Abstract
Commensal interactions between the enteric microbiota and distal intestine play important roles in regulating human health. Short-chain fatty acids (SCFAs), such as butyrate, produced through anaerobic microbial metabolism represent a major energy source for the host colonic epithelium and enhance epithelial barrier function through unclear mechanisms. Separate studies revealed that the epithelial anti-inflammatory interleukin-10 receptor α-subunit (IL-10RA) is also important for barrier formation. Based on these findings, we examined if SCFAs promote epithelial barrier through IL-10RA-dependent mechanisms. Using human intestinal epithelial cells (IECs), we discovered that SCFAs, particularly butyrate, enhanced IEC barrier formation, induced IL10RA mRNA, IL-10RA protein, and transactivation through activated Stat3 and HDAC inhibition. Loss and gain of IL10RA expression directly correlates with IEC barrier formation and butyrate represses permeability-promoting claudin-2 (Cldn2) tight-junction protein expression through an IL-10RA-dependent mechanism. Our findings provide a novel mechanism by which microbial-derived butyrate promotes barrier through IL-10RA-dependent repression of Cldn2.
Keywords: Short-chain fatty acids, butyrate, interleukin-10 receptor, IL-10RA, claudin-2, intestinal epithelial cell, barrier
INTRODUCTION
The mammalian gastrointestinal tract is home to trillions of bacteria that are separated from the host immune system by a single layer of intestinal epithelial cells (IECs). A finely regulated commensal relationship exists within the intestinal mucosa, where microbes, essential for host health, can also initiate and perpetuate mucosal disease (1). These microbes reside in the distal gut where they metabolize undigested materials and benefit the host through local synthesis of short-chain fatty acids (SCFAs). The three primary SCFAs are acetate, propionate and butyrate. Butyrate, while the least abundant of the three, is the preferred metabolic substrate for IECs and can reach luminal concentrations of 30 mM in the colon (2). Dysbiosis, in part characterized by a decrease of butyrate-producing microbial species, has been strongly associated with colonic disease, including inflammatory bowel disease (IBD) (3–5). Yet, mechanisms to explain the inverse association between butyrate and disease are not well understood.
In a healthy colon, there exists a fine balance between pro- and anti-inflammatory mediators. Pro-inflammatory cytokines, including tumor necrosis factor-alpha (TNF-α and interleukin-1beta (IL-1β) have been implicated in the pathogenesis of IBD (6, 7). However, this inflammation is normally limited by anti-inflammatory cytokines, including interleukin-10 (IL-10). IL-10 produced by IECs and leukocytes inhibits the secretion of TNF-α and interferon-gamma (IFN-γ) (8). The IL-10 receptor α subunit (IL-10RA) is expressed on numerous cell types, including IECs (9, 10). The functional IL-10 receptor is comprised of the ligand-binding alpha subunit (IL-10RA) and a beta subunit (IL-10RB) that is shared with other IL-10 receptor family members (11). The binding of IL-10 to IL-10R results in activation of the JAK-STAT signaling pathway, and transactivation of latent transcription factors. Numerous studies have shown that STAT3 activation is critical to the ligand’s anti-inflammatory activity (12, 13). Mice deficient in IL-10 or IL-10RA develop severe colitis and mice conditionally lacking intestinal epithelial IL-10RA show increased susceptibility to colitis (14–16).
While the host has developed defense mechanisms to counter threats from prokaryotes in the gut, bacterial pathogens and commensals have also evolved countermeasures to modulate the host intestinal epithelium. Recently, we have shown that microbial-derived butyrate increases mitochondrial-dependent oxygen consumption in IECs, stabilizes hypoxia-inducible factor (HIF), and induces expression of HIF-target genes that augment barrier function (17). Further, butyrate is a well-known histone deacetylase inhibitor and, through this mechanism, down-regulates pro-inflammatory mediator expression by macrophages and increases regulatory T cell differentiation (18, 19).
Given the role of barrier dysfunction in colonic disease, we sought to define mechanisms of SCFA regulation of barrier. Our studies reveal that butyrate promotes epithelial barrier formation through IL-10RA-mediated repression of permeability-promoting claudin-2 (Cldn2). Cldn2, one of 27 mammalian claudins that regulate barrier, forms a paracellular channel for small cations and water and is upregulated in IBD, contributing to diarrhea via a leak-flux mechanism (20). These findings provide a mechanistic link between host-microbial crosstalk within the mucosa and describe a mutualism between butyrate and intestinal homeostasis.
MATERIALS and METHODS
Cell culture
Both Caco2 (ATCC# HTB-37) and T84 (ATCC# CCL-248) human epithelial cell lines were obtained from ATCC (Manassas, VA) and maintained in 95% air with 5% CO2 at 37°C according to instructions provided by ATCC. Where indicated, cells were cultured on polyester transwell inserts (Costar Corp., Cambridge MA). Acetic acid, propionic acid, and butyric acid from Sigma-Aldrich (St. Louis, MO) were added to sterile filtered Hank’s balanced salt solution (Sigma) with the addition of 10 mM HEPES and pH adjusted with sodium hydroxide to pH 7.4. Transepithelial resistances (TEER) were monitored using an EVOM2 Voltohmmeter (World Precision Instruments). Cytokines were purchased from R&D Systems (Minneapolis, MN) and used at indicated concentrations. To generate shIL-10RA and oeIL10-RA cell lines, lentiviruses encoding shRNA targeting IL-10RA or IL-10RA ORF were used (MISSION™ TRC shRNA or CCSB-Broad, University of Colorado Functional Genomics Facility). We transduced T84 cells using previously described protocols (21). Stable integration was maintained by puromycin selection (3ug/ml). Knockdown and overexpression were confirmed by PCR analysis.
Gene Expression
TRIzol from Invitrogen (Grand Island, NY) was used to isolate RNA from cultured cells and tissues exposed to SCFA, TSA, and Stattic (Sigma). iScript cDNA Synthesis Kit from Bio-Rad Laboratories (Hercules, CA) and SYBR Green from Applied Biosystems (Warrington, UK) were used for cDNA synthesis and real-time PCR analysis (Applied Biosystems 7900HT) using the following mouse and human forward (Fwd) and reverse (Rev) primer sequences:
hIL10RA Fwd:CCCTGTCCTATGACCTTACCG, Rev:CACACTGCCAACTGTCAGAGT;
hSOCS3 Fwd:GGCCACTCTTCAGCATCTC, Rev:ATCGTACTGGTCCAGGAACTC; hCLDN1 Fwd:CCAGTCAATGCCAGGTACGAAT, Rev:TTGGTGTTGGGTAAGAGGTTGTT; hCLDN2 Fwd:CTCCTGGGATTCATTCCTGTT, Rev:TCAGGCACCAGTGGTGAGTAGA; hCLDN3 Fwd:CCACGCGAGAAGAAGTACA, Rev:GTAGTCCTTGCGGTCGTAGC; hCLDN7 Fwd:AATGTACGACTCGGTGCTCG, Rev:AATCTGATGGCCATACCAGG; hOCLN Fwd:GCTACGGAAGTGGCTATGG, Rev:GCGGCAATGAAACAAAAG;
hECAD Fwd:GCCCATTTCCTAAAAACCTG, Rev:CTCTGTCACCTTCAGCCATC;
hJAMA Fwd:CCTGGGAATCTTGGTTTTTG, Rev:GGAATGACGAGGTCTGTTTG;
hTJP1 Fwd:TGGTGTCCTACCTAATTCAACTCA, Rev:CGCCAGCTACAAATATTCCAACA;
hIL-10 Fwd: AATAAGGTTTCTCAAGGG, Rev:AGAACCAAGACCCAGACA;
hACTB Fwd:CCTGGCACCCAGCACAAT, Rev:GCCGATCCACACGGAGTACT;
mIl10ra Fwd:CCCATTCCTCGTCACGATCTC, Rev: TCAGACTGGTTTGGGATAGGTTT;
mActb Fwd: TACGGATGTCAACGTCACAC, Rev: AAGAGCTATGAGCTGCCTGA
Protein Analysis and Immunofluorescence
Cells and whole tissue were extracted into Tris lysis buffer with protease inhibitor (Roche), disrupted by sonication, and quantified for normalization using Pierce BCA protein assay kit (Thermo Scientific). Antibodies were used at manufacturer recommended concentrations and included: anti-IL-10RA (rabbit polyclonal), anti-Cldn2 (rabbit polyclonal), and anti–β-actin from Abcam; IL-10 (human), anti-acH3K9 (C5B11, rabbit monoclonal), anti-Stat3 (79D7, rabbit monoclonal), and anti-pStat3 (Y705, rabbit polyclonal) from Cell Signaling; and anti-IL10RA (rabbit polyclonal) from ThermoFisher. The Western blotting antibodies, peroxidase Goat Anti-Rabbit IgG and peroxidase Goat Anti-Mouse IgG, were purchased from Jackson Laboratories. Western blotting substrates, Pierce ECL and SuperSignal West Femto, were purchased from ThermoFisher.
To localize IL-10RA, T84 cells were exposed to butyrate or buffer control, fixed and processed for microscopy as described (16). Cells were localized with anti-IL-10RA followed by AlexaFluor 488 secondary Ab and counter-stained with AlexaFluor 546 (Invitrogen). Fluorescence images were obtained using an AxioCam MRc5 attached to an AxioImager A1 microscope (Zeiss, Oberkochen, Germany).
Transfection
An IL-10RA-luciferase reporter plasmid and empty vector control (Switchgear Genomics) were transfected into Caco-2 cells using Lipofectamine 3000 transfection reagent (Invitrogen) using the manufacturer’s recommended protocol. The day following transfection, cells were treated with the designated dose of butyrate. Cells were lysed the day after treatment and luciferase (Promega) was measured and normalized to protein by BCA. Lightswitch (Active Motif) were used to quantify reporters from Switchgear Genomics.
Animal Studies
Wild-type (WT) C57/B6 mice were purchased from the Jackson Laboratory (Bar Harbor, ME). Mice were housed in accordance with guidelines from the American Association for Laboratory Animal Care and Research Protocols, and experiments were approved by the Institutional Animal Care and Use Committee of the University of Colorado.
Abx treatment consisted of 3 days of 200ul/mouse oral gavage of Abx cocktail ampicillin (1mg/ml), gentamicin (1mg/ml), metronidazole (1mg/ml), neomycin (1mg/ml), and vancomycin (0.5mg/ml) from Sigma-Aldrich.
Original germ-free (GF) C57Bl/6 breeding stocks were obtained from the National Gnotobiotic. Rodent Resource Center at the University of North Carolina. Mice were bred and maintained in flexible vinyl positive pressure isolators within the Gnotobiotic Facility at the University of Colorado on a 12-hour light cycle and given free access to autoclaved water and autoclaved chow (Teklad Global Soy Protein-Free Extruded Rodent Diet, 2020sx, Harlan Laboratories, Indianapolis, IN). Animal tissues were extracted into Tris lysis buffer followed by sonication, and protein homogenates were stored at −80C until use. RNA extraction was performed as described above.
Statistics
GraphPad Prism 5 software produced by GraphPad (La Jolla, CA) was used to create figures and perform statistical analyses, including one-way ANOVA, Tukey’s post hoc test, and Student’s t-test. Error bars represent SEM unless otherwise stated, and significance was determined at p<0.05.
RESULTS
Microbial-derived Butyrate Enhances Intestinal Epithelial Barrier Formation
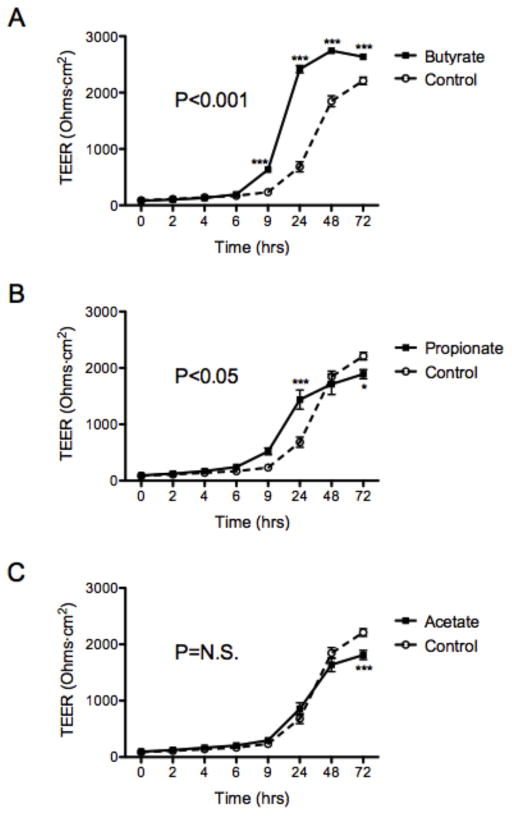
Guided by our recent findings that butyrate (BA) decreases the flux of labeled tracers across intestinal epithelial cell monolayers (17), we profiled the influence of SCFAs on intestinal barrier formation. As shown in Figure 1, T84 intestinal epithelial cells were plated on transwell inserts and exposed to physiologically-relevant concentrations of three major SCFAs: butyrate (5mM), propionate (20mM), and acetate (20mM). We monitored barrier formation, as measured by transepithelial electrical resistance (TEER), over 72h. We found that butyrate, but neither acetate nor propionate, significantly enhanced both the kinetics and magnitude of barrier formation over 72h (Figure 1A–C, p<0.001), suggesting that butyrate selectively promotes intestinal barrier formation.
Figure 1. Butyrate enhances intestinal epithelial cell barrier formation.
T84 cells were treated with HHBS (Hanks’ Buffer with 10mM HEPES) at time 0h ± (A) butyrate (5mM), (B) propionate (20mM), or (C) acetate (20mM). Transepithelial electrical resistance (TEER) was measured and analyzed over 72h. Data are representative of at least two independent experiments. Error bars represent mean ± SEM. * p<0.05; ** p<0.01; *** p<0.001; n.s., not significant (unpaired Student’s t test and ANOVA).
Butyrate Increases Interleukin-10 Receptor Expression and Function
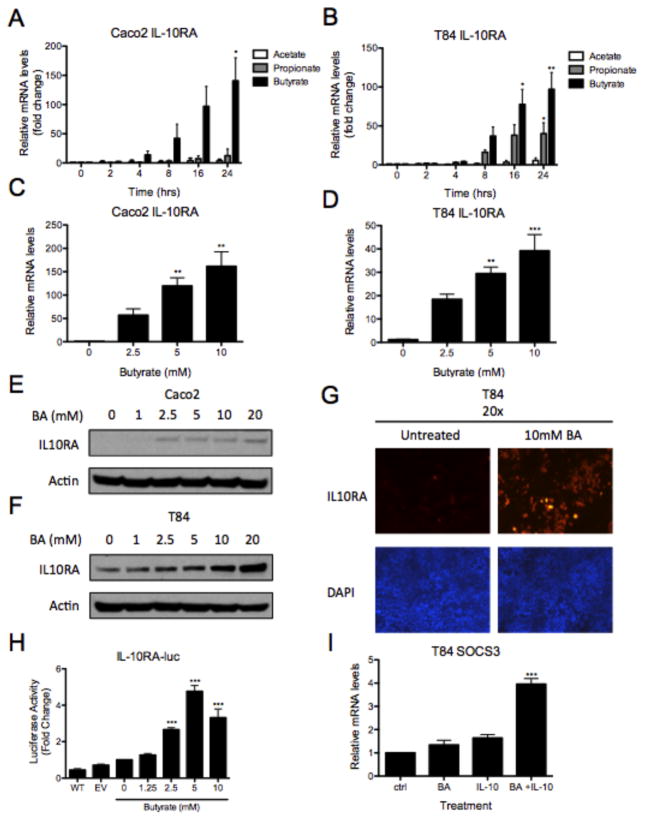
Our previous work demonstrated that the α subunit of the interleukin-10 receptor (IL-10RA) is central to the formation of epithelial barrier and that knockdown of IL-10RA results in a loss of barrier formation (16). Based on these findings, we hypothesized that butyrate-enhanced barrier formation is mediated by induction of functional IL-10RA. To test this hypothesis, human intestinal epithelial cells (Caco-2 and T84 cells) were exposed to acetate, propionate, and butyrate for up to 24 hours and IL10RA transcript was measured by quantitative PCR (qPCR). IL10RA transcript increased 140.8 ± 39.3-fold in Caco-2 cells (Figure 2A, p<0.05) and 97.0 ± 21.5-fold in T84 cells (Figure 2B, p<0.01) after 24hrs of 5mM butyrate treatment. Propionate also increased IL10RA expression by 40.0 ± 14.1-fold in T84 cells (Figure 2B, p<0.05). Further, we observed 118.6 ± 6.2-fold IL10RA mRNA induction Caco-2 cells treated with 5mM butyrate for 18 hours (Figure 2C, p<0.01). We saw a 28.3 ± 7.3-fold dose-response in T84 cells (Figure 2D, p<0.01) with the same exposure and chose to focus on butyrate for ongoing studies.
Figure 2. Butyrate upregulates interleukin-10 receptor α (IL-10RA).
(A) Caco-2 and (B) T84 cells were treated with HHBS ± acetate (20mM), propionate (20mM), or butyrate (5mM) for up to 24h. RNA was isolated, cDNA was synthesized and IL-10RA mRNA was quantitated by qPCR (data normalized to β-actin). (C, D) In a separate experiment, cells were treated with HHBS ± butyrate (0–10mM) for 18h and IL10RA mRNA was analyzed by qPCR (data normalized to β-actin). (E) Caco-2 and (F) T84 cells were treated for 24h with HHBS ± butyrate (0–20mM). Whole-cell lysates were collected and probed for IL-10RA by immunoblot (β-actin was used as loading control). (G) Representative photographs of immunofluorescence staining of IL-10RA (red) in T84 cells following HHBS ± butyrate (10mM) for 24h. Nuclei stained with DAPI (blue). (H) Caco-2 cells were transfected with empty vector control (EV) and an IL10RA promoter reporter (IL10RA-luc). After 24h, cells were treated with HHBS ± butyrate (0–10mM) for another 24h and luciferase activity was quantified. (I) T84 cells were treated with HHBS ± butyrate (10mM) for 24h and stimulated with PBS ± IL-10 (20ng/ml) for 2h. SOCS3 mRNA measured by qPCR (data normalized to β-actin). Data are representative of two or three independent experiments. Error bars represent mean ± SEM.
To further characterize the relationship between IL-10RA and butyrate, we exposed Caco-2 and T84 cells to increasing concentrations of butyrate and examined IL-10RA protein by immunoblot (Figure 2E and 2F). Our results revealed a dose-dependent induction of IL10-RA by butyrate. As shown in Figure 2G, analysis by immunofluorescence microscopy also revealed increased IL-10RA protein following exposure to 10mM butyrate treatment in T84 cells. We next evaluated whether butyrate could transactivate the IL10RA promoter. We transfected an IL10RA-luciferase reporter construct in Caco-2 cells, treated the cells with increasing concentrations of butyrate, and discovered 1.6-fold increased IL10RA-luciferase reporter activity with 2.5mM butyrate (Figure 2H, p<0.001). Additionally, butyrate-elicited IL10RA was shown to be functional based on IL-10-mediated 3.0-fold induction of suppressor of cytokine signaling 3 (SOCS3), a well-documented downstream target gene in intestinal epithelia (Figure 2I, p<0.001) (16, 22). Finally, we determined whether exogenous IL-10 would further influence barrier formation in butyrate treated monolayers. As shown in Supplemental Figure 1, pretreatment of T84 cells with butyrate (5mM, 24h) followed by IL-10 (10ng/ml) resulted in a significant increase in barrier formation compared to both media alone (p<0.001) and butyrate alone (p<0.05). The addition of anti-IL-10 (p<0.05), but not non-specific IgG (p = not significant), decreased the rate of barrier formation compared to butyrate alone. These results identify the induction of functional IL-10RA by butyrate.
Butyrate Regulates IL-10RA Expression via STAT3 Activation and Histone Deacetylase Inhibition
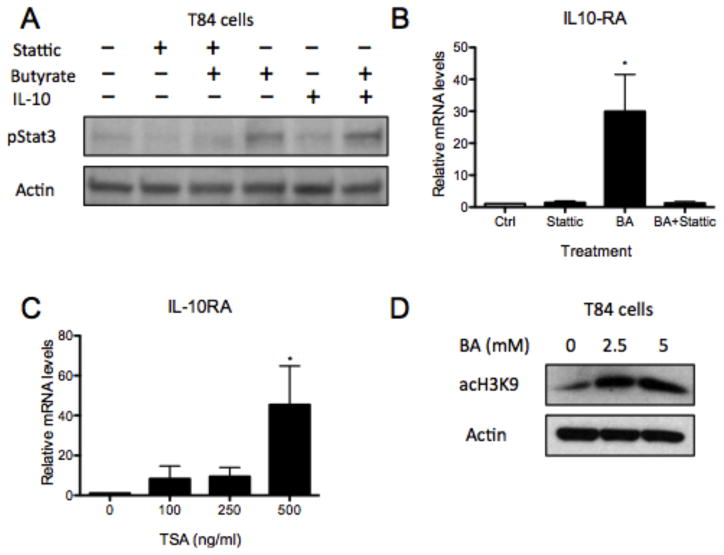
We next examined mechanisms of butyrate induction of IL-10RA. Here, we analyzed if signal transducer and activation of transcription 3 (STAT3), a downstream target of IL-10RA, could regulate IL-10RA through feed forward mechanisms, similar to IL-10 (23). T84 cells were treated with butyrate for 18h followed by IL-10 for 1h before harvest. Immunoblot of lysates revealed that butyrate alone and butyrate with IL-10 cytokine both phosphorylate STAT3 (Figure 3A). Further, pretreatment with Stattic, a STAT3 activation inhibitor, inhibited butyrate-induced STAT3 activation. In addition, stattic followed by butyrate abolished butyrate-mediated induction of IL-10RA mRNA (Figure 3B, p<0.05) (24). These results indicate that butyrate regulates IL-10RA through STAT3 activation.
Figure 3. Butyrate regulates IL-10RA through Stat3 activation and histone deacetylase inhibition.
(A) T84 cells were pretreated with DMSO ± Stattic (20mM) for 30min followed by HHBS ± butyrate (10mM) for 24h. Cells were stimulated with PBS ± IL-10 (20ng/ml) for 1h prior to harvesting the whole-cell lysates and probing for pStat3 by immunoblotting (β-actin was used as loading control). (B) T84 cells were pretreated with DMSO ± Stattic (20mM) for 30min followed by HHBS ± butyrate (5mM) for 6h. IL10RA mRNA was measured by qPCR (data normalized to β-actin). (C) T84 cells were stimulated with DMSO control or TSA (100, 250, or 500ng/ml) for 18h. RNA was isolated, cDNA was synthesized, and IL10RA mRNA was quantitated by qPCR (data normalized to β-actin). (D) T84 cells were treated with HHBS ± butyrate (2.5, 5mM) for 24h, then whole-cell lysates were collected and acH3K9 was probed by immunoblot (β-actin was used as loading control). Data are representative of at least three independent experiments. Error bars represent mean ± SEM.
Additionally, butyrate is a potent histone deacetylase inhibitor (HDACi) (19). Herein, we investigated whether butyrate mediates IL-10RA expression through HDAC inhibition. Exposure of T84 cells to trichostatin A (TSA), a well-known HDAC inhibitor, at 500ng/ml increased IL-10RA mRNA 44.4 ± 4.3-fold by qPCR (Figure 3C, p<0.05) (25). The addition of butyrate also led to significantly increased acH3K9, an active chromatin mark, in T84 cells (Figure 3D) (26). These results are supported by a previous analysis that demonstrated increased IL-10RA expression from array investigating the role of Warburg effect on butyrate-mediated colonocyte proliferation (GEO accession GSE 41113; URL: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE41113) (27). Together these findings support the HDAC inhibitory function of butytrate in induction of IL-10RA.
Recently, activation of the butyrate-binding G-protein-coupled receptor 109a (GPR109a) in the colon was found to suppress colonic inflammation and promote differentiation of Treg cells and IL-10-producing T cells (28). GPR109a is a Gi/Go protein-coupled signaling receptor that is inhibited with pertussis toxin (PTX) (29). To determine if G-protein coupling contributes to the induction of IL-10RA, we pretreated Caco-2 and T84 cells with PTX, followed by butyrate, and measured IL-10RA mRNA by qPCR. As shown in Supplemental Figure 2, IL10RA expression was not changed by PTX pretreatment, suggesting that butyrate induction of IL-10RA is independent of GPR109a.
Butyrate-Induced IL-10RA is Essential for Barrier Formation
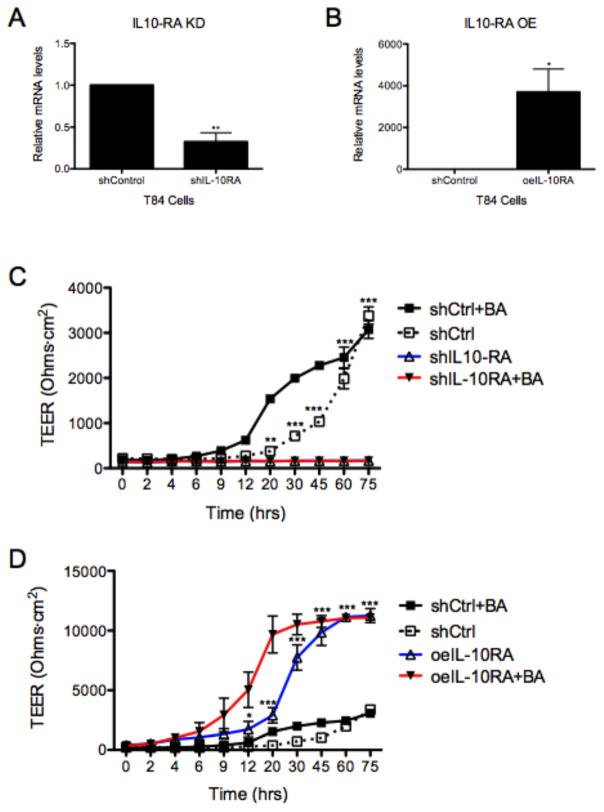
We next investigated how changes in IL-10RA expression, as in the case with butyrate levels, influenced barrier formation. For these purposes, we generated cell lines with stable loss and gain of IL-10RA using lentiviral approaches. T84 cells were transduced with shRNA-encoding lentivirus against IL-10RA (knock-down) or IL-10RA lentiORF (over-expression). The shRNA targeting IL-10RA (shIL-10RA) and lentiORF (oeIL-10RA) resulted in significantly attenuated (0.68 ± 0.11-fold) and enhanced (3703 ± 1101-fold) mRNA relative to shControl, respectively (Figure 4A and 4B, p<0.01 and p<0.05). Functionally, IL-10RA knockdown practically abolished barrier formation over 72h both in the absence and presence of butyrate (Figure 4C, p<0.001). Conversely, IL-10RA over-expression significantly enhanced barrier formation over 72h as measured by TEER and the kinetics are further enhanced in the presence of butyrate (Figure 4D, p<0.001). To define if these results paralleled an increase in IL-10, we examined the induction of IL-10 by butyrate. As shown in Supplemental Figure 3, analysis of IL-10 expression revealed that butyrate induces epithelial IL10 mRNA by as much as 30±2.5-fold (p<0.001). This induction, however, was not reflected as measurable protein is cell culture supernatants from butyrate-treated cells (0.02±.001 vs 0.01±0.001 pg/ml and 0.03±0.003 vs 0.02±0.003 pg/ml for media vs butyrate in Caco2 and T84 cells, respectively, p = not significant). These results indicate that barrier formation is dependent on epithelial expression of IL-10RA.
Figure 4. IL-10RA plays a pivotal role in formation and integrity of the intestinal epithelial cell barrier.
(A) Relative IL10RA mRNA in T84 cells transduced with lentivirus shRNA targeting IL10RA (shIL10RA) v. shControl by qPCR. (B) IL10RA gene expression, by qPCR, in T84 cells transduced with IL10RA ORF lentivirus (LentiORF; oeIL10-RA) v. shControl cells (data normalized to β-actin). (C) T84 shControl (shCtrl) and shIL10RA were treated with HHBS ± butyrate (5mM) at 0h. TEER was measured and analyzed over 75h. (D) T84 shCtrl and oeIL10RA were treated with HHBS ± butyrate (5mM) at 0h. TEER was recorded over 75h. Data are representative of three independent experiments. (A, B) Error bars represent mean ± SEM. (C, D) Error bars represent mean ± SD.
Butyrate Represses Claudin-2 in an IL-10RA-dependent Manner
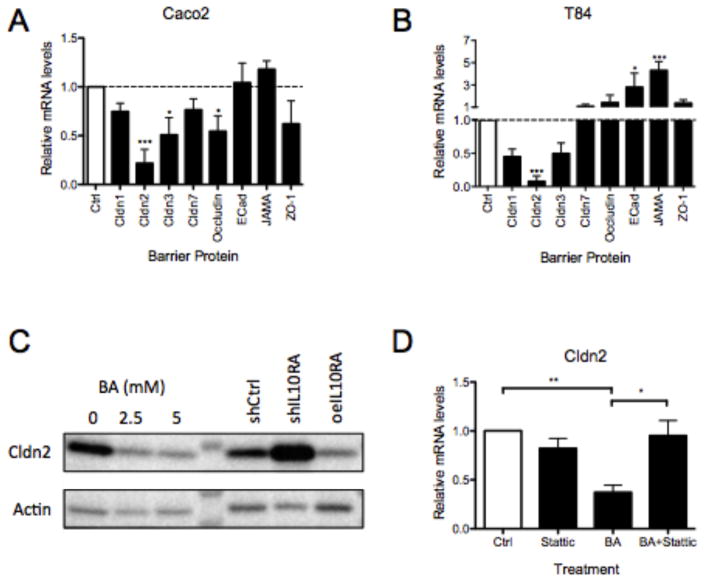
We next examined the mechanism of barrier regulation by butyrate alone and butyrate-induced IL-10RA. To this end, we exposed Caco-2 and T84 cells to butyrate and profiled the expression of a panel of junctional proteins known to be important in barrier function: Cldn1, Cldn2, Cldn3, Cldn7, occludin, E-cadherin, JAM-A and ZO-1 (30). We have used this approach successfully in the past to define targets for barrier regulation by hypoxia-inducible factor (31). Our studies revealed that butyrate preferentially repressed the expression of Cldn2 relative to control in both Caco2 (0.22 ± 0.14) and T84 cells (0.08 ± 0.07) (Figure 5A and B, p<0.001). This observation extended to the protein level, where Cldn2 protein decreased >70% by densitometry in response to physiologic concentrations of butyrate in T84 cells (Figure 5C).
Figure 5. Butyrate represses Claudin-2 through IL-10RA and pStat3.
(A) Caco-2 and (B) T84 cells were stimulated with HHBS ± butyrate (5mM) for 18h. cDNA generated from RNA isolation was analyzed by qPCR and data was normalized to β-actin. (C) Claudin-2 (Cldn2) was probed by immunoblot in T84 cells treated with HHBS ± butyrate (2.5 or 5mM) for 24h and also T84 shCtrl, shIL10RA, and oeIL10RA cell lines (β-actin was loading control). (D) T84 cells were pretreated with DMSO ± Stattic (20mM) for 30min followed by HHBS ± butyrate (5mM) for 6h. Cldn2 mRNA was measured by qPCR (data normalized to actin). Data are representative of at least three independent experiments. Error bars represent mean ± SEM.
Interestingly, we found that Cldn2 repression is dependent on IL-10RA. As shown in Figure 5C, analysis of Cldn2 protein in shIL10RA and oeIL10RA revealed a reverse correlation between Cldn2 and IL10RA. Moreover, inhibition of STAT3 with Stattic reversed the butyrate-mediated repression of Cldn2, suggesting that IL-10RA signaling through STAT3 mediates this response (Figure 5D). These results demonstrate that butyrate enhances barrier through IL-10RA-mediated repression of Cldn2 and reveals a novel mechanism of barrier regulation through host-microbe crosstalk.
Induction of Il10ra In Vivo is Microbiota-dependent
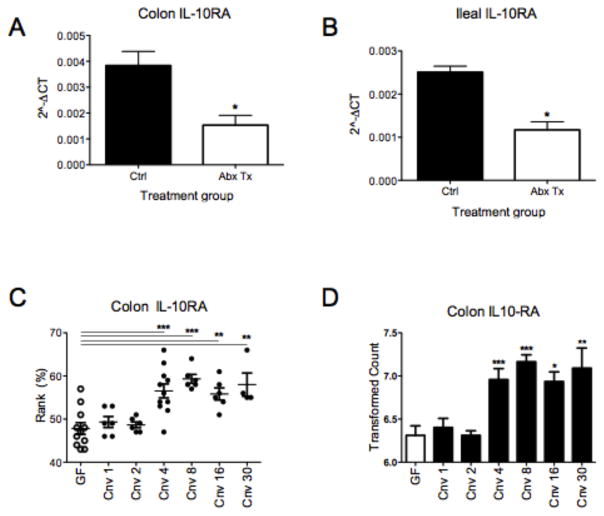
Finally, we determined the physiologic relevance of these findings in vivo. Herein, we employed antibiotic (Abx)-treated and germ-free (GF) mice to determine the impact of the microbiota on Il10ra expression. We have previously demonstrated that this Abx cocktail resulted in a 92±4% decrease in fecal butyrate levels (17). Administration of broad-spectrum Abx by oral gavage resulted in a 63±7.2% and 55±8.5 decrease in colonic and ileal Il10ra, respectively, (Figure 6A and 6B, p<0.05 for each). Examination of Il10ra mRNA in colon scrapings enriched in epithelial cells from GF mice revealed attenuated expression of Il10ra, but did not reach statistical significance (Supplemental Figure 4, p = 0.2). Likewise, as shown in Supplemental Figure 4, the administration of a butyrate supplement (tributyrin) to Abx-treated mice, as we have done in the past (17), did not significantly change the expression of either Cldn2 or IL10ra (p = 0.17 and p = 0.13 for Cldn2 and Il10ra, respectively).
Figure 6. Microbial-derived butyrate enhances Il10ra in an in vivo system.
C57BL/6 mice were gavaged with broad-spectrum antibiotics for 3d and euthanized. (A) Colon and (B) ileal tissue collected from the control and antibiotic treated mice were lysed, RNA was isolated, cDNA was synthesized, and Il10ra mRNA was analyzed by qPCR. (C and D) Data collected from Geo Profiles of both Rank % and transformed counts of germ free mice colonic Il10ra during conventionalization were graphed. Data are representative of three independent experiments (A, B). Error bars represent mean ± SEM.
We also sought to test this hypothesis by mining publicly available data sets. We discovered a time-dependent increase in colonic Il10ra rank and mRNA from array analyses investigating immune responses during conventionalization of GF mice (GEO accession GDS4319) (Figure 6C and D) (32–34). Taken together, these findings confirm in vitro results and implicate luminal-derived signals, such as butyrate, as a significant regulator of Il10ra expression in vivo.
DISCUSSION
An understanding of the interactions between the enteric microbiota and distal gut is an area of intense investigation. It is clear that microbial-derived factors contribute significantly to human health and that co-evolution has benefited both the host and the microbe (35). It has also become evident that changes to microbial community structures can shift homeostasis and contribute to a broad spectrum of diseases, including IBD (36). Recent studies have revealed that IBD is associated with significant depletion of butyrate-producing species. Given that IBD is also strongly associated with barrier dysfunction (37), we sought to understand how butyrate regulates epithelial barrier function. We report here that physiologic concentrations of butyrate repress Cldn2 expression in an IL-10RA-dependent manner to enhance epithelial barrier function, which may serve as a mechanism to prevent unwanted host immune responses against beneficial butyrate-producing microbes in the gut.
Butyrate, an end product of bacterial metabolism, typically constitutes up to 20% of SCFA in the human colon with absolute concentrations above 10mM in human feces (2). Butyrate has been demonstrated to benefit the host in a number of ways. First, it is the preferential metabolic substrate in IECs and through oxidative phosphorylation depletes local oxygen to the extent that HIF is stabilized and enhances IEC barrier via HIF (2, 17). Second, butyrate activates the GPR109a receptor to suppress colonic inflammation and carcinogenesis (28). GPR109a signaling promotes anti-inflammatory properties in colonic macrophages and dendritic cells and enables differentiation of Treg cells and IL-10-producing T cells. Third, through its actions as an HDAC inhibitor, butyrate stimulates anti-inflammatory mechanisms that may also promote the restoration of mucosal barrier function through decreased inflammatory cytokine production (38, 39). Thus, a role for butyrate in mucosal homeostasis is well established. However, the molecular details of butyrate actions on epithelial barrier function remain incompletely understood.
We confirmed previously published findings that butyrate promotes barrier formation (17). This action of butyrate on increasing both the rate and the magnitude of barrier formation was reminiscent of recent work implicating epithelial IL-10RA signaling (16). Extensions of these initial findings revealed that butyrate, and to a lesser extent propionate, prominently induces IL-10RA. Mice deficient in IL-10 or IL-10R develop spontaneous severe colitis and mutations in either IL-10 or IL-10R have been shown to result in severe cases of human IBD (40–42). Previous studies by Kominsky et al. looking at this receptor on IECs revealed a number of surprising features (16). First, IL-10RA expression was discovered based on a screen of IFN-inducible genes. This was unexpected given that IFN is one of the more predominant inducers of barrier dysfunction (43). Second, these studies revealed that IL-10RA expression is polarized to the apical surface of IECs in vitro and in vivo. Third, conditional deletion of IL-10RA in IECs revealed both increased baseline permeability and a high susceptibility to DSS colitis, suggesting an important role for IL-10RA in barrier function and recovery following insult. Along with our findings that decreased IL-10RA correlates with decreased luminal butyrate (via antibiotic treatment or germ-free conditions), we have identified an additional role for butyrate in the homeostatic maintenance of IECs via IL-10RA.
Butyrate transmits signals to the mucosa through a number of different mechanisms, including surface G-protein coupled receptor (GPCR) stimulation (28), HDAC inhibition (39), and HIF stabilization (17). The primary butyrate signaling receptor is GPR109A, a Gai coupled GPCR that also functions as the niacin receptor (28). Based on inhibition of Gαi using cholera toxin, we found no role for GPCR signaling in the induction of IL-10RA. Based on direct evidence of H3K9 acetylation and the use of TSA, we concluded that one signaling mechanism for the induction of IL-10RA was HDACi, a function for which butyrate is well established (19). It is interesting to note that propionate and acetate also increased IL-10RA mRNA, which could be due to their ability to inhibit HDACs to a lesser degree, although propionate and acetate are far less potent than butyrate as HDAC inhibitors (44, 45). We did not find an induction of IL-10RA with hypoxia treatment nor HIF stabilization (data not shown).
A surprising observation was the absolute dependence of IL-10RA expression for epithelial barrier formation. Knockdown and over-expression of IL-10RA using lentiviral approaches revealed a direct correlation between IL-10RA expression and the magnitude of barrier formation. While the addition of exogenous IL-10 enhanced the butyrate response, IL-10RA also promoted barrier formation in the absence of added IL-10. For a number of reasons, this observation was perplexing. We were not able to measure appreciable amounts of IL-10 in cell culture supernatants. Butyrate readily induced IL10 mRNA, but no increase in protein was observed. While we and others have shown that epithelial cells can make and release IL-10 in response to a number of stimuli (46–48), it is possible that butyrate also induces IL-10 mRNA degradation. IL10 mRNA, for example, is known to be degraded through processing at the 3′UTR by live probiotic bacteria including Lactobacillus paracasei (49), a known butyrate producer (50). It is also possible that the low concentrations of IL-10 protein measured in tissue culture bulk flow are sufficient at the cell surface to activate butyrate-induced IL-10RA. Additional evidence was provided using the small molecule STAT3 inhibitor, Stattic, which decreased STAT3 phosphorylation and attenuated butyrate-induced IL-10RA induction. It is also notable that the addition of butyrate to cells overexpressing IL-10RA further enhanced barrier formation, suggesting that butyrate may act through additional mechanisms to increase IEC barrier formation. While we do not know the exact nature of this additional signaling, it is possible that HDACi activity of butyrate could be involved. Butyrate was recently shown to promote barrier in T84 cells via HDAC inhibition and induction of TWIK-related potassium channel-1 (Trek1) (51).
As an extension of these results demonstrating IL-10-dependent increases in barrier induction by butyrate, we employed an unbiased screen of epithelial junction proteins to identify mechanisms of barrier development and identified butyrate-mediated repression of Cldn2 in both Caco2 and T84 cells (52). Among the claudin family of tight junction proteins, Cldn2 is considered a “leaky” claudin. An understanding of the regulatory pathways of Cldn2 expression is of significant interest because its expression is increased in IBD, celiac disease, and HIV enteropathy and colonic cancer (20). It is noteworthy that butyrate has been previously mentioned in the regulation of Cldn2 (38, 53), without insight into mechanism. The present studies add significantly to this work and identify a role for butyrate-dependent IL-10R signaling in the repression of Cldn2. Given the reciprocal relationship between loss of Cldn2 and compensation by “tight” claudin expression (54), these studies implicate an important role for butyrate in maintaining the repression of Cldn2 and maintenance of barrier function through this mechanism (20).
In a final set of experiments, we examined the role of butyrate on IL-10RA expression in vivo. To this end, IL-10RA expression was examined in mice exposed to broad-spectrum antibiotics, conditions that we have shown to result in nearly unmeasurable colonic butyrate (17). These conditions were found to significantly decrease epithelial IL-10RA expression in mice. Parallel studies in germ-free mice supported the results with antibiotics. Data mined from Geo Profiles similarly revealed increased colonic IL-10RA mRNA during conventionalization of germ-free mice. IL-10RA has already been shown to play a central role in control of homeostatic barrier function as IEC-specific IL-10RA−/− mice have significant increases in baseline permeability and systemic inflammatory responses. These results are further evidence that microbial-derived butyrate enhances IL-10RA to promote barrier and dampen immune responses.
Taken together, our results provide insight into how butyrate regulates IEC barrier and identifies a new mechanism by which microbes promote homeostasis in the distal gut. We demonstrate that butyrate, through Stat3 activation and HDAC inhibition, represses Cldn2 in an IL-10RA-dependent manner. This suggests that certain commensal bacteria may be signaling that they are non-pathogenic by producing butyrate, which leads to increased IL-10RA signaling and dampening of the host immune system. In the absence of butyrate-producing species, as occurs in IBD, barrier may be significantly compromised. These results further suggest that administration of butyrate (e.g. tributyrin) and/or promoting butyrate-producing bacteria may be of therapeutic benefit (55).
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health grants (DK103639, DK50189, DK95491) to SPC, the US Department of Veterans Affairs as well as UL1 TR001082 and T32 AI 52066 to LZ
The authors acknowledge Yaoxing Li for his contributions to performing the research presented here.
Footnotes
The authors declare no conflicts of interest.
References
- 1.Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R. Diversity, stability and resilience of the human gut microbiota. Nature. 2012;489:220–230. doi: 10.1038/nature11550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hamer HM, Jonkers D, Venema K, Vanhoutvin S, Troost FJ, Brummer RJ. Review article: the role of butyrate on colonic function. Aliment Pharmacol Ther. 2008;27:104–119. doi: 10.1111/j.1365-2036.2007.03562.x. [DOI] [PubMed] [Google Scholar]
- 3.Eeckhaut V, Machiels K, Perrier C, Romero C, Maes S, Flahou B, Steppe M, Haesebrouck F, Sas B, Ducatelle R, Vermeire S, Van Immerseel F. Butyricicoccus pullicaecorum in inflammatory bowel disease. Gut. 2013;62:1745–1752. doi: 10.1136/gutjnl-2012-303611. [DOI] [PubMed] [Google Scholar]
- 4.Machiels K, Joossens M, Sabino J, De Preter V, Arijs I, Eeckhaut V, Ballet V, Claes K, Van Immerseel F, Verbeke K, Ferrante M, Verhaegen J, Rutgeerts P, Vermeire S. A decrease of the butyrate-producing species Roseburia hominis and Faecalibacterium prausnitzii defines dysbiosis in patients with ulcerative colitis. Gut. 2014;63:1275–1283. doi: 10.1136/gutjnl-2013-304833. [DOI] [PubMed] [Google Scholar]
- 5.Sokol H, Seksik P, Furet JP, Firmesse O, Nion-Larmurier I, Beaugerie L, Cosnes J, Corthier G, Marteau P, Dore J. Low counts of Faecalibacterium prausnitzii in colitis microbiota. Inflamm Bowel Dis. 2009;15:1183–1189. doi: 10.1002/ibd.20903. [DOI] [PubMed] [Google Scholar]
- 6.Atreya I, Atreya R, Neurath MF. NF-kappaB in inflammatory bowel disease. J Intern Med. 2008;263:591–596. doi: 10.1111/j.1365-2796.2008.01953.x. [DOI] [PubMed] [Google Scholar]
- 7.Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol. 2009;9:313–323. doi: 10.1038/nri2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moore KW, de Waal Malefyt R, Coffman RL, O’Garra A. Interleukin-10 and the interleukin-10 receptor. Annu Rev Immunol. 2001;19:683–765. doi: 10.1146/annurev.immunol.19.1.683. [DOI] [PubMed] [Google Scholar]
- 9.Bourreille A, Segain JP, Raingeard de la Bletiere D, Siavoshian S, Vallette G, Galmiche JP, Blottiere HM. Lack of interleukin 10 regulation of antigen presentation-associated molecules expressed on colonic epithelial cells. Eur J Clin Invest. 1999;29:48–55. doi: 10.1046/j.1365-2362.1999.00410.x. [DOI] [PubMed] [Google Scholar]
- 10.Denning TL, Campbell NA, Song F, Garofalo RP, Klimpel GR, Reyes VE, Ernst PB. Expression of IL-10 receptors on epithelial cells from the murine small and large intestine. Int Immunol. 2000;12:133–139. doi: 10.1093/intimm/12.2.133. [DOI] [PubMed] [Google Scholar]
- 11.Commins S, Steinke JW, Borish L. The extended IL-10 superfamily: IL-10, IL-19, IL-20, IL-22, IL-24, IL-26, IL-28, and IL-29. J Allergy Clin Immunol. 2008;121:1108–1111. doi: 10.1016/j.jaci.2008.02.026. [DOI] [PubMed] [Google Scholar]
- 12.Riley JK, Takeda K, Akira S, Schreiber RD. Interleukin-10 receptor signaling through the JAK-STAT pathway. Requirement for two distinct receptor-derived signals for anti-inflammatory action. J Biol Chem. 1999;274:16513–16521. doi: 10.1074/jbc.274.23.16513. [DOI] [PubMed] [Google Scholar]
- 13.El Kasmi KC, Holst J, Coffre M, Mielke L, de Pauw A, Lhocine N, Smith AM, Rutschman R, Kaushal D, Shen Y, Suda T, Donnelly RP, Myers MG, Jr, Alexander W, Vignali DA, Watowich SS, Ernst M, Hilton DJ, Murray PJ. General nature of the STAT3-activated anti-inflammatory response. J Immunol. 2006;177:7880–7888. doi: 10.4049/jimmunol.177.11.7880. [DOI] [PubMed] [Google Scholar]
- 14.Chaudhry A, Samstein RM, Treuting P, Liang Y, Pils MC, Heinrich JM, Jack RS, Wunderlich FT, Bruning JC, Muller W, Rudensky AY. Interleukin-10 signaling in regulatory T cells is required for suppression of Th17 cell-mediated inflammation. Immunity. 2011;34:566–578. doi: 10.1016/j.immuni.2011.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuhn R, Lohler J, Rennick D, Rajewsky K, Muller W. Interleukin-10-deficient mice develop chronic enterocolitis. Cell. 1993;75:263–274. doi: 10.1016/0092-8674(93)80068-p. [DOI] [PubMed] [Google Scholar]
- 16.Kominsky DJ, Campbell EL, Ehrentraut SF, Wilson KE, Kelly CJ, Glover LE, Collins CB, Bayless AJ, Saeedi B, Dobrinskikh E, Bowers BE, MacManus CF, Muller W, Colgan SP, Bruder D. IFN-gamma-mediated induction of an apical IL-10 receptor on polarized intestinal epithelia. J Immunol. 2014;192:1267–1276. doi: 10.4049/jimmunol.1301757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kelly CJ, Zheng L, Campbell EL, Saeedi B, Scholz CC, Bayless AJ, Wilson KE, Glover LE, Kominsky DJ, Magnuson A, Weir TL, Ehrentraut SF, Pickel C, Kuhn KA, Lanis JM, Nguyen V, Taylor CT, Colgan SP. Crosstalk between Microbiota-Derived Short-Chain Fatty Acids and Intestinal Epithelial HIF Augments Tissue Barrier Function. Cell Host Microbe. 2015;17:662–671. doi: 10.1016/j.chom.2015.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Furusawa Y, Obata Y, Fukuda S, Endo TA, Nakato G, Takahashi D, Nakanishi Y, Uetake C, Kato K, Kato T, Takahashi M, Fukuda NN, Murakami S, Miyauchi E, Hino S, Atarashi K, Onawa S, Fujimura Y, Lockett T, Clarke JM, Topping DL, Tomita M, Hori S, Ohara O, Morita T, Koseki H, Kikuchi J, Honda K, Hase K, Ohno H. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013;504:446–450. doi: 10.1038/nature12721. [DOI] [PubMed] [Google Scholar]
- 19.Davie JR. Inhibition of histone deacetylase activity by butyrate. J Nutr. 2003;133:2485S–2493S. doi: 10.1093/jn/133.7.2485S. [DOI] [PubMed] [Google Scholar]
- 20.Luettig J, Rosenthal R, Barmeyer C, Schulzke JD. Claudin-2 as a mediator of leaky gut barrier during intestinal inflammation. Tissue Barriers. 2015;3:e977176. doi: 10.4161/21688370.2014.977176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Glover LE, Bowers BE, Saeedi B, Ehrentraut SF, Campbell EL, Bayless AJ, Dobrinskikh E, Kendrick AA, Kelly CJ, Burgess A, Miller L, Kominsky DJ, Jedlicka P, Colgan SP. Control of creatine metabolism by HIF is an endogenous mechanism of barrier regulation in colitis. Proc Natl Acad Sci U S A. 2013;110:19820–19825. doi: 10.1073/pnas.1302840110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Suzuki A, Hanada T, Mitsuyama K, Yoshida T, Kamizono S, Hoshino T, Kubo M, Yamashita A, Okabe M, Takeda K, Akira S, Matsumoto S, Toyonaga A, Sata M, Yoshimura A. CIS3/SOCS3/SSI3 plays a negative regulatory role in STAT3 activation and intestinal inflammation. J Exp Med. 2001;193:471–481. doi: 10.1084/jem.193.4.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Staples KJ, Smallie T, Williams LM, Foey A, Burke B, Foxwell BM, Ziegler-Heitbrock L. IL-10 induces IL-10 in primary human monocyte-derived macrophages via the transcription factor Stat3. J Immunol. 2007;178:4779–4785. doi: 10.4049/jimmunol.178.8.4779. [DOI] [PubMed] [Google Scholar]
- 24.Schust J, Sperl B, Hollis A, Mayer TU, Berg T. Stattic: a small-molecule inhibitor of STAT3 activation and dimerization. Chem Biol. 2006;13:1235–1242. doi: 10.1016/j.chembiol.2006.09.018. [DOI] [PubMed] [Google Scholar]
- 25.Yoshida M, Horinouchi S, Beppu T. Trichostatin A and trapoxin: novel chemical probes for the role of histone acetylation in chromatin structure and function. Bioessays. 1995;17:423–430. doi: 10.1002/bies.950170510. [DOI] [PubMed] [Google Scholar]
- 26.Bartova E, Krejci J, Harnicarova A, Galiova G, Kozubek S. Histone modifications and nuclear architecture: a review. J Histochem Cytochem. 2008;56:711–721. doi: 10.1369/jhc.2008.951251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Donohoe DR, Collins LB, Wali A, Bigler R, Sun W, Bultman SJ. The Warburg effect dictates the mechanism of butyrate-mediated histone acetylation and cell proliferation. Mol Cell. 2012;48:612–626. doi: 10.1016/j.molcel.2012.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Singh N, Gurav A, Sivaprakasam S, Brady E, Padia R, Shi H, Thangaraju M, Prasad PD, Manicassamy S, Munn DH, Lee JR, Offermanns S, Ganapathy V. Activation of Gpr109a, receptor for niacin and the commensal metabolite butyrate, suppresses colonic inflammation and carcinogenesis. Immunity. 2014;40:128–139. doi: 10.1016/j.immuni.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mangmool S, Kurose H. G(i/o) protein-dependent and -independent actions of Pertussis Toxin (PTX) Toxins (Basel) 2011;3:884–899. doi: 10.3390/toxins3070884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Groschwitz KR, Hogan SP. Intestinal barrier function: molecular regulation and disease pathogenesis. J Allergy Clin Immunol. 2009;124:3–20. doi: 10.1016/j.jaci.2009.05.038. quiz 21–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saeedi BJ, Kao DJ, Kitzenberg DA, Dobrinskikh E, Schwisow KD, Masterson JC, Kendrick AA, Kelly CJ, Bayless AJ, Kominsky DJ, Campbell EL, Kuhn KA, Furuta GT, Colgan SP, Glover LE. HIF-dependent regulation of claudin-1 is central to intestinal epithelial tight junction integrity. Mol Biol Cell. 2015;26:2252–2262. doi: 10.1091/mbc.E14-07-1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.El Aidy S, van Baarlen P, Derrien M, Lindenbergh-Kortleve DJ, Hooiveld G, Levenez F, Dore J, Dekker J, Samsom JN, Nieuwenhuis EE, Kleerebezem M. Temporal and spatial interplay of microbiota and intestinal mucosa drive establishment of immune homeostasis in conventionalized mice. Mucosal Immunol. 2012;5:567–579. doi: 10.1038/mi.2012.32. [DOI] [PubMed] [Google Scholar]
- 33.El Aidy S, Derrien M, Merrifield CA, Levenez F, Dore J, Boekschoten MV, Dekker J, Holmes E, Zoetendal EG, van Baarlen P, Claus SP, Kleerebezem M. Gut bacteria-host metabolic interplay during conventionalisation of the mouse germfree colon. ISME J. 2013;7:743–755. doi: 10.1038/ismej.2012.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.El Aidy S, Merrifield CA, Derrien M, van Baarlen P, Hooiveld G, Levenez F, Dore J, Dekker J, Holmes E, Claus SP, Reijngoud DJ, Kleerebezem M. The gut microbiota elicits a profound metabolic reorientation in the mouse jejunal mucosa during conventionalisation. Gut. 2013;62:1306–1314. doi: 10.1136/gutjnl-2011-301955. [DOI] [PubMed] [Google Scholar]
- 35.Garrett WS, Gordon JI, Glimcher LH. Homeostasis and inflammation in the intestine. Cell. 2010;140:859–870. doi: 10.1016/j.cell.2010.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Honda K, Littman DR. The microbiome in infectious disease and inflammation. Annu Rev Immunol. 2012;30:759–795. doi: 10.1146/annurev-immunol-020711-074937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McGuckin MA, Eri R, Simms LA, Florin TH, Radford-Smith G. Intestinal barrier dysfunction in inflammatory bowel diseases. Inflamm Bowel Dis. 2009;15:100–113. doi: 10.1002/ibd.20539. [DOI] [PubMed] [Google Scholar]
- 38.Ploger S, Stumpff F, Penner GB, Schulzke JD, Gabel G, Martens H, Shen Z, Gunzel D, Aschenbach JR. Microbial butyrate and its role for barrier function in the gastrointestinal tract. Ann N Y Acad Sci. 2012;1258:52–59. doi: 10.1111/j.1749-6632.2012.06553.x. [DOI] [PubMed] [Google Scholar]
- 39.Chang PV, Hao L, Offermanns S, Medzhitov R. The microbial metabolite butyrate regulates intestinal macrophage function via histone deacetylase inhibition. Proc Natl Acad Sci U S A. 2014;111:2247–2252. doi: 10.1073/pnas.1322269111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fraga MF, Herranz M, Espada J, Ballestar E, Paz MF, Ropero S, Erkek E, Bozdogan O, Peinado H, Niveleau A, Mao JH, Balmain A, Cano A, Esteller M. A mouse skin multistage carcinogenesis model reflects the aberrant DNA methylation patterns of human tumors. Cancer research. 2004;64:5527–5534. doi: 10.1158/0008-5472.CAN-03-4061. [DOI] [PubMed] [Google Scholar]
- 41.Glocker EO, Kotlarz D, Boztug K, Gertz EM, Schaffer AA, Noyan F, Perro M, Diestelhorst J, Allroth A, Murugan D, Hatscher N, Pfeifer D, Sykora KW, Sauer M, Kreipe H, Lacher M, Nustede R, Woellner C, Baumann U, Salzer U, Koletzko S, Shah N, Segal AW, Sauerbrey A, Buderus S, Snapper SB, Grimbacher B, Klein C. Inflammatory bowel disease and mutations affecting the interleukin-10 receptor. The New England journal of medicine. 2009;361:2033–2045. doi: 10.1056/NEJMoa0907206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lees CW, Barrett JC, Parkes M, Satsangi J. New IBD genetics: common pathways with other diseases. Gut. 2011;60:1739–1753. doi: 10.1136/gut.2009.199679. [DOI] [PubMed] [Google Scholar]
- 43.Xavier RJ, Podolsky DK. Unravelling the pathogenesis of inflammatory bowel disease. Nature. 2007;448:427–434. doi: 10.1038/nature06005. [DOI] [PubMed] [Google Scholar]
- 44.Waldecker M, Kautenburger T, Daumann H, Busch C, Schrenk D. Inhibition of histone-deacetylase activity by short-chain fatty acids and some polyphenol metabolites formed in the colon. The Journal of nutritional biochemistry. 2008;19:587–593. doi: 10.1016/j.jnutbio.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 45.Marshall S, Duong T, Wu T, Hering MA, Yada J, Higgins S, Orbus RJ, Yan ZH, Rumberger JM. Enhanced expression of uridine diphosphate-N-acetylglucosaminyl transferase (OGT) in a stable, tetracycline-inducible HeLa cell line using histone deacetylase inhibitors: kinetics of cytosolic OGT accumulation and nuclear translocation. Anal Biochem. 2003;319:304–313. doi: 10.1016/s0003-2697(03)00329-4. [DOI] [PubMed] [Google Scholar]
- 46.Colgan SP, Hershberg RM, Furuta GT, Blumberg RS. Ligation of intestinal epithelial CD1d induces bioactive IL-10: critical role of the cytoplasmic tail in autocrine signaling. Proc Natl Acad Sci U S A. 1999;96:13938–13943. doi: 10.1073/pnas.96.24.13938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nagalakshmi ML, Rascle A, Zurawski S, Menon S, de Waal Malefyt R. Interleukin-22 activates STAT3 and induces IL-10 by colon epithelial cells. International immunopharmacology. 2004;4:679–691. doi: 10.1016/j.intimp.2004.01.008. [DOI] [PubMed] [Google Scholar]
- 48.Olszak T, Neves JF, Dowds CM, Baker K, Glickman J, Davidson NO, Lin CS, Jobin C, Brand S, Sotlar K, Wada K, Katayama K, Nakajima A, Mizuguchi H, Kawasaki K, Nagata K, Muller W, Snapper SB, Schreiber S, Kaser A, Zeissig S, Blumberg RS. Protective mucosal immunity mediated by epithelial CD1d and IL-10. Nature. 2014;509:497–502. doi: 10.1038/nature13150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Demont A, Hacini-Rachinel F, Doucet-Ladeveze R, Ngom-Bru C, Mercenier A, Prioult G, Blanchard C. Live and heat-treated probiotics differently modulate IL10 mRNA stabilization and microRNA expression. J Allergy Clin Immunol. 2016;137:1264–1267. doi: 10.1016/j.jaci.2015.08.033. [DOI] [PubMed] [Google Scholar]
- 50.Moens F, Verce M, De Vuyst L. Lactate- and acetate-based cross-feeding interactions between selected strains of lactobacilli, bifidobacteria and colon bacteria in the presence of inulin-type fructans. Int J Food Microbiol. 2017;241:225–236. doi: 10.1016/j.ijfoodmicro.2016.10.019. [DOI] [PubMed] [Google Scholar]
- 51.Huang H, Liu JQ, Yu Y, Mo LH, Ge RT, Zhang HP, Liu ZG, Zheng PY, Yang PC. Regulation of TWIK-related potassium channel-1 (Trek1) restitutes intestinal epithelial barrier function. Cell Mol Immunol. 2016;13:110–118. doi: 10.1038/cmi.2014.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Anderson JM, Van Itallie CM. Physiology and function of the tight junction. Cold Spring Harb Perspect Biol. 2009;1:a002584. doi: 10.1101/cshperspect.a002584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Valenzano MC, DiGuilio K, Mercado J, Teter M, To J, Ferraro B, Mixson B, Manley I, Baker V, Moore BA, Wertheimer J, Mullin JM. Remodeling of Tight Junctions and Enhancement of Barrier Integrity of the CACO-2 Intestinal Epithelial Cell Layer by Micronutrients. PLoS One. 2015;10:e0133926. doi: 10.1371/journal.pone.0133926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Capaldo CT, Nusrat A. Claudin switching: Physiological plasticity of the Tight Junction. Semin Cell Dev Biol. 2015;42:22–29. doi: 10.1016/j.semcdb.2015.04.003. [DOI] [PubMed] [Google Scholar]
- 55.Leonel AJ, Teixeira LG, Oliveira RP, Santiago AF, Batista NV, Ferreira TR, Santos RC, Cardoso VN, Cara DC, Faria AM, Alvarez-Leite J. Antioxidative and immunomodulatory effects of tributyrin supplementation on experimental colitis. Br J Nutr. 2013;109:1396–1407. doi: 10.1017/S000711451200342X. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.