Abstract
Each day, approximately 830 women and 7,400 newborns die from complications during pregnancy and childbirth. Improving maternal and neonatal health will require bringing rapid diagnosis and treatment to the point of care in low-resource settings. However, to date there are few diagnostic tools available that can be used at the point of care to detect the leading causes of maternal and neonatal mortality in low-resource settings. Here we review both commercially available diagnostics and technologies that are currently in development to detect the leading causes of maternal and neonatal mortality, highlighting key gaps in development where innovative design could increase access to technology and enable rapid diagnosis at the bedside.
Graphical Abstract
We present diagnostic technologies available to detect the leading causes of maternal and neonatal mortality, highlighting key gaps in development.
INTRODUCTION
Two of the eight Millennium Development Goals (MDGs) adopted by world leaders to reduce global poverty, hunger, and disease focused on improving maternal health and reducing child mortality by the year 2015.1 Falling short of the goals, in 2015 the estimated rates for maternal and neonatal mortality were 216 deaths per 100,000 live births2 and 19 deaths per 1,000 live births,3 respectively. In other words, 830 women2 and 7,400 newborns3 die every day from complications in pregnancy, childbirth, and the postnatal period, amounting to an estimated 303,000 maternal deaths and 2.87 million newborn deaths per year with an additional 2.6 million stillbirths per year.4 The vast majority of these deaths occur in low-resource areas in Africa and Southeast Asia.2 To address the unmet MDGs, in 2015 the World Health Organization (WHO) developed the Sustainable Development Goals (SDGs), a set of 17 global goals to achieve by year 2030.5 The third SDG aims to ensure healthy lives across the globe and in part aims to decrease maternal mortality to less than 70 deaths per 100,000 live births and neonatal mortality to less than 12 deaths per 1,000 live births.5
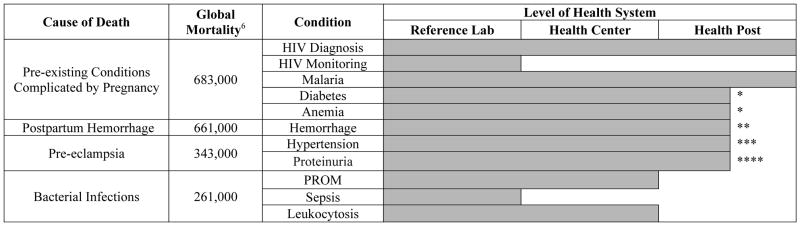
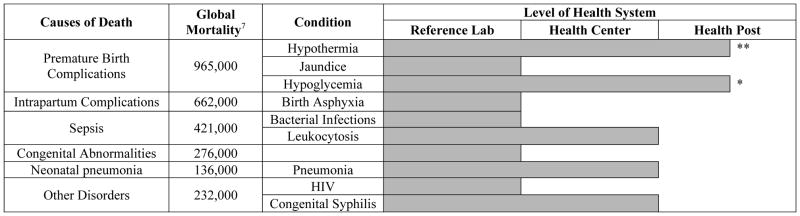
Achieving SDG three will require targeted investments to strengthen health centers in low-resource settings. In particular, there is an important need to enable rapid diagnosis and appropriate treatment of maternal and neonatal health conditions in low-resource health centers and health posts. Many of these facilities lack sophisticated laboratory infrastructure and do not have the resources to transport clinical specimens to central laboratories. Where available, point-of-care (POC) diagnostics can provide a solution to this challenge. However, as shown in Tables 1 and 2, only a limited number of POC diagnostic tools are available for use at health centers and health posts to detect the conditions that account for the majority of maternal and neonatal deaths.6,7 For many of these conditions, early detection and rapid initiation of treatment is key to reducing morbidity and mortality and achieving SDG three.8,9
Table 1.
Causes of maternal mortality globally with commercially available diagnostic tools. Global mortality column represents annual mortality rates. Level of health system indicates the level at which commercially available diagnostics can be deployed, taking into account the need for electrical power, refrigeration, consumable reagents, device and consumable costs, and necessary human resources for use.375
Available at health posts but limited by a lack of affordable consumables.
Technology exists for measuring blood loss but not for predicting those at risk.
Available at health posts but limited by a lack of human resources.
Available at health posts but limited by a lack of sensitivity.
Table 2.
Causes of neonatal mortality globally with commercially available diagnostic tools. Global mortality column represents annual mortality rates. Level of health system indicates the level at which commercially available diagnostics can be deployed, taking into account the need for electrical power, refrigeration, consumable reagents, device and consumable costs, and necessary human resources for use.375
Available at health posts but limited by a lack of affordable consumables.
Available at health posts but limited by a lack of human resources.
Currently available diagnostic tools often face barriers to implementation at the POC. Many diagnostic techniques can only be performed in laboratory facilities with access to constant power, water, and trained staff. For example, polymerase chain reaction, a standard method for diagnosing HIV in neonates, requires the use of expensive thermocycling equipment and highly trained technicians. Additionally, reagents used in many diagnostic tests have special storage or transportation requirements, such as cold transportation of antibodies used in ELISA testing to detect biomarkers of many diseases. Consumables, such as test strips or specialized cartridges, can be difficult to supply and lead to higher per-test costs. Instrumentation cost and associated maintenance costs also prevent some diagnostic technologies from being implemented in low-resource settings. The time-to-result associated with some tests limits their utility in both low- and high-resource settings. For example, bacterial culture is the gold standard to diagnosis sepsis, but the technique requires 24 to 48 hours to complete,10 preventing diagnosis-directed treatment during the effective treatment window.11 Finally, insufficient human resources can limit the efficacy of diagnostics for some conditions that require continuous monitoring, such as neonatal hypothermia. While low-cost thermometers exist to measure a neonate’s temperature, the human resources required for constant monitoring present a barrier. All of these barriers must be considered when developing a useful POC test for low-resource settings that can be appropriately implemented.
To address the shortcomings of existing diagnostics for low-resource settings, the WHO introduced a list of criteria for the ideal point-of-care test, known as ASSURED (Affordable, Sensitive, Specific, User-friendly, Robust & Rapid, Equipment-free, and Deliverable).12 Diagnostic tests targeted for use at the POC in low-resource settings should be designed with these criteria in mind to minimize barriers for successful implementation. However, there has been some criticism of the ASSURED criteria as being subjective and not sufficiently comprehensive for new technologies. Additionally, meeting all of the ASSURED criteria does not necessarily mean that a technology is appropriate for use at the POC. As Pai et al. stated in 2012, “…the technology as such does not define a POC test nor determine its use at the POC. Rather, it is the successful use at the POC that defines a diagnostic process as POC testing,”13 underscoring the importance of testing technologies at the POC and performing rigorous usability studies in the field. It is important to consider the final context in which a technology will be used during the design process and to determine how this context will affect the definitions used for and the relative importance of each of the ASSURED criteria. For example, the use of smartphones in POC diagnostics, which has previously been reviewed,14 has allowed for implementation of detection methods that previously required inaccessible equipment, changing our understanding of the criterion “equipment free”. Despite these shortcomings, the ASSURED criteria are frequently used in discussing the ability of a newly developed technology to be deployed at the POC.
When coupled with effective treatment strategies, low-cost POC diagnostics that can be administered in low-resource settings have the potential to reduce both neonatal and maternal mortality. Tables 3 and 4 summarize representative diagnostic technologies that are commercially available to meet maternal and neonatal health needs, respectively, highlighting a need to develop further commercialized technologies that reduce per-test cost, improve accuracy, and move away from reliance on power and benchtop analyzers.
Table 3.
Representative commercially available technologies available to diagnose the leading causes of maternal mortality. Notably, many of the listed technologies are lacking robust usability, scalability, and field performance studies.
| Condition | Representative Technology | Analyte Measured | Core Technology | Input Sample | Diagnostic Performance | Cost | Power* | Health System Use Level | Advantages | Limitations | Image |
|---|---|---|---|---|---|---|---|---|---|---|---|
| HIV Diagnosis | Alere Determine HIV-1/2 Ag/Ab Combo376,377 | HIV-1/2 antibodies and free HIV-1 p24 antigen | Lateral Flow | Whole Blood | Sensitivity: 99.9% Specificity: 98.3%–99.9% †376 |
$18.22 | None | Heatlh Post | Results in 20 minutes; approved for fingerstick use; excellent sensitivity | Unsuitable for acute infection and treatment monitoring |

|
| HIV Monitoring | Xpert HIV-1 Viral Load24,378,379 | HIV RNA | Automated MicrobiologyAnalyzer | Whole Blood | R2=0.92; limit of detection 40 cp/mL †**378 | $17,000 instrument $9.98/test | Mod | Reference Lab | Minimal hands-on time; 92-minute run time; high throughput; integrated sample preparation | High cost |

|
| Malaria | SD BIOLINE Malaria Ag Pf60,380,381 | Plasmodium falciparum histidine-rich protein II (PfHRP-II) | Lateral Flow | Whole Blood | Sensitivity: 95% Specificity: 97.1% † 60 |
$0.70 | None | Health Post | Low-cost; minimal training requirements; results in 30 minutes; excellect sensitivity | Counterfeit tests on market; limit-of-detection unsuitable for submicroscopic infections |

|
| Diabetes | LifeScan OneTouch Ultra 298,382 | Glucose | Lateral Flow with Reader | Whole Blood | AUC: 0.778 95% CI: 0.726–0.829 † 97 | $19.99 instrument $1.45–1.60/test | Low | Health Post | Widely available; integrated sample processing | High per-test cost |

|
| Anemia | HemoCue383–385 | Hemoglobin | Microfluidics | Whole Blood | R2=0.85–0.995 AUC: 0.96† 383 |
$449 instrument $1.53–$1.86/test | Low | Health Post | Integrated sample processing | High per-test cost |

|
| Hemorrhage | BRASS-V drape146,148,386 | Blood Loss | Blood Collection and Volume Measurement | Whole Blood | R=0.928 †† 148 | $4/test | None | Health Post | Accurate; minimal training requirements | High per-test cost |

|
| Pre-eclampsia | Alere Triage PlGF185,186,387 | Placental Growth Factor | Lateral Flow with Reader | Plasma | Sensitivity: 77% Specificity: 95% † 186 | $1750 instrument $50/test |
Mod | Health Center | Results in 20 minutes | High per test cost; requires plasma separation from whole blood |
|
| Bacterial Infection | Blood Culture388–391 | Bacterial Load | Laboratory Microbiology | Sterile Whole Blood | Sensitivity: 73% † 388 | $28 | High | Reference Lab | Prone to contamination and false positives |
|
|
| PROM | Amnisure ROM205,219,392 | Placental alpha-microglobulin 1 (PAMG-1) | Lateral Flow | Vaginal Swab | Sensitivity: 96.8% Specificity: 98.3% † 205 |
$60/test | None | Health Center | Excellent sensitivity, minimal training requirements, results in 10 minutes | High per test cost, blood contamination can give false positives |
|
Power can be battery based, with low indicating small, easily replaceable batteries, moderate indicating larger, more expensive batteries, and high indicating dependency on electrical power grid.
Reported diagnostic performance is idenpendently verified.
Diagnostic performance is manufacturer reported.
Reported diagnostic performance was achieved using plasma samples.
HIV Diagnosis image provided courtesy Alere Inc.
HIV Monitoring image provided courtesy of Cepheid.
Malaria image provided courtesy Alere Inc.
Hemorrhage image reproduced from Reference 386 under the terms of Creative Commons Attribution License.
Pre-eclampsia image provided courtesy Alere Inc.
Bacterial Infection image reproduced from Reference 391 under the terms of Creative Commons Attribution License.
PROM image provided courtesy of Qiagen.
Table 4.
Representative commercially available technologies available to diagnose the leading causes of neonatal mortality. Notably, many of the listed technologies are lacking robust usability, scalability, and field performance studies.
| Condition | Representative Technology | Analyte Measured | Core Technology | Sample Purity | Diagnostic Performance | Cost | Power* | Health System Use Level | Advantages | Limitations | Image |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Hypothermia | ThermoSpot252,254 | Body Temperature | Trans-cutaneous Sensor | N/A | Sensitivity: 88% Specificity: 97% † 252 |
$0.11 | None | Health Post | Low cost; minimal training requirements; noninvasive | Can fall off of neonate; only provides temperature range |

|
| Jaundice | BiliChek393–395 | Bilirubin | Trans-cutaneous Optical Sensor | N/A | R2=0.86 When TSB > 14 mg/dL: AUC: 0.95 95% CI: 0.92–0.97 † 393 |
$3,900 instrument $7/test | Low | Health Center | Noninvasive | High cost; may require additional confirmatory lab tests |

|
| Hypoglycemia | Nova StatStrip276,396 | Glucose | Lateral Flow with Reader | Whole Blood | Sensitivity: 90–100% Specificity: 97.7–99.4% † 276 | $250 instrument $0.30/test | Low | Health Post | Accurate with neonatal samples; integrated sample preparation | Higher instrument cost than other glucometers |

|
| Birth Asphyxia | Moyo301,397 | Fetal Heart Rate | Trans-cutaneous Doppler Sensor | N/A | Accuracy: +/−5 bpm in the range of 50–200 bpm†† 301 | $198 | Low | Heatlh Center | Noninvasive; portable | Not as sensitive for asphyxia as gold standard |

|
| Sepsis | Blood Culture388–391 | Bacterial Load | Laboratory Microbiology | Sterile Whole Blood | Sensitivity: 73% † 388 | $28 | High | Reference Lab | Prone to contamination and false positives |

|
|
| HIV | Alere™ q HIV-1/2 Detect328,398 | HIV RNA | Automated Microbiology Analyzer | Whole Blood | Sensitivity: 99.3%, Specificity: 100.0% † 328 | $25,000 instrument, $25.00/test | Mod | Reference Lab | High diagnostic performance; integrated sample preparation | Maximum of 8 samples per day, high cost |

|
| Congenital Syphilis | SD Bioline Syphilis 3.0344,399,400 | Antibodies to Treponema pallidum | Lateral Flow | Whole Blood | Sensitivity: 84.5% Specificity: 97.9% † 344 |
$0.90 | Low | Health Post | Low-cost; minimal training requirements, results in 20 minutes | Only used to diagnose mother during pregnancy |
|
| Pneumonia | Counting Beads321 | Respiratory Rate | Clinical Observation | N/A | Sensitivity for fast breathing: 68% † 321 | Provided by International Organizations | Low | Heatlh Post | Power-based equipment-free; minimal training requirements | Requires training and significant time by staff |

|
Power can be battery based, with low indicating small, easily replaceable batteries, moderate indicating larger, more expensive batteries, and high indicating dependency on electrical power grid.
Reported diagnostic performance is idenpendently verified.
Diagnostic performance is manufacturer reported.
Hypothermia image reproduced from Reference 254 under the terms of Creative Commons Attribution License.
TBS: Total Serum Bilirubin
Jaundice provided courtesy of Philips.
Hypoglycemia image provided courtesy of Nova Biomedical.
Birth Asphyxia figure from Laerdal Global Hleath AS. All rights reserved.
Sepsis image reproduced from Reference 391 under the terms of Creative Commons Attribution License.
HIV image provided courtesy Alere Inc.
Congenital Syphilis image provided courtesy Alere Inc.
Pneumonia image reproduced from Reference 321 under the terms of Creative Commons Attribution License.
Here, we review key innovations in POC diagnostic tools to detect the leading causes of maternal and neonatal death in low-resource settings. We review both commercially available technologies and technologies that are currently in development. We begin by reviewing diagnostic formats, including types of biomarkers detected in many diagnostic tests. We then focus on technologies that are available or in development to address maternal and neonatal health needs. The included figures illustrate the form and/or function of selected, representative diagnostic technologies. Finally, we discuss key unmet needs in maternal and neonatal health where further innovation in POC diagnostics is desired.
DIAGNOSTIC FORMAT
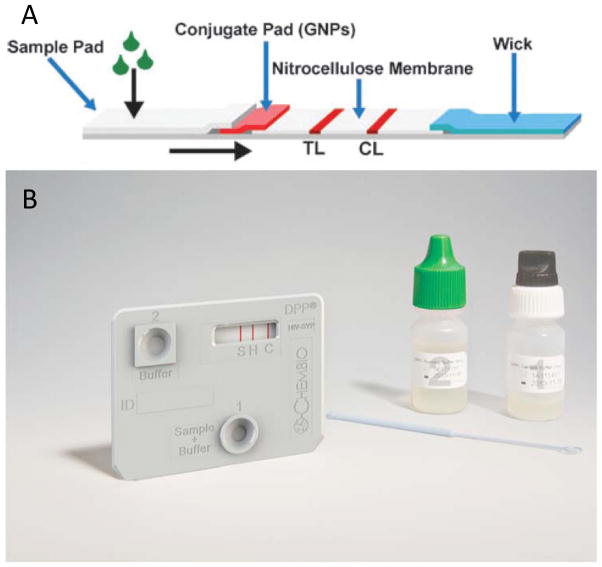
In high-resource settings, diagnostic tests are typically performed in laboratory settings by highly-trained technicians using expensive equipment. Because of this, field-appropriate benchtop analyzers have been developed to miniaturize and simplify some of the technologies found in lab instruments. Although these devices are not suitable for bedside use, they can be used in some low-resource settings that have consistent power and lab technicians. To detect biomarkers quickly and accurately at the bedside, equipment-free POC tests have been developed; one of the most common formats for these tests is the lateral flow assay (LFA), sometimes referred to as a dipstick test15 (Figure 1). While the per-test cost of LFAs is often higher than that of high-throughput laboratory instruments, the lack of instrumentation cost makes these tests extremely attractive for use in low-resource settings15,16. LFAs generally meet all of the ASSURED criteria, and are appropriate for use at health centers.15 However, one of the challenges of developing LFAs is in achieving clinically relevant sensitivity and specificity.
Figure 1.
Some examples of lateral flow tests to diagnose neonatal and maternal conditions. (a) Generalized depiction of a lateral flow device with a sample pad, conjugate pad, nitrocellulose membrane, and wicking pad. Figure reproduced from Reference 401 with the permission of The Royal Society of Chemistry.401 (b) The ChemBio DPP HIV-Syphilis Assay detects antibodies for both syphilis (first “S” line) and HIV (second “H” line) from a drop of whole blood. The patented Dual Path Platform enables separate delivery of sample and detection reagents and improves sensitivity. Figure from Chembio Diagnostic Systems, Inc. All rights reserved.
Choice of biomarker can greatly affect sensitivity and specificity of a test; therefore, biomarker selection is one of the most important considerations in the development of any diagnostic. Proteomics, metabolomics, and genomics research identifies biomarkers associated with various maternal and neonatal diseases, and verified disease biomarkers can be targeted in point-of-care diagnostics.17 Here, we briefly introduce the most commonly targeted biomarkers for diagnostics, including patient antibodies, other proteins and small molecules, and pathogen nucleic acid before discussing diagnostic technologies available for maternal and neonatal health in low-resource settings.
Antibody Detection
In response to a pathogenic threat, IgM antibodies are produced quickly, while IgG antibodies are produced later and circulate in the bloodstream longer.18,19 Most antibody-based diagnostics use recombinant proteins to detect both types of antibodies in a patient sample in order to estimate infection duration and level of exposure.18,19 However, IgG antibodies are inappropriate for detection of many neonatal conditions because maternal antibodies can be transferred to the fetus during pregnancy and persist for 12–18 months after birth.20–22 IgM antibodies can detect acute infection in neonates.23 Additionally, some antibody-based tests are unsuitable for determining treatment efficacy, as antibody levels remain elevated longer than other biomarkers.18,19
Protein and Small Molecule Biomarker Detection
Many proteins and small molecules already present in bodily fluids deviate from normal physiological levels in response to disease. Therefore, protein- and small molecule-based tests need to be optimized to detect biomarker levels that differentiate between normal and diseased states. Analyte selection is important to consider because the choice of a biomarker that is modulated in multiple conditions may result in poor diagnostic specificity for a given condition. Proteins and small molecules are often present at concentrations that can be detected without the need for amplification.15
Nucleic Acid Detection
For conditions that have low pathogen concentrations, require viral load measurement, or need high specificity against related viruses, nucleic acid testing is the standard of care. Nucleic acid tests (NATs) detect specific encoded sequences within the genetic material of pathogens. NATs may detect and amplify very few copies of the target nucleic acid, making them highly sensitive and specific. Unlike antibody tests, NATs can detect a pathogen as soon as it is present. NATs typically accomplish three goals: sample preparation, amplification of target, and detection of amplicon. Real-time PCR has allowed amplification and detection to be performed simultaneously, and several completely automated platforms have been developed for use at the POC.24
Sample Preparation
Sample preparation can be a challenge in detecting any biomarker, but it provides a significant challenge for NATs, as nucleic acid amplification is inhibited by components of bodily fluids.25,26 Sample preparation for POC tests typically consists of collection, separation, extraction, and concentration of nucleic acids. Commercialized paper-based sample collection and plasma separation technologies are commonly used for sample collection.27 Novel approaches to sample preparation, including microfluidic separation techniques28,29 and extraction and concentration techniques,29 have been previously reviewed. However, challenges remain before unprocessed blood or other clinical samples can be applied directly to amplification and detection assays at the POC.
Amplification of target
The standard method for amplification of genomic material is polymerase chain reaction (PCR), which requires expensive thermal cycling equipment, reliable electricity, trained personnel to prepare the reactions, and several hours to complete. To perform amplification at the POC, promising technologies employ isothermal enzymatic amplification methods that require a shorter incubation period and eliminate the need for thermal cycling.30
Detection of amplicon
Nucleic acid amplicons may be detected either during the amplification process (real-time detection) or at the end of the reaction (endpoint detection). The presence of amplified nucleic acids is measured via an optical or electrical signal. Real-time detection uses fluorescently labeled probes to bind to the amplicon, and instrumentation to quantify the signal.31 For end-point detection, amplification products may be tagged with reporter molecules to create a visible signal indicating the presence of the amplicon and be detected in equipment-free formats.31 Real-time detection techniques are commonly used in quantitative assays and are highly sensitive and specific, while end-point detection typically requires fewer resources and may suffice for less sensitive applications.24
The biomarker types and detection schemes discussed above each have advantages and disadvantages, and the most appropriate test for use at the POC depends on the condition and available infrastructure. We now discuss in detail technologies for each leading cause of maternal and neonatal mortality as well as areas for potential diagnostic development.
DIAGNOSTICS FOR MATERNAL HEALTH
Pre-existing Conditions Complicated by Pregnancy
HIV
HIV is a virus that attacks the body’s immune system, most notably CD4+ white blood cells that fight off infections. In 2014, an estimated 1.5 million women living with HIV gave birth, and HIV-positive women are 6–8 times more likely to die giving birth than those who are HIV negative.32 POC devices that make maternal HIV diagnosis as rapid and simple as possible are of high importance and have been the target of much funding and research over the past 20 years.33 With effective diagnosis, women with HIV can receive appropriate prophylactic strategies to reduce viral load, both for their health and the health of the neonate. Ninety percent of HIV infections in children are due to mother-to-child transmission.32 However, the prevention of mother-to-child-transmission (PMTCT) of HIV is one of the great public health successes of the past 20 years.33 The vertical transmission rate for HIV-positive pregnant women with no intervention is 25–42%, but this rate has been reduced to 1% or less when all prophylactic strategies are implemented.33 Precautions including antiretroviral therapy (ART) during pregnancy, labor and delivery, and the postnatal period to the infant, as well as elective cesarean delivery, have contributed to this success.
HIV is now widely diagnosed in adults using affordable antibody dipstick tests. However, antibody concentrations reach a peak concentration during the early stage of HIV infection and decrease after acute infection.34 Thus, antibody/antigen tests are useful for screening but not treatment monitoring. To diagnose and confirm treatment failure, the WHO strongly recommends viral load monitoring.35 Current guidelines encourage viral load testing at six and 12 months after initiating ART, and every 12 months thereafter if the patient is stable on ART. Virological failure is defined by two consecutive viral load measurements exceeding 1000 copies/mL within a 3-month interval, with adherence support between measurements, after at least six months of a new ART regimen. Despite these recommendations, where viral load testing is not available, CD4+ count and clinical symptoms are still used to diagnose treatment failure.24 Here we discuss antibody/antigen tests as well as CD4+ testing for HIV; solutions for point-of-care viral load testing are discussed in the Neonatal HIV section.
Antibody Detection
Antibody assays to diagnose HIV infection in adults detect antibodies against the HIV viral envelope proteins gp41 and gp36. Developed tests include the ChemBio sure check HIV 1/2,36 the UniGold Recombigen HIV 1/2,37 the VIKIA HIV 1/2,38 the OraQuick in-home test,39 the INSTI,40 and the Alere Determine HIV 1/2 Ab + Ag.41 The mChip is a microfluidic ELISA that detects antibodies against HIV viral envelope proteins as well as syphilis.42 While first-generation HIV tests were able to detect antibodies about a month after infection, second- and third-generation tests improved the sensitivity to allow earlier detection. Fourth-generation tests now screen for the p24 antigen in addition to antibodies, allowing detection as soon as 18 days after infection, although the p24 antigen portion has shown variable performance thus far.43 Antibody/antigen tests are the easiest and most affordable option for HIV diagnosis and have seen success with use in clinical settings. These technologies meet ASSURED criteria and are available in many low-resource settings.
In late 2016, the WHO issued a strong recommendation that HIV self-testing should be offered as an additional approach to HIV testing services.44 This recommendation was based on studies indicating that compared to standard HIV testing, HIV self-testing can result in identifying an equivalent or greater proportion of HIV-positive people. As of July 2017, there is one WHO prequalified HIV self-test, the OraQuick® HIV Self-Test (OraSure Technologies Inc., USA).45 An additional four HIV self-testing products are on the market and have been registered and approved by a founding member of the Global Harmonization Task Force: the autotest VIH® (AAZ Labs, France), the BioSURE HIV Self Test (BioSURE, United Kingdom), the OraQuick® In-Home HIV Test (OraSure Technologies Inc., USA), and the INSTI HIV Self Test (bioLytical Laboratories, Canada).45 The per-test cost of these self-tests vary widely based on implementation setting and change rapidly; for example, the WHO prequalified OraQuick® HIV Self-Test has a retail price of $9.50 in low and middle-income countries, but a June 2017 agreement between OraSure and the Bill & Melinda Gates Foundation will offer the test to public-sector buyers in 50 countries for $2.00. Retail prices for the other four products on the market currently range from $22–48 per test, though several are also available in select instances at under $10. Finally, four HIV self-tests are available in some private-sector markets, while ten self-tests that use whole blood, oral fluid, or urine are currently in the pipeline.45
Viral Load Testing
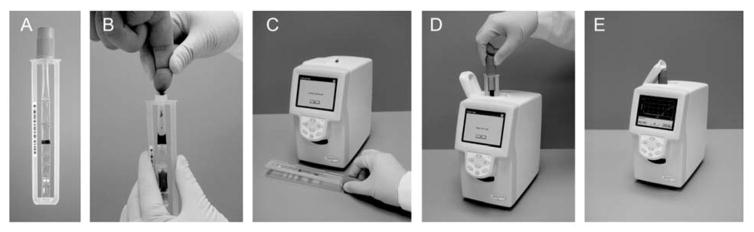
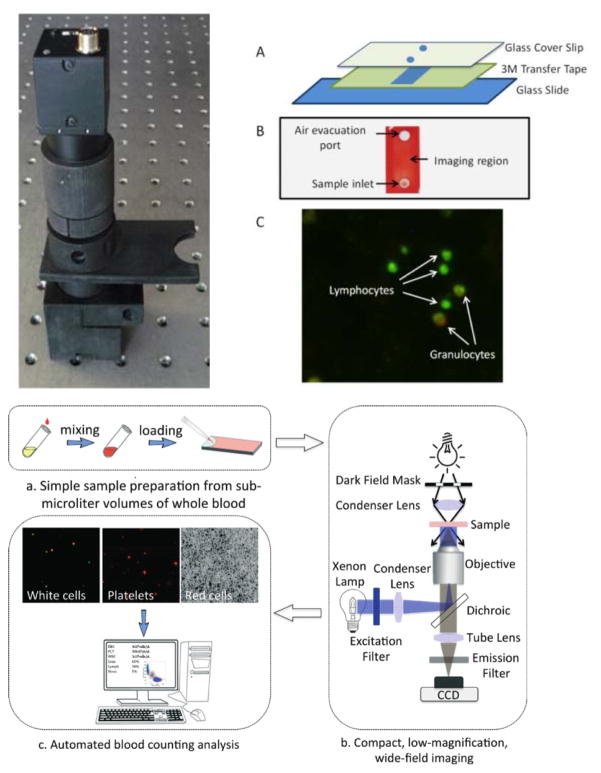
Several sample-to-answer NATs are on the market and in the pipeline for viral load testing. The SAMBA II distinguishes between viral loads above and below 1000 copies/mL in 90 minutes. In a study conducted in London, Malawi, and Uganda, the SAMBA semi-Q was 97.3% concordant with the gold standard test, the Roche TaqMan v2.46 Although easy to use, the system requires electricity and benchtop equipment, limiting its use to settings with significant infrastructure. In addition, it is only used for monitoring and not for diagnosis and is not currently able to detect HIV-2. Figure 5 illustrates the ease-of-use afforded by a rapid sample-to-answer system, including sample collection, automated analysis, and reporting of results. The LIAT Analyzer (Roche), shown in Figure 5, is a portable reverse-transcriptase PCR (RT-PCR) system that uses whole blood as a sample. Recent studies have validated a very low limit of detection of 57 copies/mL.47 The system detects multiple HIV subtypes as well as HIV-2. However, sample preprocessing is required. Another technology that is highly effective but still costly is the Gene Xpert HIV-1 Viral Load (Cepheid), a NAT that detects HIV-1 viral load in a less than two hours with only one minute of hands-on time. While this test provides quick results and requires minimal training to run, its unit cost of $17,000 USD and POC per-test cost of $9.98 limit its utility.48 Other viral load tests currently in the pipeline include the EOSCAPE-HIV Rapid RNA Assay (Wave 80 Biosciences), the TrueLab Real Time micro PCR (MolBio Diagnostics), the Bioluminescent Assay in Real Time technology (Lumora), the ExaVir Load, and the NWGHF Savanna HIV VL test, though none of these have yet been validated.49
Figure 5.
The complete workflow of the Liat Analyzer. The Liat Analyzer is an example of an automated sample-to-answer NAT platform that performs nucleic acid extraction, purification, reverse transcription, PCR amplification and real-time detection. A sample such as whole blood (shown) or plasma, is collected directly into a Liat Tube (a and b). After the tube is capped, the analyzer scans the tube barcode (c), and the tube is inserted into the analyzer (d). Then, the analyzer automatically performs all the nucleic acid testing steps and reports results in 1 hour (e). The mechanism for measurement and assay components are not depicted here. Reproduced from S. Tanriverdi, L. Chen and S. Chen, “A Rapid and Automated Sample-to-Result HIV Load Test for Near-Patient Application.” J. Infect. Dis., 2010, 201(s1), S52–S58, by permission of Oxford University Press.47
CD4 Count
Although viral load testing has been shown to be more accurate at indicating therapy failure than CD4+ testing, many low-resource environments still rely on CD4+ lymphocyte counting to monitor HIV treatment efficacy.50 Similar to antibody detection, a CD4+ lymphocyte count indicates the state of an HIV patient’s immune system. A low CD4+ count indicates a severe level of infection and correlates with poor clinical outcomes in the first year of infection. However, this amount can be affected by other conditions, and thus cannot be used as a direct measure of HIV. Additionally, baseline CD4+ counts have not been shown to be indicative of mortality after 5 years of treatment.51 However, CD4+ counts are still used as a clinical measure of immune system function and progression to AIDS, especially in areas that do not have access to viral load testing.
Commercially available and in-the-pipeline CD4 testing platforms vary widely in terms of their throughput, infrastructure requirements, and cost. Although there is no official gold standard for CD4 counting, many experts consider the high-throughput BD FACSCalibur™ (BD Biosciences) system to be a suitable reference. This platform uses flow cytometry, a technology able to take quantitative measurements of multiple features of large numbers of cells.24). The instrumentation for this platform costs $75,000, and thus is only suitable for central and national reference laboratories. The most widely used platform in low-resource settings is the BD FACSCount™ system (BD Biosciences). The FACSCount™ uses a whole blood sample, and can calculate both CD4 count and CD4 percentage, which is useful for young children. This system has been used for over a decade, and its performance has been well-validated by several independent studies.52,53 With a $30,000 instrumentation cost and a per-test cost of $3.50–10.00, this system is well suited to district hospital settings. Two additional and similar medium-throughput CD4 platforms are the Aquios CL™ (Beckman Coulter) and the Apogee Auto40 Flow Cytometer (Apogee).
The flow cytometers described above are not suitable for true POC testing due to high costs and staffing requirements. Thus, other methods of CD4 counting have been developed, and several less expensive technologies for low-throughput POC CD4 testing are on the market. The imaging-based Pima™ Analyser (Alere) introduces fluorescently labeled antibodies to a sample, then acquires and analyzes fluorescence images in an imaging chamber. This system is able to service only three tests per hour, but is suitable for all levels of healthcare and has been shown to positively impact patient retention and ART initiation.54 The CyFlow® CD4 miniPOC (Sysmex Partec) is an alernative imaging-based CD4 platform that is compact, rugged, and can be operated with a battery pack for up to 5 hours. Finally, the BD FACSPresto™ (BD Biosciences) is another imaging-based technology that has been WHO prequalified since 2014. This instrument costs less than $10,000 and each test costs less than $10, making the FACSPresto™ a more affordable option than flow cytometry for some resource-limited settings.
While the CyFlow® CD4 miniPOC can be powered temporarily with a battery pack, all of the fully quantitative CD4 tests described above currently require continuous electricity, and costs remain high.55 Large CD4 equipment also suffers from high infrastructure requirements, while smaller analyzers are limited by low throughput. For example, while a Pima™ CD4 test takes only 20 minutes, the system can only process one sample at a time, leading to long waits for patients.56,57 The only rapid, disposable, and equipment-free CD4 test currently available is the Visitect® CD4, a semiquantitative test developed by the Burnet Institute and currently licensed to Omega Diagnostics Ltd (United Kingdom). Visitect® CD4 measures the amount of CD4 protein on T-cells rather than directly measuring CD4 cells. The Visitect® CD4 uses a simple lateral flow device to capture the CD4 protein on T-cells rather than directly measuring CD4 cells, and shows a readout that indicates whether a patient’s fingerprick sample contains lymphocytes above or below a threshold of 350 cells/uL.57 An automatic battery-powered reader interprets results in less than 40 minutes, and Omega Diagnostics has also developed a smartphone application for the assay.
Finally, detection of cells in a microfluidic channel has been difficult due to the high shear stress generated in flow-based ELISA, but alternative microfluidic platforms for CD4 testing are being studied in several academic groups. One approach moves captured CD4+ lymphocytes through different aqueous phases as opposed to moving the aqueous phases across the substrate, allowing cells to be captured without experiencing high shear stress.58 It also integrates image processing using a smartphone, lowering the equipment requirements of the system. A second novel strategy carefully optimizes shear stress for lymphocyte capture, and employs only electrical methods to interrogate whole blood samples. This eliminates the need for image-based detection and makes the system more robust to environmental challenges.59 This microfluidic device was shown to be at least as accurate as the Pima CD4, and with modifications to the capture chamber, could provide over twice the testing throughput of the Pima CD4.
As of 2016, the WHO supports stopping routine CD4 count testing where viral load testing is available, limiting the use of these tests to prioritizing patients for urgent linkage to care.35 Further, ART initiation is now recommended for all adult and adolescent patients regardless of CD4 cell counts and disease stage. These recommendations indicate a decreasing importance of CD4 testing and a shift towards viral load testing as the central focus of HIV care. Accordingly, a number of CD4 technologies that were on the market or in the pipeline within the last five years have been discontinued. These include the Daktari™ CD4 Counter (Daktari Diagnostics Inc.), the MBio CD4 System (MBio Diagnostics Inc.), and the CD4 Test (Zyomyx Inc.). Daktari appears to have shifted its focus towards development of a viral load test in accordance with market shift, MBio Diagnostics is no longer marketing their CD4 system, and Zyomyx Inc. appears to no longer be in business.24 Despite this shift in priorities, CD4+ cell count remains the best indicator of a patient’s immune and clinical status, risk of opportunistic infections, and supporting the management of patients with advanced HIV disease. Furthermore, viral load testing for treatment monitoring is still inaccessible to a large portion of those affected by HIV. Therefore, innovative and cost-effective POC solutions for CD4 testing are still needed.
Malaria
Malaria, a mosquito-borne parasitic disease, affects between 150 and 300 million people per year.60 While malaria infection typically leads to flu-like symptoms, pregnancy increases susceptibility to infection and severity of disease.61,62 Complications from malaria, such as anemia, hypertension, and low birthweight, cause 10,000 maternal deaths and 200,000 neonatal deaths in Africa annually.62 In the absence of pregnancy, malaria is typically detected with blood smear microscopy or LFAs. Blood smear microscopy is a widely used method of diagnosing malaria in which trained technicians look for parasites within a blood sample using a microscope. Benefits include the ability to detect multiple species of parasite along with parasitemia, or level of infection, although appropriate training and staffing is required. Additionally, many malaria LFAs available on the market are appropriate for use at a bedside or rural health center, and over 314 million rapid tests were sold in 2014.60 Some LFAs detect only Plasmodium falciparum, the parasite species with the most severe outcomes, while other LFAs detect multiple species including P. vivax, P. malariae, and P. ovale. The WHO selection criteria for malaria LFAs include a sensitivity of 75% at 200 parasites/μL in all transmission settings, a false positive rate less than 10%, and an invalid rate less than 5%.63 The product should also be thermally stable, able to easily store, and easy-to-use. A list of reviewed LFAs that meet these criteria was recently published by the WHO.63–66
However, these tests may perform differently in cases of malaria in pregnancy. Malaria caused by Plasmodium falciparum is more difficult to diagnose during pregnancy because the parasites sequester in the placenta and therefore can have low concentrations in peripheral maternal blood.62 The number of parasites in the blood varies depending on treatment with antimalarial drugs, whether it is a mother’s first pregnancy, and other complicating factors.61,62 It is unclear from recent studies whether low parasite concentrations are associated with adverse maternal or neonatal outcomes.62,67–73 If future studies indicate little correlation between submicroscopic infections and adverse outcomes, then commonly used malaria diagnostics like blood smear microscopy or LFAs could be used for testing pregnant women at the POC. However, if future studies do show a correlation between submicroscopic infections (< 50 parasites/μL) and adverse outcomes, more complex and sensitive diagnostics will be needed to diagnose malaria in pregnancy.62,74,75
To detect submicroscopic malaria infections, several commercially available or in-the-pipeline NATs have been previously reviewed, although not validated for malaria in pregnancy.76–82 The Nanomal (QuantuMDx), Accutas (Aquila Diagnostic Systems Inc.), and DiscoGnosis LabDisk system (IMTEK) all require minimal infrastructure and sample preparation and can be used at the POC. However, cost may be prohibitive for low-resource settings, with the lowest estimated per-test cost at $2–$4 and high equipment costs.82 Other promising inexpensive malaria NATs are being developed which integrate sample preparation to reduce potential contamination, do not require cold chain storage, and have limits of detection appropriate for highly sensitive testing.76,79,81 In addition, two commercially available imaging-based tests, the Rapid Assessment of Malaria (RAM) Device (Disease Diagnostic Group Inc.) and Magneto-optical Device (MOD) (Meditopian LLC), detect submicroscopic infections in under one minute for less than $1 per test, with minimal sample preparation and rechargeable batteries to reduce infrastructure requiremnents, although the equipment cost for RAM is high.82 Finally, a few highly sensitive protein-based methods for submicroscopic malaria detection have also been reviewed.76,82–85 These tests are low-cost, easy-to-use, and do not require electricity to run, making them suitable for use at the POC. While several of the above tests may be appropriate for submicroscopic detection in low-resource settings, their diagnostic performance for malaria in pregnancy has yet to be fully validated.
Gestational Diabetes
Diabetes is a metabolic disease in which a patient experiences prolonged periods with high blood sugar levels. Diabetes developed over the course of a pregnancy is known as gestational diabetes. Gestational diabetes affects an estimated 10–25% of all pregnancies globally86,87 and can lead to serious maternal and neonatal health consequences, including pre-eclampsia, infections, obstructed labor, postpartum, hemorrhage, preterm births, stillbirths, congenital anomalies, birth injuries, and death.88,89
The WHO criteria for gestational diabetes diagnosis includes one or more of the following: fasting plasma glucose between 92–125 mg/dL, venous plasma glucose one hour after ingestion of 75 g oral glucose load above 180 mg/dL, or venous plasma glucose two hours after ingestion of 75 g oral glucose load between 153–199 mg/dL.86 The use of this diagnostic criteria requires the patient to present for the test after fasting overnight and to stay for 1–2 hours after ingesting glucose, a process known as an oral glucose tolerance test. Bhavadharini et al. report one of the greatest patient-related barriers to screening and diagnosis of gestational diabetes in low- and middle-income countries to be patients coming for checkups in the fasting state.90
Diabetes can alternatively be diagnosed through the measurement of glycated hemoglobin (HbA1C), a measure of the 3-month average glucose concentration in the patient’s blood. HbA1C testing circumvents the need for an oral glucose tolerance test. However, the utility of HbA1C tests in screening for gestational diabetes is still being investigated, as HbA1C levels are higher during pregnancy. Recent studies have shown very low sensitivity (7–81% depending on chosen cutoff value) in HbA1C tests for gestational diabetes screening and suggest confirmatory screenings with oral glucose tolerance tests.91–93 HbA1C tests are currently not readily available worldwide, are unaffordable in low-and middle-income countries, and have the potential to be adversely affected by hemoglobinopathies.94 Because of the limitations of HbA1C testing for gestational diabetes, only direct glucose measurement is discussed here.
Gold standard glucose tests are run on clinical chemistry analyzers and test the glucose in plasma. Plasma glucose tests generally require centrifugation to separate red blood cells from plasma due to glycolysis by red blood cells, a process that rapidly degrades glucose in a blood sample. However, plasma separation by centrifugation is not always available in resource-limited settings. A more detailed discussion of low-resource centrifugation methods can be found in the Hematocrit section.
Glucometers intended for self-monitoring of blood glucose (SMBG) are considered one of the founding technologies of the POC testing era,95 and SMBG is the largest market segment of POC testing.96 POC glucometers have been shown to be effective for gestational diabetes screening when venous plasma glucose measurements are not available.97 Generally, POC glucometers generate an enzymatic reaction with glucose and measure the output through photometric or amperometric detection. Commonly used enzymes include glucose oxidase (GOX) and glucose-1-dehydrogenase (GDH), and GDH modified with pyrroloquinoline quinone (GDH-PQQ) has been used recently as well. More comprehensive descriptions of the enzymatic reactions and detection methods have previously been described,98 and multiple groups have previously reviewed commercialized glucometers.96,98
While glucometers are relatively inexpensive and testing requires no sample preparation, the cost of compatible test strips can be prohibitive in low-resource settings (approximately $1.50 per test).99,100 Additional limitations of commercially available glucometers include designs that are not robust to humidity and temperature fluctuations101 as well as inaccurate conversions from whole blood glucose values to plasma glucose values.86 Conversion to plasma glucose values is required for comparison to diagnostic cutoffs; many glucometers perform this conversion by increasing the whole blood glucose value by roughly 11%, though varying hematocrit levels will determine the exact patient-specific conversion. Accurate plasma correlation remains a challenge.102
Several reviews discuss recent glucose sensing innovations that attempt to circumvent issues faced by commercially available POC glucometers. Approaches include electrochemical detection,103 sensors based on carbon nanomaterials104 and nanostructured metal-oxides,105 nonenzymatic sensors,106 non-invasive monitoring technology,107,108 and emerging technology more generally.109
Anemia
Anemia, a condition characterized by insufficient hemoglobin leading to diminished oxygen carrying capacity in the blood, affects an estimated 32 million pregnant women worldwide.110 Severe anemia is strongly associated with maternal mortality, and progress toward decreasing morbidity and mortality associated with maternal anemia has been slow over the last 20 years.111 Anemia can be diagnosed by measuring the amount of hemoglobin, the oxygen-carrying protein in red blood cells (RBCs), or hematocrit, the fraction of RBCs in the blood. RBCs are often counted as part of a complete blood count (CBC). Hemoglobin concentration, hematocrit, and RBC counts are generally correlated, though the relationship may be altered in the presence of some hematological disorders, including hemoglobinopathies.112 An important consideration for developing blood count tests is the variability of hemoglobin, WBCs, and platelets observed from one fingerprick drop to the next; drop volumes used should be sufficiently large for clinical correlation to well-mixed venous blood.113 For more information on blood counts, see the section on Bacterial Infections and Puerperal Sepsis.
Hemoglobin and hematocrit measures can be integrated into diagnostic tests for additional analytes in order to increase accuracy or to provide additional relevant diagnostic information. For example, as described in the Gestational Diabetes section, glucometers often convert whole blood glucose values to plasma glucose values inaccurately; direct hematocrit measurement incorporated into whole blood glucose measurement can allow for more accurate plasma glucose reporting. Additionally, when clinically relevant, anemia diagnostics can be multiplexed with detection of additional analytes, such as HIV antibodies as demonstrated by Guo et al.114 Here, we discuss a variety of approaches toward hemoglobin and hematocrit detection.
Hemoglobin
The gold standard of hemoglobin diagnosis, which is used in hematology analyzers in high-resource settings, relies on the conversion of hemoglobin to cyanmethemoglobin, a stable molecule that absorbs light at 540 nm.115 Absorption measurements require a spectrometer, limiting the use of this method at the POC. Other methods have been developed to quantify hemoglobin for low-resource settings. The WHO Haemoglobin Color Scale (HCS) is a semi-qualitative method of hemoglobin measurement. A drop of blood is applied to paper and is compared to a color scale by visual interpretation. The WHO HCS has a very low per-test cost (approximately $0.02 per test in Malawi) but suffers from low accuracy (sensitivity between 76–96%, specificity between 33–86%116), particularly with inadequate training or lighting,117 and especially in cases of severe anemia.118 In the Sahli method of anemia detection, hemoglobin is converted to acid hematin and compared visually to a solid glass color standard. The Sahli method has been considered the standard practice in many low-resource settings for decades. Per-test cost is reportedly higher than the WHO HCS, but sensitivity and specificity are both reported to be 85%.116 Commercially, the HemoCue system has been developed to measure hemoglobin for POC applications with higher accuracy than the WHO HCS or Sahli method, with reported sensitivity and specificity of 85–100% and 94%, respectively.116 Blood is drawn into a plastic cuvette, hemoglobin is converted to azidemethemolobin, and the sample is inserted into a spectrophotometric reader for an absorbance measurement.116 The HemoCue system was found to be the most appropriate hemoglobin measurement device in Malawi, though the per-test cost remains prohibitively high for widespread use (approximately $1.00 per cuvette in Malawi).115,119 A more in-depth look at several features of the commercially available technologies as well as a couple of technologies in development can be found in the PATH landscape report on anemia.116
More recently, the development of an alternative paper-based, colorimetric hemoglobin test was reported.120 In this approach, hemoglobin is converted to cyanmethemoglobin with Drabkin’s reagent, and the sample is applied to a microfluidic paper-based device, allowed to dry for 25 minutes, and imaged on a flatbed scanner. The use of a scanner circumvents decreases in accuracy due to ambient light conditions. The reported 95% limits of agreement between the paper-based assay and the reference assay were 1.30 and 1.18 g/dL, and the test performed with relatively high quantitative accuracy (R2=0.96). However, there is a need for sample pre-processing and a cost associated with the flatbed scanner (reported as $44 for a refurbished scanner). Tyburski et al. developed a fully disposable hemoglobin color scale test. The device has two components: a sample tube that collects 5 μL of blood after a fingerprick and a component that is pre-loaded with a color-changing reagent. The group reports an optional smartphone quantification application, as well. In laboratory evaluation, the device performed with high sensitivity and specificity by visual interpretation in cases of both severe (<7 g/dL) and mild (<11 g/dL) anemia (sensitivity: 90% and 90.2%, specificity: 94.6% and 83.7%, respectively), but relatively low quantitative accuracy (R2=0.864). The per-test cost (US $0.50) is also much higher than the WHO HCS, which may prove to be prohibitive in low-resource settings.121
Other groups have developed alternative systems that aim to bring per-test cost down while maintaining high accuracy. A low cost spectrophotometric hemoglobin detection system was developed with the use of chromatography paper as the matrix for sample deposition and hemoglobin measurement, shown in Figure 2. Absorbance at two wavelengths (528 and 656 nm) is used to calculate hemoglobin concentrations. Ninety-five percent of samples tested with this system were within 2 g/dL of HemoCue readings, and the per-test cost (< US $0.01/test) is projected to be significantly cheaper than the HemoCue system.115,122
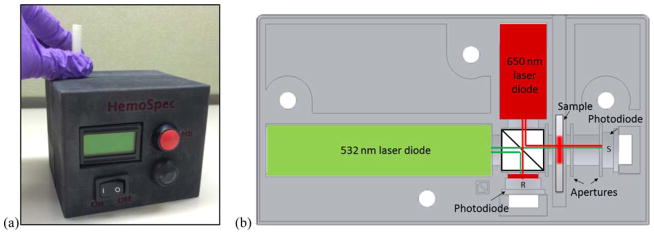
Figure 2.
Photo (a) and optical diagram (b) emphasizing the ease of use of HemoSpec, a portable device that optically measures hemoglobin concentration from blood spotted onto chromatography paper. The form, user interface, and included optical components are shown, but sample loading is not depicted. © 2014 IEEE. Reprinted, with permission, from M. Bond, J. Mvula, E. Molyneux and R. Richards-Kortum, presented in part at 2014 IEEE Healthcare Innovation Conference (HIC), 2014.122
Hematocrit
Hematocrit can also be used for anemia diagnosis. Traditionally, commercially available centrifuges, such as the Zipocrit Hematocrit Centrifuge (LW Scientific) are used in conjunction with capillary tubes loaded with a patient’s blood and sealed with wax. Centrifugation results in layers of packed RBCs, WBCs, and plasma; the height of the RBC layer can be compared to standardized charts that quantitatively produce hematocrit values for a given capillary tube fill height. While this method is cheap and relatively simple, it requires a centrifuge, which is expensive. Several approaches have recently been shown to lower the cost and complexity of centrifuges. Thompson et al. reported the development of a rotation-driven microdevice (RDM) for hematocrit determinations. The RDM is estimated to cost less than US $0.50, runs 12 samples at once, accepts less than 3 μL of whole blood, and produces a result in less than 8 minutes. Rotation is generated by a CD player motor, and a cell phone is used for quantification. Laboratory samples were comparable to clinical lab determinations, though extensive characterization has not yet been reported.123 Drawing inspiration from the whirligig, an ancient toy that generates high centrifugal force, Bhamla et al. developed the human-powered “paperfuge”. The paperfuge has an estimated cost of US $0.20, is lightweight, and can separate plasma from whole blood in less than 90 seconds with comparable performance to a commercial centrifuge.124 The paperfuge cost is insignificant compared to a commercial centrifuge, but data on usability and robustness of the paperfuge is necessary to understand potential clinical utility. The paperfuge is among other innovative approaches toward hand-powered centrifuge development,125 including repurposed salad spinners126 and egg beaters.127 Unconventional approaches for centrifugation may improve access to hematocrit measurement, but the clinical utility of these approaches must be demonstrated. Of note, centrifugation is useful in many diagnostic assays beyond hematocrit measurement.125
In addition to rotational separation, technologies have been developed to quantify hematocrit through impedance spectroscopy128,129 and paper-based plasma separation.130 Impedance spectroscopy utilizes the principle that higher hematocrit levels will increase the current flow path between reference and working electrodes. With impedance spectroscopy, results could be achieved very quickly, and the reported cost is less than US $12. Additionally, it is unclear whether sample pre-processing is required and whether non-RBC components of blood would falsely elevate hematocrit measurements. Further evaluation of complex samples and of clinical utility is required.129 The paper-based approach utilizes the natural fluid-wicking properties of paper to differentially separate blood cells from plasma. The paper-based devices are low-cost (US $0.03), though time-to-results (30 min) is much longer than with centrifugation. The upper hematocrit limit is 57%, which is below the range of some neonatal blood samples, limiting the utility of the device in its current form. The quantitative performance and reproducibility of this paper-based test has yet to be established.130 Other methods of plasma separation have been previously reviewed.28
Hemorrhage
Hemorrhage (severe bleeding) is the leading single cause of maternal mortality, accounting for 27.1% of maternal deaths worldwide, and 659,000 women die from hemorrhage per year in low-resource settings.6 Researchers have identified known risk factors such as Caesarian sections and prolonged third stage of labor, and preventative guidelines have been established.6,131 However, disease burden is still high, and low-resource diagnostics are needed to supplement the preventative guidelines. A bleeding disorder such as HELLP (hemolysis, elevated liver enzymes, low platelets) or von Willebrand disease may be a causative factor for some women who develop hemorrhage.132 Several platelet aggregation assays with or without Ristocetin have been developed to diagnose these conditions, either prior to hemorrhage or at its onset.133,134 Typically, in the presence of large amounts of blood loss or drop in blood pressure, complete blood counts and coagulation assays are performed.132,135 High resource settings particularly quantify change in peripartum hemoglobin, since it accounts for internal hematomas and hemolysis as well as external blood loss.136 Most of these tests require high infrastructure and are performed at central hospitals. For more on platelet aggregation and complete blood counts in low-resource settings, see the White Blood Cell Count and Differential Section. LFAs for coagulation have also been developed, by assessing how far 100μL of blood travels on nitrocellulose.137
POC devices have been developed to measure the amount of blood lost during hemorrhage for diagnosis and management. Quantifying blood loss is important for preventing hypoxia, heart attack, organ failure, and death.136 Typically, in low-resource settings, blood loss is estimated visually, is highly variable depending on staff training, and can be underestimated by as much as 75%.138 The use of bedpans, blood collection pads, and gravimetric sponges allow for more accurate blood loss estimates.136,139 Hemorrhage is diagnosed as blood loss over 500 mL, and many pads or cloths, including “Quaiyum’s mat” developed in Bangladesh140 (approximately $0.50 per mat)141 and a kanga system in Tanzania ($6–7 for two kanga),142,143 utilize this value to absorb only 500 mL. Other devices like Kelly’s pad in India (about $25)139,144,145 and BRASS-V drape in Nigeria (about $4 per sterilized drape)139,146–148 funnel blood to accumulate and measure it. The SAPHE mat has multiple squares of super absorbent polymer, each holding 50 mL of blood, and blood loss is estimated by counting filled squares.149 The SAPHE pad has a 0.96 Pearson’s correlation for volume blood loss, but at $0.50 to $2.50 per pad, it is not a cost-effective strategy, given the cost of misoprostol and other uterine contraction agents to prevent hemorrhage is as low as $1. Kelly’s pad can be washed and sterilized, while most other pads or cloths must be decontaminated and disposed increasing the per-use cost. An area for further innovation is the development of a more quantitative blood loss measurement technique for an extremely low per-use cost.
In addition to these blood loss techniques, a rapid diagnostic test for fibrinogen was developed to predict the severity of hemorrhage during intrapartum complications.150,151 Fibrinogen is a protein essential for coagulation and has a 99.3% specificity for predicting severe hemorrhage.150 However, the assay has a sensitivity of only 12.4%. The test may be useful for triaging, but a more sensitive biomarker is needed for accurate diagnosis.
Pre-eclampsia
Pre-eclampsia is the second largest direct cause of maternal mortality worldwide, second only to hemorrhage. The WHO estimates that 14% of maternal deaths in low-resource settings, about 341,000 per year, are caused by pre-eclampsia.6 Pre-eclampsia is a disorder characterized by high blood pressure and proteinuria, or excess protein in a 24-hour urine sample, and it can lead to severe morbidities such as seizure, placental abruption, hemorrhage, and preterm delivery. A test for proteinuria is the gold standard for pre-eclampsia, but blood pressure, blood-based biomarker tests, and algorithms can also be used for diagnosis.152,153 However, the diagnostic accuracy of these methods is debated.154 A 2005 survey in the United Kingdom claims that 45% of women with eclampsia displayed neither proteinuria nor high blood pressure in the week leading up to seizure.154,155 Additionally, while dipsticks can be used in low-resource settings to determine proteinuria with a tetrabromophenol blue colorimetric agent, urine dipsticks are often unavailable in many low-resource settings. Urine dipsticks for proteinuria do not detect key misfolded biomarkers associated with pre-eclampsia,156 vary with urine concentration,154 and can have sensitivities for pre-eclampsia as low as 55%.157 Because proteinuria is not reliably seen in all pre-eclampsia cases and the dipsticks have low sensitivity for detecting proteinuria, dipsticks are often not used when available.154,156,158–160 As such, alternative methods to diagnose pre-eclampsia in low-resource settings, including low-cost blood pressure devices and both blood- and urine-based POC diagnostics, are discussed here.
Blood Pressure Monitors
Many low resource settings do not measure blood pressure because of a lack of appropriate measuring devices or lack of trained healthcare workers.154,158,161–163 For example, 36% of health centers providing antenatal care in Malawi did not have blood pressure measurement equipment according to the Malawi Demographic Health Survey 2013–2014.154,164 However, a few blood pressure monitors have recently been developed for low-resource settings. Most notably, the Omron HEM-SOLAR and Microlife 3AS1-2 have been validated for use with pregnant women in low-resource settings.154,162,165 Omron HEM-SOLAR, a semi-automated device that uses solar energy to recharge, is slightly more accurate (cost about $30),162,166–168 whereas the Microlife 3AS1-2 is relatively cost-effective ($19) and easy to use by untrained workers.154,167,169–171 The Microlife 3AS1-2 can be manually inflated, requires rechargeable batteries, and uses a “traffic light” system to alert healthcare workers of abnormal pressures, which has received positive feedback from both trained and untrained users.169 The Nissei DS-400 (Nissei Japan Precision Instruments) has similar features of manual inflation and rechargeable batteries, although the cost is slightly higher ($30),172 and the test has only been validated on pregnant women in high-resource settings.173 The LifeSource One-Step Monitor measures blood pressure accurately with automatic inflation and rechargeable batteries, but the price is greater than other POC blood pressure monitors.174–178 Additionally, the LifeSource One-Step Monitor needs to be evaluated in a pregnant population.178 Other low-cost blood pressure monitors are in development and may be appropriate for use at the POC.154,179,180 However, as discussed above, the diagnostic accuracy of blood pressure for pre-eclampsia is debated.154
Blood-based POC diagnostics for pre-eclampsia
Several other biomarkers are also associated with increased risk for pre-eclampsia, and some of these biomarkers have been integrated into benchtop diagnostics or LFAs.181,182 Multiple benchtop readers have been developed to measure serum-based biomarkers of pre-eclampsia, specifically Placental Growth Factor (PlGF), soluble fms-like tyrosine kinase-1 (sFlt-1), and glycosylated fibronectin (GlyFn).183,184 These biomarkers have all shown promise for diagnosis of pre-eclampsia in the third trimester with area under the receiver operator curves (AUROCs) of 0.94 for PlGF, 0.96 for sFlt-1, 0.98 for sFlt-1/PlGF ratio, and 0.99 for GlyFn.184
The Triage PlGF test (Alere Inc) uses fluorescence immunoassay detection and the Triage MeterPro POC analyzer to quantitatively determine PlGF levels in blood plasma samples and returns a result in 15 minutes. Sensitivity and specificity of the Alere Triage PlGF test vary depending on the cutoff value used; between 20 and 34 weeks of gestation, the test has a 96% sensitivity and 56% specificity for pre-eclampsia using a cutoff value of <100 pg/mL and a 63% sensitivity and 90% specificity using a cutoff value of <12 pg/mL.185 When gestational age is known, a cutoff of <5th percentile for normal gestational-age dependent range improves test performance with 100% sensitivity and 96% specificity for early onset pre-eclampsia and 77% sensitivity and 95% specificity for all gestational ages.186 However, with all cutoff values, sensitivity dramatically decreases after week 34 of gestation.185
The Elecsys immunoassay sFlt-1/PlGF ratio test (Roche Diagnostics Limited) measures the relative amount of PlGF to sFlt-1 in serum samples using two separate electrochemiluminescence immunoassays with the Roche Elecsys benchtop analyzer. Similar to the Alere Triage test, Roche Elecsys performance varies on the designated cut-off value during weeks 24 through 36 of gestation. At a ratio of 23, the test has a 92% sensitivity and 81% specificity, while at a ratio of 85, the test has a 56% sensitivity and 97% specificity.185 Two additional tests have been developed, the DELFIA Xpress PlGF 1-2-3 test (PerkinElmer), which quantitatively measures PlGF levels in serum samples using a solid-phase, two-site fluoroimmunometric sandwich assay, and the BRAHMS sFlt-1 Kryptor/BRAHMS PlGF plus Kryptor PE ratio test (Thermo Fisher Scientific GmbH) which detects sFLT-1/PlGF ratio with two immunofluorescent sandwich assays.183,185,187 However, both of the latter tests must be validated in a clinical setting to determine sensitivity and specificity for pre-eclampsia diagnosis.185
Lastly, glycosylated fibronectin is a promising biomarker, and DiabetOmics currently employs a cassette reader for a fluorescent immunoassay with high sensitivity and specificity for pre-eclampsia (97% and 93%, respectively) in the third trimester.182,184 DiabetOmics test is only available in select markets, but the company has licensed the clinical test to Becton, Dickinson and Company for integration into their BD Veritor device,188 which is costly for low-resource settings.189 Alternatively, an equipment-free GlyFn test is possible, with a cutoff value of 176.4 μg/mL, although this test needs to be validated for clinical use.184
The detection of PlGF and sFlt-1 at the POC demonstrate an area for further innovation. The tests discussed above, including the Triage PlGF (Alere Inc), Elecsys sFlt1/PlGF (Roche Diagnostics Limited), and the DELFIA Xpress PIGF 1-2-3 (PerkinElmer), all have high per-test costs (>$40).185 Additionally, the clinically relevant detection ranges of PlGF (pg/mL) and sFlt-1 (ng/mL) are currently not detectable by tests appropriate for low resource settings such as LFAs. Recent innovations in developing a paper-based ELISA platform have demonstrated limits of protein detection at 100 pg/mL,83 which could be applied for detection of these protein targets.
Urine-based POC diagnostics for pre-eclampsia
While urine samples are easy to obtain and require little or no sample preparation, limitations to the commercially available dipsticks still exist, as discussed above, due to limitations in tetrabromophenol blue detection of proteinuria. Two new low-cost tests (<$0.10) for proteinuria include a proteinuria pen developed by researchers at John Hopkins and a proteinuria stamp developed by Diagnostics for All, although both are undergoing test validation and not commercially available.154 Both devices add colorimetric reagent followed by a drop of urine to paper-based platform, and a color change occurs in the presence of proteinuria. However, these tests will likely have performances equivalent to dipstick tests for proteinuria, as they function in a similar manner.160
A recent review article lists many additional promising biomarkers for pre-eclampsia, but notes that only a few of them have POC tests.182 Urine-based rapid diagnostic tests have been developed for pre-eclampsia using urinary adipsin and amyloid aggregates (GestVision).152,156,182,190 However, urinary adipsin as a biomarker has a poor specificity (70%) unless combined with diastolic blood pressure readings.182 In contrast, a GestVision diagnostic uses Congo Red Dot (CRD) to detect protein misfolding in urine, which can predict pre-eclampsia before symptoms appear.152,156 Recent articles have shown that this protein misfolding may have greater predictive value for pre-eclampsia than other biomarkers such as PlGF and sFlt-1,156,182 and the GestVision test uses amyloidophilic Congo Red dye that binds to misfolded, unfolded, or amyloid-like proteins to detect misfolded proteins quickly.191 The test is appropriate for low-resource settings due to its rapid time-to-result (<30 minutes), low cost ($0.30), high sensitivity (86%), and high specificity (85%). A healthcare worker simply spots urine onto nitrocellulose and then adds CRD to note misfolding. Additionally, while the test primarily benefits from high diagnostic performance during the third trimester, recent reports suggest it may also predict pre-eclampsia as early as the first trimester. A prospective cohort found that in the first trimester, CRD predicts 33.3%, 16.1%, and 20% of early, late, and all PE cases.152,191 The GestVision CRD test shows great promise for use at the POC in low-resource settings, but it is not yet commercially available as the company is performing final validation studies before releasing the product.
Bacterial Infections and Puerperal Sepsis
Puerperal sepsis is a leading cause of maternal death, with an estimated 5 million cases and 62,000 deaths worldwide annually. Puerperal sepsis is an infection of the genital tract occurring between the time of rupture of the amniotic membranes and the 42nd day postpartum. The infection must be accompanied by fever and at least one of the following: pelvic pain, abnormal vaginal discharge, discharge odor, and delayed reduction in size of the uterus.192 In high-resource areas, puerperal sepsis accounts for 2.1% of all maternal deaths, while in low-resource settings it is estimated to cause at least 11.6%. Further, the relative risk of mortality due to puerperal sepsis is much higher than other causes of maternal death, with a 10% mortality rate in high-income countries, and a 33% mortality rate in low-income countries.193
Protein-based Tests for Sepsis
Procalcitonin (PCT) has been demonstrated to be a consistent marker of inflammation caused by severe infections in both neonates and pregnant women194 and can be useful in assessing both infection severity and response to antibiotic treatment.195 BioMerieux’s VIDAS B.R.A.H.M.S. PCT system has demonstrated consistent measurement of PCT in both reference standards and in clinical samples.196 The VIDAS system is a benchtop reader system that performs enzyme-linked fluorescent immunoassay and reports a detection limit of 0.09 ng/mL within 20 minutes.196 However, recent meta-analysis data has shown varied results in the reported sensitivity and specificity of PCT to diagnose sepsis.197 Presepsin, on the other hand, has been demonstrated as a specific marker of sepsis that is induced by the phagocystosis of bacteria rather than the presence of inflammation, an important consideration when distinguishing sepsis from other causes of inflammation.198 Mitsubishi Chemical Corporation recently introduced the PATHFAST Presepsin for the rapid detection of presepsin. The PATHFAST system uses a chemiluminescent enzyme immunoassay to quantitatively measure presepsin in whole blood via a benchtop analyzer. The results of the PATHFAST system have shown a strong correlation of measured signal with presepsin concentration across a wide range of concentrations in both whole blood and plasma samples. Additionally, the system has been used to demonstrate a significant increase in presepsin levels in septic patients.199 Despite the progress in development of these benchtop analyzers, both of these biomarkers have clinically relevant limits of detection that have potential to be detected in an equipment-free, LFA format. Developing more POC-friendly LFAs for these biomarkers would allow detection in health posts where the infrastructure required for the VIDAS B.R.A.H.M.S. PCT system and PATHFAST Presepsin is not available, thereby allowing earlier detection of suspected sepsis cases.
In addition, blood-based LFAs have been developed to detect bacterial infections and sepsis using C-reactive protein (CRP).200 However, the concentration of CRP is very high in the blood, and LFAs to detect CRP suffer from the Hook effect, in which excess protein present in the sample binds to capture and detection antibodies separately. This prevents the formation of a sandwich at the detection line and creates a false negative result. In samples with high concentrations of target protein, the sample must be diluted prior to detection, adding user steps, or alternative techniques such as competitive assays must be used to overcome the Hook effect. For example, a near-infrared (NIR) LFA by SRI International utilizes a competitive assay, with CRP spotted onto nitrocellulose.201 Any CRP present in the blood of a neonate or mother binds to the NIR antibodies, and the antibodies do not bind to the CRP on the nitrocellulose. In this case, a lack of signal represents a positive test result. Antibodies conjugated to NIR dye improve the signal-to-background ratio and are detected with an emission scanner, limiting the readout of this assay in low-resource settings. As an alternative to the SRI International LFA, the InfectCheck CRP LFA detects CRP in a semi-quantitative ladder assay with multiple detection lines, allowing for rapid assessment of CRP level in a POC setting, although the assay does lack specificity.202
Protein-based Tests for PROM
Premature rupture of membranes (PROM), or breakage of the amniotic sac prior to labor, is associated with an increased risk of puerperal sepsis,193 as well as increased risk of preterm delivery, neonatal sepsis, and other neonatal complications.203 An array of biomarkers has been identified as promising candidates to predict PROM and preterm delivery, and from those biomarkers, several effective vaginal swab-based POC diagnostics have been developed for diagnosing PROM; they include the Amnisure ROM test (AmniSure),203–205 Actim PROM (Medix Biochimic),203,205,206 AMNI Check (MAST Diagnostica),203,207 ROM Plus (Clinical Innovations),208 AmnioQuick Duo+ (Meridian Healthcare),209 QuickLine IL-6 (Milenia),210 and Lactate Pro (Arkray).203,211 All of these tests work rapidly, with high sensitivity and specificity.
The two most commonly reviewed tests are the Amnisure ROM test, which detects the biomarker placental alpha-microglobulin 1 (PAMG-1), and the Actim PROM tests which identifies the presence of Insulinlike growth factor binding protein 1 (IGFBP-1).205,212 Many studies note that the two tests are comparable in performance, running in 5–10 minutes with a pooled sensitivity and specificity of 96.8% and 98.3% for Amnisure ROM and 92.1% and 90.5% for Actim PROM.205 Amnisure ROM is slightly more accurate in the absence of significant blood since PAMG-1 has a 1000 to 10,000 fold difference between amniotic fluid and normal cervicovaginal secretions.203 However, PAMG-1 levels in maternal blood can cause false positives for Amnisure ROM, while Actim PROM is less susceptible to false positives from blood contamination. Not only are IGFBP-1 levels in blood lower than the Actim PROM limit-of-detection, but IGFBP-1 largely exists in a phosphorylated form in blood with a lower affinity for the Actim PROM antibody.205
Two other common PROM LFAs, AmnioQuick Duo+ and ROM Plus, test both IGFBP-1 for detection of PROM in the first trimester, along with the protein alpha-fetoprotein (AFP) which significantly decreases in the third trimester.213 AmnioQuick Duo+ works in 10 minutes with 94.1% sensitivity and 87.5% specificity, and ROM Plus uses a combination of monoclonal and polyclonal antibodies with 99% sensitivity and 91% specificity in 5 to 20 minutes.208,214 Both tests are susceptible to false positives from blood contamination due to the AFP, and all women with hemorrhage were excluded from diagnostic analysis.213 The Lactate Pro test determines the amount of lactate in vaginal fluid using an electrochemical strip powered by a battery.215 Results appear in one minute with 85% sensitivity and 91% specificity. Although the Lactate Pro does require consumable batteries and lactate is not the most sensitive PROM biomarker, with a per-test cost of $1 to $3, it is relatively low cost compared to other PROM tests.203,211,216–218
While these LFAs are easy to use at the POC, issues with specificity in the presence of blood and cost have limited their uptake this far. The Amnisure ROM test, Actim PROM, and ROM plus test all have a high per-test costs (>$30).219–222 An area for further innovation is to create a truly low-cost method for diagnosing PROM in low-resource settings. Urea and creatinine have shown promise in multiple studies for highly sensitive and specific detection of PROM with close to 100% sensitivity and 100% specificity, although the optimal cutoff values have varied between studies.203,216,217,223–229 In these studies, the urea and creatinine are measured from vaginal washes, where 3 to 5 mL of saline is injected into the posterior vaginal fornix and subsequently aspirated with the same syringe.226 Urea is detected with a spectrometer and creatinine with an enzymatic based assay, but a ladder-based LFA for creatinine was developed in the laboratory which runs within 20 minutes and has 90% agreement with the traditional Jaffe method.230 Such technologies show promise for an affordable POC PROM test appropriate for low-resource settings.
Additionally, a few diagnostics can predict the risk of preterm delivery while the membrane in still intact through cervical and vaginal swab-based tests. The QuikCheck fFN test (Hologic) and Actim Partus (Medix Biochemic) predict preterm labor for women with intact membranes by measuring concentrations of fetal fibronectin (fFN) and the phosphorylated isoform of insulin-like growth factor binding protein 1 (phIGFBP-1), respectively. The QuikCheck rapid test runs in 10 minutes with 90% sensitivity and 64.8% specificity for preterm birth within 7 days.231 As a biomarker, fFN is not specific to preterm labor, which contributes to the low specificity of the QuikCheck test. In addition, the QuikChek test may be less reproducible than its US-based counterpart, the Rapid fFN test with the TLIIQ System (Hologic); the Hologic system includes internal controls and a reader to interpret test results, and quantitative fFN values can provide more information, especially for preterm risk at different gestational ages.231–234 Contamination of the vaginal swab by other fluids such as blood or semen may also affect results.231–233 Likewise, the Actim Partus test (Medix Biochimic) detects phIGFBP-1, a biomarker released from the decidua and potentially signifies labor, with 78.3% sensitivity and 89.3% specificity for preterm labor within 7 days.231 Both the QuikCheck fFN and Actim Partus test have a per-test cost that is prohibitive in low-resource settings.232 Other biomarkers such as matrix metalloproteinase-8 have been developed into a rapid test (SK Pharma Co, Ltd) for preterm delivery with high specificity (>97%) over a range of gestational ages but poor sensitivity.235 Areas for potential development include creating rapid tests with new biomarkers or a combination of the above biomarkers, which can provide both high sensitivity and specificity over a range of gestational ages, as well as a reduced per-test cost for effective use in low-resource settings.
Bacterial Culture at the Point of Care
Biomarker and nucleic acid tests can detect the presence of bacteria. However, they may cause false positives after an infection has resolved, since bacterial DNA can persist in a patient’s blood stream after the bacteria are rendered non-viable by the immune system or antibiotic treatment.236,237 Additionally, many protein biomarkers of infection are elevated in a number of inflammatory conditions further creating potential for false positives in these cases.238 Due to these limitations, some groups have begun working on techniques to perform a POC bacterial culture. These techniques aim to be faster than traditional culture, which can have a diagnostic turnaround time of 24–48 hours.236 Rather than growing bacteria as in traditional culture, POC tests aim to detect bacterial viability as well as drug sensitivity. Thus, these platforms are amenable to detecting not only viable bacteria in a sample, but also rapid screening for antibiotic resistance.239–241
Funes-Huacca et al. reported in 2012 a portable, self-contained culture device for bacteria. While their device is capable of culturing bacteria with similar detection limits to standard culture techniques, they also show rapid color change resulting from paper impregnated with a viability dye, resazurin. When viable bacteria are added to the paper and plastic device, the paper pad changes from blue to red, indicating the presence of live bacteria in less than 5 hours. The color change is visible by eye and can be quantified as number of colony forming units (CFU) with a hand-held reader, shown in Figure 3. The device demonstrated the ability to detect concentrations down to 10 CFU/mL, however these lower CFU concentrations required longer overnight incubation times.239 Additionally, methods using metabolic monitoring have been proposed to rapidly detect bacteria. In these techniques, rather than detecting the bacteria, small volumes of sample are monitored using an oxygen-sensitive fluorophore. Any viable bacteria present in a sample metabolize oxygen, and fluorescence intensity decreases. Because of the small sample volume used, changes in fluorescence can be detected in less than one hour; however, the device has only been tested in a lab with E. coli concentrations from 104 to 108 CF/mL.240 Roche recently procured a technology known as Smarticles: small particles with an embedded genetic sequence. When added to a sample, the particles specifically bind to the surface of any living target bacteria present, and its embedded genetic sequence is inserted into the bacterial cell, creating a luminescent response. This technique has the ability to be multiplexed with a panel of Smarticles specific to various bacterial species, each encoding a different luminescent response, but the approach has yet to be demonstrated in a clinical setting.237,241
Figure 3.
Schematic of paper-based bacterial culture device. This work illustrates a novel, low-cost platform for performing bacterial culture at the POC. (a) Devices are fabricated out of wax patterned paper, tape, PDMS, and a dialysis membrane. (b) When the device is folded shut, the lysogeny broth (LB) medium reservoir is brought into contact with the bacterial growth zone for bacterial culture. Because of the dialysis membrane and oxygen flow through the PDMS (c), bacteria are able to grow within the device. (d) When the device is impregnated with viability dye resazurin (PrestoBlue™), a visual color change occurs during incubation, allowing quantification of bacterial load within the sample. Reproduced from Reference 239 with permission of the Royal Society of Chemistry. (e) Antibiotics can be added to the paper disks to allow for antibiotic susceptibility testing, in which growth will be stunted surrounding the antibiotic areas in susceptible bacterial strains, whereas it will not be in antibiotic resistant strains. Reproduced from Reference 242 with permission of the Royal Society of Chemistry. The images presented here depict the platform components, assay overview, and selected results, but does not provide information on workflow and use of the platform.
Each of these techniques also has the potential to be used to rapidly assess antibiotic resistance. Because the outputs of each test are only responsive to living bacteria, comparing the response of antibiotic-free samples to those of samples that have been impregnated with antibiotics can quickly assess if the bacteria respond to the antibiotic through a quenched output.239–242
To date, there are no POC-appropriate technologies to detect bacterial infections and perform strain differentiation and antibiotic resistance testing. Further, devices based on the principle of bacterial culture are limited by sample collection challenges. First, due to low bacteremia loads in many septic patients, a larger volume of blood is required in order to collect sufficient bacteria for culture detection. Second, these devices require a sterile blood draw in order to avoid sample contamination. As such, POC detection of bacterial infections presents an opportunity for further development.
White Blood Cell Count and Differential
A WBC count with differential is frequently used in diagnosing bacterial and viral infections. In addition, a WBC count can be used to assess disease severity and monitor the effectiveness of a treatment regimen.243 The differential provides important information in distinguishing viral, bacterial, fungal, and parasitic infections, in addition to providing information about inflammation severity and autoimmune diseases.244 As noted in the section on Anemia, drop-to-drop variability of WBCs, hemoglobin, and platelets must be accounted for in WBC count test development.113
HemoCue recently developed the HemoCue WBC DIFF, a portable imaging system for performing a 5-part WBC count and differential at the POC. Similar to HemoCue’s hemoglobin test, blood is drawn into a cuvette that is then inserted into the device for measurement.245 However, the per-test cost for the cuvettes is high ($3.12/cuvette, CliaWaived.com price, February 2017).
In addition to HemoCue’s commercially available test, several in-development approaches have been reported. Majors et al. developed a digital fluorescence microscopy system to perform a three-part WBC differential that has been demonstrated in laboratory settings. The low-cost microscope is fabricated from plastic components by 3D printing and diamond turning techniques. Whole blood samples of less than 15 μL are added in a novel disposable cartridge design, removing the need for sample processing.246 Acquired images are analyzed with an automated algorithm to report the white blood cell count and 3-part differential, which differentiates lymphocytes, monocytes, and granulocytes. All samples measured in this study fell within 20% of the gold standard measurements, falling just outside the required accuracy of ±15% for CLIA waiver; the projected cost of the microscope when produced in quantities of 10,000 is approximately $613 with a per-test cost of less than $0.25, shown in Figure 4 (top).247 Similarly, Smith et al. developed a CBC measurement system for use with sub-microliter volumes of blood. In this method, a blood sample is stained and loaded into a low-cost microscope, and an automated cell counting analysis is performed. The system is able to count RBCs, WBCs, platelets, granulocytes, lymphocytes, and monocytes (Figure 4, bottom).248 Using slightly larger volumes of blood (10 μL), a cell phone-based WBC, RBC, and hemoglobin measurement system has been demonstrated. LEDs are used for excitation of the sample, and the cell phone camera is modified and used for emission detection. Automated cell counting is then performed directly on the cell phone.249 Small-volume tests must ensure drop-to-drop variability does not hinder diagnostic performance.113
Figure 4.
Technologies in development for performing a WBC count. All-plastic microscope (Top left) used with disposable slides (Top right: A, B) for complete blood count measurements. Sample diagnostic images obtained of lymphocytes and granulocytes are shown (Top right: C). The chosen images depict the size of the microscope, the relatively small number of components required to build the system and slides, and a representative image produced by the system. However, the figure does not illustrate the workflow, including sample loading. (Bottom) Method of imaging and analyzing blood count diagnostics using sub-microliter volumes of blood. The images presented here show the workflow and representative images produced by the system, but do not provide information on the size or form of the microscope. Figure Permissions: (Top left) Figure reprinted from Reference 247 with permission from the Optical Society of America; (Top right) © 2014 IEEE. Reprinted, with permission, from C. E. Majors, M. E. Pawlowski, T. Tkaczyk and R. R. Richards-Kortum, presented in part at 2014 IEEE Healthcare Innovation Conference (HIC), Seattle, WA, 2014.246 Figure reproduced from Reference 248 with permission from the Royal Society of Chemistry.
DIAGNOSTICS FOR NEONATAL HEALTH
Premature Birth Complications
Hypothermia
Neonatal hypothermia, defined as body temperature below 36.5°C, is a significant threat to neonatal survival and is also strongly correlated with premature birth, birth asphyxia, and infection.250 Traditionally, an axillary, rectal, or skin (forehead, abdomen, or foot) temperature is used to define and diagnose hypothermia in neonates. However, low nursing staff-to-patient ratios prohibit regular temperature monitoring, and thermometers that continuously and automatically measure lower temperatures to detect hypothermia are expensive and difficult to obtain.251 When low temperatures are sensed, skin-to-skin contact, swaddling, or placement in a warmer must be initiated quickly. In cases where the mother requires attention following delivery, skin-to-skin contact cannot be immediately initiated. Therefore, to ensure that other warming methods are employed when necessary, continuous temperature monitoring devices are needed for unstable and low-birth-weight infants in low-resource settings. Several promising continuous monitoring technologies are described.
The ThermoSpot is a liquid crystal thermometer in the form of a 12-mm diameter disk that is stuck to the skin via an adhesive. When placed in the axilla or on the upper abdomen, the liquid crystal disc is green for temperatures above 36.5°C, brown for temperatures 35.5–36.4°C, and black for temperatures below 35.5°C. ThermoSpot has shown a sensitivity of 88–100% in community and hospital settings for detecting hypothermia in neonates, costs only $0.11 per spot, and has been used successfully by illiterate users.252,253 One drawback to the ThermoSpot is rashes due to the adhesive. In a study involving 43 mothers, 16% reported rashes on their newborns. However, in all of these cases, transparent tape had been used to apply a non-adherent ThermoSpot, and mothers were still willing to use ThermoSpot.254 Recently, Blue Spark Technologies developed the TempTraq, a thin, disposable, flexible and battery-powered temperature skin patch for continuous monitoring of skin temperature. An initial study in adults showed no significant difference between TempTraq measurements and oral and axillary measurements (p = 0.25, 0.33), and plans are in place to test the device in a pediatric population.255 The device is commercially available as a 24-hour, single-use monitor, but the high cost of $19.99 currently prevents its accessibility in low-resource settings.
The Bempu Bracelet is a temperature-monitoring bracelet made of a silicone band and a thermistor metal cup. The bracelet blinks blue light when the neonate is normothermic, and sounds an alarm with flashing red light when the neonate is hypothermic. In a recent study, the Bempu bracelet achieved a sensitivity and specificity of 98.5% and 95%, respectively, when compared to axillary temperatures in a sample size of 2,424 alarm events.256 The Bempu is commercially available, but at $29.76 purchase price at the time of publication, is not affordable in low-resource settings. In addition, its current battery lasts for one month and is non-replaceable.
Hypothermia is closely associated with severe infection, prematurity, and asphyxia, necessitating a reliable monitoring strategy for neonates at risk.250 While ThermoSpot, Temptraq, and the Bempu bracelet, are all easy to interpret, the TempTraq only lasts 24 hours and is cost-prohibitive for low-resource settings. ThermoSpot and the Bempu bracelet show promise as technologies that can be taken home and used by a caregiver to monitor neonates in the first weeks of life. However, the Bempu must be offered at an affordable price and should be equipped with a rechargeable battery to allow reuse. Also, the qualitative results of the ThermoSpot and Bempu bracelet may be useful to an untrained caregiver, but staff may require a more exact temperature reading in a hospital setting. In conclusion, a device that is usable for at-home caregivers, like ThermoSpot, but also provides precise temperature information to nurses that is inexpensive, reusable, and easily visible in a crowded ward would be ideal for use in neonatal intensive care units and in Kangaroo Mother Care.
Jaundice
Over 60% of all newborns develop neonatal jaundice, most commonly due to elevated bilirubin levels. While many cases are benign, development of neonatal jaundice into severe hyperbilirubinemia is associated with severe morbidities and mortality. Severe hyberbilirubinemia is preventable with early diagnosis, continued monitoring, and treatment with phototherapy.257 Traditional laboratory approaches use either chemical determination, such as the diazo method, or spectrophotometry on blood samples to measure total serum bilirubin concentration.
Additionally, several non-invasive transcutaneous devices for measuring bilirubin have been developed and are commercially available. Skin reflectance is measured and normalized based on skin tone and hemoglobin, and reflected light at 455 nm and 575 nm is detected to calculate serum bilirubin.258 Because the accuracy of transcutaneous bilirubinometry can be affected by various factors, including prematurity, use during phototherapy,259 and skin color,260,261 there is debate on whether transcutaneous bilirubinometers can fully replace serum bilirubin testing for guiding treatment decisions.259,260,262,263 Guidelines for the management of late-preterm and term infants with hyperbilirubinemia in low-resource settings outline that the use of transcutaneous bilirubinometry is appropriate, but confirmatory serum bilirubin measurements should be used when transcutaneous measurements are above 12 mg/dL because of increased accuracy in serum bilirubin tests.264 Several benchtop laboratory bilirubin instruments and transcutaneous bilirubinometers have been previously described by Carceller-Blanchard et al.265
BiliStick is a newer development that aims to bring serum bilirubin measurements closer to the POC. Whole blood is applied to a plasma-separating membrane, and reflectance of blue and green light is measured from the plasma on the nitrocellulose membrane by a handheld reader. The difference in wavelength measurements is used to determine bilirubin concentration due to the signature absorbance spectrum of bilirubin in plasma. Initial clinical studies have shown 95% of BiliStick measurements falling within -2.22 mg/dL and +3.43 mg/dL of the values measured by the reference laboratory. Further, BiliStick measurements had a mean bias of 0.6 mg/dL and a correlation level of 0.961 compared to standard measurements. The cost was estimated to be less than US $160 for the instrument and a few cents for each test strip.266 In a follow-up clinical study, BiliStick was shown to have comparable diagnostic performance to the JM-103 transcutaneous bilirubinometer. Compared to clinical laboratory results, the limits of agreement were -5.8 to +3.3 mg/dL and -5.4 to +6.0 mg/dL for BiliStick and JM-103, respectively.267
Hypoglycemia
Hypoglycemia is common among neonates and can lead to neurodevelopmental disorders, visual and hearing impairments, and disorders of the central nervous system, among other morbidities.268,269 Blood glucose monitors for adults are prevalent and well-developed;98,270,271 see the Gestational Diabetes section of this review for more information. However, most commercially available POC glucometers are not optimized in the neonatal blood glucose range, which is lower than the adult glucose range. Plasma glucose values should be above 30 mg/dL in the first 24 hours of life and above 45 mg/dL for the remainder of the neonatal period. Devices therefore need to be accurate below 30 mg/dL and above 180 mg/dL, which are important values for clinical decision-making. These values lie at the limits of accurate detection for most POC glucose meters. The International Organization of Standardization (ISO) guideline ISO 15197-2013, which defines requirements for POC glucometer accuracy, outlines that 95% of samples have to fall within ±15 mg/dL for blood glucose values <100 mg/dL and within ±15% for blood glucose values >100 mg/dL.272 Further, high hematocrit levels in neonates can interfere with commercial blood glucose meters optimized for adults, leading to low glucose readings.273
The Nova StatStrip, a commercially available glucometer, has been optimized for use with neonatal samples. The StatStrip utilizes a modified glucose oxidase-based amperometric test system and is able to correct for hematocrit levels in samples of 1.2 μL.274 Studies have found the StatStrip to correlate well with laboratory-based equipment in neonatal patient samples.275,276 The StatStrip, like other glucometers, has relatively expensive test strips and an above-average meter cost (around US $250). Devices such as the Elite XL and the EML105 have been designed for use with neonatal samples, but they have not been found reliable in diagnosing neonatal hypoglycemia.273
While a POC glucometer capable of use with neonatal samples is important, pain caused by continual with heel sticks in neonates has been associated with long-term morbidities.277 Due to the need for frequent glucose monitoring in certain neonatal populations (e.g. premature neonates and infants of diabetic mothers), it would be beneficial to move toward noninvasive neonatal glucose monitoring. Research to create transdermal glucose monitors has not yet resulted in a clinical use for adults, and there are no devices for use with neonates.273 Many groups have worked toward developing continuous glucose monitors,278–281 but the need for continuous glucose monitoring in neonates is debated.282 The need for and approaches toward achieving accurate and noninvasive glucose monitoring in neonates has previously been reviewed.283
Multianalyte detection and monitoring
Multianalyte systems detect small molecules in blood; three commercially available systems include the Abbott i-Stat, Abaxis Piccolo® xpress, and Alere epoc® Reader.284 The cartridges in all three systems provide the capability to integrate sample preparation and to measure many analytes within one test run. The Abbott i-Stat uses electrochemical methods within disposable cartridges to detect a wide range of small molecules, including glucose, carbon dioxide, oxygen, potassium, chloride, sodium, lactate dehydrogenase, hematocrit, and more. 65–100 μL of blood is drawn into the cartridges without preprocessing. The detection of specific analytes includes potentiometry, amperometry, and conductive measurement, in some cases following an enzymatic reaction with the analyte of interest.285 Abaxis designed a compact disk-based approach, which utilizes centrifugal and capillary forces for sample preparation and analyte analysis.286 Alere designed a chip that utilizes electroosmotic flow and pneumatic pumps for fluid actuation with the capability for high degrees of multiplexing.284 The system is comprised of sensor containing test cards, a wireless card reader, and a mechanism for wireless transmission of data to a computer.287 The systems presented here have prohibitively high per-test costs for use in resource-limited settings (the Abbott i-Stat has a per test cost of approximately $25), but provide solutions for emergency care in higher-resource settings.
Multianalyte blood chemistry diagnostics continue to incorporate new technology developments to decrease cost. Microfluidic paper-based analytical devices (μPADs) developed by the Whitesides group are low-cost, easy-to-use platforms for bioanalysis. μPADs have shown great promise in multiplexing bioanalytical tests for urinalysis semi-quantitatively288 and quantitatively.289 μPADs for quantitative glucose, cholesterol, lactate, and alcohol testing in human blood or urine were designed to be compatible with commercial glucometers.290 Furthermore, the Whitesides group developed the universal Mobile Electrochemical Detector (uMED), a POC technology that expands the functionality of glucometers, which only perform amperometry, to include more capabilities found in a benchtop potentiostat, including cyclic voltammetry, square wave voltommetry, and potentiometry.291 The developments of μPADs and uMED increase the sophistication of POC electrochemistry in an accessible format for resource-limited settings. The number of tests that can be multiplexed into one assay has not yet matched the capabilities of the commercially available cartridge-based systems, which may be acceptable depending on the diagnostic context. Additional work toward the development of paper-based electroanalytical devices for medical diagnostics has been previously reviewed by Maxwell et al.292
Birth Asphyxia
Birth asphyxia is the third leading cause of neonatal mortality, following pre-term complications and infections, and results in 662,000 deaths per year.293,294 Birth asphyxia is characterized by a lack of oxygen supply to the neonate and can lead to mortality or lifelong morbidities, including severe organ damage, cognitive impairment, neurodegenerative diseases, epilepsies, and other chronic illnesses.295,296 Asphyxia is associated with prolonged labor, maternal infections, pre-eclampsia, hypertension, hemorrhage, prematurity, multiple births, and certain medications given to the mother.295,296
In high resource settings, asphyxia is predicted before birth using cardiotocography, which monitors fetal heartbeats and uterine contractions.297,298 In low resource settings, abnormal fetal heart rate can be detected, although this alone is not as sensitive as cardiotocography.9,299 The company Laerdal developed a fetal heart rate monitor, Moyo, for low-resource settings; Moyo is sold for less than $200.300,301 This device may help indicate birth asphxyia prior to birth and allow for more timely obstetric responses. Additionally, Jhiego has developed the e-Partogram, a handheld device to assess progression of labor. Similarly to Moyo, this device can allow for rapid detection of complications in labor and allow earlier referrals to address prolonged or obstructed labor.302
After birth, asphyxia is diagnosed immediately by testing the pH of the umbilical cord blood.297 A lack of oxygen and increase in carbon dioxide leads to metabolic acidosis, which results in low pH in cases of asphyxia.298 In addition to pH, the base excess of umbilical cord blood provides an indication of prolonged asphyxia, and the APGAR score determines severity.297,298,303,304 APGAR is a symptomatic-based scoring system based on Appearance, Pulse, Grimace, Activity, and Respiration.296 A pH value less than 7, a base deficit greater than 12 mmol/L, and an APGAR score from 0–3 indicate asphyxia.298 In low-resource settings, the APGAR symptomatic-based scoring system is most commonly used to diagnose asphyxia; however, APGAR scores and other symptomatic-based algorithms are not as sensitive as conventional methods, and delayed assessment or understaffing can result in lack of diagnosis.305–308 Recently, lactate testing for severity of asphyxia has been shown to be a valid biomarker as well but has not yet been integrated into standard clinical practice.296,304
These symptomatic-based algorithms are used for diagnosis in low-resource settings because few quantitative diagnostics for asphyxia are appropriate for the point-of-care. While several POC blood analyte monitors detect pH and lactate with handheld devices, most have not been validated for diagnosing asphyxia. A recent review lists several of these devices, along with need for consumables, parameters tested, and amount of blood required.309 Chin et al. also provide a useful review of POC microdevices for clinical chemistry.310 Additional tests capable of measuring pH and lactate have been previously discussed in the section on Multianalyte Detection and Monitoring.
A few paper-based tests have also been developed for measuring pH and lactate, including paper-fluidic electrochemical pH and lactate strips, although these tests require consumable batteries.211,311 Most equipment-free colorimetric pH strips use urine or saliva as a sample, not blood, and thus cannot be used for umbilical cord blood testing. In low-resource settings, low-cost quantitative tools to document birth asphyxia could be helpful in monitoring and evaluating efforts to improve quality of care during labor and delivery to reduce birth asphyxia. Potential solutions include the device-based and paper-based pH and lactate diagnostics mentioned above. However, clinical validation of these tests in cases of asphyxia at the POC is necessary to determine their diagnostic value.
Sepsis
Neonatal sepsis, a systematic infection occurring within the first 28 days of life, is a leading cause of neonatal morbidity and mortality.10 Each year, an estimated 421,000 infants die within the first month of life due to neonatal sepsis.7 This accounts for over 5% of childhood mortality293 and 15% of neonatal mortality.312 Further, 99% of all neonatal sepsis deaths occur in developing regions11 where there is a lack of adequate sepsis diagnostics. The immaturity of the neonatal immune system, particularly in that of premature neonates, puts them at high risk of sepsis and can complicate diagnosis.10 Fortunately, many of the diagnostic techniques discussed above in the Bacterial Infections and Puerperal Sepsis section are also applicable to neonatal sepsis. Both CRP and PCT are well-studied and characterized over the course of a neonatal bacterial infection. CRP has demonstrated a sensitivity range from 41 to 96% and a specificity range from 72 to 100%, with a value to 10 mg/L as the most commonly reported diagnostic cutoff. In contrast, PCT has reported sensitivities ranging from 62 to 100% and specificities ranging from 82 to 96%313–315 and a diagnostic cut off of 0.1 ng/mL.10 Appropriate methods to detect both of these biomarkers have been previously described. WBC counts and differentials are used in typical screening for neonatal sepsis, but have shown limited success in identifying the septic infants. Of most promise for the rapid detection of neonatal sepsis are the POC bacterial culture discussed above and molecular testing methods to detect bacterial DNA. However, to date the detection of bacterial DNA in blood has been largely limited to PCR performed in centralized laboratories.10
Pneumonia
Neonatal pneumonia, an infection of the lungs, is a leading cause of neonatal mortality world-wide and is the leading cause of respiratory failure in neonates.316 Neonatal pneumonia can be classified as early-onset, in which the infection is transmitted to the neonate during birth by aspiration of infection amniotic fluid or colonization of the birth canal, and late-onset, in which the disease in brought on by a hospital acquired infection.317 The clinical presentation of pneumonia is often non-specific, complicating diagnosis, and traditional diagnostic criteria of neonatal pneumonia has been based on the combination of chest radiographic findings316 and clinical presentation of sepsis.317 However, guidelines have been in place to detect pneumonia based on rapid breathing since the 1970s,318 and the WHO has set the respiratory rate threshold indicating neonatal pneumonia as greater than 60 breathes per minute.319
To measure respiratory rate in low-resource settings, manual counting of breaths has been shown to be a reliable measurement of respiratory rate in neonates.320 To assist community health workers in this counting, a number of international health organizations have undergone studies of using beads and small timers as counting aids to measure respiratory rate in children. In this technique, community health workers of varying levels of literacy and numeracy count the number of breaths taken by a child or neonate over the course of one minute with the use of color-coded beads, shown in Figure 6. One bead is counted per breath, and if the community health worker reaches the red beads during the one-minute period, the child or neonate is classified as having fast breathing (different strands of color-coded beads are used for measuring children in different age classifications). When measured by primarily illiterate community health workers in Uganda and South Sudan, it was found the rate of correctly classifying fast breathing in children increased from 27% to 68% when using the counting beads.321 While counting techniques have shown great improvements in the ability to accurately detect fast breathing, automated devices that decrease the time burden on health care workers could further improve pneumonia detection. Some wearable, continuous infant monitoring devices have been developed (for example, the Rest Devices’ Mimo and Snuza® HeroMD), but these devices remain prohibitively expensive for use in low-resource settings, as the per device costs range from$150 - $300.322
Figure 6.
Counting beads used by various international organizations. (a) Bead strands used by Save the Children, which employs two age-specific, color-coded strands that can be distinguished by bead size and colors. (b) Bead strands used by the Malaria Consortium and the International Rescue Committee, which also employs two age-specific, color-coded strands that are distinguished by colored beads that match the age specific amoxicillin packaging. (b) Bead strand used by UNICEF that is non-specific for ages 0–5 years and each color is made up of 10 beads for ease of counting. Reprinted from Reference 321 under the terms of Creative Commons Attribution License.
HIV
Although the mother-to-child-transmission (MTCT) rate of HIV has declined from 25–42% to 1% or less in settings where a full array of prophylactic strategies can be implemented, pediatric HIV infection remains an ongoing epidemic in less advantaged settings.323 Although new pediatric HIV infections decreased by over 50% in the 21 UNAIDS Global Plan countries in sub-Saharan Africa from 2009 to 2015, an estimated 110,000 cases were still reported in 2015, most of them from MTCT.324 In the absence of diagnosis and treatment, 50% of HIV-infected infants will die before their second birthday. Antibody based tests used for adult populations are unsuitable for neonates because maternal antibodies may persist for 12–18 months after birth.324 As such, the standard of care for early infant diagnostics (EID) of HIV is nucleic acid testing, and should be accomplished within 8 weeks of birth to ensure prompt treatment and infant survival.325 However, in 2015, only 51% of HIV-exposed infants in 21 priority African countries received a virological test within the first two months of life due to lack of diagnostic tools in low-resource settings.324 Unlike the quantitative NATs used for viral load monitoring, qualitative nucleic acid testing (a yes/no test for the presence of HIV virus) is sufficient for EID.35 Here we describe approaches toward making EID more accessible. In addition to the technologies described below, quantitative viral load technologies discussed earlier may also be used.
Although NATs are the most sensitive and specific method available for diagnosing and monitoring infectious disease, no commercially available NAT currently meets the ASSURED criteria. So far, all NAT tests require some form of instrumentation, failing to meet the ‘equipment-free’ and ‘deliverable’ criteria. Two laboratory platforms are currently used for EID: the Roche Molecular Diagnostics COBAS® HIV-1 Qualitative Test, and the Abbott RealTime Qualitative HIV-1 Test.24 Both of these instruments accept either plasma or dried blood spots as samples, and perform real-time PCR to identify the presence of HIV-1 virus. Most often, 4–5 drops of whole blood are spotted onto filter paper cards to create dried blood spot specimens, which are then transferred to a reference laboratory.326 Although these platforms are well-validated and have excellent sensitivity and specificity, they have extremely high costs and infrastructure requirements. Access to this type of testing is a critical barrier that limits access to ART in HIV-infected infants in low-resource settings.324 Where available, the time required to obtain results in a high loss-to-follow-up and low ART initiation rates.323
Sample-to-answer NATs for EID requiring fewer resources than the gold standard include the SAMBA platform (Diagnostics for the Real World Ltd), the Alere™ q HIV 1/2 Detect (Alere™), and the GeneXpert® System (Cepheid).24 These technologies may be suitable for district hospitals, but cannot be employed in more remote settings. The SAMBA platform has several assays, one of which is a fully automated sample-to-answer system that accepts whole blood for EID and has been validated in several countries including Kenya, Uganda, and Zimbabwe. The Alere™ q HIV-1/2 Detect amplifies HIV-1 and HIV-2 RNA in 52 minutes from 25 μL of blood. A 2014 study on 827 infant samples from primary health clinics in Mozambique reported 98.5% sensitivity and 99.9% sensitivity when compared to the Roche Cobas Taqman/Ampliprep instrument.327 This device recently received WHO prequalification, making it available for public sector procurement.328,329 Alere has several antibody tests available for HIV diagnostics, but the q HIV-1/2 Detect is their only test currently suitable for EID. The GeneXpert® System performs sample preparation, amplification, and detection all in a single cartridge. It is simple to operate, and has been powered successfully in mobile laboratories as well as with solar panels. A more portable version of the GeneXpert® was recently released; known as the GeneXpert Omni®, this system is highly portable at only 1 kg, designed for rugged conditions, and can be powered on a rechargeable battery.24 Finally, an emerging approach toward EID detects the p24 antigen in a lateral flow assay. The LYNX HIV p24 Antigen Assay (Northwestern Global Health Foundation) runs in less than 50 minutes. It has shown a low sensitivity of 71.9% compared to laboratory-based NATs, but may provide test results to up to 81% more patients compared to laboratory-based testing.326 This test is expected to cost only $700–$2000.
Promising approaches in development toward equipment-free EID employ isothermal nucleic acid amplification technologies.330 These approaches use enzymes to perform the strand separation that would normally be achieved by heating, allowing these reactions to be incubated in a heat block. The amplicon can then be detected on a lateral flow strip, or by a low-cost fluorescence reader. Several proof-of-concept studies have been performed to perform isothermal reactions in paper, incubate the reaction using body heat, or detect amplified HIV DNA on lateral flow strips.331–337 However, the complexities of biological samples and the need for high sensitivity again limit the current usefulness of these technologies. In a thorough review by Craw & Balachandran,330 the authors conclude that although several isothermal nucleic acid amplification techniques have been extensively validated, the limiting factor is the integration of upstream sample preparation and nucleic acid extraction with downstream detection techniques.330
Because sample preparation remains a barrier for many commercial devices, several groups have begun investigating low-cost, simple-to-use solutions for sample preparation and integration with NATs. For example, Rodriguez et al. developed a device made entirely of paper and plastic with polyethersulfone (PES) filter paper used as a sample port. An absorbent pad makes contact with the bottom of the PES sample port, and a lateral flow detection strip is initially separated from the PES sample port by a hydrophobic barrier. DNA is precipitated onto the PES sample port and washed. The absorbent pad and the hydrophobic barrier to the lateral flow detection strip are then removed, which could potentially introduce contamination, but allows for elution of the immobilized DNA down the strip. This assay was validated with HPV DNA from cervical swab samples; amplification is not required in this assay due to the abundance of HPV DNA in the swab.338 The multiplexable autonomous disposable nucleic acid amplification test (MAD NAAT) is another fully integrated sample-to-answer nucleic acid testing platform. The device accepts a nasal swab sample and produces a lateral flow result in 60 minutes, though in some samples, detection time was noted to be as fast as five minutes. The MAD NAAT is comprised of a reusable plastic housing, which facilitates heating and reagent dispensing, as well as disposable components, including fluid storage components, shown in Figure 7. Timed reagent dispensing is accomplished through melting wax barriers.339
Figure 7.
MAD NAAT is a fully integrated sample-to-answer nucleic acid testing platform. The sample processing, amplification, and detection schemes are depicted. Reagent delivery is timed by melting wax barriers (valves). The components shown here are housed in a reusable plastic cassette with heaters included, which are not depicted here, but can be seen in the original source along with figures showing user workflow and timing. The representative image shown here shows the novel integration of multiple sample preparation steps for nucleic acid testing through the MAD NAAT’s reagent delivery scheme. Figure reproduced from Reference 339 with permission from the Royal Society of Chemistry.
Technological innovation in the field of POC HIV diagnostics is still urgently needed, particularly with enclosed sample-to-answer viral load tests and with POC genotypic resistance tests. HIV-1 has a high rate of mutation, and the WHO reports that levels of HIV drug resistance in countries scaling up ART have been slowly increasing. In some areas, including East Africa, resistance rates to non-nucleoside reverse transcriptase inhibitors (NRTIs) are above 10%. Furthermore, between 10% and 30% of people receiving a first-line ART regimen will develop virological failure at some point during their treatment. Where resistance testing is not available, WHO guidelines recommend reliance on viral load monitoring to inform treatment switches. Specifically, an immediate adherence intervention is recommended when virological failure is detected, followed by a repeated viral load test three months later. If the second viral load test confirms virological failure, a switch to second-line ART is recommended. Despite this recommendation, in practice, HIV care providers often do not switch patients immediately, due to concerns that virological failure resulted from non-adherence. Excluding non-adherence as a cause of high viral load is challenging; existing adherence measurement tools rely on self-reporting measures, which are inaccurate. Therefore, POC genotypic resistance tests are urgently needed to determine the cause of virological failure and empower healthcare providers to make informed treatment decisions. Meanwhile, viral load testing platforms should continue to be made cheaper and more accessible. Innovative and cost-effective diagnostic assays may bridge the treatment gap between neonates and adults, reduce the time between infection and treatment of HIV, and control the spread of drug resistance.340
Congenital Syphilis
Syphilis is a sexually transmitted bacterial infection (Treponema pallidum) that can be transmitted from mother to child in utero.341 If syphilis is diagnosed while a woman is pregnant and she is appropriately treated early in gestation with penicillin, there is little risk of congenital syphilis in the neonate.342 However, syphilis left untreated in a pregnant woman leads to increased rates of transmission to the neonate and more adverse outcomes, including stillbirth, neonatal death, and neonatal morbidities such as visceral or neurologic damage.341,342 As such, rapid and inexpensive diagnostics in maternal populations are necessary to decrease mortality and morbidity associated with congenital syphilis. If a mother infected with syphilis does not receive appropriate treatment while pregnant, the neonate must undergo more rigorous diagnostic and monitoring to determine if they are infected, including testing of long bone radiographical examination to identify characteristic bone lesions and cerebrospinal fluid testing.342
Antibody-based Syphilis Tests for Detection in Pregnant Women
Syphilis is detected in a pregnant woman using both treponemal and nontreponemal antibody-based LFA tests, where treponemal tests detect antibodies generated directly against the causative bacterial agent Treponema pallidum and nontreponemal tests detect antibodies associated with the host response to the infection. Treponemal tests use Treponema pallidum antigens spotted onto the paper membrane to target antibodies, but these tests cannot distinguish between past and present infections.343–345 The most commonly reviewed LFAs for treponemal syphilis include Determine (Alere), SD Bioline (Standard Diagnostics Inc.), Syphicheck (Qualpro), and Visitect (Omega).343–345 In general, these tests meet the ASSURED criteria, as they are specific (> 95% in whole blood), low-cost (<$1), user-friendly, rapid (<15 minutes), robust, and equipment-free.344,346 However, sensitivity varies depending on the test and sample type with whole blood sensitivities of 86.3%, 84.5%, 74.5%, and 74.26% for Determine, SD Bioline, Syphicheck, and Visitect respectively.344 On the other hand, diagnostics for nontreponemal syphilis detect anti-cardiolipin antibodies or other non-specific antibodies generated against host reactions to Treponema pallidum. Known as the rapid plasma reagin (RPR) test, this test is inexpensive ($0.15 – $0.23 per test) and quick (< 10 minutes),346 but it detects biomarkers that are present in many other diseases, including autoimmune diseases, and therefore may produce false positive results.343–345 A few diagnostics, such as the DPP Screen and Confirm assay (ChemBio), multiplex both types of syphilis screens onto the same strip.343–345 The DPP Screen and Confirm assay (ChemBio) is reported to have reduced sensitivity compared to some treponemal-only tests like Determine (Alere) with plasma sensitivities of 89.8% and 97.3% respectively.343 However, the multiplexed treponemal and nontreponemal antibodies can help distinguish past and active infections.343 Additionally, in a recent pilot study in Rwanda, a microfluidic-based syphilis and HIV multiplexed diagnostic detected treponemal syphilis with an AUC of 0.90 and nontreponemal syphilis with an AUC of 0.92347. This device costs $34 per test, but is easy-to-use and appropriate for use at the POC with a smartphone to power the microfluidic component and provide quantitative readout.
Syphilis Tests for Detection in Neonates
Screening for syphilis in newborns is difficult, since IgG antibodies transferred from mother to child can circulate in an infant for 15 months after birth.348 While IgM antibodies do not cross the placenta, the CDC does not recommend IgM tests due to poor test performance, as commercially available IgM tests like Capita Syphilis-M EIA (Trinity BioTech) have sensitivities ranging from 64% in early latent infection to 93% in primary infection.342,348,349 Instead nontreponomal tests that detect both IgG and IgM antibodies are used for diagnosting neonates, although they have a high false positive rate.348
Other methods for diagnosing congenital syphilis include darkfield microscopy and nucleic acid testing, which both have high sensitivities. However, darkfield microscopy and real time PCR machines like Rotor-Gene (Corbett Research) and the iCycler (Bio-Rad) are inappropriate for use at the POC in low-resource settings due to infrastructure requirements, need for trained personnel, and high costs.350 Other low-cost and easy-to-use methods of nucleic acid amplification could be applied to the detection of congenital syphilis at the POC. Likewise, new methods for detecting congenital syphilis in a neonate at the POC in the absence of diagnosis and treatment in the mother are needed to rapidly screen neonates and initiate appropriate treatment.
EMERGING INFECTIONS
Zika Virus
While Zika virus (ZIKV) is not a leading cause of maternal or neonatal mortality, it has lately been recognized as an emerging threat to both neonatal and maternal health and has been the subject of accelerated diagnostic innovation. Zika virus is a mosquito-borne flavivirus with a relatively mild clinical manifestation resembling dengue fever and chikungunya, including fever, headache, myalgia, and rash.351 However, Zika virus drew attention in early 2015 with a widespread outbreak in Brazil that illuminated the relationship between prenatal infection and poor pregnancy outcomes, including congenital Zika transmission to the neonate and microcephaly.352 Because of the non-specific nature of the clinical manifestation, differential diagnosis can be difficult in pregnant women,351 leading to the rapid development of new diagnostics for Zika virus.
Antibody-based Zika Tests
To differentiate between acute and past Zika infection, the Biocan Zika test uses a combination of the ZIKV NS1 and envelope proteins to detect both IgG and IgM antibodies from whole blood, achieving results from a whole blood sample within 10 minutes.19 The test also claims 99.5% specificity, but validation studies need to verify this claim. Typically, serological cross-reactivity between flaviviruses such as Zika, dengue, West Nile, and yellow fever limits the specificity of these antibody-based tests, and the Center for Disease Control (CDC) recommends a plaque reduction neutralization test for diagnosing Zika in order to avoid this limitation.19
NATs for Zika
Because of the cross-reactivity issues present in antibody-based tests, NATs present an emerging solution to distinguish between flaviviruses. The CDC developed a Zika virus assay comprised of two one-step real-time RT-PCR reactions detecting the premembrane gene and envelope gene. The specificity of the assay was confirmed by testing with RNA from a variety of other flaviviruses, and no cross-reactivity was seen. Additionally, the analystical sensitivity was shown to be as low as 100 copies. This assay was used demonstrate a relatively short duration of detectable viremia following the onset of clinical symptoms (less than 3 days). It was also used to demonstrate detection in sanples other than plasma and serum, including saliva, urine, and amniotic fluid.353 This RT-PCR assay demonstrates the potential for NATs to provide rapid and specific diagnosis of Zika virus, but lacks development into a POC-friendly test format.
Recently, several portable, low-cost platforms for molecular diagnosis of Zika have been demonstrated. Chan et al. developed a platform that utilizes reverse transcriptase recombinase polymerase amplification to perform RT-PCR isothermally from urine samples. A modified 3D printer was used for magnetic particle-based nucleic acid extraction, and the included heating unit within the printer can be used to heat samples for the RPA reaction. Fluorescence monitoring was performed using a smartphone camera for nucleic acid quantification following amplification, and clinical relevant sensitivity was demonstrated (5 plaqye-forming units/mL).354 Song et al. developed an instrument-free POC platform for the molecular detection of Zika virus. In their platform, reverse-transcription loop-mediated isothermal amplification is performed in a disposable cassette that is chemically heated in a reaction cup without the need for electrical power. This platform has demonstrated a limit of detection of 5 plaque-forming units from oral samples, and the authors report a per test cost of $2.00.355 Additionally, Pardee et al. demonstrated a test that can identify single-base differences between viral strains of Zika using only a drop of blood applied to paper discs. This test employs isothermal RNA amplification, toehold switch RNA sensors, and CRISPR/Cas-9 technology to achieve a sample-to-answer result in less than three hours.356 The toe-hold switches use a synthetic biology technique that resembles a RNA hairpin; the ribosome binding site (RBS) is contained in the loop of the hairpin, and a trigger RNA sequence unfolds the hairpin structure to expose the RBS and allow translation to a protein structure to occur. This technique has demonstrated excellent orthogonality when tested against similar flaviviruses and is illustrated in Figure 8. The test was demonstrated using a handheld reader with an estimated cost of $250 and an estimated per-test cost of $0.10–$1.00.357
Figure 8.
A novel detection technique for Zika virus that incorporates toe-hold switches and CRISPR-Cas9 technology capable of detecting single-nucleotide differences in RNA strands. (Top) Sensors for specific RNA strands are developed using a novel detection technique known as toe-hold. In short, these toe-hold switches (shown as the RNA hairpin complex) only unfold in the presence of the target RNA strand, revealing a ribosome binding site; this, in combination with a cell-free system dried into paper discs, results in the translation of proteins that cause a visual color change. (Bottom left) The toe-hold switches are combined with an isothermal RNA amplification technique to detect this color change in samples with clinically relevant RNA levels. (Bottom right) CRISPR-Cas9 is used to detect strain mutations; in the American ZIKV strain, the target RNA contains a PAM site (protospacer adjacent motif) generated by the strain mutation at which CRISPR-Cas9 binds and cleaves the target RNA, preventing the downstream translation of color-change proteins. As such, color change will only occur in the African ZIKV strain. This figure provides an overview of the molecular detection components of the system, but does not show the size of the system, required equipment, or workflow to perform the assay. Figure reprinted from Cell, 165, K. Pardee, A. A. Green, M. K. Takahashi, D. Braff, G. Lambert, J. W. Lee, T. Ferrante, D. Ma, N. Donghia, M. Fan, N. M. Daringer, I. Bosch, D. M. Dudley, D. H. O’Connor, L. Gehrke and J. J. Collins, “Rapid, Low-Cost Detection of Zika Virus Using Programmable Biomolecular Components,” 1255–1266, Copyright (2016), with permission from Elsevier.356
There is a great need for further development of diagnostic tests for Zika virus that have been field validated. Currently available diagnostic methods are limited to laboratory settings, preventing the effiecient monitoring and diagnosis of Zika at the POC. Further, tests that multiplex Zika with other common flaviviruses such as dengue and chikungunya have the potential to be of great utility when distriguishing flaviviruses at the POC.353
DISCUSSION
While great progress has been made to decrease maternal and neonatal mortality, there are still major gaps in the availability of POC tools that aim to diagnose the leading causes of maternal and neonatal death in low-resource settings, as shown in Tables 1 and 2. Only two maternal conditions, HIV and malaria, have readily available POC diagnostics that do not face major barriers for effective implementation at the bedside. Hemorrhage, pre-eclampsia, and bacterial infections combined account for over 50% of global maternal mortality, yet there are no available POC diagnostics to identify women at risk for developing these conditions.6 Furthermore, no neonatal conditions discussed here have POC diagnostics that are appropriate for use at a health post. The only diagnostic tools available for these and other conditions face numerous barriers to being implemented at the POC, including infrastructure requirements, supply chain concerns, consumable use, per-test and instrument costs, time-to-results, and human resource requirements. Many of these challenges to implementation are summarized in Tables 3 and 4. New developments should focus on making existing technologies less dependent on staffing and infrastructure, and bridging the gaps that currently exist, especially in biomarker validation, monitoring technologies, and sample preparation. Finally, affordable platform technologies and devices that link diagnostic results with clinical action have the potential to connect more people with improved health care while using fewer resources.
With advancements in proteomics and metabolomics, several biomarkers have been identified as potential targets for future maternal and neonatal POC diagnostics. Panels of biomarkers have been listed for preterm labor,358 pre-eclampisa,152,181–183,187,359–364 PROM,203 congenital syphilis,365 bacterial infection,313,366,367 and inflammation markers associated with neonatal pneumonia.313,366,367 Many are currently performed in high-resource platforms such as Western Blots, ELISAs, or benchtop analyzers, and have potential for development as RDTs.183,187 Protein biomarkers such as PCT, presepsin, and PlGF/sFlt-1 currently require small benchtop readers, but have clinically relevant limits of detection that suggest they could be implemented in microfluidic and LFA platforms. Potential biomarkers should be evaluated for sensitivity and specificity of diagnosing a certain disease before development of POC-friendly diagnostics that meet ASSURED criteria. These biomarkers present a major opportunity to move detection away from centralized laboratory facilities and towards implementation at the bedside.
In some cases, uptake of POC diagnostics is limited by a lack of human resources rather than a lack of infrastructure. Here, effective and automatic monitoring technologies are needed to allow effective implementation. For example, appropriate technologies to measure temperature and respiratory rate of preterm babies exist, but healthcare providers must manually evaluate neonates on a regular basis; inadequate staffing levels often lead to infrequent monitoring. While 37% of global health care providers work in the Americas to serve 10% of the global disease burden, only 4% of global health care providers work in sub-Saharan Africa to serve 25% of the global disease burden.368 Low-resource settings would benefit greatly from affordable, robust technologies that constantly monitor temperature or analyte values and alert healthcare providers when values are no longer within the normal range.
A third major gap in development exists in the lack of adequate sample preparation techniques. The technical requirements of preanalytical procedures to extract and concentrate a biomarker prevent many in-development tests, most notably NATs,29,31 from being implemented into POC clinical settings. Existing preanalytical procedures are also a major source of errors in laboratory diagnostics.369 Therefore, there is a need for further research into streamlined and integrated sample preparation modules that allow tests to accept unprocessed patient samples and produce diagnostic results with minimal user steps. Well-designed sample preparation techniques can increase the sensitivity and specificity of existing POC diagnostics tools, reduce human error associated with existing techniques, and allow access to platform technologies that use nucleic acid detection for applications such as neonatal HIV.
Given the existing health system constraints, novel technology platforms should be designed strategically in order to best implement them in low-resource settings. For example, assays that require more resources should be equipped to handle multiple disease targets. Platform technologies, such as GeneXpert, have the potential to revolutionize POC disease testing by detecting numerous disease targets with interchangeable test cartridges that contain pre-dried reagents and a code containing identifying information. Although high-throughput platform technologies may only be suitable for large, high-resource laboratories, they may have a more significant impact on disease burden than multiple tests that require fewer resources but also report with lower sensitivity. Another strategy to use existing resources efficiently at the POC is multiplexing. However, there have been significant challenges in implementing multiplexed assays at the POC, most notably a decrease in analyte sensitivity as additional test targets are added. For example, a platform to detect the presence of three intestinal protozoa using isothermal amplification and lateral flow detection was demonstrated in 2016; however, the limit of detection was approximately an order of magnitude higher for each target than it was when detected in a singleplex assay.370 Additionally, many commercially available multiplexed NAATs, such as GeneXpert, are only capable of a low-throughput of samples, limiting their usability in populations with high burdens of disease.371 There is an urgent need to continue progress towards developing platforms capable of multiplexing disease targets into a single test without sacrificing test sensitivity.372
To overcome barriers to access, technologies themselves should help strengthen linkage to care after diagnosis whenever possible. For example, recent developments in smartphone usage at the POC suggest that smartphones could be used to connect test results to electronic medical records systems.14 Also, barcodes are commonly used in platform technologies to identify kits, reagents, and expiration dates, reducing the level of training required of healthcare workers and the chance of user error.24 Finally, the recent use of quick response (QR) codes in lateral flow strips indicates that QR codes could be used to communicate test results among healthcare workers, to transfer data in ways similar to barcodes, or as anti-counterfeit measures for diseases that face many counterfeit tests, such as malaria.373 Imaging platforms that allow smartphones to image a printed code could be useful for a wide range of applications including mobile health initiatives and surveillance efforts for other infectious diseases.
The past few decades have witnessed major declines in child and maternal mortality and progress in the fight against HIV and malaria in developing countries. In 2014, the number of new HIV infections had decreased 20% since the peak of the global epidemic in 1998.34 Due to the cooperation of several international agencies, significant funding, and the timely innovation of sensitive and specific point-of-care diagnostic tools, 1.2 million new HIV child infections have been avoided in the 21 most affected countries in sub-Saharan Africa, and over 2 million more pregnant women have started receiving lifesaving antiretroviral therapy.324 Similar interventions allowed malaria rapid diagnostic test sales to increase from 46 million in 2008 to 314 million in 2014, and have contributed to an estimated 60% worldwide decrease in malaria mortality rates over the past 16 years.374 The successes of adult HIV and malaria diagnostics illustrate the potential for bringing effective, inexpensive, and life-saving technologies to the POC. However, progress in other areas is still urgently needed. With strategic design, new technologies will continue to expand access to quality diagnostic tools to the areas most affected by the leading causes of maternal and neonatal mortality.
Supplementary Material
Acknowledgments
This work was supported by Grant #2013138 from the Doris Duke Charitable Foundation
Research reported in this publication was supported by the National Cancer Institute of the National Institutes of Health under Award Number R01CA186132. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of health.
This work was supported by Grant #AID-GH-F-15-00012 from the U.S. Agency for International Development.
References
- 1.World Health Organization. Health in 2015: From MDGs to SDGs. 2015 [Google Scholar]
- 2.World Health Organization, UNICEF, UNFPA, World Bank Group and UN. Trends in Maternal Mortality: 1990 to 2015. 2015 [Google Scholar]
- 3.UNICEF. Levels & Trends in Child Mortality; Report 2015. 2015 [Google Scholar]
- 4.De Bernis L, Kinney MV, Stones W, Ten Hoope-Bender P, Vivio D, Leisher SH, Bhutta ZA, Gülmezoglu M, Mathai M, Belizán JM, Franco L, McDougall L, Zeitlin J, Malata A, Dickson KE, Lawn JE. Lancet. 2016;387:703–716. doi: 10.1016/S0140-6736(15)00954-X. [DOI] [PubMed] [Google Scholar]
- 5.United Nations General Assembly. Transforming our world: the 2030 Agenda for Sustainable Development. 2015 [Google Scholar]
- 6.Say L, Chou D, Gemmill A, Tunçalp Ö, Moller AB, Daniels J, Gülmezoglu AM, Temmerman M, Alkema L. Lancet Glob Heal. 2014;2:e323–e333. doi: 10.1016/S2214-109X(14)70227-X. [DOI] [PubMed] [Google Scholar]
- 7.Liu L, Oza S, Hogan D, Perin J, Rudan I, Lawn JE, Cousens S, Mathers C, Black RE. Lancet. 2015;385:430–440. doi: 10.1016/S0140-6736(14)61698-6. [DOI] [PubMed] [Google Scholar]
- 8.Black RE, Laxminarayan R, Temmerman M, Walker N. Intergovernmental Panel on Climate Change, editor. Climate Change 2013 - The Physical Science Basis. Cambridge University Press; Cambridge: 2016. pp. 1–30. [Google Scholar]
- 9.Institute of Medicine (US) Committee on Improving Birth Outcomes. Improving Birth Outcomes: Meeting the Challenge in the Developing World. National Academies Press; Washington, D.C: 2003. [PubMed] [Google Scholar]
- 10.Simonsen KA, Anderson-Berry AL, Delair SF, Davies HD. Clin Microbiol Rev. 2014;27:21–47. doi: 10.1128/CMR.00031-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ganatra HA, Stoll BJ, Zaidi AKM. Clin Perinatol. 2010;37:501–523. doi: 10.1016/j.clp.2010.02.004. [DOI] [PubMed] [Google Scholar]
- 12.Peeling RW, Holmes KK, Mabey D, Ronald A. Sex Transm Infect. 2006;82:v1–v6. doi: 10.1136/sti.2006.024265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pai NP, Vadnais C, Denkinger C, Engel N, Pai M. PLoS Med. 2012:9. doi: 10.1371/journal.pmed.1001306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu X, Akay A, Wei H, Wang S, Pingguan-Murphy B, Erlandsson BE, Li X, Lee W, Hu J, Wang L, Xu F. Proc IEEE. 2015;103:236–247. [Google Scholar]
- 15.Wong R, Tse H, editors. Lateral Flow Immunoassay. Humana Press; Totowa, NJ: 2009. [Google Scholar]
- 16.St John A, Price CP. Clin Biochem Rev. 2013;34:61–74. [PMC free article] [PubMed] [Google Scholar]
- 17.Drucker E, Krapfenbauer K. EPMA J. 2013;4:7. doi: 10.1186/1878-5085-4-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Charrel RN, Leparc-Goffart I, Pas S, de Lamballerie X, Koopmans M, Reusken C. Bull World Health Organ. 2016;94:574–584D. doi: 10.2471/BLT.16.171207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rabe IB, Staples JE, Villanueva J, Hummel KB, Johnson JA, Rose L, Hills S, Wasley A, Fischer M, Powers AM. Morb Mortal Wkly Rep. 2016;65:543–546. doi: 10.15585/mmwr.mm6521e1. [DOI] [PubMed] [Google Scholar]
- 20.Anderson DA, Crowe SM, Garcia M. Curr HIV/AIDS Rep. 2011;8:31–37. doi: 10.1007/s11904-010-0067-z. [DOI] [PubMed] [Google Scholar]
- 21.Moncada PA, Montoya JG. Expert Rev Anti Infect Ther. 2012;10:815–828. doi: 10.1586/eri.12.58. [DOI] [PubMed] [Google Scholar]
- 22.Neto EC, Rubin R, Schulte J, Giugliani R. Emerg Infect Dis. 2004;10:1069–1073. doi: 10.3201/eid1006.030830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bennett JE, Dolin R, Blaser MJ. Mandell, Douglas, and Bennett’s principles and practice of infectious diseases. 8. Elsevier Health Sciences; 2014. [Google Scholar]
- 24.UNITAID. HIV/AIDS Diagnostic Technology Landscape. 5. 2015. [Google Scholar]
- 25.Schrader C, Schielke A, Ellerbroek L, Johne R. J Appl Microbiol. 2012;113:1014–1026. doi: 10.1111/j.1365-2672.2012.05384.x. [DOI] [PubMed] [Google Scholar]
- 26.Kersting S, Rausch V, Bier F, von Nickisch-Rosenegk M. Malar J. 2014;13:99. doi: 10.1186/1475-2875-13-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tang RH, Yang H, Choi JR, Gong Y, Feng SS, Pingguan-Murphy B, Huang QS, Shi JL, Mei QB, Xu F. Crit Rev Biotechnol. 2017;37:411–428. doi: 10.3109/07388551.2016.1164664. [DOI] [PubMed] [Google Scholar]
- 28.Kersaudy-Kerhoas M, Sollier E. Lab Chip. 2013;13:3323. doi: 10.1039/c3lc50432h. [DOI] [PubMed] [Google Scholar]
- 29.Cui F, Rhee M, Singh A, Tripathi A. Annu Rev Biomed Eng. 2015;17:267–286. doi: 10.1146/annurev-bioeng-071114-040538. [DOI] [PubMed] [Google Scholar]
- 30.Li J, Macdonald J. Biosens Bioelectron. 2015;64:196–211. doi: 10.1016/j.bios.2014.08.069. [DOI] [PubMed] [Google Scholar]
- 31.Niemz A, Ferguson TM, Boyle DS. Trends Biotechnol. 2011;29:240–250. doi: 10.1016/j.tibtech.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Elizabeth Glaser Pediatric AIDS Foundation. Maternal, newborn and child health: The Foundation to a Successful Response to Pediatric HIV. 2017 [Google Scholar]
- 33.Hurst SA, Appelgren KE, Kourtis AP. Expert Rev Anti Infect Ther. 2015;13:169–181. doi: 10.1586/14787210.2015.999667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Haleyur Giri Setty MK, Hewlett IK. AIDS Res Treat. 2014;2014:1–20. doi: 10.1155/2014/497046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.World Health Organization. 2016 [Google Scholar]
- 36.Jaspard M, Le Moal G, Saberan-Roncato M, Plainchamp D, Langlois A, Camps P, Guigon A, Hocqueloux L, Prazuck T. PLoS One. 2014;9:e101148. doi: 10.1371/journal.pone.0101148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shafiee H, Jahangir M, Inci F, Wang S, Willenbrecht RBM, Giguel FF, Tsibris AMN, Kuritzkes DR, Demirci U. Small. 2013;9:2553–2563. doi: 10.1002/smll.201202195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gomes C, Azevedo-Pereira JM. J Virol Methods. 2011;173:353–356. doi: 10.1016/j.jviromet.2011.03.009. [DOI] [PubMed] [Google Scholar]
- 39.Rosenberg NE, Kamanga G, Phiri S, Nsona D, Pettifor A, Rutstein SE, Kamwendo D, Hoffman IF, Keating M, Brown LB, Ndalama B, Fiscus SA, Congdon S, Cohen MS, Miller WC. J Infect Dis. 2012;205:528–34. doi: 10.1093/infdis/jir789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Singh AE, Lee B, Fenton J, Preiksaitis J. Expert Opin Med Diagn. 2013;7:299–308. doi: 10.1517/17530059.2013.774370. [DOI] [PubMed] [Google Scholar]
- 41.Brauer M, De Villiers JC, Mayaphi SH. J Virol Methods. 2013;189:180–183. doi: 10.1016/j.jviromet.2013.01.017. [DOI] [PubMed] [Google Scholar]
- 42.Chin CD, Laksanasopin T, Cheung YK, Steinmiller D, Linder V, Parsa H, Wang J, Moore H, Rouse R, Umviligihozo G, Karita E, Mwambarangwe L, Braunstein SL, van de Wijgert J, Sahabo R, Justman JE, El-Sadr W, Sia SK. Nat Med. 2011;17:1015–1019. doi: 10.1038/nm.2408. [DOI] [PubMed] [Google Scholar]
- 43.Duong YT, Mavengere Y, Patel H, Moore C, Manjengwa J, Sibandze D, Rasberry C, Mlambo C, Li Z, Emel L, Bock N, Moore J, Nkambule R, Justman J, Reed J, Bicego G, Ellenberger DL, Nkengasong JN, Parekh BS. J Clin Microbiol. 2014;52:3743–3748. doi: 10.1128/JCM.01989-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.World Health Organization. Guidelines on HIV self-testing and partner notification: supplement to consolidated guidelines on HIV testing services. 2016 [PubMed] [Google Scholar]
- 45.UNITAID. HIV rapid diagnostics for self-testing. 3. 2017. [Google Scholar]
- 46.Lee HH, Dineva MA, Chua YL, Ritchie AV, Ushiro-Lumb I, Wisniewski CA. J Infect Dis. 2010;201:S65–S72. doi: 10.1086/650385. [DOI] [PubMed] [Google Scholar]
- 47.Tanriverdi S, Chen L, Chen S. J Infect Dis. 2010;201:S52–S58. doi: 10.1086/650387. [DOI] [PubMed] [Google Scholar]
- 48.Schnippel K, Meyer-Rath G, Long L, MacLeod W, Sanne I, Stevens WS, Rosen S. Trop Med Int Heal. 2012;17:1142–1151. doi: 10.1111/j.1365-3156.2012.03028.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shafiee H, Wang S, Inci F, Toy M, Henrich TJ, Kuritzkes DR, Demirci U. Annu Rev Med. 2015;66:387–405. doi: 10.1146/annurev-med-092112-143017. [DOI] [PubMed] [Google Scholar]
- 50.Hoffmann CJ, Maritz J, van Zyl GU. Trop Med Int Heal. 2016;21:219–223. doi: 10.1111/tmi.12639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.May MT, Vehreschild J-J, Trickey A, Obel N, Reiss P, Bonnet F, Mary-Krause M, Samji H, Cavassini M, Gill MJ, Shepherd LC, Crane HM, d’Arminio Monforte A, Burkholder GA, Johnson MM, Sobrino-Vegas P, Domingo P, Zangerle R, Justice AC, Sterling TR, Miró JM, Sterne JAC. Clin Infect Dis. 2016;62:1571–1577. doi: 10.1093/cid/ciw183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lopez A, Caragol I, Candeias J, Villamor N, Echaniz P, Ortuno F, Sempere A, Strauss K, Orfao A. Cytometry. 1999;38:231–237. doi: 10.1002/(sici)1097-0320(19991015)38:5<231::aid-cyto5>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 53.Strauss K, Hannet I, Engels S, Shiba A, Ward DM, Ullery S, Jinguji MG, Valinsky J, Barnett D, Orfao A, Kestens L. Cytometry. 1996;26:52–59. doi: 10.1002/(SICI)1097-0320(19960315)26:1<52::AID-CYTO8>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 54.Jani IV, Sitoe NE, Alfai ER, Chongo PL, Quevedo JI, Rocha BM, Lehe JD, Peter TF. Lancet. 2011;378:1572–1579. doi: 10.1016/S0140-6736(11)61052-0. [DOI] [PubMed] [Google Scholar]
- 55.Stevens W, Gous N, Ford N, Scott LE. BMC Med. 2014;12:173. doi: 10.1186/s12916-014-0173-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Scott L. South African Med J. 2013;103:883–4. doi: 10.7196/samj.7167. [DOI] [PubMed] [Google Scholar]
- 57.Glynn MT, Kinahan DJ, Ducrée J. Lab Chip. 2013;13:2731. doi: 10.1039/c3lc50213a. [DOI] [PubMed] [Google Scholar]
- 58.Wang S, Tasoglu S, Chen PZ, Chen M, Akbas R, Wach S, Ozdemir CI, Gurkan UA, Giguel FF, Kuritzkes DR, Demirci U. Sci Rep. 2014;4:3796. doi: 10.1038/srep03796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Watkins NN, Hassan U, Damhorst G, Ni H, Vaid A, Rodriguez W, Bashir R. Sci Transl Med. 2013;5:214ra170–214ra170. doi: 10.1126/scitranslmed.3006870. [DOI] [PubMed] [Google Scholar]
- 60.World Health Organization. Malaria Rapid Diagnostic Test Performance: Summary results of WHO product testing of malaria RDTs: rounds 1–6 (2008–2015) 2015 [Google Scholar]
- 61.Uneke CJ. Yale J Biol Med. 2007;80:39–50. [PMC free article] [PubMed] [Google Scholar]
- 62.Fried M, Muehlenbachs A, Duffy PE. Expert Rev Anti Infect Ther. 2012;10:1177–1187. doi: 10.1586/eri.12.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Global Malaria Programme. Recommended selection criteria for procurement of malaria rapid diagnostic tests. 2016 [Google Scholar]
- 64.World Health Organization. Malaria Rapid Diagnostic Test Performance: Results of WHO product testing of malaria RDTs: round 6 (2014–2015) 2015 [Google Scholar]
- 65.World Health Organization. World Health Statistics: 2016. 2016 [Google Scholar]
- 66.Wilson ML. Clin Infect Dis. 2012;54:1637–1641. doi: 10.1093/cid/cis228. [DOI] [PubMed] [Google Scholar]
- 67.Williams JE, Cairns M, Njie F, Laryea Quaye S, Awine T, Oduro A, Tagbor H, Bojang K, Magnussen P, ter Kuile FO, Woukeu A, Milligan P, Chandramohan D, Greenwood B. Clin Infect Dis. 2016;62:837–844. doi: 10.1093/cid/civ1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cohee LM, Kalilani-Phiri L, Boudova S, Joshi S, Mukadam R, Seydel KB, Mawindo P, Thesing P, Kamiza S, Makwakwa K, Muehlenbachs A, Taylor TE, Laufer MK. Malar J. 2014;13:274. doi: 10.1186/1475-2875-13-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rantala A-M, Taylor SM, Trottman PA, Luntamo M, Mbewe B, Maleta K, Kulmala T, Ashorn P, Meshnick SR. Malar J. 2010;9:269. doi: 10.1186/1475-2875-9-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mayor A, Moro L, Aguilar R, Bardaji A, Cistero P, Serra-Casas E, Sigauque B, Alonso PL, Ordi J, Menendez C. Clin Infect Dis. 2012;54:1561–1568. doi: 10.1093/cid/cis236. [DOI] [PubMed] [Google Scholar]
- 71.Saute F, Menendez C, Mayor A, Aponte J, Gomez-Olive X, Dgedge M, Alonso P. Trop Med Int Health. 2002;7:19–28. doi: 10.1046/j.1365-3156.2002.00831.x. [DOI] [PubMed] [Google Scholar]
- 72.Mockenhaupt FP, Rong B, Till H, Eggelte TA, Beck S, Gyasi-Sarpong C, Thompson WN, Bienzle U. Trop Med Int Heal. 2000;5:167–73. doi: 10.1046/j.1365-3156.2000.00532.x. [DOI] [PubMed] [Google Scholar]
- 73.Mockenhaupt FP, Bedu-Addo G, von Gaertner C, Boyé R, Fricke K, Hannibal I, Karakaya F, Schaller M, Ulmen U, Acquah PA, Dietz E, Eggelte TA, Bienzle U. Malar J. 2006;5:119. doi: 10.1186/1475-2875-5-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tegegne B, Getie S, Lemma W, Mohon AN, Pillai DR. Malar J. 2017;16:34. doi: 10.1186/s12936-017-1692-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kyabayinze DJ, Tibenderana JK, Nassali M, Tumwine LK, Riches C, Montague M, Counihan H, Hamade P, Van Geertruyden J-P, Meek S. Malar J. 2011;10:306. doi: 10.1186/1475-2875-10-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Oriero EC, Van Geertruyden J-P, Nwakanma DC, D’Alessandro U, Jacobs J. Expert Rev Mol Diagn. 2015;15:1419–1426. doi: 10.1586/14737159.2015.1090878. [DOI] [PubMed] [Google Scholar]
- 77.Safavieh M, Kanakasabapathy MK, Tarlan F, Ahmed MU, Zourob M, Asghar W, Shafiee H. ACS Biomater Sci Eng. 2016;2:278–294. doi: 10.1021/acsbiomaterials.5b00449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Giuffrida MC, Spoto G. Biosens Bioelectron. 2017;90:174–186. doi: 10.1016/j.bios.2016.11.045. [DOI] [PubMed] [Google Scholar]
- 79.Oriero EC, Jacobs J, Van Geertruyden J-P, Nwakanma D, D’Alessandro U. J Antimicrob Chemother. 2015;70:2–13. doi: 10.1093/jac/dku343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Roth JM, Korevaar DA, Leeflang MMG, Mens PF. Crit Rev Clin Lab Sci. 2016;53:87–105. doi: 10.3109/10408363.2015.1084991. [DOI] [PubMed] [Google Scholar]
- 81.Britton S, Cheng Q, McCarthy JS. Malar J. 2016;15:88. doi: 10.1186/s12936-016-1158-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.UNITAID. Malaria Diagnostics Technology and Market Landscape. 3. 2016. [Google Scholar]
- 83.Grant BD, Smith CA, Karvonen K, Richards-Kortum R. Anal Chem. 2016;88:2553–2557. doi: 10.1021/acs.analchem.5b03999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bauer WS, Kimmel DW, Adams NM, Gibson LE, Scherr TF, Richardson KA, Conrad JA, Matakala HK, Haselton FR, Wright DW. Analyst. 2017;142:1569–1580. doi: 10.1039/c7an00278e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Anderson CE, Shah KG, Yager P. 2017:383–411. doi: 10.1016/bs.mie.2017.01.018. [DOI] [PubMed] [Google Scholar]
- 86.World Health Organization. Global Report on Diabetes. 2016 [Google Scholar]
- 87.Zhu Y, Zhang C. Curr Diab Rep. 2016;16:7. doi: 10.1007/s11892-015-0699-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Coustan DR. Clin Chem. 2013;59:1310–1321. doi: 10.1373/clinchem.2013.203331. [DOI] [PubMed] [Google Scholar]
- 89.Kanguru L, Bezawada N, Hussein J, Bell J. Glob Health Action. 2014;7:23987. doi: 10.3402/gha.v7.23987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bhavadharini B, Uma R, Saravanan P, Mohan V. Clin Diabetes Endocrinol. 2016;2:13. doi: 10.1186/s40842-016-0031-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Renz PB, Cavagnolli G, Weinert LS, Silveiro SP, Camargo JL. PLoS One. 2015;10:e0135989. doi: 10.1371/journal.pone.0135989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Khalafallah A, Phuah E, Al-Barazan AM, Nikakis I, Radford A, Clarkson W, Trevett C, Brain T, Gebski V, Corbould A. BMJ Open. 2016;6:e011059. doi: 10.1136/bmjopen-2016-011059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ye M, Liu Y, Cao X, Yao F, Liu B, Li Y, Wang Z, Xiao H. Diabetes Res Clin Pract. 2016;114:43–49. doi: 10.1016/j.diabres.2016.02.007. [DOI] [PubMed] [Google Scholar]
- 94.World Health Organization. Use of glycated haemoglobin (HbA1c) in the diagnosis of diabetes mellitus. 2011 [PubMed] [Google Scholar]
- 95.Drain PK, Hyle EP, Noubary F, Freedberg KA, Wilson D, Bishai WR, Rodriguez W, Bassett IV. Lancet Infect Dis. 2014;14:239–249. doi: 10.1016/S1473-3099(13)70250-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.St John A, Price CP. Clin Biochem Rev. 2014;35:155–67. [PMC free article] [PubMed] [Google Scholar]
- 97.Bhavadharini B, Mahalakshmi MM, Maheswari K, Kalaiyarasi G, Anjana RM, Deepa M, Ranjani H, Priya M, Uma R, Usha S, Pastakia SD, Malanda B, Belton A, Unnikrishnan R, Kayal A, Mohan V. Acta Diabetol. 2016;53:91–97. doi: 10.1007/s00592-015-0761-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Rebel A, Rice MA, Fahy BG. J Diabetes Sci Technol. 2012;6:396–411. doi: 10.1177/193229681200600228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.World Health Organization. Medical Devices: Managing the Mismatch. 2010. [Google Scholar]
- 100.Mendis S, Al Bashir I, Dissanayake L, Varghese C, Fadhil I, Marhe E, Sambo B, Mehta F, Elsayad H, Sow I, Algoe M, Tennakoon H, Truong LD, Lan LTT, Huiuinato D, Hewageegana N, Fahal NAW, Mebrhatu G, Tshering G, Chestnov O. Int J Hypertens. 2012;2012:1–7. doi: 10.1155/2012/584041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Weigl BH, Drake JK. Point Care J Near-Patient Test Technol. 2013;12:33–40. [Google Scholar]
- 102.Vučić Lovrenčić M, Radišić Biljak V, Božičević S, Pape-Medvidović E, Ljubić S. Int J Endocrinol. 2013;2013:1–6. doi: 10.1155/2013/206309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Chen C, Xie Q, Yang D, Xiao H, Fu Y, Tan Y, Yao S. RSC Adv. 2013;3:4473. [Google Scholar]
- 104.Zhu Z, Garcia-Gancedo L, Flewitt AJ, Xie H, Moussy F, Milne WI. Sensors. 2012;12:5996–6022. doi: 10.3390/s120505996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Rahman MM, Ahammad AJS, Jin J-H, Ahn SJ, Lee J-J. Sensors. 2010;10:4855–4886. doi: 10.3390/s100504855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Tian K, Prestgard M, Tiwari A. Mater Sci Eng C. 2014;41:100–118. doi: 10.1016/j.msec.2014.04.013. [DOI] [PubMed] [Google Scholar]
- 107.Bandodkar AJ, Wang J. Trends Biotechnol. 2014;32:363–371. doi: 10.1016/j.tibtech.2014.04.005. [DOI] [PubMed] [Google Scholar]
- 108.Vashist SK. Anal Chim Acta. 2012;750:16–27. doi: 10.1016/j.aca.2012.03.043. [DOI] [PubMed] [Google Scholar]
- 109.Oliver NS, Toumazou C, Cass AEG, Johnston DG. Diabet Med. 2009;26:197–210. doi: 10.1111/j.1464-5491.2008.02642.x. [DOI] [PubMed] [Google Scholar]
- 110.Stevens GA, Finucane MM, De-Regil LM, Paciorek CJ, Flaxman SR, Branca F, Peña-Rosas JP, Bhutta ZA, Ezzati M. Lancet Glob Heal. 2013;1:e16–e25. doi: 10.1016/S2214-109X(13)70001-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.New S, Wirth M. BJOG An Int J Obstet Gynaecol. 2015;122:166–169. doi: 10.1111/1471-0528.13225. [DOI] [PubMed] [Google Scholar]
- 112.Kohne E. Dtsch Arztebl Int. 2011;108:532–40. doi: 10.3238/arztebl.2011.0532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Bond MM, Richards-Kortum RR. Am J Clin Pathol. 2015;144:885–894. doi: 10.1309/AJCP1L7DKMPCHPEH. [DOI] [PubMed] [Google Scholar]
- 114.Guo TW-A, Patnaik R, Kuhlmann K, Rai AJ, Sia SK. Lab Chip. 2015;15:3514–3520. doi: 10.1039/c5lc00609k. [DOI] [PubMed] [Google Scholar]
- 115.Bond M, Elguea C, Yan JS, Pawlowski M, Williams J, Wahed A, Oden M, Tkaczyk TS, Richards-Kortum R. Lab Chip. 2013;13:2381. doi: 10.1039/c3lc40908b. [DOI] [PubMed] [Google Scholar]
- 116.PATH. Maternal anemia rapid landscape analysis document. 2012 [Google Scholar]
- 117.Darshana LGT, Uluwaduge DI. Anemia. 2014;2014:1–4. doi: 10.1155/2014/531670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Marn H, Critchley JA. Lancet Glob Heal. 2016;4:e251–e265. doi: 10.1016/S2214-109X(16)00005-X. [DOI] [PubMed] [Google Scholar]
- 119.Medina Lara A, Mundy C, Kandulu J, Chisuwo L, Bates I. J Clin Pathol. 2005;58:56–60. doi: 10.1136/jcp.2004.018366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Yang X, Piety NZ, Vignes SM, Benton MS, Kanter J, Shevkoplyas SS. Clin Chem. 2013;59:1506–1513. doi: 10.1373/clinchem.2013.204701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Tyburski EA, Gillespie SE, Stoy WA, Mannino RG, Weiss AJ, Siu AF, Bulloch RH, Thota K, Cardenas A, Session W, Khoury HJ, O’Connor S, Bunting ST, Boudreaux J, Forest CR, Gaddh M, Leong T, Lyon LA, Lam WA. J Clin Invest. 2014;124:4387–4394. doi: 10.1172/JCI76666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Bond M, Mvula J, Molyneux E, Richards-Kortum R. presented in part at 2014 IEEE Healthcare Innovation Conference (HIC); 2014. [Google Scholar]
- 123.Thompson BL, Gilbert RJ, Mejia M, Shukla N, Haverstick DM, Garner GT, Landers JP. Anal Chim Acta. 2016;924:1–8. doi: 10.1016/j.aca.2016.04.028. [DOI] [PubMed] [Google Scholar]
- 124.Bhamla MS, Benson B, Chai C, Katsikis G, Johri A, Prakash M. Nat Biomed Eng. 2017;1:9. [Google Scholar]
- 125.Bond M, Richards-Kortum R. Nat Biomed Eng. 2017;1:17. [Google Scholar]
- 126.Brown J, Theis L, Kerr L, Zakhidova N, O’Connor K, Uthman M, Oden ZM, Richards-Kortum R. Am J Trop Med Hyg. 2011;85:327–332. doi: 10.4269/ajtmh.2011.10-0399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Wong AP, Gupta M, Shevkoplyas SS, Whitesides GM. Lab Chip. 2008;8:2032–7. doi: 10.1039/b809830c. [DOI] [PubMed] [Google Scholar]
- 128.Trebbels D, Hradetzky D, Zengerle R. presented in part at 2009 Annual International Conference of the IEEE Engineering in Medicine and Biology Society; 2009. [DOI] [PubMed] [Google Scholar]
- 129.Punter-Villagrasa J, Cid J, Páez-Avilés C, Rodríguez-Villarreal I, Juanola-Feliu E, Colomer-Farrarons J, Miribel-Català P. Sensors. 2015;15:4564–4577. doi: 10.3390/s150204564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Berry SB, Fernandes SC, Rajaratnam A, DeChiara NS, Mace CR. Lab Chip. 2016;16:3689–3694. doi: 10.1039/c6lc00895j. [DOI] [PubMed] [Google Scholar]
- 131.Ngwenya S. Int J Womens Health. 2016;8:647–650. doi: 10.2147/IJWH.S119232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.James A, Cooper DL, Paidas MJ. Int J Womens Health. 2015 doi: 10.2147/IJWH.S89573. [DOI] [Google Scholar]
- 133.Just S. Semin Thromb Hemost. 2016;43:075–091. doi: 10.1055/s-0036-1592164. [DOI] [PubMed] [Google Scholar]
- 134.Ettel M, Nardi MA, McVoy L. Int J Lab Hematol. 2017 doi: 10.1111/ijlh.12622. [DOI] [PubMed] [Google Scholar]
- 135.Royal College of Obstetricians and Gynaecologists. Prevention and management of postpartum haemorrhage. 2009 [Google Scholar]
- 136.Atukunda EC, Mugyenyi GR, Obua C, Atuhumuza EB, Musinguzi N, Tornes YF, Agaba AG, Siedner MJ. PLoS One. 2016;11:e0152408. doi: 10.1371/journal.pone.0152408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Li H, Han D, Pauletti GM, Steckl AJ. Lab Chip. 2014;14:4035–4041. doi: 10.1039/c4lc00716f. [DOI] [PubMed] [Google Scholar]
- 138.Hancock A, Weeks AD, Lavender DT. BMC Pregnancy Childbirth. 2015;15:230. doi: 10.1186/s12884-015-0653-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.PATH. Blood Loss Measurement. 2013 [Google Scholar]
- 140.Prata N, Quaiyum MA, Passano P, Bell S, Bohl DD, Hossain S, Azmi AJ, Begum M. Soc Sci Med. 2012;75:2021–2027. doi: 10.1016/j.socscimed.2012.06.028. [DOI] [PubMed] [Google Scholar]
- 141.The Economist Newspaper Limited. Econ. 2013 [Google Scholar]
- 142.Prata N, Mbaruku G, Campbell M. Int J Gynecol Obstet. 2005;89:49–50. doi: 10.1016/j.ijgo.2005.01.019. [DOI] [PubMed] [Google Scholar]
- 143.Spangler SA. Med Anthropol Q. 2011;25:479–498. doi: 10.1111/j.1548-1387.2011.01181.x. [DOI] [PubMed] [Google Scholar]
- 144. [accessed March 2017];Kelly’s Pad. http://www.healthklin.com/Kellys-Pad.
- 145. [accessed March 2017];Kelly’s Pad. http://maternova.org/kellys-pad.
- 146. [accessed March 2017];Calibrated obstetric drape (case of 100) https://maternova.net/products/calibrated-obstetric-drape.
- 147.Lertbunnaphong T, Lapthanapat N, Leetheeragul J, Hakularb P, Ownon A. Singapore Med J. 2015;57:325–328. doi: 10.11622/smedj.2016107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Patel A, Goudar SS, Geller SE, Kodkany BS, Edlavitch SA, Wagh K, Patted SS, Naik VA, Moss N, Derman RJ. Int J Gynecol Obstet. 2006;93:220–224. doi: 10.1016/j.ijgo.2006.02.014. [DOI] [PubMed] [Google Scholar]
- 149.Wilcox L, Ramprasad C, Gutierrez A, Oden M, Richards-Kortum R, Sangi-Haghpeykar H, Gandhi M. Matern Child Health J. 2017;21:516–523. doi: 10.1007/s10995-016-2135-5. [DOI] [PubMed] [Google Scholar]
- 150.Cortet M, Deneux-Tharaux C, Dupont C, Colin C, Rudigoz R-C, Bouvier-Colle M-H, Huissoud C. Br J Anaesth. 2012;108:984–989. doi: 10.1093/bja/aes096. [DOI] [PubMed] [Google Scholar]
- 151.Dudek MM, Lindahl TL, Killard AJ. Anal Chem. 2010;82:2029–2035. doi: 10.1021/ac902763a. [DOI] [PubMed] [Google Scholar]
- 152.Nwanodi O. Healthcare. 2016;4:26. doi: 10.3390/healthcare4020026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.National Institute for Health and Care Excellence. PlGF-based testing to help diagnose suspected pre-eclampsia (Triage PlGF test, Elecsys immunoassay sFlt-1/PlGF ratio, DELFIA Xpress PlGF 1-2-3 test, and BRAHMS sFlt-1 Kryptor/BRAHMS PlGF plus Kryptor PE ratio) 2016 [Google Scholar]
- 154.Magee LA, Von Dadelszen P, Stones W, Mathai M, editors. The FIGO textbook of pregnancy hypertension: An evidence-based guide to monitoring, prevention and management. The Global Library of Women’s Medicine; London: 2016. [Google Scholar]
- 155.Knight M. Obstet Anesth Dig. 2008;28:145–146. [Google Scholar]
- 156.Buhimschi IA, Nayeri UA, Zhao G, Shook LL, Pensalfini A, Funai EF, Bernstein IM, Glabe CG, Buhimschi CS. Sci Transl Med. 2014;6:245ra92–245ra92. doi: 10.1126/scitranslmed.3008808. [DOI] [PubMed] [Google Scholar]
- 157.Khashia KM, Willett MJ, Elgawly RM. J Obstet Gynaecol (Lahore) 2007;27:388–389. doi: 10.1080/01443610701327602. [DOI] [PubMed] [Google Scholar]
- 158.Herrick TM, Harner-Jay CM, Levisay AM, Coffey PS, Free MJ, LaBarre PD. BMC Pregnancy Childbirth. 2014;14:10. doi: 10.1186/1471-2393-14-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Osungbade KO, Ige OK. J Pregnancy. 2011;2011:1–6. doi: 10.1155/2011/481095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. [accessed March 2017];Proteinuria testing: Technology Solutions Landscape. http://sites.path.org/mnhtech/assessment/preeclampsia-and-eclampsia/proteinuria/technology-solution/
- 161.Villar J, Say L, Shennan A, Lindheimer M, Duley L, Conde-Agudelo A, Merialdi M. Int J Gynecol Obstet. 2004;85:S28–S41. doi: 10.1016/j.ijgo.2004.03.009. [DOI] [PubMed] [Google Scholar]
- 162.Baker EC, Hezelgrave N, Magesa SM, Edmonds S, de Greeff A, Shennan A. Trop Doct. 2012;42:101–103. doi: 10.1258/td.2011.110352. [DOI] [PubMed] [Google Scholar]
- 163.Oiyemhonlan B, Udofia E, Punguyire D. Afr J Reprod Health. 2013;17:129–40. [PubMed] [Google Scholar]
- 164.Malawi Ministry of Health and ICF International. Malawi Service Provision Assessment 2013–14. Lilongwe, Malawi: 2014. [Google Scholar]
- 165.de Greef A, Ashraf D, Saad L, Hezelgrave NL, Shennan AH. Arch Dis Child - Fetal Neonatal Ed. 2011;96:Fa105-Fa105. [Google Scholar]
- 166.Parati G, Kilama MO, Faini A, Facelli E, Ochen K, Opira C, Mendis S, Wang J, Atkins N, O’Brien E. Hypertension. 2010;56:1047–1053. doi: 10.1161/HYPERTENSIONAHA.110.160408. [DOI] [PubMed] [Google Scholar]
- 167.Kewalbansing PV, Ishwardat ARPP, Brewster LM, Oehlers G, van Montfrans GA. Blood Press Monit. 2013;18:78–84. doi: 10.1097/MBP.0b013e32835ebafe. [DOI] [PubMed] [Google Scholar]
- 168.Carey D. Omron HEM-Solar(HEM-4500-SOLE) UBM; 2011. [Google Scholar]
- 169.de Greeff A, Nathan H, Stafford N, Liu B, Shennan AH. Blood Press Monit. 2008;13:342–348. doi: 10.1097/MBP.0b013e32830fd07c. [DOI] [PubMed] [Google Scholar]
- 170.Nathan HL, de Greeff A, Hezelgrave NL, Chappell LC, Shennan AH. Blood Press Monit. 2015;20:299–302. doi: 10.1097/MBP.0000000000000134. [DOI] [PubMed] [Google Scholar]
- 171.Nathan H, El Ayadi A, Hezelgrave N, Seed P, Butrick E, Miller S, Briley A, Bewley S, Shennan A. BJOG An Int J Obstet Gynaecol. 2015;122:268–275. doi: 10.1111/1471-0528.13206. [DOI] [PubMed] [Google Scholar]
- 172. [accessed March 2017];Nissei DS-400 Semi-Automatic Upper Arm BPM. http://www.white-medical.co.uk/Nissei-DS-400-Semi-Automatic-Upper-Arm-BPM.
- 173.de Greeff A, Shennan AH. Trop Doct. 2015;45:168–173. doi: 10.1177/0049475515581542. [DOI] [PubMed] [Google Scholar]
- 174.Casiglia E, Tikhonoff V, Albertini F, Palatini P. Hypertension. 2016;68:896–903. doi: 10.1161/HYPERTENSIONAHA.116.07961. [DOI] [PubMed] [Google Scholar]
- 175.Rogoza AN, Pavlova TS, Sergeeva MV. Blood Press Monit. 2000;5:227–31. doi: 10.1097/00126097-200008000-00006. [DOI] [PubMed] [Google Scholar]
- 176.Davis PD, Dennis JL, Railton R. J Hum Hypertens. 2004 doi: 10.1038/sj.jhh.1001804. [DOI] [PubMed] [Google Scholar]
- 177. [accessed July 2007];LifeSource UA-767 Blood Pressure Monitor Review. http://bloodpressuremonitorguide.com/lifesource-ua-767-review.
- 178.A&D Medical. Advanced/Deluxe One Step Auto-Inflation: Instruction Guide- Model UA-767 Plus. 2009 [Google Scholar]
- 179.Golparvar M, Naddafnia H, Saghaei M. Anesth Analg. 2002;95:1686–90. doi: 10.1097/00000539-200212000-00040. [DOI] [PubMed] [Google Scholar]
- 180.Arteta C, Domingos JS, Pimentel MAF, Santos MD, Chiffot C, Springer D, Raghu A, Clifford GD. In: Wireless Mobile Communication and Healthcare. MobiHealth 2011. Lecture Notes of the Institute for Computer Sciences, Social Informatics and Telecommunications Engineering. Nikita KS, Lin JC, Fotiadis DI, Arredondo Waldmeyer MT, editors. Vol. 83. Springer; Berlin, Heidelberg: 2012. pp. 335–342. [Google Scholar]
- 181.PATH. Candidate blood based biomarkers for preeclampsia testing. 2014 [Google Scholar]
- 182.Acestor N, Goett J, Lee A, Herrick TM, Engelbrecht SM, Harner-Jay CM, Howell BJ, Weigl BH. Clin Chem Lab Med. 2016;54:17–27. doi: 10.1515/cclm-2015-0069. [DOI] [PubMed] [Google Scholar]
- 183.Andersen LB, Frederiksen-Møller B, Work Havelund K, Dechend R, Jørgensen JS, Jensen BL, Nielsen J, Lykkedegn S, Barington T, Christesen HT. J Am Soc Hypertens. 2015;9:86–96. doi: 10.1016/j.jash.2014.11.008. [DOI] [PubMed] [Google Scholar]
- 184.Rasanen J, Quinn MJ, Laurie A, Bean E, Roberts CT, Nagalla SR, Gravett MG. Am J Obstet Gynecol. 2015;212:82.e1–82.e9. doi: 10.1016/j.ajog.2014.07.052. [DOI] [PubMed] [Google Scholar]
- 185.Frampton GK, Jones J, Rose M, Payne L. Health Technol Assess (Rockv) 2016;20:1–160. doi: 10.3310/hta20870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Benton SJ, Hu Y, Xie F, Kupfer K, Lee S-W, Magee LA, von Dadelszen P. Am J Obstet Gynecol. 2011;205:469.e1–469.e8. doi: 10.1016/j.ajog.2011.06.058. [DOI] [PubMed] [Google Scholar]
- 187.Diagnostics Assessment Programme. Pre-eclampsia: The Triage PlGF test, Elecsys immunoassay sFlt-1/PlGF ratio, DELFIA Xpress PlGF 1-2-3 test and BRAHMS sFlt-1 Kryptor/PlGF plus Kryptor PE ratio to aid the assessment of suspected pre-eclampsia. 2015 [Google Scholar]
- 188.Kirkpatrick T, Dolecki MN. [accessed March 2017];BD Licenses Technology to Advance Testing for Preeclampsia, Gestational Diabetes: Tests from DiabetOmics to be Integrated into BD Veritor™ Point-of-Care Diagnostic Platform. https://www.bd.com/press/2016/BD-Licenses-Technology-to-Advance-Testing-for-Preeclampsia-Gestational-Diabetes.aspx.
- 189. [accessed March 2017];BD Veritor™ System for the Rapid Detection of Flu A+B. https://www.fishersci.com/shop/products/bd-veritor-system-the-rapid-detection-flu-a-b-3/p-4322335.
- 190.Wang T, Zhou R, Gao L, Wang Y, Song C, Gong Y, Jia J, Xiong W, Dai L, Zhang L, Hu H. Hypertension. 2014;64:846–851. doi: 10.1161/HYPERTENSIONAHA.113.02688. [DOI] [PubMed] [Google Scholar]
- 191.Sammar M, Syngelaki A, Sharabi-Nov A, Nicolaides K, Meiri H. Fetal Diagn Ther. 2017;41:23–31. doi: 10.1159/000444450. [DOI] [PubMed] [Google Scholar]
- 192.Seale AC, Mwaniki M, Newton CR, Berkley Ja. Lancet Infect Dis. 2009;9:428–438. doi: 10.1016/S1473-3099(09)70172-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Arulkumaran N, Singer M. Best Pract Res Clin Obstet Gynaecol. 2013;27:893–902. doi: 10.1016/j.bpobgyn.2013.07.004. [DOI] [PubMed] [Google Scholar]
- 194.Meisner M. Clin Chim Acta. 2002;323:17–29. doi: 10.1016/s0009-8981(02)00101-8. [DOI] [PubMed] [Google Scholar]
- 195.Meisner M, Tschaikowsky K, Palmaers T, Schmidt J. Crit Care. 1999;3:45. doi: 10.1186/cc306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Kim K-E, Han J-Y. Korean J Lab Med. 2010;30:153. doi: 10.3343/kjlm.2010.30.2.153. [DOI] [PubMed] [Google Scholar]
- 197.Tang BMP, Eslick GD, Craig JC, McLean AS. Lancet Infect Dis. 2007;7:210–7. doi: 10.1016/S1473-3099(07)70052-X. [DOI] [PubMed] [Google Scholar]
- 198.Zou Q, Wen W, Zhang X-C. World J Emerg Med. 2014;5:16–9. doi: 10.5847/wjem.j.issn.1920-8642.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Okamura Y, Yokoi H. Clin Chim Acta. 2011;412:2157–2161. doi: 10.1016/j.cca.2011.07.024. [DOI] [PubMed] [Google Scholar]
- 200.Volanakis J. Mol Immunol. 2001;38:189–197. doi: 10.1016/s0161-5890(01)00042-6. [DOI] [PubMed] [Google Scholar]
- 201.Swanson C, D’Andrea A. Clin Chem. 2013;59:641–8. doi: 10.1373/clinchem.2012.200360. [DOI] [PubMed] [Google Scholar]
- 202.Leung W, Chan CP, Rainer TH, Ip M, Cautherley GWH, Renneberg R. J Immunol Methods. 2008;336:30–36. doi: 10.1016/j.jim.2008.03.009. [DOI] [PubMed] [Google Scholar]
- 203.Caughey AB, Robinson JN, Norwitz ER. Rev Obstet Gynecol. 2008;1:11–22. [PMC free article] [PubMed] [Google Scholar]
- 204.Chhabra S, Kumar N, Kalra P. J Basic Clin Reprod Sci. 2015;4:29. [Google Scholar]
- 205.Palacio M, Kühnert M, Berger R, Larios CL, Marcellin L. BMC Pregnancy Childbirth. 2014;14:183. doi: 10.1186/1471-2393-14-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Rutanen E-M, Kärkkäinen TH, Lehtovirta J, Uotila JT, Hinkula MK, Hartikainen A-L. Clin Chim Acta. 1996;253:91–101. doi: 10.1016/0009-8981(96)80001-e. [DOI] [PubMed] [Google Scholar]
- 207.Morris PG, Jain K. J Obstet Gynaecol (Lahore) 1998;18:33–36. doi: 10.1080/01443619868235. [DOI] [PubMed] [Google Scholar]
- 208.Rogers LC, Scott L, Block JE. Clin Med Insights Reprod Heal. 2016;10:15. doi: 10.4137/CMRH.S38386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Eleje GU, Ezugwu EC, Eke AC, Ikechebelu JI, Obiora CC, Ojiegbe NO, Ezebialu IU, Ezeama CO, Nwosu BO, Udigwe GO, Okafor CI, Ezugwu FO. J Perinat Med. 2017:45. doi: 10.1515/jpm-2016-0204. [DOI] [PubMed] [Google Scholar]
- 210.Chaemsaithong P, Romero R, Korzeniewski SJ, Martinez-Varea A, Dong Z, Yoon BH, Hassan SS, Chaiworapongsa T, Yeo L. J Matern Neonatal Med. 2016;29:349–359. doi: 10.3109/14767058.2015.1006620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Wiberg-Itzel E, Pettersson H, Cnattingius S, Nordstrom L. BJOG An Int J Obstet Gynaecol. 2006;113:1426–1430. doi: 10.1111/j.1471-0528.2006.01088.x. [DOI] [PubMed] [Google Scholar]
- 212.Liang D, Qi H, Luo X, Xiao X, Jia X. J Obstet Gynaecol Res. 2014;40:1555–1560. doi: 10.1111/jog.12381. [DOI] [PubMed] [Google Scholar]
- 213.Abdelazim IA, Al-Sherbeeny MM, Ibrahim MEM, Fahmy AA, Rabei NH, Aziz Khalifa AA. Acta Medial Int. 2016;3:69–74. [Google Scholar]
- 214.McQuivey RW, Block JE. Med Devices Evid Res. 2016;6:69–74. doi: 10.2147/MDER.S106106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Wiberg-Itzel E, Cnattingius S, Nordström L. BJOG An Int J Obstet Gynaecol. 2005;112:754–758. doi: 10.1111/j.1471-0528.2004.00521.x. [DOI] [PubMed] [Google Scholar]
- 216.El-Messidi A, Cameron A. J Obstet Gynaecol Canada. 2010;32:561–569. doi: 10.1016/S1701-2163(16)34525-X. [DOI] [PubMed] [Google Scholar]
- 217.Desai Shyam V, Tank P. Handbook on Preterm Prelabor Rupture of Membranes in a Low Resource Setting. 1. Jaypee Brothers Medical Publishers (P) Ltd; New Delhi: 2012. [Google Scholar]
- 218. [accessed January 2017];Lactate Pro 2 Test Strips 25/Box. http://www.habdirect.co.uk/lactate-pro-2-test-strips.
- 219. [accessed January 2017];AmniSure ROM Test (Rupture of [fetal] Membranes test) https://www.qiagen.com/us/shop/sample-technologies/protein/protein-preparation/amnisure-rom-test-10-min-us/#orderinginformation.
- 220. [accessed January 2017];AmniSure® ROM Test - box of 10 tests. http://www.medshop.co.nz/Amnisure.
- 221.South Eastern Sydney Local Health District Obstetric Clinical Guidelines Group. Actim PROM: Qualitative diagnosis of preterm premature rupture of membranes guideline. Sydney, Australia: 2009. [Google Scholar]
- 222. [accessed January 2017];ROM Plus Fetal Membranes Rupture Test. http://www.superiormedical.com/product/rom-plus-fetal-membranes-rupture-test/
- 223.Osman OM, Elghazaly M. Open J Obstet Gynecol. 2014;4:967–972. [Google Scholar]
- 224.Kafali H, Öksüzler C. Arch Gynecol Obstet. 2007;275:157–160. doi: 10.1007/s00404-006-0240-1. [DOI] [PubMed] [Google Scholar]
- 225.Mohamed AMM, Mostafa WAI. Kasr Al-Aini J Obstet Gynecol. 2011;2:41–47. [Google Scholar]
- 226.Hanfy A. Med J Cairo Univ. 2010;78:313–317. [Google Scholar]
- 227.Zanjani MS, Haghighi L. J Obstet Gynaecol Res. 2012;38:505–508. doi: 10.1111/j.1447-0756.2011.01692.x. [DOI] [PubMed] [Google Scholar]
- 228.Tigga M, Malik S. Int J Reprod Contraception, Obstet Gynecol. 2015;4:1070–1075. [Google Scholar]
- 229.Gezer C, Ekin A, Golbasi C, Kocahakimoglu C, Bozkurt U, Dogan A, Solmaz U, Golbasi H, Taner CE. J Matern Neonatal Med. 2016 doi: 10.1080/14767058.2016.1188072. [DOI] [PubMed] [Google Scholar]
- 230.Fung K-K, Chan CP-Y, Renneberg R. Anal Bioanal Chem. 2009;393:1281–1287. doi: 10.1007/s00216-008-2551-5. [DOI] [PubMed] [Google Scholar]
- 231.Bruijn MMC, Vis JY, Wilms FF, Oudijk MA, Kwee A, Porath MM, Oei G, Scheepers HCJ, Spaanderman MEA, Bloemenkamp KWM, Haak MC, Bolte AC, Vandenbussche FPHA, Woiski MD, Bax CJ, Cornette JMJ, Duvekot JJ, Nij Bijvank BWA, van Eyck J, Franssen MTM, Sollie KM, van der Post JAM, Bossuyt PMM, Opmeer BC, Kok M, Mol BWJ, van Baaren G-J. Eur J Obstet Gynecol Reprod Biol. 2016;206:220–224. doi: 10.1016/j.ejogrb.2016.09.018. [DOI] [PubMed] [Google Scholar]
- 232.Corabian P. The Actim™ Partus versus the TLiIQ® System as rapid response tests to aid in diagnosing preterm labour in sympotmatic women. Edmonton AB Canada; 2008. [Google Scholar]
- 233.Abbott DS, Radford SK, Seed PT, Tribe RM, Shennan AH. Am J Obstet Gynecol. 2013;208:122.e1–122.e6. doi: 10.1016/j.ajog.2012.10.890. [DOI] [PubMed] [Google Scholar]
- 234.Bruijn M, Vis J, Wilms F, Oudijk M, Kwee A, Porath M, Oei G, Scheepers H, Spaanderman M, Bloemenkamp K, Haak M, Bolte A, Vandenbussche F, Woiski M, Bax C, Cornette J, Duvekot J, Nij Bijvanck B, van Eyck J, Franssen M, Sollie K, van der Post J, Bossuyt P, Opmeer B, Kok M, Mol B, van Baaren G-J. BJOG An Int J Obstet Gynaecol. 2016;123:1965–1971. doi: 10.1111/1471-0528.13752. [DOI] [PubMed] [Google Scholar]
- 235.Nien JK, Yoon BH, Espinoza J, Kusanovic JP, Erez O, Soto E, Richani K, Gomez R, Hassan S, Mazor M, Edwin S, Bahado-Singh R, Romero R. Am J Obstet Gynecol. 2006;195:1025–1030. doi: 10.1016/j.ajog.2006.06.054. [DOI] [PubMed] [Google Scholar]
- 236.Klouche M, Schröder U. Clin Chem Lab Med. 2008;46:888–908. doi: 10.1515/CCLM.2008.157. [DOI] [PubMed] [Google Scholar]
- 237.Aellen S, Que Y-A, Guignard B, Haenni M, Moreillon P. Antimicrob Agents Chemother. 2006;50:1913–1920. doi: 10.1128/AAC.00869-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Singer M, Deutschman CS, Seymour CW, Shankar-Hari M, Annane D, Bauer M, Bellomo R, Bernard GR, Chiche J-D, Coopersmith CM, Hotchkiss RS, Levy MM, Marshall JC, Martin GS, Opal SM, Rubenfeld GD, van der Poll T, Vincent J, Angus DC. JAMA. 2016;315:801. doi: 10.1001/jama.2016.0287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Funes-Huacca M, Wu A, Szepesvari E, Rajendran P, Kwan-Wong N, Razgulin A, Shen Y, Kagira J, Campbell R, Derda R. Lab Chip. 2012;12:4269. doi: 10.1039/c2lc40391a. [DOI] [PubMed] [Google Scholar]
- 240.Ayyash S, Wu Wen-I, Ravi Selvaganapathy P. presented in part at 2014 IEEE Healthcare Innovation Conference (HIC); 2014. [Google Scholar]
- 241.Smarticles Technology. [accessed June 2016]; https://www.rochemicrobiologytests.com/healthcare-associated-infections/innovative-solutions.html.
- 242.Deiss F, Funes-Huacca ME, Bal J, Tjhung KF, Derda R. Lab Chip. 2014;14:167–71. doi: 10.1039/c3lc50887k. [DOI] [PubMed] [Google Scholar]
- 243.Houwen B. Lab Hematol. 2001;7:89–100. [Google Scholar]
- 244.Laudicina RJ, Simonian Y. In: Clinical Laboratory Hematology. 2. McKenzie SB, Williams JL, editors. Pearson; Boston: 2010. pp. 104–145. [Google Scholar]
- 245.Russcher H, Van Deursen N, Ermens T, De Jonge R. Ned Tijdschr voor Klin Chemie en Lab. 2013;38:140–141. [Google Scholar]
- 246.Majors CE, Pawlowski ME, Tkaczyk T, Richards-Kortum RR. presented in part at 2014 IEEE Healthcare Innovation Conference (HIC); Seattle, WA. 2014. [Google Scholar]
- 247.Forcucci A, Pawlowski ME, Majors C, Richards-Kortum R, Tkaczyk TS. Biomed Opt Express. 2015;6:4433. doi: 10.1364/BOE.6.004433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Smith ZJ, Gao T, Chu K, Lane SM, Matthews DL, Dwyre DM, Hood J, Tatsukawa K, Heifetz L, Wachsmann-Hogiu S. Lab Chip. 2014;14:3029–36. doi: 10.1039/c4lc00567h. [DOI] [PubMed] [Google Scholar]
- 249.Zhu H, Sencan I, Wong J, Dimitrov S, Tseng D, Nagashima K, Ozcan A. Lab Chip. 2013;13:1282. doi: 10.1039/c3lc41408f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Lunze K, Bloom DE, Jamison DT, Hamer DH. BMC Med. 2013;11:24. doi: 10.1186/1741-7015-11-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Kennedy N, Gondwe L, Morley DC. Lancet. 2000;355:1364. doi: 10.1016/s0140-6736(05)72592-7. [DOI] [PubMed] [Google Scholar]
- 252.Green DA. Arch Dis Child - Fetal Neonatal Ed. 2006;91:F96–F98. doi: 10.1136/adc.2005.078410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Pejaver RK, Nisarga R, Gowda B. Indian J Pediatr. 2004;71:795–6. doi: 10.1007/BF02730715. [DOI] [PubMed] [Google Scholar]
- 254.Mole TB, Kennedy N, Ndoya N, Emond A. PLoS One. 2012;7:e45823. doi: 10.1371/journal.pone.0045823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Stoner K, Frisone J. Use of a Skin Patch Device to Obtain Temperature Measurements (Adult Study) 2015 [Google Scholar]
- 256.Tanigasalam V, Bhat V, Adhisivam B. Novel method of detecting hypothermia in low birth weight neonates in resource limited settings using wristband (Bempu Bracelet) device, a descriptive study. Pondicherry: 2016. [Google Scholar]
- 257.Greco C, Arnolda G, Boo N-Y, Iskander IF, Okolo AA, Rohsiswatmo R, Shapiro SM, Watchko J, Wennberg RP, Tiribelli C, Coda Zabetta CD. Neonatology. 2016;110:172–180. doi: 10.1159/000445708. [DOI] [PubMed] [Google Scholar]
- 258.World Health Organization. Bilirubinometer. 2011. [Google Scholar]
- 259.Rubio A, Epiard C, Gebus M, Deiber M, Samperiz S, Genty C, Ego A, Debillon T. Neonatology. 2017;111:1–7. doi: 10.1159/000447736. [DOI] [PubMed] [Google Scholar]
- 260.Rylance S, Yan J, Molyneux E. Paediatr Int Child Health. 2014;34:101–107. doi: 10.1179/2046905513Y.0000000050. [DOI] [PubMed] [Google Scholar]
- 261.Olusanya BO, Imosemi DO, Emokpae AA. Pediatrics. 2016;138:e20160907–e20160907. doi: 10.1542/peds.2016-0907. [DOI] [PubMed] [Google Scholar]
- 262.Murli L, Thukral A, Sankar MJ, Vishnubhatla S, Deorari AK, Paul VK, Sakariah A, Dolma, Agarwal R. J Perinatol. 2017;37:182–187. doi: 10.1038/jp.2016.189. [DOI] [PubMed] [Google Scholar]
- 263.Mansouri M, Mahmoodnejad A, Sarvestani RT, Gharibi F. Int J Pediatr. 2015;3:633–641. [Google Scholar]
- 264.Olusanya BO, Ogunlesi TA, Kumar P, Boo N-Y, Iskander IF, de Almeida MFB, Vaucher YE, Slusher TM. BMC Pediatr. 2015;15:39. doi: 10.1186/s12887-015-0358-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Grohmann K. Pediatrics. 2006;117:1174–1183. doi: 10.1542/peds.2005-0590. [DOI] [PubMed] [Google Scholar]
- 266.Coda Zabetta CD, Iskander IF, Greco C, Bellarosa C, Demarini S, Tiribelli C, Wennberg RP. Neonatology. 2013;103:177–181. doi: 10.1159/000345425. [DOI] [PubMed] [Google Scholar]
- 267.Greco C, Iskander IF, Akmal DM, El Houchi SZ, Khairy DA, Bedogni G, Wennberg RP, Tiribelli C, Zabetta CDC. Nat Publ Gr. 2017 doi: 10.1038/jp.2017.94. [DOI] [PubMed] [Google Scholar]
- 268.Su J, Wang L. Transl Pediatr. 2012;1:108–15. doi: 10.3978/j.issn.2224-4336.2012.04.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Sweet CB, Grayson S, Polak M. J Pediatr Pharmacol Ther. 2013;18:199–208. doi: 10.5863/1551-6776-18.3.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Bergenstal RM. Insulin. 2008;3:5–14. [Google Scholar]
- 271.Tack C, Pohlmeier H, Behnke T, Schmid V, Grenningloh M, Forst T, Pfützner A. Diabetes Technol Ther. 2012;14:330–337. doi: 10.1089/dia.2011.0170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Klonoff DC. Diabetes Spectr. 2014;27:174–179. doi: 10.2337/diaspect.27.3.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Beardsall K. Early Hum Dev. 2010;86:263–267. doi: 10.1016/j.earlhumdev.2010.05.005. [DOI] [PubMed] [Google Scholar]
- 274.Vanavanan S, Santanirand P, Chaichanajarernkul U, Chittamma A, DuBois JA, Shirey T, Heinz M. Clin Biochem. 2010;43:186–192. doi: 10.1016/j.clinbiochem.2009.09.010. [DOI] [PubMed] [Google Scholar]
- 275.Lockyer MG, Fu K, Edwards RM, Collymore L, Thomas J, Hill T, Devaraj S. Clin Biochem. 2014;47:840–843. doi: 10.1016/j.clinbiochem.2014.01.004. [DOI] [PubMed] [Google Scholar]
- 276.Makaya T, Memmott A, Bustani P. J Paediatr Child Health. 2012;48:342–346. doi: 10.1111/j.1440-1754.2011.02253.x. [DOI] [PubMed] [Google Scholar]
- 277.Ranger M, Chau CMY, Garg A, Woodward TS, Beg MF, Bjornson B, Poskitt K, Fitzpatrick K, Synnes AR, Miller SP, Grunau RE. PLoS One. 2013;8:e76702. doi: 10.1371/journal.pone.0076702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Beardsall K. Arch Dis Child - Fetal Neonatal Ed. 2005;90:F307–f310. doi: 10.1136/adc.2004.051979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Lodwig V, Kulzer B, Schnell O, Heinemann L. J Diabetes Sci Technol. 2014;8:397–402. doi: 10.1177/1932296814525825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.DeSalvo D, Buckingham B. Curr Diab Rep. 2013;13:657–662. doi: 10.1007/s11892-013-0398-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Rodbard D. Diabetes Technol Ther. 2016;18:S2-3–S2-13. doi: 10.1089/dia.2018.0092. [DOI] [PubMed] [Google Scholar]
- 282.Harris DL, Battin MR, Weston PJ, Harding JE. J Pediatr. 2010;157:198–202.e1. doi: 10.1016/j.jpeds.2010.02.003. [DOI] [PubMed] [Google Scholar]
- 283.Woo HC, Tolosa L, El-Metwally D, Viscardi RM. Arch Dis Child Fetal Neonatal Ed. 2014;99:F153–7. doi: 10.1136/archdischild-2013-304682. [DOI] [PubMed] [Google Scholar]
- 284.Chin CD, Linder V, Sia SK. Lab Chip. 2012;12:2118. doi: 10.1039/c2lc21204h. [DOI] [PubMed] [Google Scholar]
- 285.Papadea C, Foster J, Grant S, Ballard SA, Cate JC, Southgate WM, Purohit DM. Ann Clin Lab Sci. 2002;32:231–43. [PubMed] [Google Scholar]
- 286.Gorkin R, Park J, Siegrist J, Amasia M, Lee BS, Park J-M, Kim J, Kim H, Madou M, Cho Y-K. Lab Chip. 2010;10:1758. doi: 10.1039/b924109d. [DOI] [PubMed] [Google Scholar]
- 287.Nichols JH, Rajadhyaksha A, Rodriguez M. Point Care J Near-Patient Test Technol. 2008;7:7–11. [Google Scholar]
- 288.Martinez AW, Phillips ST, Whitesides GM, Carrilho E. Anal Chem. 2010;82:3–10. doi: 10.1021/ac9013989. [DOI] [PubMed] [Google Scholar]
- 289.Nie Z, Nijhuis CA, Gong J, Chen X, Kumachev A, Martinez AW, Narovlyansky M, Whitesides GM. Lab Chip. 2010;10:477–483. doi: 10.1039/b917150a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Nie Z, Deiss F, Liu X, Akbulut O, Whitesides GM. Lab Chip. 2010;10:3163. doi: 10.1039/c0lc00237b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Nemiroski A, Christodouleas DC, Hennek JW, Kumar AA, Maxwell EJ, Fernandez-Abedul MT, Whitesides GM. Proc Natl Acad Sci. 2014;111:11984–11989. doi: 10.1073/pnas.1405679111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Maxwell EJ, Mazzeo AD, Whitesides GM. MRS Bull. 2013;38:309–314. [Google Scholar]
- 293.Liu L, Johnson HL, Cousens S, Perin J, Scott S, Lawn JE, Rudan I, Campbell H, Cibulskis R, Li M, Mathers C, Black RE. Lancet. 2012;379:2151–2161. doi: 10.1016/S0140-6736(12)60560-1. [DOI] [PubMed] [Google Scholar]
- 294. [accessed March 2017];Newborn death and illness. http://www.who.int/pmnch/media/press_materials/fs/fs_newborndealth_illness/en/
- 295.Chikuse B, Chirwa E, Maluwa A, Malata A, Odland J. Open J Nurs. 2012;2:351–357. [Google Scholar]
- 296.Golubnitschaja O, Yeghiazaryan K, Cebioglu M, Morelli M, Herrera-Marschitz M. EPMA J. 2011;2:197–210. doi: 10.1007/s13167-011-0087-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Bogdanovic G, Babovic A, Rizvanovic M, Ljuca D, Grgic G, DjuranovicMilicic J. Med Arch. 2014;68:102. doi: 10.5455/medarh.2014.68.102-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Pirnareva E, Tsankova M. Ultrasound Obstet Gynecol. 2016;48:322. [Google Scholar]
- 299.Chaturvedi P, Shah N. Indian J Pediatr. 1991;58:63–67. doi: 10.1007/BF02810413. [DOI] [PubMed] [Google Scholar]
- 300. [accessed March 2017];MOYO- Improved Fetal Heart Rate Monitoring for Safer Births. https://savinglivesatbirth.net/summaries/294.
- 301. [accessed March 2017];Moyo Fetal Heart Rate Monitor. http://www.laerdalglobalhealth.com/doc/2555/Moyo-Fetal-Heart-Rate-Monitor.
- 302.Jhpiego. Addressing Labor Complications: A Low-Cost Solution for a Persistent Global Health Challenge | Childbirth. Baltimore: 2011. [Google Scholar]
- 303.Gilstrap LC, Leveno KJ, Burris J, Williams ML, Little BB. Am J Obstet Gynecol. 1989;161:825–30. doi: 10.1016/0002-9378(89)90410-9. [DOI] [PubMed] [Google Scholar]
- 304.Malin GL, Morris RK, Khan KS. BMJ. 2010;340:c1471–c1471. doi: 10.1136/bmj.c1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Ghosh D, Bhakoo ON, Narang A, Dhall K. J Trop Pediatr. 1997;43:108–11. doi: 10.1093/tropej/43.2.108. [DOI] [PubMed] [Google Scholar]
- 306.Wall SN, Lee AC, Niermeyer S, English M, Keenan WJ, Carlo W, Bhutta ZA, Bang A, Narayanan I, Ariawan I, Lawn JE. Int J Gynecol Obstet. 2009;107:S47–S64. doi: 10.1016/j.ijgo.2009.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.World Health Organization. Guidelines on Basic Newborn Resuscitation. 2012. [PubMed] [Google Scholar]
- 308.Leach A, McArdle TF, Banya WA, Krubally O, Greenwood AM, Rands C, Adegbola R, de Francisco A, Greenwood BM. Ann Trop Paediatr. 1999;19:33–43. doi: 10.1080/02724939992617. [DOI] [PubMed] [Google Scholar]
- 309.Stoot LJ, Cairns NA, Cull F, Taylor JJ, Jeffrey JD, Morin F, Mandelman JW, Clark TD, Cooke SJ. Conserv Physiol. 2014;2:cou011-cou011. doi: 10.1093/conphys/cou011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Chin CD, Chin SY, Laksanasopin T, Sia SK. In: Point-of-Care Diagnostics on a Chip. Issadore D, Westervelt RM, editors. Springer-Verlag; Berlin, Heidelberg: 2013. pp. 3–21. [Google Scholar]
- 311.Yang J, Kwak TJ, Zhang X, McClain R, Chang W-J, Gunasekaran S. ACS Sensors. 2016;1:1235–1243. doi: 10.3791/53339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.You D, Hug L, Chen Y, Newby H, Wardlaw T, Mathers C, Boerma T, Hogan D, Ho J, Suzuki E, Pelletier F, Andreev K, Gerland P, Gu D, Li N, Sawyer C, Bay G, Miller T, Faijer DJ, Hill K. Levels & Trends in Child Mortality: Report 2014. 2014 [Google Scholar]
- 313.Meem M, Modak JK, Mortuza R, Morshed M, Islam MS, Saha SK. J Glob Health. 2011;1:201–9. [PMC free article] [PubMed] [Google Scholar]
- 314.Bersani I, Auriti C, Ronchetti MP, Prencipe G, Gazzolo D, Dotta A. Biomed Res Int. 2015;2015:1–10. doi: 10.1155/2015/253520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Kocabaş E, Sarikçioğlu A, Aksaray N, Seydaoğlu G, Seyhun Y, Yaman A. Turk J Pediatr. 2007;49:7–20. [PubMed] [Google Scholar]
- 316.Liu J, Liu F, Liu Y, Wang H-W, Feng Z-C. Chest. 2014;146:383–388. doi: 10.1378/chest.13-2852. [DOI] [PubMed] [Google Scholar]
- 317.Duke T. Arch Dis Child - Fetal Neonatal Ed. 2005;90:F211–f219. doi: 10.1136/adc.2003.048108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Simoes EAF, Cherian T, Chow J, Shahid-Salles Sa, Laxminarayan R, John TJ. Acute Respiratory Infections in Children. 2006 [Google Scholar]
- 319.Redd SC, Vreuls R, Metsing M, Mohobane PH, Patrick E, Moteetee M. Bull World Health Organ. 1994;72:113–8. [PMC free article] [PubMed] [Google Scholar]
- 320.Singhi S, Bhalla AK, Bhandari A, Narang A. Ann Trop Paediatr. 2003;23:135–8. doi: 10.1179/027249303235002206. [DOI] [PubMed] [Google Scholar]
- 321.Noordam AC, Barbera Lainez Y, Sadruddin S, van Heck PM, Chono AO, Acaye GL, Lara V, Nanyonjo A, Ocan C, Kallander K. Health Policy Plan. 2015;30:696–704. doi: 10.1093/heapol/czu047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.King D. BMJ. 2014;349:g6639–g6639. doi: 10.1136/bmj.g6639. [DOI] [PubMed] [Google Scholar]
- 323.World Health Organization. Consolidated guidelines on the use of antiretroviral drugs for treating and preventing HIV infection: recommendations for a public health approach. 2013 [PubMed] [Google Scholar]
- 324.UNAIDS. On the fast track to an AIDS-free generation. 2016 [Google Scholar]
- 325.Woelk GB, Kieffer MP, Walker D, Mpofu D, Machekano R. Trials. 2016;17:88. doi: 10.1186/s13063-016-1202-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Meggi B, Bollinger T, Mabunda N, Vubil A, Tobaiwa O, Quevedo JI, Loquiha O, Vojnov L, Peter TF, Jani IV. PLoS One. 2017;12:e0169497. doi: 10.1371/journal.pone.0169497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Jani IV, Meggi B, Mabunda N, Vubil A, Sitoe NE, Tobaiwa O, Quevedo JI, Lehe JD, Loquiha O, Vojnov L, Peter TF. JAIDS J Acquir Immune Defic Syndr. 2014;67:e1–e4. doi: 10.1097/QAI.0000000000000250. [DOI] [PubMed] [Google Scholar]
- 328.World Health Organization. WHO Prequalification of In Vitro Diagnostics PUBLIC REPORT. 2016 [Google Scholar]
- 329.Lustig J, Cunningham J. Alere q HIV-1 / 2 Detect Point-of-Care Molecular HIV Assay Receives WHO Prequalification. 2016 [Google Scholar]
- 330.Craw P, Balachandran W. Lab Chip. 2012;12:2469. doi: 10.1039/c2lc40100b. [DOI] [PubMed] [Google Scholar]
- 331.Rohrman BA, Richards-Kortum RR. Lab Chip. 2012;12:3082–8. doi: 10.1039/c2lc40423k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Rohrman BA, Leautaud V, Molyneux E, Richards-Kortum RR. PLoS One. 2012;7:e45611. doi: 10.1371/journal.pone.0045611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Rodriguez NM, Linnes JC, Fan A, Ellenson CK, Pollock NR, Klapperich CM. Anal Chem. 2015;87:7872–7879. doi: 10.1021/acs.analchem.5b01594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Choi JR, Hu J, Tang R, Gong Y, Feng S, Ren H, Wen T, Li X, Wan Abas WAB, Pingguan-Murphy B, Xu F. Lab Chip. 2016;16:611–621. doi: 10.1039/c5lc01388g. [DOI] [PubMed] [Google Scholar]
- 335.Lillis L, Lehman D, Singhal MC, Cantera J, Singleton J, Labarre P, Toyama A, Piepenburg O, Parker M, Wood R, Overbaugh J, Boyle DS. PLoS One. 2014;9:e108189. doi: 10.1371/journal.pone.0108189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Crannell ZA, Rohrman B, Richards-Kortum R. PLoS One. 2014;9:e112146. doi: 10.1371/journal.pone.0112146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Crannell ZA, Rohrman B, Richards-Kortum R. Anal Chem. 2014;86:5615–5619. doi: 10.1021/ac5011298. [DOI] [PubMed] [Google Scholar]
- 338.Rodriguez NM, Wong WS, Liu L, Dewar R, Klapperich CM. Lab Chip. 2016;16:753–763. doi: 10.1039/c5lc01392e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Lafleur LK, Bishop JD, Heiniger EK, Gallagher RP, Wheeler MD, Kauffman P, Zhang X, Kline EC, Buser JR, Kumar S, Byrnes SA, Vermeulen NMJ, Scarr NK, Belousov Y, Mahoney W, Toley BJ, Ladd PD, Lutz BR, Yager P. Lab Chip. 2016;16:3777–3787. doi: 10.1039/c6lc00677a. [DOI] [PubMed] [Google Scholar]
- 340.Mauk M, Song J, Bau HH, Gross R, Bushman FD, Collman RG, Liu C. Lab Chip. 2017;17:382–394. doi: 10.1039/c6lc01239f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Wijesooriya NS, Rochat RW, Kamb ML, Turlapati P, Temmerman M, Broutet N, Newman LM. Lancet Glob Heal. 2016;4:e525–e533. doi: 10.1016/S2214-109X(16)30135-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.Arnold SR, Ford-Jones EL. Paediatr Child Health. 2000;5:463–9. doi: 10.1093/pch/5.8.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.Causer LM, Kaldor JM, Fairley CK, Donovan B, Karapanagiotidis T, Leslie DE, Robertson PW, McNulty AM, Anderson D, Wand H, Conway DP, Denham I, Ryan C, Guy RJ. PLoS One. 2014;9:e91504. doi: 10.1371/journal.pone.0091504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344.Jafari Y, Peeling RW, Shivkumar S, Claessens C, Joseph L, Pai NP. PLoS One. 2013;8:e54695. doi: 10.1371/journal.pone.0054695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Kay NS, Peeling RW, Mabey DC. Expert Rev Anti Infect Ther. 2014;12:63–73. doi: 10.1586/14787210.2014.860356. [DOI] [PubMed] [Google Scholar]
- 346.Kleutsch L, Harvey SA, Rennie W. Rapid syphilis tests in Tanzania: A long road to adoption. Case Study. Bethesda, MD: 2009. [Google Scholar]
- 347.Laksanasopin T, Guo TW, Nayak S, Sridhara AA, Xie S, Olowookere OO, Cadinu P, Meng F, Chee NH, Kim J, Chin CD, Munyazesa E, Mugwaneza P, Rai AJ, Mugisha V, Castro AR, Steinmiller D, Linder V, Justman JE, Nsanzimana S, Sia SK. Sci Transl Med. 2015;7:273re1–273re1. doi: 10.1126/scitranslmed.aaa0056. [DOI] [PubMed] [Google Scholar]
- 348.Morshed MG, Singh AE. Clin Vaccine Immunol. 2015;22:137–147. doi: 10.1128/CVI.00681-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349.CDC. [accessed July 2017];Congenital Syphilis. https://www.cdc.gov/std/tg2015/congenital.htm.
- 350.Koek AG, Bruisten SM, Dierdorp M, van Dam AP, Templeton K. Clin Microbiol Infect. 2006;12:1233–1236. doi: 10.1111/j.1469-0691.2006.01566.x. [DOI] [PubMed] [Google Scholar]
- 351.Mlakar J, Korva M, Tul N, Popović M, Poljšak-Prijatelj M, Mraz J, Kolenc M, Resman Rus K, Vesnaver Vipotnik T, Fabjan Vodušek V, Vizjak A, Pižem J, Petrovec M, Avšič Županc T. N Engl J Med. 2016;374:951–958. doi: 10.1056/NEJMoa1600651. [DOI] [PubMed] [Google Scholar]
- 352.Rasmussen SA, Jamieson DJ, Honein MA, Petersen LR. N Engl J Med. 2016;374:1981–1987. doi: 10.1056/NEJMsr1604338. [DOI] [PubMed] [Google Scholar]
- 353.Waggoner JJ, Pinsky A. J Clin Microbiol. 2016;54:860–867. doi: 10.1128/JCM.00279-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354.Chan K, Weaver SC, Wong P-Y, Lie S, Wang E, Guerbois M, Vayugundla SP, Wong S. Sci Rep. 2016;6:38223. doi: 10.1038/srep38223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 355.Song J, Mauk MG, Hackett BA, Cherry S, Bau HH, Liu C. Anal Chem. 2016;88:7289–7294. doi: 10.1021/acs.analchem.6b01632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356.Pardee K, Green AA, Takahashi MK, Braff D, Lambert G, Lee JW, Ferrante T, Ma D, Donghia N, Fan M, Daringer NM, Bosch I, Dudley DM, O’Connor DH, Gehrke L, Collins JJ. Cell. 2016;165:1255–1266. doi: 10.1016/j.cell.2016.04.059. [DOI] [PubMed] [Google Scholar]
- 357.Pardee K, Green AA, Ferrante T, Cameron DE, DaleyKeyser A, Yin P, Collins JJ. Cell. 2014;159:940–954. doi: 10.1016/j.cell.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 358.Heng YJ, Liong S, Permezel M, Rice GE, Di Quinzio MKW, Georgiou HM. Front Physiol. 2015;6:151. doi: 10.3389/fphys.2015.00151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359.Dusse LM, Rios DRA, Pinheiro MB, Cooper AJ, Lwaleed BA. Clin Chim Acta. 2011;412:17–21. doi: 10.1016/j.cca.2010.09.030. [DOI] [PubMed] [Google Scholar]
- 360.Kazmi R, Cooper A, Lwaleed B. Semin Thromb Hemost. 2011;37:131–136. doi: 10.1055/s-0030-1270339. [DOI] [PubMed] [Google Scholar]
- 361.Kenny LC, Broadhurst DI, Dunn W, Brown M, North RA, McCowan L, Roberts C, Cooper GJS, Kell DB, Baker PN. Hypertension. 2010;56:741–749. doi: 10.1161/HYPERTENSIONAHA.110.157297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 362.Blankley RT, Fisher C, Westwood M, North R, Baker PN, Walker MJ, Williamson A, Whetton AD, Lin W, McCowan L, Roberts CT, Cooper GJS, Unwin RD, Myers JE. Mol Cell Proteomics. 2013;12:3148–3159. doi: 10.1074/mcp.M112.026872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 363.Dawonauth L, Rademacher L, L’Omelette AD, Jankee S, Lee Kwai Yan MY, Jeeawoody RB, Rademacher TW. J Reprod Immunol. 2014;101–102:148–152. doi: 10.1016/j.jri.2013.06.001. [DOI] [PubMed] [Google Scholar]
- 364.Williams PJ, Gumaa K, Scioscia M, Redman CW, Rademacher TW. Hypertens (Dallas, Tex 1979) 2007;49:84–9. doi: 10.1161/01.HYP.0000251301.12357.ba. [DOI] [PubMed] [Google Scholar]
- 365.Marangoni A, Foschi C, Capretti MG, Nardini P, Compri M, Corvaglia LT, Faldella G, Cevenini R. Clin Vaccine Immunol. 2016;23:410–416. doi: 10.1128/CVI.00032-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 366.Lubell Y, Blacksell SD, Dunachie S, Tanganuchitcharnchai A, Althaus T, Watthanaworawit W, Paris DH, Mayxay M, Peto TJ, Dondorp AM, White NJ, Day NPJ, Nosten F, Newton PN, Turner P. BMC Infect Dis. 2015;15:511. doi: 10.1186/s12879-015-1272-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 367.Mussap M, Puxeddu E, Puddu M, Ottonello G, Coghe F, Comite P, Cibecchini F, Fanos V. Clin Chim Acta. 2015;451:65–70. doi: 10.1016/j.cca.2015.07.025. [DOI] [PubMed] [Google Scholar]
- 368.Castillo-Laborde C. Hum Resour Health. 2011;9:4. doi: 10.1186/1478-4491-9-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 369.Plebani M. 2012;33:85–88. [PMC free article] [PubMed] [Google Scholar]
- 370.Crannell Z, Castellanos-Gonzalez A, Nair G, Mejia R, White AC, Richards-Kortum R. Anal Chem. 2016;88:1610–1616. doi: 10.1021/acs.analchem.5b03267. [DOI] [PubMed] [Google Scholar]
- 371.Zumla A, Al-Tawfiq JA, Enne VI, Kidd M, Drosten C, Breuer J, Muller MA, Hui D, Maeurer M, Bates M, Mwaba P, Al-Hakeem R, Gray G, Gautret P, Al-Rabeeah AA, Memish ZA, Gant V. Lancet. 2014 doi: 10.1016/S1473-3099(14)70827-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 372.Dincer C, Bruch R, Kling A, Dittrich PS, Urban GA. Trends Biotechnol. 2017 doi: 10.1016/j.tibtech.2017.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 373.Scherr TF, Gupta S, Wright DW, Haselton FR. Lab Chip. 2017:17. doi: 10.1039/c6lc01580h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 374.WHO. [accessed March 2017];Fact Sheet: World Malaria Day. 2016 http://www.who.int/malaria/media/world-malaria-day-2016/en/
- 375.World Health Organization. Interagency list of medical devices for essential interventions for reproductive, maternal, newborn and child health. 2015 [Google Scholar]
- 376.CDC. Alere Determine™ HIV-1/2 Ag/Ab Combo Information Sheet for Testing Programs. 2016 [Google Scholar]
- 377.Hoenigl M, Graff-Zivin J, Little SJ. Clin Infect Dis. 2016;62:501–11. doi: 10.1093/cid/civ912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 378.Moyo S, Mohammed T, Wirth KE, Prague M, Bennett K, Holme MP, Mupfumi L, Sebogodi P, Moraka NO, Boleo C, Maphorisa CN, Seraise B, Gaseitsiwe S, Musonda RM, van Widenfelt E, Powis KM, Gaolathe T, Tchetgen Tchetgen EJ, Makhema JM, Essex M, Lockman S, Novitsky V. J Clin Microbiol. 2016;54:3050–3055. doi: 10.1128/JCM.01594-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 379.Cepheid. [accessed April 2017];GeneXpert IV. http://www.cepheid.com/en/cepheid-solutions-uk/systems/genexpert-systems/genexpert-iv.
- 380. [accessed February 2017];SD Bioline Malaria Ag RDT Series. http://www.standardia.com/en/home/product/Rapid_Diagnostic_Test/Malaria_Ag_Pf-HRP2.html.
- 381. [accessed March 2017];A guide for the selection of malaria RDTs to be purchased with 3DF grants. http://www.threediseasesfund.org/images/stories/pdf/procurement/SOPs_and_Guidelines/Malaria-RDTs-Guideline-3DF-v1.0.pdf.
- 382. [accessed March 2017];OneTouch Ultra®2 Meter. http://www.shoponetouch.com/product/35580.
- 383.Sanchis-Gomar F, Cortell-Ballester J, Pareja-Galeano H, Banfi G, Lippi G. J Lab Autom. 2013;18:198–205. doi: 10.1177/2211068212457560. [DOI] [PubMed] [Google Scholar]
- 384. [accessed March 2017];CLIA waived: HemoCue. http://www.cliawaived.com/web/HemoCue.htm.
- 385. [accessed March 2017];Hemoglobin test - HemoCue® Hb 201+ System. http://www.hemocue.us/en-us/solutions/hematology/hemocue-hb-201plus-system.
- 386.Boopathi A, Nayak SR, Rao A, Rao B. Open J Obstet Gynecol. 2014;4:666–671. [Google Scholar]
- 387.Alere. The Alere Triage System. Jouy en Josas; France: 2013. [Google Scholar]
- 388.Lee A, Mirrett S, Reller LB, Weinstein MP. J Clin Microbiol. 2007;45:3546–3548. doi: 10.1128/JCM.01555-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 389.Marik PE. Crit Care. 2014;18:529. doi: 10.1186/s13054-014-0529-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 390.Schuur JD, Lin M. Blood Culture Testing: Send Samples Selectively to Lower Costs. [accessed March 2017];Medico-Legal Risk. http://www.acepnow.com/article/blood-culture-testing-send-samples-selectively-lower-costs-medico-legal-risk/
- 391.Ali MJ, Ayyar A, Motukupally SR, Sharma S, Naik MN. 2014 doi: 10.1186/s12348-014-0027-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 392. [accessed March 2017];AmniSure ROM Test Overview. http://www.amnisure.com/care/test.
- 393.Raimondi F, Lama S, Landolfo F, Sellitto M, Borrelli AC, Maffucci R, Milite P, Capasso L. BMC Pediatr. 2012;12:625. doi: 10.1186/1471-2431-12-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 394.Robertson A, Kazmierczak S, Vos P. J Perinatol. 2002;22:12–14. doi: 10.1038/sj.jp.7210592. [DOI] [PubMed] [Google Scholar]
- 395.Philips BiliChek System. [accessed April 2017]; http://www.usa.philips.com/healthcare/product/HC989805644871/bilichek-bilirubinometer.
- 396. [accessed April 2017];Nova Biomedical: StatStrip. http://www.novabio.us/statstrip-glu/
- 397. [accessed March 2017];Moyo-Fetal Heart Rate Monitor. http://designawards.core77.com/Commercial-Equipment/49815/Moyo-Fetal-Heart-Rate-Monitor.
- 398.Elizabeth Glaser Pediatric AIDS Foundation and UNITAID. Point-of-Care Early Infant Diagnosis. 2016 [Google Scholar]
- 399.World Health Organization. Diagnostics Evaluation Series No. 1- Laboratory-based evaluation of rapid syphilis diagnostics. 2003 [Google Scholar]
- 400. [accessed March 2017];SD BIOLINE Syphilis 3.0. http://www.standardia.com/en/home/product/Rapid_Diagnostic_Test/SyphilisTest.html.
- 401.Wilson R. Chem Soc Rev. 2008;37:2028. doi: 10.1039/b712179m. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.