Abstract
Currently, there are no effective therapeutic strategies targeting Kras driven cancers, and therefore, identifying new targeted therapies and overcoming drug resistance have become paramount for effective long-term cancer therapy. We have found that reducing expression of the palmitoyl transferase DHHC20 increases cell death induced by the EGFR inhibitor gefitinib in Kras and EGFR mutant cell lines, but not MCF7 cells harboring wildtype Kras. We show that the increased gefitinib sensitivity in cancer cells induced by DHHC20 inhibition is mediated directly through loss of palmitoylation on a previously identified cysteine residue in the C-terminal tail of EGFR. We utilized an EGFR point mutant in which the palmitoylated cysteine 1025 is mutated to alanine (EGFRC1025A), that results in receptor activation. Expression of the EGFR mutant alone in NIH3T3 cells does not increase sensitivity to gefitinib-induced cell death. However, when EGFRC1025A is expressed in cells expressing activated KrasG12V, EGFR inhibitor induced cell death is increased. Surprisingly, lung cancer cells harboring the EGFR inhibitor resistant mutation, T790M, become sensitive to EGFR inhibitor treatment when DHHC20 is inhibited. Finally, the small molecule, 2-bromopalmitate, which has been shown to inhibit palmitoyl transferases, acts synergistically with gefitinib to induce cell death in the gefitinib resistant cell line NCI-H1975.
Keywords: Palmitoylation, Receptor Tyrosine Kinase Signaling, oncogenic signaling, Kras, EGFR
Introduction
EGFR and other members of the ErbB family of receptor tyrosine kinases (RTKs) play critical roles in reacting to extracellular growth cues by initiating downstream signaling cascades through various effector pathways (1–3). Mutations in EGFR, leading to its constant activation and subsequent uncontrolled cell growth, are detectable in 10% to 30% of tumors from patients with non–small cell lung cancer (NSCLC)(4). Ligand (EGF) binding to EGFR induces receptor dimerization and subsequent auto-phosphorylation of specific tyrosine residues on the C-terminal tail. Activating EGFR point mutations in exon 21, such as L858R, and deletions of exon 19 are often predictors of response to EGFR tyrosine kinase inhibitor (TKI) therapy with gefitinib or erlotinib (5). However, the majority of patients with NSCLCs harboring these activating EGFR mutations relapse within 10 to 16 months of treatment with EGFR TKIs (5–6). In over half of these patients, resistance to EGFR TKI therapy is associated with the acquisition of a secondary T790M mutation in the EGFR TK domain, which alters interaction of reversible TKIs with the ATP-binding pocket (4,7). It is therefore critical to develop strategies to overcome drug resistance. We have recently uncovered a previously unknown regulation of EGFR through EGFR palmitoylation.
Protein palmitoylation is the reversible covalent attachment of a 16-carbon saturated fatty acid palmitate onto cysteine residues. Addition of the large hydrophobic palmitate facilitates association of proteins at the plasma membrane (PM) influencing formation of cell signaling complexes.(8–12) Palmitoylation is mediated by a family of 23 protein acyl-transferases containing a conserved DHHC (aspartic acid, histidine, histidine, cysteine) motif essential for catalysis (13–15). DHHC20 palmitoylates the epidermal growth factor receptor (EGFR) on specific cysteine residues on the C-terminal tail, suppressing its activation. Unexpectedly, inhibition of DHHC20 in both breast and lung cancer cells increases induction of cell death in response to the EGFR inhibitor gefitinib, despite the fact the cells harbor activated Kras mutations and wild type EGFR (16). Mutations in Kras, predominantly an amino acid substitution at codon 12 or 13, lead to upregulation of the Ras/MAPK signaling and ERK activation, ultimately driving cell division. While mutations in Ras are common in cancer it has been challenging to therapeutically target Ras directly because of the nucleotide-binding pocket’s very high affinity for GTP.
Here we report that using a palmitoylation defective EGFR mutant we demonstrate the increased response to gefitinib is mediated by the palmitoylated cysteine residue 1025, but only when in combination with activated Kras. Alternatively, inhibition of DHHC20 also increases sensitivity to gefitinib-induced cell death in cancer cells harboring not only activating EGFR mutations, but also the gefitinib resistant T790M mutation, independent of activated Kras. These results demonstrate a previously unreported mechanism to overcome mutation driven drug resistance by targeting a recently identified modification of EGFR.
Materials and Methods
Cell culture
MDA-MB-231, MCF7, and NIH3T3 cells (ATCC) were cultured in DMEM containing 10% FBS. NCI-H1975 cells (ATCC) were maintained in RPMI supplemented with 10% FBS. Cells were treated with gefitinib (Selleck Chemicals) and 2-bromopalmitate (2-BP) (Sigma).
Immunoblot analysis
Cell lysates were prepared in 1% Triton-X-100 buffer, including Tris-HCl (pH 7.5) and sodium chloride solution (NaCl). Lysates were analyzed by immunoblotting with the following antibodies: Anti-DHHC20 (HPA014702) and anti-FLAG M2 antibodies were purchased from Sigma-Aldrich. Anti-pY1068-EGFR, EGFR-XP, pERK, ERK, pS473-AKT, pT308-AKT, AKT, β-actin were obtained from Cell Signaling Technologies. Anti-HA antibodies were purchased from Biolegend. Immune complexes were detected with horseradish peroxidase-conjugated secondary antibodies and enhanced chemiluminescence (ECL) (Thermo Scientific).
Silencing of Human DHHC20
The oligonucleotides for shControl and shDHHC20 constructs were synthesized (Integrated DNA Technologies) and inserted into the pLKO.1 vector. shControl encodes the non-targeting sequence of SHC002 (Sigma); the shRNA target sequence of human DHHC20 is 50-GAGCTCTGCGTGTTTACTATT-30. MDA-MB-231, MCF7 and NCI-H1975 cells were transduced with lentivirus encoding shControl or shDHHC20 and selected by puromycin treatment (1 mg/ml) for several passages.
Cell Viability
Cells were treated with gefitinib (5 µM) and/or 2-BP (500 nM) for 72 hr, and viability was measured by trypan blue staining. Quantification was done using a 1-way ANOVA with Tukey’s multiple comparison post-test.
Determination of IC50 Values and Isobologram analysis
Cells were seeded on a 96-well plate in 100 µl growth media at a density of 1500 cells per well. After 24 hours post-seeding, the cells were treated with gefitinib and/or 2-BP for an additional 72 hours. Cell viability was assessed using the alamar blue viability assay (Invitrogen). Triplicate wells for each experiment were analyzed and the experiment was performed three times. The IC50 values were determined by a non-linear regression of the dose-response effect data using Prism for MacOSX (GraphPad Software). Cells were exposed to 1:1 ratios of the respective IC50s for gefitinib and 2-BP at ¼ xIC50, ½ xIC50, IC50, 2 xIC50, and 4 xIC50. The assessment of synergy was performed using CalcuSyn software (Biosoft). The combination index (CI) was evaluated to assess synergism (CI<1), additive effect (CI~1) or antagonism (CI>>1).
Plasmids and generation of stable cell lines
To generate inducible cell lines, wildtype EGFR and EGFR C1025A cDNA was first subcloned into the inducible pTRIPZ backbone with a puromycin resistance marker and FLAG tag. Empty pTRIPZ, which expresses the rtTA3, puromycin resistance marker, and FLAG tag was used as a negative control. Virus production was performed by transfecting HEK293T cells with the pTRIPZ constructs, psPAX2 and pMD2.G plasmids (Addgene) using TransIT-LT1 (Mirus) according to the manufacturer’s instructions. MDA-MB-231 and NIH3T3 were infected with pTRIPZ virus using polybrene and incubated for 24 hours. Post-infection, fresh media was added on infected cells and incubated for an additional 48 hours before selection. Cells infected with the pTRIPZ constructs were selected with 1 µg/ml puromycin for several passages.
Expression of EGFR cDNA was induced with 1µg/ml doxycycline. Lentivirus of human mutant Kras4B(G12V) in pLenti-PGK-hygromycin resistance with an HA tag (Addgene plasmid #35633, [Singh et al. Cell, 2012]) was generated using HEK293T cells, Gag, VSVG and Rev. NIH3T3 cells infected with pTRIPZ constructs and selected with puromycin were subsequently infected with Kras4B(G12V)-HA. NIH3T3 pTRIPZ-plenti-Kras4B(G12V) cells were selected with puromycin (1 µg/ml) and hygromycin (500 µg/ml) together for several passages.
Results
Increased gefitinib-induced cell death is mediated by the palmitoylation site C1025 on the C-terminal tail of EGFR
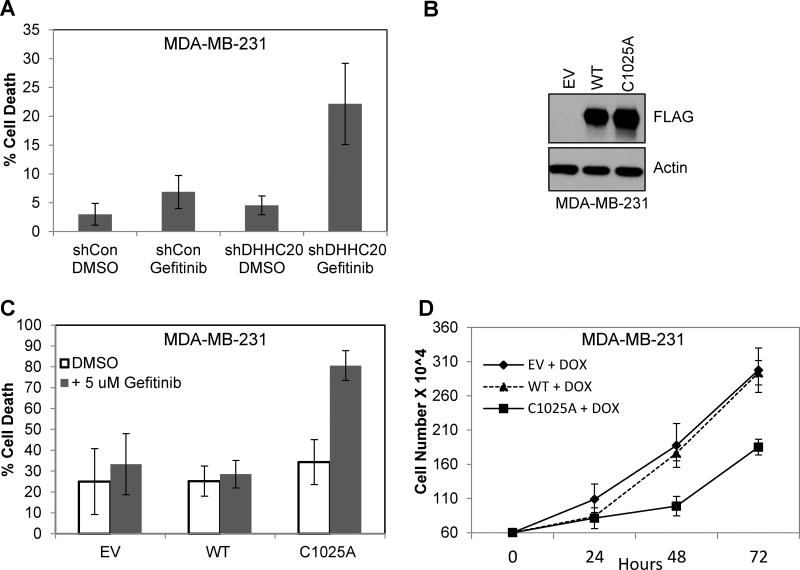
We reported previously that inhibition of DHHC20 increased the sensitivity of the triple negative breast cancer cell line, MDA-MB-231, to TKI induced cell death (Fig. 1A) (16). Although inhibition of DHHC20 by shRNA leads to an increase in sensitivity to gefitinib-induced cell death, it is still unclear if this effect was directly mediated by inhibiting EGFR palmitoylation or if it is the effect of another unknown target of DHHC20. We previously showed that EGFR is palmitoylated on the C-terminal tail and that mutating one of the palmitoylated cysteine residues 1025 to alanine is sufficient to reduce receptor palmitoylation, induce receptor autophosphorylation, adaptor binding and downstream signaling to AKT and ERK when transiently expressed in NIH3T3 cells (16). To test if blocking EGFR palmitoylation directly increases gefitinib sensitivity of human cancer cells, we developed a conditional system for expressing wild type EGFR (EGFRWT) or palmitoylation defective EGFR with cysteine 1025 mutated to alanine (EGFRC1025A) in MDA-MB-231 cells with a tetracycline-inducible promoter. Induction of the cells with doxycycline for 72 hours resulted in protein levels of EGFRC1025A is slightly higher than EGFRWT consistent with previous findings (Fig. 1B) (16). After 24 hours of doxycycline inductions, cells were treated with gefitinib (5µM) for 72 hours. The percentage of dead cells expressing EGFRWT were similar to cells infected with an empty vector control treated with gefitinib (Fig. 1C). Cells expressing the EGFRC1025A mutant and treated with DMSO showed similar percentages of cell death as EGFRWT expressing cells (Fig. 1C). However, when the EGFRC1025A mutant cells were treated with gefitinib, cell death increased to 80.6% compared to the 32.4% in gefitinib treated EGFRWT expressing cells (Fig. 1C). One potential damaging outcome when blocking EGFR palmitoylation is increased tumor growth caused by activated EGFR. However, expression of EGFRC1025A did not increase growth of the breast cancer cells and in fact significantly reduced cell proliferation compared to cells overexpressing EGFRWT (Fig 1D). These results indicate that blocking EGFR palmitoylation at cysteine residue 1025 is sufficient to induce gefitinib sensitivity in triple negative breast cancer cells. This confirms that the increased sensitivity to gefitinib-induced cell death imposed by DHHC20 inhibition via shRNA is caused by loss of EGFR palmitoylation at cysteine residue 1025.
Figure 1. Expression of palmitoylation defective EGFRC1025A mediates gefitinib-induced cytotoxicity.
(A) Gefitinib increases cytotoxicity in DHHC20 silenced Kras mutant cells MDA-MB-231. (B) MDA-MB-231 (KrasG13D) cells stably expressing inducible EGFRWT or EGFRC1025A or empty vector control (EV) were treated with doxocycline (1µg/ml) for 15 hours. Immunoblotting with anti-FLAG shows induced expression of FLAG tagged EGFRWT and EGFRC1025A. (C) Cells were treated with doxocycline (1µg/ml) every 24 hours for 72 hours and with DMSO or 5 µM gefitinib at 24 hours post-seeding. Expression of EGFRC1025A induced sensitivity to gefitinib. Cell viability was measured by Trypan Blue staining at 72 hours post-treatment (mean +/−StDev). (D) Induced expression of EGFRC1025A in MDA-MB-231 cells reduces cell growth.
DHHC20 silencing does not increase gefitinib sensitivity in wild type Kras expressing MCF7 breast cancer cells
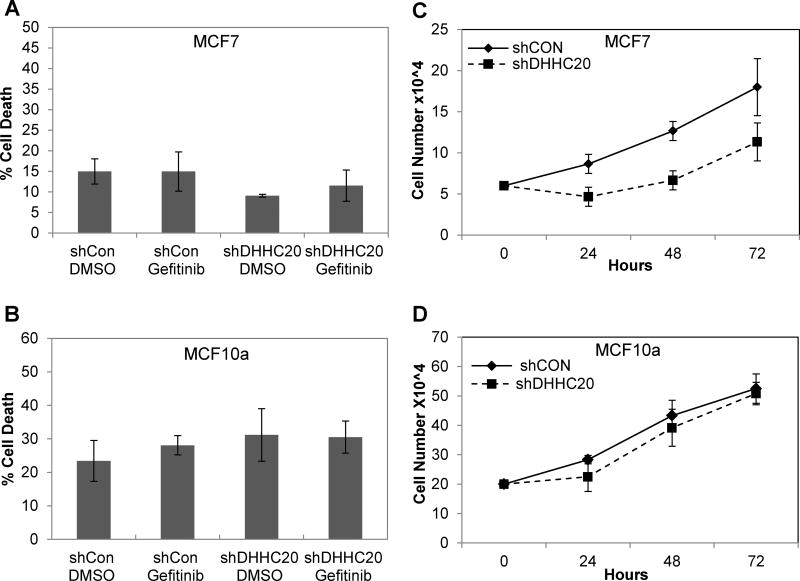
The MDA-MB-231 breast cancer cells express wild type EGFR, but harbor an activating mutation in Kras (KrasG12D). We asked if a similar sensitivity to gefitinib is observed in cancer cells expressing wild type Kras upon DHHC20 silencing. Silencing DHHC20 in the breast cancer cell line MCF7 harboring a mutation in the PI3K pathway (PIK3CAE545K) had no effect on gefitinib-induced cell death consistent with a requirement for oncogenic Kras (Fig. 1A). However, similar to the slowed growth upon expression of EGFRC1025A in MDA-MB-231, inhibiting DHHC20 in MCF7 cells did slow cell growth, suggesting that DHHC20 plays a role in proliferation in the presence of a PIK3CA mutation (Fig. 1B). Inhibiting DHHC20 did modestly increase phosphorylation of both ERK and AKT, suggesting that the loss of DHHC20 promotes the signaling of the constitutively active mutant PIK3CA (Supp. Fig. 1B). We next examined the gefitinib sensitivity of downstream EGFR signaling in shControl and shDHHC20 expressing cells. Treatment of MCF7 cells with 5 µM gefitinib decreased pERK only when DHHC20 was silenced by shRNA, but pAKT was only slightly inhibited by gefitinib in the MCF7 shDHHC20 cells (Supp. Fig. 1B).
Since MCF7 cells were still not sensitive to gefitinib-induced cell death after DHHC20 inhibition we asked if non-transformed cells were also resistant. When DHHC20 was inhibited in MCF10A cells, there was no increase in the sensitivity to gefitinib-induced cell death after 72 hours of treatment (Fig. 2C). Furthermore, inhibition of DHHC20 in MCF10a cells had no effect on cell growth (Fig. 2D), indicating that DHHC20 is not essential for normal cell growth. We also examined the gefitinib sensitivity of downstream EGFR signaling in shControl and shDHHC20 expressing MCF10a cells. Treatment with 5 µM gefitinib decreased pEGFR and pERK in both shControl and shDHHC20 cells as expected because gefitinib will inhibit EGFR in these non-transformed cells (Supp. Fig. 1C). These results thus far indicate that inhibition of DHHC20 only increases sensitivity to gefitinib-induced cell death in cancer cells with activating mutations in Kras.
Figure 2. DHHC20 inhibition increases gefitinib sensitivity in cells with activating mutations and resistance mutations in EGFR.
MCF7 and MCF10A are resistant to gefitinib cytotoxicity in response to DHHC20 silencing. (A, B) shControl and shDHHC20 cells were treated with DMSO or 5 µM gefitinib. Cell viability was measured by Trypan Blue staining at 72 hours. (C) Knockdown of DHHC20 in MCF7 (PIK3CAE545K) cells slows growth. (D) Knockdown of DHHC20 in the non-transformed MCF10a cells has no effect on cell growth.
Increased gefitinib-induced cell death mediated by EGFRC1025A requires mutant KrasG12V
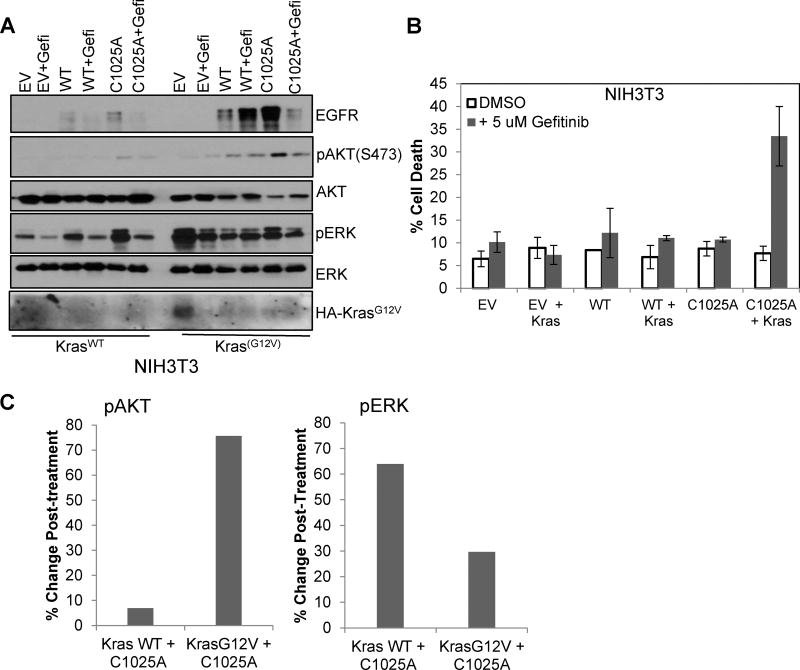
To determine the requirements of gefitinib-induced cell death in shDHHC20 cells, NIH3T3 cells were stably infected with tetracycline-inducible EGFRWT or palmitoylation defective EGFRC1025A. The level of EGFRWT expression was lower than EGFRC1025A after 12 hours of induction (Fig. 3A). After 72 hours of induced expression of EGFRWT or EGFRC1025A, there was no effect on cell viability after 72 hours of gefitinib treatment (Fig. 3B). Since expression of EGFRC1025A alone was not sufficient to induce gefitinib sensitivity we asked if oncogenic Kras is required. The mutant KrasG12V was stably expressed in NIH3T3 cells together with tetracycline-inducible EGFRWT or EGFRC1025A. Treatment with gefitinib increased the percentage of cell death to 33.4% in KrasG12V cells expressing EGFRC1025A compared to 11.0% in cells expressing EGFRWT and KrasG12V (Fig. 3B). This indicates the combination of oncogenic Kras with palmitoylation defective EGFRC1025A is required and sufficient to increase gefitinib-induced cell death. When downstream signaling was examined, we found that cells expressing both KrasG12V and EGFRC1025A had higher levels of pAKT compared to those with KrasWT. This increase in pAKT is inhibited by gefitinib treatment whereas the change in EGFRC1025A KrasWT is minimal (Fig. 3A). However, the EGFRC1025A and KrasG12V condition had lower levels of pERK compared to EGFRC1025A and KrasWT, and were reduced upon gefitinib treatment to similar levels in both conditions (Fig. 3A). Therefore, the increase in cell death may be more dependent on the gefitinib-induced changes in AKT activation than ERK.
Figure 3. Mutation of the palmitoylated cysteine 1025 of EGFR in combination with KrasG12V increases gefitinib sensitivity.
(A) NIH3T3 cells stably expressing inducible full length EGFRWT or EGFRC1025A or empty vector control (EV) with KrasWT (left) or KrasG12V (right). Immunoblotting with EGFR shows induced expression of EGFRWT and EGFRC1025A, and immunoblotting with HA shows expression of HA-tagged KrasG12V. (B) Induced expression of EGFRC1025A in NIH3T3 stably expressing KrasG12V increased gefitinib-induced cell death. Cells were treated with doxocycline (1µg/ml) every 24 hours for 72 hours and with DMSO or 5 µM Gefitinib at 24 hours post-seeding. Cell viability was measured by Trypan Blue staining at 72 hours post-treatment (mean +/−StDev).
DHHC20 inhibition increases gefitinib sensitivity in cells with the activating and resistance mutations in EGFR
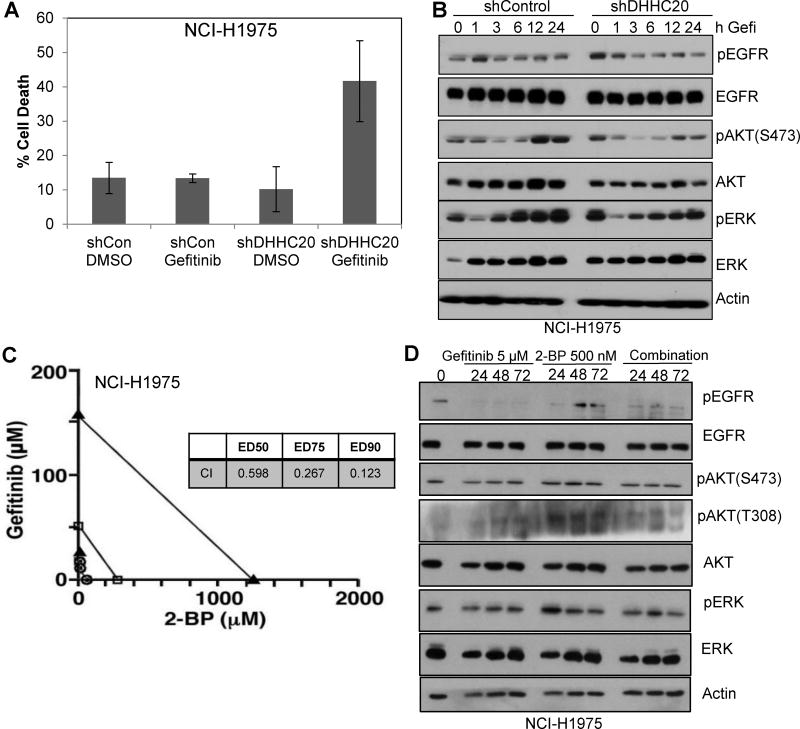
Upon examination of the KrasWT lung cancer cell line, NCI-H1975, inhibiting DHHC20 increased gefitinib-induced cell death (41.7% vs. 13.4%) (Fig. 4A). The increase of sensitivity of the NCI-H1975 shDHHC20 cell line to gefitinib is quite unexpected since this line harbors an activating mutation L858R and the acquired secondary mutation, T790M, in the kinase domain imparting gefitinib resistance.
Figure 4. Palmitoyl transferase inhibitor 2-bromopalmitate synergizes with gefitinib in inducing cell death in gefitinib resistant cells.
(A) shControl and shDHHC20 NCI-H1975 cells were treated with DMSO or 5 µM gefitinib. Cell viability was measured by Trypan Blue staining at 72 hours. Gefitinib increases cytotoxicity in H1975 DHHC20 silenced cells. All graphs show mean +/−StDev. (B) Treatment of NCI-H1975 DHHC20 silenced cells with gefitinib decreases EGFR, AKT and ERK phosphorylation. (C) NCI-H1975 cells were treated with (i) fixed IC50 ratios of gefitinib alone at 24 hours post-seeding, (ii) fixed IC50 ratios of 2-BP alone at 24 hours post-seeding, or (iii) Gefitinib in combination with 2-BP. The multiple effect-level isobologram analyses at 72 hours post-treatment are shown for the ED50 (open circle), ED75 (closed square) and ED90 (closed triangle) values. The combination of Gefitinib and 2-BP is synergistic in NCI-H1975 cells. (D) NCI-H1975 cells were treated with 5 µM Gefitinib, 500 nm 2-BP or both at 24 hours post-seeding. Cells were harvested after the indicated treatment time points and lysates were immunoblotted with indicated antibodies.
To address if the increase in cell death in the gefitinib resistant line is associated with an increase in EGFR sensitivity to gefitinib, we examined EGFR signaling in the shDHHC20 cells. When DHHC20 was silenced in NCI-H1975, cells exhibited increased EGFR, AKT and ERK phosphorylation (Fig. 4B). The increase in signaling suggests that the palmitoyl transferase inhibits the mutant EGF receptor. After treatment of NCI-H1975 shDHHC20 cells with gefitinib, the elevated levels of pEGFR were reduced within 1 hour, suggesting that the increase in EGFR phosphorylation upon DHHC20 inhibition was not caused by decreased dephosphorylation. The simplest explanation for the increased EGFR autophosphorylation upon DHHC20 silencing is the high basal kinase activity of the activating mutation. Similarly, levels of pERK and pAKT decreased after 1 hour and remain low for 24 hours after treatment (Fig. 4B). In the NCI-H1975 shControl cells, the gefitinib treatment increased pAKT and pERK levels as well as total levels of AKT and ERK at 6–24 hours of treatment through a mechanism that is still unclear (Fig. 4B). This suggests that reduction of DHHC20 by shRNA increases the sensitivity of these gefitinib resistant cells to inhibitor treatment.
Palmitate analog 2-bromopalmitate synergizes with gefitinib to induce cell death in gefitinib resistant cells
Silencing DHHC20 by shRNA causes chronic inhibition of DHHC20 and constitutively elevated levels of EGFR signaling. The immediate effects of acute DHHC20 inhibition through pharmacologic inhibition would provide greater insight into the utility of DHHC20 as a therapeutic target. Although currently there is not a specific inhibitor to palmitoyl transferases, the palmitate analog 2-bromopalmitate (2-BP) has been shown to inhibit DHHC domain containing palmitoyl transferases at micromolar concentrations (17). Our previous study showed that 2-BP was sufficient to increase sensitivity of MDA-MB-231 cells to gefitinib-induced cell death (16). We wanted to examine the effect of 2-BP on gefitinib sensitivity in greater detail, specifically on gefitinib resistant cells.
We asked if acute treatment of cancer cells with 2-BP is sufficient to sensitize the gefitinib resistant NCI-H1975 cells to gefitinib-induced cell death in and if the sensitivity is comparable to the wild type EGFR expressing MDA-MB-231 cells. The combination treatment increased cell death compared to either gefitinib or 2-BP alone in both MDA-MB-231 and NCI-H1975 cells. Based on these responses to treatment with gefitinib and 2-BP, we assessed the effects of combining 2-BP and gefitinib using the Chou-Talalay method (18). With half-maximal inhibitory concentrations of gefitinib (IC50 = 3.5 µM for MDA-MB-231 and 12.67 µM for NCI-H1975) and 2-BP (IC50 = 11.74 µM for MDA-MB-231 and 11.46 µM for H1975), these compounds were tested alone for effects on MDA-MB-231 and NCI-H1975 cell growth at 1/8X, 1/4X, 1/2X, 1X, 2X, and 4X the IC50 values and at equipotent concentrations at the same ratios in combination. Isobologram analysis of the data at ED50, ED75 and ED90 values showed an additive effect of the gefitinib/2-BP combination in MDA-MB-231 cells (data not shown), but showed synergy of the gefitinib/2-BP combination in NCI-H1975 cells with CI values of less than 1 (Fig. 4C). Upon examination of downstream signaling in NCI-H1975, we found 2-BP treatment increased pEGFR, pAKT(T308) and pERK consistent with the shDHHC20 condition (Fig. 4D). With gefitinib treatment in combination with 2-BP, pEGFR and pAKT(T308) notably reduced and pERK is modestly reduced in contrast to the MDA-MB-231 in which there was no detectable change between treatment groups. Therefore, the signaling mechanism behind the observed synergy between drugs may be through regulation of combined AKT(T308) and ERK phosphorylation. This shows that targeting DHHC20 may be an effective approach to overcoming EGFR TKI resistance in NSCLC.
Discussion
We observe increased gefitinib sensitivity in cells expressing EGFR harboring both activating L858R and a resistance mutation T790M, an acquired secondary mutation that imparts resistance to EGFR TKIs, such as gefitinib. Our results also reveal a requirement for oncogenic Kras for increased inhibitor sensitivity mediated by blocking EGFR palmitoylation in cells with wild type EGFR. Expression of a mutant form of EGFR that is resistant to palmitoylation at cysteine 1025 leads to increased sensitivity to gefitinib-induced cell death, but only in the presence of oncogenic KrasG12V. Expression of activated PIK3CA was insufficient to induce sensitivity to gefitinib, confirming the selectivity for KrasG12V (Supp. Fig. 1D).
The alterations in EGFR signaling that lead to increased gefitinib-induced cell death are still not entirely clear. Inhibition of DHHC20 in EGFR mutant background increases both pERK and pAKT levels and are both effectively inhibited by gefitinib. One possibility it is the change in signaling from the artificially high levels induced by DHHC20 inhibition or EGFRC1025A expression down to the gefitinib inhibited levels that causes the cells to crisis and die. Cells with activated Kras become sensitized to gefitinib with DHHC20 inhibition, but the resulting increase in pERK caused by DHHC20 inhibition is not reduced by gefitinib. It is therefore unclear why the MDA-MB-231 cells become sensitive to gefitinib when DHHC20 is knocked down.
Furthermore, the combined effect of DHHC20 and EGFR inhibition is greater in the cells with mutant EGFR than cells with mutant Kras. The synergy between gefitinib and 2-bromopalmitate (2-BP) in the NCI-H1975 cells compared to the additive effect in MDA-MB-231 cells is particularly surprising. Gefitinib targets EGFR harboring the L858R activating mutation, which increases the affinity of the drug for activated EGFR relative to ATP, in NCI-H1975. It is therefore not surprising that the gefitinib/2-BP combination treatment is more effective in this cell line than the EGFR wild type cell line, MDA-MB-231. What is surprising is that these cells are resistant to gefitinib because of the T790M secondary acquired resistance mutation. The T790M mutation increases the affinity of the ATP binding pocket for ATP over gefitinib. Due to toxicity, we have been unable to express EGFRC1025A in the NCI-H1975 cells or express EGFRL858R/T790M also harboring the C1025A mutation in any cell type. We therefore can’t conclusively demonstrate that the increased gefitinib sensitivity in the NCI-H1975 cells is through EGFR palmitoylation directly.
Our results reveal two potential vulnerabilities in the EGFR/MAPK pathway mediated by DHHC20 inhibition. First is the gefitinib-induced sensitivity of Kras mutant cancers to inhibition of DHHC20 or blocking EGFR palmitoylation. Oncogenic mutation in Kras is one of the most common mutations in cancer and yet targeting Kras therapeutically has been elusive. Therefore, inducing sensitivity of Kras mutant cells to EGFR inhibitor therapy is an unprecedented alternative approach. The second is the re-sensitization of gefitinib resistant cancer cells by inhibition of DHHC20. While we have not yet shown that this effect is through EGFR, the synergistic effect between 2-BP and gefitinib is striking. The fact we have been unable to express the C1025A mutant with the activating L858R mutant suggests there may be a form of synthetic lethality possibly from hyperactivation of the pathway. Future studies will determine how palmitoylation effects the gate-keeper mutation in the kinase domain and if it is through the C-terminal tail.
Supplementary Material
Highlights.
The effect of inhibiting the palmitoyl transferase DHHC20 on EGFR activation and increased sensitivity to the EGFR inhibitor gefitinib requires activating mutations in either Kras or EGFR itself.
Treatment of inhibitor resistant lung cancer cells harboring the activating EGFR mutation L858R and the drug resistance mutation, T790M, with a palmitoyl-transferase inhibitor combined with the EGFR TKI, gefitinib, causes synergistic induction of cell death.
The palmitoyl transferase DHHC0 may be an attractive therapeutic target for Kras driven and gefitinib resistant cancers.
Acknowledgments
We would like to thank Donita Brady for useful discussion and advice regarding the experiments and manuscript. We would also like to thank Kris Wood for the PIK3CA (H1047R) lentiviral plasmid.
Funding
This work was supported by NIH Grant R01CA181633 (to ESW) and by ACS Grant RSG-15-027-01 (to E.S.W).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures of Potential Conflicts of Interest
No potential conflict of interests were disclosed by the authors of this manuscript.
References
- 1.Roskoski R., Jr ErbB/HER protein-tyrosine kinases: Structures and small molecule inhibitors. Pharmacol Res. 2014;87:42–59. doi: 10.1016/j.phrs.2014.06.001. [DOI] [PubMed] [Google Scholar]
- 2.Walton GM, Chen WS, Rosenfeld MG, Gill GN. Analysis of deletions of the carboxyl terminus of the epidermal growth factor receptor reveals self-phosphorylation at tyrosine 992 and enhanced in vivo tyrosine phosphorylation of cell substrates. J Biol Chem. 1990;265(3):1750–4. [PubMed] [Google Scholar]
- 3.Lemmon MA, Schlessinger J, Ferguson KM. The EGFR Family: Not So Prototypical Receptor Tyrosine Kinases. Cold Spring Harb Perspect Biol. 2014;6.4:a020768. doi: 10.1101/cshperspect.a020768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Normanno N, De Luca A, Bianco C, Strizzi L, Mancino M, Maiello MR, Carotenuto A, De Feo G, Caponigro F, Salomon DS. Epidermal growth factor receptor (EGFR) signaling in cancer. Gene. 2006;366:2–16. doi: 10.1016/j.gene.2005.10.018. [DOI] [PubMed] [Google Scholar]
- 5.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. Ca Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 6.Chen Z, Fillmore C, Hammerman P, Kim C, Wong K-K. Non-small-cell lung cancers: a heterogeneous set of diseases. Nat Rev Cancer. 2014;14:535–546. doi: 10.1038/nrc3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jorissen R, et al. Epidermal growth factor receptor: mechanisms of activation and signalling. Exp Cell Res. 2003;284:31–53. doi: 10.1016/s0014-4827(02)00098-8. [DOI] [PubMed] [Google Scholar]
- 8.Fukata Y, Fukata M. Protein palmitoylation in neuronal development and synaptic plasticity. Nat Rev Neuro. 2010;11:161–175. doi: 10.1038/nrn2788. [DOI] [PubMed] [Google Scholar]
- 9.Linder M, Deschenes R. Palmitoylation: policing protein stability and traffic. Nat Rev Mol Cell Bio. 2007;8:74–84. doi: 10.1038/nrm2084. [DOI] [PubMed] [Google Scholar]
- 10.Young F, Butland S, Sanders S, Sutton L, Hayden M. Putting proteins in their place: Palmitoylation in Huntington disease and other neuropsychiatric diseases. Prog Neurobiol. 2012;97:220–238. doi: 10.1016/j.pneurobio.2011.11.002. [DOI] [PubMed] [Google Scholar]
- 11.Aicart-Ramos C, Valero R, Rodriguez-Crespo I. Protein palmitoylation and subcellular trafficking. Biochim Biophys Acta. 2011;1808:2981–94. doi: 10.1016/j.bbamem.2011.07.009. [DOI] [PubMed] [Google Scholar]
- 12.Wang W, Runkle K, Terkowski S, Ekaireb R, Witze E. Protein Depalmitoylation Is Induced by Wnt5a and Promotes Polarized Cell Behavior. J Biol Chem. 2015;290:15707–15716. doi: 10.1074/jbc.M115.639609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Conibear E, Davis NG. Palmitoylation and depalmitoylation dynamics at a glance. J Cell Sci. 2010;123(Pt 23):4007–10. doi: 10.1242/jcs.059287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Greaves J, Chamberlain L. DHHC palmitoyl transferases: substrate interactions and (patho)physiology. Trends Biochem Sci. 2011;36:245–253. doi: 10.1016/j.tibs.2011.01.003. [DOI] [PubMed] [Google Scholar]
- 15.McCormick PJ, Dumaresq-Doiron K, Pluviose AS, Pichette V, Tosato G. Palmitoylation controls recycling in lysosomal sorting and trafficking. Lefrancois S. Traffic. 2008;9:1984–97. doi: 10.1111/j.1600-0854.2008.00814.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Runkle K, Kharbanda A, Stypulkowski E, Cao X-J, Wang W, Garcia BA, Witze E. Inhibition of DHHC20-Mediated EGFR Palmitoylation Creates a Dependence on EGFR Signaling. Mol Cell. 2016;62:385–396. doi: 10.1016/j.molcel.2016.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jennings BC, Nadolski MJ, Ling Y, Baker MB, Harrison ML, Deschenes RJ, Linder ME. 2-Bromopalmitate and 2-(2-hydroxy-5-nitro-benzylidene)-benzo[b]thiophen-3-one inhibit DHHC-mediated palmitoylation in vitro. J Lipid Res. 2009;50(2):233–42. doi: 10.1194/jlr.M800270-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chou TC. Drug combination studies and their synergy quantification using the Chou- Talalay method. Cancer Res. 2010;70(2):440–6. doi: 10.1158/0008-5472.CAN-09-1947. Chou TC. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.