Abstract
The endocannabinoid system (eCBs), named after the plant Cannabis sativa, comprises cannabinoid receptors, endogenous ligands known as “endocannabinoids”, and enzymes involved in the biosynthesis and degradation of these ligands, as well as putative transporters for these ligands. ECBs proteins and small molecules have been detected in early embryonic stages of many vertebrate models. As a result, cannabinoid receptors and endogenous as well as exogenous cannabinoids influence development and behavior in many vertebrate species. Understanding the precise mechanisms of action for the eCBs will provide an invaluable guide towards elucidation of vertebrate development and will also help delineate how developmental exposure to marijuana might impact health and cognitive and executive functioning in adulthood. Here we review the developmental roles of the eCBs in vertebrates, focusing our attention on the zebrafish model. Since little is known regarding the eCBs in zebrafish, we provide new data on the expression profiles of eCBs genes during development and in adult tissue types of this model organism. We also highlight exciting areas for future investigations, including the synaptic regulation of eCBs, its role in reward and addiction, and in nervous system development and plasticity.
Keywords: Endocannabinoid System, Cannabinoid Receptors, AEA, 2-AG, Zebrafish, Development, THC
Graphical abstract
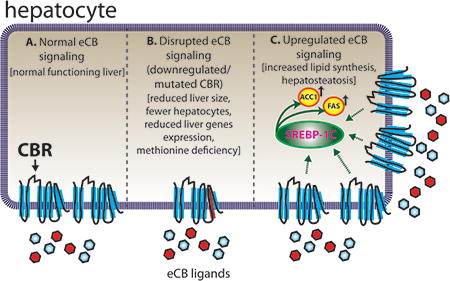
Proper liver function in zebrafish appears to be dependent on a normal, functioning eCBs.
Introduction
After the first cannabinoid receptor CB1 was identified as a binding site for psychotropic cannabinoids and cloned for further localization studies (Herkenham et al 1990, Matsuda et al 1990), many laboratories started to investigate the phenomenon previously associated with the consumption of cannabinoids, including the feeling of happiness, excitement, dissociation of ideas, spatiotemporal errors, mood fluctuation, illusion, and hallucinations (Moreau 1973). Researchers explored the physiological roles of this receptor, and in turn discovered Anandamide (AEA) (Devane et al 1992) and 2-arachidonoylglycerol (2-AG) (Mechoulam et al 1996) as endogenous ligands for CB1. Both ligands specifically interact with CB1 leading to the inhibition of adenylyl cyclase (Howlett et al 2010). In addition, a second cannabinoid receptor CB2, which shared 48% identity with CB1, was identified. CB2 is mostly expressed in the spleen, suggesting a role in the immune system (Munro et al 1993). The set of cannabinoid receptors, the two endocannabinoids and the enzymes responsible for their synthesis and degradation are together known as the endocannabinoid system (eCBs).
The eCBs has received considerable attention from the research community. More than 16000 papers can be found on the NCBI website using keyword searches for “cannabinoid receptor” and “endocannabinoid system”. Despite these advances, the diversity of actions characterizing the stimulation of the two receptors by endogenous and exogenous ligands remains incompletely understood. In particular, the roles of the eCBs in reward and addiction and their impact on embryonic and postnatal development await further investigations.
In this article, we will first highlight the eCBs for its known synaptic actions and role in reward and addiction. Evolutionary considerations will then be given by discussing the eCBs in mammals and amphibians. Due to the focus on zebrafish, the current state of knowledge of the eCBs will be subsequently discussed in greater detail in this model organism. Finally, we present new data on the spatiotemporal expression profiles of the eCBs in zebrafish to bridge this knowledge gap.
eCBs signalling at the synapse
The two major cannabinoid receptors, CB1 and CB2, belong to the large family of seven transmembrane-spanning G-protein-coupled receptors (GPCRs) (Matsuda et al 1990, Munro et al 1993). Genes encoding orthologs of the mammalian CB1 are found throughout vertebrates including chicken, turtle, frog, and fish (Elphick & Egertova 2001). Within the central nervous system (CNS), the two endocannabinoids are synthesized and released “on demand” into the synaptic cleft, where they work as retrograde synaptic messengers through binding to the CB receptors on the presynaptic terminal of neurons (Chevaleyre et al 2006, Elphick & Egertova 2005). The activation of cannabinoid receptors in turn inhibits the release of many neurotransmitters (e. g. serotonin, glycine, gamma-aminobutyric acid, glutamate, cholecystokinin). Specific catabolic enzymes are then responsible for the degradation of the ligands. The eCB signalling pathway at the synapse is described in greater detail in Figure 1A and a schematic representation showing gene relationship and function is shown in Figure 1B. Moreover, Table 1 summarizes eCBs' gene names and function.
Figure 1. eCBs signaling in neurons.
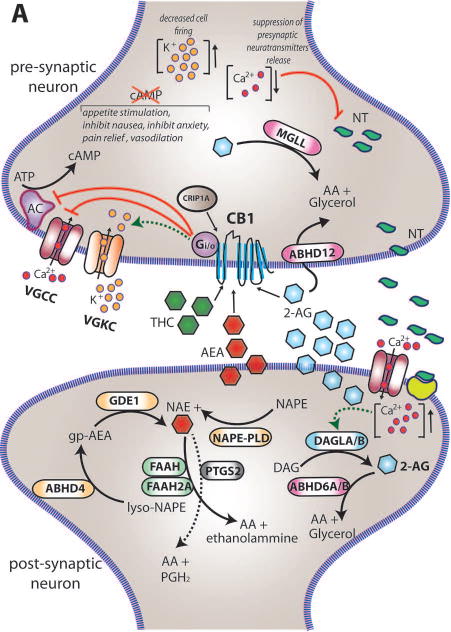
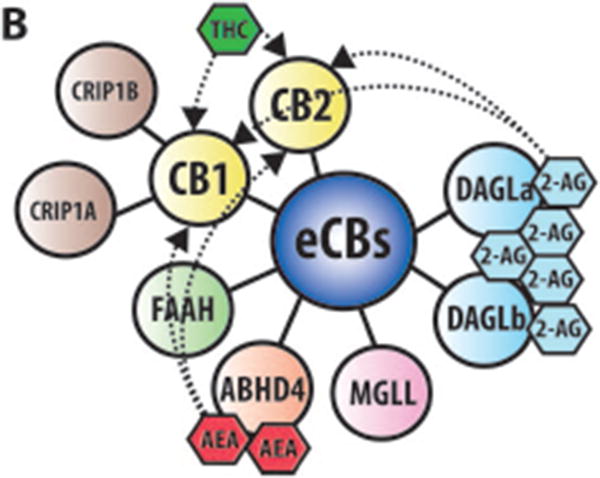
(A) Within the brain the endocannabinoids AEA and 2-AG are biosynthesized from different membrane phospholipid families, both esterified with arachidonic acid (AA). Several possible biosynthetic routes for the formation of AEA in the post-synaptic neuron have been suggested with multiple enzymes implicated: Nacylphosphatidylethanolamine specific phospholipase D (NAPE-PLD), αβ-Hydrolase domain-containing 4 (ABHD4), and glycerophosphodiesterase-1 (GDE1). The biosynthetic precursors of 2-AG are converted to 2-AG by the action of sn-1-diacylglycerol lipases α and β (DAGLα and DAGLβ). Endogenous AEA and 2-AG and exogenous Δ9-THC (THC) activate the CB1 receptor, exposed on the pre-synaptic neuron, causing (1) G-protein mediated inactivation of voltage-gated calcium channels (VGCC) which results in a transient reduction of neurotransmitter (NT) release, (2) G-protein mediated activation of voltage-gated potassium channels (VGKC), which decreases cell firing, and (3) G-protein mediated inhibition of adenylate cyclase (AC) with consequent reduction of cAMP levels. 2-AG is then degraded by three serine hydrolases, MGLL (monoglyceride lipase), ABHD6 (αβ-Hydrolase domain-containing 6) and ABHD12 (αβ-Hydrolase domain-containing 12), that account for approximately 99% of 2-AG hydrolysis in the CNS (Savinainen et al 2012). MGLL is responsible for approx. 85% of 2-AG hydrolysis and co-localizes with CB1 in axon terminals (Savinainen et al 2012). ABHD6 accounts for approx. 4% of brain 2-AG hydrolase activity; in neurons it resides post-synaptically, often juxtaposed with CB1 where it regulates intracellular pools of 2-AG at the site of generation. ABHD12 is highly expressed in microglia and accounts for approx. 9% of total brain 2-AG hydrolysis. AEA is generally degraded by the fatty acid amide hydrolases FAAH and FAAH2A. ProstaglandinEndoperoxide Synthase 2 PTGS2 (COX-2) possesses the capacity to metabolize AEA in vivo and can compete with FAAH for AEA in several brain regions (Glaser & Kaczocha 2010). The cannabinoid receptor interacting protein CRIP1A, transiently interacting with CB1 can stabilize and regulate the inactive state of the receptor (Niehaus et al 2007). In contrast with this conclusion, Guggenhuber and colleagues proposed that CRIP1A regulates CB1 activity in an agonist-dependent manner (Guggenhuber et al 2016). (B) Schematic representation of the main proteins belonging to the eCBs. CB1 and CB2 receptors get activated by endogenous (2-AG and AEA) and exogenous (THC) cannabinoids; 2-AG is synthesised by DAGLα and DAGLβ and degraded by MGLL; AEA is mainly synthesised by ABHD4 and degraded by FAAH. CRIP1A and CRIP1B are known interaction partners of CB1; Levels of 2-AG in the brain are higher than those of AEA.
Table 1.
Description of protein names, standard abbreviations, conventional zebrafish names (https://zfin.org) and function of endocannabinoid related genes investigated in this study.
| Protein Name | Standard Abbreviation | ZFIN Gene Name | Function |
|---|---|---|---|
| cannabinoid receptor 1 | CB1 | cnr1 | G-protein coupled receptor located primarily in the CNS; activated by endogenous and exogenous cannabinoids |
| cannabinoid receptor 2 | CB2 | cnr2 | G-protein coupled receptor located primarily in peripheral organs of the immune system and in the brain; activated by endogenous and exogenous cannabinoids |
| G protein-coupled receptor 55a | GPR55A | gpr55a | G- protein coupled receptor widely expressed in the brain; recently found to be activated by endogenous and exogenous cannabinoids; its activation leads to stimulation of rhoA, cdc4 and rac1 |
| cannabinoid receptor interacting protein 1a | CRIP1A | cnrip1a | CB1 receptor interacting protein that interacts with the distance C-terminus of CB1 altering/modulating CB1 interactions with G-protein |
| monoglyceride lipase | MGLL (MAGL) | mgll | member of the serine hydrolases superfamily; it catalyzes the hydrolysis of 2-AG to AA and Glycerol |
| αβ-hydrolase domain containing 6b | ABHD6B | abhd6b | member of the serine hydrolases superfamily; itcatalizes the hydrolysis of 2-AG to AA and Glycerol |
| αβ-hydrolase domain containing 12 | ABHD12 | abhd12 | member of the serine hydrolases superfamily; it catalyzes the hydrolysis of 2-AG to AA and Glycerol in the CNS |
| diacylglycerol lipase, alpha | DAGLα | dagla | diacylglycerol lipase; it catalyzes the hydrolysis of diacylglycerol (DAG) to the most abundantendocannabinoid 2-AG |
| diacylglycerol lipase, beta | DAGLβ | daglb | diacylglycerol lipase; it catalyzes the hydrolysis of diacylglycerol (DAG) to the most abundant endocannabinoid 2-AG |
| fatty acid amide hydrolase | FAAH | faah | fatty acid amide hydrolase with a single N-terminal transmembrane domain; principal catabolic enzyme for a class of lipids known as fatty acid amides (FAAs) of which AEA belongs to |
| fatty acid amide hydrolase 2a | FAAH2A | faah2a | fatty acid amide hydrolase; it degrades bioactive fatty acid amides, including AEA (AEA = arachidonic acid + ethanolamine) |
| N-acyl phosphatidylethanolamine phospholipase D | NAPE-PLD | napepld | member of the metallo-beta-lactamase family with phosphodiesterase activity; it releases NAE from NAPE to for AEA |
| αβ-hydrolase domain containing 4 | ABHD4 | abhd4 | hydrolase that acts on either NAPE or lyso-NAPE to generate the glycerophospho-arachidonoyl ethanolamide which is subsequently cleavaged to give AEA |
| glycerophosphodiester phosphodiesterase 1 | GDE1 (MIR16) | gde1 | phosphodiesterase with glycerophospho-NAE phosphodiesterase activity; ithydrolyses the phosphodiesterase bond of GP-NArE to release free AEA |
| prostaglandin-endoperoxide synthase 2 a | PTGS2A (COX-2; COX-2A) | ptgs2a | cycloxigenase involved in AEA (and 2-AG?) metabolism |
| N-acylsphingosine amidohydrolase (acid ceramidase) 1a | NAAA1A (ASAH1A) | asah1a | member of the choloylglycine hydrolase family with similar structure to faah. Unlike faah, naaa1a operates in acidic environments (pH 4.5) |
| protein tyrosine phosphatase, non-receptor type 22 | PTPN22 | ptpn22 | protein tyrosine phosphase highly expressed in the immune system; it dephosphorylates pAEA to yield AEA |
| peroxisome proliferator-activated receptor alpha b | PPARαβ | pparab | nuclear receptor transcription factor protein suggested as a binding target of endocannabinoids |
| peroxisome proliferator-activated receptor gamma | PPARγ (ARF6) | pparg | nuclear receptor transcription factors protein suggested as a binding target of endocannabinoids |
Role of the eCBs in reward and addiction
The role of the eCBs in reward processing and motivated behaviors has been extensively studied. The ventral tegmental area (VTA) and the nucleus accumbens (NAc) play central roles in the processing of rewarding stimuli and in drug addiction. The VTA also contains at least two additional neuronal phenotypes that are not dopaminergic (DA) (Cameron et al 1997). DA neurons produce endogenous cannabinoids to regulate their own activity through the interaction with the afferent neurons: current data support a CB1 receptor-mediated increase in dopamine neuron activity, due to induction of local disinhibitory mechanisms, such as depolarization-induced suppression of inhibition or depolarization-induced suppression of excitation (DSE) at inhibitory (i.e., GABAergic) or excitatory (i.e., glutamatergic) synapses, respectively (Zlebnik & Cheer 2016). Previous findings on addiction showed that AEA and its synthetic analog methanandamide are effective reinforcers of intravenous self-administration behavior in squirrel monkeys, an animal model of human drug abuse and suggests that medications that promote the actions of endogenously released cannabinoids could also activate brain reward processes and have the potential for abuse (Justinova et al 2005). It is important to understand how marijuana can mediate these effects. There is evidence that cannabis is addictive: around 9% of users become dependent on the drug, showing signs of addiction such as developing tolerance or experiencing withdrawal symptoms (Cressey 2015). It has been demonstrated that the psychoactive component of marijuana, Δ9-THC, alters the activity of central reward pathways in a manner that is consistent with other abused drugs but the cellular mechanisms through which this occurs rely upon the combined regulation of several afferent pathways to the VTA (Lupica et al 2004). The precise mechanism by which the eCBs facilitates DA burst firing in vivo is yet to be fully understood.
The evolution of the eCBs
The eCBs is widely conserved across organisms, although the patterns of evolution for each protein vary. CB receptor genes appear to be present only in chordates (Elphick 2012). It is believed that CB1 and CB2 arose from a gene duplication event of a common ancestral gene. Remarkably, duplicate CB receptor genes have been found in teleosts. For example, zebrafish have two cb2 genes, and puffer fish have two cb1 genes (Elphick & Egertova 2001). This may be evidence of a second gene duplication event in a common ancestor of these fish, followed by loss of a gene copy in subsequent families. Unlike CB receptors, DAGLs (2-AG synthesis enzymes) are more widely conserved among bacteria, fungi, plants and animals (Yuan et al 2016). However, DAGL substrate selectivity across organisms differs. For example, mammalian DAGL specifically hydrolyzes DAGs, while bacterial DAGL can catalyze hydrolysis of DAG, MAG, and glycerol. It is worth noting that the two isoforms of DAGL, DAGLα and DAGLβ, have distinct evolutionary patterns. Yuan et al compiled a thorough account of the similarities and differences between the evolutions of each isoform. As with DAGL, 2-AG is also largely conserved. 2-AG has been found in animals as primitive as fresh water polyps (De Petrocellis et al 1999). The 2-AG degradation enzyme MGLL is also largely conserved across many different phyla; it is found in animals such as placozoans and cnidarians. However, several insect species like Drosophila lost this gene (Elphick 2012). ABHD4 is one of the proteins suggested to be involved in AEA biosynthesis and is also highly conserved, having orthologues in a wide variety of species from fruit flies and lizards to mammals (Ensembl Genome Browser). FAAH and FAAH2, enzymes involved in the degradation of AEA, likely underwent a gene duplication event in an ancestral animal preceding organisms with nervous systems. Though these genes are prevalent across species, certain lineages lost one of these genes. For example, rodents lack FAAH2, and insect species like Drosophila lack FAAH (Elphick 2012). Lastly, the cannabinoid receptor interacting protein CRIP1A is thought to originate in the first organisms with nervous systems; this protein is ubiquitous and has been found in species such as N. vectenses (cnidaria) and C. elegans (nematoda). Even though CRIP1A interacts with CB1, its origins significantly precede CB receptor appearance, suggesting other functions for this protein in addition to interacting with CB receptors (Elphick 2012).
The eCBs in developing mammals
The eCBs with its metabolic enzymes, receptors, and secondary messenger cascades play a major role in development/neurodevelopment. Understanding of the signaling pathways will help reveal the basis of developmental defects that are associated with prenatal drug abuse. The presence of CB1 receptor at early developmental stages suggests that the eCBs contributes to CNS development, such as axonal elongation, myelination, migration, cell proliferation, and synaptogenesis (Fernandez-Ruiz et al 2000). Multiple studies have shown that mRNA expression of the CB1 receptor is distributed in both the fetal and neonatal rat brain (Berrendero et al 1998, Berrendero et al 1999, Romero et al 1997). CB1 activity and receptor binding can be identified as early as 14 gestational days old, which overlap with the expression of most neurotransmitters (Insel 1995). In mammals, the CB1 receptor plays a major role in neural progenitor proliferation and survival. The proliferation of neural progenitor cells has been associated with the dependence of the activation of the CB1 receptor in areas such as the cerebellum and hippocampus (Trazzi et al 2010). Studies using knockout mice demonstrated that inactivation of both the CB1 and CB2 receptor impairs neural progenitor cell proliferation (Aguado et al 2005, Palazuelos et al 2006). Also, reduced CB1 function in vivo is frequently linked to alterations with regards to hippocampal and cortical development (Aguado et al 2005, Zurolo et al 2010). It is shown that mice lacking CB1 receptors have suppressed cortical progenitor proliferation (Aguado et al 2005, Mulder et al 2008).
AEA and 2-AG are present throughout prenatal development, but fluctuate and vary with a wide range (Berrendero et al 1999, Fernandez-Ruiz et al 2000). In mice, AEA has been associated with the activation of embryo implantation inside the uterus during days 4-6 of pregnancy (Paria et al 2001). Throughout perinatal development, AEA levels are low at mid-gestation but increase gradually until adulthood (Berrendero et al 1999). However, fetal 2-AG levels are approximately the same concentration in young and adult rat brains, with only a distinguishable surge of 2-AG immediately after birth (Berrendero et al 1999, Fernandez-Ruiz et al 2000). Overall, it has been observed that in adult brains, the concentrations of 2-AG are much greater than the levels of AEA, a difference of 2000-8000 pmol/g of tissue versus 3-6 pmol/g of tissue, respectively (Berrendero et al 1999, Fernandez-Ruiz et al 2000).
In humans, the CB1 receptor is saturated in the cerebellum, hippocampus, caudate nucleus, and cerebral cortex and is evident in the brain as early as prenatal development. At just gestational week 9, the CB1 receptor can be found in Cajal–Retzius cells and in the sub-ventricular zone (Zurolo et al 2010). During the second trimester of gestation, CB1 receptors can be traced in the hippocampal CA region (Wang et al 2003). However, the high concentrations of the CB1 receptor in fibre-enriched areas are only detected during prenatal development and are almost non-existent in the adult brain (Mato et al 2003). Thus, the expression of the CB1 receptor during early development of the nervous system suggests that the endocannabinoid system plays a role on neural development in humans, which ultimately can be associated with possible neuropsychiatric disorders (Galve-Roperh et al 2009, Jutras-Aswad et al 2009).
Alterations of the endocannabinoid receptor signalling during early human development can result in changes of the developing brain, for instance, impairment of neuronal maturation, connectivity, or migration, which could play a direct role in adult brain dysfunction (Pang et al 2008). Indeed, genetic polymorphisms in eCBs genes have been associated with functional differences. A missense mutation in the FAAH gene has been associated with problematic drug use (Hariri et al 2009, Sipe et al 2002). In 2010, further studies have shown that mutations in the ABHD12 gene (functioning in degradation of AEA) cause the neurodegenerative disease PHARC, with symptoms including polyneuropathy, hearing loss, ataxia, retinitis pigmentosa, and cataracts (Fiskerstrand et al 2010). Additionally, the inhibition of the CB1 receptor during cortical neurogenesis resulted in deficits to subcortical projections that impaired proper motor function in adulthood (Diaz-Alonso et al 2012). Alteration of eCBs signalling also affects the way the brain processes emotion, reward, and threat (Galve-Roperh et al 2009, Jutras-Aswad et al 2009). Reduction or enhancement of G-protein mediated signalling due to genetic polymorphisms in the CB1 gene has been linked to psychiatric disorders such as schizophrenia, depression, and psychosis (Ballon et al 2006). Similarly, polymorphisms in the CB2 gene has also been linked to symptoms of depression and schizophrenia (Onaivi et al 2008).
The eCBs in amphibians
Under the classification of amphibians, CB1 receptors have been identified within species such as Taricha granulosa (Soderstrom et al 2000), Xenopus laevis (Cottone et al 2003), and Rana esculenta (Meccariello et al 2007). However, Xenopus tropicalis is one of the few in which CB2 gene is present (Elphick & Egertova 2001). In situ hybridization experiments conducted on Xenopus granulosa have revealed that CB1 mRNA expression can be detected early during development in the telencephalon, specifically in the nucleus amygdalae, dorso lateralis, and stria terminalis. Additionally, expression of the CB1 mRNA can be found in the cerebellum, preoptic region, stratum griseum of the hindbrain, and thalamus (Hollis et al 2006). In Xenopus laevis, there has been detection of CB1 mRNA in the embryos at stage 28. Upon reaching stage 41, CB1 mRNA is detected in the rhombencephalon and olfactory bulb (Migliarini et al 2006). During adulthood, Xenopus laevis has CB1 mRNA-positive cells in regions such as amygdala, hypothalamus, cerebellum, spinal cord, mesencephalic tegmentum, dorsal/medial pallium, and cells of the pituitary gland such as the thyrotrophs, lactotrophs, and gonadotrophs (Cesa et al 2002, Cesa et al 2001, Cottone et al 2003, Salio et al 2002).
Similarly, CB1 immunostaining in neurons has been revealed in Rana esculenta at high level, specifically in the pre-optic regions, hindbrain, hypothalamus, and telencephalic hemispheres (Cottone et al 2008, Meccariello et al 2008). There are postulations that gonadal activity is influenced by the eCBs due to fluctuations in CB1 mRNA expression in regions of the brain associated with the sexual cycle (Meccariello et al 2006, Meccariello et al 2008). During the frog sexual cycle, gonadotropin-releasing hormone I (GnRH-I) mRNA and CB1 levels have an inverse relationship of expression in the diencephalon and telencephalon (Chianese et al 2012, Meccariello et al 2008). In these regions, AEA acts as an antagonist to the synthesis of GnRH-I and GnRH-II, which will trigger an increase in CB1 transcription, suggesting a relationship between GnRH and the eCBs (Chianese et al 2011, Chianese et al 2012, Meccariello et al 2008). Additionally, there are speculations of endocannabinoid-mediated responses such as anxiety, stress, and fear due to the CB1 mRNA in situ hybridization staining of the amygdaloid complex in Taricha granulosa (Cottone et al 2003).
The eCBs in Zebrafish
As a non-mammalian vertebrate, the zebrafish (Danio rerio) is evolutionarily more distant from humans than rodent models but evolutionarily closer to humans than other invertebrate models, such as worms (C. elegans) or fruit flies (D. melanogaster). Indeed, the eCBs is highly conserved between mammals and zebrafish but not the aforementioned invertebrate model organisms (Elphick 2012). Zebrafish development occurs externally and the transparency of its embryos through larval stages makes it an ideal model to understand the role played by eCBs in development.
Neural development
Watson et al showed that knockdown of cb1 gene activity by morpholino antisense oligonucleotides resulted in defects of axonal growth and fasciculation (Watson et al 2008). More recently, Martella et al showed that 2-AG plays a key role in axonal growth and fasciculation, and that the eCBs is critical for the development of functional vision and locomotion (Martella et al 2016a).
Anxiety
The eCBs is involved in modulating anxiety across various animal models (Krug & Clark 2015). Low level stimulation of CB receptors commonly causes anxiolytic effects, while high level stimulation is anxiogenic (Viveros et al 2005). A light/dark preference testing arena was utilized to see the effects of CB receptor stimulation on fish behavior (Connors et al 2014). In this assay, zebrafish were placed into a tank with both light and dark regions. Normally, adult zebrafish have a significantly higher preference for dark areas (Serra et al 1999), but exposure to anxiolytic drugs can increase the time spent in light areas (Guo 2004). Connors et al showed that supplementing zebrafish food with the potent synthetic CB receptor agonist WIN55212-2 (1 μg/day for 7 days) increases the time spent in light areas, suggesting anxiolytic effects. In contrast, a spatial tank test revealed anxiogenic properties of Δ9-THC, a CB receptor agonist (Stewart & Kalueff 2014). Zebrafish pre-exposed for 20 minutes with 30 mg/L or 50 mg/L THC spent less time in the upper half of the tank, suggesting an increase in anxiety compared to control fish. Though CB receptor agonists were used in both the light/dark preference assay and the spatial tank test, opposing effects on anxiety are likely due to different amounts of drug dosed, corroborating the idea that the relationship between CB receptor stimulation and anxiety is dosage-dependent and/or due to different routes of administration. Another method of evaluating anxiety involves escape response. Ruhl et al measured the response of the fish to a threatening visual stimulus and did not have significant evidence of THC at 100 nM being anxiolytic (Ruhl et al 2014). However, even though THC at 100 nM did not produce any change in behavioral performance, the same concentration severely impaired spatial memory (Ruhl et al 2014). Along with differences in concentration, discrepancies in anxiolytic effects across studies may lie in the fact that different cannabinoids were administered; WIN55212-2 is a full agonist, while THC is a partial agonist. Interestingly, a more recent threat experiment using the same concentration of THC as Ruhl et al yielded different results; THC administration appeared to have some anxiolytic properties (THC treated fish spent less time at the bottom of the tank), yet did not reduce other behaviors indicative of anxiety (freezing and erratic movements) (Ruhl et al 2016). The fear stimulus in this experiment was a pheromone, and a possible explanation for the conflicting results may be that CB stimulation distinctly affects different sensory inputs. Lastly, social interactions may also be indicative of nervous behavior. Fish treated with 1 mg/L WIN55212-2 swam longer with a stranger fish than controls, suggesting anxiolytic activity (Barba-Escobedo & Gould 2012). Though most evidence from zebrafish experiments agree with conclusions from other animal models, some conflicting evidence calls for more experiments to have a clearer understanding of the connection between the eCBs and anxiety in zebrafish.
Lipid Homeostasis and Appetite
Zebrafish have become a common and relevant model for the study of lipid homeostasis, and the eCBs has been associated with changes in lipid homeostasis and food intake (Krug & Clark 2015). In rodent models, CB1 stimulation in the liver has been shown to induce fatty acid synthesis, increase appetite, and promote obesity, while CB1 downregulation produces opposite effects (Gary-Bobo et al 2007, OseiHyiaman et al 2005, Wiley et al 2005). Similar results were seen in zebrafish. Liu et al reported the importance of the eCBs in the liver; cb1 and cb2 double mutant fish have impaired liver development and function (Liu et al 2016). Specifically the authors showed that inhibition of CB receptor activity disrupts liver development and metabolic function in zebrafish, impacting hepatic differentiation and liver size due to fewer hepatocytes and reduced liver-specific gene expression and proliferation (Liu et al 2016). In contrast, when cb1 is overexpressed in zebrafish liver, the expression levels of genes involved in the fatty acid production, transport, and storage are consequently increased, resulting in hepatosteatosis (Pai et al 2013). Addition of Cb1 antagonist AM251 rescued this phenotype, suggesting the role of Cb1 in stimulating lipid accumulation. Additionally, AEA administration to zebrafish has been shown to increase expression of Srebp, a transcription factor involved in sterol synthesis (Migliarini & Carnevali 2008). In another study, Fraher et al showed that alteration of the activity of the eCBs and Retinoic Acid (RA) pathways have additive function in lipid abundance during zebrafish development (zebrafish embryos were exposed to chemical treatments WIN55212-2, Rimonabant, 4-diethylaminobenzaldehyde, BMS 753, BMS 614, BMS 961, CD 2665, oleamide, AM 630, bisphenol A diglycidyl ether, and Rosiglitazone) (Fraher et al 2015).
A recent study by Martella et al determined that bisphenol A (BPA) stimulates hepatosteatosis in zebrafish via upregulation of the eCBs (Martella et al 2016b). While stimulation of CB receptors appears to increase lipid synthesis, not all cannabinoids facilitate this process. A study showed that phytocannabinoids cannabidiol (CBD) and Δ9-tetrahydrocannabivarin (THCV) reduce lipid levels in zebrafish, agreeing with studies done in rodent models (Silvestri et al 2015). Unlike THC, THCV is a Cb1 receptor antagonist, and CBD has minimal affinity for either CB receptor. Therefore, the effects on lipid homeostasis by these particular cannabinoids likely occur through either Cb1 downregulation or interaction with receptors outside of the eCB system. It is worth noting that this experiment measured lipid metabolism in vivo by quantifying the amount of yolk in zebrafish embryos over time. Therefore, further studies must be done to see if these phytocannabinoids can protect against hepatosteatosis in adult fish. CB receptors have also been shown to modulate fish appetite. In an experiment done by Piccinetti et al, the administration of melatonin reduced zebrafish food intake. These melatonin-treated fish consequently had reduced Cb1 expression, suggesting that Cb1 has a role in stimulating hunger in zebrafish (Piccinetti et al 2010). Additionally, downregulation of Cb1 in zebrafish has been shown to reduce appetite in a dose dependent manner (Shimada et al 2012). Nishio et al conclude that Cb1 down-regulates the expression of cocaine- and amphetamine-related transcript (CART)-3 to induce hunger in zebrafish (Nishio et al 2012).
Immune System, and Neuroinflammation
The eCBs is also associated with immune system processes such as inflammation. As in mammals, zebrafish Cb2 receptors are highly expressed in white blood cells (Krug & Clark 2015). Administration of various Cb2 agonists in zebrafish reduced leukocyte migration to a tail wound (Liu et al 2013). Conversely, knocking out cb2 resulted in increased leukocyte migration compared to control fish. These studies suggest that Cb2 is a critical component in the modulation of inflammation responses, agreeing with other animal model studies. Cb2 activation is thought to inhibit leukocyte migration by down-regulating arachidonate 5-lipoxygenase (Alox5) through the JNK/c-Jun/Alox5 pathway. Additionally, Cb2 plays a role in neuroinflammation; in experimental allergic encephalomyelitis (EAE), a mouse model of brain inflammation, Cb2 is upregulated 200-fold in resting microglial cells (Maresz et al 2005). The reduction of Cb2 receptors on invading T-cells is shown to facilitate neurodegenerative disease progression (Maresz et al 2007). Experiments with neurodegenerative zebrafish models are needed to test whether Cb2 has a similar role in regulating neuroinflammatory disease progression in zebrafish.
Results
CB1 and CRIP1A proteins are conserved between zebrafish and mammalian orthologues
Zebrafish Cb1 protein (from transcript ENSDART00000011283.6) is made up of 475 aa compared to the 472 aa of human CB1 (ENST00000369501.2) and 473 aa of its mouse counterparts (ENSMUST00000057188.6). Different from human and mouse transcripts, which have a single exon, the zebrafish transcript has two exons. Comparison of human and mouse CB1 amino acid sequence with corresponding zebrafish orthologue Cb1 using ClustalW sequence alignment software revealed sequence identity around 70% relative to the human and mouse proteins (Figure S2). Human and mouse amino acid sequences share 97% identity.
CRIP1A, encoded by the CNRIP1a gene, was recently identified to interact with the C terminal region of CB1 (Ahmed et al 2014). Zebrafish Crip1A protein sequence was well conserved and similar in length when compared to the human and mouse proteins. In particular, zebrafish Crip1A (ENDART00000130828.2) protein contains 162 aa compared to the 164 aa of the human (ENST00000263655.3) and mouse (ENSMUST00000058159.5) counterparts. In all three species, the open reading frame (ORF) spanned three exons. ClustalW sequence alignment revealed sequence identity of around 60% relative to the human and mouse proteins (Figure S3). Human and mouse CRIP1A amino acid sequence identity is 97%.
Zebrafish cb1 and cnrip1a transcripts are detected in the developing zebrafish brain
Whole-mount in situ hybridization revealed enriched cb1 expression in discrete brain regions of zebrafish embryos. Similarly to what was previously described in (Lam et al 2006), cb1 transcript was detected in the preoptic area at 30 hpf (Figure 3 A,B) and extends to other areas of the telencephalon, diencephalon, and midbrain at 50 hpf (Figure 3 C,D). At 72 hpf, cb1 transcript was detected in the olfactory bulb and weakly in the midbrain (Figure 3 E,F).
Figure 3. Whole mount in situ hybridization analysis of cb1 and cnrip1a genes.
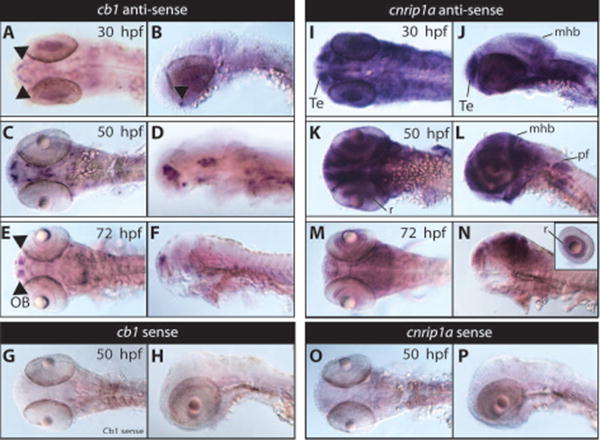
cb1 (A-H) and cnrip1a (I-P) expression in developing zebrafish (30, 50 and 72 hpf). Cb1 expression was examined using whole-mount in situ hybridization in wild type embryos at the 30 hpf (A, B; dorsal and lateral respectively), 50 hpf (C,D; dorsal and lateral respectively), and 72 hpf (E,F dorsal and lateral respectively) stages. By 30 hpf, cb1 expression is highly restricted to the pre-optic area (A,B). At 50 hpf, cb1 expression was prominent in very specific areas of the brain including the olfactory bulbs, telencephalon, optic tectum and hypothalamus (C,D). At 72 hpf, cb1 expression was further restricted to the telencephalon in the olfactory bulbs and weakly in the midbrain area (E,F). No signal was detected using cb1 sense probes at all stages, 50 hpf is shown (G,H). cnrip1a expression was examined using whole-mount in situ hybridization in wild type embryos at the 30 hpf (I, J; dorsal, lateral respectively), 50 hpf (K, L; dorsal, lateral respectively), and 72 hpf (M, N; dorsal, lateral respectively) stages. By 30 hpf, cnrip1a expression is highly expressed in the head region showing an enrichment in the telencephalon, midbrain, midbrain-hindbrain boundary and eyes (I,J). At 50 hpf, strong cnrip1a expression was prominent in specific areas of the brain, eyes retina, pectoral fins, and potentially in digestive organs (K,L). At 72 hpf, cnrip1a expression was further restricted to the retina, telencephalon, midbrain, midbrain-hindbrain boundary (M, N). No signal was detected using cnrip1a sense probes at all stages, 50 hpf is shown (O,P). Te, telencephalon; pf, pectoral fins; r, retina; mhb, midbrain-hindbrain boundary, OB, olfactory bulbs.
We next analyzed the expression pattern of cnrip1a transcript. cnrip1a transcript is highly expressed in the head region and in the brain at 30 hpf (Figure 3, I,J) showing an enrichment in the telencephalon, midbrain, hindbrain, and eyes. At 50 hpf cnrip1a transcript is detected in the brain, retina, pectoral fins, and potentially in digestive organs which they are not easily demarcated due to the strong signal of the probe (Figure 3 K,L). At 72 hpf cnrip1a expression is reduced but was still detectable in the mid- hind-brain regions (Figure 3 M,N). For both cb1 and cnrip1a there was no detectable staining with the sense control probes at all stages analyzed (50 hpf stages are shown as an example in Figure 3 G,H,O,P).
Expression profile of zebrafish eCBs genes during embryogenesis
Next we employed qPCR analysis to investigate the expression profiles of zebrafish eCBs genes during embryogenesis using cDNAs prepared from ten zebrafish embryonic developmental stages (between 1 hour and 120 hours post fertilization). cb1 expression was low during development although a very clear maternal-zygotic transition phase was detected (Figure 4, A). These data were consistent with in situ hybridization analysis shown in Figure 3. Similar low expression was seen for orphan receptor gpr55a (Figure 4, A). cb2 was expressed at higher levels and its expression seemed to be paired with the onset of peripheral organogenesis (Figure 3, A). Expression analysis of cnrip1a showed high level of expression starting at 24 hpf in concomitance with brain development (Figure 4, B). High levels of expression were detected in later stages, too. Its maternal expression also suggested a role at very early stages (Figure 4, B). The expression levels of genes encoding Mgll and Abhd6a and b (enzymes involved in 2-AG degradation) were relatively low during zebrafish development (Figure 4, C) while abhd12 showed high maternal mRNA levels, which continues throughout organogenesis (Figure 4, C). dagla showed a similar expression profile to the daglb, even though daglb seemed required during initial phases of embryonic development (Figure 4, D). faah and faah2a gene expression levels were very similar during development and fairly weak (Figure 4, E). A similar expression profile was seen for nape-pld and gde1 (Figure 4, F) but not for abhd4 of which the maternal expression was higher (Figure 4, F), suggesting a role during embryo early stages and a potential requirement for AEA synthesis during these stages. Expression of other eCBs related genes is shown in Figure 4G. Of these genes naaa1a showed elevated relative expression mRNA levels before 24 hpf (Figure 4, G).
Figure 4. Developmental expression of zebrafish eCBs genes.
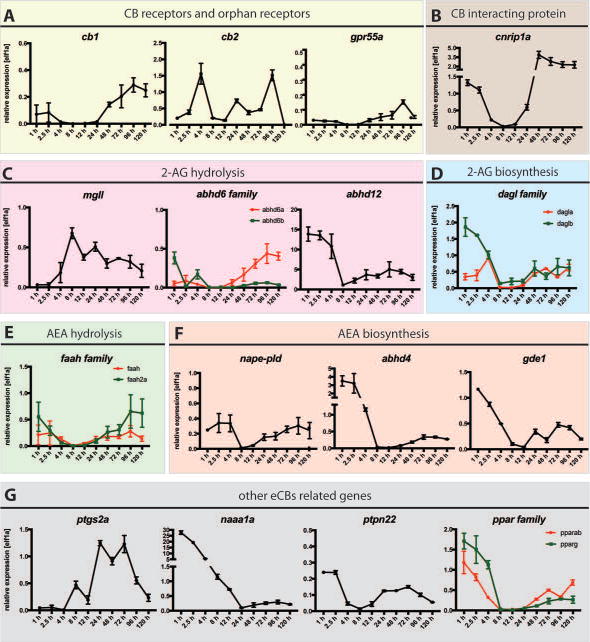
qPCR analysis of mRNA prepared at ten embryonic developmental stages of WT embryos (X axis: 1-120 hours post fertilization, hpf) using primers against (A) Cannabinoid Receptors, cb1 and cb2 and putative orphan receptor gpr55a (B) cannabinoid receptor interacting proteins, cnrip1a, (C) enzymes responsible for 2-AG hydrolysis mgll, abhd6a and abhd6b and abhd12, (D) enzymes responsible for 2-AG synthesis, dagla and daglb, (E) enzymes responsible for AEA hydrolysis faah and faah2a (F) enzymes responsible for AEA synthesis nape-pld, abhd4 and gde1 (G) genes associated with the eCBs, ptgs2a, naaa1a (asah1A), ptpn22, pparab and pparg. Elf1α was used as internal control to determine the relative mRNA expression. Relative average expression ± SEM (qPCR results are representative of two experimental repeats, 2 repeats/experiment). GraphPad Prism 7 software was used for statistical analysis.
Expression profiles of zebrafish eCBs genes in adult tissue types
We also analyzed the expression of zebrafish eCBs genes in adult tissues by qPCR. cb1 mRNA was present at very high levels in the brain while very little is detected in the eyes and testis (Figure 5, A); cb2 was detected in the brain, kidney, spleen and testis (Figure 5, A); grp55A was predominant in the brain, spleen and testis (Figure 5, A). The expression of cnrip1a was extremely high in the brain, eyes and testis (Figure 5, B). The highest expression for mgll was detected in the brain, kidney, spleen and eyes (Figure 5, C). While abhd6b appeared to be expressed at levels barely detectable, abhd6a was mostly expressed in the intestine, liver and testis (Figure 5, C). abhd12 showed variable expression within different tissues, and it was found more abundant in the brain, muscles, eyes and reproductive organs with a lower level in kidney, heart and intestine (Figure 5, C). The different expression levels of these serine hydrolases in the brain are explainable by their respective activity in the brain: indeed MGLL accounts for 85% of 2-AG hydrolysis, ABHD6 accounts for approx. 4% of brain 2-AG hydrolase and ABHD12 for 9% (Savinainen et al 2012). dagla and daglb showed a similar pattern of expression in the brain, muscles, kidney, eyes and testis; in the spleen dagla showed higher levels of expression compared to daglb (Figure 5, D). faah and faah2a were expressed at comparable levels in the brain (Figure 5, E); faah was also moderately expressed in the skin and testis (Figure 5, E) while faah2a was also detected intestine, eyes and testis (Figure 5, E). napepld, gde1 and abhd4 were widely expressed in almost all organs with similar mRNA levels in the brain (Figure 5, F); abhd4 was found more abundant in the spleen and testis (Figure 5, F). ptgs2a was predominantly expressed in the skin, spleen and eyes (Figure 5, G); naaa1a was greatly expressed in the reproductive organs (Figure 5, G), this was consistent with its high levels of maternal mRNA (Figure 4, G). pparab and pparg showed similar expression in muscles and spleen (Figure 5, G); pparab expression was greater in the brain, heart and eyes while pparg seemed to be enriched in the testis (Figure 5, G).
Figure 5. WT zebrafish adult tissue types expression of zebrafish eCBs genes.
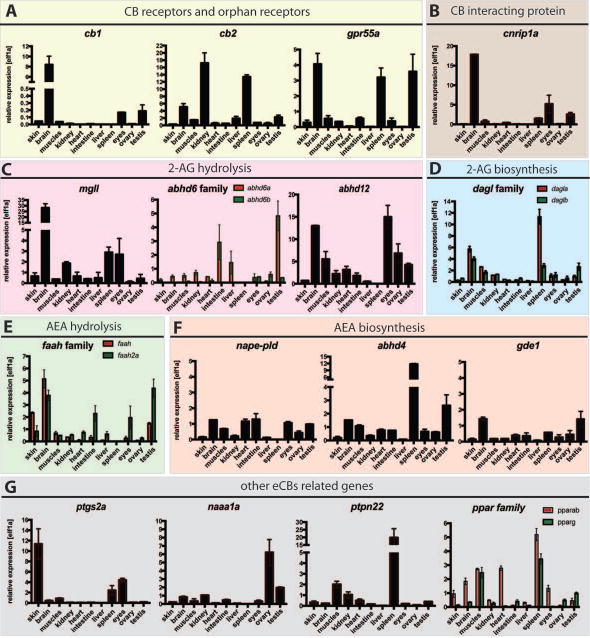
qPCR detection of zebrafish eCBs genes in zebrafish tissues (X axis: skin, brain, muscles, kidney, heart, intestine, liver, spleen, eyes, ovary, testis) using primers against (A) Cannabinoid Receptors, cb1 and cb2 and putative orphan receptor gpr55a (B) cannabinoid receptor interacting proteins, cnrip1a, (C) enzymes responsible for 2-AG hydrolysis mgll, abhd6a and abhd6b and abhd12, (D) enzymes responsible for 2-AG synthesis, dagla and daglb, (E) enzymes responsible for AEA hydrolysis faah and faah2a (F) enzymes responsible for AEA synthesis nape-pld, abhd4 and gde1 (G) genes associated with the eCBs, ptgs2A, naaa1a (asah1a), ptpn22, pparab and pparg. Elf1α was used as internal control to determine the relative mRNA expression. Relative average expression ± SEM (qPCR results are representative of two experimental repeats and 2 repeats/experiment). GraphPad Prism 7 software was used for statistical analysis.
Discussion
Although the eCBs has been studied in various animal models and humans, there are still many unknown features of this system. For example, it is not clear how activity regulates the availability of endogenous eCB ligands to specific synapses. The circuit mechanisms underlying eCBs's role in reward, addiction, and anxiety remain to be elucidated. How endogenous and exogenous CB ligands impact nervous system development and plasticity is also an important question for which deep mechanistic insights can be gained.
Zebrafish (Danio rerio) has become a prominent vertebrate model organism to study biological processes in vivo (Hill et al 2005, Santoriello & Zon 2012, Stern & Zon 2003). This is due to a combination of salient properties for elucidating embryonic development, physiology and diseases. Though a vertebrate, it has the strengths of invertebrate model systems, such as small size, high fecundity, and a relatively short generation time (Lieschke & Currie 2007). Moreover, its rapid and synchronous embryonic development greatly facilitates phenotypic analysis and high throughput experimental approaches. Its transparent and easily accessible embryos and larvae make zebrafish ideally suited for cell-type specific gene activity alterations and subsequent in vivo observations.
Despite the clear advantages in using this model, limited functional studies of the eCBs have been carried out using zebrafish. Here we present expression profiles of all genes known to be involved in endocannabinoid signaling in zebrafish at different developmental stages and in individual adult organs. Our observational gene expression studies contribute to the existing data about the endocannabinoid system and emphasize the benefit of this model in providing new insights. We found that zebrafish Cb1 and Crip1a are highly conserved with their human and mouse counterparts in terms of sequence identity at the amino acid level (Figure S2 and S3). Using in situ hybridization and qPCR to assess spatial and temporal expression in zebrafish embryos respectively, we found that cb1 transcript is restricted to very specific areas of the brain while cnrip1a is highly and widely expressed within the CNS; it is possible that Crip1A has an independent role from Cb1, which has already been suggested by Guggenhuber and colleagues (Elphick 2012, Guggenhuber et al 2016). Consequently, despite the initial exclusive characterization of Crip1A protein as Cb1 interacting protein it would be interesting to investigate further. Another interesting finding is that, despite the proposed role for Cb1 in the liver, in our studies zebrafish cb1 does not show detectable levels of mRNA expression in the adult liver. This is consistent with what has been previously stated in Alswat et al: “in the normal liver, the expression of CB1 and CB2 receptors is modest, which probably explains why the focus of research on the role of eCBs in the liver pathophysiology has come only recently. Indeed, early studies of brain CB1 receptors used the liver as a negative control” (Alswat 2013, Galiegue et al 1995).
The manipulation and analysis of the eCBs in zebrafish (i.e. the creation of zebrafish knock-out models) could bridge existing knowledge gaps on their function. Zebrafish studies could also contribute to a better understanding of the toxicological effects of exogenous cannabinoids. Through the employment of knockout models and well controlled assays, the effects of acute and chronic phytocannabinoid administration on development and behavior can be assessed.
The eCBs has the potential to be utilized in various therapeutic strategies. Previous studies have given evidence that activation or inhibition of the eCBs could alleviate the symptoms of various disease states, including multiple sclerosis, Alzheimer's disease, Parkinson's disease, Huntington's disease, obesity, anxiety, and depression (Di Marzo et al 2015, Krug & Clark 2015). On the other hand, exposure to exogenous CB ligands such as marijuana may have unwanted consequences on development and health. Further exploration of the eCBs, in particular by harvesting the strength of model organisms such as zebrafish, will allow for its exploitation in therapeutic contexts while avoiding the side effects of modulating eCB signaling.
Materials and Methods
Zebrafish husbandry
Wild type of the AB strain adult (1 y old) and larval zebrafish (Danio rerio, of either sex) were used in this study. The animals were raised at the University of California, San Francisco zebrafish facility at 28°C under a 14/10 hour light/dark cycle in accordance with National Institutes of Health and University of California, San Francisco guidelines.
Quantitative polymerase chain reaction (qPCR) analysis
Total RNA was prepared from isolated adult tissues (skin, brain, muscles, kidney, heart, intestine, liver, spleen, eyes, ovary, testis) and representative developmental embryo stages (1 hpf to 120 hpf dechorionated embryos) of zebrafish using TRIzol reagent (Invitrogen) by homogenization and purified using RNeasy Mini Kit (Qiagen). cDNAs were synthesized from 500 ng of purified RNA using qScript™ cDNA SuperMix (Quanta Biosciences) and used as templates. qPCR was performed using Applied Biosystems SYBR Green PCR Master Mix and the ABI7900HT machine. Forward and Reverse primers, listed in Table S1, were designed using ncbi primer blast software with exon-exon junction parameters and Danio rerio RefSeq for off targets. elf1α primers were used as standard CT (McCurley & Callard 2008) to generate Ct values.
Whole mount in situ hybridization
cb1 and cnrip1a sense and anti-sense in situ probes were created by cloning PCR products from zebrafish embryos' cDNA template into commercial TOPO vector pCR™4-TOPO®cloning kit (Invitrogen). After checking the directionality of the PCR products by sequencing (Quintara Biosciences), linearization of the vector by specific restriction enzymes was performed (details shown in Figure S1). Digoxigenin labeled RNA probes for in situ hybrydization (ISH) were generated using the DIG labeling kit (Roche) according to the manufacturer's instructions using either T3 or T7 RNA polymerases (depending on directionality). DNA template was removed using DNase. Hybridization of embryos collected at 30, 50 and 72 hpf (incubation step performed at 68°C) and detection with anti-digoxigenin was done as previously described (Guo et al 1999). After staining, embryos were cleared with glycerol, and whole-mounted for viewing. Images were taken using Zeiss Axioskop 2 plus microscope, Canon EOS DS126431 camera and MicroManager software.
Supplementary Material
Figure 2. The eCBs in Zebrafish Liver.
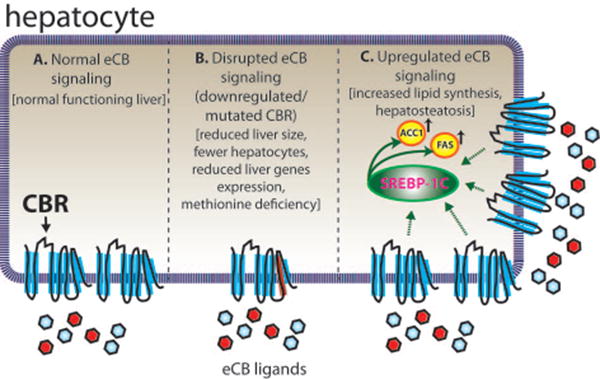
Proper liver function in zebrafish appears to be dependent on a normal, functioning eCBs. (A) Zebrafish liver functions regularly when eCB signaling is unaltered. (B) Mutation of cb1 and cb2 impair liver development and function (Liu et al 2016). Loss of CB1 and CB2 function results in a smaller liver with less hepatocytes and reduced expression of liver-specific genes in zebrafish. Additionally, methionine levels are irregular, which is known to cause a variety of metabolic problems, including hepatosteatosis. (C) Overexpression of CB1 results in fish with hepatosteatosis (Pai et al 2013). Increased CB1 receptor signaling stimulates SREBP-1c, a transcription factor which upregulates the expression of ACC1 and FAS (genes involved in fatty acid synthesis). ACC1, acetyl coenzyme-A carboxylase-1; CBR, cannabinoid receptor; FAS, fatty acid synthase; SREBP-1c, sterol regulatory element-binding protein 1c.
Acknowledgments
We would like to thank the Guo lab members for the helpful discussions and Michael Munchua, Hongbin Yuan, and Xingnu Zhai for excellent fish care.
Fundings: These studies were supported by Federal NIH fundings (Grant number R21DA038447 and R01 DA035680).
Footnotes
Competing interests: The authors declare that they have no competing interests.
References
- Aguado T, Monory K, Palazuelos J, Stella N, Cravatt B, et al. The endocannabinoid system drives neural progenitor proliferation. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2005;19:1704–6. doi: 10.1096/fj.05-3995fje. [DOI] [PubMed] [Google Scholar]
- Ahmed MH, Kellogg GE, Selley DE, Safo MK, Zhang Y. Predicting the molecular interactions of CRIP1a-cannabinoid 1 receptor with integrated molecular modeling approaches. Bioorg Med Chem Lett. 2014;24:1158–65. doi: 10.1016/j.bmcl.2013.12.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alswat KA. The role of endocannabinoids system in fatty liver disease and therapeutic potentials. Saudi J Gastroenterol. 2013;19:144–51. doi: 10.4103/1319-3767.114505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballon N, Leroy S, Roy C, Bourdel MC, Charles-Nicolas A, et al. (AAT)n repeat in the cannabinoid receptor gene (CNR1): association with cocaine addiction in an African-Caribbean population. Pharmacogenomics J. 2006;6:126–30. doi: 10.1038/sj.tpj.6500352. [DOI] [PubMed] [Google Scholar]
- Barba-Escobedo PA, Gould GG. Visual social preferences of lone zebrafish in a novel environment: strain and anxiolytic effects. Genes Brain Behav. 2012;11:366–73. doi: 10.1111/j.1601-183X.2012.00770.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berrendero F, Garcia-Gil L, Hernandez ML, Romero J, Cebeira M, et al. Localization of mRNA expression and activation of signal transduction mechanisms for cannabinoid receptor in rat brain during fetal development. Development. 1998;125:3179–88. doi: 10.1242/dev.125.16.3179. [DOI] [PubMed] [Google Scholar]
- Berrendero F, Sepe N, Ramos JA, Di Marzo V, Fernandez-Ruiz JJ. Analysis of cannabinoid receptor binding and mRNA expression and endogenous cannabinoid contents in the developing rat brain during late gestation and early postnatal period. Synapse. 1999;33:181–91. doi: 10.1002/(SICI)1098-2396(19990901)33:3<181::AID-SYN3>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- Cameron DL, Wessendorf MW, Williams JT. A subset of ventral tegmental area neurons is inhibited by dopamine, 5-hydroxytryptamine and opioids. Neuroscience. 1997;77:155–66. doi: 10.1016/s0306-4522(96)00444-7. [DOI] [PubMed] [Google Scholar]
- Cesa R, Guastalla A, Cottone E, Mackie K, Beltramo M, Franzoni MF. Relationships between CB1 cannabinoid receptors and pituitary endocrine cells in Xenopus laevis: an immunohistochemical study. Gen Comp Endocrinol. 2002;125:17–24. doi: 10.1006/gcen.2001.7720. [DOI] [PubMed] [Google Scholar]
- Cesa R, Mackie K, Beltramo M, Franzoni MF. Cannabinoid receptor CB1-like and glutamic acid decarboxylase-like immunoreactivities in the brain of Xenopus laevis. Cell Tissue Res. 2001;306:391–8. doi: 10.1007/s004410100461. [DOI] [PubMed] [Google Scholar]
- Chevaleyre V, Takahashi KA, Castillo PE. Endocannabinoid-mediated synaptic plasticity in the CNS. Annu Rev Neurosci. 2006;29:37–76. doi: 10.1146/annurev.neuro.29.051605.112834. [DOI] [PubMed] [Google Scholar]
- Chianese R, Ciaramella V, Fasano S, Pierantoni R, Meccariello R. Anandamide modulates the expression of GnRH-II and GnRHRs in frog, Rana esculenta, diencephalon. Gen Comp Endocrinol. 2011;173:389–95. doi: 10.1016/j.ygcen.2011.07.001. [DOI] [PubMed] [Google Scholar]
- Chianese R, Ciaramella V, Scarpa D, Fasano S, Pierantoni R, Meccariello R. Anandamide regulates the expression of GnRH1, GnRH2, and GnRH-Rs in frog testis. Am J Physiol Endocrinol Metab. 2012;303:E475–87. doi: 10.1152/ajpendo.00086.2012. [DOI] [PubMed] [Google Scholar]
- Connors KA, Valenti TW, Lawless K, Sackerman J, Onaivi ES, et al. Similar anxiolytic effects of agonists targeting serotonin 5-HT1A or cannabinoid CB receptors on zebrafish behavior in novel environments. Aquat Toxicol. 2014;151:105–13. doi: 10.1016/j.aquatox.2013.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottone E, Guastalla A, Mackie K, Franzoni MF. Endocannabinoids affect the reproductive functions in teleosts and amphibians. Mol Cell Endocrinol. 2008;286:S41–5. doi: 10.1016/j.mce.2008.01.025. [DOI] [PubMed] [Google Scholar]
- Cottone E, Salio C, Conrath M, Franzoni MF. Xenopus laevis CB1 cannabinoid receptor: molecular cloning and mRNA distribution in the central nervous system. J Comp Neurol. 2003;464:487–96. doi: 10.1002/cne.10808. [DOI] [PubMed] [Google Scholar]
- Cressey D. The cannabis experiment. Nature. 2015;524:280–3. doi: 10.1038/524280a. [DOI] [PubMed] [Google Scholar]
- De Petrocellis L, Melck D, Bisogno T, Milone A, Di Marzo V. Finding of the endocannabinoid signalling system in Hydra, a very primitive organism: possible role in the feeding response. Neuroscience. 1999;92:377–87. doi: 10.1016/s0306-4522(98)00749-0. [DOI] [PubMed] [Google Scholar]
- Devane WA, Hanus L, Breuer A, Pertwee RG, Stevenson LA, et al. Isolation and structure of a brain constituent that binds to the cannabinoid receptor. Science. 1992;258:1946–9. doi: 10.1126/science.1470919. [DOI] [PubMed] [Google Scholar]
- Di Marzo V, Stella N, Zimmer A. Endocannabinoid signalling and the deteriorating brain. Nat Rev Neurosci. 2015;16:30–42. doi: 10.1038/nrn3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Alonso J, Aguado T, Wu CS, Palazuelos J, Hofmann C, et al. The CB(1) cannabinoid receptor drives corticospinal motor neuron differentiation through the Ctip2/Satb2 transcriptional regulation axis. J Neurosci. 2012;32:16651–65. doi: 10.1523/JNEUROSCI.0681-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elphick MR. The evolution and comparative neurobiology of endocannabinoid signalling. Philos Trans R Soc Lond B Biol Sci. 2012;367:3201–15. doi: 10.1098/rstb.2011.0394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elphick MR, Egertova M. The neurobiology and evolution of cannabinoid signalling. Philos Trans R Soc Lond B Biol Sci. 2001;356:381–408. doi: 10.1098/rstb.2000.0787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elphick MR, Egertova M. The phylogenetic distribution and evolutionary origins of endocannabinoid signalling. Handb Exp Pharmacol. 2005:283–97. doi: 10.1007/3-540-26573-2_9. [DOI] [PubMed] [Google Scholar]
- Fernandez-Ruiz J, Berrendero F, Hernandez ML, Ramos JA. The endogenous cannabinoid system and brain development. Trends Neurosci. 2000;23:14–20. doi: 10.1016/s0166-2236(99)01491-5. [DOI] [PubMed] [Google Scholar]
- Fiskerstrand T, H'Mida-Ben Brahim D, Johansson S, M'Zahem A, Haukanes BI, et al. Mutations in ABHD12 cause the neurodegenerative disease PHARC: An inborn error of endocannabinoid metabolism. Am J Hum Genet. 2010;87:410–7. doi: 10.1016/j.ajhg.2010.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraher D, Ellis MK, Morrison S, McGee SL, Ward AC, et al. Lipid Abundance in Zebrafish Embryos Is Regulated by Complementary Actions of the Endocannabinoid System and Retinoic Acid Pathway. Endocrinology. 2015;156:3596–609. doi: 10.1210/EN.2015-1315. [DOI] [PubMed] [Google Scholar]
- Galiegue S, Mary S, Marchand J, Dussossoy D, Carriere D, et al. Expression of central and peripheral cannabinoid receptors in human immune tissues and leukocyte subpopulations. Eur J Biochem. 1995;232:54–61. doi: 10.1111/j.1432-1033.1995.tb20780.x. [DOI] [PubMed] [Google Scholar]
- Galve-Roperh I, Palazuelos J, Aguado T, Guzman M. The endocannabinoid system and the regulation of neural development: potential implications in psychiatric disorders. Eur Arch Psychiatry Clin Neurosci. 2009;259:371–82. doi: 10.1007/s00406-009-0028-y. [DOI] [PubMed] [Google Scholar]
- Gary-Bobo M, Elachouri G, Gallas JF, Janiak P, Marini P, et al. Rimonabant reduces obesity-associated hepatic steatosis and features of metabolic syndrome in obese Zucker fa/fa rats. Hepatology. 2007;46:122–9. doi: 10.1002/hep.21641. [DOI] [PubMed] [Google Scholar]
- Glaser ST, Kaczocha M. Cyclooxygenase-2 mediates anandamide metabolism in the mouse brain. J Pharmacol Exp Ther. 2010;335:380–8. doi: 10.1124/jpet.110.168831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guggenhuber S, Alpar A, Chen R, Schmitz N, Wickert M, et al. Cannabinoid receptor-interacting protein Crip1a modulates CB1 receptor signaling in mouse hippocampus. Brain Struct Funct. 2016;221:2061–74. doi: 10.1007/s00429-015-1027-6. [DOI] [PubMed] [Google Scholar]
- Guo S. Linking genes to brain, behavior and neurological diseases: what can we learn from zebrafish? Genes Brain Behav. 2004;3:63–74. doi: 10.1046/j.1601-183x.2003.00053.x. [DOI] [PubMed] [Google Scholar]
- Guo S, Wilson SW, Cooke S, Chitnis AB, Driever W, Rosenthal A. Mutations in the zebrafish unmask shared regulatory pathways controlling the development of catecholaminergic neurons. Developmental biology. 1999;208:473–87. doi: 10.1006/dbio.1999.9204. [DOI] [PubMed] [Google Scholar]
- Hariri AR, Gorka A, Hyde LW, Kimak M, Halder I, et al. Divergent effects of genetic variation in endocannabinoid signaling on human threat- and reward-related brain function. Biol Psychiatry. 2009;66:9–16. doi: 10.1016/j.biopsych.2008.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herkenham M, Lynn AB, Little MD, Johnson MR, Melvin LS, et al. Cannabinoid receptor localization in brain. Proc Natl Acad Sci U S A. 1990;87:1932–6. doi: 10.1073/pnas.87.5.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill AJ, Teraoka H, Heideman W, Peterson RE. Zebrafish as a model vertebrate for investigating chemical toxicity. Toxicological sciences : an official journal of the Society of Toxicology. 2005;86:6–19. doi: 10.1093/toxsci/kfi110. [DOI] [PubMed] [Google Scholar]
- Hollis DM, Coddington EJ, Moore FL. Neuroanatomical distribution of cannabinoid receptor gene expression in the brain of the rough-skinned newt, Taricha granulosa. Brain Behav Evol. 2006;67:135–49. doi: 10.1159/000090978. [DOI] [PubMed] [Google Scholar]
- Howlett AC, Blume LC, Dalton GD. CB(1) cannabinoid receptors and their associated proteins. Curr Med Chem. 2010;17:1382–93. doi: 10.2174/092986710790980023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Insel T. The Fourth Generation of Progress. Raven Press; New York: 1995. pp. 683–94. [Google Scholar]
- Justinova Z, Solinas M, Tanda G, Redhi GH, Goldberg SR. The endogenous cannabinoid anandamide and its synthetic analog R(+)-methanandamide are intravenously self-administered by squirrel monkeys. J Neurosci. 2005;25:5645–50. doi: 10.1523/JNEUROSCI.0951-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jutras-Aswad D, DiNieri JA, Harkany T, Hurd YL. Neurobiological consequences of maternal cannabis on human fetal development and its neuropsychiatric outcome. Eur Arch Psychiatry Clin Neurosci. 2009;259:395–412. doi: 10.1007/s00406-009-0027-z. [DOI] [PubMed] [Google Scholar]
- Krug RG, 2nd, Clark KJ. Elucidating cannabinoid biology in zebrafish (Danio rerio) Gene. 2015;570:168–79. doi: 10.1016/j.gene.2015.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam CS, Rastegar S, Strahle U. Distribution of cannabinoid receptor 1 in the CNS of zebrafish. Neuroscience. 2006;138:83–95. doi: 10.1016/j.neuroscience.2005.10.069. [DOI] [PubMed] [Google Scholar]
- Lieschke GJ, Currie PD. Animal models of human disease: zebrafish swim into view. Nat Rev Genet. 2007;8:353–67. doi: 10.1038/nrg2091. [DOI] [PubMed] [Google Scholar]
- Liu LY, Alexa K, Cortes M, Schatzman-Bone S, Kim AJ, et al. Cannabinoid receptor signaling regulates liver development and metabolism. Development. 2016;143:609–22. doi: 10.1242/dev.121731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YJ, Fan HB, Jin Y, Ren CG, Jia XE, et al. Cannabinoid receptor 2 suppresses leukocyte inflammatory migration by modulating the JNK/c-Jun/Alox5 pathway. J Biol Chem. 2013;288:13551–62. doi: 10.1074/jbc.M113.453811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupica CR, Riegel AC, Hoffman AF. Marijuana and cannabinoid regulation of brain reward circuits. Br J Pharmacol. 2004;143:227–34. doi: 10.1038/sj.bjp.0705931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maresz K, Carrier EJ, Ponomarev ED, Hillard CJ, Dittel BN. Modulation of the cannabinoid CB2 receptor in microglial cells in response to inflammatory stimuli. J Neurochem. 2005;95:437–45. doi: 10.1111/j.1471-4159.2005.03380.x. [DOI] [PubMed] [Google Scholar]
- Maresz K, Pryce G, Ponomarev ED, Marsicano G, Croxford JL, et al. Direct suppression of CNS autoimmune inflammation via the cannabinoid receptor CB1 on neurons and CB2 on autoreactive T cells. Nat Med. 2007;13:492–7. doi: 10.1038/nm1561. [DOI] [PubMed] [Google Scholar]
- Martella A, Sepe RM, Silvestri C, Zang J, Fasano G, et al. Important role of endocannabinoid signaling in the development of functional vision and locomotion in zebrafish. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2016a;30:4275–88. doi: 10.1096/fj.201600602R. [DOI] [PubMed] [Google Scholar]
- Martella A, Silvestri C, Maradonna F, Gioacchini G, Allara M, et al. Bisphenol A Induces Fatty Liver by an Endocannabinoid-Mediated Positive Feedback Loop. Endocrinology. 2016b;157:1751–63. doi: 10.1210/en.2015-1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mato S, Del Olmo E, Pazos A. Ontogenetic development of cannabinoid receptor expression and signal transduction functionality in the human brain. Eur J Neurosci. 2003;17:1747–54. doi: 10.1046/j.1460-9568.2003.02599.x. [DOI] [PubMed] [Google Scholar]
- Matsuda LA, Lolait SJ, Brownstein MJ, Young AC, Bonner TI. Structure of a cannabinoid receptor and functional expression of the cloned cDNA. Nature. 1990;346:561–4. doi: 10.1038/346561a0. [DOI] [PubMed] [Google Scholar]
- McCurley AT, Callard GV. Characterization of housekeeping genes in zebrafish: male-female differences and effects of tissue type, developmental stage and chemical treatment. BMC Mol Biol. 2008;9:102. doi: 10.1186/1471-2199-9-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meccariello R, Chianese R, Cacciola G, Cobellis G, Pierantoni R, Fasano S. Type-1 cannabinoid receptor expression in the frog, Rana esculenta, tissues: a possible involvement in the regulation of testicular activity. Mol Reprod Dev. 2006;73:551–8. doi: 10.1002/mrd.20434. [DOI] [PubMed] [Google Scholar]
- Meccariello R, Chianese R, Cobellis G, Pierantoni R, Fasano S. Cloning of type 1 cannabinoid receptor in Rana esculenta reveals differences between genomic sequence and cDNA. FEBS J. 2007;274:2909–20. doi: 10.1111/j.1742-4658.2007.05824.x. [DOI] [PubMed] [Google Scholar]
- Meccariello R, Franzoni MF, Chianese R, Cottone E, Scarpa D, et al. Interplay between the endocannabinoid system and GnRH-I in the forebrain of the anuran amphibian Rana esculenta. Endocrinology. 2008;149:2149–58. doi: 10.1210/en.2007-1357. [DOI] [PubMed] [Google Scholar]
- Mechoulam R, Ben Shabat S, Hanus L, Fride E, Vogel Z, et al. Endogenous cannabinoid ligands--chemical and biological studies. J Lipid Mediat Cell Signal. 1996;14:45–9. doi: 10.1016/0929-7855(96)01507-6. [DOI] [PubMed] [Google Scholar]
- Migliarini B, Carnevali O. Anandamide modulates growth and lipid metabolism in the zebrafish Danio rerio. Mol Cell Endocrinol. 2008;286:S12–6. doi: 10.1016/j.mce.2008.01.021. [DOI] [PubMed] [Google Scholar]
- Migliarini B, Marucci G, Ghelfi F, Carnevali O. Endocannabinoid system in Xenopus laevis development: CB1 receptor dynamics. FEBS Lett. 2006;580:1941–5. doi: 10.1016/j.febslet.2006.02.057. [DOI] [PubMed] [Google Scholar]
- Moreau J. Hashish and Mental Illness. Raven Press; New York: 1973. [Google Scholar]
- Mulder J, Aguado T, Keimpema E, Barabas K, Ballester Rosado CJ, et al. Endocannabinoid signaling controls pyramidal cell specification and long-range axon patterning. Proc Natl Acad Sci U S A. 2008;105:8760–5. doi: 10.1073/pnas.0803545105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro S, Thomas KL, Abu-Shaar M. Molecular characterization of a peripheral receptor for cannabinoids. Nature. 1993;365:61–5. doi: 10.1038/365061a0. [DOI] [PubMed] [Google Scholar]
- Niehaus JL, Liu Y, Wallis KT, Egertova M, Bhartur SG, et al. CB1 cannabinoid receptor activity is modulated by the cannabinoid receptor interacting protein CRIP 1a. Mol Pharmacol. 2007;72:1557–66. doi: 10.1124/mol.107.039263. [DOI] [PubMed] [Google Scholar]
- Nishio S, Gibert Y, Berekelya L, Bernard L, Brunet F, et al. Fasting induces CART down-regulation in the zebrafish nervous system in a cannabinoid receptor 1-dependent manner. Mol Endocrinol. 2012;26:1316–26. doi: 10.1210/me.2011-1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onaivi ES, Ishiguro H, Gong JP, Patel S, Meozzi PA, et al. Brain neuronal CB2 cannabinoid receptors in drug abuse and depression: from mice to human subjects. PLoS One. 2008;3:e1640. doi: 10.1371/journal.pone.0001640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osei-Hyiaman D, DePetrillo M, Pacher P, Liu J, Radaeva S, et al. Endocannabinoid activation at hepatic CB1 receptors stimulates fatty acid synthesis and contributes to diet-induced obesity. J Clin Invest. 2005;115:1298–305. doi: 10.1172/JCI23057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pai WY, Hsu CC, Lai CY, Chang TZ, Tsai YL, Her GM. Cannabinoid receptor 1 promotes hepatic lipid accumulation and lipotoxicity through the induction of SREBP-1c expression in zebrafish. Transgenic Res. 2013;22:823–38. doi: 10.1007/s11248-012-9685-0. [DOI] [PubMed] [Google Scholar]
- Palazuelos J, Aguado T, Egia A, Mechoulam R, Guzman M, Galve-Roperh I. Nonpsychoactive CB2 cannabinoid agonists stimulate neural progenitor proliferation. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2006;20:2405–7. doi: 10.1096/fj.06-6164fje. [DOI] [PubMed] [Google Scholar]
- Pang T, Atefy R, Sheen V. Malformations of cortical development. Neurologist. 2008;14:181–91. doi: 10.1097/NRL.0b013e31816606b9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paria BC, Song H, Wang X, Schmid PC, Krebsbach RJ, et al. Dysregulated cannabinoid signaling disrupts uterine receptivity for embryo implantation. J Biol Chem. 2001;276:20523–8. doi: 10.1074/jbc.M100679200. [DOI] [PubMed] [Google Scholar]
- Piccinetti CC, Migliarini B, Olivotto I, Coletti G, Amici A, Carnevali O. Appetite regulation: the central role of melatonin in Danio rerio. Horm Behav. 2010;58:780–5. doi: 10.1016/j.yhbeh.2010.07.013. [DOI] [PubMed] [Google Scholar]
- Romero J, Garcia-Palomero E, Berrendero F, Garcia-Gil L, Hernandez ML, et al. Atypical location of cannabinoid receptors in white matter areas during rat brain development. Synapse. 1997;26:317–23. doi: 10.1002/(SICI)1098-2396(199707)26:3<317::AID-SYN12>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- Ruhl T, Prinz N, Oellers N, Seidel NI, Jonas A, et al. Acute administration of THC impairs spatial but not associative memory function in zebrafish. Psychopharmacology (Berl) 2014;231:3829–42. doi: 10.1007/s00213-014-3522-5. [DOI] [PubMed] [Google Scholar]
- Ruhl T, Zeymer M, von der Emde G. Cannabinoid modulation of zebrafish fear learning and its functional analysis investigated by c-Fos expression. Pharmacol Biochem Behav. 2016;153:18–31. doi: 10.1016/j.pbb.2016.12.005. [DOI] [PubMed] [Google Scholar]
- Salio C, Cottone E, Conrath M, Franzoni MF. CB1 cannabinoid receptors in amphibian spinal cord: relationships with some nociception markers. J Chem Neuroanat. 2002;24:153–62. doi: 10.1016/s0891-0618(02)00040-6. [DOI] [PubMed] [Google Scholar]
- Santoriello C, Zon LI. Hooked! Modeling human disease in zebrafish. J Clin Invest. 2012;122:2337–43. doi: 10.1172/JCI60434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savinainen JR, Saario SM, Laitinen JT. The serine hydrolases MAGL, ABHD6 and ABHD12 as guardians of 2-arachidonoylglycerol signalling through cannabinoid receptors. Acta Physiol (Oxf) 2012;204:267–76. doi: 10.1111/j.1748-1716.2011.02280.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serra EL, Medalha CC, Mattioli R. Natural preference of zebrafish (Danio rerio) for a dark environment. Braz J Med Biol Res. 1999;32:1551–3. doi: 10.1590/s0100-879x1999001200016. [DOI] [PubMed] [Google Scholar]
- Shimada Y, Hirano M, Nishimura Y, Tanaka T. A high-throughput fluorescence-based assay system for appetite-regulating gene and drug screening. PLoS One. 2012;7:e52549. doi: 10.1371/journal.pone.0052549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silvestri C, Paris D, Martella A, Melck D, Guadagnino I, et al. Two nonpsychoactive cannabinoids reduce intracellular lipid levels and inhibit hepatosteatosis. J Hepatol. 2015;62:1382–90. doi: 10.1016/j.jhep.2015.01.001. [DOI] [PubMed] [Google Scholar]
- Sipe JC, Chiang K, Gerber AL, Beutler E, Cravatt BF. A missense mutation in human fatty acid amide hydrolase associated with problem drug use. Proc Natl Acad Sci U S A. 2002;99:8394–9. doi: 10.1073/pnas.082235799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soderstrom K, Leid M, Moore FL, Murray TF. Behaviroal, pharmacological, and molecular characterization of an amphibian cannabinoid receptor. J Neurochem. 2000;75:413–23. doi: 10.1046/j.1471-4159.2000.0750413.x. [DOI] [PubMed] [Google Scholar]
- Stern HM, Zon LI. Cancer genetics and drug discovery in the zebrafish. Nature reviews. Cancer. 2003;3:533–9. doi: 10.1038/nrc1126. [DOI] [PubMed] [Google Scholar]
- Stewart AM, Kalueff AV. The behavioral effects of acute Delta(9)-tetrahydrocannabinol and heroin (diacetylmorphine) exposure in adult zebrafish. Brain Res. 2014;1543:109–19. doi: 10.1016/j.brainres.2013.11.002. [DOI] [PubMed] [Google Scholar]
- Trazzi S, Steger M, Mitrugno VM, Bartesaghi R, Ciani E. CB1 cannabinoid receptors increase neuronal precursor proliferation through AKT/glycogen synthase kinase-3beta/beta-catenin signaling. J Biol Chem. 2010;285:10098–109. doi: 10.1074/jbc.M109.043711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viveros MP, Marco EM, File SE. Endocannabinoid system and stress and anxiety responses. Pharmacol Biochem Behav. 2005;81:331–42. doi: 10.1016/j.pbb.2005.01.029. [DOI] [PubMed] [Google Scholar]
- Wang X, Dow-Edwards D, Keller E, Hurd YL. Preferential limbic expression of the cannabinoid receptor mRNA in the human fetal brain. Neuroscience. 2003;118:681–94. doi: 10.1016/s0306-4522(03)00020-4. [DOI] [PubMed] [Google Scholar]
- Watson S, Chambers D, Hobbs C, Doherty P, Graham A. The endocannabinoid receptor, CB1, is required for normal axonal growth and fasciculation. Mol Cell Neurosci. 2008;38:89–97. doi: 10.1016/j.mcn.2008.02.001. [DOI] [PubMed] [Google Scholar]
- Wiley JL, Burston JJ, Leggett DC, Alekseeva OO, Razdan RK, et al. CB1 cannabinoid receptor-mediated modulation of food intake in mice. Br J Pharmacol. 2005;145:293–300. doi: 10.1038/sj.bjp.0706157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan D, Wu Z, Wang Y. Evolution of the diacylglycerol lipases. Prog Lipid Res. 2016;64:85–97. doi: 10.1016/j.plipres.2016.08.004. [DOI] [PubMed] [Google Scholar]
- Zlebnik NE, Cheer JF. Drug-Induced Alterations of Endocannabinoid-Mediated Plasticity in Brain Reward Regions. J Neurosci. 2016;36:10230–38. doi: 10.1523/JNEUROSCI.1712-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurolo E, Iyer AM, Spliet WG, Van Rijen PC, Troost D, et al. CB1 and CB2 cannabinoid receptor expression during development and in epileptogenic developmental pathologies. Neuroscience. 2010;170:28–41. doi: 10.1016/j.neuroscience.2010.07.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


