Abstract
Parkinson’s disease (PD) is the second most common age-related neurodegenerative disorder after Alzheimer’s disease. To date, the clinical diagnosis of PD is primarily based on the late onset of motor impairments. Unfortunately, at this stage, most of the dopaminergic neurons may have already been lost, leading to the limited clinical benefits of current therapeutics. Therefore, early identification of PD, especially at the prodromal stage, is still a main challenge in the diagnosis and management of this disease. Recently, microRNAs (miRNAs) in cerebrospinal fluid or peripheral blood have been proposed as putative biomarkers to assist in PD diagnosis and therapy. In this review, we systematically summarize the changes of miRNA expression profiles in PD patients, and highlight their putative roles in the diagnosis and treatment of this devastating disease.
Keywords: Parkinson’s disease, MicroRNA, Biomarker, Diagnosis
Introduction
Parkinson’s disease (PD) is the second most prevalent neurodegenerative disorder affecting ~1% of the population >55 years old [1]. Pathologically, PD is characterized by the progressive deterioration of dopamine (DA) neurons and the presence of Lewy bodies composed of α-synuclein in the substantia nigra pars compacta (SNc), as well as the loss of DA signaling in the striatum. Numerous genes have been identified as contributors to the etiology of PD, including two dominantly-inherited genes [α-synuclein or SNCA and leucine-rich repeat kinase 2 (LRRK2)], three recessive genes (Parkin, DJ-1, and PINK1) [2], and other PD-associated genes (UCHL1, ATP13A2, GIGYF2, HTRA2, PLA2G6, FBXO7, VPS35, EIF4G1, DNAJC16, and GBA) [3, 4]. These genes can interact with environmental risk factors to initiate the main pathogenic mechanisms involved in PD, including apoptosis, autophagy, inflammation, mitochondrial dysfunction, and oxidative stress [5], and finally generate PD pathologies. Clinically, PD is primarily diagnosed by the late onset of motor disorders including resting tremor, muscle rigidity, stiffness of the limbs, bradykinesia, and postural instability [6]. However, when these manifestations are observed, a significant loss (60%–70%) of DA neurons has already occurred, leading to limited clinical benefits of current therapeutics [7, 8]. Therefore, the early identification of PD, especially at the prodromal stage, is a main challenge for early diagnosis and therapy. Non-motor symptoms, such as sleep disturbances, sensory abnormalities, autonomic dysfunction, cognitive deficits, and mood disorders, are sometimes present before motor symptoms and have been recently reported to be beneficial for the early diagnosis of PD [9, 10]. However, non-motor symptoms also occur frequently in individuals without PD as a part of normal aging. Previous studies have suggested that 68%–88% of normal, comparably aged individuals experience at least one non-motor symptom. Thus, developing reliable clinical diagnostic methods that are feasible, objective, measurable, specific, and sensitive for PD is essential to improve the diagnosis and management of the disease.
MicroRNAs (miRNAs) are small non-coding RNA molecules 18–25 nucleotides long [11]. Since miRNAs can be released into peripheral fluids from both the central nervous system (CNS) and peripheral organs in a cell-free manner, they can bind with protein components and be encapsulated by microvesicles (for example, exosomes) to cross the blood-brain barrier [12–15]. Therefore, miRNAs can be detected in fluids such as plasma, serum, tears, saliva, breast milk, urine, and cerebrospinal fluid (CSF) [16]. Intracellular miRNAs can interact with the RNA-induced silencing complex (RISC) and target the 3’-untranslated region (UTR) of mRNAs, causing subsequent degradation or translational modulation of target genes [17, 18]. miRNAs are believed to regulate the expression of ~30% of human genes, and each miRNA is estimated to regulate ~200 target genes [19]. Furthermore, miRNA-gene-related regulation may change homeostasis and molecular functions in cells, which may cause the abnormal expression of miRNAs in the CNS [12]. More importantly, the expression profiles of brain miRNAs change during the course of brain development, thus these tiny molecules may act as mighty regulators to restore imbalanced or dysregulated pathophysiological pathways at the onset and very early stages of PD. Moreover, with high stability and quantifiability, lower cost, and shorter time of assay development, as well as their diversity and potential to target multiple downstream genes, miRNAs may have putative roles in the diagnosis and treatment of PD [20].
miRNAs and PD
miRNAs play critical roles in the biology of DA neurons, the predominant cell type affected by neurodegeneration in PD [1]. miRNAs have recently been found to regulate gene expression in the cortex of patients with PD, and the cortical expression patterns of miRNAs accurately distinguish PD brains from non-diseased brains [21, 22]. Previous studies have further addressed the roles of miRNAs in PD pathogenesis by focusing on the PD-related genes, such as α-synuclein or LRRK2, and have identified miRNAs that specifically regulate their expressions [23, 24]. More recently, miRNAs have been easily quantified in various body fluids as cell-free or exosome-derived molecules, suggesting that miRNAs might be used as specific and objective biomarkers for PD diagnosis at premotor or prodromal phases. Several reports have shown that altered miRNAs might be associated with the premotor stage. For instance, down-regulated expression of miR-34b and c have been found in the amygdala and frontal cortex of PD patients at premotor stages, and not related to drug treatment [25]. Decreased expression of miR-19b has been found in patients with idiopathic rapid eye movement sleep behavior disorder 4.67 ± 2.61 years before a diagnosis of PD or dementia with Lewy bodies [26]. It is conceivable that the altered expression patterns of miRNAs in the brain, CSF, and peripheral blood of PD patients may provide valuable information to help the early diagnosis and management of PD. This will require a large number of samples at different stages of PD and carefully matched healthy and disease controls, for confirmation and eventual use in clinical practice.
Expression Patterns of miRNAs in Body Fluids of PD Patients
Differential Expression of miRNAs in PD Patients and Controls
In recent years, the altered expression of miRNAs in various body-fluid samples from PD patients has been reported. For instance, the expression levels of miR-335, miR-374a/b, miR-199, miR-126, miR-151-5p, miR-29b/c, miR-147, miR-28-5p, miR-30b/c, miR-301a, and miR-26a in peripheral blood mononuclear cells (PBMCs) from PD patients are lower than those from healthy controls [27]. Plasma-based circulating miRNAs including miR-1826, miR450b-3p, miR-626 and miR-505 are differentially expressed in PD patients and controls, and may serve as potential biomarkers for PD diagnosis [7]. Since CSF is directly associated with the CNS, it has been considered as a reliable source for investigating biomarkers of PD. Previous studies have demonstrated that let-7-3p, miR-128, miR-433, miR-485-5p, miR-132-5p, and miR-212-3p from CSF are differentially expressed in PD patients and controls [28]. Moreover, miR-1 and miR-19b-3p in CSF exosomes are significantly reduced in PD patients, while miR-153, miR-409-3p, miR-10a-5p, and let-7g-3p are significantly up-regulated, demonstrating that CSF exosomal RNA molecules might be reliable biomarkers with fair robustness in regard to specificity and sensitivity in differentiating PD from healthy controls [29]. Previous studies have also demonstrated changes of miRNAs expression in serum from patients with PD. For example, miR-338-3p, miR-30e-3p, and miR-30a-3p are up-regulated, whereas miR-16-2-3p and miR-1294 are down-regulated in the serum of PD patients [30]. miR-29c, miR-146a, miR-214, and miR-221 in PD patients are also significantly lower than in healthy controls. In addition, positive correlations between miR-221 and UPDRS-III, as well as between miR-221 and UPDRS-V in PD patients have been demonstrated, suggesting that miRNAs can be used as biomarkers for detecting the stages of PD [31]. Table 1 summarizes the alterations of miRNAs in various body-fluids from PD patients assessed using different methods. Generally, these studies used small samples and simple technologies. Future studies with larger sample size and samples from multiple clinical sites should be encouraged.
Table 1.
miRNAs in PD patients and controls.
| Technology | Source | Numbers | Altered miRNAs | Changes | Reference | |
|---|---|---|---|---|---|---|
| PD | Controls | |||||
| Microarrays and RT-PCR | PMBCs | 19 | 13 | miR-335, miR-374a/b, miR-199, miR-126, miR-151-5p, miR-29b/c, miR-147, miR-28-5p, miR-30b/c, miR-301a, miR-26a | Decreased | Martins et al. [27] |
| Microarrays | Plasma | 32 | 32 | miR-1826, miR450b-3p, miR-626, miR-505 | Khoo et al. [7] | |
| Taqman miRNA arrays | Plasma | 31 | 25 | miR-181c, miR-331-5p, miR-193a-3p, miR-196b, miR-454, miR-125a-3p, miR-137 | Increased | Cardo et al. [66] |
| RT-PCR | Serum | 138 | 112 | miR-29c, miR-146a, miR-214, and miR-221 | Decreased | Ma et al. [31] |
| Solexa sequencing and RT-PCR | Serum | 169 | 180 | miR-141, miR-214, miR-146b-5p, miR-193a-3p | Decreased | Dong et al. [67] |
| Solexa sequencing and RT-PCR | Serum | 106 | 91 | miR-185, miR-15b, miR-221 and miR-181a | Decreased | Ding et al. [68] |
| miR-195 | Increased | |||||
| NGS RNA sequencing | Serum | 50 | 62 | miR-16-2-3p, miR-1294 | Decreased | Burgos et al. [28] |
| miR-338-3p, miR-30e-3p, and miR-30a-3p | Increased | |||||
| CSF | 67 | 78 | let-7, miR-128, miR-433, miR-485-5p, miR-132, miR-212 | Different expression | ||
| Taqman miRNA arrays | CSF | 47 | 27 | miR-1, miR-19b-3p | Decreased | Gui et al. [29] |
| miR-153, miR-409-3p, miR-10a-5p, let-7g-3p | Increased | |||||
| RT-PCR | CSF | 44 | 42 | miR-144-5p, miR-200a-3p and miR-542-3p | Increased | Mo et al. [38] |
PBMCs peripheral blood mononuclear cells, CSF cerebrospinal fluid, NGS next generation small RNA sequencing.
The actions of these differentially-expressed miRNAs on PD-related genes in the CNS have been gradually revealed. For instance, pathogenic LRRK2 antagonizes the repression of E2F1/DP expression by let-7 or miR-184*; conversely, an increased level of let-7 or miR-184* attenuates the pathogenic effects of LRRK2 [32]. Moreover, miR-433 binds to the promoter region of fibroblast growth factor 20, indirectly causing the overexpression of α-synuclein [33], a characteristic pathological marker of PD. miR-128 targeted to transcription factor EB acts indirectly against the accumulation of α-synuclein; and miR-7 and miR-153 can bind to the 3’-UTR of SNCA and thus directly inhibit the expression of α-synuclein [24, 34]. Moreover, our previous study demonstrated that miR-132 represses the differentiation of DA neurons and that increased miR-132/miR-212 expression down-regulates the Nurr1 level [35]. These findings suggest that different miRNAs play either protective or harmful roles, keeping the balance between PD-risk/causal factors and protective factors. Further studies are needed to uncover the detailed networks underlying the complicated interactions between miRNAs and PD, and thus provide experimental evidence to support the beneficial roles of miRNAs in early diagnosis and management.
miRNA Expression in PD Patients Carrying Different Gene Mutations
The altered expression profiles of miRNAs in the body-fluids of PD patients may predict the impact of these miRNAs on the expression of PD-related genes and subsequently the functions of these genes and their downstream targets. Moreover, interactions between miRNAs and PD-related genes have also been identified. Apart from the antagonistic interactions between LRRK2 and let-7 or miR-184* [32], the expression of miR-29a, miR-29c, and miR-19b in serum from PD patients carrying LRRK2 mutations is also significantly reduced [36]. Our recent study indicated that the miR-29 family (miR-29a, b, and c) directly repress the expression of Tet1 [37], suggesting that this family may provide potential biomarkers for Tet1-associated PD. Moreover, dramatic up-regulation of miR-144-5p, miR-200a-3p, and miR-542-3p has been detected in both α-synuclein A53T mutant mice and the CSF of PD patients [38]. Interestingly, the changes of miRNA expression have strong correlations with the severity of PD, suggesting that the differentially-expressed miRNAs may be useful biomarkers to distinguish different stages of PD [38].
The detection of miRNAs in body-fluids from PD patients carrying PD-related gene mutations is not widely used because of the limited number of samples from PD patients with gene mutations. Several studies have reported changes in miRNA profiles in animal models or cell lines transfected with PD-related genes [39, 40]. For example, deficiency of PINK1 might decrease the expression of mir-326, mir-330, and mir-3099, affecting the development of mouse brain and the differentiation of neural stem cells [39]. Moreover, since the alterations of miRNA expression in PD patients carrying different gene mutations are not the same, miRNAs may also serve as biomarkers for the differential diagnosis of PD patients carrying different genetic variants.
Differential Expression of miRNAs in Treated and Untreated PD Patients
Previous studies have indicated altered miRNA expression profiles in PD patients with pharmacotherapy or deep brain stimulation (DBS) compared with untreated controls. Margis et al. [41] analyzed the expression patterns of miRNAs in peripheral blood from untreated PD patients, levodopa/carbidopa-treated PD patients, early-onset PD patients, and healthy controls. They found that miR-1, miR-22*, and miR-29 in total peripheral blood are able to distinguish untreated PD from healthy individuals, while miR-16-2*, miR-26a-2*, and miR-30a can differentiate treated from untreated patients [41]. Alieva et al. found that the levels of miR-7, miR-9-3p, miR-9-5p, miR-129, and miR-132 in peripheral blood lymphocytes are higher in treated than in untreated PD patients [42]. Moreover, Serafin et al. demonstrated that miR-30b-5p, miR-103a-3p, and miR-29a-3p in PBMCs are significantly higher in levodopa-treated PD patients than in controls, while there is no statistically significant difference between drug-naïve patients and controls [43].
Soreq et al. analyzed the miRNA expression in blood leukocytes from DBS-treated PD patients and healthy controls [44]. All patients were given bilateral subthalamic nucleus electrode implantation, and the blood samples were collected at one day pre-DBS and post-DBS (6–18 weeks), ON stimulus and OFF stimulus. They found that DBS induced a stimulus-dependent reversal of miRNAs in the leukocytes from PD patients, as evidenced by the increased expression of hsa-miR-20a and decreased hsa-miR-378 after DBS treatment. Moreover, they also found decreased expressions of miR-652, miR-15a*, miR-29c, miR-29a, miR-376c, and miR-143, together with increased expressions of miR-423, miR-365, miR-486, miR-1260, and miR-218, after cessation of electrical stimulation [44]. The different expression profiles of miRNAs between anti-PD drug/DBS-treated and untreated individuals as well as different stages of DBS-treated PD suggest that miRNAs may be useful for predicting the therapeutic outcome of PD [44]. Furthermore, as the miRNAs differentially expressed between treated and untreated PD patients have been identified, further analysis of the target genes of these miRNAs may be useful for a better understanding of the mechanisms underlying the efficacy of PD therapy.
miRNA Detection in Brain Samples from PD Patients
Although the measurement of peripheral miRNAs provides a convenient and feasible method for the diagnosis and management of PD, the detection of miRNAs in brain tissues is still of most importance for advancing our understanding of the on-going biology of the disease. One recent analysis of postmortem cerebral cortex, midbrain, and cerebellum from PD patients and unaffected controls found that miR-133b, which regulates the maturation and function of midbrain DA neurons, is deficient in the midbrain of patients with PD [45]. Furthermore, a negative feedback loop between miR-133b and paired-like homeodomain transcription factor Pitx3 is also revealed [45]. Another study analyzed the miRNA expression in differently-affected brain regions from idiopathic PD patients at either premotor stages (Braak stages 1 to 3) or motor stages (Braak stages 4 and 5) [25]. They found that the expressions of miR-34b and miR-34c are decreased at the motor stages in several affected brain regions of PD patients, including the amygdala, frontal cortex, substantia nigra, and cerebellum. More interestingly, a significant reduction in the expression of both miR-34b and miR-34c has been reported in the amygdala of patients at pre-motor stages who received no treatment related to PD [25]. Moreover, the downregulation of miR-34b and c is coupled with reduced expression of the important PD genes DJ-1 and Parkin [25, 46, 47].
Other studies have also demonstrated the significant up-regulation of miR-21, miR-224, miR-373*, miR-26b, miR-106a*, and miR-301b, which target the chaperone-mediated autophagy pathway in the SNc and amygdala of PD patients [48]. Moreover, a significant differential expression of miR-135b in the SNc between PD patients and healthy controls has been reported [49]. In addition, the expression level of miR-205, a suppressor of LRRK2 expression, is significantly lower in the frontal cortex and striatum of PD patients [23].
Different Expression of miRNAs in Patients with PD and Other Neurological Diseases
miRNAs may be valuable biomarkers for distinguishing PD from other neurological diseases. For instance, the expression of miR-10b-5p is significantly decreased in PD patients, especially in the later onset stage, whereas this miRNA is increased in the brains of patients with Huntington’s disease, especially in the early stage [22]. miRNAs may also be helpful in the differential diagnosis of PD from the C subtype of multiple system atrophy (MSA). While miR-223*, miR-324-3p, and miR-24 are up-regulated and miR-339-5p is down-regulated in sera from both PD and MSA patients, the levels of miR-24, miR-34b, and miR-148b are significantly higher in MSA than in PD patients [50]. Moreover, the down-regulation of miR-19b in patients with idiopathic rapid eye movement sleep behavior disorder facilitates the diagnosis of PD or dementia with Lewy bodies [26].
In summary, alterations of miRNAs in different stages of PD have been reported. It may be useful to find the changes in the early stage of disease, as this may help early diagnosis and predict the disease progression. Furthermore, following the changes of miRNAs with treatment may permit monitoring the progress of the disease and uncover the therapeutic mechanisms and pathways. However, these studies are limited by the small number of samples and the lack of verification and pathway identification.
So far, only a handful miRNAs have shown consistent results in different studies on body-fluids or brain tissues from PD patients. For instance, decreased expression of miR-29c has been reported in CSF, serum, and PBMCs in several studies comparing late-stage PD with controls [26, 27, 29, 36]. miR-29a and c in serum from PD patients carrying LRRK2 mutations have also been reported to be significantly reduced [36]. Blood miR-29a is higher in L-dopa-treated patients with PD versus controls. Besides, miR-29b in PBMCs from PD patients is lower than in healthy controls [27]. The miR-29 family (miR-29a, b, and c) in peripheral blood can be used to distinguish untreated PD from healthy individuals [41]. We found a regulatory interaction between the miR-29 family and the DNA demethylase Tet1 [37], implying its potential application for Tet-related human diseases. Therefore, we consider the miR-29 family to be the most promising candidates for biomarkers of PD.
Moreover, the miRNAs that act on PD-related genes may have more potential value for further network studies. For example, miR-205, let-7, and miR-184* are associated with LRRK2 [23, 32]; miR-433, miR-7, and miR-153 with α-synuclein [24, 33, 34]; miR-132 with Nurr1 [35]; miR-34b and c with DJ-1 and Parkin [25, 46, 47]; and miR-133b with Pitx3 [45]. So, these miRNAs are worth further investigation to assess their value as biomarkers of PD.
miRNA-Based Therapy for PD
To date, the clinical diagnosis of PD is primarily based on the late onset of motor impairments. Unfortunately, at this stage, the majority of DA neurons may have already been lost, leading to limited clinical benefits of current therapeutics. Therefore, the symptomatic therapy currently used for PD lacks long-lasting efficacy and fails to reverse the pathological progression of the disease. Moreover, other therapeutic strategies, including DBS [51], cell transplantation [52], and gene therapy [53], can improve the motor symptoms, but they are expensive and their applications are still far from generalized. Considering the important roles of miRNAs in PD pathogenesis and progression, these small RNA molecules appear to be promising tools in PD therapy, via predicting more potential molecular targets and providing biomarkers for better or more individualized therapeutic strategies.
miRNA Mimic Application
In 2007, small double-stranded RNA molecules that mimic miRNAs and can serve as gain-of-function tools for specific miRNAs were developed [54]. miRNA mimics downregulate certain target proteins by binding to the 3’-UTR of the mRNA of a specific target gene. miRNA mimics have been reported to regulate PD-related risk genes and unravel proteins associated with PD. For instance, miR-205 targets LRRK2 in the brains of sporadic PD patients, and miR-153 targets α-synuclein in rodents’ primary neuron culture [23, 24]. However, this strategy also carries the potential risk of off-target effects and the probability of unwanted interference with other genes [34, 55].
Anti-miRNA Oligonucleotides and Antagomir
Anti-miRNA oligonucleotides (AMOs) and antagomir can downregulate miRNA through binding to the mature guide strand and induce degradation or stoichiometric duplex formation [56]. Compared with conventional inhibitors, antagomir is more stable and has a stronger inhibitory effect in vivo and in vitro. Antagomir does not need a transfection reagent in cell experiments, avoiding the complex packaging process. In animal experiments, the systemic or local injection, inhalation or oral administration of these therapeutics has been developed to prolong the therapeutic effects for several weeks.
AMOs enable the identification of miRNAs involved in the pathophysiological process of PD and interfere with molecular pathways, mainly those involved in apoptosis, autophagy, inflammation, mitochondrial dysfunction and oxidative stress, neurite outgrowth, dendritic spine morphology, neuronal differentiation, and synaptic plasticity [5, 34, 54].
Other therapeutic intervention strategies have also been established. miRNA sponges are able to target an entire miRNA family sharing the same seed sequence [55]; and miRNA-mask, a single stranded 2’-O-methyl-modified antisense oligonucleotide, binds to the 3’-UTR of the target mRNA of an miRNA, thereby masking the miRNA binding site and then de-repressing the target genes [56]. However, these two approaches, to date, have not been applied to PD.
Fortunately, following the rapid research progress in nanotechnology and the development of novel and efficient nanovehicles for siRNA and miRNA delivery, the miRNA delivery challenges in vivo seem to have been overcome. For instance, various techniques, such as the covalent coupling of RNAs to cholesterol, lipids, peptides, and antibodies, as well as chemical nucleotide modifications in the RNA structure, have been established in order to increase the RNA protection and response sensitivity [57, 58]. Moreover, the nanoparticulate polyethylenimines seem to be a promising tool for the delivery of small RNA molecules [59].
Strategy for miRNA Target Gene Prediction
miRNAs have been demonstrated to play key roles in many biological processes, including development, cell differentiation, proliferation, apoptosis, and tumor metastasis, but the specific function of each miRNA as well as its target genes is not yet clearly understood. Accordingly, it is important to predict miRNA target genes in order to analyze and reveal the functions of miRNAs that are involved in biological processes. Bioinformatics and biological tools are the main methods that are useful in current studies on miRNA target genes.
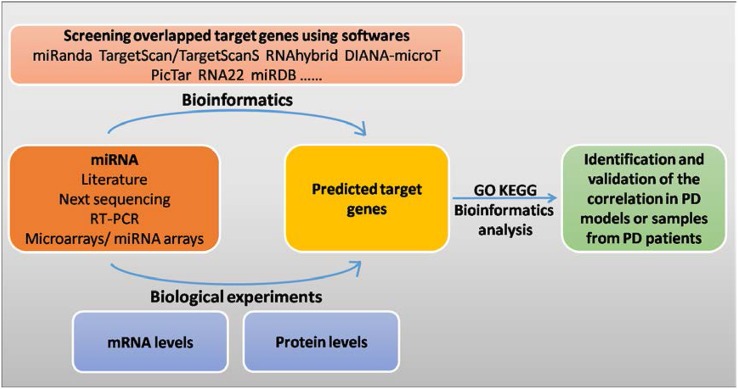
Bioinformatics tools are based on algorithms to grade and screen target genes from samples, and provide reference information to researchers, so the results can be validated by experiments. Moreover, with the increasing number of miRNAs in miRBase, identifying and keeping track of links between miRNAs and their targets are becoming increasingly complex. However, researchers can use miRNA target-prediction tools such as MiRanda, TargetScan, RNAhybrid, DIANA-microT, and RNA22, which have been designed with different algorithms to explore interesting miRNAs obtained from the literature, experiments, or databases [60]. Considering certain limitations of computational approaches in predicting miRNA target genes, many investigators have conducted biological experiments to directly find the miRNA target genes from the levels of mRNAs and their related proteins. For instance, gene chip analysis has been used for assessing the changes of mRNAs after modifying the expression levels of miRNAs in cells [61], while RISC immunopurification [62] has been used to isolate miRNA target genes. Considering that the actions of miRNAs are mediated through post-transcriptional inhibition and cause changes in the levels of proteins, several research groups have used proteomics to identify miRNA target genes [63, 64].
The reported research processes for finding miRNA target genes are illustrated in Fig. 1. While the main line of study is clear, more newly-developed technologies and analytical methods will further help to provide a better understanding of the network of interactions between miRNAs and their target genes.
Fig. 1.
Bioinformatics and experiments are used to screen for miRNA target genes. GO gene ontology, KEGG kyoto encyclopedia of genes and genomes.
Conclusions
Among many methods available to detect miRNAs, two of the most frequently used are reverse transcription real-time quantitative PCR (RT-qPCR) and next-generation small RNA sequencing [65]. By using these traditional methods, various miRNA expression profiles have been generated, and comparative analysis has been performed to detect the potential benefits of miRNAs for the early diagnosis and treatment of PD. Since the miRNAs differentially expressed between PD patients and healthy individuals and between treated and untreated PD patients have been reported, we conclude that measurement of certain miRNAs in body-fluids may aid the diagnosis of PD. Furthermore, application of miRNA mimics to interfere with the target genes may have the potential to develop new drugs for the treatment of PD. However, the specificity of miRNAs and the reproducibility of their effects are still concerned due to small sample sizes and the heterogeneity (age, sex, and severity of disease) of recruited patients. To acquire consistent and reliable miRNA biomarkers, the following points should be considered: (1) establishing standard operating protocols for sample collection, preservation, and detection from different labs and research centers; (2) obtaining comprehensive information on enrolled PD patients, especially the severity and duration of disease; (3) re-validating potential biomarkers found using different screening approaches by RT-qPCR assays, and the process should be double-blind in different and larger cohorts with larger sample size. Besides, multi-unit cooperation to collect human samples for miRNA research and new trace-detecting technologies that can measure subtle changes in miRNAs in body-fluids and brain tissues are urgently needed.
Acknowledgements
This review was supported by the National Natural Science Foundation of China (81430021 and 81370470).
Footnotes
Ying Wang and Zhaofei Yang have contributed equally.
References
- 1.Grasso M, Piscopo P, Crestini A, Confaloni A, Denti MA. Circulating microRNAs in neurodegenerative diseases. EXS. 2015;106:151–169. doi: 10.1007/978-3-0348-0955-9_7. [DOI] [PubMed] [Google Scholar]
- 2.Hamza TH, Zabetian CP, Tenesa A, Laederach A, Montimurro J, Yearout D, et al. Common genetic variation in the HLA region is associated with late-onset sporadic Parkinson’s disease. Nat Genet. 2010;42:781–785. doi: 10.1038/ng.642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.De Rosa P, Marini ES, Gelmetti V, Valente EM. Candidate genes for Parkinson disease: Lessons from pathogenesis. Clin Chim Acta. 2015;449:68–76. doi: 10.1016/j.cca.2015.04.042. [DOI] [PubMed] [Google Scholar]
- 4.Klein C, Westenberger A. Genetics of Parkinson’s disease. Cold Spring Harb Perspect Med. 2012;2:a008888. doi: 10.1101/cshperspect.a008888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Saghazadeh A, Rezaei N. MicroRNA machinery in Parkinson’s disease: a platform for neurodegenerative diseases. Expert Rev Neurother. 2015;17:1–27. doi: 10.1586/14737175.2015.1114886. [DOI] [PubMed] [Google Scholar]
- 6.Moustafa AA, Chakravarthy S, Phillips JR, Gupta A, Keri S, Polner B, et al. Motor symptoms in Parkinson’s disease: A unified framework. Neurosci Biobehav Rev. 2016;68:727–740. doi: 10.1016/j.neubiorev.2016.07.010. [DOI] [PubMed] [Google Scholar]
- 7.Khoo SK, Petillo D, Kang UJ, Resau JH, Berryhill B, Linder J, et al. Plasma-based circulating MicroRNA biomarkers for Parkinson’s disease. J Parkinsons Dis. 2012;2:321–331. doi: 10.3233/JPD-012144. [DOI] [PubMed] [Google Scholar]
- 8.Hughes AJ, Daniel SE, Ben-Shlomo Y, Lees AJ. The accuracy of diagnosis of parkinsonian syndromes in a specialist movement disorder service. Brain. 2002;125:861–870. doi: 10.1093/brain/awf080. [DOI] [PubMed] [Google Scholar]
- 9.Chaudhuri KR, Healy DG, Schapira AH. Non-motor symptoms of Parkinson’s disease: diagnosis and management. Lancet Neurol. 2006;5:235–245. doi: 10.1016/S1474-4422(06)70373-8. [DOI] [PubMed] [Google Scholar]
- 10.Copped F. Genetics and epigenetics of Parkinson’s disease. Sci World J. 2012;2012:489830. doi: 10.1100/2012/489830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garcia-Gimenez JL, Sanchis-Gomar F, Lippi G, Mena S, Ivars D, Gomez-Cabrera MC, et al. Epigenetic biomarkers: A new perspective in laboratory diagnostics. Clin Chim Acta. 2012;413:1576–1582. doi: 10.1016/j.cca.2012.05.021. [DOI] [PubMed] [Google Scholar]
- 12.Kumar S, Vijayan M, Bhatti JS, Reddy PH. MicroRNAs as peripheral biomarkers in aging and age-related diseases. Prog Mol Biol Transl Sci. 2017;146:47–94. doi: 10.1016/bs.pmbts.2016.12.013. [DOI] [PubMed] [Google Scholar]
- 13.Cao X, Yeo G, Muotri AR, Kuwabara T, Gage FH. Noncoding RNAs in the mammalian central nervous system. Annu Rev Neurosci. 2006;29:77–103. doi: 10.1146/annurev.neuro.29.051605.112839. [DOI] [PubMed] [Google Scholar]
- 14.Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 15.Magdalinou N, Lees AJ, Zetterberg H. Cerebrospinal fluid biomarkers in parkinsonian conditions: an update and future directions. J Neurol Neurosurg Psychiatry. 2014;85:1065–1075. doi: 10.1136/jnnp-2013-307539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Etheridge A, Gomes CP, Pereira RW, Galas D, Wang K. The complexity, function and applications of RNA in circulation. Front Genet. 2013;4:115. doi: 10.3389/fgene.2013.00115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 18.Qiu L, Tan EK, Zeng L. microRNAs and neurodegenerative diseases. Adv Exp Med Biol. 2015;888:85–105. doi: 10.1007/978-3-319-22671-2_6. [DOI] [PubMed] [Google Scholar]
- 19.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 20.Goodall EF, Heath PR, Bandmann O, Kirby J, Shaw PJ. Neuronal dark matter: the emerging role of microRNAs in neurodegeneration. Front Cell Neurosci. 2013;7:178. doi: 10.3389/fncel.2013.00178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mouradian MM. MicroRNAs in Parkinson’s disease. Neurobiol Dis. 2012;46:279–284. doi: 10.1016/j.nbd.2011.12.046. [DOI] [PubMed] [Google Scholar]
- 22.Hoss AG, Labadorf A, Beach TG, Latourelle JC, Myers RH. microRNA Profiles in Parkinson’s Disease Prefrontal Cortex. Front Aging Neurosci. 2016;8:36. doi: 10.3389/fnagi.2016.00036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cho HJ, Liu G, Jin SM, Parisiadou L, Xie C, Yu J, et al. MicroRNA-205 regulates the expression of Parkinson’s disease-related leucine-rich repeat kinase 2 protein. Hum Mol Genet. 2013;22:608–620. doi: 10.1093/hmg/dds470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Doxakis E. Post-transcriptional regulation of alpha-synuclein expression by mir-7 and mir-153. J Biol Chem. 2010;285:12726–12734. doi: 10.1074/jbc.M109.086827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Minones-Moyano E, Porta S, Escaramis G, Rabionet R, Iraola S, Kagerbauer B, et al. MicroRNA profiling of Parkinson’s disease brains identifies early downregulation of miR-34b/c which modulate mitochondrial function. Hum Mol Genet. 2011;20:3067–3078. doi: 10.1093/hmg/ddr210. [DOI] [PubMed] [Google Scholar]
- 26.Fernandez-Santiago R, Iranzo A, Gaig C, Serradell M, Fernández M, Tolosa E, et al. MicroRNA association with synucleinopathy conversion in rapid eye movement behavior disorder. Ann Neurol. 2015;77:895–901. doi: 10.1002/ana.24384. [DOI] [PubMed] [Google Scholar]
- 27.Martins M, Rosa A, Guedes LC, Fonseca BV, Gotovac K, Violante S, et al. Convergence of miRNA expression profiling, alpha-synuclein interacton and GWAS in Parkinson’s disease. PLoS One. 2011;6:25443. doi: 10.1371/journal.pone.0025443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Burgos K, Malenica I, Metpally R, Courtright A, Rakela B, Beach T, et al. Profiles of extracellular miRNA in cerebrospinal fluid and serum from patients with Alzheimer’s and Parkinson’s diseases correlate with disease status and features of pathology. PLoS One. 2014;9:94839. doi: 10.1371/journal.pone.0094839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gui Y, Liu H, Zhang L, Lv W, Hu X. Altered microRNA profiles in cerebrospinal fluid exosome in Parkinson disease and Alzheimer disease. Oncotarget. 2015;6:37043–37053. doi: 10.18632/oncotarget.6158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Filatova EV, Alieva AKh, Shadrina MI, Slominsky PA. MicroRNAs: possible role in pathogenesis of Parkinson’s disease. Biochemistry (Mosc) 2012;77:813–819. doi: 10.1134/S0006297912080020. [DOI] [PubMed] [Google Scholar]
- 31.Ma W, Li Y, Wang C, Xu F, Wang M, Liu Y. Serum miR-221 serves as a biomarker for Parkinson’s disease. Cell Biochem Funct. 2016;34:511–515. doi: 10.1002/cbf.3224. [DOI] [PubMed] [Google Scholar]
- 32.Gehrke S, Imai Y, Sokol N, Lu B. Pathogenic LRRK2 negatively regulates microRNA-mediated translational repression. Nature. 2010;466:637–641. doi: 10.1038/nature09191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang G, van der Walt JM, Mayhew G, Li YJ, Züchner S, Scott WK, et al. Variation in the miRNA-433 binding site of FGF20 confers risk for Parkinson disease by overexpression of alpha-synuclein. Am J Hum Genet. 2008;82:283–289. doi: 10.1016/j.ajhg.2007.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Junn E, Mouradian MM. MicroRNAs in neurodegenerative diseases and their therapeutic potential. Pharmacol Ther. 2012;133:142–150. doi: 10.1016/j.pharmthera.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang D, Li T, Wang Y, Tang Y, Cui H, Tang Y, et al. miR-132 regulates the differentiation of dopamine neurons by directly targeting Nurr1 expression. J Cell Sci. 2012;125:1673–1682. doi: 10.1242/jcs.086421. [DOI] [PubMed] [Google Scholar]
- 36.Botta-Orfila T, Morató X, Compta Y, Lozano JJ, Falgàs N, Valldeoriola F, et al. Identification of blood serum micro-RNAs associated with idiopathic and LRRK2 Parkinson’s disease. J Neurosci Res. 2014;92:1071–1077. doi: 10.1002/jnr.23377. [DOI] [PubMed] [Google Scholar]
- 37.Cui Y, Li T, Yang D, Li S, Le W. miR-29 regulates Tet1 expression and contributes to early differentiation of mouse ESCs. Oncotarget. 2016;7:64932–64941. doi: 10.18632/oncotarget.10751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mo M, Xiao Y, Huang S, Cen L, Chen X, Zhang L, et al. MicroRNA expressing profiles in A53T mutant alpha-synuclein transgenic mice and Parkinsonian. Oncotarget. 2017;8:15–28. doi: 10.18632/oncotarget.13905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Choi I, Woo JH, Jou I, Joe EH. PINK1 Deficiency decreases expression levels of mir-326, mir-330, and mir-3099 during brain development and neural stem cell differentiation. Exp Neurobiol. 2016;25:14–23. doi: 10.5607/en.2016.25.1.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Amrita Datta Chaudhuri Doo Chul Choi, Savan Kabaria, Alan Tran, Eunsung Junn. MicroRNA-7 regulates the function of mitochondrial permeability transition pore by targeting VDAC1 expression. J Biol Chem. 2016;291:6483–6493. doi: 10.1074/jbc.M115.691352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Margis R, Margis R, Rieder CR. Identification of blood microRNAs associated to Parkinsonis disease. J Biotechnol. 2011;152:96–101. doi: 10.1016/j.jbiotec.2011.01.023. [DOI] [PubMed] [Google Scholar]
- 42.Alieva AK, Filatova EV, Karabanov AV, IIIarioshkin SN, Limborska SA, Shadrina MI, et al. miRNA expression is highly sensitive to a drug therapy in Parkinson’s disease. Parkinsonism Relat Disord 2015, 21: 72–74. [DOI] [PubMed]
- 43.Serafin A, Foco L, Zaniqni S, Blankenburg H, Picard A, Zanon A, et al. Overexpression of blood microRNAs 103a, 30b, and 29a in L-dopa-treated patients with PD. Neurology. 2015;84:645–653. doi: 10.1212/WNL.0000000000001258. [DOI] [PubMed] [Google Scholar]
- 44.Soreq L, Salomonis N, Bronstein M, Greenberg DS, Israel Z, Bergman H, et al. Small RNA sequencing-microarray analyses in Parkinson leukocytes reveal deep brain stimulation-induced splicing changes that classify brain region transcriptomes. Front Mol Neurosci. 2013;6:10. doi: 10.3389/fnmol.2013.00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim J, Inoue K, Ishii J, Vanti WB, Voronov SV, Murchison E, et al. A microRNA feedback circuit in midbrain dopamine neurons. Science. 2007;317:1220–1224. doi: 10.1126/science.1140481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shapshak P. Molecule of the month: miRNA and Parkinson’s disease protein PARK2. Bioinformation. 2013;9:381–382. doi: 10.6026/97320630009381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Saito Y, Saito H. MicroRNAs in cancers and neurodegenerative disorders. Front Genet. 2012;3:194. doi: 10.3389/fgene.2012.00194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Alvarez-Erviti L, Seow Y, Schapira AH, Rodriguez-Oroz MC, Obeso JA, Cooper JM. Influence of microRNA deregulation on chaperone-mediated autophagy and alpha-synuclein pathology in Parkinson’s disease. Cell Death Dis. 2013;4:545. doi: 10.1038/cddis.2013.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cardo LF, Coto E, Ribacoba R, Menéndez M, Moris G, Suárez E, et al. MiRNA profile in the substantia nigra of Parkinson’s disease and healthy subjects. J Mol Neurosci. 2014;54:830–836. doi: 10.1007/s12031-014-0428-y. [DOI] [PubMed] [Google Scholar]
- 50.Vallelunga A, Raqusa M, Di Mauro S, Iannitti T, Pilleri M, Biundo R, et al. Identification of circulating microRNAs for the differential diagnosis of Parkinson’s disease and Multiple System Atrophy. Front Cell Neurosci. 2014;8:156. doi: 10.3389/fncel.2014.00156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Muthuraman M, Deuschl G, Koirala N, Riedel C, Volkmann J, Groppa S. Effects of DBS in parkinsonian patients depend on the structural integrity of frontal cortex. Sci Rep. 2017;7:43571. doi: 10.1038/srep43571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kumar A, Narayanan K, Chaudhary RK, Mishra S, Kumar S, Vinoth KJ, et al. Current perspective of stem cell therapy in neurodegenerative and metabolic diseases. Mol Neurobiol. 2016 doi: 10.1007/s12035-016-0217-4. [DOI] [PubMed] [Google Scholar]
- 53.Valdés P, Schneider BL. Gene Therapy: A Promising approach for neuroprotection in Parkinson’s disease? Front Neuroanat. 2016;10:123. doi: 10.3389/fnana.2016.00123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Basak I, Patil KS, Aives G, Larsen JP, Møller SG, et al. microRNAs as neuroregulators, biomarkers and therapeutic agents in neurodegenerative diseases. Cell Mol Life Sci. 2016;73:811–827. doi: 10.1007/s00018-015-2093-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Roshan R, Ghosh T, Scaria V, Pillai B. MicroRNAs: novel therapeutic targets in neurodegenerative diseases. Drug Discov Today. 2009;14:1123–1129. doi: 10.1016/j.drudis.2009.09.009. [DOI] [PubMed] [Google Scholar]
- 56.Boutla A, Delidakis C, Tabler M. Developmental defects by antisense-mediated inactivation of micro-RNAs 2 and 13 in Drosophila and the identification of putative target genes. Nucleic Acids Res. 2003;31:4973–4980. doi: 10.1093/nar/gkg707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Molasy M, Walczak A, Szaflik J, Szaflik JP, Majsterek I. MicroRNAs in glaucoma and neurodegenerative diseases. J Hum Genet. 2017;62:105–112. doi: 10.1038/jhg.2016.91. [DOI] [PubMed] [Google Scholar]
- 58.Conde J, Edelman ER, Artzi N. Target-responsive DNA/RNA nanomaterials for microRNA sensing and inhibition: the jack-of-all-trades in cancer nanotheranostics? Adv Drug Deliv Rev. 2015;81:169–183. doi: 10.1016/j.addr.2014.09.003. [DOI] [PubMed] [Google Scholar]
- 59.Höbel S, Aigner A. Polyethylenimines for siRNA and miRNA delivery in vivo. Wiley Interdiscip Rev Nanomed Nanobiotechnol. 2013;5:484–501. doi: 10.1002/wnan.1228. [DOI] [PubMed] [Google Scholar]
- 60.Marz M, Ferracin M, Klein C. MicroRNAs as biomarker of Parkinson disease? Small but mighty. Neurology. 2015;84:636–638. doi: 10.1212/WNL.0000000000001275. [DOI] [PubMed] [Google Scholar]
- 61.Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 62.Easow G, Teleman AA, Cohen SM. Isolation of microRNA targets by miRNP immunopurification. RNA. 2007;13:1198–1204. doi: 10.1261/rna.563707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Selbach M, Schwanhäusser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 64.Baek D, Villén J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Batistela MS, Josviak ND, Sulzbach CD, de Souza RL. An overview of circulating cell-free microRNAs as putative biomarkers in Alzheimer’s and Parkinson’s Diseases. Int J Neurosci. 2017;127:547–558. doi: 10.1080/00207454.2016.1209754. [DOI] [PubMed] [Google Scholar]
- 66.Cardo LF, Coto E, de Mena L, Ribacoba R, Moris G, Menéndez M, et al. Profile of microRNAs in the plasma of Parkinson’s disease patients and healthy controls. J Neurol. 2013;260:1420–1422. doi: 10.1007/s00415-013-6900-8. [DOI] [PubMed] [Google Scholar]
- 67.Dong H, Wang C, Lu S, Yu C, Huang L, Feng W, et al. A panel of four decreased serum microRNAs as a novel biomarker for early Parkinson’s disease. Biomarkers. 2016;21:129–137. doi: 10.3109/1354750X.2015.1118544. [DOI] [PubMed] [Google Scholar]
- 68.Ding H, Huang Z, Chen M, Wang C, Chen X, Chen J, et al. Identification of a panel of five serum miRNAs as a biomarker for Parkinson’s disease. Parkinsonism Relat Disord. 2016;22:68–73. doi: 10.1016/j.parkreldis.2015.11.014. [DOI] [PubMed] [Google Scholar]