Abstract
Parkinson’s disease (PD) is a neurodegenerative disease with a long preclinical phase. The continuous loss of dopaminergic (DA) neurons is one of the pathogenic hallmarks of PD. Diagnosis largely depends on clinical observation, but motor dysfunctions do not emerge until 70%–80% of the nigrostriatal nerve terminals have been destroyed. Therefore, a biomarker that indicates the degeneration of DA neurons is urgently needed. Transcription factors are sequence-specific DNA-binding proteins that regulate RNA synthesis from a DNA template. The precise control of gene expression plays a critical role in the development, maintenance, and survival of cells, including DA neurons. Deficiency of certain transcription factors has been associated with DA neuron loss and PD. In this review, we focus on some transcription factors and discuss their structure, function, mechanisms of neuroprotection, and their potential for use as biomarkers indicating the degeneration of DA neurons.
Keywords: Parkinson’s disease, Transcription factor, Biomarker, Cell death, Dopaminergic neuron
Introduction
Parkinson’s disease (PD) is the most common neurodegenerative disorder accompanied by movement dysfunction. Pathologically, PD is characterized by the presence of cytoplasmic inclusions in dopaminergic (DA) neurons termed Lewy bodies (LBs) and the continuous loss of DA neurons in the substantia nigra pars compacta (SNc) [1]. Decreased dopamine levels in the dorsal striatum trigger the classical clinical manifestations following the selective loss of DA neurons. Great accomplishments have been achieved in past decades regarding our understanding of the pathological mechanisms underlying PD; however, the etiology of DA neuron loss in PD still remains to be elucidated. The death of DA neurons is considered a signal of symptom onset. The drug management generally used today aims to relieve the symptoms by improving the dopamine levels in the midbrain or by stimulating dopamine receptors. However, these motor dysfunctions do not emerge until 70%–80% of nigrostriatal nerve terminals have been destroyed. And the loss of DA neurons cannot be rescued or reversed by drug administration [2]. It seems that early intervention is the most effective measure for coping with PD. Sensitive and specific diagnostic methods are urgently needed to predict the death of DA neurons in the preclinical phase. Fortunately, several biomarkers have been suggested as potential candidates for detecting the death of DA neurons even before the emergence of motor dysfunction.
A biomarker is a measurable indicator reflecting normal or abnormal biological processes [3]. Comprehensive information about the nature of any particular disease can be acquired from specific changes in pathological, biochemical, and genetic processes, in addition to clinical observation. An ideal biomarker should be sensitive, reproducible, fully validated, easy to measure, and cheap [4]. Biological fluids are the most common material used for biomarker testing. Among these, cerebrospinal fluid (CSF) may be the most sensitive source for evaluating PD pathology, due to its close association with the degeneration process of neurons. Different CSF markers are used for routine differential diagnosis in amyotrophic lateral sclerosis and Alzheimer’s disease. Other fluids like plasma, urine, and saliva that are easier to obtain are also promising candidates for biomarker testing, since their sampling is more likely to be accepted by patients.
As for PD in particular, the optimal biomarker should reflect the degeneration of DA neurons. Transcription factors are sequence-specific DNA-binding proteins that regulate RNA synthesis from a DNA template. The precise control of gene expression is vital for the development, maintenance, and survival of neurons. Moreover, apart from modulating transcription processes, transcription factors have additional functions that are also of critical significance for regulating neuron survival. An imbalance in the levels of transcription factors can be induced by oxidative stress, mitochondrial dysfunction, neuroinflammation, and neurotoxins, leading to the dysregulation of gene expression and degeneration of DA neurons in PD [5]. In this review, we focus on several critical transcription factors including myocyte enhancer factor 2 family (MEF2), nuclear receptor related 1 (Nurr1), paired like homeodomain 3 (Pitx3), and engrailed homeobox 1/2 (En1/2). We discuss their structure, function, mechanisms of neuroprotection, and their potential for use as biomarkers to indicate the degeneration of DA neurons. The dysregulation of signaling pathways in PD is also summarized for a better understanding of the death of DA neurons.
Transcription Factors Involved in Maintaining the Function and Survival of DA Neurons
There have been great achievements in recent years regarding our understanding of the genetic and signaling networks that control the generation, maintenance, and survival of DA neurons. Transcription factors play a crucial role in illuminating the processes underlying the degeneration of DA neurons. It is therefore important to elucidate the mechanisms behind these transcription factors during development and later stages in DA neurons. These mechanisms can provide novel insights into PD pathogenesis and suggest clinical biomarker utilization.
Mature DA neurons express elevated levels of tyrosine hydroxylase (TH), dopamine transporter (DAT), vesicular monoamine transporter 2 (VMAT2), dopa decarboxylase, among others. They are thought of as phenotypic markers that determine the identity of the mature DA neuron. Transcription factors are implicated in the expression of phenotypic markers. The number of SNc neurons is greatly reduced and they no longer express TH in Pitx3-deficient mice [6]. When the Nurr1 gene is deleted (Nurr−/−), mice have poor motor function and histopathological examination shows that DA neurons are decreased in the midbrain. TH and DAT, as well as aromatic amino-acid carboxylase, are absent from the nigrostriatal pathway [7, 8]. There is also an indication that En1/2 participates in the acquisition of DA neuronal identity by interacting with other factors [9].
Neurotrophic factors (NTFs) are peptides essential for the growth, maturation, and survival of neurons and axons. The levels and functions of these factors have been associated with PD. It has been reported in a genome-wide analysis that brain-derived neurotrophic factor (BDNF) is one of the transcription targets of MEF2s [10], which differently regulate the transcription of BDNF in response to neuronal depolarization [11]. In addition, Nurr1 has been shown to regulate the transcription of BDNF in rat midbrain primary cultures. BDNF mRNA and protein are down-regulated following the decreased expression of Nurr1. Reporter gene assay experiments have shown that BDNF is a direct target gene of Nurr1 [12], which also modulates the expression of the glial cell line-derived neurotrophic factor (GDNF) receptor Ret. GDNF signaling is disrupted in nigral DA neurons when Nurr1 is down-regulated by treatment with α-synuclein [13]. Pitx3 can increase the production of BDNF and GDNF in transfected astrocytes. This has also been confirmed in SH-SY5Y cells and primary ventral mesencephalic cultures [14, 15]. Moreover, BDNF has been reported to be regulated by GDNF, and Pitx3 is also involved in this regulation of BDNF [16].
The etiology of DA neuronal degeneration is not fully understood; however, stress is thought to be one of its major causes. Mitochondrial toxins have been identified to induce animal models of PD including those using MPTP (1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine), 6-OHDA (6-hydroxydopamine), and paraquat. Apoptosis of DA neurons is also attributed to endoplasmic reticulum (ER) stress in vivo and in vitro. MEF2s have been shown to be post-translationally modified when exposed to certain kinds of stimuli. Nitrosative stress-induced dysfunction in MEF2-PGC1α (peroxisome proliferator-activated receptor gamma coactivator 1-alpha) transcription accounts for apoptotic cell death in the isogenic human induced pluripotent stem cell model of PD [17]. In addition, Nurr1 has been reported to play a role in the resistance to oxidative stress, as revealed by microarray analyses. Nurr1 can shuttle between the cytosol and the nucleus when exposed to oxidative stress [18]. Nrf1 and certain genes for antioxidant enzymes are direct targets of Pitx2/3. Reactive oxygen species levels are excessively upregulated in double conditional Pitx2/3 mouse mutants, leading to DNA damage and apoptosis of differentiating cells [19]. Engrailed homeoprotein is a neuroprotective transcription factor in PD. Injection of engrailed-2 can restore heterochromatin markers, decrease DNA strand breaks, and defend DA neurons against oxidative stress [20].
Certain transcription factors are also involved in other mechanisms that protect DA neurons. MEF2D has been shown to regulate the production of interleukin-10 to protect DA neurons [21]. Nurr1 has an important function in sustaining respiratory activity. Transcription factors have a close relationship with PD and they maintain the function and survival of DA neurons via multiple mechanisms.
Transcription Factors and Parkinson’s Disease
Defective Transcription Factors in DA Neuronal Degeneration
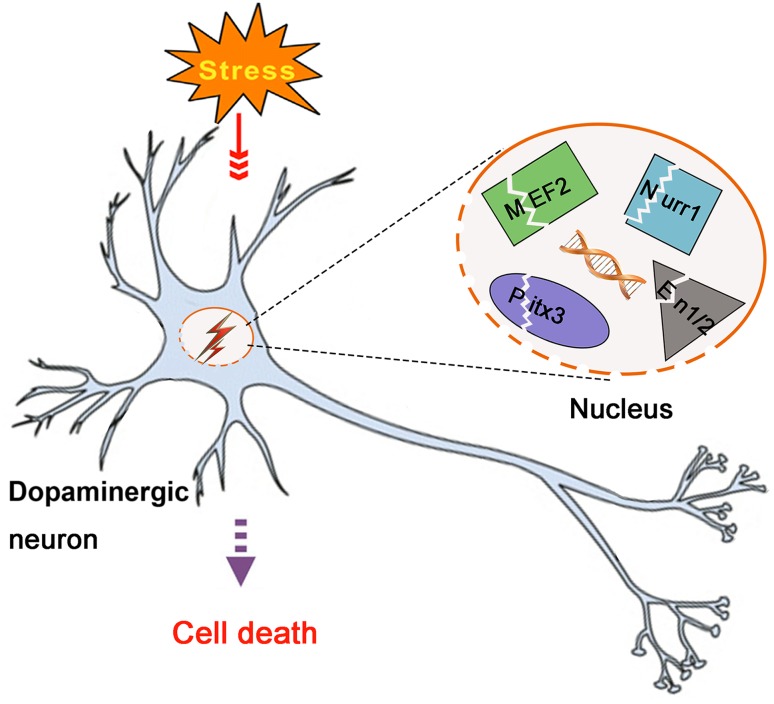
Several transcription factors maintain the survival of neurons via canonical functions to regulate gene expression and additional functions including translation modulation, oxidative stress resistance, and others. MEF2s, Nurr1, Pitx3, and En1/2 are prominent transcription factors that play vital roles in the development, maintenance, and survival of DA neurons. They accomplish these functions by interacting with different kinds of signaling pathways to maintain the homeostasis of mitochondria, decrease ER stress, regulate neuroinflammation, and modulate other biological processes. Studies have shown that they are defective and tend to be suppressed in PD animal models and patients with PD (Fig. 1). These transcription factors are directly involved in the survival of DA neurons and therefore have the potential to be biomarkers indicating the death of DA neurons and indicating PD even in the preclinical phase.
Fig. 1.
Involvement of nuclear transcription factors in PD pathogenesis. In PD, all kinds of stresses (neurotoxin, aging, oxidative stress, etc.) lead to the dysregulation of the nuclear transcription factors MEF2s, Nurr1, Pitx3 and En1/2 and thereby disrupt their functions, contributing to the death of dopaminergic neurons.
MEF2 Proteins
The MEF2 proteins are members of the MADS (MCM1-agamous-deficiens-serum response factor) family of transcription factors, composed of MEF2-A, -B, -C, and -D proteins [22]. The first 56 amino-acids at the N terminus, termed the MADS-box, generally bind to A/T-rich DNA sequences. The MEF2 domain, adjacent to the MADS-box, binds preferentially to the consensus sequence 5’-CC(A/T)(T/A)AAATAG-3’ [23]. The C-terminal portion of MEF2 proteins contains the transcriptional activation domain, as well as a number of regulatory minidomains including a nuclear localization sequence and multiple phosphorylation motifs [24]. MEF2 family members regulate the expression of many genes regulated by neuronal activity, including ARC (activity-regulated cytoskeleton-associated protein), BDNF, and HOMER1A (Homer protein homolog 1) [25]. As noted earlier, BDNF is a neurotrophic factor for DA neurons [26]. In addition, MEF2 family members are regulated by several kinases and post-translational regulatory mechanisms, which are critical for the control of their function [27–32].
Studies have implied that MEF2A/C/D-knockout (KO) mice have early postnatal lethality with increased neuronal apoptosis, indicating a pivotal role of the MEF2 factors in neuronal survival [33, 34]. The first data revealing the role of MEF2s in neuronal survival were provided by research studying the mechanisms of neuronal activity-dependent survival. The authors found that phosphorylation of MEF2C by p38 activates its function and promotes neuronal survival [35]. MEF2A has been suggested to contain a p38 MAPK (mitogen-activated protein kinase) docking site in its crystal structure. The p38-MEF2 pathway has been shown to be important for the development, proliferation, and survival of neurons. The p38-MEF2 pathway has been shown to mediate the anti-apoptotic effect in neurodegenerative disease. MEF2s are also involved in different cellular activities and functions that help the cell to resist various stimuli including oxidative stress and neurotoxins [36]. Results from earlier studies have shown that the function of MEF2D is maintained partly by adequate CMA (chaperone-mediated autophagy) activity while α-synuclein disturbs this degradation process. MEF2D is a neuronal survival factor in PD, and deregulation of the CMA-MEF2D pathway may be a cause of PD [37]. Gao et al. implied that oxidization of the survival factor MEF2D is induced by 6-OHDA, and that it plays a critical role in oxidative stress-induced DA neuronal death [30]. MEF2D directly regulates the expression of the ND6 gene, a component of mitochondrial complex I. When treated with rotenone, the DNA-binding of MEF2D is weakened, resulting in decreased levels of ND6 protein and reduced ATP production [38]. Interestingly, MEF2D can also maintain cell survival via regulating interleukin-10 production in microglia [21]. The PD animal models used now are induced by different kinds of neurotoxins, including MPTP, 6-OHDA, and rotenone [39]. Motor syndromes and DA neuronal loss are seen after the injection of the toxin. Adequate evidence suggests that proteins and mRNAs of MEF2s are destabilized after using neurotoxin [40–43]; therefore, there is a close correlation between MEF2 levels and the death of DA neurons.
NR4A2 (Nurr1) Nuclear Receptor Subfamily 4 Group A Member 2
Nurr1 (also named NR4A2) belongs to an NR4A subgroup of the nuclear receptor superfamily that is part of the ligand-activated transcription factor family. The transcription factor is composed of four parts: the modulator domain in the N-terminal, the DNA-binding domain, the ligand-binding domain, and the C-terminus named transactivation-dependent activation function 2. Nurr1 can work in the form of a monomer or a homodimer. Moreover, it can function effectively by binding with retinoid X receptors to form heterodimers. The first time Nurr1 was associated with PD was for its critical role in the development of midbrain DA neurons [44]. Its significant function has been validated in Nurr1-deficient mice, in which the number of DA neurons is greatly decreased [45]. In addition, the expression of Nurr1 seems to regulate the expression of TH, of aromatic amino-acid carboxylase, of the DA transporter, and of the vesicular monoamine transporter, which are the phenotypic markers of DA neurons [46]. Nurr1 expression tends to decrease in midbrain DA neurons in an age-dependent manner; this process may help to clarify the pathogenesis of PD. Nurr1 regulates the transcription of many target substrates and it is also precisely controlled by related signaling pathways. MAPK pathways are involved in the activation and induction of Nurr1. The apoptosis signal-regulating kinase 1-p38 pathway mediates the cytoplasmic translocation of Nurr1 on exposure to oxidative stress [47]. The phosphorylation of Nurr1 by extracellular signal-regulated kinases 1/2 upregulates the transcription of TH.
Research has shown that Nurr1 regulates axon genesis and maintains the fiber integrity of mesencephalic DA (mDA) neurons. Moreover, Nurr1 may play an anti-inflammatory role in maintaining the survival of neurons. Liu et al. showed that Nurr1 restrains the expression of C-C motif chemokine ligand 2, leading to decreased release of TNF-α and interleukin-1β in PD models both in vivo and in vitro [48]. Interestingly, Nurr1 can recruit the CoREST corepressor complex to clear nuclear factor-κB (NFkB)-p65 in microglia and astrocytes in order to repress exaggerated inflammation and protect DA neurons [49, 50]. Mitochondrial dysfunction is thought to be an important contributor to the pathology of PD. Nurr1 is also involved in the survival of DA neurons by regulating mitochondrial function. When adult mice are treated with tamoxifen to induce conditional ablation of Nurr1, the expression of 90% of all genes concerned with oxidative phosphorylation is down-regulated in Nurr1-ablated DA neurons, implying a critical role of Nurr1 in respiratory processes [51]. In addition, evidence indicates an interplay between Nurr1 and α-synuclein. Nurr1 is involved in the function of GDNF via regulation of its receptors [52]. However, conditional expression of α-synuclein disrupts this function and promotes the degradation of Nurr1. Conversely, decreased Nurr1 expression previously reported in patients with PD may transcriptionally induce the expression of α-synuclein [53]. DA neurons in heterozygous Nurr1-deficient (Nurr1 +/–) mice have been found to be more sensitive to toxic stress, including exposure to lactacystin or MPTP [54]. Mutations in the Nurr1 gene are associated with familial and sporadic PD [55]. Taken together, Nurr1 assists in the development of midbrain DA neurons, maintaining normal respiratory chains, supporting neurotrophic signal propagation, resisting exaggerated inflammation, and decreasing the susceptibility to environmental stimuli and neurotoxins.
Pitx3
Pitx3 is one of the homeodomain-containing transcription factors. The Pitx3 protein comprises 302 amino-acids with 98% identity in the mouse and human. An OAR domain is situated at the C-terminal of the protein, which has been reported to restrain cartilage homeoprotein 1 protein transcriptional activity by binding to DNA [56].
Pitx3 defines a pathway for the development and survival of midbrain DA neurons. This pathway has been investigated in young adult Pitx3-null mice (Pitx3−/−) that show decreased TH staining and display impaired motor coordination, implying a critical role of Pitx3 in DA neuronal development. Evidence shows that Pitx3 directly binds to the promoter of the TH gene and upregulates its expression [57]. Neurotrophic factors like BDNF and GDNF play a critical role in protecting DA neurons [58]. In SH-SY5Y cells and primary cultures, Pitx3 modulates the transcription and translation of both BDNF and GDNF. Astrocytes overexpressing Pitx3 produce more neurotrophic factor and protect DA neurons from rotenone-induced injury. MiR-133b is specifically enriched in the midbrain and deficient in patients with PD. Pitx3 has been reported to regulate the transcription of precursor-miR-133b. Pitx3 directly binds to the promoter of miR-133b sequences and modulates the expression of miR-133b precursors [59]. In addition, activation of the mTOR pathway increases striatal dopamine concentrations and the mRNA levels of Pitx3, En1, and Nurr1, which protect DA neurons in genetic and neurotoxin-induced (MPTP or 6-OHDA) mouse models of PD.
En1/2
The homeoproteins En1/2 not only serve as a survival factor for mDA neurons during development, but continue to play a neuroprotective role in adult DA neurons [60]. There are five distinct subregions within the protein, designated EH1–5. EH4 is a homeodomain composed of a highly conserved stretch of ~60 amino-acids. The EH1 and EH5 domains function to repress transcriptional activity. The EH2/3 domains determine the affinity of the protein to DNA. The phosphorylation of a common serine-rich site in EH2 increases the DNA binding of recombinant engrailed several-fold [61].
En1/2 play a vital role in the development of mDA neurons. En1/2 are initially expressed on embryonic day E8 in a cluster of cells in the midbrain. Mice deficient in both genes (En1 -/-; En2 -/-) die at birth and exhibit a loss of DA neurons, while the loss of one En1 allele (En1 -/+) results in the progressive loss of mDA neurons at 6 weeks after birth [62]. In addition, En1 assists in the protection of DA neurons from death in PD animal models induced by 6-OHDA, MPTP, and A30P a-synuclein, indicating its critical role in the survival and maintenance of DA neurons during adulthood. It has been reported that En1/2 survival activity is mediated through both the activation of the Erk1/2 MAPK survival pathway and suppression of the pro-apoptotic activity of p75 neurotrophin receptor [63]. En2 has also been reported to activate the phosphatidylinositol 3-kinase/AKT pathway and inhibit PTEN [64]. Mitochondrial dysfunction is considered to be a cause of PD, and En1/2 have been reported to regulate the activity of mitochondria. Transduction of En1/2 proteins stimulates the translation of nuclear-encoded mitochondrial mRNAs of Ndufs1/Ndufs3, subunits of complex I, leading to elevated complex I activity through the mTOR pathway [65, 66].
Signaling and Dysregulation of Transcription Factors in Neuronal Degeneration
Cell death is often imposed on DA neurons by extracellular or intracellular stimuli including oxidative stress, microglia activation, chronic inflammation, exposure to neurotoxins, and DNA damage [67]. Although the mechanisms leading to survival or death are not fully understood, it is clear that a growing number of pathways are dysregulated and involved in linking these stimuli with the cell-death machinery. Moreover, these pathways interplay with transcription factors. They can regulate or be modulated by transcription factors. The dysfunction of certain signaling pathways is always found in animal models and in patients with PD. Testing the critical molecules in these signaling pathways may help to elucidate how transcription factor biomarkers reflect the degeneration of DA neurons more sensitively and precisely.
MAPK Pathways and Neuronal Death
MAPKs are serine–threonine protein kinases that regulate cell differentiation, inflammation, innate immunity, apoptosis, and survival. MAPKs comprise three types: p38 MAPK, c-Jun NH2-terminal kinase (JNK), and extracellular signal-regulated kinase (ERK) [68]. MAPKs mediate the activation of microglia and neuroinflammation, maintain the homeostasis of mitochondria, regulate ER stress, and phosphorylate and regulate the expression of Bcl-2 protein family members. Increasing experimental evidence shows that neurotoxins (such as 6-OHDA, MPTP, and rotenone) selectively phosphorylate p38 MAPK in DA neurons. Phosphorylated p38 MAPK affects apoptosis by phosphorylating neuronal survival factor MEF2s, inducing the translocation of Nurr1, and activating NF-κB [47, 69]. Modulating ERK signaling in patients with PD may protect DA neurons and potentially have therapeutic effects on motor dysfunction [70].
Role of the PI3K/AKT/mTOR Pathway in Neuronal Degeneration
PI3Ks are responsible for propagating signals arising from cytokines, growth factors, and hormones into intracellular communication by generating phospholipids. AKT, a crucial signaling mediator downstream of PI3K, is a serine/threonine protein kinase regulating multiple cellular processes including proliferation, metabolism, autophagy, and cell survival. The central hubs of this network are PI3K/AKT and mTOR signaling. PI3K-AKT signaling is defective in PD and is associated with loss of DA neurons. In addition, the PI3K/AKT signaling pathway has been reported to regulate Leucine-rich repeat kinase2 (LRRK2), PTEN-induced putative kinase 1 (PINK1), and DJ-1 whose mutations are thought to be a cause of familial PD. This pathway could also modulate the autophagic activity that regulates clearance of the protein aggregates occurring in neurodegeneration, to protect DA neurons. Moreover, certain transcription factors are modulated by this pathway. It has been revealed that activated PI3K/AKT/mTOR pathways increase the mRNA levels of Pitx3 and En1/2 [71].
Other Pathways Associated with DA Neurons
NF-κB is involved in inflammation pathways and plays an important role in neurodegenerative disorders. TNF-α is one of the most powerful activators of the pathway. Once activated, NF-κB transcription factors translocate to the nucleus and regulate the transcription of target genes [72]. Other pathways, like Wnt and CREB signaling pathways, also regulate the transcription of genes or the modification of proteins, resulting in the death or survival of DA neurons.
Transcription Factors as Potential Biomarkers Reflecting the Death of DA Neurons
PD has a long preclinical period, during which motor dysfunction is inconspicuous while DA neurons are starting to degenerate [73]. With traditional diagnostic methods, early diagnosis and timely therapeutic intervention are difficult to achieve. The loss of DA neurons signifies the onset of PD, which can occur decades before classical symptoms emerge. Methods that allow the detection of degenerating DA neurons in the preclinical phase are therefore urgently needed. Biomarkers seem to be one solution to this problem, and great efforts have been devoted to the selection of potential candidates reflecting the degeneration of neurons. Encouragingly, several kinds of biomarkers have been examined and reported, including markers for non-motor dysfunctions, genetic mutations, and biochemical and metabolic molecules [74]. Emerging evidence has shown that transcription factors have significant functions in the maintenance and survival of DA neurons. They can be detected in biological fluids in humans and are positively associated with an increased risk for PD. They may serve to mirror the degeneration of DA neurons, help diagnosis in the preclinical phase, track disease progression, and assess the effect of therapeutic interventions [75, 76]. Transcription factors are therefore one of the potential candidates for biomarkers in the diagnosis of PD.
It has been revealed that each MEF2 isoform has a unique distribution pattern in a different region of the brain [77]. Considering the critical role of MEF2s in the survival of DA neurons and their abundant expression in neurons, it seems that MEF2s could be used as a biomarker, reflecting the death of neurons and indicating PD. The biomarker research regarding MEF2s is still lacking, and great efforts are needed in order to make possible the clinical use of MEF2s in the diagnosis of PD.
Studies have revealed that a decrease of Nurr1 tends to be coupled with the loss of DA neurons in PD, suggesting its potential as a biomarker for neuronal death. A research team recently measured NURR1 and PITX3 gene expression in 255 PD patients and 211 age- and gender-matched healthy controls. The results, measured in human peripheral blood lymphocytes (PBLs), indicated decreased NURR1 and PITX3 gene expression in Chinese patients with PD [78]. Another study showed that 60-year-old or older female patients with PD show lower levels of NURR1 expression in PBLs [79]. When a research team recently measured the expression levels of genes in the NR4A subfamily in patients with PD and AD, they found that, similar to the results from PBLs, the levels of not only Nurr1 but all subfamily members were decreased in patients with PD [80]. Decreased expression of the NURR1 gene in PBLs may help differentiate individuals with PD from patients with other disorders associated with impaired central nervous system function. Unfortunately, no study has yet reported the testing and analysis of Nurr1 in CSF. A validation of the results from PBLs in CSF would be convincing and useful for further research.
Pitx3 is expressed in a group of mDA neurons in the ventral tegmental area and SNc. In addition, evidence has shown that several single-nucleotide polymorphisms of the PITX3 gene are associated with a higher risk or early onset of PD [81]. The gene expression of PITX3 was measured in human PBLs from 255 Chinese patients with PD and 211 healthy controls using a quantitative real-time PCR technique. Surprisingly, the expression of the PITX3 gene is reduced in the PBLs of Chinese patients with PD [78]. In males and older individuals, decreased PITX3 gene expression is associated with an increased risk for PD, indicating the possible systemic involvement of this gene in PD. There is no research on Pitx3 in other biological fluids. Even though Pitx3 seems to be promising as a biomarker for PD, further validation is required.
The expression of En1/2 starts early, at embryonic day E8, and becomes restricted to mDA neurons in the adult. Diana et al. have reported that En1 protein is expressed in solid-type salivary gland adenoid cystic carcinoma and correlates with a significantly lower survival rate, indicating its potential as a biomarker for this cancer [82]. In addition, En2 protein can be detected in urine by ELISA, and its average levels are higher in patients with prostate and bladder cancer [83, 84]. As for the crucial role of En1/2 in the development, maintenance, and survival of DA neurons, there is a possibility for the protein to be a biomarker indicating the death of DA neurons.
Regrettably the transcription factors mentioned above have not been used in clinical diagnosis, great efforts are needed to promote the application of them. Promising results need to be validated in more biological materials, including CSF, plasma, urine, and saliva, to increase the accuracy of diagnosis. Moreover, the levels of biomarkers in different groups of PD patients are controversial. Standards for biomarker research are urgently needed to eliminate irrelevant factors. Furthermore, sample sizes should be expanded in future studies that aim to transfer the promising laboratory results to clinical testing.
Conclusions
In the past decades, α-synuclein, Aβ, and other molecules have been examined and linked to the diagnosis of PD [85]. However, most biomarkers have not achieved routine clinical use. Certain transcription factors are directly involved in the survival of DA neurons. Alterations in the levels of these factors indicate the death of DA neurons. These factors therefore have been suggested to reflect disease progression and to help diagnosing PD in the preclinical phase. It is almost impossible to deduce the death of DA neurons from one molecule only. Transcription factor biomarkers play a crucial role in predicting the death of DA neurons, and can play an important role in diagnosing PD more precisely and sensitively, in combination with other biomarkers.
Acknowledgements
This review was supported by the National Key Research and Development Program of China (2016YFC1306603) and the National Natural Science Foundation of China (31671060).
References
- 1.Yuan L, Song Z, Deng X, Yang Z, Yang Y, Guo Y, et al. Genetic analysis of FBXO2, FBXO6, FBXO12, and FBXO41 variants in Han Chinese patients with sporadic Parkinson’s disease. Neurosci Bull. 2017 doi: 10.1007/s12264-017-0122-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bernheimer H, Birkmayer W, Hornykiewicz O, Jellinger K, Seitelberger F. Brain dopamine and the syndromes of Parkinson and Huntington. Clinical, morphological and neurochemical correlations. J Neurol Sci. 1973;20:415–455. doi: 10.1016/0022-510X(73)90175-5. [DOI] [PubMed] [Google Scholar]
- 3.Rachakonda V, Pan TH, Le WD. Biomarkers of neurodegenerative disorders: how good are they? Cell Res. 2004;14:347–358. doi: 10.1038/sj.cr.7290235. [DOI] [PubMed] [Google Scholar]
- 4.van Dijk KD, Teunissen CE, Drukarch B, Jimenez CR, Groenewegen HJ, Berendse HW, et al. Diagnostic cerebrospinal fluid biomarkers for Parkinson’s disease: a pathogenetically based approach. Neurobiol Dis. 2010;39:229–241. doi: 10.1016/j.nbd.2010.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blaudin de The FX, Rekaik H, Prochiantz A, Fuchs J, Joshi RL. Neuroprotective transcription factors in animal models of Parkinson disease. Neural Plast. 2016;2016:6097107. doi: 10.1155/2016/6097107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Le W, Zhang L, Xie W, Li S, Dani JA. Pitx3 deficiency produces decreased dopamine signaling and induces motor deficits in Pitx3(-/-) mice. Neurobiol Aging. 2015;36:3314–3320. doi: 10.1016/j.neurobiolaging.2015.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zetterstrom RH, Solomin L, Jansson L, Hoffer BJ, Olson L, Perlmann T. Dopamine neuron agenesis in Nurr1-deficient mice. Science. 1997;276:248–250. doi: 10.1126/science.276.5310.248. [DOI] [PubMed] [Google Scholar]
- 8.Baffi JS, Palkovits M, Castillo SO, Mezey E, Nikodem VM. Differential expression of tyrosine hydroxylase in catecholaminergic neurons of neonatal wild-type and Nurr1-deficient mice. Neuroscience. 1999;93:631–642. doi: 10.1016/S0306-4522(99)00124-4. [DOI] [PubMed] [Google Scholar]
- 9.Veenvliet JV, Dos Santos MT, Kouwenhoven WM, von Oerthel L, Lim JL, van der Linden AJ, et al. Specification of dopaminergic subsets involves interplay of En1 and Pitx3. Development. 2013;140:3373–3384. doi: 10.1242/dev.094565. [DOI] [PubMed] [Google Scholar]
- 10.Flavell SW, Kim TK, Gray JM, Harmin DA, Hemberg M, Hong EJ, et al. Genome-wide analysis of MEF2 transcriptional program reveals synaptic target genes and neuronal activity-dependent polyadenylation site selection. Neuron. 2008;60:1022–1038. doi: 10.1016/j.neuron.2008.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lyons MR, Schwarz CM, West AE. Members of the myocyte enhancer factor 2 transcription factor family differentially regulate Bdnf transcription in response to neuronal depolarization. J Neurosci. 2012;32:12780–12785. doi: 10.1523/JNEUROSCI.0534-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Volpicelli F, Caiazzo M, Greco D, Consales C, Leone L, Perrone-Capano C, et al. Bdnf gene is a downstream target of Nurr1 transcription factor in rat midbrain neurons in vitro. J Neurochem. 2007;102:441–453. doi: 10.1111/j.1471-4159.2007.04494.x. [DOI] [PubMed] [Google Scholar]
- 13.Decressac M, Kadkhodaei B, Mattsson B, Laguna A, Perlmann T, Bjorklund A. α-Synuclein-induced down-regulation of Nurr1 disrupts GDNF signaling in nigral dopamine neurons. Sci Transl Med. 2012;4:163ra156. doi: 10.1126/scitranslmed.3004676. [DOI] [PubMed] [Google Scholar]
- 14.Peng C, Fan S, Li X, Fan X, Ming M, Sun Z, et al. Overexpression of pitx3 upregulates expression of BDNF and GDNF in SH-SY5Y cells and primary ventral mesencephalic cultures. FEBS Lett. 2007;581:1357–1361. doi: 10.1016/j.febslet.2007.02.054. [DOI] [PubMed] [Google Scholar]
- 15.Yang D, Peng C, Li X, Fan X, Li L, Ming M, et al. Pitx3-transfected astrocytes secrete brain-derived neurotrophic factor and glial cell line-derived neurotrophic factor and protect dopamine neurons in mesencephalon cultures. J Neurosci Res. 2008;86:3393–3400. doi: 10.1002/jnr.21774. [DOI] [PubMed] [Google Scholar]
- 16.Peng C, Aron L, Klein R, Li M, Wurst W, Prakash N, et al. Pitx3 is a critical mediator of GDNF-induced BDNF expression in nigrostriatal dopaminergic neurons. J Neurosci. 2011;31:12802–12815. doi: 10.1523/JNEUROSCI.0898-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ryan SD, Dolatabadi N, Chan SF, Zhang X, Akhtar MW, Parker J, et al. Isogenic human iPSC Parkinson’s model shows nitrosative stress-induced dysfunction in MEF2-PGC1alpha transcription. Cell. 2013;155:1351–1364. doi: 10.1016/j.cell.2013.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garcia-Yague AJ, Rada P, Rojo AI, Lastres-Becker I, Cuadrado A. Nuclear import and export signals control the subcellular localization of Nurr1 protein in response to oxidative stress. J Biol Chem. 2013;288:5506–5517. doi: 10.1074/jbc.M112.439190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.L’Honore A, Commere PH, Ouimette JF, Montarras D, Drouin J, Buckingham M. Redox regulation by Pitx2 and Pitx3 is critical for fetal myogenesis. Dev Cell. 2016;39:756. doi: 10.1016/j.devcel.2016.12.011. [DOI] [PubMed] [Google Scholar]
- 20.Rekaik H, Blaudin de The FX, Fuchs J, Massiani-Beaudoin O, Prochiantz A, Joshi RL. Engrailed homeoprotein protects mesencephalic dopaminergic neurons from oxidative stress. Cell Rep. 2015;13:242–250. doi: 10.1016/j.celrep.2015.08.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang S, Gao L, Lu F, Wang B, Gao F, Zhu G, et al. Transcription factor myocyte enhancer factor 2D regulates interleukin-10 production in microglia to protect neuronal cells from inflammation-induced death. J Neuroinflammation. 2015;12:33. doi: 10.1186/s12974-015-0258-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu YT, Breitbart RE, Smoot LB, Lee Y, Mahdavi V, Nadal-Ginard B. Human myocyte-specific enhancer factor 2 comprises a group of tissue-restricted MADS box transcription factors. Genes Dev. 1992;6:1783–1798. doi: 10.1101/gad.6.9.1783. [DOI] [PubMed] [Google Scholar]
- 23.Black BL, Olson EN. Transcriptional control of muscle development by myocyte enhancer factor-2 (MEF2) proteins. Annu Rev Cell Dev Biol. 1998;14:167–196. doi: 10.1146/annurev.cellbio.14.1.167. [DOI] [PubMed] [Google Scholar]
- 24.Potthoff MJ, Olson EN. MEF2: a central regulator of diverse developmental programs. Development. 2007;134:4131–4140. doi: 10.1242/dev.008367. [DOI] [PubMed] [Google Scholar]
- 25.Dietrich JB. The MEF2 family and the brain: from molecules to memory. Cell Tissue Res. 2013;352:179–190. doi: 10.1007/s00441-013-1565-2. [DOI] [PubMed] [Google Scholar]
- 26.Allen SJ, Watson JJ, Shoemark DK, Barua NU, Patel NK. GDNF, NGF and BDNF as therapeutic options for neurodegeneration. Pharmacol Ther. 2013;138:155–175. doi: 10.1016/j.pharmthera.2013.01.004. [DOI] [PubMed] [Google Scholar]
- 27.Yin Y, She H, Li W, Yang Q, Guo S, Mao Z. Modulation of neuronal survival factor MEF2 by kinases in Parkinson’s disease. Front Physiol. 2012;3:171. doi: 10.3389/fphys.2012.00171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.She H, Yang Q, Mao Z. Neurotoxin-induced selective ubiquitination and regulation of MEF2A isoform in neuronal stress response. J Neurochem. 2012;122:1203–1210. doi: 10.1111/j.1471-4159.2012.07860.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ke K, Shen J, Song Y, Cao M, Lu H, Liu C, et al. CDK5 contributes to neuronal apoptosis via promoting MEF2D phosphorylation in rat model of intracerebral hemorrhage. J Mol Neurosci. 2015;56:48–59. doi: 10.1007/s12031-014-0466-5. [DOI] [PubMed] [Google Scholar]
- 30.Gao L, She H, Li W, Zeng J, Zhu J, Jones DP, et al. Oxidation of survival factor MEF2D in neuronal death and Parkinson’s disease. Antioxid Redox Signal. 2014;20:2936–2948. doi: 10.1089/ars.2013.5399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chan SF, Sances S, Brill LM, Okamoto S, Zaidi R, McKercher SR, et al. ATM-dependent phosphorylation of MEF2D promotes neuronal survival after DNA damage. J Neurosci. 2014;34:4640–4653. doi: 10.1523/JNEUROSCI.2510-12.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang Q, Mao Z. Dysregulation of autophagy and Parkinson’s disease: the MEF2D link. Apoptosis. 2010;15:1410–1414. doi: 10.1007/s10495-010-0475-y. [DOI] [PubMed] [Google Scholar]
- 33.Akhtar MW, Kim MS, Adachi M, Morris MJ, Qi X, Richardson JA, et al. In vivo analysis of MEF2 transcription factors in synapse regulation and neuronal survival. PLoS One. 2012;7:e34863. doi: 10.1371/journal.pone.0034863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Latchney SE, Jiang Y, Petrik DP, Eisch AJ, Hsieh J. Inducible knockout of Mef2a, -c, and -d from nestin-expressing stem/progenitor cells and their progeny unexpectedly uncouples neurogenesis and dendritogenesis in vivo. FASEB J. 2015;29:5059–5071. doi: 10.1096/fj.15-275651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mao Z, Bonni A, Xia F, Nadal-Vicens M, Greenberg ME. Neuronal activity-dependent cell survival mediated by transcription factor MEF2. Science. 1999;286:785–790. doi: 10.1126/science.286.5440.785. [DOI] [PubMed] [Google Scholar]
- 36.Burton TR, Dibrov A, Kashour T, Amara FM. Anti-apoptotic wild-type Alzheimer amyloid precursor protein signaling involves the p38 mitogen-activated protein kinase/MEF2 pathway. Brain Res Mol Brain Res. 2002;108:102–120. doi: 10.1016/S0169-328X(02)00519-3. [DOI] [PubMed] [Google Scholar]
- 37.Yang Q, She H, Gearing M, Colla E, Lee M, Shacka JJ, et al. Regulation of neuronal survival factor MEF2D by chaperone-mediated autophagy. Science. 2009;323:124–127. doi: 10.1126/science.1166088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Szczepanek K, Lesnefsky EJ, Larner AC. Multi-tasking: nuclear transcription factors with novel roles in the mitochondria. Trends Cell Biol. 2012;22:429–437. doi: 10.1016/j.tcb.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sonia Angeline M, Sarkar A, Anand K, Ambasta RK, Kumar P. Sesamol and naringenin reverse the effect of rotenone-induced PD rat model. Neuroscience. 2013;254:379–394. doi: 10.1016/j.neuroscience.2013.09.029. [DOI] [PubMed] [Google Scholar]
- 40.Zhang Q, Xie H, Ji Z, He R, Xu M, He Y, et al. Cdk5/p25 specific inhibitory peptide TFP5 rescues the loss of dopaminergic neurons in a sub-acute MPTP induced PD mouse model. Neurosci Lett. 2016;632:1–7. doi: 10.1016/j.neulet.2016.08.023. [DOI] [PubMed] [Google Scholar]
- 41.Wang B, Cai Z, Lu F, Li C, Zhu X, Su L, et al. Destabilization of survival factor MEF2D mRNA by neurotoxin in models of Parkinson’s disease. J Neurochem. 2014;130:720–728. doi: 10.1111/jnc.12765. [DOI] [PubMed] [Google Scholar]
- 42.She H, Mao Z. Regulation of myocyte enhancer factor-2 transcription factors by neurotoxins. Neurotoxicology. 2011;32:563–566. doi: 10.1016/j.neuro.2011.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim MK, Kim SC, Kang JI, Hyun JH, Boo HJ, Eun SY, et al. 6-Hydroxydopamine-induced PC12 cell death is mediated by MEF2D down-regulation. Neurochem Res. 2011;36:223–231. doi: 10.1007/s11064-010-0309-x. [DOI] [PubMed] [Google Scholar]
- 44.Perlmann T, Jansson L. A novel pathway for vitamin A signaling mediated by RXR heterodimerization with NGFI-B and NURR1. Genes Dev. 1995;9:769–782. doi: 10.1101/gad.9.7.769. [DOI] [PubMed] [Google Scholar]
- 45.Le W, Conneely OM, Zou L, He Y, Saucedo-Cardenas O, Jankovic J, et al. Selective agenesis of mesencephalic dopaminergic neurons in Nurr1-deficient mice. Exp Neurol. 1999;159:451–458. doi: 10.1006/exnr.1999.7191. [DOI] [PubMed] [Google Scholar]
- 46.Jankovic J, Chen S, Le WD. The role of Nurr1 in the development of dopaminergic neurons and Parkinson’s disease. Prog Neurobiol. 2005;77:128–138. doi: 10.1016/j.pneurobio.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 47.Watanabe T, Sekine S, Naguro I, Sekine Y, Ichijo H. Apoptosis signal-regulating kinase 1 (ASK1)-p38 pathway-dependent cytoplasmic translocation of the orphan nuclear receptor NR4A2 is required for oxidative stress-induced necrosis. J Biol Chem. 2015;290:10791–10803. doi: 10.1074/jbc.M114.623280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu W, Gao Y, Chang N. Nurr1 overexpression exerts neuroprotective and anti-inflammatory roles via down-regulating CCL2 expression in both in vivo and in vitro Parkinson’s disease models. Biochem Biophys Res Commun. 2017;482:1312–1319. doi: 10.1016/j.bbrc.2016.12.034. [DOI] [PubMed] [Google Scholar]
- 49.Saijo K, Winner B, Carson CT, Collier JG, Boyer L, Rosenfeld MG, et al. A Nurr1/CoREST pathway in microglia and astrocytes protects dopaminergic neurons from inflammation-induced death. Cell. 2009;137:47–59. doi: 10.1016/j.cell.2009.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bensinger SJ, Tontonoz P. A Nurr1 pathway for neuroprotection. Cell. 2009;137:26–28. doi: 10.1016/j.cell.2009.03.024. [DOI] [PubMed] [Google Scholar]
- 51.Kadkhodaei B, Alvarsson A, Schintu N, Ramskold D, Volakakis N, Joodmardi E, et al. Transcription factor Nurr1 maintains fiber integrity and nuclear-encoded mitochondrial gene expression in dopamine neurons. Proc Natl Acad Sci U S A. 2013;110:2360–2365. doi: 10.1073/pnas.1221077110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lin X, Parisiadou L, Sgobio C, Liu G, Yu J, Sun L, et al. Conditional expression of Parkinson’s disease-related mutant alpha-synuclein in the midbrain dopaminergic neurons causes progressive neurodegeneration and degradation of transcription factor nuclear receptor related 1. J Neurosci. 2012;32:9248–9264. doi: 10.1523/JNEUROSCI.1731-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang YX, Latchman DS. Nurr1 transcriptionally regulates the expression of alpha-synuclein. Neuroreport. 2008;19:867–871. doi: 10.1097/WNR.0b013e3282ffda48. [DOI] [PubMed] [Google Scholar]
- 54.Pan T, Zhu W, Zhao H, Deng H, Xie W, Jankovic J, et al. Nurr1 deficiency predisposes to lactacystin-induced dopaminergic neuron injury in vitro and in vivo. Brain Res. 2008;1222:222–229. doi: 10.1016/j.brainres.2008.05.022. [DOI] [PubMed] [Google Scholar]
- 55.Tan EK, Chung H, Zhao Y, Shen H, Chandran VR, Tan C, et al. Genetic analysis of Nurr1 haplotypes in Parkinson’s disease. Neurosci Lett. 2003;347:139–142. doi: 10.1016/S0304-3940(03)00539-1. [DOI] [PubMed] [Google Scholar]
- 56.Brouwer A, ten Berge D, Wiegerinck R, Meijlink F. The OAR/aristaless domain of the homeodomain protein Cart1 has an attenuating role in vivo. Mech Dev. 2003;120:241–252. doi: 10.1016/S0925-4773(02)00416-1. [DOI] [PubMed] [Google Scholar]
- 57.Cazorla P, Smidt MP, O’Malley KL, Burbach JP. A response element for the homeodomain transcription factor Ptx3 in the tyrosine hydroxylase gene promoter. J Neurochem. 2000;74:1829–1837. doi: 10.1046/j.1471-4159.2000.0741829.x. [DOI] [PubMed] [Google Scholar]
- 58.Tang T, Li Y, Jiao Q, Du X, Jiang H. Cerebral dopamine neurotrophic factor: a potential therapeutic agent for Parkinson’s disease. Neurosci Bull. 2017 doi: 10.1007/s12264-017-0123-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kim J, Inoue K, Ishii J, Vanti WB, Voronov SV, Murchison E, et al. A microRNA feedback circuit in midbrain dopamine neurons. Science. 2007;317:1220–1224. doi: 10.1126/science.1140481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rekaik H, Blaudin de The FX, Prochiantz A, Fuchs J, Joshi RL. Dissecting the role of Engrailed in adult dopaminergic neurons–Insights into Parkinson disease pathogenesis. FEBS Lett. 2015;589:3786–3794. doi: 10.1016/j.febslet.2015.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Simon HH, Thuret S, Alberi L. Midbrain dopaminergic neurons: control of their cell fate by the engrailed transcription factors. Cell Tissue Res. 2004;318:53–61. doi: 10.1007/s00441-004-0973-8. [DOI] [PubMed] [Google Scholar]
- 62.Kordower JH, Olanow CW, Dodiya HB, Chu Y, Beach TG, Adler CH, et al. Disease duration and the integrity of the nigrostriatal system in Parkinson’s disease. Brain. 2013;136:2419–2431. doi: 10.1093/brain/awt192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Alavian KN, Sgado P, Alberi L, Subramaniam S, Simon HH. Elevated P75NTR expression causes death of engrailed-deficient midbrain dopaminergic neurons by Erk1/2 suppression. Neural Dev. 2009;4:11. doi: 10.1186/1749-8104-4-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li Y, Liu H, Lai C, Su Z, Heng B, Gao S. Repression of engrailed 2 inhibits the proliferation and invasion of human bladder cancer in vitro and in vivo. Oncol Rep. 2015;33:2319–2330. doi: 10.3892/or.2015.3858. [DOI] [PubMed] [Google Scholar]
- 65.Alvarez-Fischer D, Fuchs J, Castagner F, Stettler O, Massiani-Beaudoin O, Moya KL, et al. Engrailed protects mouse midbrain dopaminergic neurons against mitochondrial complex I insults. Nat Neurosci. 2011;14:1260–1266. doi: 10.1038/nn.2916. [DOI] [PubMed] [Google Scholar]
- 66.Stettler O, Joshi RL, Wizenmann A, Reingruber J, Holcman D, Bouillot C, et al. Engrailed homeoprotein recruits the adenosine A1 receptor to potentiate ephrin A5 function in retinal growth cones. Development. 2012;139:215–224. doi: 10.1242/dev.063875. [DOI] [PubMed] [Google Scholar]
- 67.Tao K, Wang B, Feng D, Zhang W, Lu F, Lai J, et al. Salidroside protects against 6-hydroxydopamine-induced cytotoxicity by attenuating ER stress. Neurosci Bull. 2016;32:61–69. doi: 10.1007/s12264-015-0001-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jha SK, Jha NK, Kar R, Ambasta RK, Kumar P. p38 MAPK and PI3K/AKT signalling cascades inParkinson’s disease. Int J Mol Cell Med. 2015;4:67–86. [PMC free article] [PubMed] [Google Scholar]
- 69.Han J, Molkentin JD. Regulation of MEF2 by p38 MAPK and its implication in cardiomyocyte biology. Trends Cardiovasc Med. 2000;10:19–22. doi: 10.1016/S1050-1738(00)00039-6. [DOI] [PubMed] [Google Scholar]
- 70.Jacobsen KX, MacDonald H, Lemonde S, Daigle M, Grimes DA, Bulman DE, et al. A Nurr1 point mutant, implicated in Parkinson’s disease, uncouples ERK1/2-dependent regulation of tyrosine hydroxylase transcription. Neurobiol Dis. 2008;29:117–122. doi: 10.1016/j.nbd.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 71.Domanskyi A, Geissler C, Vinnikov IA, Alter H, Schober A, Vogt MA, et al. Pten ablation in adult dopaminergic neurons is neuroprotective in Parkinson’s disease models. FASEB J. 2011;25:2898–2910. doi: 10.1096/fj.11-181958. [DOI] [PubMed] [Google Scholar]
- 72.Mincheva-Tasheva S, Soler RM. NF-kappaB signaling pathways: role in nervous system physiology and pathology. Neuroscientist. 2013;19:175–194. doi: 10.1177/1073858412444007. [DOI] [PubMed] [Google Scholar]
- 73.Li S, Wang Y, Wang F, Hu LF, Liu CF. A new perspective for Parkinson’s disease: circadian rhythm. Neurosci Bull. 2017;33:62–72. doi: 10.1007/s12264-016-0089-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Xiao Q, Chen S, Le W. Hyposmia: a possible biomarker of Parkinson’s disease. Neurosci Bull. 2014;30:134–140. doi: 10.1007/s12264-013-1390-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Eller M, Williams DR. Biological fluid biomarkers in neurodegenerative parkinsonism. Nat Rev Neurol. 2009;5:561–570. doi: 10.1038/nrneurol.2009.135. [DOI] [PubMed] [Google Scholar]
- 76.Parnetti L, Castrioto A, Chiasserini D, Persichetti E, Tambasco N, El-Agnaf O, et al. Cerebrospinal fluid biomarkers in Parkinson disease. Nat Rev Neurol. 2013;9:131–140. doi: 10.1038/nrneurol.2013.10. [DOI] [PubMed] [Google Scholar]
- 77.Lin X, Shah S, Bulleit RF. The expression of MEF2 genes is implicated in CNS neuronal differentiation. Brain Res Mol Brain Res. 1996;42:307–316. doi: 10.1016/S0169-328X(96)00135-0. [DOI] [PubMed] [Google Scholar]
- 78.Liu H, Wei L, Tao Q, Deng H, Ming M, Xu P, et al. Decreased NURR1 and PITX3 gene expression in Chinese patients with Parkinson’s disease. Eur J Neurol. 2012;19:870–875. doi: 10.1111/j.1468-1331.2011.03644.x. [DOI] [PubMed] [Google Scholar]
- 79.Le W, Pan T, Huang M, Xu P, Xie W, Zhu W, et al. Decreased NURR1 gene expression in patients with Parkinson’s disease. J Neurol Sci. 2008;273:29–33. doi: 10.1016/j.jns.2008.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Montarolo F, Perga S, Martire S, Navone DN, Marchet A, Leotta D, et al. Altered NR4A subfamily gene expression level in peripheral blood of Parkinson’s and Alzheimer’s disease patients. Neurotox Res. 2016;30:338–344. doi: 10.1007/s12640-016-9626-4. [DOI] [PubMed] [Google Scholar]
- 81.Bergman O, Hakansson A, Westberg L, Nordenstrom K, Carmine Belin A, Sydow O, et al. PITX3 polymorphism is associated with early onset Parkinson’s disease. Neurobiol Aging. 2010;31:114–117. doi: 10.1016/j.neurobiolaging.2008.03.008. [DOI] [PubMed] [Google Scholar]
- 82.Bell D, Bell A, Roberts D, Weber RS, El-Naggar AK. Developmental transcription factor EN1–a novel biomarker in human salivary gland adenoid cystic carcinoma. Cancer. 2012;118:1288–1292. doi: 10.1002/cncr.26412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Morgan R, Bryan RT, Javed S, Launchbury F, Zeegers MP, Cheng KK, et al. Expression of Engrailed-2 (EN2) protein in bladder cancer and its potential utility as a urinary diagnostic biomarker. Eur J Cancer. 2013;49:2214–2222. doi: 10.1016/j.ejca.2013.01.019. [DOI] [PubMed] [Google Scholar]
- 84.Marszall MP, Sroka W, Adamowski M, Slupski P, Jarzemski P, Siodmiak J, et al. Engrailed-2 protein as a potential urinary prostate cancer biomarker: a comparison study before and after digital rectal examination. Eur J Cancer Prev. 2015;24:51–56. doi: 10.1097/CEJ.0000000000000046. [DOI] [PubMed] [Google Scholar]
- 85.Henchcliffe C, Dodel R, Beal MF. Biomarkers of Parkinson’s disease and dementia with Lewy bodies. Prog Neurobiol. 2011;95:601–613. doi: 10.1016/j.pneurobio.2011.09.002. [DOI] [PubMed] [Google Scholar]