Abstract
Dynamic changes of the post-translational O-GlcNAc modification (O-GlcNAcylation) are controlled by O-linked β-N-acetylglucosamine (O-GlcNAc) transferase (OGT) and the glycoside hydrolase O-GlcNAcase (OGA) in cells. O-GlcNAcylation often occurs on serine (Ser) and threonine (Thr) residues of the specific substrate proteins via the addition of O-GlcNAc group by OGT. It has been known that O-GlcNAcylation is not only involved in many fundamental cellular processes, but also plays an important role in cancer development through various mechanisms. Recently, accumulating data reveal that O-GlcNAcylation at histones or non-histone proteins can lead to the start of the subsequent biological processes, suggesting that O-GlcNAcylation as ‘protein code’ or ‘histone code’ may provide recognition platforms or executive instructions for subsequent recruitment of proteins to carry out the specific functions. In this review, we summarize the interaction of O-GlcNAcylation and epigenetic changes, introduce recent research findings that link crosstalk between O-GlcNAcylation and epigenetic changes, and speculate on the potential coordination role of O-GlcNAcylation with epigenetic changes in intracellular biological processes.
Keywords: O-GlcNAcylation, post-translational modification, histone modification, epigenetics
Introduction
A large number of intracellular proteins often undergo post-translational modifications. As one of the highly dynamic, inducible, and reversible post-translational modification, O-GlcNAc on Ser/Thr residues of nuclear, cytoplasmic, and mitochondrial proteins is ubiquitous in eukaryotic cells (Wells and Hart, 2003; Bullen et al., 2014). Dynamic changes of O-GlcNAcylation are controlled by OGT and OGA (Gao et al., 2001). The sugar nucleotide UDP-GlcNAc serves as a donor for O-GlcNAc addition to nucleocytoplasmic proteins whereas OGA removes the O-GlcNAc group from proteins (Mailleux et al., 2016). Numerous studies have demonstrated that O-GlcNAcylation is involved in many fundamental cellular processes, including gene transcription (Ha and Lim, 2015), cell signaling (Cassey, 1995), and apoptosis (Butkinaree et al., 2008). More than those, many transcriptional factors, tumor suppressors, oncogenes, and cell receptors have been found to be tightly associated with O-GlcNAcylation in tumorigenesis (Bond and Hanover, 2015; Charoensuksai et al., 2015). Recently, accumulating data reveal that O-GlcNAcylation is frequently related to epigenetic changes. For example, O-GlcNAcylation at serine 112 site of histone H2B (H2BS112) by OGT stimulates H2B at lysine 120 (H2BK120) ubiquitination that further activates gene transcription such as ring finger protein 20 (RNF20) (Fujiki et al., 2011; Nakamura et al., 2011), suggesting the existence of complicated coordinative role between O-GlcNAcylation and the other histone post-translational modifications. In this review, we focus on the O-GlcNAcylation by OGT, summarize the current understanding of the crosstalk between O-GlcNAcylation and epigenetic changes in cells.
Intracellular O-GlcNac modification is regulated by OGT and OGA
As is well known, O-GlcNAcylation is the process of adding O-GlcNAc via an O-linkage to Ser or Thr residues of intracellular proteins. Thousands of O-GlcNAcylated proteins have been found to implicate in the regulation of core processes including cellular metabolism, growth, and other important function (Copeland et al., 2008; Hanover et al., 2012). In cells, O-GlcNAcylation is often referred to as a nutrient sensor. The sensitivity of nutrient is based on the levels of UDP-GlcNAc, the donor substrate for protein O-GlcNAcylation, while UDP-GlcNAc is the end product of the hexosamine biosynthetic pathway (HBP). Thus, the levels of UDP-GlcNAc in cells are closely associated with flux through the HBP (Marshall et al., 1991). According to the current reports, OGT and OGA are the only two enzymes to control intracellular O-GlcNAcylation.
OGT is highly conserved from Caenorhabditis elegans to human. Functional domain studies have been clarified that full length of OGT is composed of two important regions: an N-terminal tetratricopeptide-repeat (TPR) super-helical structure and a C-terminal catalytic domain (Jinek et al., 2004; Kreppel and Hart, 1999). Wherein, TPR domain is involved in substrate recognition through serving as protein:protein docking sites for substrate targeting proteins (Lubas and Hanover, 2000), and C-terminal catalytic domain binds to donor UDP-GlcNAc and is responsible for catalyzing the substrate targeting proteins by adding O-GlcNAc group (Lazarus et al., 2011). In cells, three different transcripts encoding nucleocytoplasmic isoform (ncOGT, 116 kDa), the mitochondrial isoform (mOGT, 103 kDa), and the short isoform (sOGT, 75 kDa) are produced by alternative gene splicing. C-terminal of these isoforms is completely the same, and only differs in the number of N-terminal TPRs (Hanover et al., 2003). As shown in Fig. 1, 13.5, 9, and 3 TPRs are contained in ncOGT, mOGT, and sOGT, respectively. The different localizations and the number of TPRs in each isoform suggest that each OGT isoform possesses its own substrate specificity and function (Lazarus et al., 2011; Love et al., 2003; Trapannone et al., 2016; Sacoman et al., 2017; Shin et al., 2011; Riu et al., 2008).
Figure 1.
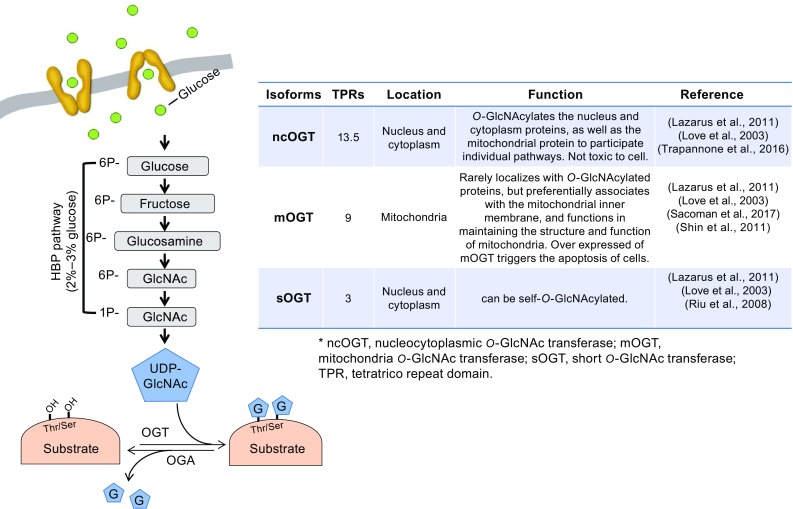
The process of O -GlcNAc modification and the molecular structure of OGT isoforms. A range of 2%–3% glucose uptake by the cells goes through the HBP pathway, experiences a series of modification and synthesize UDP-GlcNAc, the substrates of O-GlcNAc reaction. OGT/OGA are the enzymes in charge of add/remove UDP-GlcNAc at Ser or Thr residues respectively to control the balance of intracellular O-GlcNAcylation level. The molecular structure of 3 isoforms of OGT is highly conserved. By alternative splicing, 3 isoforms of OGT including nucleocytoplasmic isoform (ncOGT, 116 kDa), the mitochondrial isoform (mOGT, 103 kDa), and the short isoform (sOGT, 75 kDa) are produced.
Another important enzyme that affects intracellular O-GlcNAcylation is O-GlcNAc hydrolase OGA. OGA was initially isolated from crude cellular extract, and it catalyzes hydrolytic cleavage of O-GlcNAc from proteins (Gao et al., 2001). Full-length OGA is composed of N-terminal N-acetyl-β-D-glucosamindase domain and C-terminal with acetyltransferase-like (AT) domain (Toleman et al., 2004). Alternative gene splicing results in two different lengths of OGA isoforms: one isoform has a 916 amino acid (OGA-L) and predominantly localizes in the cytoplasm, while the other only has a 677 amino acid (OGA-S), and localizes in nuclear and lipid-droplet (Comtesse et al., 2001).
In view of the OGT and OGA are the only enzymes that involved in O-GlcNAc post-translational modification in cells, it is not difficult to understand that both enzymes are essential to cell growth, ontogeny, and survival in mammals. As reported, OGT modulates the somatic cell function and embryo viability through O-GlcNAc modification of X-chromosome-linked proteins in mice (O’Donnell et al., 2004). Furthermore, global O-GlcNAcylation of rat brain protein is relatively high at early development stage, and gradually decreases during the development, suggesting the coordinative role between OGT and OGA in cell growth and differentiation (Liu et al., 2012).
Mode of OGT in cells
Although the expression level of OGT in the pancreas and brain is higher than other tissues (Kreppel et al., 1997), OGT is ubiquitously expressed in all tissues, showing the essential role of OGT in cells. In line with this view, numerous studies have confirmed the broad presence of O-GlcNAc modification involved in many fundamental cellular processes such as cell morphogenesis, cell signaling, apoptosis, and transcription (Lazarus et al., 2006). It is worth noting that, in addition to free form, OGT can also be assembled in complex to participate in intracellular biological processes.
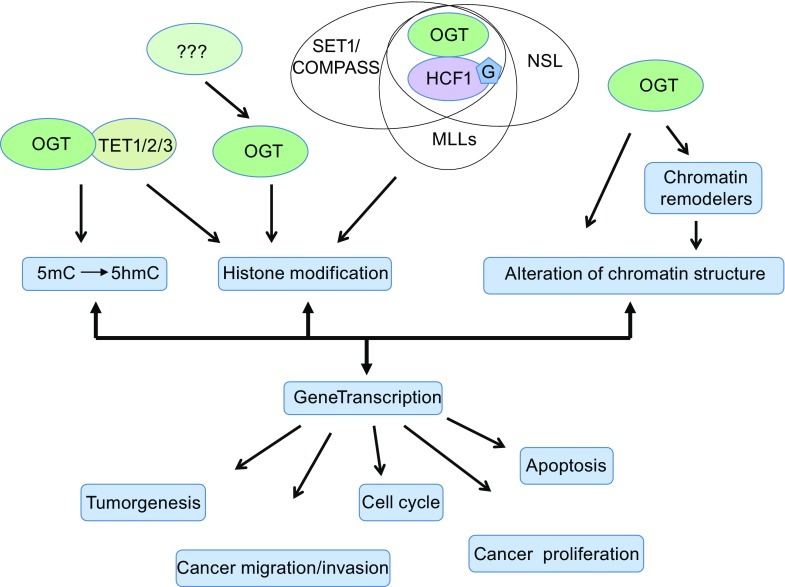
Ten-eleven translocation (TET) family enzymes including TET1, TET2, TET3 catalyze the conversion of 5-methylcytosine (mC) to 5-hydroxymethylation (hmC) (Ito et al., 2011). Recent studies have found that each enzyme of TET family (TETs) can be complexed with OGT to play a cellular function. The complexes of TET2/TET3 with OGT can O-GlcNAcylate the host cell factor 1 (HCF1), a shared subunit of the methyltransferase SET1/COMPASS and histone acetyltransferase NSL complexes, therefore impact the histone H3K4 tri-methylation (H3K4me3) via SET1/COMPASS (Deplus et al., 2013). In-depth study demonstrated that the TET3-OGT complex enhances the recruitment of OGT to chromatin through its stabilization (Ito et al., 2014). TET1, being no exception, can interact with OGT, and the TET1-OGT complex regulates the recruitment of OGT to chromatin and TET1 activity in embryonic stem cells (Vella et al., 2013). Thus, we speculate that the TETs-OGT crosstalk not only help OGT to raise in chromatin, but also regulates gene transcription by affecting histone modification and chromatin structure. In addition to TETs proteins, the function of OGT is also connected to other intracellular proteins. For example, a heterotrimeric complex formed by URI, OGT, and PP1γ regulates cellular O-GlcNAcylation in response to metabolic stress (Burén et al., 2016). However, in C2C12 skeletal muscle cells, OGT and AMPK (AMP-activated protein kinase) cooperatively regulate nutrient-sensitive intracellular metabolism, growth, and proliferation (Bullen et al., 2014). Furthermore, a stable complex of MLL5 (mixed lineage leukemia 5), OGT, and USP7 (ubiquitin specific protease 7) was verified by Zhang Y’s group (Ding et al., 2015), and the coordinative expression between three proteins was observed in primary cervical adenocarcinomas. The above results suggest that OGT participates in intracellular different biological processes via forming different protein complexes.
We previously described a second human MOF (males absent on the first)-containing histone acetyltransferases NSL (non-specific lethal) complex, which comprises 9 subunits and can acetylate histone H4 at lysine 16 (K16), 5 (K5), and 8 (K8) (Cai et al., 2010). Although the function of OGT in NSL complex is unclear, the coordinative role of OGT and HCF1 has been reported. For instance, O-GlcNAcylation by OGT to the specific repeats region of HCF-1 may provide instructions for the HCF-1 proteolysis (Capotosti et al., 2011). In addition, the interaction of OGT/O-GlcNAcylation and HCF1 stabilizes PGC-1α, a master regulator of gluconeogenesis. Knockdown both OGT and HCF1 significantly decreases the expression of gluconeogenic genes induced by PGC-1α in fed mice, suggesting the OGT/HCF1 sub-complex-mediated cooperative regulation in gluconeogenesis (Ruan et al., 2012). Also, OGT and HCF1 were characterized as major cellular binding partner of THAP1, a sequence-specific DNA binding factor (Mazars et al., 2010). Besides those stable complexes, OGT can also form a temporary complex with OGA, PP1γ, and Aurora B to regulate the post-translational status of vimentin under the coordination of O-GlcNAcylation and phosphorylation (Slawson et al., 2008), indicating the manifold functions of OGT assembled in complexes (Fig. 2).
Figure 2.
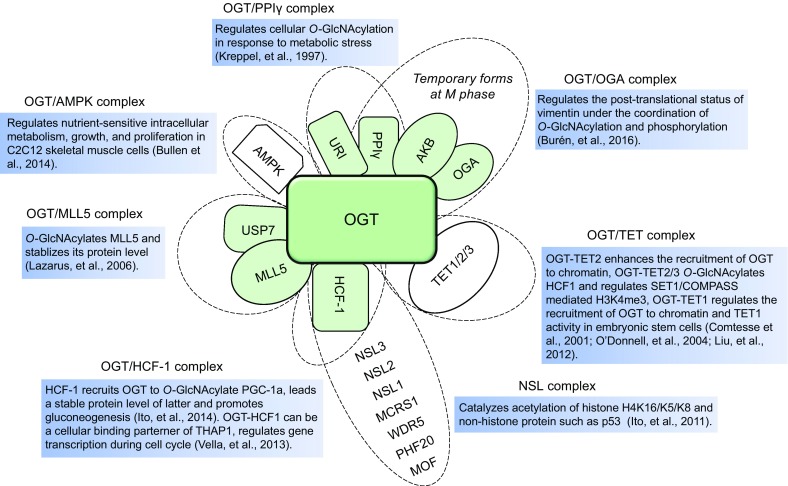
Regulation and function of OGT-mediated protein complexes
Crosstalk between O-GlcNAcylation and epigenetic mechanisms
Changes in chromatin structure are mainly caused by N-terminal tails of histone modification and ATP-dependent chromatin remodelers (Gerhold and Gasser, 2014; Längst and Manelyte, 2015; Jin et al., 2005). These complexes regulate the gene expression by epigenetic mechanism in most cellular biological processes (Bannister and Kouzarides, 2011). Recent research evidence shows that O-GlcNAcylation by OGT is often implicated in chromatin-mediated processes in a coordinated manner (Fig. 3). According to OGT is an important and indispensable enzyme in cell metabolic pathway, unbalanced O-GlcNAc modification on substrate proteins leads to various kinds of diseases, such as diabetes, neurologic disorders, cardiovascular disease, and cancer (Gao et al., 2001). Especially, high O-GlcNAc modification has been found in breast cancer (Caldwell et al., 2010; Gu et al., 2010; Krzeslak et al., 2012), prostate cancer (Lynch et al., 2012; Kamigaito et al., 2014; Gu et al., 2014), lung and colon cancer (Mi et al., 2011), esophageal cancer (Qiao et al., 2012), and hepatocellular carcinoma (Zhu et al., 2012). More importantly, numerous tumor-related proteins are also modified and regulated by OGT, such as oncoprotein c-Myc (Chou et al., 1995; Itkonen et al., 2013) and tumor suppressor gene p53 (Yang et al., 2006; de Queiroz et al., 2016). The role of OGT/O-GlcNAcylation in some important signal transduction pathways cannot be ignored. In cholangiocarcinoma (CCA) cells, matrix metalloproteinases (MMP)-mediated migration and invasion of CCA cells are regulated by O-GlcNAcylation through affecting the nuclear translocation of NFκB (Phoomak et al., 2016).
Figure 3.
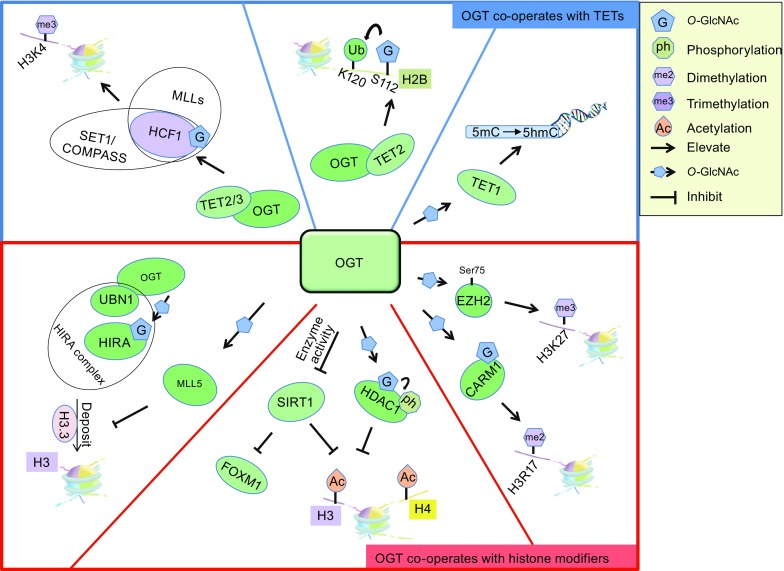
Crosstalk between O -GlcNAcylation and chromatin remodelers. Extensive connections of O-GlcNAcylation with chromatin modifiers are detected in cells
Coordination of O-GlcNAcylation and TET proteins
Given that TETs implicated in controlling genome-wide DNA methylation and cellular differentiation, the impact of TETs proteins in tumorigenesis has been investigated (Shi et al., 2013; Fu et al., 2013). Subsequent research found that TET1 is a fusion partner of mixed-lineage leukemia (MLL)-rearranged acute myeloid leukemia (AML) (Ono et al., 2002; Lorsbach et al., 2003). More importantly, TET genes, especially TET2 are frequently mutated in various cancers, including myelodysplastic syndrome (MDS), cohort of chronic myelomonocytic leukemia (CMML), primary and secondary AML, blastic plasmacytoid dendritic neoplasm (Scourzic et al., 2015; Delhommeau et al., 2009; Langemeijer et al., 2009; Tefferi et al., 2009). However, transcriptional regulation of TETs might be associated with its interplay with histone modifications. Supporting this speculation, OGT-dependent H2BS112 O-GlcNAcylation is regulated by TET2 in embryonic stem cells (Chen et al., 2013). While genome-wide analysis confirms that O-GlcNAcylation of H2BS112 is widely distributed over chromosomes, and is frequently co-localizes with H2BK120 mono-ubiquitination (Fujiki et al., 2011), suggesting the complexity among TET2, O-GlcNAcylation, and H2BK120 ubiquitination.
As we mentioned earlier, TETs proteins function to recruit OGT to chromatin. Thus, it is not difficult to understand that knocking down TETs proteins declines the O-GlcNAcylation of chromatin proteins. In mouse embryonic stem (ES) cells, TET1-OGT interaction links to multiple chromatin regulators including Sin3A and NuRD complexes (Shi et al., 2013). Follow-up studies clarify that TET1 with those chromatin regulators together commands the level of local 5hmC to regulate the expression of target genes (Wu et al., 2011; Williams et al., 2011; Yildirim et al., 2011), suggesting the existence of crosstalk between TET1-OGT and TET1-chromatin remodelers in gene transcription. Consistent with this conjecture, knocking down either TET1 or OGT in mouse ES cells leads to reduced genomic targeting of both Ezh2 (enhancer of Zeste 2 polycomb repressive complex 2 subunit, a subunit of PRC2 complex) and Sin3A, indicating the importance of TET1-OGT coordination in repressing developmental genes (Shi et al., 2013). In drosophila, polycomb group gene super sex comb (sxc) that regulates the bithorax complex is turned out to encode OGT, and play essential role in polycomb repression (Gambetta et al., 2009; Ingham, 1984). Confirming to this conclusion, TET1-OGT complex represses gene transcription through interaction with Sin3A/HDAC1/Sirt1 and PCR2/Ezh2 complexes (Hardivillé and Hart, 2012; Williams et al., 2012). Similar effects are reflected in TET2-OGT or TET3-OGT interaction. They can target OGT to transcriptional start site (TSS), and in collaboration with HCF1 recruit SET1/COPASS complexes, therefore activate gene transcription through histone H3K4me3 at target promoters (Deplus et al., 2013). TETs-OGT/O-GlcNAcylation is involved in some cancer development. In 5-fluorouracilresistant colon cancer cells (SNUC5/5-FUR), highly expressed OGT binds to TET1 and recruits to the Nrf2 (nuclear factor erythroid 2-related factor 2) promoter region, suggesting the role of OGT in TET1-mediated Nrf2 expression (Kang et al., 2016). Taken together, TETs proteins in collaboration with OGT play a critical role in regulating chromatin structure and gene transcription.
Coordination of O-GlcNAcylation and other modifiers on histones
Intracellular biological processes are extremely complex, and are often regulated by two or more histone modifiers in a co-ordinated manner. O-GlcNAcylation-mediated mechanism is no exception, often in collaboration with other histone modifications to regulate the biological processes in cells. Besides all four histones (H2A, H2B, H3, and H4) can be O-GlcNAcylated by OGT (Zhang et al., 2011; Sakabe et al., 2010) (Table 1), the accurate modified sites are also identified gradually. Genome-wide studies confirm that O-GlcNAcyaltion of histone H2A at Ser40 is dramatically changed during the differentiation in mouse trophoblast stem cells (Hirosawa et al., 2016). However, O-GlcNAcylation on histone H2A at Thr101 can relax chromatin structure through destabilizing H2A-H2B dimer (Lercher et al., 2015). On the other hand, occurrence of O-GlcNAcylation on histone variant H2AX at Ser139 is often detected in DNA damage foci (Chen and Yu, 2016). Obviously, O-GlcNAcylation on histone H2A at different sites is tightly associated with different intracellular functions. In cells, O-GlcNAcylation on histone H2BS112 may preserve a stable chromatin at the early stage of adipocyte differentiation, thus repressing gene transcription in cell fate (Ronningen et al., 2015). And H2BS112-O-GlcNAcylation facilitates H2B at K120 ubiquitination, the latter further acts as a platform recruiting the SET1/COMPASS complex binding to histone H3, thereby activates gene transcription through histone H3K4me3 (Deplus et al., 2013).
Table 1.
O-GlcNAcylation sites and functions of histone tails. O-GlcNAc modification is observed in all four histones (H2A, H2B, H3 and H4) as well as histone variants H3.3 at indicated sites.
| Histones | O-GlcNAcylated sites | Functions | References |
|---|---|---|---|
| H2A | Ser40 | Tightly relates with the differentiation in mouse trophoblast stem cells | (Hirosawa et al., 2016) |
| Thr101 | Destabilizes H2A-H2B dimer, further relaxes the structure of chromatin | (Lercher et al., 2015) | |
| H2AX | Ser139 | Co-localizes with DNA damage foci, may function in DNA damage repair | (Chen and Yu 2016) |
| H2B | Ser112 | Preserves a stable chromatin and represses gene transcription at the early stage of adipocyte differentiation Promotes H2BK120 ubiquitination, participates the regulation of H3K4me3 and gene transcription |
(Ronningen et al., 2015) (Deplus et al., 2013) |
| Ser36 | May be a part of the histone code | (Sakabe et al., 2010) | |
| H3 | Thr32 | Increases the phosphorylation of Thr32, Ser28, and Ser10, which are the specific mark of mitosis | (Zhang et al., 2011) (Fong et al., 2005) |
| Ser10 | Competitively reduces the levels of H3S10 phosphorylation, therefore regulates the pathway that H3S10P involved in, such as passing the G2-M phase check point, regulating the H4K16ac | (Zhang et al., 2011) | |
| H4 | Ser47 | May be a part of the histone code | (Sakabe et al., 2010) |
Ser40/139/112/10/36/47, serine residues 40/139/112/10/36/47; Thr101/32, threonine residue 101/32; H2BK120, H2B lysine 120; H3K4me3, H3 lysine 4 tri-methylation; H4K16ac, H4 lysine 16 acetylation
Interestingly, competitive modification between O-GlcNAcylation and phosphorylation on Ser/Thr residues of substrate proteins may be closely related with functional switch. For example, higher O-GlcNAcylated H3 at Thr32 is observed during interface than mitosis. Further research demonstrates that O-GlcNAcylated Thr32 reduces mitosis-specific phosphorylation of Thr32, Ser28, and Ser10 on H3, suggesting the switching function of O-GlcNAcylation-mediated Thr32 in mitosis (Zhang et al., 2011; Fong et al., 2005). Importantly, according to O-GlcNAcylation of H3 can competitively reduce the level of H3S10 phosphorylation, but removal of O-GlcNAc from H3S10 is required for entering mitosis during the G2-M transition phase (Zhang et al., 2011), H3S10 has been considered as a molecular checkpoint for entering mitosis (Van Hooser et al., 1998). Further study confirmed that H3S10 phosphorylation provides a binding platform for the phospho-binding 14-3-3 proteins and histone acetyltransferase MOF to trigger acetylation of histone H4 at lysine 16 (H4K16ac), and H3S10 phosphorylation and H4K16ac further coordinatively regulate the binding site for bromodomain protein BRD4 (Zippo et al., 2009). Moreover, in addition to up-regulation of H3S10 phosphorylation in hepatocellular carcinoma and primary lung cancer (Zhu et al., 2016), there is sufficient evidence to prove that H3S10 phosphorylation is responsible for neoplastic cell transformation and oncogene c-fos/c-Jun activation (Choi et al., 2005), suggesting the important coordinative role between O-GlcNAcylation and other histone modifications in tumor development.
Coordination of O-GlcNAcylation and the other chromatin remodelers
In addition to direct glycosylation of histones, OGT can also affect other histones modifications by crosslinking other chromatin remodelers. In line with this view, overexpression of OGT raises global H3K9ac and H3K27me3 level in cells, suggesting that OGT may collaborate with other chromatin remodelers to regulate histone modification (Sakabe and Hart, 2010). Histone H3 is a well-known specific substrate of CARM1 (a co-activator-associated argine methyltrnsferse 1), however the arginine methylation at H3R17 was regulated by O-GlcNAc modification of CARM1 (Charoensuksai et al., 2015; Sakabe and Hart, 2010; Schurter et al., 2001). Interestingly, the crosstalk between OGT and chromatin remodelers also exists in DNA damage repair pathway. O-GlcNAc-modified H2AX and MDC1 (mediator of DNA damage check point 1) are enriched at DNA damage foci, but suppress the expansion of DNA damage-produced phosphorylation at H2AX Ser139 site on the chromatin (Chen and Yu, 2016). What happens on histone variant H3.3 is another example. OGT regulates histone chaperone HIRA complex via O-GlcNAc modification, and subsequently deposits histone variant H3.3 to genic regions, then further governs H3.3 nucleosome assembly and cell senescence (Lee and Zhang, 2016; Ricketts and Marmorstein, 2016). Although the mechanism of targeting of histone H3.3 deposition at specific loci in whole genome remains largely unclear, histone H3.3 has been considered as epigenetic regulator that is dispensable for proper expression of genes (Kato et al., 2015). Furthermore, the function of histone variant H3.3 is also regulated by O-GlcNAcylated MLL5 which is a specific histone H3K4 tri-methylase (Gallo et al., 2015). In cells, O-GlcNAcylated MLL5 represses histone H3.3 expression and facilitates the self-renewal of adult glioblastoma (Ding et al., 2015; Sebastian et al., 2009). On the other hand, histone H3.3 substitution and mutation are found in several tumors such as bone tumors and glioblastoma (Kato et al., 2015). However, whether substituted and mutated H3.3 is related to OGT-mediated O-GlcNAcylation remains to be further investigated.
Crosstalk of O-GlcNAcylation and epigenetics in cancer
Intracellular unbalanced epigenetic changes or abnormal O-GlcNAcylation levels can lead to various diseases. However, the evidence of crosslink between epigenetic changes and O-GlcNAcylation in tumorigenesis is gradually increased. For example, in addition to modulating the activity of deacetylase SIRT1 in an AMPK-dependent manner in breast cancer cells (Ferrer et al., 2017), OGT-mediated O-GlcNAcylation can also affect cancer cell growth and invasion through regulating the oncogenic transcription factor FOXM1 (Caldwell et al., 2010), suggesting the interplay of O-GlcNAcylation and SIRT1 in cancer cells. Another deacetylase HDAC1 also can be O-GlcNAc modified to promote its phosphorylation level, and simultaneously enhances its enzyme activity. This O-GlcNAc modified HDAC1 further functions in regulating the global histone acetylation level, and therefore influences the cell proliferation, invasion, and migration by regulating target genes in HepG2 cells (Zhu et al., 2016), suggesting the potential coordinative role in hepatic carcinoma. Given that abnormal histone acetylation has been implicated in tumorigenesis, the crosslink mechanism of O-GlcNAcylation and histone acetylation might provide a new direction for cancer therapy. On the other hand, OGT can modulate the integrity of multi-protein complex through O-GlcNAcylation-mediated subunit protein stability. For instance, OGT-mediated O-GlcNAcylation of Ezh2 at Ser75 stabilizes the PCR2 complex, which further promotes histone H3K27 tri-methylation in breast cancer MCF-7 cells (Chu et al., 2014). Taken together, the collaborative effects of OGT with histones and other non-histone proteins have been involved in gene transcription and tumorigenesis. Therefore, figuring out the mechanism of the network between OGT and epigenetic changes in tumorigenesis will provide a novel theoretical basis for cancer research.
Conclusion and perspectives
In summary, as the only intracellular mono-O-GlcNAc transferase, OGT has been implicated in fundamental intracellular biological functions through regulating cellular metabolism, chromatin structure, and gene transcription by cooperating with epigenetic mechanism. Although the precise regulation mechanism of network between OGT-mediated O-GlcNAcylation and epigenetic changes still remains largely unclear and to be further investigated, increasing evidence has proven that OGT-mediated O-GlcNAcylation and epigenetic changes form a complex network to further control the gene expression and signaling pathways. This coordination between O-GlcNAcylation and epigenetic changes contributes a critical role in initiation and progression of many human diseases (Fig. 4).
Figure 4.
Schematic diagram of coordination between OGT-mediated O -GlcNAcylation and chromatin remodelers in intracellular fundamental functions
ACKNOWLEDGEMENTS
This work was supported by the National Natural Science Foundation of China (Grant No. 31571316) and the Project of Jilin Province Science and Technology Development Program (No. 20140414057GH).
ABBREVIATIONS
AMPK, AMP-activated protein kinase; CARM1, a co-activator-associated argine methyltransferse 1; Ezh2, enhancer of Zeste 2; ESC, mouse embryonic stem cells; H2BK120, histone H2B at lysine 120; H2BS112, histone H2B at serine 112; H3K4me3, histone H3K4 tri-methylation; H4K16ac, H4K16 acetylation; HATs, histone acetyltransferases; HBP, hexosamine biosynthetic pathway; HCF1, host cell factor 1; HDACs, histone deacetylases; MLL5, mixed lineage leukemia 5; MOF, males absent on the first; mOGT, the mitochondrial isoform of OGT; MSL, male-specific lethal; ncOGT, nucleocytoplasmic isoform of OGT; Nrf2, nuclear factor erythroid 2-related factor 2; NSL, non-specific lethal; OGA, glycoside hydrolase O-GlcNAcase; O-GlcNAcylation, O-GlcNAc modification; OGT, β-N-acetylglucosamine (O-GlcNAc) transferase; sOGT, short isoform of OGT; TET, ten-eleven translocation; TPR, tetratricopeptide-repeat; USP7, ubiquitin specific protease 7.
COMPLIANCE WITH ETHICS GUIDELINES
Donglu Wu, Yong Cai, and Jingji Jin declare that they have no conflict of interest. This article does not contain any studies with human or animal subjects performed by any of the authors.
AUTHORS CONTRIBUTIONS
Donglu Wu, Yong Cai, and Jingji Jin participated in writing, editing and making figures. All authors read and approved the final manuscript.
References
- Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res. 2011;21:381–395. doi: 10.1038/cr.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond MR, Hanover JA. A little sugar goes a long way: the cell biology of O-GlcNAc. J Cell Biol. 2015;208:869–880. doi: 10.1083/jcb.201501101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullen JW, Balsbaugh JL, Chanda D, Shabanowitz J, Hunt DF, Neumann D, Hart GW. Cross-talk between two essential nutrient-sensitive enzymes: O-GlcNAc transferase (OGT) and AMP-activated protein kinase (AMPK) J Biol Chem. 2014;289:10592–10606. doi: 10.1074/jbc.M113.523068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burén S, Gomes AL, Teijeiro A, Fawal MA, Yilmaz M, Tummala KS, Perez M, Rodriguez-Justo M, Campos-Olivas R, Megías D, Djouder N. Regulation of OGT by URI in response to glucose confers c-MYC-dependent survival mechanisms. Cancer Cell. 2016;30:290–307. doi: 10.1016/j.ccell.2016.06.023. [DOI] [PubMed] [Google Scholar]
- Butkinaree C, Cheung WD, Park S, Park K, Barber M, Hart GW. Characterization of beta-N-acetylglucosaminidase cleavage by caspase-3 during apoptosis. J Biol Chem. 2008;283:23557–23566. doi: 10.1074/jbc.M804116200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y, Jin J, Swanson SK, Cole MD, Choi SH, Florens L, Washburn MP, Conaway JW, Conaway RC. Subunit composition and substrate specificity of a MOF-containing histone acetyltransferase distinct from the male-specific lethal (MSL) complex. J Biol Chem. 2010;285:4268–4672. doi: 10.1074/jbc.C109.087981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldwell SA, Jackson SR, Shahriari KS, Lynch TP, Sethi G, Walker S, Vosseller K, Reginati MJ. Nutrient sensor O-GlcNAc transferase regulates breast cancer tumorigenesis through targeting of the oncogenic transcription factor FoxM1. Oncogene. 2010;29:2831–2842. doi: 10.1038/onc.2010.41. [DOI] [PubMed] [Google Scholar]
- Capotosti F, Guernier S, Lammers F, Waridel P, Cai Y, Jin J, Conaway JW, Conaway RC, Herr W. O-GlcNAc transferase catalyzes site-specific proteolysis of HCF-1. Cell. 2011;144:376–388. doi: 10.1016/j.cell.2010.12.030. [DOI] [PubMed] [Google Scholar]
- Cassey PJ. Protein lipidation in cell signaling. Science. 1995;268:221–225. doi: 10.1126/science.7716512. [DOI] [PubMed] [Google Scholar]
- Charoensuksai P, Kuhn P, Wang L, Sherer N, Xu W. O-GlcNAcylation of co-activator-associated arginine methyltransferase 1 regulates its protein substrate specificity. Biochem. J. 2015;466:587–599. doi: 10.1042/BJ20141072. [DOI] [PubMed] [Google Scholar]
- Chen Q, Yu X. OGT restrains the expansion of DNA damage signaling. Nucleic Acids Res. 2016;44:9266–9278. doi: 10.1093/nar/gkw663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Chen Y, Bian C, Fujiki R, Yu X. TET2 promotes histone O-GlcNAcylation during gene transcription. Nature. 2013;493:561–564. doi: 10.1038/nature11742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi HS, Choi BY, Cho YY, Mizuno H, Kang BS, Bode AM, Dong Z. Phosphorylation of Histone H3 at Serine 10 is Indispensable for Neoplastic Cell Transformation. Cancer Res. 2005;65:5818–5827. doi: 10.1158/0008-5472.CAN-05-0197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou TY, Hart GW, Dang CV. c-Myc is glycosylated at threonine 58, a known phosphorylation site and a mutational hot spot in lymphomas. J Biol Chem. 1995;270:18961–18965. doi: 10.1074/jbc.270.32.18961. [DOI] [PubMed] [Google Scholar]
- Chu CS, Lo PW, Yeh YH, Hsu PH, Peng SH, Teng YC, Kang ML, Wong CH, Juan LJ. O-GlcNAcylation regulates EZH2 protein stability and function. Proc Natl Acad Sci USA. 2014;111:1355–1360. doi: 10.1073/pnas.1323226111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comtesse N, Maldener E, Meese E. Identification of a nuclear variant of MGEA5: a cytoplasmic hyaluronidase and a beta-N-acetylglucosaminidase. Biochem Biophys Res Commun. 2001;283:634–640. doi: 10.1006/bbrc.2001.4815. [DOI] [PubMed] [Google Scholar]
- Copeland RJ, Bullen JW, Hart GW. Cross-talk between GlcNAcylation and phosphorylation: roles in insulin resistance and glucose toxicity. Am J Physiol Endocrinol Metab. 2008;295:E17–E28. doi: 10.1152/ajpendo.90281.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Queiroz RM, Madan R, Chien J, Dias WB, Slawson C. Changes in O-Linked N-Acetylglucosamine (O-GlcNAc) Homeostasis Activate the p53 Pathway in Ovarian Cancer Cells. J Biol Chem. 2016;291:18897–18914. doi: 10.1074/jbc.M116.734533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delhommeau F, Dupont S, Della Valle V, James C, Trannoy S, Massé A, Kosmider O, Le Couedic JP, Robert F, Alberdi A, Lécluse Y, Plo I, Dreyfus FJ, Marzac C, Casasevall N, Lacombe C, Romana SP, Dessen P, Soulier J, Viquié F, Fontenay M, Vainchenker W, Bernard OA. Mutation in TET2 in myeloid cancers. N Engl J Med. 2009;360:2289–2301. doi: 10.1056/NEJMoa0810069. [DOI] [PubMed] [Google Scholar]
- Deplus R, Delatte B, Schwinn MK, Defrance M, Méndez J, Murphy N, Dawson MA, Volkmar M, Putmans P, Calonne E, Shih AH, Levine RL, Bernard O, Mercher T, Solary E, Urh M, Daniels DL, Fuks F. TET2 and TET3 regulate GlcNAcylation and H3K4 methylation through OGT and SET1/COMPASS. EMBO J. 2013;32:645–655. doi: 10.1038/emboj.2012.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding X, Jiang W, Zhou P, Liu L, Wan X, Yuan X, Wang X, Chen M, Chen J, Yang J, Kong C, Li B, Peng C, Wong CC, Hou F, Zhang Y. Mixed Lineage Leukemia 5 (MLL5) Protein stability is cooperatively regulated by O-GlcNAc transferase (OGT) and ubiquitin specific protease 7 (USP7) PLoS ONE. 2015;10:e0145023. doi: 10.1371/journal.pone.0145023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrer CM, Lu TY, Bacigalupa ZA, Katsetos CD, Sinclair DA, Reginato MJ. O-GlcNAcylation regulates breast cancer metastasis via SIRT1 modulation of FOXM1 pathway. Oncogene. 2017;36:559–569. doi: 10.1038/onc.2016.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong JJ, Nguyen BL, Bridger R, Medrano EE, Wells L, Pan S, Sifers RN. Beta-N-acetylglucosamine (O-GlcNAc) is a novel regulator of mitosis-specific phosphorylations on histone H3. J Biol Chem. 2005;287:12195–12203. doi: 10.1074/jbc.M111.315804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X, Jin L, Wang X, Luo A, Hu J, Zheng X, Tsark WM, Riggs AD, Ku HT, Huang W. MicroRNA-26a targets ten eleven translocation enzymes and is regulated during pancreatic cell differentiation. Proc Natl Acad Sci USA. 2013;110:17892–17897. doi: 10.1073/pnas.1317397110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiki R, Hashiba W, Sekine H, Yokoyama A, Chikanishi T, Ito S, Imai Y, Kim J, He HH, Igarashi K, Kanno J, Ohtake F, Kitagawa H, Roeder RG, Brown M, Kato S. GlcNAcylation of histone H2B facilitates its monoubiquitination. Nature. 2011;480:557–560. doi: 10.1038/nature10656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallo M, Coutinho FJ, Vanner RJ, Gayden T, Mack SC, Murison A, Remke M, Li R, Takayama N, Desai K, Lee L, Lan X, Park NI, Barsyte-Lovejoy D, Smil D, Sturm D, Kushida MM, Head R, Cusimano MD, Bernstein M, Clarke ID, Dick JE, Pfister SM, Rich JN, Arrowsmith CH, Taylor MD, Jabado N, Bazett-Jones DP, Lupien M, Dirks PB. MLL5 orchestrates a cancer self-renewal state by repressing the histone variant H3.3 and globally reorganizing chromatin. Cancer Cell. 2015;28:715–729. doi: 10.1016/j.ccell.2015.10.005. [DOI] [PubMed] [Google Scholar]
- Gambetta MC, Oktaba K, Müller J. Essential role of the glycosyltransferase sxc/Ogt in polycomb repression. Science. 2009;325:93–96. doi: 10.1126/science.1169727. [DOI] [PubMed] [Google Scholar]
- Gao Y, Wells L, Comer FI, Parker GJ, Hart GW. Dynamic O-glycosylation of nuclear and cytosolic proteins: cloning and characterization of a neutral, cytosolic beta-N-acetylglucosaminidase from human brain. J Biol Chem. 2001;276:9838–9845. doi: 10.1074/jbc.M010420200. [DOI] [PubMed] [Google Scholar]
- Gao Y, Wells L, Comer FI, Parker GJ, Hart GW. Dynamic O-glycosylation of nuclear and cytosolic proteins: cloning and characterization of a neutral, cytosolic beta-N-acetylglucosaminidase from human brain. J Biol Chem. 2001;276:9838–9845. doi: 10.1074/jbc.M010420200. [DOI] [PubMed] [Google Scholar]
- Gerhold CB, Gasser SM. INO80 and SWR complexes: relating structure to function in chromatin remodeling. Trends Cell Biol. 2014;24:619–631. doi: 10.1016/j.tcb.2014.06.004. [DOI] [PubMed] [Google Scholar]
- Gu Y, Mi W, Ge Y, Liu H, Fan Q, Han C, Yang J, Han F, Lu X, Yu W. GlcNAcylation plays an essential role in breast cancer metastasis. Cancer Res. 2010;70:6344–6351. doi: 10.1158/0008-5472.CAN-09-1887. [DOI] [PubMed] [Google Scholar]
- Gu Y, Gao J, Han C, Zhang X, Liu H, Ma L, Sun X, Yu W. O-GlcNAcylation is increased in prostate cancer tissues and enhances malignancy of prostate cancer cells. Mol. Med. Rep. 2014;10:897–904. doi: 10.3892/mmr.2014.2269. [DOI] [PubMed] [Google Scholar]
- Ha C, Lim K. O-GlcNAc modification of Sp3 and Sp4 transcription factors negatively regulates their transcriptional activities. Biochem Biophys Res Commun. 2015;467:341–347. doi: 10.1016/j.bbrc.2015.09.155. [DOI] [PubMed] [Google Scholar]
- Hanover JA, Yu S, Lubas WB, Shin SH, Ragano-Caracciola M, Kochran J, Love DC. Mitochondrial and nucleocytoplasmic isoforms of O-linked GlcNAc transferase encoded by a single mammalian gene. Arch Biochem Biophys. 2003;409:287–297. doi: 10.1016/S0003-9861(02)00578-7. [DOI] [PubMed] [Google Scholar]
- Hanover JA, Krause MW, Love DC. Post-translational modifications: bittersweet memories: linking metabolism to epigenetics through O-GlcNAcylation. Nat Rev Mol Cell Biol. 2012;13:312–321. doi: 10.1038/nrm3334. [DOI] [PubMed] [Google Scholar]
- Hardivillé S, Hart GW. Nutrient regulation of gene expression by O-GlcNAcylation of chromatin. Curr Opin Chem Biol. 2012;33:88–94. doi: 10.1016/j.cbpa.2016.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirosawa M, Hayakawa K, Yoneda C, Arai D, Shiota H, Suzuki T, Tanaka S, Dohmae N, Shiota K. Novel O-GlcNAcylation on Ser(40) of canonical H2A isoforms specific to viviparity. Sci. Rep. 2016;6:31785. doi: 10.1038/srep31785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingham PW. A gene that regulates the bithorax complex differentially in larval and adult cells of Drosophila. Cell. 1984;37:815–823. doi: 10.1016/0092-8674(84)90416-1. [DOI] [PubMed] [Google Scholar]
- Itkonen HM, Minner S, Guldvik IJ, Sandmann MJ, Tsourlakis MC, Berge V, Svindland A, Schlomm T, Mills IG. O-GlcNAc transferase integrates metabolic pathways to regulate the stability of c-MYC in human prostate cancer cells. Cancer Res. 2013;73:5277–5287. doi: 10.1158/0008-5472.CAN-13-0549. [DOI] [PubMed] [Google Scholar]
- Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, He C, Zhang Y. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333:1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito R, Katsura S, Shimada H, Tsuchiya H, Hada M, Okumura T, Sugawara A, Yokoyama A. TET3-OGT interaction increases the stability and the presence of OGT in chromatin. Genes Cells. 2014;19:52–65. doi: 10.1111/gtc.12107. [DOI] [PubMed] [Google Scholar]
- Jin J, Cai Y, Li B, Conaway RC, Workman JL, Conaway JW, Kusch T. In and out: histone variant exchange in chromatin. Trends Biochem Sci. 2005;30:680–687. doi: 10.1016/j.tibs.2005.10.003. [DOI] [PubMed] [Google Scholar]
- Jinek M, Rehwinkel J, Lazarus BD, Izaurralde E, Hanover JA, Conti E. The superhelical TPR-repeat domain of O-linked GlcNAc transferase exhibits structural similarities to importin alpha. Nat Struct Mol Biol. 2004;11:1001–1007. doi: 10.1038/nsmb833. [DOI] [PubMed] [Google Scholar]
- Kamigaito T, Okaneya T, Kawakubo M, Shimojo H, Nishizawa O, Nakayama J. Overexpression of O-GlcNAc by prostate cancer cells is significantly associated with poor prognosis of patients. Prostate Cancer Prostatic Dis. 2014;17:18–22. doi: 10.1038/pcan.2013.56. [DOI] [PubMed] [Google Scholar]
- Kang KA, Piao MJ, Ryu YS, Kang HK, Chang WY, Keum YS, Hyun JW. Interaction of DNA demethylase and histone methyltransferase upregulates Nrf2 in 5-fluorouracil-resistant colon cancer cells. Oncotarget. 2016;7:40594–40620. doi: 10.18632/oncotarget.9745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato S, Ishii T, Kouzmenko A (2015) Point mutations in an epigenetic factor lead to multiple types of bone tumors: role of H3.3 histone variant in bone development and disease. Bonekey. Rep. 4:715 [DOI] [PMC free article] [PubMed]
- Kreppel LK, Hart GW. Regulation of a cytosolic and nuclear O-GlcNAc transferase. Role of the tetratricopeptide repeats. J Biol Chem. 1999;274:32015–32022. doi: 10.1074/jbc.274.45.32015. [DOI] [PubMed] [Google Scholar]
- Kreppel LK, Blomberg MA, Hart GW. Dynamic glycosylation of nuclear and cytosolic proteins. Cloning and characterization of a unique O-GlcNAc transferase with multiple tetratricopeptide repeats. J Biol Chem. 1997;272:9308–9315. doi: 10.1074/jbc.272.14.9308. [DOI] [PubMed] [Google Scholar]
- Krzeslak A, Forma E, Bernaciak M, Romanowicz H, Brys M. Gene expression of O-GlcNAc cycling enzymes in human breast cancers. Clin Exp Med. 2012;12:61–65. doi: 10.1007/s10238-011-0138-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langemeijer SM, Kuiper RP, Berends M, Knops R, Aslanyan MG, Massop M, Stevens-Linders E, van Hoogen P, van Kessel AG, Raymakers RA, Kamping EJ, Verhoef GE, Verburgh E, Hagemeijer A, Vandenberghe P, de Witte T, van der Reijden BA, Jansen JH. Acquired mutations in TET2 are common in myelodysplastic syndromes. Nat. Genet. 2009;41:838–842. doi: 10.1038/ng.391. [DOI] [PubMed] [Google Scholar]
- Längst G, Manelyte L. Chromatin remodelers: from function to dysfunction. Genes. 2015;6:299–324. doi: 10.3390/genes6020299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarus BD, Love DC, Hanover JA. Recombinant O-GlcNAc transferase isoforms: identification of O-GlcNAcase, yes tyrosine kinase, and tau as isoform-specific substrates. Glycobiology. 2006;16:415–421. doi: 10.1093/glycob/cwj078. [DOI] [PubMed] [Google Scholar]
- Lazarus MB, Nam Y, Jiang J, Sliz P, Walker S. Structure of human O-GlcNAc transferase and its complex with a peptide substrate. Nature. 2011;469:564–567. doi: 10.1038/nature09638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JS, Zhang Z. O-linked N-acetylglucosamine transferase (OGT) interacts with the histone chaperone HIRA complex and regulates nucleosome assembly and cellular senescence. Proc Natl Acad Sci USA. 2016;113:E3213–E3220. doi: 10.1073/pnas.1600509113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lercher L, Raj R, Patel NA, Price J, Mohammed S, Robinson CV, Schofield CJ, Davis BG. Generation of a synthetic GlcNAcylated nucleosome reveals regulation of stability by H2A-Thr101 GlcNAcylation. Nat. Commun. 2015;6:7978. doi: 10.1038/ncomms8978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Li X, Yu Y, Shi J, Liang Z, Run X, Li Y, Dai CL, Grundke-Iqbal I, Iqbal K, Liu F, Gong CX. Developmental regulation of protein O-GlcNAcylation, O-GlcNAc transferase, and O-GlcNAcase in mammalian brain. PLoS ONE. 2012;7:43724. doi: 10.1371/journal.pone.0043724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorsbach RB, Moore J, Mathew S, Raimondi SC, Mukatira ST, Downing JR. TET1, a member of a novel protein family, is fused to MLL in acute myeloid leukemia containing the t (10;11)(q22;q23) Leukemia. 2003;17:637–641. doi: 10.1038/sj.leu.2402834. [DOI] [PubMed] [Google Scholar]
- Love DC, Kochan J, Cathey RL, Shin SH, Hanover JA. Mitochondrial and nucleocytoplasmic targeting of O-linked GlcNAc transferase. J Cell Sci. 2003;116:647–654. doi: 10.1242/jcs.00246. [DOI] [PubMed] [Google Scholar]
- Lubas WA, Hanover JA. Functional expression of O-linked GlcNAc transferase. Domain structure and substrate specificity. J Biol Chem. 2000;275:10983–10988. doi: 10.1074/jbc.275.15.10983. [DOI] [PubMed] [Google Scholar]
- Lynch TP, Ferrer CM, Jackson SR, Shahriari KS, Vosseller K, Reginato MJ. Critical role of O-linked beta-Nacetylglucosamine transferase in prostate cancer invasion, angiogenesis, and metastasis. J Biol Chem. 2012;287:11070–11081. doi: 10.1074/jbc.M111.302547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mailleux F, Gélinas R, Beauloye C, Horman S, Bertrand L. O-GlcNAcylation, enemy or ally during cardiac hypertrophy development? Biochim Biophys Acta. 2016;1862:2232–2243. doi: 10.1016/j.bbadis.2016.08.012. [DOI] [PubMed] [Google Scholar]
- Marshall S, Bacote V, Traxinger RR. Discovery of a metabolic pathway mediating glucose-induced desensitization of the glucose transport system. Role of hexosamine biosynthesis in the induction of insulin resistance. J Biol Chem. 1991;266:4706–4712. [PubMed] [Google Scholar]
- Mazars R, Gonzalez-de-Peredo A, Cayrol C, Lavigne AC, Vogel JL, Ortega N, Lacroix C, Gautier V, Huet G, Ray A, Monsarrat B, Kristie TM, Girard JP. The THAP-Zinc Finger Protein THAP1 Associates with Coactivator HCF-1 and O-GlcNAc Transferase, a link between DYT6 and DYT3 dystonias. J Biol Chem. 2010;285:13364–13371. doi: 10.1074/jbc.M109.072579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi W, Gu Y, Han C, Liu H, Fan Q, Zhang X, Cong Q, Yu W. O-GlcNAcylation is a novel regulator of lung and colon cancer malignancy. Biochim Biophys Acta. 2011;1812:514–519. doi: 10.1016/j.bbadis.2011.01.009. [DOI] [PubMed] [Google Scholar]
- Nakamura K, Kato A, Kobayashi J, Yanagihara H, Sakamoto S, Oliveira DV, Shimada M, Tauchi H, Suzuki H, Tashiro S, Zou L, Komatsu K. Regulation of homologous recombination by RNF20-dependent H2B ubiquitination. Mol Cell. 2011;41:515–528. doi: 10.1016/j.molcel.2011.02.002. [DOI] [PubMed] [Google Scholar]
- O’Donnell N, Zachara NE, Hart GW, Marth JD. Ogt-dependent X-chromosome-linked protein glycosylation is a requisite modification in somatic cell function and embryo viability. Mol Cell Biol. 2004;224:1680–1690. doi: 10.1128/MCB.24.4.1680-1690.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono R, Taki T, Taketani T, Taniwaki M, Kobayashi H, Hayashi Y. LCX, leukemia-associated protein with a CXXC domain, is fused to MLL in acute myeloid leukemia with trilineage dysplasia having t(10;11)(q22;q23) Cancer Res. 2002;62:4075–4080. [PubMed] [Google Scholar]
- Phoomak C, Vaeteewoottacharn K, Sawanyawisuth K, Seubwai W, Wongkham C, Silsirivanit A, Wongkhamb S. Mechanistic insights of O-GlcNAcylation that promote progression of cholangiocarcinoma cells via nuclear translocation of NF-κB. Sci. Rep. 2016;6:27853. doi: 10.1038/srep27853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao Z, Dang C, Zhou B, Li S, Zhang W, Jiang J, Zhang J, Kong R, Ma Y. O-linked N-acetylglucosamine transferase (OGT) is overexpressed and promotes O-linked protein glycosylation in esophageal squamous cell carcinoma. J. Biomed Res. 2012;26:268–273. doi: 10.7555/JBR.26.20110121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricketts MD, Marmorstein R. A molecular prospective for HIRA complex assembly and H3.3-specific histone chaperone. J Mol Biol. 2016 doi: 10.1016/j.jmb.2016.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riu IH, Shin IS, Do SI. Sp1 modulates ncOGT activity to alter target recognition and enhanced thermotolerance in E. coli. Biochem Biophys Res Commun. 2008;372:203–209. doi: 10.1016/j.bbrc.2008.05.034. [DOI] [PubMed] [Google Scholar]
- Ronningen T, Shah A, Oldenburg AR, Vekterud K, Delbarre E, Moskaug JO, Collas P. Prepatterning of differentiation-driven nuclear lamin A/C-associated chromatin domains by GlcNAcylated histone H2B. Genome Res. 2015;25:1825–1835. doi: 10.1101/gr.193748.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan HB, Han X, Li MD, Singh JP, Qian K, Azarhoush S, Zhao L, Bennett AM, Samuel VT, Wu J, Yates JR, Yang X. O-GlcNAc transferase/host cell factor C1 complex regulates gluconeogenesis by modulating PGC-1α stability. Cell Metab. 2012;16:226–237. doi: 10.1016/j.cmet.2012.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacoman JL, Dagda RY, Burnham-Marusich AR, Dagda RK, Berninsone PM. Mitochondrial O-GlcNAc transferase (mOGT) regulates mitochondrial structure, function and survival in HeLa cells. J Biol Chem. 2017;292:4499–4518. doi: 10.1074/jbc.M116.726752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakabe K, Hart GW. O-GlcNAc Transferase Regulates Mitotic Chromatin Dynamics. J Biol Chem. 2010;285:34460–34468. doi: 10.1074/jbc.M110.158170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakabe K, Wang Z, Hart GW. Beta-N-acetylglucosamine (O-GlcNAc) is part of the histone code. Proc Natl Acad Sci USA. 2010;107:19915–19920. doi: 10.1073/pnas.1009023107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schurter BT, Koh SS, Chen D, Bunick GJ, Harp JM, Hanson BL, Henschen-Edman A, Mackay DR, Stallcup MR, Aswad DW. Methylation of histone H3 by coactivator-associated arginine methyltransferase 1. Biochemistry. 2001;40:5747–5756. doi: 10.1021/bi002631b. [DOI] [PubMed] [Google Scholar]
- Scourzic L, Mouly E, Bernard OA. TET proteins and the control of cytosine demethylation in cancer. Genome Med. 2015;7:9. doi: 10.1186/s13073-015-0134-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebastian S, Sreenivas P, Sambasivan R, Cheedipudi S, Kandalla P, Pavlath GK, Dhawan J. MLL5, a trithorax homolog, indirectly regulates H3K4 methylation, represses cyclin A2 expression, and promotes myogenic differentiation. Proc Natl Acad Sci USA. 2009;106:4719–4724. doi: 10.1073/pnas.0807136106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi FT, Kim H, Lu W, He Q, Liu D, Goodell MA, et al. Ten-eleven transloca tion 1 (Tet1) is regulated by O-linked N-acetylglucosamine transferase (Ogt) for target gene repression in mouse embryonic stem cells. J Biol Chem. 2013;288:20776–20784. doi: 10.1074/jbc.M113.460386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin SH, Love DC, Hanover JA. Elevated O-GlcNAc-dependent signaling through inducible mOGT expression selectively triggers apoptosis. Amino Acids. 2011;40:885–893. doi: 10.1007/s00726-010-0719-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slawson C, Lakshmanan T, Knapp S, Hart GW. A mitotic GlcNAcylation/phosphorylation signaling complex alters the posttranslational state of the cytoskeletal protein vimentin. Mol Biol Cell. 2008;19:4130–4140. doi: 10.1091/mbc.E07-11-1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tefferi A, Pardanani A, Lim KH, Abdel-Wahab O, Lasho TL, Patel J, Gangat N, Finke CM, Schwager S, Mullally A, Li CY, Hanson CA, Mesa R, Bernard O, Delhommeau F, Vainchenker W, Gilliland DG, Levine RL. TET2 mutations and their clinical correlates in polycythemia vera, essential thrombocythemia and myelofibrosis. Leukemia. 2009;23:905–911. doi: 10.1038/leu.2009.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toleman C, Paterson AJ, Whisenhunt TR, Kudlow JE. Characterization of the histone acetyltransferase (HAT) domain of a bifunctional protein with activable O-GlcNAcase and HAT activities. J Biol Chem. 2004;279:53665–53673. doi: 10.1074/jbc.M410406200. [DOI] [PubMed] [Google Scholar]
- Trapannone R, Mariappa D, Ferenbach AT, van Aalten DM. Nucleocytoplasmic human O-GlcNAc transferase is sufficient for O-GlcNAcylation of mitochondrial proteins. Biochem. J. 2016;473:1693–1702. doi: 10.1042/BCJ20160092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Hooser A, Goodrich DW, Allis CD, Brinkley BR, Mancini MA. Histone H3 phosphorylation is required for the initiation, but not maintenance, of mammalian chromosome condensation. J Cell Sci. 1998;111:3497–3506. doi: 10.1242/jcs.111.23.3497. [DOI] [PubMed] [Google Scholar]
- Vella P, Scelfo A, Jammula S, Chiacchiera F, Williams K, Cuomo A, Roberto A, Christensen J, Bonaldi T, Helin K, Pasini D. Tet proteins connect the O-linked N -acetylglucosamine transferase Ogt to chromatin in embryonic stem cells. Mol Cell. 2013;49:645–656. doi: 10.1016/j.molcel.2012.12.019. [DOI] [PubMed] [Google Scholar]
- Wells L, Hart GW. O-GlcNAc turns twenty: functional implications for post-translational modification of nuclear and cytosolic proteins with a sugar. FEBS Lett. 2003;546:154–158. doi: 10.1016/S0014-5793(03)00641-0. [DOI] [PubMed] [Google Scholar]
- Williams K, Christensen J, Pedersen MT, Johansen JV, Cloos PA, Rappsilber J, Helin K. TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature. 2011;473:343–348. doi: 10.1038/nature10066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams K, Christensen J, Helin K. DNA methylation: TET proteins-guardians of CpG islands? EMBO Rep. 2012;13:28–35. doi: 10.1038/embor.2011.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, D’Alessio AC, Ito S, Xia K, Wang Z, Cui K, Zhao K, Sun YE, Zhang Y. Dual functions of Tet1 in transcriptional regulation in mouse embryonic stem cells. Nature. 2011;473:389–393. doi: 10.1038/nature09934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang WH, Kim JE, Nam HW, Ju JW, Kim HS, Kim YS, Cho JW. Modification of p53 with O-linked N-acetylglucosamine regulates p53 activity and stability. Nat Cell Biol. 2006;8:1074–1083. doi: 10.1038/ncb1470. [DOI] [PubMed] [Google Scholar]
- Yildirim O, Li R, Hung JH, Chen PB, Dong X, Ee LS, Weng Z, Rando OJ, Fazzio TG. Mbd3/NURD complex regulates expression of 5-hydroxymethylcytosine marked genes in embryonic stem cells. Cell. 2011;147:1498–1510. doi: 10.1016/j.cell.2011.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Roche K, Nasheuer HP, Lowndes NF. Modification of histones by sugar β-N-acetylglucosamine (GlcNAc) occurs on multiple residues, including histone H3 serine 10, and is cell cycle-regulated. J Biol Chem. 2011;286:37483–37495. doi: 10.1074/jbc.M111.284885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q, Zhou L, Yang Z, Lai M, Xie H, Wu L, Xing C, Zhang F, Zheng S. O-GlcNAcylation plays a role in tumor recurrence of hepatocellular carcinoma following liver transplantation. Med Oncol. 2012;29:985–993. doi: 10.1007/s12032-011-9912-1. [DOI] [PubMed] [Google Scholar]
- Zhu X, Li D, Zhang Z, Zhu W, Li W, Zhao J, Xing X, He Z, Wang S, Wang F, Ma L, Bai Q, Zeng X, Li J, Gao C, Xiao Y, Wang Q, Chen L, Chen W. Persistent phosphorylation at specific H3 serine residues involved in chemical carcinogen-induced cell transformation. Mol: Carcinog; 2016. [DOI] [PubMed] [Google Scholar]
- Zhu G, Tao T, Zhang D, Liu X, Qiu H, Han L, Xu Z, Xiao Y, Cheng C, Shen A. O-GlcNAcylation of histone deacetylase-1 in hepatocellular carcinoma promotes cancer progression. Glycobiology. 2016;26:820–833. doi: 10.1093/glycob/cww025. [DOI] [PubMed] [Google Scholar]
- Zippo A, Serafini R, Rocchigiani M, Pennacchini S, Krepelova A, Oliviero S. Histone crosstalk between H3S10ph and H4K16ac generates a histone code that mediates transcription elongation. Cell. 2009;138:1122–1136. doi: 10.1016/j.cell.2009.07.031. [DOI] [PubMed] [Google Scholar]