Abstract
The mechanistic target of rapamycin (mTOR) is a central regulator of growth and proliferation and mTOR inhibition is a promising therapy for a variety of diseases and disorders. Inhibition of mTOR complex I (mTORC1) with rapamycin delays aging and increases healthy longevity in laboratory animals and is used clinically at high doses to prevent organ transplant rejection and to treat some forms of cancer. Clinical use of rapamycin is associated with several unwanted side effects, however, and several strategies are being taken to identify mTORC1 inhibitors with fewer side effects. We describe here a yeast-based growth assay that can be used to screen for novel inhibitors of mTORC1. By testing compounds using a wild-type strain and isogenic cells lacking either TOR1 or FPR1, we can resolve not only whether a compound is an inhibitor of mTORC1 but also whether the inhibitor acts through a mechanism similar to rapamycin by binding Fpr1. Using this assay, we show that rapamycin derivatives behave similarly to rapamycin, while caffeine and the ATP competitive inhibitors Torin 1 and GSK2126458 are mTORC1 inhibitors in yeast that act independently of Fpr1. Some mTOR inhibitors in mammalian cells do not inhibit mTORC1 in yeast, and several nutraceutical compounds were not found to specifically inhibit mTOR but resulted in a general inhibition of yeast growth. Our screening method holds promise as a means of effectively assaying drug libraries for mTOR-inhibitory molecules in vivo that may be adapted as novel treatments to fight diseases and extend healthy longevity.
Electronic supplementary material
The online version of this article (doi:10.1007/s11357-017-9988-4) contains supplementary material, which is available to authorized users.
Keywords: mTOR, Saccharomyces cerevisiae, Yeast
Introduction
The budding yeast, Saccharomyces cerevisiae, is a premier model system for identifying conserved genetic and pharmacological interventions that extend life span (Longo et al. 2012; Kaeberlein 2010). A particularly good example of this is the mechanistic target of rapamycin (mTOR) pathway that was first genetically implicated in aging in yeast (Fabrizio et al. 2001) and has since emerged as an important target for delaying aging in multicellular invertebrates and mice (Johnson et al. 2013a, 2015). Likewise, the small molecule mTOR inhibitor, rapamycin, was first shown to extend life span in yeast (Powers et al. 2006) and has since been shown to have similar pro-longevity effects in nematodes (Robida-Stubbs et al. 2012), fruit flies (Bjedov et al. 2010), and mice (Harrison et al. 2009).
In addition to increased life span, rapamycin maintains organismal health during aging in mice, evidenced by decreased occurrence of multiple age-related diseases (Johnson et al. 2013b). Rapamycin treatment in aging mice reduces cancer (Anisimov et al. 2011; Popovich et al. 2014), prevents cognitive dysfunction (Halloran et al. 2012; Majumder et al. 2012), attenuates declining renal and hepatic function (Neff et al. 2013), improves muscle and visual performance (Neff et al. 2013), and reverses cardiac (Flynn et al. 2013; Dai et al. 2014) and immune decline (Chen et al. 2009). Improved cardiac function from rapamycin treatment has recently been similarly observed in middle-aged companion dogs (Urfer et al. 2017a, b), while improved immune function has been observed in healthy elderly people treated with the rapamycin derivative everolimus (RAD001) (Mannick et al. 2014). Transient rapamycin treatment regimens lasting as few as 12 weeks and initiated late in life are also effective at increasing life span and healthspan in mice (Bitto et al. 2016). These findings place rapamycin and other mTOR inhibitors among the leading candidates for translational interventions to promote healthy aging in people and companion animals (Blagosklonny 2010; Kaeberlein et al. 2015, 2016).
The mTOR is a nutrient and growth factor responsive kinase that functions in two distinct protein complexes: mTOR complex 1 (mTORC1) and mTOR complex 2 (mTORC2) (Loewith et al. 2002; Wullschleger et al. 2006). Rapamycin binds to the FK506 binding protein FKPB12 (Fpr1 in yeast), and the FKBP12-rapamycin complex inhibits the activity of mTORC1 by disrupting the physical interaction between the mTOR protein and a second mTORC1 component, raptor (Kog1 in yeast) (Heitman et al. 1991; Lorenz and Heitman 1995). Deletion of FPR1 confers resistance to rapamycin in yeast (Heitman et al. 1991; Benton et al. 1994). The mTORC1 complex regulates a variety of downstream cellular processes including messenger RNA (mRNA) translation, autophagy, and mitochondrial metabolism, all of which are affected by rapamycin treatment (Saxton and Sabatini 2017). Unlike mTORC1, mTORC2 activity is not directly inhibited by rapamycin (Jacinto et al. 2004). The mTORC2 complex is less well characterized than mTORC1, but is similarly involved in regulating a variety of cellular processes including cytoskeleton organization and regulation of metabolism. Long-term treatment with rapamycin in mammals is reported to cause indirect inhibition of mTORC2, which is implicated in metabolic defects including insulin resistance and glucose intolerance (Lamming et al. 2012). Both mTOR complexes are essential for viability in yeast and multicellular eukaryotes.
Budding yeast contain two genes that encode the mTOR kinase: TOR1 and TOR2. Tor1 functions exclusively in mTORC1, while Tor2 functions in both complexes (Helliwell et al. 1994). Consistent with this, deletion of TOR2 leads to inviability due to the complete lack of mTORC2 activity (Kunz et al. 1993), while deletion of TOR1 results in viable cells that are long lived and sensitive to rapamycin, due to reduced mTORC1 activity (Heitman et al. 1991; Kaeberlein et al. 2005a). Despite their sensitivity to rapamycin, tor1Δ yeast cells do not show a substantial reduction in mRNA translation or doubling time (McCormick et al. 2015; Beaupere et al. 2017), indicating that Tor2 provides sufficient mTORC1 activity for relatively normal growth in rich medium.
Multiple pharmaceutical mTORC1 and general mTOR inhibitors are used as chemotherapeutic agents (Folkes et al. 2008; Chresta et al. 2010; Knight et al. 2010; O’Donnell et al. 2017; Liu et al. 2012). Additionally, several natural products and natural product mixtures are reported to inhibit mTOR signaling in cell culture and rodent models, including: curcumin (Beevers et al. 2006, 2009), green tea extract (Zhang et al. 2006), epigallocatechin-3-gallate (Zhang et al. 2006; Van Aller et al. 2011), caffeine (Saiki et al. 2011; Reinke et al. 2006), genistein (Anastasius et al. 2009), lycopene and eicosapentaenoic acid (Liu et al. 2012), sulforaphane (Wiczk et al. 2012), alpha-lipoic acid (Xie et al. 2012; Li et al. 2014), glucosamine (Jiang et al. 2014), quercetin (Meng et al. 2015; Lu et al. 2015), berberine (Fan et al. 2015), and resveratrol (Park et al. 2016). In most cases, the mechanistic basis for inhibition of mTOR via these natural products is unknown, and it remains unclear whether these compounds act via direct inhibition of mTOR, mTORC1, or through indirect effects on components of the mTOR/nutrient response network.
To facilitate identification of new small molecule mTOR inhibitors in vivo, we have developed a simple yeast-based assay that quantifies differential growth inhibition in rapamycin-sensitized and rapamycin-resistant genetic backgrounds using a Bioscreen C MBR plate reader/shaker/incubator. We have previously optimized the Bioscreen C MBR machine to obtain high-resolution growth curves of budding yeast cells for chronological life span analysis (Murakami et al. 2008; Murakami and Kaeberlein 2009) and assessing differential sensitivities of yeast strains to different chemical and environmental stressors (Delaney et al. 2013). Here, we extend this method by assessing the impact of rapamycin and several other small molecules on doubling time and outgrowth in rich media for wild-type BY4742 cells and isogenic tor1Δ and fpr1Δ cells. Our method relies on the fact that deletion of TOR1 confers sensitivity to rapamycin due to diminished mTORC1 activity, while deletion of FPR1 confers resistance to rapamycin due to the necessary role of Fpr1 in rapamycin-mediated mTORC1 inhibition. Given this, we predict that mTORC1 inhibitors with mechanisms similar to rapamycin will display similar differential growth inhibition across the three genotypes, while compounds that inhibit mTORC1 by a mechanism distinct from rapamycin will have a greater inhibitory effect on growth of tor1Δ cells relative to WT or fpr1Δ cells, but that WT and fpr1Δ cells will show similar inhibition of growth at a given drug concentration. Drugs that do not inhibit mTORC1 will either have no effect on growth in any of the genotypes or will similarly inhibit growth across all three genotypes. We report the validation of this assay using rapamycin, rapamycin derivatives, and mTOR catalytic inhibitors. We also report the effect of several natural product compounds on mTORC1 inhibition and outgrowth, finding that among the natural products tested, only caffeine displays the outgrowth profile expected for an in vivo inhibitor of mTORC1 in yeast.
Results
TOR1 mutants are hypersensitive, and FPR1 mutants are resistant to rapamycin and rapalogs
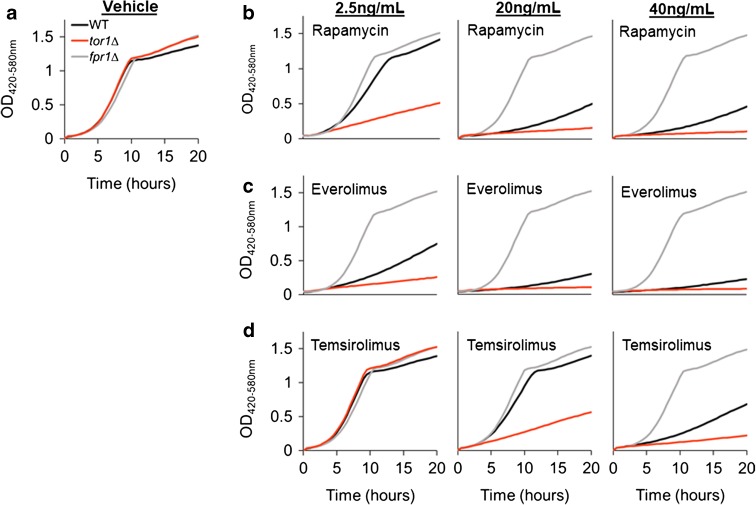
We tested the effect of known mTORC1 inhibitors on growth kinetics in three haploid yeast strains: wild-type (WT) BY4742 and isogenic tor1Δ, and fpr1Δ single-gene deletion mutants. We began by analyzing dose responses for rapamycin and two rapalogs, everolimus and temsirolimus. As expected, all concentrations of rapamycin tested reduced tor1Δ outgrowth to a greater extent than WT cells, while fpr1Δ cells were resistant to growth inhibition (Figs. 1 and 2). Everolimus and rapamycin produced remarkably similar growth inhibitory responses, while higher concentrations of temsirolimus were necessary to achieve growth inhibition comparable to the other two drugs.
Fig. 1.
Differential growth of wild-type (WT), tor1Δ, and fpr1Δ strains in the presence of rapamycin and rapalogs. tor1Δ mutants are hypersensitive, and fpr1Δ mutants are resistant to rapamycin and rapalogs. Representative growth curves of WT (black), tor1∆ (red), and fpr1∆ (gray) yeast grown in YPD with (a) 1.5% DMSO (vehicle) or 2.5–40 ng/mL (left to right) (b) rapamycin, (c) everolimus, or (d) temsirolimus (color figure online)
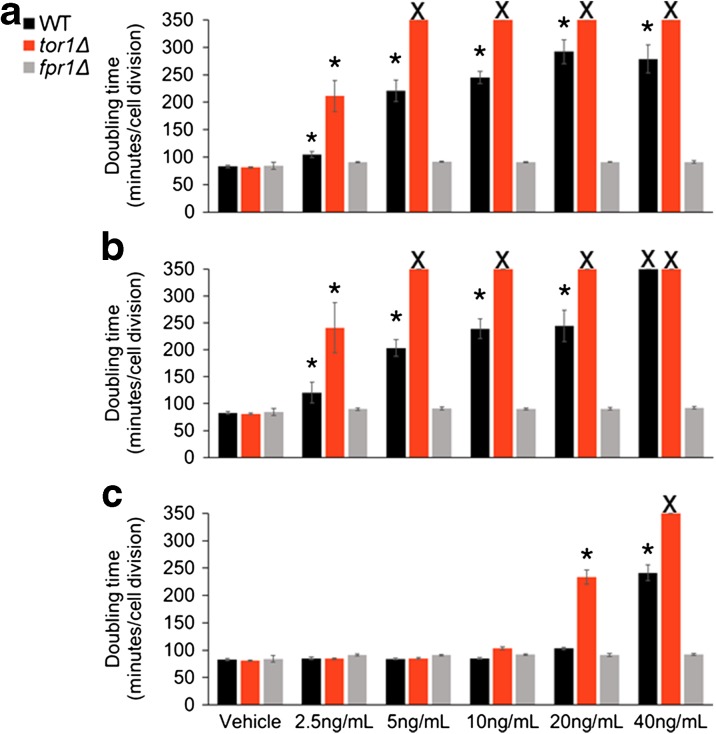
Fig. 2.
Effect of rapamycin and rapalogs on maximal doubling time of wild-type (WT), tor1Δ, and fpr1Δ yeast cells. Maximal doubling times of wild-type (black), tor1∆ (red), and fpr1∆ (gray) in vehicle or 2.5–40 ng/mL (a) rapamycin, (b) everolimus, or (c) temsirolimus (n = 3–5 biological replicates for each treatment). Error bars = SEM. Asterisk means significantly different from 1.5% DMSO control (vehicle) (p < 0.05, Welch’s t test). X = doubling time could not be calculated due to no growth, indicated by OD ≤0.3 after 20 h (color figure online)
Sensitivity of the tor1Δ strain to mTORC1 inhibitors can been seen at 2.5 ng/mL rapamycin or everolimus and at 20 ng/mL temsirolimus, where deletion of TOR1 results in a significantly greater increase in doubling time relative to WT (Fig. 2 and Supplemental Table 1). Resistance of the fpr1Δ strain is most evident at the highest concentration of each drug tested, where both WT and tor1Δ strains have doubling times exceeding 200 min, while the fpr1Δ cells are still growing as rapidly as in the vehicle control (doubling time 80–90 min).
One consequence of diminished mTORC1 signaling is diminished protein translation (Steffen et al. 2008; Beretta et al. 1996). To determine if diminished protein translation is sufficient to recapitulate rapamycin-like patterns of growth inhibition, we tested concentrations of the general protein translation inhibitor cycloheximide (CHX) ranging from 1 to 500 ng/mL. Cycloheximide treatment potently and similarly inhibits growth of all three strains at concentrations of 50 ng/mL and greater (Supplemental Fig. 1), demonstrating that general translation inhibition does not recapitulate the differential effects of rapamycin on growth of these three yeast strains.
ATP competitive inhibitors of mTORC1 produce a distinct growth inhibitory profile compared to rapamycin treatment
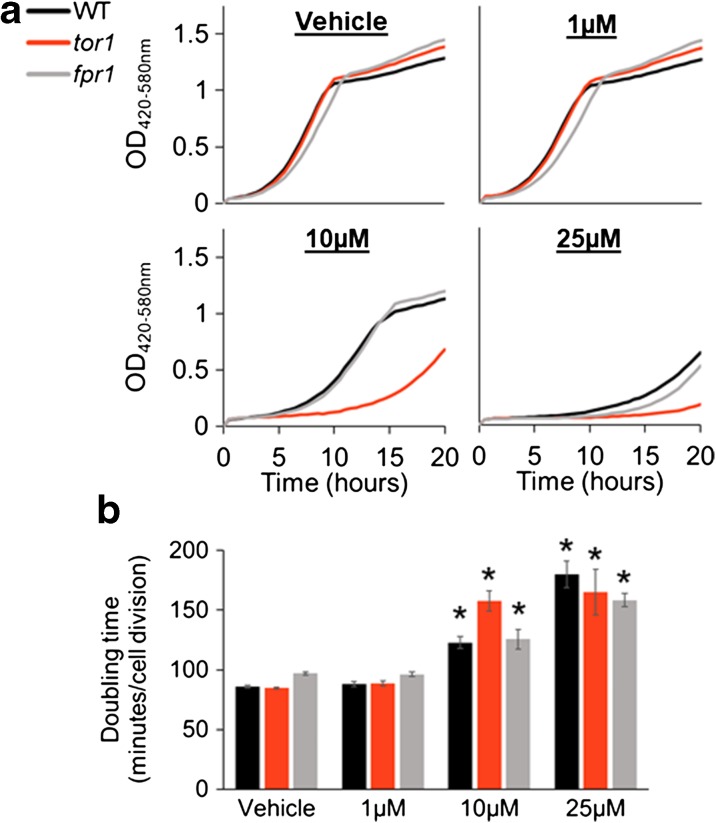
In addition to rapamycin-like compounds that inhibit mTORC1 signaling via interaction with Fpr1, several drugs have been developed that act as ATP-competitive inhibitors of mTOR in mammalian cells. To understand how these compounds differentially impact growth in our yeast strains, we tested dose responses for Torin 1, GSK2126458, GDC-0941, and AZD8055 (Fig. 3, Supplemental Fig. 2, and Supplemental Table 2). All of the ATP-competitive inhibitors required micromolar concentrations to inhibit growth, compared to nanomolar concentrations when using rapamycin and rapalogs. At 10 μM Torin 1, tor1Δ cells were strongly growth inhibited, while WT cells or fpr1Δ cells were less strongly affected (Fig. 3 and Supplemental Table 2). At 25 μM Torin 1, growth of all three strains was severely impacted, with WT and fpr1Δ cells showing similar reductions in growth rate, as expected for a catalytic inhibitor of mTOR that does not act through Fpr1.
Fig. 3.
The catalytic mTOR inhibitor Torin 1 produces a distinct growth inhibitory profile relative to rapamycin. tor1Δ mutants are hypersensitive to Torin 1, and fpr1Δ mutants are not resistant. a Representative growth curves of WT, tor1Δ, and fpr1Δ outgrown in the presence of 2.5% DMSO (vehicle) (top left), 1 μM Torin 1 (top right), 10 μM Torin 1 (bottom left), or 25 μM Torin 1 (bottom right). b Maximal doubling time (minutes/cell division) for WT, tor1Δ, and fpr1Δ strains grown in either 2.5% DMSO (vehicle control) (n = 9 biological replicates for WT and fpr1Δ, and biological replicates n = 10 for tor1Δ) or 1–25 μM Torin 1 (n = 4 biological replicates for each strain in 1 μM Torin 1 and n = 5 biological replicates for each strain in 10 and 25 μM Torin 1). Asterisk means significantly different from vehicle (p < 0.05, Welch’s t test). Error bars = SEM
GSK2126458, another catalytic inhibitor of mTOR, only weakly impacted yeast growth (Supplemental Fig. 2 and Supplemental Table 2). Doubling time was modestly increased by 100 μM GSK2126458 in all three strains, while growth of only the tor1Δ cells was impacted by lower concentrations. Interestingly, neither GDC-0941 nor AZD8055 produced a measurable change in growth in any yeast strains (Supplemental Fig. 2 and Supplemental Table 2).
Among several putative mTORC1 inhibitory nutraceuticals, caffeine shows specificity for mTORC1 in yeast
Many nutraceutical compounds are described as having an mTORC1 inhibitory effect, particularly in the context of human cancer cell culture models. To identify and validate mTORC1-modulating nutraceuticals in yeast, we tested a subset of these compounds in our Bioscreen C MBR assay (Table 1). Most of the tested nutraceuticals produced no effect on growth at concentrations up to 100 μg/mL (Table 1). Quercetin significantly inhibited WT and tor1Δ doubling time by 23 and 25%, respectively, at the highest concentration tested (p < 0.01, Welch’s t test), but did not produce a significant difference in fpr1Δ doubling time (p = 0.24, Welch’s t test) even though a trend toward increased doubling time is seen in the strain. Two compounds tested, berberine and lycopene, inhibited growth in a genotype-independent manner. Lycopene only modestly impacted growth, while berberine strongly inhibited growth at 100 μg/mL.
Table 1.
Maximum doubling time of wild-type (WT), tor1Δ, and fpr1Δ cells grown in rich YPD medium supplemented with the indicated neutraceuticals. Doubling time (DT) (minutes/cell division), standard error of the mean (SEM), percent change, and number of biological replicate cultures tested (n) for putative mTOR-inhibitory neutraceutical compounds
| Treatment | WT | tor1Δ | fpr1Δ | ||||||
|---|---|---|---|---|---|---|---|---|---|
| DT (SEM) | % change | n | DT (SEM) | % change | n | DT (SEM) | % change | n | |
| 1% DMSO | 83.3 (0.9) | – | 13 | 82.8 (0.9) | – | 13 | 90.8 (1.0) | – | 13 |
| 100 μg/mL Alpha-lipoic acid | 85.4 (2.3) | 2.6 | 3 | 85.2 (1.2) | 2.9 | 3 | 91.7 (2.1) | −6.2 | 3 |
| 100 μg/mL Broccoli concentrate | 83.3 (1.1) | 0.0 | 3 | 82.7 (2.8) | −0.2 | 3 | 89.2 (1.9) | −1.8 | 3 |
| 100 μg/mL Glucosamine | 81.0 (2.5) | −2.8 | 4 | 81.2 (2.1) | −2.0 | 4 | 87.5 (2.6) | −3.6 | 4 |
| 100 μg/mL Lycopene | 95.8 (0.3) | 15.0* | 3 | 101.1 (2.8) | 22.2* | 3 | 107.7 (0.7) | 18.6* | 3 |
| 100 μg/mL Quercetin | 104.1 (3.1) | 25.0* | 4 | 102.0 (6.0) | 23.2* | 4 | 105.4 (10.2) | 16.1 | 4 |
| 100 μg/mL Resveratrol | 82.7 (0.8) | −0.7 | 3 | 82.4 (0.5) | −0.5 | 3 | 87.0 (1.5) | −4.2 | 3 |
| 1 μg/mL Berberine | 80.2 (2.4) | −3.7 | 3 | 79.7 (2.6) | −3.7 | 3 | 87.0 (4.2) | −4.2 | 3 |
| 10 μg/mL Berberine | 102.2 (1.9) | 22.7* | 3 | 95.2 (1.8) | 14.9* | 3 | 101.8 (2.3) | 12.1* | 3 |
| 100 μg/mL Berberine | 227.1 (6.7) | 172.6* | 3 | 228.6 (2.9) | 176.1* | 3 | 238.1 (2.5) | 162.3* | 3 |
*p < 0.05, compared to 1% DMSO, Welch’s (unequal variance) t test
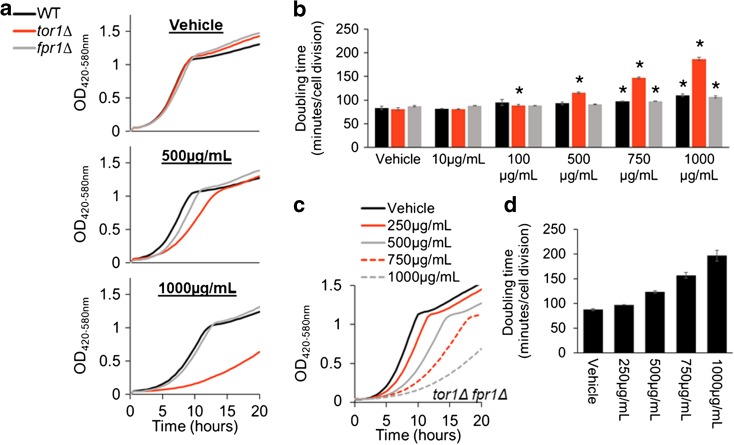
Among the nutraceuticals tested, caffeine was the only compound that showed growth inhibition consistent with an mTOR inhibitory effect, increasing doubling time specifically in the tor1Δ mutant cells at 100 μg/mL (Fig. 4). Caffeine also increased WT and fpr1Δ doubling times at concentrations greater than 750 μg/mL. To assess epistatic relationships between Fpr1 and mTORC1, we constructed a tor1Δ fpr1Δ double mutant. Growth inhibitory profiles in tor1Δ fpr1Δ were identical to those of tor1Δ in our caffeine dose response (Fig. 3c), confirming that Fpr1 is not required for caffeine-mediated growth inhibition in yeast.
Fig. 4.
Caffeine inhibits yeast growth in tor1Δ cells independently of Fpr1. a Representative growth curves of 6.67% H2O (vehicle) (top left), 500 μg/mL (middle), and 1000 μg/mL (bottom). b Maximal doubling time (minutes/cell division) for WT, tor1Δ, and fpr1Δ yeast grown in vehicle or 10–1000 μg/mL caffeine (n = 3–5 independent cultures for each strain-treatment tested). Asterisk means significantly different from vehicle control (p < 0.05, Welch’s t test). Error bars = SEM. c tor1Δ fpr1Δ are sensitive to caffeine, indicating that caffeine-mediated growth inhibition in tor1Δ cells is independent of Fpr1. d Peak doubling times for tor1Δ fpr1Δ grown in vehicle or 250–1000 μg/mL caffeine (n = 6 biological replicate cultures for each strain-treatment tested). Error bars = SEM
Discussion
Rapamycin and other mTOR inhibitors have emerged as one of the most promising classes of molecules for treating a variety of diseases and promoting healthy longevity (Kaeberlein 2013). The largest barrier to clinical utilization of these compounds for such purposes is the perceived risk of adverse side effects. While it remains unclear how significant an issue this is in relatively healthy people at lower doses, the perception that mTOR inhibitors are risky drugs remains a significant challenge. Thus, there is utility in developing or identifying novel mTOR inhibitors that are effective in vivo and which may have fewer side effects. Of particular interest are natural product “nutraceutical” mTOR inhibitors.
By utilizing yeast strains with differential sensitivity to mTORC1 inhibition, we developed an in vivo screening platform that allows us to preliminarily categorize compounds as growth inhibitory, mTOR inhibitory, and/or rapamycin mimetics. Compounds that inhibit WT, tor1Δ, and fpr1Δ cells similarly are general growth inhibitors, but are unlikely to be either direct or indirect mTOR inhibitors, at least in yeast. Compounds that induce growth inhibition preferentially or specifically in tor1Δ cells are candidate mTOR inhibitors, and if fpr1Δ cells are resistant to these compounds, then they are likely inhibiting mTOR by a mechanism similar to rapamycin. The assay system performed as expected here for rapamycin, the rapalogs everolimus and temsirolimus, the catalytic mTOR inhibitors Torin 1 and GSK2126458, and the natural product mTOR inhibitor caffeine. Therefore, we conclude that this assay system is likely to be suitable for identification of novel, unknown pharmaceutical and natural product mTOR inhibitors.
Our analyses also confirmed previously observed cases of differential sensitivity between yeast and mammals with regard to the catalytic mTOR inhibitors AZD8055 and GDC-0941, which may be due to sequence differences between the mammalian and yeast proteins (Wu et al. 2015). It is also likely that equivalent doses of some compounds will yield different intracellular concentrations in yeast cells relative to mammalian cells, and that yeast may be able to clear or otherwise detoxify certain compounds more or less effectively. Thus, it is important to recognize that any hits identified in the yeast-based screening system described here will need to be validated in mammalian cells to confirm similar mTOR-inhibitory effects, and that failure to detect mTOR inhibition in this system for a given compound does not rule out the possibility that the compound could inhibit mTOR in mammalian cells. Nonetheless, given the biochemical and mechanistic similarities between yeast and mammalian mTORC1 and mTORC2, we anticipate that many compounds will behave similarly in both systems, as is the case for several examples reported here.
It is of particular interest that, among the nutraceutical compounds tested, only caffeine demonstrated growth kinetics consistent with mTORC1 inhibition in our yeast assay. This supports prior studies of caffeine on mTOR activity in budding yeast (Reinke et al. 2006; Wanke et al. 2008) and fission yeast (Rallis et al. 2013), and is of interest in light of numerous reports that caffeine can extend life span in invertebrate models (Sutphin et al. 2012; Bridi et al. 2015) and that coffee consumption is associated with reduced mortality in people (Loftfield et al. 2015; Je and Giovannucci 2014). It is intriguing to speculate that these effects could be related to the caffeine’s mTOR inhibitory activities.
The absence of effects from the other nutraceutical compounds tested suggests that they may not be true mTOR inhibitors. One potential explanation is that some of these compounds, which were generally reported to inhibit mTOR in cancer cell culture models, act via indirect mechanisms through targets that are not present or not similarly affected in yeast. Another possibility is that some of these compounds inhibit mammalian cell growth or nutrient uptake, perhaps specifically in the context of cancer cell culture models, which could have indirect effects on mTOR signaling in response. One interesting example is resveratrol, which is reported to have numerous targets in mammalian cells and to inhibit mTOR through both direct and indirect mechanisms. In one study, resveratrol was found to enhance the physical interaction between mTOR and its inhibitor DEPTOR (Liu et al. 2010), which has no obvious yeast ortholog. Resveratrol has also been suggested to inhibit mTOR by indirect mechanisms including regulation of phosphoinositide 3-kinase (PI3K), Akt (Brito et al. 2009), and AMP-activated protein kinase (AMPK) (Tillu et al. 2012). We found no evidence here to support an mTOR-inhibitory role for resveratrol in yeast, nor any effect on growth at all, consistent with prior work showing that resveratrol does not impact yeast growth or replicative life span (Kaeberlein et al. 2005b).
Overall, the system described here represents a sensitive, high-throughput, and inexpensive approach to identify growth inhibitory molecules with specificity for mTOR in vivo. It may be possible to further optimize this system, for example, by screening compounds in a drug sensitized background or by humanizing the system through expression of human proteins in the mTOR signaling pathway in yeast. Additionally, this method is easily adapted for testing drug interactions by adding a mixture of two or more compounds, as well as for investigating the impact of genetic diversity on mTORC1 inhibition through use of large-scale genetic libraries or wild isolate yeast strains (Fay and Benavides 2005; Liti et al. 2009). The identification of new mTOR inhibitors and genetic variants that impact sensitivity to mTOR inhibition will facilitate the development of new therapies and personalized approaches for a breadth of conditions where mTOR signaling is perturbed.
Methods
Yeast strains and culture conditions
All yeast strains used were in the BY4742 genetic background (MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0). BW1121 (BY4742) was purchased from Thermo Fisher Scientific (Waltham, MA). GS668 (fpr1Δ::kanMX4) and GS983 (tor1Δ::kanMX4) were obtained from the haploid MATα yeast deletion collection (Winzeler et al. 1999). PCR was used to confirm knockout identity. For GS983, 5′-TTGAATCCTAATTTCTTGCTCAATC-3′ and 5′-AAGGCATATATTGATGCTCAAAAAG-3′ primers were used to confirm knockout. For GS668, 5′-GTTACTTGATGATATTAAGCACGGG-3′ and 5′-ACAAAAATGAACCATTAGCAAAGAG-3′ primers were used to confirm knockout. The tor1Δ fpr1Δ double deletion strain was constructed by mating tor1Δ::URA3 (Kaeberlein et al. 2005a) and fpr1Δ::kanMX4 and selecting for Ura+ G418r haploid spores after sporulation and tetrad dissection. PCR was used to verify presence of gene deletions. For overnight culture and growth analysis, yeast extract peptone dextrose (YPD) (1% w/v Bacto™ yeast extract (BD), 2% w/v Bacto™ peptone (BD), 2% w/v dextrose) media were used. Yeast were cultured at 30 °C for all experiments. All experiments were repeated at least three times with biological replicates. The number of biological replicates is indicated in each figure legend. Error bars in each figure represent standard error of the mean (SEM).
Growth analysis using Bioscreen C MBR
Growth analysis of maximal growth rate in yeast strains was performed using a Bioscreen C CMB (Growth Curves USA, Piscataway, NJ, USA) as previously described (Murakami et al. 2008, 2011; Burtner et al. 2009). Briefly, colonies outgrown from frozen stocks were inoculated into 5 mL YPD and incubated at 30 °C in a roller drum for 12–16 h. Two microliters of outgrown culture was used to inoculate 148 μL YPD into 100-well honeycomb plates for growth analysis. At least three colonies per strain were analyzed in triplicate for each drug treatment. Doubling times were calculated identifying the slope of the inflection point along growth curves using the online web tool Yeast Outgrowth Data Analyzer (YODA) (Olsen et al. 2010). Welch’s (unequal variance) t test was used to assess statistical significance.
Drug preparation and suppliers
All drugs except caffeine and cycloheximide were suspended in DMSO (these drugs were suspended in H2O). Rapamycin, everolimus, and temsirolimus were purchased from LC Laboratories (Woburn, MA, USA). Cycloheximide and berberine were purchased from Sigma-Aldrich (St. Louis, MO, USA). Torin 1 was purchased from Cayman Chemical (Ann Arbor, MI, USA). AZD8055, GDC-0941, and GSK2126458 were kind gifts from Jason Pitt. Caffeine was purchased from MP Biomedicals (Santa Ana, CA, USA). Resveratrol was purchased from AstaTech Inc. (Bristol, PA, USA). Alpha-lipoic acid, broccoli concentrate, glucosamine, lycopene, and quercetin were provided by USANA Health Sciences, Inc. (Salt Lake City, UT, USA).
Electronic supplementary material
(PDF 162 kb).
(PDF 306 kb).
(PDF 44 kb).
(PDF 45 kb).
Acknowledgments
This work was supported by the University of Washington Nathan Shock Center of Excellence in the Basic Biology of Aging Invertebrate Longevity and Healthspan Core (P30AG013280) and a grant to MK from USANA Health Sciences. MBL was supported by the Howard Hughes Medical Institute (HHMI) Gilliam Fellowship for Advanced Study, the National Institutes of Health (NIH) Cellular and Molecular Biology training grant (T32GM7270-39), and the University of Washington Graduate Opportunities and Minority Achievement Program (UW GO-MAP) Bank of America Fellowship.
Footnotes
Electronic supplementary material
The online version of this article (doi:10.1007/s11357-017-9988-4) contains supplementary material, which is available to authorized users.
References
- Anastasius N, Boston S, Lacey M, Storing N, Whitehead SA. Evidence that low-dose, long-term genistein treatment inhibits oestradiol-stimulated growth in MCF-7 cells by down-regulation of the PI3-kinase/Akt signalling pathway. J Steroid Biochem Mol Biol. 2009;116(1–2):50–55. doi: 10.1016/j.jsbmb.2009.04.009. [DOI] [PubMed] [Google Scholar]
- Anisimov VN, et al. Rapamycin increases lifespan and inhibits spontaneous tumorigenesis in inbred female mice. Cell Cycle. 2011;10(24):4230–4236. doi: 10.4161/cc.10.24.18486. [DOI] [PubMed] [Google Scholar]
- Beaupere C, et al. CAN1 arginine permease deficiency extends yeast replicative lifespan via translational activation of stress response genes. Cell Rep. 2017;18(8):1884–1892. doi: 10.1016/j.celrep.2017.01.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beevers CS, Li F, Liu L, Huang S. Curcumin inhibits the mammalian target of rapamycin-mediated signaling pathways in cancer cells. Int J Cancer. 2006;119(4):757–764. doi: 10.1002/ijc.21932. [DOI] [PubMed] [Google Scholar]
- Beevers CS, et al. Curcumin disrupts the mammalian target of rapamycin-raptor complex. Cancer Res. 2009;69(3):1000–1008. doi: 10.1158/0008-5472.CAN-08-2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton BM, Zang JH, Thorner J. A novel FK506- and rapamycin-binding protein (FPR3 gene product) in the yeast Saccharomyces cerevisiae is a proline rotamase localized to the nucleolus. J Cell Biol. 1994;127(3):623–639. doi: 10.1083/jcb.127.3.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beretta L, Gingras AC, Svitkin YV, Hall MN, Sonenberg N. Rapamycin blocks the phosphorylation of 4E-BP1 and inhibits cap-dependent initiation of translation. EMBO J. 1996;15(3):658–664. [PMC free article] [PubMed] [Google Scholar]
- Bitto A, et al. Transient rapamycin treatment can increase lifespan and healthspan in middle-aged mice. elife. 2016;5:e16351. doi: 10.7554/eLife.16351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjedov I, et al. Mechanisms of life span extension by rapamycin in the fruit fly Drosophila melanogaster. Cell Metab. 2010;11(1):35–46. doi: 10.1016/j.cmet.2009.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blagosklonny MV. Increasing healthy lifespan by suppressing aging in our lifetime: preliminary proposal. Cell Cycle. 2010;9(24):4788–4794. doi: 10.4161/cc.9.24.14360. [DOI] [PubMed] [Google Scholar]
- Bridi JC, et al. Lifespan extension induced by caffeine in Caenorhabditis elegans is partially dependent on adenosine signaling. Front Aging Neurosci. 2015;7:220. doi: 10.3389/fnagi.2015.00220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brito PM, et al. Resveratrol inhibits the mTOR mitogenic signaling evoked by oxidized LDL in smooth muscle cells. Atherosclerosis. 2009;205(1):126–134. doi: 10.1016/j.atherosclerosis.2008.11.011. [DOI] [PubMed] [Google Scholar]
- Burtner CR, Murakami CJ, Kennedy BK, Kaeberlein M. A molecular mechanism of chronological aging in yeast. Cell Cycle. 2009;8(8):1256–1270. doi: 10.4161/cc.8.8.8287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Liu Y, Zheng P. mTOR regulation and therapeutic rejuvenation of aging hematopoietic stem cells. Sci Signal. 2009;2(98):ra75. doi: 10.1126/scisignal.2000559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chresta CM, et al. AZD8055 is a potent, selective, and orally bioavailable ATP-competitive mammalian target of rapamycin kinase inhibitor with in vitro and in vivo antitumor activity. Cancer Res. 2010;70(1):288–298. doi: 10.1158/0008-5472.CAN-09-1751. [DOI] [PubMed] [Google Scholar]
- Dai DF, et al. Altered proteome turnover and remodeling by short-term caloric restriction or rapamycin rejuvenate the aging heart. Aging Cell. 2014;13(3):529–539. doi: 10.1111/acel.12203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaney JR, et al. Stress profiling of longevity mutants identifies Afg3 as a mitochondrial determinant of cytoplasmic mRNA translation and aging. Aging Cell. 2013;12(1):156–166. doi: 10.1111/acel.12032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabrizio P, Pozza F, Pletcher SD, Gendron CM, Longo VD. Regulation of longevity and stress resistance by Sch9 in yeast. Science. 2001;292(5515):288–290. doi: 10.1126/science.1059497. [DOI] [PubMed] [Google Scholar]
- Fan X, et al. Berberine alleviates ox-LDL induced inflammatory factors by up-regulation of autophagy via AMPK/mTOR signaling pathway. J Transl Med. 2015;13:92. doi: 10.1186/s12967-015-0450-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay JC, Benavides JA. Evidence for domesticated and wild populations of Saccharomyces cerevisiae. PLoS Genet. 2005;1(1):66–71. doi: 10.1371/journal.pgen.0010005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn JM, et al. Late-life rapamycin treatment reverses age-related heart dysfunction. Aging Cell. 2013;12(5):851–862. doi: 10.1111/acel.12109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folkes AJ, et al. The identification of 2-(1H-indazol-4-yl)-6-(4-methanesulfonyl-piperazin-1-ylmethyl)-4-morpholin-4-yl-t hieno[3,2-d]pyrimidine (GDC-0941) as a potent, selective, orally bioavailable inhibitor of class I PI3 kinase for the treatment of cancer. J Med Chem. 2008;51(18):5522–5532. doi: 10.1021/jm800295d. [DOI] [PubMed] [Google Scholar]
- Halloran J, et al. Chronic inhibition of mammalian target of rapamycin by rapamycin modulates cognitive and non-cognitive components of behavior throughout lifespan in mice. Neuroscience. 2012;223:102–113. doi: 10.1016/j.neuroscience.2012.06.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison DE, et al. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature. 2009;460(7253):392–395. doi: 10.1038/nature08221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitman J, Movva NR, Hall MN. Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science. 1991;253(5022):905–909. doi: 10.1126/science.1715094. [DOI] [PubMed] [Google Scholar]
- Helliwell SB, et al. TOR1 and TOR2 are structurally and functionally similar but not identical phosphatidylinositol kinase homologues in yeast. Mol Biol Cell. 1994;5(1):105–118. doi: 10.1091/mbc.5.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacinto E, et al. Mammalian TOR complex 2 controls the actin cytoskeleton and is rapamycin insensitive. Nat Cell Biol. 2004;6(11):1122–1128. doi: 10.1038/ncb1183. [DOI] [PubMed] [Google Scholar]
- Je Y, Giovannucci E. Coffee consumption and total mortality: a meta-analysis of twenty prospective cohort studies. Br J Nutr. 2014;111(7):1162–1173. doi: 10.1017/S0007114513003814. [DOI] [PubMed] [Google Scholar]
- Jiang L, Jin Y, Wang H, Jiang Y, Dong J. Glucosamine protects nucleus pulposus cells and induces autophagy via the mTOR-dependent pathway. J Orthop Res. 2014;32(11):1532–1542. doi: 10.1002/jor.22699. [DOI] [PubMed] [Google Scholar]
- Johnson SC, Rabinovitch PS, Kaeberlein M. mTOR is a key modulator of ageing and age-related disease. Nature. 2013;493(7432):338–345. doi: 10.1038/nature11861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson SC, Martin GM, Rabinovitch PS, Kaeberlein M. Preserving youth: does rapamycin deliver? Sci Transl Med. 2013;5(211):211fs240. doi: 10.1126/scitranslmed.3007316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson SC, Sangesland M, Kaeberlein M, Rabinovitch PS. Modulating mTOR in aging and health. Interdiscip Top Gerontol. 2015;40:107–127. doi: 10.1159/000364974. [DOI] [PubMed] [Google Scholar]
- Kaeberlein M. Lessons on longevity from budding yeast. Nature. 2010;464(7288):513–519. doi: 10.1038/nature08981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein M. mTOR inhibition: from aging to autism and beyond. Scientifica (Cairo) 2013;2013:849186. doi: 10.1155/2013/849186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein M, et al. Regulation of yeast replicative life span by TOR and Sch9 in response to nutrients. Science. 2005;310(5751):1193–1196. doi: 10.1126/science.1115535. [DOI] [PubMed] [Google Scholar]
- Kaeberlein M, et al. Substrate-specific activation of sirtuins by resveratrol. J Biol Chem. 2005;280(17):17038–17045. doi: 10.1074/jbc.M500655200. [DOI] [PubMed] [Google Scholar]
- Kaeberlein M, Rabinovitch PS, Martin GM. Healthy aging: the ultimate preventative medicine. Science. 2015;350(6265):1191–1193. doi: 10.1126/science.aad3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein M, Creevy KE, Promislow DE. The dog aging project: translational geroscience in companion animals. Mamm Genome. 2016;27(7–8):279–288. doi: 10.1007/s00335-016-9638-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight SD, et al. Discovery of GSK2126458, a highly potent inhibitor of PI3K and the mammalian target of rapamycin. ACS Med Chem Lett. 2010;1(1):39–43. doi: 10.1021/ml900028r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunz J, et al. Target of rapamycin in yeast, TOR2, is an essential phosphatidylinositol kinase homolog required for G1 progression. Cell. 1993;73(3):585–596. doi: 10.1016/0092-8674(93)90144-F. [DOI] [PubMed] [Google Scholar]
- Lamming DW, et al. Rapamycin-induced insulin resistance is mediated by mTORC2 loss and uncoupled from longevity. Science. 2012;335(6076):1638–1643. doi: 10.1126/science.1215135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Dungan CM, Carrier B, Rideout TC, Williamson DL. Alpha-lipoic acid supplementation reduces mTORC1 signaling in skeletal muscle from high fat fed, obese Zucker rats. Lipids. 2014;49(12):1193–1201. doi: 10.1007/s11745-014-3964-x. [DOI] [PubMed] [Google Scholar]
- Liti G, et al. Population genomics of domestic and wild yeasts. Nature. 2009;458(7236):337–341. doi: 10.1038/nature07743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, et al. Resveratrol inhibits mTOR signaling by promoting the interaction between mTOR and DEPTOR. J Biol Chem. 2010;285(47):36387–36394. doi: 10.1074/jbc.M110.169284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, et al. Selective ATP-competitive inhibitors of TOR suppress rapamycin-insensitive function of TORC2 in Saccharomyces cerevisiae. ACS Chem Biol. 2012;7(6):982–987. doi: 10.1021/cb300058v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewith R, et al. Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol Cell. 2002;10(3):457–468. doi: 10.1016/S1097-2765(02)00636-6. [DOI] [PubMed] [Google Scholar]
- Loftfield E, et al. Association of coffee consumption with overall and cause-specific mortality in a large US prospective cohort study. Am J Epidemiol. 2015;182(12):1010–1022. doi: 10.1093/aje/kwv146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longo VD, Shadel GS, Kaeberlein M, Kennedy B. Replicative and chronological aging in Saccharomyces cerevisiae. Cell Metab. 2012;16(1):18–31. doi: 10.1016/j.cmet.2012.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz MC, Heitman J. TOR mutations confer rapamycin resistance by preventing interaction with FKBP12-rapamycin. J Biol Chem. 1995;270(46):27531–27537. doi: 10.1074/jbc.270.46.27531. [DOI] [PubMed] [Google Scholar]
- Lu Q, et al. Quercetin inhibits the mTORC1/p70S6K signaling-mediated renal tubular epithelial-mesenchymal transition and renal fibrosis in diabetic nephropathy. Pharmacol Res. 2015;99:237–247. doi: 10.1016/j.phrs.2015.06.006. [DOI] [PubMed] [Google Scholar]
- Majumder S, et al. Lifelong rapamycin administration ameliorates age-dependent cognitive deficits by reducing IL-1beta and enhancing NMDA signaling. Aging Cell. 2012;11(2):326–335. doi: 10.1111/j.1474-9726.2011.00791.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannick JB, et al. mTOR inhibition improves immune function in the elderly. Sci Transl Med. 2014;6(268):268ra179. doi: 10.1126/scitranslmed.3009892. [DOI] [PubMed] [Google Scholar]
- McCormick MA, et al. A comprehensive analysis of replicative lifespan in 4,698 single-gene deletion strains uncovers conserved mechanisms of aging. Cell Metab. 2015;22(5):895–906. doi: 10.1016/j.cmet.2015.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng FD, et al. Synergistic effects of snail and quercetin on renal cell carcinoma Caki-2 by altering AKT/mTOR/ERK1/2 signaling pathways. Int J Clin Exp Pathol. 2015;8(6):6157–6168. [PMC free article] [PubMed] [Google Scholar]
- Murakami C & Kaeberlein M (2009) Quantifying yeast chronological life span by outgrowth of aged cells. J Vis Exp. doi:10.3791/1156 [DOI] [PMC free article] [PubMed]
- Murakami CJ, Burtner CR, Kennedy BK, Kaeberlein M. A method for high-throughput quantitative analysis of yeast chronological life span. J Gerontol A Biol Sci Med Sci. 2008;63(2):113–121. doi: 10.1093/gerona/63.2.113. [DOI] [PubMed] [Google Scholar]
- Murakami CJ, Wall V, Basisty N, Kaeberlein M. Composition and acidification of the culture medium influences chronological aging similarly in vineyard and laboratory yeast. PLoS One. 2011;6(9):e24530. doi: 10.1371/journal.pone.0024530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neff F, et al. Rapamycin extends murine lifespan but has limited effects on aging. J Clin Invest. 2013;123(8):3272–3291. doi: 10.1172/JCI67674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell JS, Massi D, Teng MWL, & Mandala M (2017) PI3K-AKT-mTOR inhibition in cancer immunotherapy, redux. Semin Cancer Biol. doi:10.1016/j.semcancer.2017.04.015 [DOI] [PubMed]
- Olsen B, Murakami CJ, Kaeberlein M. YODA: software to facilitate high-throughput analysis of chronological life span, growth rate, and survival in budding yeast. BMC Bioinformatics. 2010;11:141. doi: 10.1186/1471-2105-11-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park D, et al. Resveratrol induces autophagy by directly inhibiting mTOR through ATP competition. Sci Rep. 2016;6:21772. doi: 10.1038/srep21772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popovich IG, et al. Lifespan extension and cancer prevention in HER-2/neu transgenic mice treated with low intermittent doses of rapamycin. Cancer Biol Ther. 2014;15(5):586–592. doi: 10.4161/cbt.28164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers RW, 3rd, Kaeberlein M, Caldwell SD, Kennedy BK, Fields S. Extension of chronological life span in yeast by decreased TOR pathway signaling. Genes Dev. 2006;20(2):174–184. doi: 10.1101/gad.1381406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rallis C, Codlin S, Bahler J. TORC1 signaling inhibition by rapamycin and caffeine affect lifespan, global gene expression, and cell proliferation of fission yeast. Aging Cell. 2013;12(4):563–573. doi: 10.1111/acel.12080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinke A, Chen JC, Aronova S, Powers T. Caffeine targets TOR complex I and provides evidence for a regulatory link between the FRB and kinase domains of Tor1p. J Biol Chem. 2006;281(42):31616–31626. doi: 10.1074/jbc.M603107200. [DOI] [PubMed] [Google Scholar]
- Robida-Stubbs S, et al. TOR signaling and rapamycin influence longevity by regulating SKN-1/Nrf and DAF-16/FoxO. Cell Metab. 2012;15(5):713–724. doi: 10.1016/j.cmet.2012.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiki S, et al. Caffeine induces apoptosis by enhancement of autophagy via PI3K/Akt/mTOR/p70S6K inhibition. Autophagy. 2011;7(2):176–187. doi: 10.4161/auto.7.2.14074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxton RA, Sabatini DM. mTOR signaling in growth, metabolism, and disease. Cell. 2017;168(6):960–976. doi: 10.1016/j.cell.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffen KK, et al. Yeast life span extension by depletion of 60s ribosomal subunits is mediated by Gcn4. Cell. 2008;133(2):292–302. doi: 10.1016/j.cell.2008.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutphin GL, Bishop E, Yanos ME, Moller RM, Kaeberlein M. Caffeine extends life span, improves healthspan, and delays age-associated pathology in Caenorhabditis elegans. Longev Healthspan. 2012;1:9. doi: 10.1186/2046-2395-1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillu DV, et al. Resveratrol engages AMPK to attenuate ERK and mTOR signaling in sensory neurons and inhibits incision-induced acute and chronic pain. Mol Pain. 2012;8:5. doi: 10.1186/1744-8069-8-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urfer SR, et al. A randomized controlled trial to establish effects of short-term rapamycin treatment in 24 middle-aged companion dogs. Geroscience. 2017;39(2):117–127. doi: 10.1007/s11357-017-9972-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urfer SR, et al. Asymptomatic heart valve dysfunction in healthy middle-aged companion dogs and its implications for cardiac aging. Geroscience. 2017;39(1):43–50. doi: 10.1007/s11357-016-9956-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Aller GS, et al. Epigallocatechin gallate (EGCG), a major component of green tea, is a dual phosphoinositide-3-kinase/mTOR inhibitor. Biochem Biophys Res Commun. 2011;406(2):194–199. doi: 10.1016/j.bbrc.2011.02.010. [DOI] [PubMed] [Google Scholar]
- Wanke V, et al. Caffeine extends yeast lifespan by targeting TORC1. Mol Microbiol. 2008;69(1):277–285. doi: 10.1111/j.1365-2958.2008.06292.x. [DOI] [PubMed] [Google Scholar]
- Wiczk A, Hofman D, Konopa G, Herman-Antosiewicz A. Sulforaphane, a cruciferous vegetable-derived isothiocyanate, inhibits protein synthesis in human prostate cancer cells. Biochim Biophys Acta. 2012;1823(8):1295–1305. doi: 10.1016/j.bbamcr.2012.05.020. [DOI] [PubMed] [Google Scholar]
- Winzeler EA, et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285(5429):901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- Wu TJ, et al. Identification of a non-gatekeeper hot spot for drug-resistant mutations in mTOR kinase. Cell Rep. 2015;11(3):446–459. doi: 10.1016/j.celrep.2015.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124(3):471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- Xie R, et al. Alpha-lipoic acid pre- and post-treatments provide protection against in vitro ischemia-reperfusion injury in cerebral endothelial cells via Akt/mTOR signaling. Brain Res. 2012;1482:81–90. doi: 10.1016/j.brainres.2012.09.009. [DOI] [PubMed] [Google Scholar]
- Zhang Q, et al. Green tea extract and (−)-epigallocatechin-3-gallate inhibit mast cell-stimulated type I collagen expression in keloid fibroblasts via blocking PI-3K/AkT signaling pathways. J Invest Dermatol. 2006;126(12):2607–2613. doi: 10.1038/sj.jid.5700472. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF 162 kb).
(PDF 306 kb).
(PDF 44 kb).
(PDF 45 kb).