Abstract
Pristinamycin, produced by Streptomyces pristinaespiralis, which is a streptogramin-like antibiotic consisting of two chemically unrelated components: pristinamycin I (PI) and pristinamycin II (PII), shows potent activity against many antibiotic-resistant pathogens. However, so far pristinamycin production titers are still quite low, particularly those of PI. In this study, we constructed a PI single component producing strain by deleting the PII biosynthetic genes (snaE1 and snaE2). Then, two metabolic engineering approaches, including deletion of the repressor gene papR3 and chromosomal integration of an extra copy of the PI biosynthetic gene cluster (BGC), were employed to improve PI production. The final engineered strain ΔPIIΔpapR3/PI produced a maximum PI level of 132 mg/L, with an approximately 2.4-fold higher than that of the parental strain S. pristinaespiralis HCCB10218. Considering that the PI biosynthetic genes are clustered in two main regions in the 210 kb “supercluster” containing the PI and PII biosynthetic genes as well as a cryptic polyketide BGC, these two regions were cloned separately and then were successfully assembled into the PI BGC by the transformation-associated recombination (TAR) system. Collectively, the metabolic engineering approaches employed is very efficient for strain improvement in order to enhance PI titer.
Keywords: Streptomyces pristinaespiralis, Pristinamycin I, Biosynthetic gene cluster, Metabolic engineering
1. Introduction
Pristinamycin is a streptogramin-like antibiotic consisting of a mixture of two chemically unrelated components: pristinamycin I (PI), a branched cyclic hexadepsipeptide (Fig. 1A) and pristinamycin II (PII), a polyunsaturated cyclo-peptide macrolactone [1]. PI and PII are produced by Streptomyces pristinaespiralis in a ratio of 30:70 and are synthesized by non-ribosomal peptide synthetases (NRPSs) and hybrid polyketide synthase (PKS)/NRPS enzymes, respectively [2]. The combination of the two compounds shows a strong synergistic effect, with a bactericidal activity 100-fold higher than that of the single components [3]. Pristinamycin is highly active against many antibiotic-resistant pathogens, particularly Gram-positive bacteria, including methicillin-resistant Staphylococcus aureus (MRSA), vancomycin-resistant S. aureus (VRSA) and Enterococcus faecium (VREF) [4]. Currently, pristinamycin and its derivatives are used as therapeutics to treat severe bacterial infections caused by multi-drug resistant pathogens. However, so far the production titers of pristinamycin (both PI and PII) by S. pristinaespiralis are still quite low and require to be improved for scale-up industrial production.
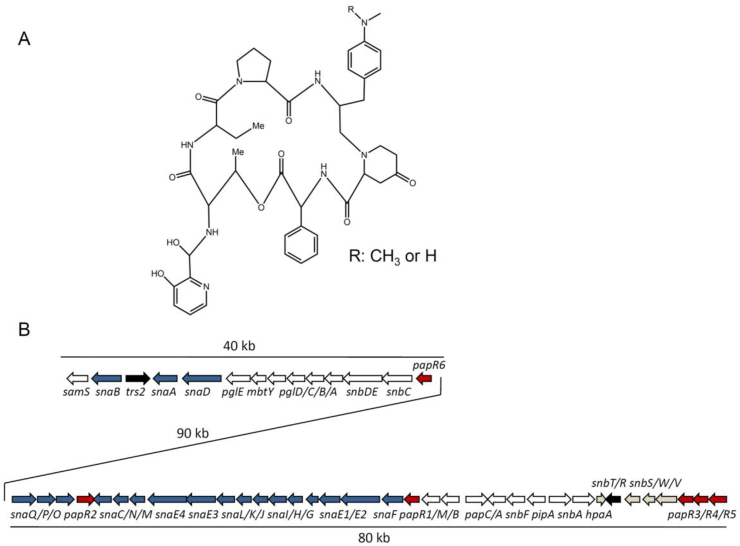
Fig. 1.
The chemical structure of pristinamycin I (PI) and organization of the pristinamycin biosynthetic gene cluster (BGC) in S. pristinaespiralis. A. The PI chemical structure. B. PI and PII biosynthetic genes are indicated as white and blue arrows, respectively. Regulatory genes are shown as red arrows. Genes for resistance and of general function are shown as black arrows. Genes of unknown function are indicated as grey arrows. PI and PII biosynthetic genes are interspersed by approximately 90 kb as indicated.
Because of the toxic effects of the synergistic active PI/PII combination on mycelial growth of S. pristinaespiralis [5], as well as mutual interference when they are purified, we aimed to generate two high single component (PI or PII) producing strains. Recently, in our research group, markedly increased PII production has been achieved by two metabolic engineering approaches [5], [6], including modification of the pathway-specific regulation pathway and multi-copy chromosomal integration of the PII biosynthetic gene cluster (BGC). For these approaches S. pristinaespiralis HCCB10218 has been used as the starting strain, which is generated from traditional physical and chemical mutagenesis of the wild-type strain ATCC25486 [7]. By adding a macroreticular resin, the final engineered strain SBJ1005 produced maximum PII titers of approximately 2.2 g/L and 2 g/L in Erlenmeyer flasks and a 5-L bioreactor, respectively, which are the highest PII titers ever reported [6]. However, so far, the construction of high PI-producing strains was not reported yet.
In the past few years, the genes responsible for the biosynthesis of pristinamycin I has been well characterized [2]. They are clustered together with the PII biosynthetic genes as well as a cryptic type II polyketide BGC in a 210 kb “supercluster” [2] (Fig. 1B). In addition, much effort has been made in our understanding of the regulation of pristinamycin biosynthesis. PapR1, R2 and R4, belong to SARP (Streptomyces antibiotic regulatory proteins) family regulators and act as activators for both PI and PII biosynthesis, while PapR3 and R5, belonging to TetR family proteins, act as repressors [2], [8]. PapR6 is a response regulator-like protein and acts as a pathway-specific regulator of PII biosynthesis [7]. SpbR, a γ-butyrolactone receptor, acts as a global player in the regulation of pristinamycin production but affects also bacterial growth and development [9]. Knowledge on the pristinamycin BGC as well as on the regulation of pristinamycin biosynthesis provides clues for strain improvement by metabolic engineering approaches. Furthermore, in Streptomyces various metabolic engineering approaches have been developed [10], [11], such as manipulating regulatory pathways, increasing the supply of specific building blocks and amplification of secondary metabolite BGCs of interest, which have been proven as efficient methods for Streptomyces strain improvement.
In this study, two PII biosynthetic genes snaE1 and E2 (encoding the deduced hybrid PKS/NRPS complex SnaE1 and the predicted PKS SnaE2, respectively) were deleted to yield a sole PI-producing strain. Then, two metabolic engineering approaches, including deletion of the repressor gene papR3 and adding an extra copy of the PI BGC, were employed to enhance PI production. The final engineered strain produced over two-fold higher PI titers than that of the parental strain S. pristinaespiralis HCCB10218, indicating that the strategy employed here is very efficient for PI overproduction.
2. Materials and methods
2.1. Bacterial strains, plasmids and culture conditions
All strains and plasmids used in this study are listed in Table 1. S. pristinaespiralis HCCB10218 (CGMCC 5486) is a pristinamycin producing strain isolated after traditional mutagenesis (physical and chemical mutations) of the wild-type strain ATCC 25486 [7]. S. pristinaespiralis strains were cultivated on RP agar at 30 °C for the preparation of spore suspensions as well as for intergeneric conjugation [5]. For the analysis of PI production, S. pristinaespiralis strains were grown in seed medium and fermentation medium as described previously [2]. E. coli strains were grown on Luria-Bertani (LB) agar or in LB broth at 37 °C. DH5α and EPI300 were used as the hosts for plasmid construction and for the propagation of the recombinant plasmid (pCAP-PI) containing the PI BGC, respectively. The methylation defective E. coli ET12567/pUZ8002 (a RK2 derivative) was used as the donor in the experiments of intergeneric conjugation between E. coli and S. pristinaespiralis [12]. Saccharomyces cerevisiae VL6-48 was grown in YPD medium [13] and was used for cloning and assembly of the PI BGC. When necessary, antibiotics, such as kanamycin (50 μg/ml), apramycin (50 μg/ml), chloramphenicol (33 μg/ml) or/and ampicillin (100 μg/ml) were added.
Table 1.
Strains and plasmids used in this study.
| Strains | Genotype | Source |
|---|---|---|
| E. coli strains | ||
| DH5α | F–Φ80lacZΔM15 Δ(lacZYA-argF) U169 deoR recA1 endA1 hsdR17(rk-mk+) phoA supE44 λ- thi-1 gyrA96 relA1 | Gibco-BRL |
| EPI300 | recA1 endA1 araD139 Δ(ara, leu)7697 λ- rpsL(Strr) trfA dhfr | Epicentre |
| ET12567/pUZ8002 | The methylation defective strain ET12567 containing the RP4 derivative plasmid pUZ8002 | [18] |
| S. cerevisiae | ||
| VL6-48 | Host strain; MAT alpha, his3-D200, trp1-D1, ura3-52, lys2, ade2-101, met14, psi + cir0 | ATCC MYA-3666 |
| S. pristinaespiralis | ||
| HCCB10218 | Parental strain | [7] |
| ΔPII | Mutant strain, HCCB10218 with deletion of the PII biosynthetic genes (snaE1 and snaE2) | This study |
| ΔPIIΔpapR3 | Mutant strain, HCCB10218 with deletion of both the PII biosynthetic genes (snaE1 and snaE2) and the repressor gene papR3 | This study |
| ΔPIIΔpapR3/PI | Mutant strain, ΔPIIΔpapR3 with chromosomal integration of one copy of the assembled PI BGC via site-specific recombination | This study |
| Plasmids | ||
| pKCcas9dO | CRISPR/Cas9 editing plasmid for deletion of the pathway-specific activator gene actII-ORF4 of ACT biosynthesis in S. coelicolor, acc(3)IV, pSG5 ori, tipAp-Scocas9, j23119p-sgRNA | [14] |
| pKCcas9dPII | CRISPR/Cas9 editing plasmid for deletion of the PII biosynthetic genes (snaE1 and snaE2), acc(3)IV, pSG5 ori, tipAp-Scocas9, j23119p-sgRNA | This study |
| pKCcas-papR3 | CRISPR/Cas9 editing plasmid for papR3 deletion, acc(3)IV, pSG5 ori, tipAp-Scocas9, j23119-sgRNA | This study |
| pCAP01 | Gene cluster capture vector; ARSH4/CEN6, pUC ori, aph(3)II, ΦC31 int/attP, oriT (RP4) | [13] |
| pCAP-spr1 | Recombinant plasmid containing the PI BGC region between pglE and snbC (28 kb) cloned in pCAP01 | This study |
| pCAP-spr2 | Recombinant plasmid containing the PI BGC region between papM and hpaA (11 kb) cloned in pCAP01 | This study |
| pCAP-PI | Recombinant plasmid containing the assembled PI BGC cloned in pCAP01 | This study |
2.2. Construction of S. pristinaespiralis mutants
The mutants ΔPII and ΔPIIΔpapR3, with the respective deletion of the PII biosynthetic genes (snaE1/E2, totally 18,781 bp), as well as both, snaE1/E2 and papR3 repressor gene deletion (1575 bp) were constructed on the basis of the parental strain HCCB10218 using the CRISPR/Cas9-mediated genome editing method as described previously [14]. Here, we present ΔPII as an example to give a brief introduction of the procedure for mutant construction: The sgRNA transcription cassette was obtained by PCR with the primer pair snaEgRNA-fw/gRNA-rev and the plasmid pKCcas9dO as the template. Two homologous arms (1921 and 1973 bp) were amplified from the genomic DNA of S. pristinaespiralis HCCB10218 using the primer pairs snaE-up-fw/rev and snaE-down-fw/rev, respectively. The three DNA fragments were ligated together by an overlapping PCR using the primers snaEsgRNA-fw/snaE-down-rev. The obtained PCR product was cloned as SpeI/HindIII–restricted fragment into the plasmid pKCcas9dO, which was pretreated with the same two restriction enzymes, resulting in pKCcas-PII. pKCcas-PII was introduced into HCCB10218 by intergeneric conjugation between ET12567/pUZ8002 and S. pristinaespiralis. The mutants with correct deletion of snaE1/E2 were identified by colony PCR with the primer pairs, JsnaE-inner-fw/rev and JsnaE-outer-fw/rev, respectively, followed by DNA sequencing. The obtained mutant was named as ΔPII. Using the ΔPII mutant as the starting strain, the ΔPIIΔpapR3 mutant was constructed similarly by deleting the repressor gene papR3. The primers used are listed in Table 2.
Table 2.
Primers used in this study.
| Primers | Sequences (5′-3′) |
|---|---|
| Amplification of two sgRNAs used for deletion of the PII biosynthetic genes (snaE1 and snaE2) and papR3, respectively (N20, guide sequences are underlined and the restriction enzyme sites are in italics) | |
| snaE-gRNA-fw | AGCTAGCTCAGTCCTAGGTATAATACTAGTTGGGTCCGTAGACGTTCCACGTTTTAGAGCTAGAAATA |
| papR3-gRNA-fw | AGCTAGCTCAGTCCTAGGTATAATACTAGTCGGCGCGTACGACGGGGTCCGTTTTAGAGCTAGAAATACTCAAAAAAAGCACCGACTCGG |
| gRNA-rev | CTCAAAAAAAGCACCGACTCGG |
| Amplification of upstream and downstream homologous arms (the sequences for overlapping PCR are underlined and the enzyme sites are in italics) | |
| snaE-up-fw | CACCGAGTCGGTGCTTTTTTTGAGGCCGTAGGCGTTGTTGAGG |
| snaE-up-rev | CCGCACCAACGGCTACACC |
| snaE-down-fw | GGCACGGTGTAGCCGTTGGTGCGGGAGGAAGGCGGGGAAGGTC |
| snaE-down-rev | CGTTGTAAAACGACGGCCAGTGCCAAGCTTGCACCCTGGACACCTGGCT |
| papR3-up-fw | AAGTGGCACCGAGTCGGTGCTTTTTTTGAGCGAAACCCGACGCACGAT |
| papR3-up-rev | CGTTCCCGTCGGCGAGTCATC |
| papR3-down-fw | CATGCGGGATGACTCGCCGACGGGAACGGCGTGGGTGGTTGCCTCTG |
| papR3-down-rev | CGTTGTAAAACGACGGCCAGTGCCAAGCTTCGAGCGGATGCTGCACGAC |
| Verification of the deletion of the PII biosynthetic genes (snaE1 and snaE2) and papR3 | |
| JsnaE-inner-fw | GTGTCAGGGGCGGGGAGGAA |
| JsnaE-inner-rev | CTGCACGGCGTCGTCCACG |
| JsnaE-outer-fw | CGGGCGGACAGGAACACCA |
| JsnaE-outer-rev | ACATCCGCACCGCCTTCG |
| JpapR3-inner-fw | ACCATCGGTGTACGGCTTCT |
| JpapR3-inner-fw | GGACGCCACCCATGTGCTGA |
| JpapR3-outer-fw | GTGTCGTCGTGGGCAGGTGT |
| JpapR3-outer-fw | CGGCTATCTGCTGAACACCTCC |
| Amplification of sgRNAs for Cas9-mediated in vitro DNA digestion (N20, guide sequences are underlined and T7 promoter sequences are in italics) | |
| spr1-gRNA1-fw | GACTGACACTGATAATACGACTCACTATAGGACGAAGGTGCAGTTGAAGTTTTAGAGCTAGAAATAGC |
| spr1-gRNA2-fw | GACTGACACTGATAATACGACTCACTATAGGTGCGCAAGGAACTGGAGTTTTAGAGCTAGAAATAGC |
| gRNA-rev | CTCAAAAAAAGCACCGACTCGG |
| Amplification of upstream and downstream homologous arms for the construction of the spr1 and spr2 capture vectors (the overlapping sequences designed for the assembly of spr1 and spr2 are underlined and the enzyme sites are in italics) | |
| spr1UP-fw | GACTAGTTACGTGCCGAGGGCCTTGAGGTA |
| spr1UP-rev | GGAATTCCATATGGAGTCCGAGGACACCGCGCGAAGC |
| spr1DOWN-fw | GGAATTCCATATGTCAGCGCAGGCCGGTCTCGAAGGCGTA |
| spr1DOWN-rev | GGGGTACCGCTCTAGACCGCCCTGTGCGAACTGCTGGAACCCGACGACCTGCTCTTAGTTCCTCGCGCAGCCACAGAT |
| spr2UP-fw | GACTAGTACCAGCTGATCGCGGTCCATCTGTGGCTGCGCGAGGAACTAAGAGCAGGTCGTCGGGTTC |
| spr2UP-rev | GGAATTCCATATGATCCTCCAGCTGCGCGAACTGA |
| spr2DOWN-fw | GGAATTCCATATGGTATGCCGTTCATCGAAGTG |
| spr2DOWN-rev | CCGCTCGAGACTACGACCGTCCCGTGCCGCGAAG |
| Verification of direct cloning of spr1 and spr2 as well as of the assembly of PI BGC | |
| Jspr1-A-fw | CCAGGAAACGGACGAAGCG |
| Jspr1-A-rev | CCTGACGGGCTTGTCTGCTC |
| Jspr1-B -fw | GAGGGAGTCGTAGGTCTGCTGC |
| Jspr1-B- rev | GAGCCCTACCAGCACATCGTC |
| Jspr2-A-fw | GGACCCGTTGGCAGGAAGCA |
| Jspr2-A-rev | ACACGGCTCCTACCAACTCG |
| Jspr2-B-fw | TGGTCCTTCAGGCACAGCA |
| Jspr2-B-rev | CGACCACATCACCCTCAAGACC |
2.3. Cloning of the PI BGC
The PI biosynthetic genes are located in two main regions, including region 1 (named as spr1, from pglE to snbC, approximately 28 kb) and region 2 (named as spr2, from papM to hpaA, approximately 11 kb), as presented in Fig. 1. spr1 and spr2 were cloned separately (2.3.1) and then assembled together (2.3.2). It should be noted that the samS gene possibly involved in PI biosynthesis, which is located upstream of the PII biosynthetic gene snaB, was not included. The primers used are listed in Table 2.
2.3.1. Cloning of two partial PI BGC (spr1 and spr2) separately
The spr1 region was cloned by the CRISPR/Cas9-mediated TAR technology [15]. Two sgRNAs directed to the upstream region of snbC and downstream region of pglE were designed. The spr1 sgRNAs transcription cassettes with T7 promoter sequence were amplified from the plasmid pKCcas9dO with the primer pairs, spr1gRNA1-fw/gRNA-rev and spr1gRNA2-fw/gRNA-rev, respectively. Then, in vitro transcription was performed using the MEGAScriptTM Kit (Ambion), followed by purification with the MEGAClear™ Kit (Ambion). Finally, CRISPR/Cas9 (Tolo Biotech., Shanghai, China)-mediated genomic DNA digestion was conducted as described previously [15]. The enzymatically fragmented genomic DNA was purified by ethanol precipitation.
The spr1 capture vector was constructed as follows. Two homologous arms (approximately 1 kb) corresponding to flanking regions of spr1 were amplified from S. pristinaespiralis HCCB10218 genomic DNA with two primer pairs, spr1up-fw/rev and spr1down-fw/rev, respectively, and introduced into the SpeI/KpnI-digested plasmid pCAP01, generating the spr1 capture plasmid pCAP01-spr1. Then, spheroplast cells of S. cerevisiae VL6-48 were transformed with 1 μg linearized pCAP01-spr1 (treated by EcoRI) and 2–3 μg Cas9-treated HCCB10218 genomic DNA. Desired transformants was screened on SD-Trp agar. The positive clones were verified by PCR. Two primer pairs, including Jspr1-A-fw/rev (amplifying the joint region between the vector and spr1) and Jspr1-B-fw/rev (amplifying the inner regions of spr1) were used. The putative positive plasmids were extracted from yeast and then transformed into E. coli EPI 300 for propagation. Direct cloning of the spr1 region was verified by restriction enzyme analysis, resulting in the plasmid pCAP-spr1.
Direct cloning of the spr2 region was carried out using traditional TAR method [13], similarly as that of the CRISPR/Cas9-mediated TAR used for spr1 cloning. The only difference is that genomic DNA of HCCB10218 was digested with XbaI and NdeI but not Cas9. The obtained plasmid was named pCAP-spr2.
2.3.2. Assembly of the PI BGC
Assembly of the PI BGC was performed by the TAR method [13]. To assemble spr1 and spr2 into the PI BGC, we designed a 80-bp overlapping region between spr1 and spr2. The capture vector for spr1 and spr2 assembly was constructed as that of pCAP-spr1. The primer pairs used for the amplification of the two homologous arms were spr1up-fw/rev and spr2down-fw/rev. pCAP-spr1 and pCAP-spr2 were digested with SpeI/KpnI and SpeI/XhoI, respectively, to isolate spr1 and spr2 large DNA fragments. Correct assembly between spr1 and spr2 was confirmed by colony PCR (using primer pairs of Jspr1-B-fw/rev and Jspr2-B-fw/rev) and restriction enzyme analysis, resulting in the plasmid pCAP-PI, which contained the expected PI BGC.
2.4. S. pristinaespiralis fermentation and quantitative analysis of PI production
S. pristinaespiralis fermentation was conducted according to the method described by Mast et al. [2]. Briefly, S. pristinaespiralis strains were cultured in 25 ml of seed medium in 250 ml flasks at 27 °C on a rotary shaker (240 rpm). After 40–44 h, cultures (2 ml each) were harvested to inoculate 3 × 25 ml of fermentation medium in 250 ml-volume flasks that were incubated at 27 °C (240 rpm). Fermentation cultures (0.5 ml each) were collected at five different time points (30, 48, 60, 72 and 96 h), followed by extraction with the same volume of acetone for 60 min. The mixtures were centrifuged at 4000 rpm for 10 min. The supernatants were analyzed for PI production by high-performance liquid chromatography (HPLC) according to the method described previously [16]. The elution time for PIA (the main component of PI) was approximately 9.9 min. Purified PIA (98%, provided by Shanghai Liyi Co. Ltd, Shanghai, China) was used to make standard curves.
3. Results
3.1. Construction of a sole pristinamycin I (PI)-producing strain
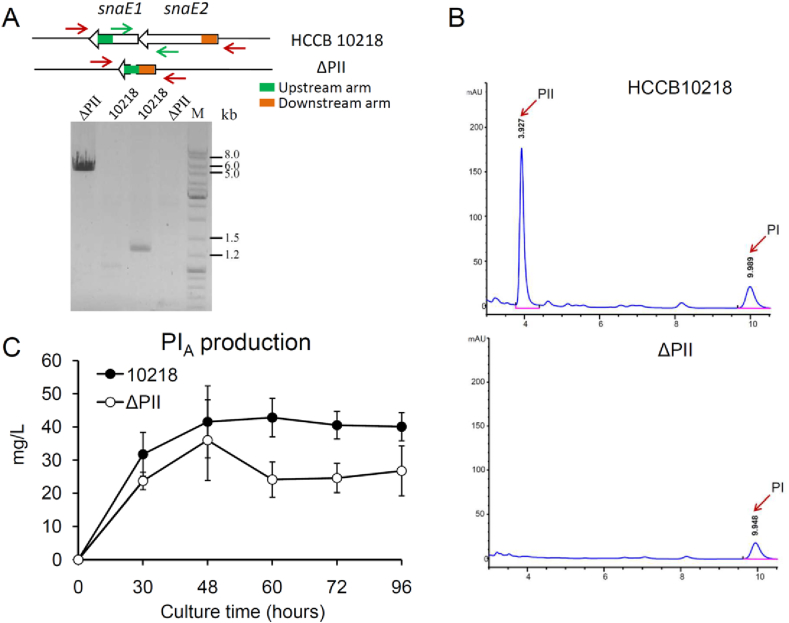
To construct a sole PI-producing strain, we generated the mutant ΔPII on the basis of the parental strain S. pristinaespiralis HCCB10218 with an in-frame deletion of two PII biosynthetic genes, snaE1 and snaE2, which encode the deduced hybrid PKS/NRPS complex SnaE1 and the predicted PKS SnaE2 [2]. The correct deletion of these two genes was verified by PCR (Fig. 2A) and DNA sequencing (data not shown). Deletion of snaE1/E2 has no effect on bacterial growth (data not shown). Fermentation cultures of HCCB10218 and the ΔPII mutant were collected at five different time points for the analysis of pristinamycin (both PI and PII) production titers by HPLC. These analyses showed that compared with HCCB10218 that could produce both, PI and PII, the ΔPII mutant only formed PI (Fig. 2B). Furthermore, to our surprise, we found that deletion of snaE1/E2 led to approximately 20–40% reduced PI production titers in comparison to the parental strain HCCB10218 (Fig. 2C).
Fig. 2.
Verification of the sole PI component producing strain ΔPII. A. Verification of the deletion of the PII biosynthetic genes (snaE1 and snaE2) by colony PCR. The primer pair located upstream of snaE1 and downstream of snaE2 (JsnaE-outer-fw/rev) is indicated as red arrows. The expected band size for the ΔPII mutant is 4152 bp. In the parental strain HCCB10218, due to too large DNA fragment (>20 kb), no band could be amplified by PCR. The primer pair located within the snaE1 and snaE2 ORF (JsnaE-inner-fw/rev) is indicated as green arrows. The expected size for the parental strain HCCB10218 is 1220 bp. For the ΔPII mutant sample, where snaE1 and snaE2 are deleted, no band was amplified by PCR. B. Pristinamycin production of the parental strain HCCB10218 and the ΔPII mutant analyzed by HPLC. C. Effect of the snaE1/E2 deletion on PI production. Fermentation cultures of the ΔPII mutant and the parental strain HCCB10218 were collected at five time points as indicated. Fermentation analysis was performed in triplicate and was repeated twice. Error bars indicate the standard deviations for three biological replicates.
3.2. Deletion of the repressor gene papR3 results in enhanced PI production
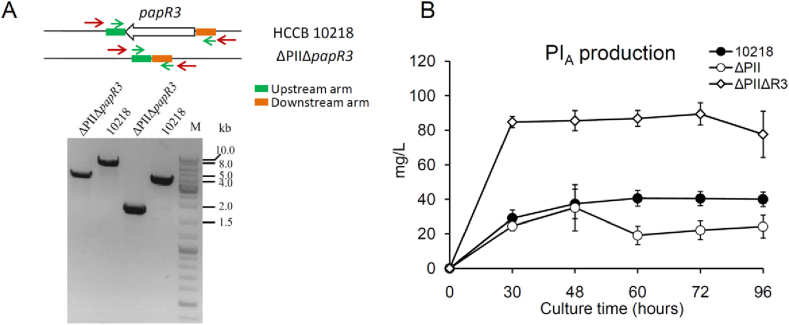
In S. pristinaespiralis, the TetR-family regulator PapR3 has been identified as repressor of PI and PII biosynthesis [2], [5], [8]. Deletion of papR3 has been shown to result in a markedly enhanced PI and PII production [5]. To further increase PI production in ΔPII, we constructed the mutant ΔPIIΔpapR3 with an in-frame deletion of papR3 on the basis of the ΔPII mutant. The correct deletion of papR3 was confirmed by colony PCR (Fig. 3A) and DNA sequencing (data not shown). Bacterial growth was not affected upon deletion of papR3 (data not shown). Fermentation cultures of HCCB10218 and the mutants ΔPII and ΔPIIΔpapR3 were collected at five different time points for PI production analysis. The results showed that deletion of papR3 on the basis of ΔPII mutant resulted in a markedly enhanced PI production. The ΔPIIΔpapR3 mutant produced a maximum PI level of 90 mg/L, showing a 1.4-fold higher than the parental strain HCCB10218 (approximately 38 mg/L) (Fig. 3B).
Fig. 3.
Verification of the ΔPIIΔpapR3 mutant with deletion of two PII biosynthetic genes snaE1/E2 and the repressor gene papR3. A. Verification of papR3 deletion by colony PCR. The primer pairs located outside and inside the two homologous arms (JpapR3-outer-fw/rev and JpapR3-inner-fw/rev) are indicated as red and green arrows, respectively. The expected band sizes amplified by red primers (outside) for the ΔPII mutant and HCCB10218 are 3287 and 4862 bp, respectively. The primer pair amplified by green primers (inside) are 1685 and 3260 bp for the ΔPII mutant and HCCB10218, respectively. B. Effect of papR3 deletion on PI production. Fermentation cultures of three strains, including HCCB10218, ΔPII and ΔPIIΔpapR3, were collected at five time points as indicated. Fermentation analysis was performed in triplicate and was repeated twice. Error bars indicate the standard deviations for three biological replicates.
3.3. Adding an extra copy of the PI BGC leads to further increase of PI production
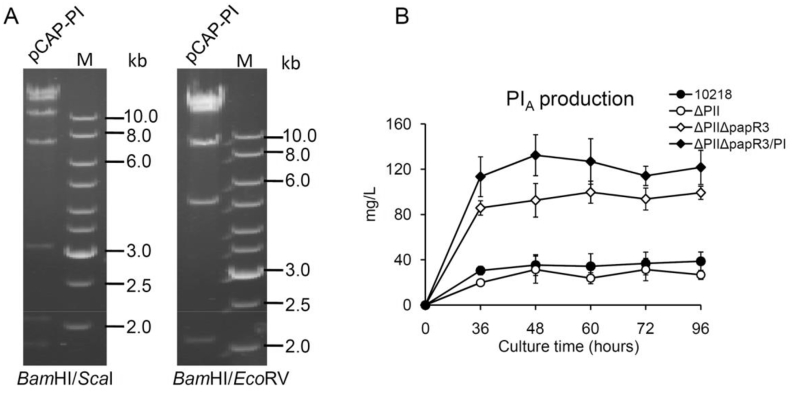
Another efficient strategy to optimize antibiotic production yield is to increase the copy number of secondary metabolite BGCs [5], [6], [10]. Thus, we applied this metabolic engineering strategy in order to increase PI production in ΔPIIΔpapR3. To achieve this, the first step is to clone the PI BGC. The PI biosynthetic genes are scattered in two main regions, namely, region 1 (named as spr1, from snbC to pglE, approximately 28 kb) and region 2 (named as spr2, from papM to hpaA, approximately 11 kb) (Fig. 1). These two regions were successfully cloned using the TAR method [13], [15] and then assembled into the PI BGC to yield the plasmid pCAP-PI. The correctness of the pCAP-PI was confirmed by PCR (data not shown) and restriction enzyme analysis using BamHI/ScaI and BamHI/EcoRV, respectively (Fig. 4A).
Fig. 4.
Effect on PI production after chromosomal integration of an extra copy of the PI BGC into ΔPIIΔpapR3. A. Verification of the plasmid pCAP-PI containing the assembled PI BGC by restriction enzyme analysis. Two groups of enzymes were used as indicated. The expected sizes after BamHI/ScaI digestion are 1780, 2008, 3013, 6917, 9666, 12771 and 16339 bp. The expected sized after BamHI/EcoRV digestion are 2008, 4793, 8447, 14809 and 22437 bp. B. Effects on PI production after chromosomal integration of an extra copy of the PI BGC into ΔPIIΔpapR3. Fermentation cultures of the four strains, including HCCB10218, ΔPII, ΔPIIΔpapR3 and ΔPIIΔpapR3/PI, were collected at five time points as indicated. Fermentation analysis was performed in triplicate and was repeated twice. Error bars indicate the standard deviations for three biological replicates.
3.3.1. Introduction of pCAP-PI into the ΔPIIΔpapR3 mutant further increases PI production yield
We introduced the plasmid pCAP-PI (containing the PI BGC) into the mutant ΔPIIΔpapR3 by the ΦC31 integrase-mediated site-specific DNA recombination. This resulted in strain ΔPIIΔpapR3/PI, where an extra copy of the PI BGC is present. The engineered strain showed similar morphological development and bacterial growth as HCCB10218 and ΔPIIΔpapR3 (data not shown). Fermentation cultures of HCCB10218, ΔPII, ΔPIIΔpapR3 andΔPIIΔpapR3 were taken at five different time points for PI production analysis. The HPLC analyses demonstrated that the introduction of an extra copy of the PI BGC led to a further increase of PI production. The ΔPIIΔpapR3/PI mutant produced a maximum PI level of 132 mg/L, showing approximately 30% and 240% higher than those of the ΔPIIΔpapR3 mutant and HCCB10218, respectively (Fig. 4B). Therefore, it could be concluded that the combination of the two metabolic engineering approaches employed is very efficient for the improvement of PI production by S. pristinaespiralis.
4. Discussion
In this study, a high pristinamycin I (PI) single component-producing strain was constructed by deleting the PII biosynthetic genes combined with two metabolic engineering approaches, including the deletion of the repressor gene papR3 and the addition of an extra copy of the PI BGC. The final engineered strain ΔPIIΔpapR3/PI produced a maximum PI level of 132 mg/L, showing an approximately 2.4-fold higher than that of the parental strain HCCB10218. However, the PI titer of ΔPIIΔpapR3/PI is still insufficient for industrial production. Therefore, other metabolic engineering strategies, which have been widely proven as efficient methods for Streptomyces strain improvement [10], such as increasing precursor supply, overexpression of activator genes and deletion of competing pathways (other secondary metabolite BGCs) could be included to further enhance PI titer.
Interestingly, we found that deletion of the PII biosynthetic genes resulted in 20–40% reduced PI production. In S. pristinaespiralis, pristinamycin biosynthesis is under complex regulation involving up to seven cluster-situated regulators, including SpbR and additional six regulators (PapR1-PapR6) [8], as well as regulators located outside of the pristinamycin BGC, such as AtrA-p or Spy1 [16], [17]. We propose that PII may act as a coactivator or an inducer for PI biosynthesis. In the presence of PII, activators could bind to the promoter region of target genes and activate PI production, whereas repressors would dissociate from target promoter and result in the derepression of PI gene expression. Accordingly, loss of PII production would lead to a downregulation of PI biosynthetic gene expression and thus decrease PI production. So far, the detailed mechanisms of pristinamycin biosynthesis control remain to be elucidated. However, a better understanding of the regulatory principles of pristinamycin biosynthesis would be greatly beneficial to perform further strain improvement for PI overproduction in S. pristinaespiralis.
Acknowledgements
This work was sponsored by the National Natural Science Foundation of China (31430004, 31421061, 31630003, 31370081 and 31570072) and the Science and Technology Commission of Shanghai Municipality (16490712100).
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Contributor Information
Wolfgang Wohlleben, Email: wolfgang.wohlleben@biotech.uni-tuebingen.de.
Weihong Jiang, Email: whjiang@sibs.ac.cn.
Yinhua Lu, Email: yhlu@sibs.ac.cn.
References
- 1.Barriere J.C., Berthaud N., Beyer D., Dutka-Malen S., Paris J.M., Desnottes J.F. Recent developments in streptogramin research. Curr Pharm Des. 1998;4:155–180. [PubMed] [Google Scholar]
- 2.Mast Y., Weber T., Golz M., Ort-Winklbauer R., Gondran A., Wohlleben W. Characterization of the ‘pristinamycin supercluster’ of Streptomyces pristinaespiralis. Microb Biotechnol. 2011;4:192–206. doi: 10.1111/j.1751-7915.2010.00213.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Di Giambattista M., Chinali G., Cocito C. The molecular basis of the inhibitory activities of type A and type B synergimycins and related antibiotics on ribosomes. J Antimicrob Chemother. 1989;24:485–507. doi: 10.1093/jac/24.4.485. [DOI] [PubMed] [Google Scholar]
- 4.Mast Y., Wohlleben W. Streptogramins - two are better than one! Int J Med Microbiol. 2014;304:44–50. doi: 10.1016/j.ijmm.2013.08.008. [DOI] [PubMed] [Google Scholar]
- 5.Li L., Zhao Y., Ruan L., Yang S., Ge M., Jiang W. A stepwise increase in pristinamycin II biosynthesis by Streptomyces pristinaespiralis through combinatorial metabolic engineering. Metab Eng. 2015;29:12–25. doi: 10.1016/j.ymben.2015.02.001. [DOI] [PubMed] [Google Scholar]
- 6.Li L., Zheng G., Chen J., Ge M., Jiang W., Lu Y. Multiplexed site-specific genome engineering for overproducing bioactive secondary metabolites in actinomycetes. Metab Eng. 2017;40:80–92. doi: 10.1016/j.ymben.2017.01.004. [DOI] [PubMed] [Google Scholar]
- 7.Dun J., Zhao Y., Zheng G., Zhu H., Ruan L., Wang W. PapR6, a putative atypical response regulator, functions as a pathway-specific activator of pristinamycin II biosynthesis in Streptomyces pristinaespiralis. J Bacteriol. 2015;197:441–450. doi: 10.1128/JB.02312-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mast Y., Guezguez J., Handel F., Schinko E. A complex signaling cascade governs pristinamycin biosynthesis in Streptomyces pristinaespiralis. Appl Environ Microbiol. 2015;81:6621–6636. doi: 10.1128/AEM.00728-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Folcher M., Gaillard H., Nguyen L.T., Nguyen K.T., Lacroix P., Bamas-Jacques N. Pleiotropic functions of a Streptomyces pristinaespiralis autoregulator receptor in development, antibiotic biosynthesis, and expression of a superoxide dismutase. J Biol Chem. 2001;276:44297–44306. doi: 10.1074/jbc.M101109200. [DOI] [PubMed] [Google Scholar]
- 10.Zhang M.M., Wang Y., Ang E.L., Zhao H. Engineering microbial hosts for production of bacterial natural products. Nat Prod Rep. 2016;33:963–987. doi: 10.1039/c6np00017g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Olano C., Lombo F., Mendez C., Salas J.A. Improving production of bioactive secondary metabolites in actinomycetes by metabolic engineering. Metab Eng. 2008;10:281–292. doi: 10.1016/j.ymben.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 12.Kieser T.B.M., Butter M., Chater K. John Innes Foundation; Norwich, England: 2000. Practical Streptomyces genetics. [Google Scholar]
- 13.Yamanaka K., Reynolds K.A., Kersten R.D., Ryan K.S., Gonzalez D.J., Nizet V. Direct cloning and refactoring of a silent lipopeptide biosynthetic gene cluster yields the antibiotic taromycin A. Proc Natl Acad Sci USA. 2014;111:1957–1962. doi: 10.1073/pnas.1319584111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang H., Zheng G., Jiang W., Hu H., Lu Y. One-step high-efficiency CRISPR/Cas9-mediated genome editing in Streptomyces. Acta Biochim Biophys Sin (Shanghai) 2015;47:231–243. doi: 10.1093/abbs/gmv007. [DOI] [PubMed] [Google Scholar]
- 15.Lee N.C., Larionov V., Kouprina N. Highly efficient CRISPR/Cas9-mediated TAR cloning of genes and chromosomal loci from complex genomes in yeast. Nucleic Acids Res. 2015;43:e55. doi: 10.1093/nar/gkv112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang W., Tian J., Li L., Ge M., Zhu H., Zheng G. Identification of two novel regulatory genes involved in pristinamycin biosynthesis and elucidation of the mechanism for AtrA-p-mediated regulation in Streptomyces pristinaespiralis. Appl Microbiol Biotechnol. 2015;99:7151–7164. doi: 10.1007/s00253-015-6638-6. [DOI] [PubMed] [Google Scholar]
- 17.Jin Q., Yin H., Hong X., Jin Z. Isolation and functional analysis of spy1 responsible for pristinamycin yield in Streptomyces pristinaespiralis. J Microbiol Biotechnol. 2012;22:793–799. doi: 10.4014/jmb.1111.11031. [DOI] [PubMed] [Google Scholar]
- 18.Gust B., Challis G.L., Fowler K., Kieser T., Chater K.F. PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc Natl Acad Sci USA. 2003;100:1541–1546. doi: 10.1073/pnas.0337542100. [DOI] [PMC free article] [PubMed] [Google Scholar]