Abstract
Background
The dissemination of extended-spectrum β-lactamase (ESBL)-producing bacteria presented a great concern worldwide. Gram-negative organisms such as Escherichia coli and Klebsiella pneumoniae are the most frequently isolated pathogens responsible for nosocomial infections.
Objectives
The aim of this study was to investigate and to follow the emergence of resistance and the characterization of Extended-Spectrum Beta-Lactamases (ESBL) among broad-spectrum beta-lactam-Escherichia coli clinical isolates recovered from the military hospital and Habib Thameur hospital in Tunisia.
Methods
A total of 113 E.coli isolates obtained during the period 2004 through 2012 showed a significant degree of multi-resistance. Among these strains, the double-disk synergy test confirmed the ESBL phenotype in 46 isolates. These included 32(70%) strains from Hospital A and 14(30%) from Hospital B.
Results
The ESBL was identified as CTX-M-15. The ESBL resistance was transferred by a 60 kb plasmid CTXM-15-producing isolates were unrelated according to the PFGE analysis and characterization of the regions surrounding the blaCTX-M-15 showed the ISEcp1 elements located in the upstream region of the bla gene and 20 of them truncated by IS26.
Conclusion
ESBL producing E. coli strains are a serious threat in the community in Tunisia and we should take into consideration any possible spread of such epidemiological resistance.
Keywords: CTX-M-15, diverse clones, ESBLs, Escherichia coli
Introduction
The production of extended-spectrum β-lactamases continues to be the important cause of resistance among gram negative bacteria.
The TEM- and SHV- are the first ESBLs type, mutant derivatives of established plasmid-mediated β-lactamases, however, a novel type of plasmid mediated ESBLs, the CTX-M enzymes, cefotaximases, emerged worldwide in the last decade, and preferentially hydrolyze cefotaxime over ceftazidime and are inhibited by clavulanic acid, sulbactam, and tazobactam1 were reported in the second half of the 1980s classified in Ambler class A and in group 2be of the Bush, Jacoby and Medeiros classification2.
More than 65 CTX-M β-lactamases were revealed worldwide. The phylogenic study clustered them in five major groups: CTXM 1,-2, -8, -9, and -25 groups3–5.
The number of reports studing the CTX-M variants continue to increase,CTX-M-28 enzyme was recently reported in Tunisa6,7.
CTX-M-9, first described in 1994 in Spain8, Guyana and the United Kingdom9 also in Tunisa10.
Also, in 2006, Boyd et al mention the first report of CTX-M-16 type-producing Enterobacteriaceae in Tunisia and in Africa11.
Another variant of CTX-M type, CTX-M-8 was detected in cefotaxime-resistant Proteus mirabilis strain in association with a plasmid mediated AmpC lactamase12.
CTX-M-15 is the most prevalent β-lactamase detected amongst the ESBL-positive K. pneumoniae and E. coli strains derived from CTX-M-3 by a substitution of Asp-240-Gly which increases its catalytic efficiency against ceftazidime13,14 first described in 200115,16
Many reports have documented the emergence of CTX-M gene9, and the first report of the CTX-M-15 in Tunisia was cited in the Charles Nicolle Hospital in 1984 and it was described in various studies in Tunisia including that of coque et al, the gene has been found in E. coli strains in a Tunisian Hospital17, France18, and Central African Republic19–25 .91% of the ESBL-producing isolates carried blaCTX-M-15 genes21.
The production of CTX-M enzymes is an emerging phenomenon that has been called ‘the CTX-M pandemic’16.
The insertion sequence ISEcp1 was found to be involved in the mobility of blaCTX-M,was located upstream the bla CTX-M-27 gene e in a neonatal ward of the maternity department of Farhat Hached Hospital, Sousse26. It has been found also upstream the CTX-M-14 producing E. coli isolated from hospitalized patients in a university Hospital of Tunisia27, and upstream the CTX-M-15 gene in Proteus mirabilis and Morganella morganii isolated at the Military Hospital of Tunis24.
ISEcp1 was located upstream of the blaCTX-M gene on E. coli isolates from food samples28.
CTX-M genes may spread through clonal dissemination or horizontal gene transfer19.
Methods
Bacterial strain
These clinical strains were isolated from samples collected in different wards, including the emergency (25, 86 %), reanimation (16.07 %), hemodialysis (4.56 %), neonatal (4.24 %), pediatrics (4.39 %), gastroenterology (13.32 %), external (12.56 %) and urology (19 %).
68% of strains were from urine, 17.8% from blood culture and 14.2% from Pus.
All the isolates were identified by the Vitek automated system (bioMérieux, Vitek 32) and API 20E system (bioMérieux, Marcy l'Etoile, France).
E. coli DH5a (recA1, F_, end A1, gyrA96, thi-1, hsdR17, rK_, mK+, supE44, relA1, DlacU69, F80lazDM15) and E. coli HB101 (F_, D(gpt-proA) 62, leuB6, supE44, ara-14, galK2, lac Y1, D(mcrc-mrr), rps, L26, Xyl-rmtl 1, thi-1, IncFI, rec AB, strr), were used respectively for the transformation and conjugation experiments.
Antimicrobial susceptibility and synergy testing
Routine antibiograms were determined by the disk diffusion method on Mueller-Hinton agar (MH, Diagnostics Pasteur) using susceptibility breakpoints as recommended by the Clinical and Laboratory Standards Institute (CLSI)29.
The double-disk synergy test was used to detect the ESBL production as previously described30,24 by using amoxicillin-clavulanate against cefotaxime, ceftriaxone, ceftazidime and aztreonam.
Minimum inhibitory concentrations (MICs) of selected anti-microbial agents were determined by using the dilution method on Mueller-Hinton agar according to CLSI guidelines29.
Table 1 shows MICs (µg/mL) of various antimicrobial agents obtained for the clinical isolate E. coli, transconjugant and transformant, and the E. coli recipients.
Table 1.
Primers used for detection of resistance genes.
| PCR Target |
Primer name |
Primer sequence | Amplicon sizes (pb) |
Annealing temperatures (°C) |
References |
| CTX-M | CTX-M-A | 5′ TTT GCG ATG TGC AGT ACC AGT AA3′ |
544 | 57°C | (Edelstein et al., 2003) |
| CTX-M-B | 5′ CGA TAT CGT TGG TGG TGC ATA3′ |
||||
| TEM | TEM-F | 5′ATGAGTATTCAACATTTCCGTG3′ | 844 | 55°C | (Yagi et al., 2000) |
| TEM-R | 5′TTACCAATGCTTAATCAGTGAG3′ | ||||
| SHV | SHV-F | 5′ ATTTGTCGCTTCTTTACTCGC3′ | 861 | 55°C | (Essack et al., 2001) |
| SHV-R | 5′ TTTATGGCGTTACCTTTGACC′ | ||||
| ISEcp1 | ISEcp1-A | 5′GCAGGTCTTTTTCTGCTCC3′ | 490 | 57 °C | Lartigue et al., 2004) |
| ISEcp1-B | 5′ATTTCCGCAGCACCGTTTGC3′ |
CTX, cefotaxime; CRO, ceftriaxone; CAZ, ceftazidime; ATM, aztreonam; TIC, ticarcillin; STR, streptomycin; IMP, imipenem; ERT, ertapenem; CHL, chloramphenicol; TET, tetracycline; OFX, ofloxacin. NM, not measured.
β-Lactamase extraction and isoelectric focusing
IEF was performed as described previously17,31, using a culture grown overnight at 37°C in Trypto-Caseine Soy broth (TCS). These exponentially growing bacteria were harvested at 17,400 _ g (Rotor F 0650, Bekman) and bacterial suspensions were prepared by sonication in UP 400 S (dv. Hielscher, Germany) five times for 45 s each time.
Crude extract was centrifuged at 17,400 _ g (Universel 32 R, Hettich) for 15 min at 4°C.
The supernatant of the sonicate was subjected to isoelectric focusing on ampholine polyacrylamide gel with a pH range of 3–10 at a voltage range of 100–300, at 4°C in a 111Mini IEF Cell (Bio-Rad). TEM-1 (pI 5.4), TEM-2 (pI 5.6), TEM-3 (pI 6.3), SHV-1 (pI 7.6) and SHV-12 (pI 8.2) were used as pI markers.
β-lactamase activities were revealed by iodemetric method using benzylpenicillin to 1 mM and cefotaxime (3 mM) as substrates in phosphate buffer (25 mM; pH 7).
β-Lactamase assay
Hydrolytic activities of crude extracts for β-lactam antibiotics were determined by the spectrophotometric method at the wave length of maximal absorbance for the β-lactam ring of each antibiotic.32
The decrease in absorbance of the antibiotics at an appropriate concentration was measured in a temperature controlled spectrophotometer (Varian R CARY 50 Bio UV-visible) at 37°C.
Specific activity is calculated on depending of Ross and O'Callaghan equation in 197533
Effect of inhibitors (IC50 determination)
Crude enzyme extract was incubated with clavulanic acid and sulbactam increasing concentrations. EDTA was used at a fixed concentration of 1 mM. Residuals β-lactamases activities were determined by the spectrophotometric method using cephalothin 1 mM as substrate. The inhibitor concentration required to inhibit 50% of enzyme activity was determined graphically (IC50)34.
Analysis of plasmids and transfer of resistance
Plasmid DNA was extracted with the alkaline lysis method, as described by Sambrook et al35.
Conjugation experiments were carried out with E. coli HB101, as previously described7,24. (31; 9;3;4).
The transconjugants were selected on LB agar supplemented with streptomycin (100 µg/ml) and ampicillin (100 µg/ml).
Transformation experiments were carried out by using E. coli DH5α as the recipient as previously described31,36.
Transformants were selected on Luria-Bertani medium agar plates supplemented with ampicillin (100 mg/ml).
Transformants were subjected to DDST to confirm the presence of ESBL genes and were examined for co-transfer of other antibiotic resistance determinants present in the donor clinical isolates by disk diffusion.
Characterization of the resistance genes using PCR method and sequencing
Primers used for amplification of resistance genes, annealing temperatures and predicted amplicon sizes are shown in Table 1. They were used to detect blaCTX-M, blaSHV, blaTEM and the sequences surrounding the bla CTX-M gene, ISEcp1and IS2637,38 also the aac(6′)-ib-cr gene.
The DNA amplification programs consisted of initial denaturation for 5 min at 94°C, followed by 30 cycles of denaturation for 30 s at 94°C, annealing temperatures differed according to the primer pair used and were for 45 s at 57°C for the blaCTX-M and blaISEcp1; 55°C for blaTEM and blaSHV. Finally, the polymerization for 5 min at 72°C.
PCR products were purified using QIAquick PCR Purification Kit (Qiagen, USA) and sequenced using the Big-Dye Terminator v.3.1 Cycle Sequence Kit and ABI Prism 310 automatic sequencer (Applied Biosystems, USA).
The nucleotide sequences were compared with those included in the GenBank database using BLAST (http:/www.ncbi.nlm.nih.gov/blast/).
Detection of ISEcp1
PCR for ISEcp1 was used to examine the region upstream of the the blaCTX-M gene with the primers shown in Table 1. An annealing temperature,57°C was used followed by sequencing of the PCR product.
Typing of isolates by MLST
Molecular typing was performed for one representative E. coli isolate by MLST by PCR amplification of the seven housekeeping loci (adk, fumC, gyrB,icd, mdh, purA and recA).
The amplicon was sequenced and compared with the MLST database. (http://mlst.ucc.ie/mlst/dbs/Ecoli)39.
Molecular typing using Pulsed-field gel electrophoresis (PFGE)
The epidemiological relationship of CTX-M-15 positive strains was studied by PFGE using XbaI as restriction enzymes (Bio-Rad Laboratories, France).
Restriction fragments of DNA were separated by electrophoresis which was performed in a 1.2% agarose gel on a CHEF DRIII apparatus (Bio-Rad Laboratories, Richmond, CA, USA) with 6V/cm for 19 h at 14°C with an initial switch time of 2.2 to 52.0 sec.
XbaI-digested DNA were compared visually based on differences in the number and mobility of bands40.
Results
Studied strains show multidrug resistance phenotype with various antibiotics. They were highly resistant to antibiotics such as penicillins, cephalosporins, aminoglycosides, quinolones and tetracycline, (MIC 512– 64µg/mL) whereas remained susceptible to imipinem (MIC <2µg/ml) and meropenem (MIC <0,5µg/ml) (Table 2 ). Similar results were observed with the transformants and transconjugants except with quinolones, phenicol and aminoglycosides, suggesting that this resistance could be in chromosome.
Table 2.
MICs (µg/mL) of various antimicrobial agents obtained for the clinical isolate E. coli, transconjugant and transformant, and the E. coli recipients.
| Isolates | Year of isolation |
Ward | Specimen | MIC (mg/l) | ||||||||||
| Cefotaxime | Ceftriaxon | Ceftazidime | Aztreonam | Ticarcillin | Streptomycin | Imipenem | ERT | CHL | Tetracyclin | OFX | ||||
| E.coli 4836 | 2011 | urology | urine | >512 | >512 | >512 | >512 | >512 | 128 | <2 | 0.5 | 128 | 64 | 64 |
|
E. coli HB101 X E. coli 4836 |
_ | 512 | 512 | 512 | 128 | 512 | 256 | <2 | 0.5 | 32 | 64 | 128 | ||
| E. coli HB101 | _ | <2 | <2 | <2 | <2 | 8 | 2 | <2 | <2 | <2 | <2 | <2 | ||
|
E. coli DH5α/ E. coli 4836 |
_ | 512 | 512 | 16 | 512 | >512 | 256 | ≤0.006 | <0.5 | 8 | 4 | <2 | ||
| E. coli DH5a | _ | <2 | <2 | <2 | <2 | <2 | <2 | ≤0.006 | <0.5 | 2 | <2 | <2 | ||
46 E. coli strains were phenotypically confirmed as ESBL with the double disc synergy showing a marked synergy between ceftazidime, céfotaxime, aztreonam, ceftriaxoneand amoxicillin-clavulanic acid on MH agar plates as well as the transformants and transconjugants suggested the presence of a class A ESBL.
The resistance to expanded-spectrum cephalosporins was successfully transferred to a recipient strains, E. coli HB101 with the frequency of transfer equivalent to 5× 10−5 transconjugant/recipient and this indicated that the gene was plasmidic transferred with an estimated molecular size of 60 kb of the transferable plasmid.
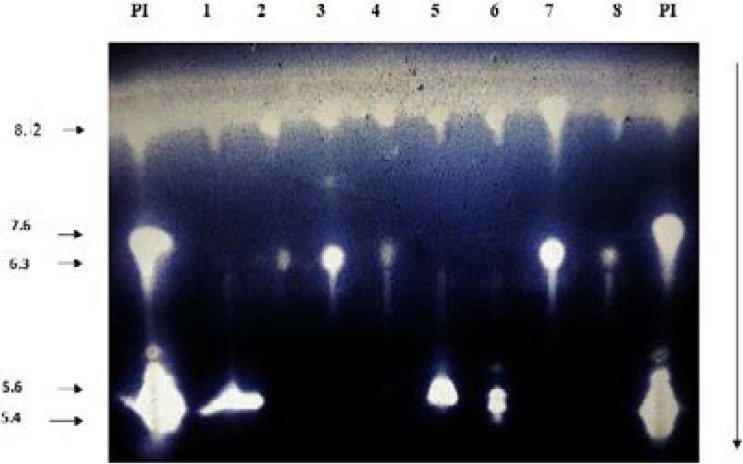
An extended-spectrum β-lactamase band with a pI 8.6 with cefotaxime as substrate was detected in the 46 ESBL strains (Figure 1), transformants and transconjugants.
Figure1.
Isoelectric focusing (7%) with cefotaxime for 1mM as substrate of the E. coli strains 1–8: E. coli strains; pI: pI markers TEM-1 (pI 5.4), TEM-2 (pI 5.6), TEM-3 (pI 6.3), SHV-1 (pI 7.6) and SHV-12 (pI 8.2)
Kinetics resuts showed that E. coli strains hydrolyze benzylpenicillin, ticarcillin with a high level of hydrolytic activity, specific activities ranged between (8.77–6.67U/mg of protein) and hydrolyze cephalosporins with a higher hydrolytic activity to cefotaxime (U/mg of protein) (Table 3).
Table 3.
Specific activities β -lactamases of E. coli strain 4836 and its transconjugant (µmol of substrate hydrolyzed/min/mg of protein)
| Antibiotics | E. coli 4836 | E. coli HB101 X E. coli 4836 |
| Benzylpenicilli | 8.77 | 2.63 |
| Ticarcillin | 6.67 | 2.43 |
| Imipenem | ND | ND |
| Cefoxitine | 0.654 | 0.755 |
| Cefotaxime | 5.16 | 2.55 |
| Ceftriaxone | 0.239 | 0.192 |
| Ceftazidime | 0 .16 | 0.116 |
| Aztreonam | 0. 455 | 0.430 |
| Cefpirome | 0.31 | 0.02 |
ND: non detected.
These enzymes are not inhibited by EDTA and are not defined as metallo-enzymes. The analysis of the IC50 showed that the clavulanic acid with IC50 of 3.5 µM was the powerful inhibitor and categorized this enzyme to class A: serine active β-lactamases (Table 4).
Table 4.
Inhibitors effect of activities β-lactamases of E. coli 4836 strain (IC50)
| E. coli 4836 | IC50 (µM) | ||
| Clavulanic acid | Sulbactam | EDTA | |
| 3.5 | 14.7 | - | |
(−): without effect
PCR analyses confirmed the presence of blaCTX-M-3-related genes in parental strain E. coli 4836, and transformant E. coli DH5α/ E.coli 4836 and transconjugant E. coli HB101 X, E.coli 4836 indicating that this gene is located on conjugative plasmid.Sequencing of the deduced amino acid, followed by BLAST searches, and confirmed blaCTX-M as blaCTX-M-15.
This enzyme preferentially hydrolyzed cefotaxime over ceftazidime.
15 strains produced the quinolone resistance determinants aac(6′)-Ib-cr whereas ISEcp1 sequence was found in CTX-M-15- producing isolates.
When using the specialized primers reversibly of IS sequences (ISEcp1 and the IS26) with blaCTX-M-15 primers, We came to the result that the ISEcp1 sequence was found upstream of the blaCTX-M-15 gene.
On the other hand, IS26 transposase region was detected upstream of the ISEcp1 sequence on 22 of our studied E. coli isolates.
According to the MLST analysis of one E. coli isolate, it showed that this belonged to ST 131, displayed specific O25 type (O 25b) and the E. coli isolate belonged to the B2 phylogenetic subgroup I.
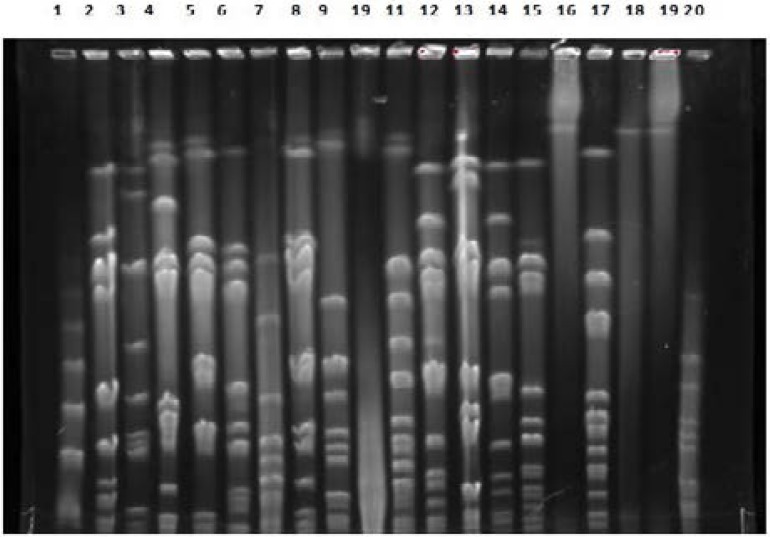
PFGE (Figure 2) showed that the 46 strains presented different profiles, and our studied strains were unrelated suggesting that the dissemination of bla CTX-M-15gene among E. coli clinical strains due to horizontal transfer of multi- resistance or/and the genetic mobile element are responsible for the dissemination of these gene.
Figure 2.
Pulsed-field gel electrophoresis (PFGE) of XbaI-digested genomic DNA of the E. coli isolates producing CTX M-15 (Lane 1–20)
Discussion
Our study investigated the genetic environment of blaCTX-M genes in fourty six Escherichia coli extended spectrum β-lactamase (ESBL)-positive isolates. Of these 46 strains, 10 E. coli isolates (24 %) produced β-lactamase activities with a varied isoelectric points (pIs) between 5.5 and 8,6 (lane 1,5 and 6, Figure 1), they carried the blaTEM gene (9 from hospital A and 1 from hospital B). Sequencing of PCR products of this latter showed that it corresponded to the TEM-1.
9 of the studied E. coli isolates (26 %) presented a basic pIs of 7.3 and 8.6 (lane 2,3,4,7 and 8, Figures 1). They carried the blaSHV gene (8 from hospital A and 1 from hospital B) and it corresponded to SHV-1 by sequencing. The presence of the insertion sequence ISEcp1upstream the gene in all the CTX-M-15 strain producers is a real concern which plays an important role of the mobilization of this latter as has been reported in several studies16,41.
According to the MLST analysis of one E. coli isolate, it showed that this belonged to ST 131, displays specific O25 type (O 25b) and the E. coli isolate belonged to the B2 phylogenetic subgroup I. This phylogroup B2 ST131 is previously described on E.coli CTX-M producers responsible for the clonal diffusing of the CTX-M-15 gene41,42 and this isolate may belong to this clone.
The presence of the blaCTX-M-15 encoding gene amongst unrelated strains argued for genetic transit of mobile elements amongst unrelated strains. Our study confirms that the CTX-M-15 gene is the most prevalent ESBL found in Tunisian hospitals and the association of the bla CTX-M-15 with the insertion sequence ISEcp1 facilitates its dissemination.
In short with reference to our insightful study, we should control the emergence of ESBL producing E. coli strains from hospitals environment which in some cases lack both the efficient surveillance and cleaning services.
This finding is alarming for healthcare providers and reinforces other previous studies in Tunisia to prevent a further spread and in the light of the current and fabulous medical breakthroughs, that we are now witnessing on a daily basis of, has to be a wake-up call for us to further enhance our efforts and better our practical measures.
Conflict of interest
There is no conflict of interest.
References
- 1.Bradford PA, Cherubin CE, Idemyor V, Rasmussen BA, Bush K. Multiply resistant Klebsiella pneumonia strains from two Chicago hospitals: identification of the extended-spectrum TEM-12 and TEM-10 ceftazidime-hydrolyzing beta-lactamases in a single isolate. Antimicrob Agents Chemother. 1994;38:761–766. doi: 10.1128/aac.38.4.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bush K, Jacoby GA, Medeiros AA. A functional classification scheme for beta-lactamases and its correlation with molecular structure. Antimicrob Agents Chemother. 1995;39:1211–1233. doi: 10.1128/aac.39.6.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Labia R. Analysis of the bla toho gene coding for Toho-2- beta-lactamase. Antimicrob Agents Chemother. 1999;43:2576–2577. doi: 10.1128/aac.43.10.2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barlow M, Reik RA, Jacobs SD, Medina M, Meyer MP, McGowan JE, Tenover FC. High Rate of Mobilization for blaCTX-Ms. Emerg Infect Dis. 2008;14:423–428. doi: 10.3201/eid1403.070405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Novais A, Canton R, Moreira R, Peixe L, Baquero F. Emergence and dissemination of Enterobacteriaceae isolates producing CTX-M-1-like enzymes in Spain are associated with IncFII (CTX-M-15) and broad-host-range (CTX-M-1, -3, and −32) plasmids. Antimicrob Agents Chemother. 2007;51:796–799. doi: 10.1128/AAC.01070-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ben Achour N, Mercuri P, Power P, Belhadj C, Ben Moussa M, Galleni M, Belhadj Omrane. First detection of CTX-M-28 in a Tunisian hospital from a cefotaxime-resistant Klebsiella pneumoniae strain. Pathol Biol. 2009a;57:343–348. doi: 10.1016/j.patbio.2008.07.016. [DOI] [PubMed] [Google Scholar]
- 7.Bourouis A, Chihi H, Mahrouki S, Ayari K, Ben Moussa M, Belhadj O. Molecular characterization of a transferable blaCTX-M-28gene in clinical isolates of Enterobacter cloacae. J Micro and Antimic. 2013;4:38–43. [Google Scholar]
- 8.Govinden U, Mocktar C, Moodley P, Sturm AW, Essack SY. Geographical evolution of the CTX-M β-lactamase-an update. Afr J Biotechnol. 2007;6:831–839. [Google Scholar]
- 9.Mamlouk K, Boutiba-Ben Boubaker I, Gautier V, Vimont S, Picard B, Ben Redjeb S. Emergence and outbreaks of CTX-M β-lactamase-producing. Escherichia coli and Klebsiella pneumoniae strains in a Tunisian hospital. J Clin Microbiol. 2006;44:4049–4056. doi: 10.1128/JCM.01076-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bourouis A, Dubois V, Coulange L, André C, Belhadj C, Ben Moussa M, Belhadj O. First report of CTX-M-9 in a clinical isolate of Enterobacter cloacae in a Tunisian hospital. Pathol Biol. 2009;59:187–191. doi: 10.1016/j.patbio.2009.03.008. [DOI] [PubMed] [Google Scholar]
- 11.Boyd DA, Tyler S, Christianson S, McGeer A, Muller MP, Willey BM, Bryce E, Gardam M, Nordmann P, Mulvey MR. Complete nucleotide sequence of a 92-kilobase plasmid harboring the CTX-M-15 extended spectrum β-lactamase involved in an outbreak in long-term-care facilities in Toronto, Canada. Antimicrob Agents Chemother. 2004;48:3758–3764. doi: 10.1128/AAC.48.10.3758-3764.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mahrouki S, Chihi H, Bourouis A, Ayari K, BEN MOUSSA M, Belhadj O. High-level cefotaxime-resistant Proteus mirabilis strain isolated from a Tunisian intensive care unit ward: CTX-M-8 extended-spectrum β-lactamase coproduced with a plasmid mediated AmpC lactamase. African J Biotech. 2013;21:3278–3282. [Google Scholar]
- 13.Poirel L, Gniadkowski M, Nordmann P. Biochemical analysis of the ceftazidime-hydrolysing extended-spectrum beta-lactamase CTX-M-15 and of its structurally related beta-lactamase CTX-M-3. J Antimicrob Chemother. 2002;50:1031–1034. doi: 10.1093/jac/dkf240. [DOI] [PubMed] [Google Scholar]
- 14.Poirel L, Kampfer P, Nordmann P. Chromosome-encoded Ambler class A beta-lactamase of Kluyvera georgiana, a probable progenitor of a subgroup of CTX-M extended-spectrum beta-lactamases. Antimicrob Agents Chemother. 2002;46:4038–4040. doi: 10.1128/AAC.46.12.4038-4040.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Karim A, Poirel L, Nagarajan S, Nordmann P. Plasmid-mediated extended-spectrum beta lactamase (CTX-M-3 like) from India and gene association with insertion sequence ISEcp1. FEMS Microbiol Lett. 2001;201:237–241. doi: 10.1111/j.1574-6968.2001.tb10762.x. [DOI] [PubMed] [Google Scholar]
- 16.Abbassi MS, Torres C, Achour W, Vinue L, Saenz Y, Costa D, Bouchami O, Ben Hassen A. Genetic characterisation of CTX-M-15-producing Klebsiella pneumoniae and Escherichia coli strains isolated from stem cell transplant patients in Tunisia. Int J Antimicrob Agents. 2008;32:308–314. doi: 10.1016/j.ijantimicag.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 17.Coque T M, Novais Â, Carattoli A, Poirel L, Pitout J, Peixe L, Nordmann P. Dissemination of clonally related Escherichia coli strains expressing extended-spectrum beta-lactamase CTX-M-15. Emerg Infect Dis. 2008;14:195–200. doi: 10.3201/eid1402.070350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mnif B, Vimont S, Boyd A, Bourit E, Picard B, Branger C, Denamur E, Arlet G. Molecular characterization of addiction systems of plasmids encoding extended-spectrum β-lactamases in Escherichia coli. J Antimicrob Chemother. 2010;65:1599–1603. doi: 10.1093/jac/dkq181. [DOI] [PubMed] [Google Scholar]
- 19.Lavollay M, Mamlouk K, Frank T, Akpabie A, Burghoffer B, Ben Redjeb S, Bercion R, Gautier V, Arlet G. Clonal dissemination of a CTX-M-15 β-lactamase-producing Escherichia coli strain in the Paris area, Tunis, and Bangui. Antimicrob Agents Chemother. 2006;50:2433–2438. doi: 10.1128/AAC.00150-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dahmen S, Bettaieb D, Mansour W, Boujaafar N, Bouallègue O, Arlet G. Characterization and molecular epidemiology of extended-spectrum β-lactamases in clinical isolates of Enterobacteriaceae in a Tunisian University Hospital. Microb Drug Resist. 2010;16:163–170. doi: 10.1089/mdr.2009.0108. [DOI] [PubMed] [Google Scholar]
- 21.Dahmen S, Poirel L, Mansour W, Bouallègue O, Nordmann P. Prevalence of plasmid-mediated quinolone resistance determinants in Enterobacteriaceae from Tunisia. Clin Microbiol Infect. 2009;16:1019–1023. doi: 10.1111/j.1469-0691.2009.03010.x. [DOI] [PubMed] [Google Scholar]
- 22.Elhani D, Bakir L, Aouni M, Passet V, Arlet G, Brisse S, Weill F-X. Molecular epidemiology of extended-spectrum β-lactamase-producing Klebsiella pneumoniae strains in a university hospital in Tunis, Tunisia, 1999–2005. Clin Microbiol Infect. 2010;16:157–164. doi: 10.1111/j.1469-0691.2009.03057.x. [DOI] [PubMed] [Google Scholar]
- 23.Ktari S, Arlet G, Mnif B, Gautier V, Mahjoubi F, Ben Jmeaa M. Emergence of multidrug-resistant Klebsiella pneumoniae isolates producing VIM-4 Metallo- β -lactamase, CTX-M-15 extended-spectrum b-lactamase, and CMY-4 AmpC b-lactamase in a Tunisian university hospital. Antimicrob Agents Chemother. 2006;50:4198–4201. doi: 10.1128/AAC.00663-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mahrouki S, Belhadj O, Chihi H, Ben Moussa M, Celenza G, Amicosante GF. Chromosomal blaCTX-M-15 associated with ISEcp1 in Proteus mirabilis and Morganella morganii isolated at military hospital of Tunis, Tunisia. J Med Microbiol. 2012;61:1286–1289. doi: 10.1099/jmm.0.039487-0. [DOI] [PubMed] [Google Scholar]
- 25.Mahrouki S, Bourouis A, Chihi H, Ouertani R, Ferjani M, Ben Moussa M. First characterisation of plasmid-mediated quinolone resistance- qnrS1 co-expressed blaCTX-M-15 and blaDHA-1 genes in clinical strain of Morganella morganii recovered from a Tunisian intensive care unit. Indian J Med Microbiol. 2012;30:437–441. doi: 10.4103/0255-0857.103765. [DOI] [PubMed] [Google Scholar]
- 26.Bouallegue OG, Ben Salem Y, Fabre L, Demartin M, Grimont PAD, Mzoughi R, Weill FX. Nosocomial outbreak caused by Salmonella enterica serotype Livingstone producing CTX-M-27 extended-spectrum β-lactamase in a neonatal unit in Sousse, Tunisia. J Clin Microbiol. 2005;34:1037–1044. doi: 10.1128/JCM.43.3.1037-1044.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lahlaoui H, BEN MOUSSA M. CTX-M-14 type β-Lactamase producing Escherichia coli isolated from hospitalized patients in Tunisia. APMIS. 2011;119:759–761. doi: 10.1111/j.1600-0463.2011.02800.x. [DOI] [PubMed] [Google Scholar]
- 28.Ben Slama K, Ben Sallem R, Jouini A, Rachid S, Moussa L, Saenz Y, Estepa V, Somalo S, Boudabous A, Torres C. Diversity of genetic lineages among CTX-M-15 and CTX-M-14 producing Escherichia coli strains in a Tunisian hospital. Curr Microbiol. 2011;62:1794–1801. doi: 10.1007/s00284-011-9930-4. [DOI] [PubMed] [Google Scholar]
- 29.Wayne PA. Performance standards for antimicrobial susceptibility testing; sixteenth informational supplement, CLSI document; M100- S16. 2006;26:44–51. [Google Scholar]
- 30.Jacoby GA, Han P. Detection of extended-spectrum β-lactamases in clinical isolates of Klebsiella pneumoniae and Escherichia coli. J Clin Microbiol. 1996;34:908–911. doi: 10.1128/jcm.34.4.908-911.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ben Achour N, Chouchani C, Bouhawala N, Amor A, Belhadj O. Clinical strain of Proteus mirabilis able to produce a plasmid mediated ceftazidimase. Pathol Biol. 2007 doi: 10.1016/j.patbio.2007.03.002. [DOI] [PubMed] [Google Scholar]
- 32.Philippon L, Nass T, Bouthors AT, Barakett V, Nordmann P. OXA-18, a class D acid-inhibited extended spectrum β-lactamase from Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1997;41:2188–2195. doi: 10.1128/aac.41.10.2188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ross GW, O'callagham CH, Hash H. β-lactamases asseys Methods in enzymology. Vol. 13. New York: Academic Press; 1975. pp. 69–85. [DOI] [PubMed] [Google Scholar]
- 34.Réjiba S, Limam F, Belhadj C, Belhadj O, Ben-Mahrez K. Biochemical characterization of a novel extended-spectrum β -lactamase from Pseudo monas aeruginosa. Microb Drug Resist. 2002;8:9–13. doi: 10.1089/10766290252913700. [DOI] [PubMed] [Google Scholar]
- 35.Sambrook J, Fritsch F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 36.Hanahan D. Technique for transformation of E.coli. DNA cloning. 1985;1:109–135. [Google Scholar]
- 37.Eckert C, Gautier V, Arlet G. DNA sequence analysis of the genetic environment of various blaCTX-M genes. J Antimicrob Chemother. 2006;57:14–23. doi: 10.1093/jac/dki398. [DOI] [PubMed] [Google Scholar]
- 38.Woodford N, Ward ME, Kaufmann ME, Turton J, Fagan EJ, James D, Johnson AP, Pike R, Warner M, Cheasty T, Pearson A, Harry S, Leach JB, Loughrey A, Lowes JA, Warren RE, Livermore DM. Community and hospital spread of Escherichia coli producing CTX-M extended-spectrum beta-lactamases in the UK. J Antimicrob Chemother. 2004;54:735–743. doi: 10.1093/jac/dkh424. [DOI] [PubMed] [Google Scholar]
- 39.Wirth T, Falush D, Lan RT, Colles F, Mensa P, Wieler LH, Karch H, Reeves PR, Maiden MCJ, Ochman H, Achtman M. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol Microbiol. 2006;60:1136–1151. doi: 10.1111/j.1365-2958.2006.05172.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tenover FC, Mohammed MJ, Gorton TS, Dembek ZF. Detection and reporting of organisms producing extended-spectrum β-lactamases: survey of laboratories in Connecticut. J Clin Microbiol. 1999;37:4065–4070. doi: 10.1128/jcm.37.12.4065-4070.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Clermont O, Lavollay M, Vimont S, Deschamps C, Forestier C, Branger C, Denamur E, Arlet G. The CTXM-15-producing Escherichia coli diffusing clone belongs to a highly virulent B2 phylogenetic subgroup. J Antimicrob Chemother. 2008;61:1024–1028. doi: 10.1093/jac/dkn084. [DOI] [PubMed] [Google Scholar]
- 42.Clermont O, Bonacorsi S, Bingen E. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl Environ Microbiol. 2000;66:4555–4558. doi: 10.1128/aem.66.10.4555-4558.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]