The recent remarkable advances in multiple myeloma (MM) therapy and outcomes have had mixed impact on patients with adverse risk genetics, many of whom continue to have inferior outcomes. This applies particularly to deletion of chromosome 17p13 [del(17p)], found in ≈10% of newly diagnosed MM (ND) and at higher prevalence in advanced disease.1, 2, 3, 4, 5 The TP53 gene is located within the minimally deleted region on 17p13, and is thought to confer the adverse risk in a haploinsufficient manner.6 The incidence of TP53 mutations in ND is ≈3% but also increases with disease progression,7, 8 and is associated with shorter survival.9 Although TP53 mutations are uncommon in the absence of del(17p), approximately one third of del(17p) MM patients are reported to have a TP53 mutation, and this increases in refractory disease to more than 50%.8, 10
The prognostic impact of TP53 mutations in del(17p) MM remains unresolved.2, 10 We therefore carried out mutational analysis of a series of MM patients who had fluorescence in situ hybridization (FISH)-identified del(17p) to correlate with clinical outcomes.
Ethical approval was obtained from National Research Ethics Service, London and informed consent in accordance with the Declaration of Helsinki. Out of 286 patients (2009–2014) tested at our centre using FISH on CD138+ bone marrow cells (Miltenyi microbead isolation), del(17p) was detected in 10 out of 98 (10.2%) ND and 42 out of 188 (22.3%) relapsed patients. An additional 6 patients were diagnosed with del(17p) at another centre. Material was available for all 16 ND and 35 relapse patients for mutational screening. In 23 patients, samples were available at more than one time point (range 2–4). FISH analysis was performed using standard probes.11 Genomic DNA was amplified by polymerase chain reaction and screened for TP53 mutations in exons 4–11 using denaturing high-performance liquid chromatography (DHPLC; Transgenomic, Glasgow, UK). Mutations were verified by Sanger sequencing. Extended mutation testing was performed on TP53-mutated samples using targeted next generation sequencing (NGS; Supplementary Materials). Clinical details including treatment and disease response (2014 IMWG criteria) were collated. Survival was estimated using Kaplan–Meier methods, and differences were assessed by log-rank test, with P-values <0.05 considered statistically significant.
Eighteen different TP53 mutations were identified in 18 del(17p) patients (35%) by DHPLC (Table 1). Consistent with other studies,10 most mutations occurred in exons 5, 6 and 8 encoding an integral part of the p53 DNA binding domain. Seventeen (94%) were single-nucleotide variants (SNVs), a similar spectrum to that reported previously,10 although others have found deletions or insertions.2 Seventeen SNVs were documented on the International Agency for Research on Cancer TP53 database; 14 (82%) were missense mutations, two nonsense mutations and one altered a splice site. Most were predicted to produce a non-functional protein with <20% transcriptional activity. The remaining mutation was a novel in-frame indel. In 83% of samples, the variant allele frequency of the TP53 mutation was >50%, consistent with the known del(17p) in a significant proportion of cells. In three patients, additional low-level TP53 mutations were detected by NGS.
Table 1. Summary of TP53 mutations identified in the cohort.
| ID | Disease status at time of mutation detection | Months since diagnosis | Del(17p) % by FISH | Mutated TP53 exon | TP53 mutation | TP53 mutation VAF (%)a | On IARC TP53 | Mutational effectb | Additional mutationsc | Additional FISH abnormalities |
|---|---|---|---|---|---|---|---|---|---|---|
| 1d | Relapse | 178 | 100 | 7 | p.C242S | 52 | Yes | Non-functional missense | DIS3 (14%), TRAF3 (1%) | t(4;14) |
| Relapse | 184 | 74 | 7 | p.C242S | 90 | Yes | Non-functional missense | DIS3 (4%), TRAF3 (13%) | t(4;14), 13q- | |
| 2 | Relapse | 73 | 78 | 6 | p.P190L | 12 | Yes | Partially functional missense | None | None |
| 3d | Relapse | 75 | 53 | Int/Ex 9 10 | c.993+2T>C p.R342Xe | 90 12 | Yes Yes | Disrupt splicing Nonsense | DIS3 (88%), FGFR3 (36%) | t(4;14), 1q+, 13q- |
| Relapse | 86 | 87 | Int/Ex 9 | c.993+2T>C | 97 | Yes | Disrupt splicing | DIS3 (99%), FGFR3 (45%) | t(4;14), 1q+,1p- | |
| 4d | Relapse | 56 | 99 | 6 | p.I195T | 97 | Yes | Non-functional missense | KRAS (48%), FAM46C (31%) | 1q+, 13q- |
| Relapse | 79 | 99 | 6 | p.I195T | 93 | Yes | Non-functional missense | KRAS (47%), FAM46C (16%) | 1q+, 13q- | |
| 5 | Relapse | 46 | 79 | 5 | p.C141Y | 66 | Yes | Non-functional missense | None | None |
| 6 | Relapse | 63 | 33 | 6 | p.H193R | 32 | Yes | Non-functional nonsense | NRAS (45%), DIS3 (41%) | Hyperdiploid |
| 7 | Relapse | 26 | 86 | 8 6 | p.R267W p.H214Re | 64 11 | Yes | Non-functional missense Non-functional missense | None | 1q+ |
| 8 | Relapse | 26 | 100 | 5 9 | p.A138_T140delinsTSGDRPA p.Q317Xe | 78 9 | No Yes | In-frame indel Nonsense | ATM (27%), DIS3 (34%), 2x NRAS (9%) (12%) | 1q+ |
| 9d | Relapse | 14 | 80 | 7 | p.Y234C | 99 | Yes | Non-functional missense | KRAS (43%) | 1p-, 13q- |
| Relapse | 19 | 80 | 7 | p.Y234C | 80 | Yes | Non-functional missense | KRAS (38%) | 1p-, 13q- | |
| 10 | Relapse | 12 | 100 | 8 | p.E268A | 97 | Yes | Non-functional missense | None | None |
| 11d | NDMM | 0 | 100 | 10 | p.L348S | 67 | Yes | Non-functional missense | PRDM1 (65%), DIS3 (11%) | 13q-, t(4;14), 1p-,Tetraploid |
| Relapse | 8 | 86 | 10 | p.L348S | 39 | Yes | Non-functional missense | PRDM1 (38%), DIS3 (24%), FAM46C (32%) | 13q-, t(4;14), 1p-, Tetraploid | |
| 12 | Relapse | 11 | 91 | 5 | p.A161T | 97 | Yes | Partially functional missense | None | t(14;16), 1q+ |
| 13 | Relapse | 12 | 100 | 8 | p.G279E | 68 | Yes | Non-functional missense | BRAF (30%) | 1q+ |
| 14 | Relapse | 10 | 100 | 8 | p.P278S | 73 | Yes | Non-functional missense | FAM46C (23%) | None |
| 15 | NDMM | 0 | 84 | 6 | p.Q192X | 88 | Yes | Missense | NRAS (36%), PIK3CA (41%), TRAF3 (45%) | t(14;16), 13q- |
| 16 | NDMM | 0 | 96 | 4 | p.E62X | 91 | Yes | Nonsense | None | None |
| 17 | NDMM | 0 | 92 | 5 | p.V172D | 98 | Yes | Non-functional missense | None | t(11;14) |
| 18 | NDMM | 0 | 77 | 4 | p.F109V | 47 | Yes | Non-functional missense | None | t(14;16), 1q+ |
Abbreviations: FISH, fluorescence in situ hybridization; Int/Ex, intron/exon boundary; NDMM, newly diagnosed multiple myeloma; NGS, next generation sequencing; VAF, variant allele frequency.
VAF as determined by targeted NGS.
Taken from the International Agency for Research on Cancer (IARC) TP53 database; proteins with <20% transcriptional activity were predicted to be non-functional and with 20–75% transcriptional activity partially functional.
Details are provided in Supplementary Table S3.
Patients with longitudinal samples.
Mutation only detected by targeted NGS.
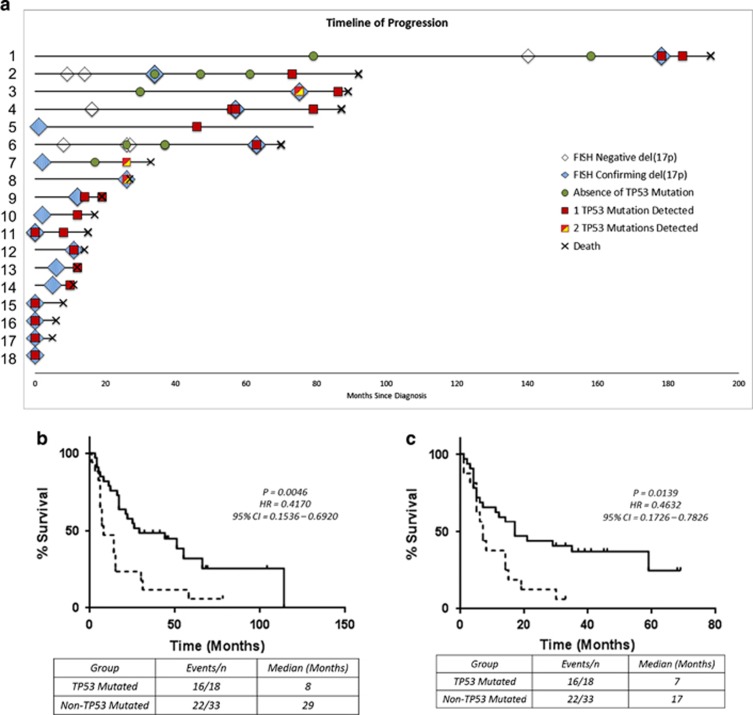
Twenty-three patients were tested more than once, including 8 patients with TP53 mutations, allowing us to explore the chronology of these genetic events. TP53 mutations were identified at multiple time points in 5 patients (Table 1). In case 1, the mutant level increased with disease progression, indicating selection of the TP53-mutated clone. Case 11 showed clonal evolution with the appearance of a FAM46C mutation at relapse. Three patients (cases 3, 4 and 9) had consistently high mutant levels at both time points. The subclonal nonsense mutation detected in case 3 was only present in the first mutated sample. Thirteen of the 18 TP53-mutated patients (72%) were mutant-positive in the first sample analyzed (Figure 1a). In five this was at diagnosis, indicating that 31% (5 of 16) of patients in this cohort presented with both del(17p) and a TP53 mutation (cases 11, 15–18). However, this frequency may be an underestimate as diagnostic samples were not available from two patients who presented with del(17p), both of whom were TP53-mutated in a subsequent relapse sample (cases 5 and 10). In six patients, it was not possible to determine the chronology of their acquisition as earlier FISH results or stored samples were not available. In cases 1, 3 and 6, TP53 mutations were only detected at relapse despite mutational testing in earlier samples. Two of these patients also only acquired the del(17p) at relapse. Of note, in both the remaining two patients (cases 2 and 7), del(17p) acquisition clearly preceded the TP53 mutation. Case 2 acquired del(17p) at first relapse 34 months post-diagnosis but only acquired a TP53 mutation at their fourth relapse 73 months post-diagnosis. Case 7 presented with del(17p), was mutant-negative at relapse 17 months later, but acquired a TP53 mutation at second relapse 26 months post-diagnosis. The chronology of FISH and mutational testing for the 33 del(17p) patients without TP53 mutations is shown in Supplementary Figure S1.
Figure 1.
Clinical impact of TP53 mutations. (a) Swimmer plot displaying the time of del(17p) FISH testing/detection and TP53 mutation screening for each TP53-mutated patient. Kaplan–Meier curves for (b) overall survival (OS) from first detection of del(17p) in patients with and without TP53 mutations, and (c) from first detection of the mutation in TP53-mutated patients compared to the last time point tested in non-mutated patients. Dotted Line, TP53-mutated patients; solid line, non-TP53 patients. CI, 95% confidence intervals; HR, hazard ratios.
There was no difference in age at diagnosis or disease isotype between TP53-mutated and non-mutated patients (Supplementary Table S1). Ten TP53-mutated patients (56%) also had at least one other high-risk cytogenetic feature detected at some point in their disease (t(4;14), t(14;16) or 1q gain; Table 1), but this frequency was not significantly different from the non-TP53-mutated patients (56 vs 49%, P=0.6). Ten TP53-mutated patients (56%) also had additional mutations in previously reported myeloma driver genes9, 12 [DIS3(5), FAM46C(3), NRAS(3), KRAS(2), TRAF3(2), FGFR3(1), PRDM1(1), ATM(1), BRAF(1) and PIK3CA(1)] and most of these patients (7, 39%) had two or more additional mutations (Table 1); in our series, the incidence of DIS3 mutations (28%) appears higher.
Median overall survival (OS) from diagnosis was significantly shorter in TP53-mutated than in non-mutated patients (19 vs 74 months, P=0.02; Supplementary Figure S2), as was median OS from the time at which del(17p) was first detected (8 vs 29 months, P<0.01; Figure 1b). Median progression-free survival (PFS) from first detection of del(17p) was similar between the two groups (7 vs 12 months, P=0.51; Supplementary Figure S3A), but the acquisition of a TP53 mutation was associated with extremely poor survival. Median PFS of patients from the time a mutation was first detected was 5 months (Supplementary Figure S3B), and median OS was 7 months (Figure 1c). In contrast, patients who remained without detectable TP53 mutation had a median PFS of 11.5 months, with median OS 17 months, calculated from the date of the last sample tested. There was thus a striking difference in survival between patients with and without TP53 mutations. These results confirm that, although homozygous deletion of TP53 in MM is uncommon,3 inactivating TP53 mutations that are likely to produce non-functional proteins are frequent in del(17p) MM. TP53 haploinsufficiency in MM causes downstream p53 pathway deregulation, altering the delivery of an apoptotic response,6 but the biological consequence of the additional functional loss conferred by mutation of the remaining allele remains unclear. In this cohort, we observed that TP53 mutation usually occurred after, or simultaneously with, allelic loss of 17p13, and conferred a significantly poorer prognosis for those patients with both abnormalities. Bi-allelic inactivation of tumor suppressor genes, including TP53, has been reported to drive relapse in MM. A recent longitudinal study similarly identified that bi-allelic events leading to complete TP53 inactivation resulted in the poorest prognosis at relapse.13 Patients with the lowest levels of TP53 mRNA have also been reported to have the worst clinical outcome.3, 14 There is also evidence of other mechanisms leading to p53 inactivation in MM, including promoter methylation6 and aberrant expression and function of p53-regulating miRNAs,15 thus investigation of TP53 mRNA and p53 protein levels may be warranted. Collectively our results confirm that, in the context of del(17p), acquisition of a TP53 mutation on the other allele confers significantly worse clinical outcomes, and suggest that allelic loss of 17p13 likely precedes such mutations. In the era of genomic medicine, there is a case for routinely screening for TP53 mutations in del(17p) MM to aid management decisions and direct these high-risk patients towards alternative treatment strategies aimed at counteracting the loss of tumor suppressor activity. Finally, the possibility of a different target gene for del(17p) in patients who do not have mutations of TP53 remains to be explored.
Acknowledgments
UCL/UCLH receives funding as an NIHR Biomedical Research Centre. We would like to acknowledge the generous support of The Snowdome Foundation.
Footnotes
Supplementary Information accompanies this paper on Blood Cancer Journal website (http://www.nature.com/bcj)
The authors declare no conflict of interest.
Supplementary Material
References
- Avet-Loiseau H, Attal M, Moreau P, Charbonnel C, Garban F, Huilin C et al. Genetic abnormalities and survival in multiple myeloma: the experience of the Intergroupe Francophone du Myélome. Blood 2007; 109: 3489–3495. [DOI] [PubMed] [Google Scholar]
- Chng WJ, Price-Troska T, Gonzalez-Paz N, Van Wier S, Jacobus S, Blood E et al. Clinical significance of TP53 mutation in myeloma. Leukemia 2007; 21: 582–584. [DOI] [PubMed] [Google Scholar]
- Boyd KD, Ross FM, Tapper WJ, Chiecchio L, Dagrada G, Konn ZJ et al. The clinical impact and molecular biology of del(17p) in multiple myeloma treated with conventional or thalidomide-based therapy. Genes Chromosomes Cancer 2011; 50: 765–774. [DOI] [PubMed] [Google Scholar]
- Fonseca R, Bergasel PL, Drach J, Shaughnessy J, Gutierrez N, Stewart AK et al. The clinical impact and molecular biology of del(17p) in multiple myeloma treated with conventional or thalidomide-based therapy. Genes Chromosomes Cancer 2011; 50: 765–774. [DOI] [PubMed] [Google Scholar]
- Bergsagel PL, Mateos M-V, Gutierrez NC, Rajkumar SV, San Miguel JF. Improving overall survival and overcoming adverse prognosis in the treatment of cytogenetically high-risk multiple myeloma. Blood 2013; 121: 884–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teoh PJ, Chung TH, Sebastian S, Choo SN, Yan J, Ng SB et al. p53 haploinsufficiency and functional abnormalities in multiple myeloma. Leukemia 2014; 28: 2066–2074. [DOI] [PubMed] [Google Scholar]
- Lionetti M, Barbieri M, Manzoni M, Fabris S, Bandini C, Todoerti K et al. Molecular spectrum of TP53 mutations in plasma cell dyscrasias by next generation sequencing: an Italian cohort study and overview of the literature. Oncotarget 2016; 7: 21353–21361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kortüm KM, Mai EK, Hanafiah NH, Shi CX, Zhu YX, Bruins L et al. Targeted sequencing of refractory myeloma reveals a high incidence of mutations in CRBN and Ras pathway genes. Blood 2016; 128: 1226–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker BA, Boyle EM, Wardell CP, Murison A, Begum DB, Dahir NM et al. Mutational spectrum, copy number changes, and outcome: results of a sequencing study of patients with newly diagnosed myeloma. J Clin Oncol 2015; 33: 3911–3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodé L, Eveillard M, Trichet V, Soussi T, Wuillème S, Richebourg S et al. Mutations in TP53 are exclusively associated with del(17p) in multiple myeloma. Haematologica 2010; 95: 1973–1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D, Stephenson C, Percy L, Lach A, Chatters S, Kempski H et al. Cohort analysis of FISH testing of CD138(+) cells in relapsed multiple myeloma: implications for prognosis and choice of therapy. Br J Haematol 2015; 171: 881–883. [DOI] [PubMed] [Google Scholar]
- Kortüm KM, Langer C, Monge J, Bruins L, Egan JB, Zhu YX et al. Targeted sequencing using a 47 gene multiple myeloma mutation panel (M3P) in -17p high risk disease. Br J Haematol 2015; 168: 507–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinhold N, Ashby C, Rasche L, Chavan SS, Stein C, Stephens OW et al. Clonal selection and double-hit events involving tumor suppressor genes underlie relapse in myeloma. Blood 2016; 128: 1735–1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong W, Wu X, Starnes S, Johnson SK, Haessler J, Wang S et al. An analysis of the clinical and biologic significance of TP53 loss and the identification of potential novel transcriptional targets of TP53 in multiple myeloma. Blood 2008; 112: 4235–4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichiorri F, Suh SS, Rocci A, De Luca L, Taccioli C, Santhanam R et al. Downregulation of p53-inducible microRNAs 192, 194 and 215 impairs the p53/MDM2 autoregulatory loop in multiple myeloma development. Cancer Cell 2010; 18: 367–381. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.