ABSTRACT
Most integral outer membrane proteins (OMPs) of Gram-negative bacteria, such as Escherichia coli, assume a β-barrel structure. The β-barrel assembly machine (Bam), a five-member complex composed of β-barrel OMP BamA and four associated lipoproteins, BamB, BamC, BamD, and BamE, folds and inserts OMPs into the outer membrane. The two essential proteins BamA and BamD interact to stabilize two subcomplexes, BamAB and BamCDE, and genetic and structural evidence suggests that interactions between BamA and BamD occur via an electrostatic interaction between a conserved aspartate residue in a periplasmic domain of BamA and a conserved arginine in BamD. In this work, we characterize charge-change mutations at these key BamA and BamD residues and nearby charged residues in BamA with respect to OMP assembly and Bam complex stability. We show that Bam complex stability does not correlate with function, that BamA and BamD must adopt at least two active conformational states during OMP assembly, and that these charged residues are not required for function. Rather, these charged residues are important for coordinating the activities of BamA and BamD to allow efficient OMP assembly. We present a model of OMP assembly wherein recognition and binding of unfolded OMP substrate by BamA and BamD induce a signaling interaction between the two proteins, causing conformational changes necessary for the assembly reaction to proceed. By analogy to signal sequence recognition by SecYEG, we believe these BamA-BamD interactions ensure that both substrate and complex are competent for OMP assembly before the assembly reaction commences.
IMPORTANCE Conformational changes in the proteins of the β-barrel assembly machine (Bam complex) are associated with the folding and assembly of outer membrane proteins (OMPs) in Gram-negative bacteria. We show that electrostatic interactions between the two essential proteins BamA and BamD coordinate conformational changes upon binding of unfolded substrate that allow the assembly reaction to proceed. Mutations affecting this interaction are lethal not because they destabilize the Bam complex but rather because they disrupt this coordination. Our model of BamA-BamD interactions regulating conformation in response to proper substrate interaction is reminiscent of conformational changes the secretory (Sec) machinery undergoes after signal sequence recognition that ensure protein quality control.
KEYWORDS: Escherichia coli, Gram-negative bacteria, membrane biogenesis, outer membrane proteins, protein-protein interactions
INTRODUCTION
The β-barrel outer membrane proteins (OMPs) play crucial roles in maintaining the integrity of the outer membrane (OM), transporting nutrients, and removing small-molecule toxins from Gram-negative bacteria (1). OMPs destined for the OM are translocated via the secretory (Sec) machinery through the inner membrane (IM), ferried across the periplasm via chaperone proteins, and then properly folded and assembled into the OM by the β-barrel assembly machine (Bam) complex (2). The Bam complex is a heteropentamer composed of BamA (itself an OMP with five N-terminal periplasmic POTRA domains) and four associated OM lipoproteins, BamBCDE (3–7).
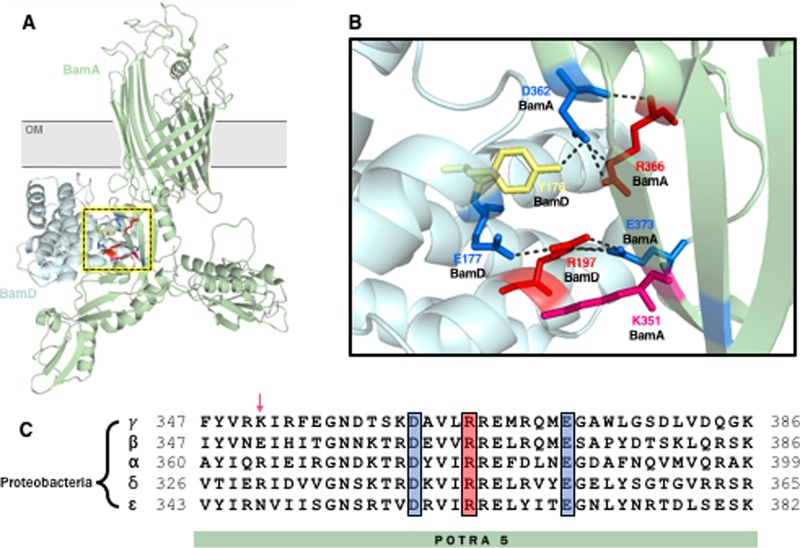
BamA and BamD are both required for viability, and each essential component forms a stable subcomplex involving the accessory lipoproteins (BamAB and BamCDE) (3–7). Structural studies show electrostatic interactions between the membrane-proximal POTRA domain (POTRA5) and the C terminus of BamD, specifically between conserved residues E373 of BamA and R197 of BamD (8–12) (Fig. 1A). A conditionally lethal mutation that perturbs this interaction (bamAE373K) is thought to prevent the formation of a critical salt bridge between these conserved residues and, consequently, to split the Bam holocomplex into BamAB and BamCDE subcomplexes (13).
FIG 1.
A network of electrostatic interactions connects BamA and BamD. (A) The interface between BamA (green) and BamD (blue) centers around a set of conserved charged residues in POTRA5 (D362, R366, and E373) that interact with one another and with Y176, E177, and R197 of BamD (PDB: 5EKQ). The BamD-POTRA5 interface is shown in the inset and in greater detail in panel B. (C) ClustalW alignment of BamA POTRA5 peptide sequences from representative proteobacterial species within Alphaproteobacteria (α) (Brucella abortus), Betaproteobacteria (β) (Neisseria gonorrhoeae), Gammaproteobacteria (γ) (Escherichia coli), Deltaproteobacteria (δ) (Myxococcus xanthus), and Epsilonproteobacteria (ε) (Campylobacter jejuni) classes. Conserved residues D362, R366, and E373 are highlighted, and the position of the variable residue corresponding to K351 in E. coli is indicated by an arrow.
The lethality conferred by bamAE373K is suppressed by a compensatory mutation in BamD (bamDR197L) that restores Bam complex function but does not restore the stable BamABCDE holocomplex (13). This suggests that bamAE373K and bamDR197L are both gain-of-function mutations that enable Bam function even in the absence of a direct physical interaction between BamA and BamD (13).
Biophysical characterization of POTRA5 revealed local conformational plasticity in the electrostatic network within this domain (which includes E373), and it has been proposed that interchanging salt bridges between charged residues in this network underlie the conformational exchange within POTRA5 observed by nuclear magnetic resonance (NMR) (9) (Fig. 1B). Consistent with this suggestion, the E373K mutation introduced as described above was found to shift the conformational equilibrium of POTRA5 (9).
In light of the observed effects of E373 mutation on BamA function, conformation, and protein-protein interactions, we sought to discern the role of this electrostatic network on Bam complex formation and function by combining POTRA5 charge-change variants with wild-type (bamD+) or gain-of-function (bamDR197L) alleles of bamD. Analysis of complex function and complex stability in this mutant panel shows that while the stability of the Bam holocomplex is sensitive to charge-change mutations in the conserved POTRA5 electrostatic network, holocomplex stability is not correlated with Bam complex function. Our results suggest that this network of electrostatic interactions is not required for Bam function per se, but that this electrostatic network evolved as a mechanism for regulating the conformational dynamics of the essential Bam components during OMP assembly. We propose that conserved POTRA5 residues enable coordination of the conformational dynamics of BamA and BamD during the OMP assembly cycle and that mutations that uncouple the intermolecular communication between BamA and BamD may stabilize BamA and BamD conformations that are incompatible and that therefore cannot productively engage in the coordinated process of membrane protein assembly.
RESULTS
Charge change mutations alter the electrostatic network in POTRA5.
To probe the importance of the dynamic electrostatic interactions between BamA residue E373 and BamD residue R197, as well as the electrostatic interactions between BamAE373 and other residues within this region of POTRA5, we generated point mutations that alter the charge of the residues in the POTRA5 electrostatic network (K351, D362, and R366) (Fig. 1B) and characterized their effects on Bam complex function and stability in bamD+ and bamDR197L backgrounds. Three of these residues (D362, R366, and E373) are highly conserved across proteobacteria, while K351 is variable (Fig. 1C). As observed previously (13), the bamAE373K allele causes lethality at physiological temperature (37°C) in bamD+ strains carrying the inducible PBAD-bamA+ construct grown in the absence of the inducer (arabinose), a phenotype that is completely suppressed upon introduction of the bamDR197L allele; the bamAE373K bamDR197L double mutant grows comparably to a wild-type (WT) strain in the absence of arabinose.
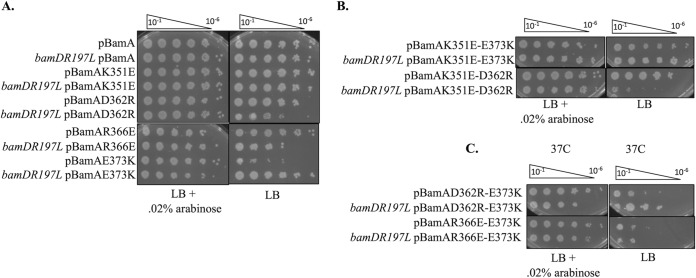
Like bamAE373K mutants, bamAK351E, bamAD362R, and bamAR366E mutants are viable in merodiploid strains in which WT bamA is expressed (i.e., in the presence of arabinose). This indicates that each of the charge change mutations is recessive to bamA+. However, when these mutant BamA variants were expressed in the absence of WT BamA (i.e., in the absence of arabinose), a variety of phenotypes were observed depending on the cognate allele of bamD carried in each strain (Fig. 2A and Table 1); these are described below. In general, these phenotypes were stronger at 37°C than at 30°C.
FIG 2.
Charge-change mutations in BamA POTRA5 cause growth defects. Strains containing an arabinose-inducible copy of bamA (JCM320) as well as pZS21::bamAmut were grown to stationary phase with arabinose, 10-fold serial dilutions (top) were spotted onto LB medium with 25 μg/ml kanamycin or LB with 25 μg/ml kanamycin and 0.2% arabinose, and plates were incubated overnight at 37°C. (A) bamAD362R and bamAR366E are synthetically lethal with bamDR197L, while bamAE373K is synthetically lethal with bamD+. bamAK351E showed no growth defects. (B) bamAK351E is an intragenic suppressor of bamAE373K but not bamAD362R. (C) Both bamAD362R and bamAR366E show reciprocal incompatibility with bamAE373K.
TABLE 1.
Phenotypes of charge change substitutions in POTRA5a
| pZS21::bamA variant | Growth at 37°C in background: |
Stable Bam complex in background: |
||
|---|---|---|---|---|
| bamD+ | bamDR197L | bamD+ | bamD197L | |
| Wild type | + | + | + | + |
| bamAK351E | + | + | + | + |
| bamAD362R | + | − | − | − |
| bamAR366E | + | − | + | + |
| bamAE373K | − | + | − | − |
| bamAK351E-E373K | + | + | − | − |
| bamAK351E-D362R | + | − | ND | ND |
| bamAD362R-E373K | − | − | − | − |
| bamAR366E-E373K | − | − | − | − |
ND, not determined.
The K351E substitution seems not to compromise Bam function, as bamAK351E had no effect on growth or Bam complex function in both the bamD+ and bamDR197L backgrounds at both 30 and 37°C (Fig. 2A; also see Fig. S1 in the supplemental material). The bamAD362R and bamAR366E alleles, on the other hand, display an inverse genetic interaction with the wild-type (bamD+) and gain-of-function (bamDR197L) alleles compared to bamAE373K: bamAD362R and bamAR366E mutants are viable in a bamD+ background but grow poorly at 37°C (Fig. 2A) and exhibit mucoidy in a bamDR197L background at 30°C (Fig. S1 in the supplemental material).
In summary, every BamA POTRA5 charge-change mutant tested was functional with at least one bamD allele tested, demonstrating that none of these conserved residues is necessary for BamA function. These mutations must not affect the ability of BamA and BamD to interact with unfolded substrate or to undergo the conformational changes necessary for normal OMP assembly. It is likely, then, that normal Bam activity (and therefore viability) is compromised by “incompatible” combinations of bamA and bamD alleles (e.g., bamAR366E bamDR197L or bamAE373K bamD+).
Stability of the Bam complex does not correlate with function.
Normally, the Bam complex exists as a stable five-member complex that can be purified intact without the use of cross-linking agents by affinity purification with any one of the members (3, 5). This complex comprises two stable subcomplexes (BamAB and BamCDE) that can form efficiently both in vivo and in vitro and can combine to form a holocomplex centered around POTRA5 (3, 5, 6). It was previously shown that the conditionally lethal E373K mutation disrupts the interaction between the BamAB and BamCDE subcomplexes and that this separation likely contributes to the lethal effects of this mutation by interfering with the ability of BamA and BamD to functionally collaborate. The bamDR197L mutation suppresses the lethal effects of bamAE373K, but it does not restore a stable BamAB-BamCDE interaction (13). This observation led to the previous suggestion that the physical separation of the subcomplexes compromises Bam activity not because a stable BamA-BamD interaction is critical for function per se but because a signaling interaction occurs between wild-type BamA and BamD proteins that regulates (and perhaps coordinates) their activities. If this is true, we should observe that perturbing additional residues in the electrostatic POTRA5 network implicated in the stable BamA-BamD interaction can influence holocomplex stability without necessarily compromising complex function. To test this assumption and to determine the effect of the other bamA charge change mutations on complex stability, we used affinity purification of His-tagged BamA to monitor the BamAB-BamCDE interaction in the presence of these mutations.
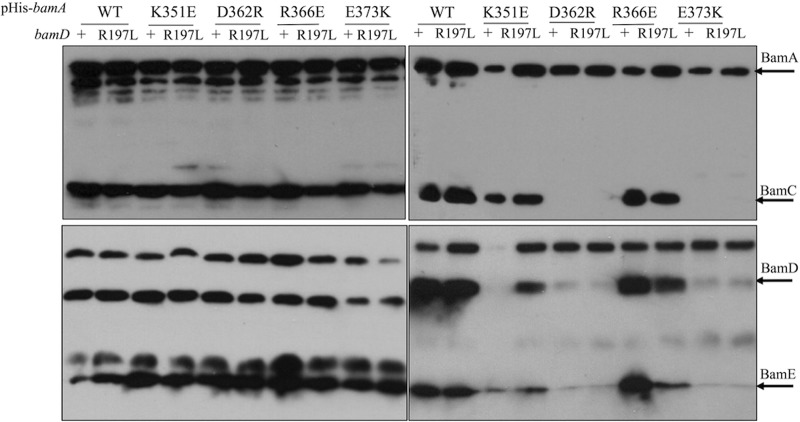
We found that while the R366E mutation has no effect on Bam complex stability (i.e., BamCDE can be copurified with BamA carrying the R366E substitution), the K351E mutation affects the interaction with BamD+ specifically, and the D362R mutation (like E373K) destabilizes the complex in both the bamD+ and bamDR197L backgrounds (Fig. 3). This result is intriguing because bamAE373K and bamAD362R exhibit opposite compatibilities with bamD+ and bamDR197L despite the fact that the E373K and D362R mutations both destabilize the Bam complex (Fig. 2A). In contrast, bamAR366E has the same compatibilities with respect to bamD as does the bamAD362R allele (Fig. 2A); yet, unlike D362R, the R366E mutation does not affect complex stability in either a bamD+ or bamDR197L background. These results clearly demonstrate that Bam complex function does not correlate with Bam complex stability and that incompatibility between bamDR197L and bamAD362R or bamAR366E is unrelated to the stable physical interaction between BamA and BamD. Furthermore, the reciprocal functional relationship between these bamA and bamD alleles cannot be explained by the presence or absence of a stable Bam complex.
FIG 3.
Complex stability does not correlate with function. Affinity purification of His-bamA and His-bamAmut was performed in both a bamD+ and bamDR197L background. Samples were run on SDS-PAGE gels and immunoblotted for BamA, BamC, BamD, and BamE. Left, sample before purification; right, samples after purification. When the complex has been destabilized, BamCDE no longer stably associate with BamA and are not eluted during purification.
An intragenic suppressor of bamAE373K does not restore complex stability.
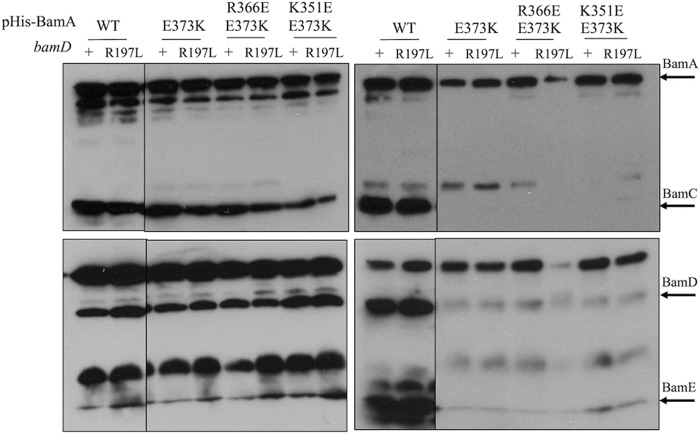
To further probe the role of the dynamic electrostatic interactions in BamA POTRA5, we constructed strains carrying multiple charge change mutations in bamA and characterized their phenotypes in both the bamD+ and the bamDR197L backgrounds. Remarkably, we discovered that K351E acts as an intragenic suppressor of bamAE373K in that it restores growth in a bamD+ background at physiological temperatures (Fig. 2B). As with the extragenic suppressor bamDR197L, which rescues the viability of a bamAE373K mutant without restoring the BamA-BamD interaction, bamAK351E also suppresses the lethality caused by the E373K mutation without restoring the stability of the Bam complex; in both the bamD+ and the bamDR197L backgrounds, the K351E E373K double mutant exhibits the same affinity purification profile as BamAE373K, indicating that the stable interaction between BamA and BamD has not been restored (Fig. 4).
FIG 4.
Double mutations do not restore complex stability even when they restore function. Affinity purification of His-BamA and His-BamAmut was performed in both a bamD+ and bamDR197L background. Samples were run on SDS-PAGE gels and immunoblotted for BamA, BamC, BamD, and BamE. Left, samples before purification; right, samples after purification. When the complex has been destabilized, BamCDE no longer stably associate with BamA and are not eluted during purification. Regardless of the double mutation combination, BamAE373K always destabilizes the Bam complex.
Interestingly, bamAK351E is not a generic suppressor of incompatible bamA and bamD alleles; it does not suppress the synthetic lethality of bamAD362R bamDR197L (Fig. 2B). On the contrary, the K351E D362R double mutant exhibits a more severe growth phenotype than the D362R single mutant in a bamDR197L background (Fig. 2 and Table 1). This demonstrates that the bamAK351E mutation is not a gain-of-function mutation that is cis dominant to other charge change mutations in POTRA5; this mutation cannot suppress all defective BamA-BamD interactions.
Reciprocal incompatibility between alleles of BamA and BamD causes synthetic lethality.
To further explore the contribution of charge POTRA5 residues to Bam function, we constructed double mutants carrying the E373K mutation and either the D362R or R366E substitution and assessed for compatibility with both wild-type bamD and bamDR197L mutant alleles (Fig. 2C). The bamAD362R-E373K double mutant is inviable at all tested temperatures in a bamD+ background and is partially suppressed by bamDR197L at 37°C (Fig. 2C and S1). Similar (if slightly stronger) phenotypes are seen in a bamAR366E-E373K double mutant, which is inviable at all temperatures in a bamD+ background and with bamDR197L at both 30°C and 37°C (Fig. 2C and S1).
While bamDR197L suppresses the lethality of bamAE373K, it does not suppress the lethality of the bamAR366E-E373K and bamAD362R-E373K double mutants, indicating that the bamDR197L mutation is incompatible with the R366E or D362R substitutions in BamA, even when the gain-of-function bamAE373K mutation is present. Furthermore, the bamAR366E and bamAD362R mutations do not suppress the lethality of bamAE373K in a bamD+ background even though each of those mutations has no defect in a bamD+ background, suggesting that both sets of mutations have cis-dominant and opposite defects with respect to interactions with different bamD alleles; that is, the bamAE373K phenotype is observed when these BamA double mutants are expressed in a bamD+ background, whereas the bamAD362R or bamAR366E phenotype is observed in a bamDR197L background. Additionally, while BamAD362R does not interact with either BamD or BamDR197L and BamAR366E interacts normally with both BamD and BamDR197L (Fig. 3), in both double mutants (bamAD362R-E373K and bamAR366E-E373K) a stable physical interaction between BamA and BamD cannot be detected (Fig. 4). Like the mutant with the bamAE373K mutation alone, the double mutants show Bam subcomplex separation in both a bamD+ and bamDR197L background. This suggests that the bamAE373K mutation is cis-dominant in its ability to destabilize the Bam complex, as no double mutants containing bamAE373K, including the bamD+-compatible allele bamAK351E-E373K, permit the formation of a stable Bam holocomplex.
We suggest that the bamAD362R-E373K and bamAR366E-E373K double mutants cannot properly coordinate with either bamD allele, rendering them unable to undergo the necessary conformational changes to proceed with OMP assembly. That neither mutation overrides the other in the double mutant is further evidence of the reciprocal incompatibility between the POTRA5 mutants.
DISCUSSION
The salt bridge between the invariant residues BamAE373 and BamDR197 is a prominent feature of all Bam complex crystal structures to date, and charge change mutations at BamA373 are known to disrupt the BamA-BamD interaction. However, several observations challenge the notion that the critical role of BamAE373, and the salt bridge it forms with BamDR197, is simply to stabilize an essential physical interaction between BamA POTRA5 and BamD. Chief among these observations is the finding that intragenic and extragenic suppressors of bamAE373K restore Bam function but fail to restore complex stability, demonstrating that the BamAE373-BamDR197 salt bridge is not critically important for Bam complex function. We argue that the dynamic electrostatic interactions among the closely linked charged residues in POTRA5 and BamD are conserved not because they are central to the function(s) of these essential factors but rather because they serve to coordinate the conformational dynamics, and therefore the activities, of BamA and BamD.
The bamAE373K mutation and its extragenic suppressor bamDR197L allow both mutant BamA and BamD proteins to function in the absence of the stable physical interaction that normally occurs between them. The simplest explanation for this suppression posits that the mutations alter the conformation of both proteins so as to bypass the requirement for a stable interaction. Presumably, then, BamAE373K cripples Bam function in the presence of wild-type BamD, not because BamA is incompetent to bind and assemble substrates but because the lack of a stable interaction prevents proper BamA-induced activation of the cognate BamD partner. This conclusion is supported by recent NMR-based characterization of the conformational dynamics of POTRA5 carrying various residues at position 373; this work suggests that the E373K mutation does not result in a novel POTRA5 conformation but rather shifts the observed two-state conformational equilibrium of POTRA5 to one extreme (9). In what follows, we use the terms of allostery to describe these conformations (14) with unfolded OMPs as the ligand. In the absence of interaction with an unfolded OMP, BamA remains in an inert, or “tense,” basal state. Because the BamAE373K variant is fully functional in the presence of a suppressor mutation and in the absence of a direct interaction with BamD, we suggest that the E373K mutation biases the conformation of BamA toward a relaxed conformation that is normally achieved only when the Bam complex engages client OMPs (the ligand) in the course of normal assembly.
It is striking that the BamAD362R and the BamAR366E mutation proteins behave in a fashion opposite that of BamAE373K, whereas only one of the two mutations affects Bam complex stability. Like E373K, the D362R substitution also disrupts complex stability, but the mutant protein can function only with wild-type BamD and not the gain-of-function BamDR197L variant, which is otherwise compatible with both BamA+ and BamAE373K. bamAR366E is also incompatible with bamDR197L, but the synthetic lethality observed must not be caused by a disruption of the physical interaction between BamAR366E and BamDR197L, as the complete holocomplex forms in the presence of both mutations. Regardless of complex stability, we suspect that BamAD362R and BamAR366E cannot properly coordinate with BamDR197L in the way that BamAE373K can. To explain these reciprocal relationships, we propose that the BamAD362R and BamAR366E mutant proteins exist in a conformation, which we designate tense, being distinct from that of BamAE373K, “relaxed,” and that the R366E and D362R substitutions drive the conformational equilibrium of POTRA5 in the opposite direction as E373K. Similarly, we argue that the R197L mutation biases the conformation of BamD toward a relaxed state that is normally precipitated by an interaction with POTRA5 and OMP substrates. One of the BamA conformations (tense, typified by BamAD362R and BamAR366E) is compatible with wild-type BamD, whereas the other (relaxed, typified by BamAE373K) is compatible with BamDR197L (itself a relaxed conformer). Clearly, each conformational state described for both BamA and BamD must be functional, but they must be paired with a complementary conformation of the partner protein to efficiently complete the OMP assembly cycle.
Underlying our mechanistic interpretation is the suspicion that BamA and BamD in the resting state complex are primed to accept incoming substrate and that the binding of a component of the substrate to the exposed nexus of charged residues at the BamA-BamD interface triggers an allosteric event that shifts the conformational equilibria of BamA and BamD from tense to relaxed states that are compatible with OMP assembly. Presumably, the conformational dynamics observed in BamA require the reorganization of electrostatic interactions involving the conserved charged residues characterized in this work, and previous structural and spectroscopic analyses of POTRA5 support this assertion (9, 15, 16).
The notion of induced coordinated conformational dynamics of BamA and BamD is consistent with the fact that both BamA and BamD interact with unfolded OMP substrates. Indeed, BamD directly interacts with the C termini of substrate OMPs (17–20), a prime candidate for the allosteric inducer of Bam complex activation. This interpretation also highlights potential mechanistic conservation between BamA and the Omp85 family transport protein FhaC, the conformation of which is regulated by binding of its substrate (filamentous hemagglutinin [FHA]) to the β-barrel-proximal POTRA domain (21).
Evidently, the stable interaction between the BamAB and BamCDE subcomplexes can be disrupted by mutation without compromising function in any detectable way. It is essentially impossible to prove that the two subcomplexes do not interact weakly and transiently; however, we think it quite possible that these complexes can function in the absence of a direct interaction. As it is highly unlikely that a folding intermediate of an OMP could diffuse from one subcomplex to another, we think it more likely that each subcomplex interacts simultaneously with the same substrate protein even though the subcomplexes do not stably interact.
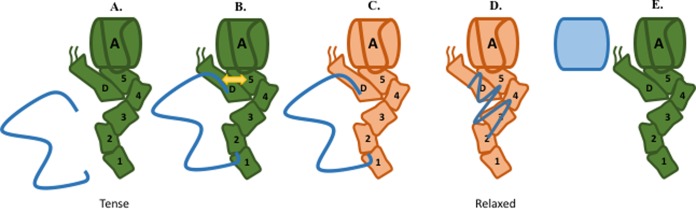
The results summarized in the preceding paragraphs have led us to propose a model wherein BamA-BamD interactions serve to coordinate conformational changes in BamA and BamD that initiate the assembly process (Fig. 5). In our model, the unfolded substrate is bound by BamA and/or BamD (Fig. 5A) (20, 22, 23). Substrate binding induces conformational changes (tense to relaxed) in both BamA and BamD (Fig. 5C) that are communicated between the two proteins via the dynamic electrostatic network between POTRA5 and BamDR197 (Fig. 5B). Relaxed BamA and BamD are then competent to continue the OMP assembly cycle (Fig. 5D). After the nascent OMP is inserted into the OM, the Bam complex returns to its initial state (tense) to accept another incoming substrate (Fig. 5E).
FIG 5.
BamA-BamD interactions coordinate BamA and BamD conformational changes during OMP assembly. (A) The wild-type Bam complex exists in an initial (tense) state that is competent to accept substrate and assemble OMPs. (B) Unfolded substrate arrives at the Bam complex and interacts with BamD and BamA. This interaction induces communication between BamA and BamD at POTRA5. (C) Proper binding of substrate by BamA and BamD triggers conformational changes that allow assembly to proceed. (D) Activated (relaxed) BamA and BamD can now continue to efficiently assemble OMPs into the outer membrane. (E) After OMP assembly, the Bam complex is recycled to its initial (tense) state.
In the absence of a stable interaction between BamA and BamD, coordination of the activities of these two proteins after substrate binding is likely impaired. Under these conditions, gain-of-function mutations that shift the conformational equilibrium without input from the partner protein bypass the need for direct communication, allowing OMP assembly to proceed. For example, bamAE373K disrupts BamA-BamD interactions and alters the conformation of BamA, but BamD remains in its initial state, awaiting coordination with BamA that is normally dependent on a protein-protein interaction between them. In this case, the bamAE373K mutation is lethal unless bamDR197L independently alters the conformational equilibrium of BamD (and both proteins bypass the need for direct interaction); here, we anticipate that BamAE373K and BamDR197L each represent the relaxed forms of BamA and BamD, respectively. Additionally, we suggest that the K351E mutation shifts the equilibrium of BamAK351E-E373K back toward the tense conformation that can properly and efficiently communicate with BamD to allow proper coordination to occur, suppressing bamAE373K lethality regardless of the allele of bamD present.
On the contrary, bamAD362R and bamAR366E are partial-loss-of-function mutations that bias the conformational equilibrium toward the tense state so as to preclude proper communication between BamA and the BamDR197L mutant, which is conformationally shifted toward the relaxed state. As BamA then fails to coordinate with BamD, OMP assembly cannot proceed efficiently. However, these BamA mutant proteins must be capable of coordinating with wild-type BamD even in the absence of a stable complex, as there are no growth defects in an otherwise wild-type background. This may well indicate that bound substrate can drive BamA and BamD coordination even if stable complex formation cannot be demonstrated.
Whether or not additional genetic, biochemical, and structural inquiries support the model, our results clearly demonstrate the existence of two different conformations of both BamA and BamD, and neither one of these conformations is inactive. The SecYEG complex, which translocates proteins into the periplasm and assembles proteins in the IM, can exist in both a closed and an open complex (24). The prl alleles of the sec genes suppress signal sequence mutations (25), even deletions that remove the entire signal sequence without noticeable effects on wild-type secreted or membrane proteins (26, 27), either by stabilizing the open complex or by destabilizing the close complex (27). We suggest that, like the Sec complex (28), the Bam complex also uses conformational changes as a proofreading step to ensure substrate quality control.
MATERIALS AND METHODS
Strain construction.
The strains used in this study are listed in Table 2. Charge change mutations were introduced into the pZS21::BamA and pET22-42::His6-BamA plasmids using site-directed mutagenesis (QuikChange), according to the manufacturer's instructions. Relevant oligonucleotides designed using PrimerX (http://bioinformatics.org/primerx/cgi-bin/DNA_1.cgi) are listed in Table S1 in the supplemental material. Strains constructed in the BamA deletion strain (5) were maintained on 0.02% arabinose with 25 μg/ml kanamycin. Strains containing the pET22-42::His6-BamA plasmid were maintained on 25 μg/ml ampicillin.
TABLE 2.
Relevant strains used
| Strain | Genotype or description | Reference or source |
|---|---|---|
| JCM158 | MC4100 arar/− | 5 |
| JCM320 | JCM158 ΔbamA Δ(λatt-lom)::bla PBAD bamA araC | 3 |
| DPR437 | JCM320/pZS21::bamA+ | 13 |
| ALM067 | JCM320 bamDR197L/pZS21::bamA+ | This study |
| FA101 | JCM320/pZS21::bamAK351E | This study |
| FA102 | JCM320 bamDR197L/pZS21::bamAK351E | This study |
| DPR1006 | JCM320/pZS21::bamAD362R | This study |
| DPR1014 | JCM320 bamDR197L/pZS21::bamAD362R | This study |
| DPR1007 | JCM320/pZS21::bamAR366E | This study |
| DPR1015 | JCM320 bamDR197L/pZS21::bamAR366E | This study |
| DPR662 | JCM320/pZS21::bamAE373K | 13 |
| DPR682 | JCM320 bamDR197L/pZS21::bamAE373K | 13 |
| FA105 | JCM320/pZS21::bamAK351E-E373K | This study |
| FA106 | JCM320 bamDR197L/pZS21::bamAK351E-E373K | This study |
| ALM430 | JCM320/pZS21::bamAK351E-D362R | This study |
| ALM431 | JCM320 bamDR197L/pZS21::bamAK351E-D362R | This study |
| ALM063 | JCM320/pZS21::bamAD362R-E373K | This study |
| ALM432 | JCM320 bamDR197L/pZS21::bamAD362R-E373K | This study |
| ALM064 | JCM320/pZS21::bamAR366E-E373K | This study |
| ALM068 | JCM320 bamDR197L/pZS21::bamAR366E-E373K | This study |
| DPR821 | JCM158/pET22-42::His6-BamA+ | 13 |
| DPR850 | JCM158 bamDR197L/pET22-42::His6-BamA+ | This study |
| ALM426 | JCM158/pET22-42::His6-BamAK351E | This study |
| ALM427 | JCM158 bamDR197L/pET22-42::His6-BamAK351E | This study |
| ALM420 | JCM158/pET22-42::His6-BamAD362R | This study |
| ALM421 | JCM158 bamDR197L/pET22-42::His6-BamAD362R | This study |
| ALM424 | JCM158/pET22-42::His6-BamAR366E | This study |
| ALM425 | JCM158 bamDR197L/pET22-42::His6-BamAR366E | This study |
| DPR822 | JCM158/pET22-42::His6-BamAE373K | 13 |
| DPR871 | JCM158 bamDR197L/pET22-42::His6-BamAE373K | This study |
| ALM410 | JCM158/pET22-42::His6-BamAK351E-E373K | This study |
| ALM411 | JCM158 bamDR197L/pET22-42::His6-BamAK351E-E373K | This study |
| ALM422 | JCM158/pET22-42::His6-BamAD362R-E373K | This study |
| ALM423 | JCM158 bamDR197L/pET22-42::His6-BamAD362R-E373K | This study |
| ALM424 | JCM158/pET22-42::His6-BamAR366E-E373K | This study |
| ALM425 | JCM158 bamDR197L/pET22-42::His6-BamAR366E-E373K | This study |
Growth assays.
Relevant strains were grown overnight at 37°C in LB containing 0.02% arabinose with 25 μg/ml kanamycin. Tenfold serial dilutions in LB without arabinose were made in 200-μl wells in a 96-well plate and then spotted onto LB plates containing 25 μg/ml kanamycin with and without arabinose. Plates were incubated at 30 and 37°C for 18 h and at 24°C for 36 h.
Affinity purification.
Cells were grown in 200 ml of LB with 25 μg/ml ampicillin at 30°C until reaching an optical density (OD) of ∼0.8. Cells were harvested by centrifugation at 5,000 × g for 10 min. Cell pellets were washed twice with 20 ml of 20 mM potassium phosphate (pH 7.2) and 150 mM NaCl. Cells were lysed in 10 ml of BugBuster solution (Novagen) containing 5 μg/ml lysozyme, 50 μg/ml DNase 1, 50 μg/ml RNase 1, and 1 μM phenylmethylsulfonyl fluoride (PMSF) by 1 h rocking at 4°C. Cellular debris was removed by centrifugation at 10,000 × g for 20 min. After 100-μl samples were removed for Western blotting, the cleared lysate (whole-cell extract) was incubated with 100 μl of nickel-nitrilotriacetic acid (Ni-NTA) beads (Qiagen) by 1 h rocking at 4°C. After incubation, beads were collected by spinning at 2,500 × g for 5 min and transferred to Eppendorf tubes. Beads were pulse-spun and washed 5 times with 1 ml of buffer containing 50 mM potassium phosphate (pH 8.0), 300 mM NaCl, and 20 mM imidazole. Two 250-μl elutions in buffer containing 50 mM potassium phosphate (pH 8.0), 300 mM NaCl, and 200 mM imidazole were pooled. Input samples were mixed with 2× SDS sample buffer, and the eluate was mixed with 5× SDS sample buffer (both without reducing agents). Samples were boiled at 100°C for 10 min. SDS-PAGE analysis was performed using 12% acrylamide gels run at 140 V. Gels were transferred to nitrocellulose membranes and blocked with a 1% (wt/vol) milk solution for 1 h before antibodies to detect BamA (1:20,000 dilution), BamC (1:40,000 dilution), BamD (1:10,000 dilution), and BamE (1:10,000 dilution) were added for incubation. A donkey anti-rabbit IgG–horseradish peroxidase (HRP) conjugate (Amersham) secondary antibody was incubated at a concentration of 1:8,000. Luminata Classico Western HRP substrate (EMD Millipore) and X-ray film (LabScientific) were used to visualize bands.
Supplementary Material
ACKNOWLEDGMENTS
We thank the Silhavy lab for critical discussion and reading of the manuscript.
This work was supported by National Institute of General Medical Sciences grant GM034821 awarded to T.J.S.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00373-17.
REFERENCES
- 1.Silhavy TJ, Kahne D, Walker S. 2010. The bacterial cell envelope. Cold Spring Harb Perspect Biol 2:a000414. doi: 10.1101/cshperspect.a000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ricci DP, Silhavy TJ. 2012. The Bam machine: a molecular cooper. Biochim Biophys Acta 1818:1067–1084. doi: 10.1016/j.bbamem.2011.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu T, Malinverni J, Ruiz N, Kim S, Silhavy TJ, Kahne D. 2005. Identification of a multicomponent complex required for outer membrane biogenesis in Escherichia coli. Cell 121:235–245. doi: 10.1016/j.cell.2005.02.015. [DOI] [PubMed] [Google Scholar]
- 4.Sklar JG, Wu T, Kahne D, Silhavy TJ. 2007. Defining the roles of the periplasmic chaperones SurA, Skp, and DegP in Escherichia coli. Genes Dev 21:2473–2484. doi: 10.1101/gad.1581007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Malinverni JC, Werner J, Kim S, Sklar JG, Kahne D, Misra R, Silhavy TJ. 2006. YfiO stabilizes the YaeT complex and is essential for outer membrane protein assembly in Escherichia coli. Mol Microbiol 61:151–164. doi: 10.1111/j.1365-2958.2006.05211.x. [DOI] [PubMed] [Google Scholar]
- 6.Hagan CL, Kim S, Kahne D. 2010. Reconstitution of outer membrane protein assembly from purified components. Science 328:890–892. doi: 10.1126/science.1188919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim S, Malinverni JC, Sliz P, Silhavy TJ, Harrison SC, Kahne D. 2007. Structure and function of an essential component of the outer membrane protein assembly machine. Science 317:961–964. doi: 10.1126/science.1143993. [DOI] [PubMed] [Google Scholar]
- 8.Bakelar J, Buchanan SK, Noinaj N. 2016. The structure of the β-barrel assembly machinery complex. Science 351:180–186. doi: 10.1126/science.aad3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sinnige T, Weingarth M, Daniëls M, Boelens R, Bonvin AMJJ, Houben K, Baldus M. 2015. Conformational plasticity of the POTRA 5 domain in the outer membrane protein assembly factor BamA. Structure 23:1317–1324. doi: 10.1016/j.str.2015.04.014. [DOI] [PubMed] [Google Scholar]
- 10.Iadanza MG, Higgins AJ, Schiffrin B, Calabrese AN, Brockwell DJ, Ashcroft AE, Radford SE, Ranson NA. 2016. Lateral opening in the intact β-barrel assembly machinery captured by cryo-EM. Nat Commun 7:12865. doi: 10.1038/ncomms12865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Han L, Zheng J, Wang Y, Yang X, Liu Y, Sun C, Cao B, Zhou H, Ni D, Lou J, Zhao Y, Huang Y. 2016. Structure of the BAM complex and its implications for biogenesis of outer-membrane proteins. Nat Struct Mol Biol 23:192–196. doi: 10.1038/nsmb.3181. [DOI] [PubMed] [Google Scholar]
- 12.Gu Y, Li H, Dong H, Zeng Y, Zhang Z, Paterson NG, Stansfeld PJ, Wang Z, Zhang Y, Wang W, Dong C. 2016. Structural basis of outer membrane protein insertion by the BAM complex. Nature 531:64–69. doi: 10.1038/nature17199. [DOI] [PubMed] [Google Scholar]
- 13.Ricci DP, Hagan CL, Kahne D, Silhavy TJ. 2012. Activation of the Escherichia coli β-barrel assembly machine (Bam) is required for essential components to interact properly with substrate. Proc Natl Acad Sci U S A 109:3487–3491. doi: 10.1073/pnas.1201362109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Monod J, Wyman J, Changeux JP. 1965. On the nature of allosteric transitions: a plausible model. J Mol Biol 12:88–118. doi: 10.1016/S0022-2836(65)80285-6. [DOI] [PubMed] [Google Scholar]
- 15.Gatzeva-Topalova PZ, Warner LR, Pardi A, Sousa MC. 2010. Structure and flexibility of the complete periplasmic domain of BamA: the protein insertion machine of the outer membrane. Structure 18:1492–1501. doi: 10.1016/j.str.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang H, Gao Z-Q, Hou H-F, Xu J-H, Li L-F, Su X-D, Dong Y-H. 2011. High-resolution structure of a new crystal form of BamA POTRA4-5 from Escherichia coli. Acta Crystallogr Sect F Struct Biol Cryst Commun 67:734–738. doi: 10.1107/S1744309111014254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Robert V, Volokhina EB, Senf F, Bos MP, Van Gelder P, Tommassen J. 2006. Assembly factor Omp85 recognizes its outer membrane protein substrates by a species-specific C-terminal motif. PLoS Biol 4:e377. doi: 10.1371/journal.pbio.0040377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Albrecht R, Zeth K. 2011. Structural basis of outer membrane protein biogenesis in bacteria. J Biol Chem 286:27792–27803. doi: 10.1074/jbc.M111.238931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sandoval CM, Baker SL, Jansen K, Metzner SI, Sousa MC. 2011. Crystal structure of BamD: an essential component of the β-barrel assembly machinery of Gram-negative bacteria. J Mol Biol 409:348–357. doi: 10.1016/j.jmb.2011.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hagan CL, Wzorek JS, Kahne D. 2015. Inhibition of the β-barrel assembly machine by a peptide that binds BamD. Proc Natl Acad Sci U S A 112:2011–2016. doi: 10.1073/pnas.1415955112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guérin J, Saint N, Baud C, Meli AC, Etienne E, Locht C, Vezin H, Jacob-Dubuisson F. 2015. Dynamic interplay of membrane-proximal POTRA domain and conserved loop L6 in Omp85 transporter FhaC. Mol Microbiol 98:490–501. doi: 10.1111/mmi.13137. [DOI] [PubMed] [Google Scholar]
- 22.Lee J, Xue M, Wzorek JS, Wu T, Grabowicz M, Gronenberg LS, Sutterlin HA, Davis RM, Ruiz N, Silhavy TJ, Kahne DE. 2016. Characterization of a stalled complex on the β-barrel assembly machine. Proc Natl Acad Sci U S A 113:8717–8722. doi: 10.1073/pnas.1604100113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wzorek JS, Lee J, Tomasek D, Hagan CL, Kahne DE. 2017. Membrane integration of an essential β-barrel protein prerequires burial of an extracellular loop. Proc Natl Acad Sci U S A 114:2598–2603. doi: 10.1073/pnas.1616576114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Van Den Berg B, Clemons WM Jr, Collinson I, Modis Y, Hartmann E, Harrison SC, Rapoport TA. 2004. X-ray structure of a protein-conducting channel. Nature 427:36–44. doi: 10.1038/nature02218. [DOI] [PubMed] [Google Scholar]
- 25.Emr SD, Hanley-Way S, Silhavy TJ. 1981. Suppressor mutations that restore export of a protein with a defective signal sequence. Cell 23:79–88. doi: 10.1016/0092-8674(81)90272-5. [DOI] [PubMed] [Google Scholar]
- 26.Flower AM, Doebele RC, Silhavy TJ. 1994. PrlA and PrlG suppressors reduce the requirement for signal sequence recognition. J Bacteriol 176:5607–5614. doi: 10.1128/jb.176.18.5607-5614.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Derman AI, Puziss JW, Bassford PJ Jr, Beckwith J. 1993. A signal sequence is not required for protein export in prlA mutants of Escherichia coli. EMBO J 12:879–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Osborne RS, Silhavy TJ. 1993. PrlA suppressor mutations cluster in regions corresponding to three distinct topological domains. EMBO J 12:3391–3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.