ABSTRACT
Outer membrane protein (OMP) biogenesis in Escherichia coli is a robust process essential to the life of the organism. It is catalyzed by the β-barrel assembly machine (Bam) complex, and a number of quality control factors, including periplasmic chaperones and proteases, maintain the integrity of this trafficking pathway. Little is known, however, about how periplasmic proteases recognize and degrade OMP substrates when assembly is compromised or whether different proteases recognize the same substrate at distinct points in the assembly pathway. In this work, we use well-defined assembly-defective mutants of LptD, the essential lipopolysaccharide assembly translocon, to show that the periplasmic protease DegP degrades substrates with assembly defects that prevent or impair initial contact with Bam, causing the mutant protein to accumulate in the periplasm. In contrast, another periplasmic protease, BepA, degrades a LptD mutant substrate that has engaged the Bam complex and formed a nearly complete barrel. Furthermore, we describe the role of the outer membrane lipoprotein YcaL, a protease of heretofore unknown function, in the degradation of a LptD substrate that has engaged the Bam complex but is stalled at an earlier step in the assembly process that is not accessible to BepA. Our results demonstrate that multiple periplasmic proteases monitor OMPs at distinct points in the assembly process.
IMPORTANCE OMP assembly is catalyzed by the essential Bam complex and occurs in a cellular environment devoid of energy sources. Assembly intermediates that misfold can compromise this essential molecular machine. Here we demonstrate distinctive roles for three different periplasmic proteases that can clear OMP substrates with folding defects that compromise assembly at three different stages. These quality control factors help ensure the integrity of the permeability barrier that contributes to the intrinsic resistance of Gram-negative organisms to many antibiotics.
KEYWORDS: assembly intermediates, β-barrel proteins, chaperones, lipopolysaccharide, outer membrane, periplasm, protease
INTRODUCTION
The Gram-negative bacterial outer membrane (OM) is an essential, selectively permeable, lipid barrier replete with integral β-barrel outer membrane proteins (OMPs) and lipoproteins (1, 2). The outer leaflet of the asymmetric OM bilayer is composed of lipopolysaccharide (LPS), which contributes to the intrinsic resistance Gram-negative bacteria exhibit against membrane perturbants and many antibiotics. Some OMPs serve as pores for nutrient diffusion, while others facilitate the assembly of β-barrel proteins and LPS into the OM (1, 3, 4). The biogenesis and assembly of OMPs is a tightly regulated process that features multiple redundant steps and quality control measures, including protein chaperoning and degradation of defective substrates (4–7).
OMPs are secreted through the inner membrane (IM) translocase (Sec) machinery before being transported across the aqueous periplasm and finally assembled into the OM by the β-barrel assembly machine (Bam) complex (4). Prior to assembly, substrates are maintained in a folding-competent state by a partially redundant network of chaperone proteins and folding factors, such as SurA, FkpA, and Skp (5, 7–9). Skp and the chaperone-protease DegP also accommodate substrates that “fall off” the SurA-Bam folding pathway during basal substrate mishandling or envelope stress. Skp is hypothesized to aid in rescuing assembly, while DegP is responsible for both chaperoning and degrading off-pathway substrates (7, 9–11).
DegP is a multimeric chaperone-protease that has long been recognized as an essential factor for the maintenance of envelope integrity in response to OMP misfolding. The deletion of degP from a surA strain results in a synthetically lethal phenotype above 25°C, and ΔdegP ΔsurA strains accumulate unfolded OMPs such as LamB and OmpA (7, 9). ΔdegP strains alone are temperature sensitive, and the degP gene is upregulated in response to envelope stress sensed by the σE stress response (10, 12). DegP forms interactions with substrates through its PDZ domain and is capable of forming a variety of multimeric structures to enclose or to degrade misfolded proteins (13, 14).
In order to ensure that the Bam complex is not compromised or to prevent the insertion of defective substrates in the OM, it stands to reason that proteases other than DegP also degrade substrates before or at the point of OMP insertion by the Bam complex. Nevertheless, few periplasmic proteases have been as well described or implicated in OMP biogenesis. Recently, Narita et al. revealed a role for the zinc metalloprotease YfgC (renamed BepA) in the biogenesis of the LPS assembly OMP LptD (15). The authors showed that BepA associates with the Bam complex and displays, like DegP, protease and chaperone functions that both contribute to the maturation of LptD. Also like DegP, BepA is regulated by the σE stress response (12). Reflecting the importance of BepA in quality control, ΔbepA mutants are sensitive to a range of antibiotics.
Weski and Ehrmann recently conducted a thorough synthetic mutant phenotypic analysis of many periplasmic quality control factors, revealing the redundancy of the OM quality control network and the importance of proteases and chaperones under suboptimal growth conditions (16). We sought to further clarify the roles of DegP and BepA in the biogenesis of OMPs while identifying and exploring the potential role(s) that other envelope proteases may play in maintaining the integrity of OMP assembly. Acknowledging the potential redundancy among the OMP quality control network and the efficiency of wild-type OMP assembly, our experimental design used defined mutants in a complex OMP substrate to explore envelope protease function (11).
We focused here on the biogenesis of the aforementioned OMP LptD, the OM component and terminal member of the LPS biogenesis pathway (3, 17–19). LptD is a unique OMP with a 26-strand C-terminal β-barrel arranged around a lipoprotein plug (LptE), a soluble periplasmic N-terminal β-jellyroll domain, and two nonconsecutive disulfide bonds (20–22). Wild-type LptD biogenesis requires that the substrate be maintained in a folding-competent state by SurA, Skp, and FkpA (23, 24) before reaching the Bam complex, where the barrel is assembled in coordination with its lipoprotein plug LptE (22, 25). When LptD maturation is stalled by limiting levels of LptE, BepA degrades the improperly folded substrate (15). After assembly, LptD requires the reorganization of two disulfide bonds to become a fully mature two-protein translocon (24, 25). Due to the reliance of LptD on multiple biogenesis factors and quality control mechanisms, we reasoned that a wide range of proteases might monitor its assembly. In using LptD mutants that have defined assembly defects, we aimed to unambiguously define the role of proteases in monitoring OMPs at specific stages in the assembly process.
Here we report genetic and biochemical evidence that supports the roles of DegP and BepA in degrading LptD at distinct points in the assembly pathway, Furthermore, we identify a novel role for the as-yet-unknown protease YcaL in the degradation of LptD after the substrate is initially engaged with the Bam complex. Specifically, we propose that YcaL degrades substrates that have engaged the Bam complex but are stalled at an early step that is distinct from the step at which DegP or BepA degrades defective β-barrels. Furthermore, we find that DegP, BepA, and YcaL may degrade substrates that vary in folding competence. These analyses support a model in which periplasmic proteases exhibit functionally segregated substrate recognition and degradation profiles, implying that proteases monitor OMPs along the entire biogenesis pathway.
RESULTS
Characterization of OM proteases using informed combinations of mutations.
In a wild-type strain, the degradation of OMP substrates is so efficient that whole-cell levels of an OMP are typically a reliable proxy for the assembled substrate. Therefore, to reveal the potential roles of various periplasmic proteases in the degradation of LptD, we selected well-characterized mutations from previous investigations in which specific steps in assembly of the LptD were demonstrated to be defective. Through the use of these well-defined mutants, we sought to determine how periplasmic proteases may degrade a substrate at different steps in assembly and thereby prevent defective OMP insertion or Bam inhibition.
In this double mutant analysis, we separately combined null mutations in 10 factors proposed to be important in periplasmic protein quality control (degP, degQ, ompT, ptrA, tsp, ycaL, ydgD, loiP, bepA, and yhiJ) (16, 26) with well-characterized mutations in the LptD assembly pathway. Levels of β-mercaptoethanol (β-ME)-treated (reduced) LptD protein are an indication of the whole-cell levels of LptD, which may include misfolded or intermediate substrates. The levels of fully oxidized LptD, however, are representative of properly matured and functional LptDE translocons (24, 27). Therefore, we determined both reduced and oxidized LptD levels as well as any temperature sensitivity phenotypes of each resulting double mutant strain. Because LptD is essential for the insertion of LPS in the OM and thus defects in LptD biogenesis cause increased OM permeability (2, 17, 28), we also examined OM permeability in each strain.
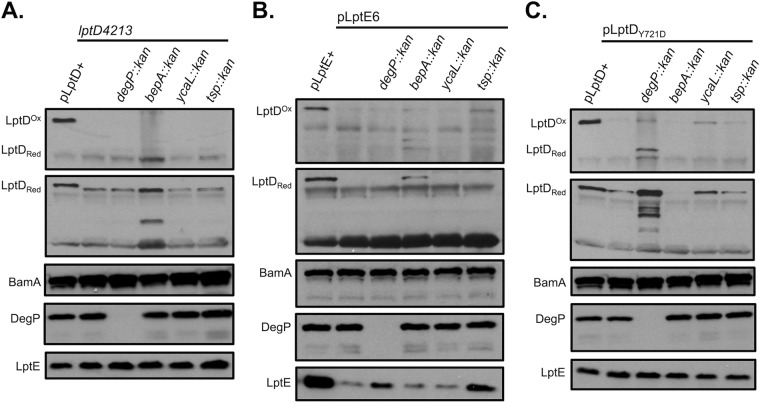
First, we assessed the role of proteases in the degradation of three LptD substrates with mutations known to impair assembly at different steps and to result in decreased LptD protein levels. Specifically, we compared the protease profiles of double mutants containing either the lptD4213, lptE6, or lptDY721D mutation and each of the protease null mutations. The lptD4213 mutation encodes a 23-amino acid deletion in LptD that results in stalled late-stage assembly when the substrate is partially folded around LptE and in contact with the Bam components BamA and BamD (28, 29). Although less extensively characterized, the lptE6 mutation similarly seems to affect later-stage assembly; the 6-bp deletion in lptE affects interactions between LptD and LptE and the assembly of the complex but does not affect assembled complex stability (22). The lptDY721D mutation affects assembly at a step prior to the stage at which LptD4213 is stalled. It does not block recognition by the Bam complex but causes slow folding (J. Lee, H. A. Sutterlin, J. S. Wzorek, C. L. Hagan, M. Grabowicz, T. J. Silhavy, and D. Kahne, submitted for publication).
BepA degrades LptD substrates that are stalled at later steps in the OMP assembly process.
The combination of many protease null mutations with lptD4213, lptE6, and lptDY721D resulted in no discernible restoration of LptD levels or synthetic permeability (Table 1) or temperature sensitivity phenotypes (data not shown). However, the deletion of bepA from the lptD4213 mutant strain resulted in striking restoration of the reduced levels of LptD4213, consistent with the previously described role of this protease in the maturation and degradation of LptD (15) (Fig. 1A). Importantly, bepA deletion did not restore the oxidized mature levels of LptD4213, indicating that the accumulated substrate remains stalled on the Bam complex or is misfolded in the membrane. Accordingly, the deletion of bepA does not improve the permeability phenotypes of the lptD4213 strain, further implying that the substrate that accumulates in the absence of this protease is nonfunctional (Table 1). LptD4213 levels were not restored by the deletion of any other protease, including DegP.
TABLE 1.
Outer membrane permeability and LptD assembly phenotypes of constructed protease mutants
| Genotype | Wild-type background |
lptD4213 background |
lptE6 background |
lptDY721D background |
||||
|---|---|---|---|---|---|---|---|---|
| OM phenotypea | LptD restorationb | OM phenotype | LptD restoration | OM phenotype | LptD restoration | OM phenotype | LptD restoration | |
| MC4100 | ++++ | NA | ++ | − | ++ | − | ++ | − |
| degP::kan | ++++ | NA | + | − | ++ | − | + | +++ |
| degQ::kan | ++++ | NA | ++ | − | ++ | − | ++ | − |
| ompT::kan | ++++ | NA | ++ | − | ++ | − | ++ | − |
| ptrA::kan | ++++ | NA | ++ | − | ++ | − | ++ | − |
| tsp::kan | ++++ | NA | ++ | − | +++ | − | ++ | − |
| ycaL::kan | ++++ | NA | ++ | − | ++ | − | +++ | ++ |
| ydgD::kan | ++++ | NA | ++ | − | ++ | − | ++ | − |
| loiP::kan | ++++ | NA | ++ | − | ++ | − | ++ | − |
| bepA::kan | ++ | NA | + | +++ | ++ | ++ | ++ | − |
| yhiJ::kan | ++++ | NA | ++ | − | ++ | − | ++ | − |
OM phenotype: ++++, growth similar to wild-type growth in the presence of antibiotics; −, no growth.
LptD restoration: NA, not applicable; ++++, restoration of LptD levels close to wild-type levels; −, no restoration of LptD levels.
FIG 1.
LptD is degraded by different proteases at discrete points in the assembly process. Relative OMP levels were determined from whole-cell lysates in wild-type strains, i.e., NR754 (A), GC169 (B), or HC340 (C), and strains containing each of the chromosomal mutations listed, in a lptD4213 (A), lptE6 (B), or lptDY721D (C) background by SDS-PAGE and immunoblotting for the proteins indicated. Red, reduced; Ox, oxidized.
In an effort to corroborate the role of BepA in the degradation of LptD, we then performed the same analyses with lptE6. Indeed, when BepA, but not DegP or other proteases examined, was deleted from the lptE6 strain, we observed a marked increase in the levels of reduced LptD (Fig. 1B). Although the deletion of degP and tsp resulted in an increase in LptE6 levels, neither protease affected the accumulation of LptD, suggesting that BepA acts directly on the LptD OMP substrate. Taken together, these data demonstrate that BepA, but not other periplasmic proteases, degrades LptD substrates that are stalled in later stages of assembly as partial barrels bound to the Bam complex.
The unknown periplasmic protease YcaL degrades an LptDY721D population that is competent to form fully functional complexes.
In stark contrast to the lptD4213 and lptE6 mutants, the deletion of bepA resulted in no increase in reduced LptDY721D levels, suggesting that BepA does not detectably degrade this mutant substrate. Instead, we observed a marked increase in LptDY721D levels when DegP, and to a lesser extent YcaL, was absent (Fig. 1C). The deletion of ycaL did not increase the level of reduced LptDY721D to nearly the extent of a degP-null strain, implying either that YcaL accesses only a fraction of DegP-susceptible substrate or that DegP and YcaL degrade distinct populations of LptDY721D. Surprisingly, the ΔdegP lptDY721D-accumulated substrate did not convert markedly to a fully functional mature complex, indicating that the fraction of LptDY721D normally degraded by DegP is a misfolded, off-pathway species residing either in the periplasm or at the Bam complex. However, a greater proportion of the small fraction of accumulated substrate in the ΔycaL strain appeared to accumulate as properly oxidized, mature LptD machinery, indicating that the substrate degraded by YcaL is still competent for assembly.
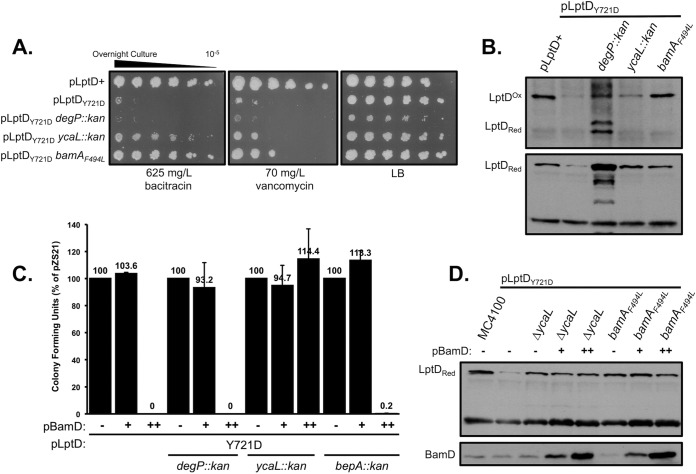
In order to confirm the apparent ameliorating effect of ycaL deletion on the assembly of a properly oxidized fraction of LptD, we examined the OM permeability defects of each double mutant. The lptDY721D mutation confers OM permeability, resulting in sensitivity to both vancomycin and bacitracin. Consistent with the finding that little LptDY721D is assembled as the fully mature form in a degP-null mutant, degP deletion failed to rescue these defects and further reduced the viability of the strain on LB agar (Fig. 2A). The deletion of ycaL, however, partially restored vancomycin and bacitracin resistance in a lptDY721D background, indicating that the accumulated substrate in this background forms a functional LptD/E translocon. The suppression of lptDY721D phenotypes by ΔycaL also appears to be specific, as ycaL deletion did not suppress the vancomycin and bacitracin sensitivities observed in other OM-compromised mutants such as ΔbamB and ΔbamE strains (see Table S2 in the supplemental material). Taken together, the increased levels of oxidized LptD and the decrease in OM permeability observed in a ΔycaL lptDY721D strain indicate that YcaL, a protease of heretofore unknown function, degrades a conformer of LptDY721D stalled at the Bam complex that is competent for assembly if the residence time on the Bam complex is increased.
FIG 2.
Deletion of ycaL improves the viability and LptD levels of a lptDY721D strain. (A) Antibiotic sensitivity was assessed by spotting serial 10-fold dilutions of stationary-phase cultures of strains containing the indicated mutations on LB agar, with or without vancomycin or bacitracin, and incubating the cultures at 37°C. (B) Relative LptD levels were determined in a wild-type strain and strains containing each of the chromosomal mutations listed, in a lptDY721D background, by SDS-PAGE and immunoblotting for the proteins indicated. (C) CFU were determined in control and lptDY721D strains containing each of the mutations listed by transformation with the indicated plasmids, i.e., pZS21 (−), pZS21::bamD (+), or pZS21::bamD O/E (++), and calculation of the resulting surviving colonies. (D) Relative protein levels were determined from whole-cell lysates from a wild-type strain and strains containing each of the chromosomal mutations and plasmids listed, in a lptDY721D background, by SDS-PAGE and immunoblotting for the proteins indicated. Red, reduced; Ox, oxidized.
During our analysis, we noted that the OM permeability suppression phenotypes of the ΔycaL lptDY721D strain strikingly resembled those of a lptDY721D spontaneous gain-of-function suppressor in bamA (Lee et al., submitted). Indeed, ΔycaL restores vancomycin and bacitracin resistance similarly to bamAF494L and increases the assembly of mature LptD, although not to the extent observed in a bamAF494L suppressor background (Fig. 2B). Therefore, we hypothesized that the ycaL-null mutation might similarly suppress defects associated with aberrant LptDY721D biogenesis.
Elsewhere, Lee et al. report that lptDY721D confers a novel synthetic phenotype with the essential Bam lipoprotein BamD. Specifically, ∼10-fold overexpression of BamD is synthetically lethal with the lptDY721D mutation, implying that LptDY721D forms aberrant interactions with BamD that are exacerbated when the Bam component is provided in excess. The bamAF494L mutation suppresses this synthetic lethality and improves LptD biogenesis in the mutant background, further indicating that the lethal interaction between LptD and BamD is alleviated and mutant substrate assembly is enhanced by an alteration in the BamA barrel that facilitates LptDY721D assembly (Lee et al., submitted). Strikingly, ycaL deletion also suppresses the lethality of BamD overexpression in the lptDY721D strain (Fig. 2C). In contrast, despite the fact that degP deletion drastically increases the level of reduced LptDY721D, we observed no suppression of lethality upon BamD overexpression in a ΔdegP lptDY721D background. In either a ΔycaL or bamAF494L background, BamD overexpression does not lead to a decrease in LptDY721D levels, implying that the substrate in a ΔycaL or bamAF494L background is stable and not further degraded by proteases such as DegP (Fig. 2D). The similar suppression observed with ΔycaL and bamAF494L supports the notion that YcaL normally degrades a conformer of LptDY721D that is stalled on the assembly machine but is capable of being assembled by the Bam complex. Furthermore, the striking lack of suppression of BamD overexpression phenotypes in a ΔdegP background implies that YcaL and DegP do not access the same pools of mutant substrate.
Periplasmic protease activity is functionally segregated to distinct steps in the OMP assembly process.
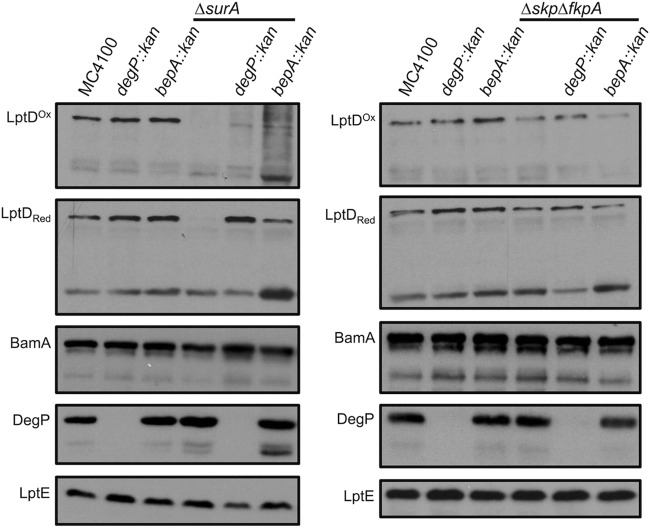
Because we observed a striking difference in the degradation of lptD mutants by BepA, DegP, and YcaL, we sought to confirm the apparent functional segregation of these proteases using mutations that clearly impair LptD biogenesis at the earliest steps of assembly. LptD assembly is known to be dependent on the periplasmic chaperones SurA, Skp, and FkpA (23). The degP deletion confers temperature sensitivity at 37°C in a Δskp ΔfkpA strain, and a ΔdegP ΔsurA strain is viable only at 25°C (9). Therefore, we introduced ycaL-, bepA-, and degP-null mutations into ΔsurA and Δskp ΔfkpA strains and characterized each mutant at the viable temperature (9). In accordance with the well-described role of DegP as the primary protease responsible for degrading misfolded periplasmic proteins, degP deletion resulted in significant accumulation of reduced LptD in a ΔsurA background and a modest increase in LptD levels in a Δskp ΔfkpA mutant background (Fig. 3). In contrast, and consistent with the findings of Weski and Ehrmann (16), bepA deletion did not significantly increase LptD levels in a Δskp ΔfkpA background and resulted in only a slight increase in LptD levels in a ΔsurA background. These results further underscore the functional segregation between DegP, which degrades substrates that undergo aberrant assembly in the periplasm or during initial contact with the Bam complex, and BepA, which degrades substrates that stall at later points in assembly, when the substrate is partially folded and engaged with the Bam complex.
FIG 3.
DegP and BepA have functionally segregated roles in degrading LptD. Relative OMP levels were determined from whole-cell lysates from a wild-type strain and strains containing each of the chromosomal mutations listed, in a ΔsurA or Δskp ΔfkpA background, by SDS-PAGE and immunoblotting for the proteins indicated. The ΔsurA ΔdegP strain and all control strains were grown at 25°C, while the Δskp ΔfkpA ΔdegP strain and all control strains were grown at 30°C. Red, reduced; Ox, oxidized.
Interestingly, these results also corroborated the fact that SurA is crucial for LptD to reach the Bam complex and become fully assembled. In a ΔsurA degP-null mutant, despite the fact that abundant LptD exists in the reduced form, very little of the substrate matures to oxidized LptD (Fig. 3). In a Δskp ΔfkpA degP-null strain, however, the accumulated LptD species readily converts to the fully oxidized form. Taken together, these results imply that SurA is necessary to direct a large fraction of LptD to the Bam complex and Skp and FkpA play a different redundant role in substrate assembly.
DISCUSSION
Our work here provides evidence that three envelope proteases, DegP, YcaL and BepA, are functionally segregated by the step at which they degrade stalled mutant LptD substrate. In this way, we reason that the three different proteases recognize defective substrates at distinct points in assembly to prevent assembly machine inhibition or membrane integration of defective OMPs. Furthermore, due to our observation that ycaL deletion suppresses rather than exacerbates the phenotypes of the lptDY721D mutant, we propose that these proteases serve separate roles in OMP biogenesis. Consistent with previous descriptions, DegP degrades misfolded OMPs within the periplasmic space that have fallen off the pathway and are no longer competent for assembly. However, BepA degrades OMPs at a different step in assembly, acting against substrates like LptD4213 that have progressed far enough in the assembly pathway to become rudimentary barrels. In this way, BepA may act as the “protease of last resort” for defective OMPs. In stark contrast to DegP and BepA, the previously unknown protease YcaL uniquely degrades substrates like LptDY721D that are stalled at an earlier step in the assembly process and are inaccessible to DegP and BepA (Fig. 4).
FIG 4.
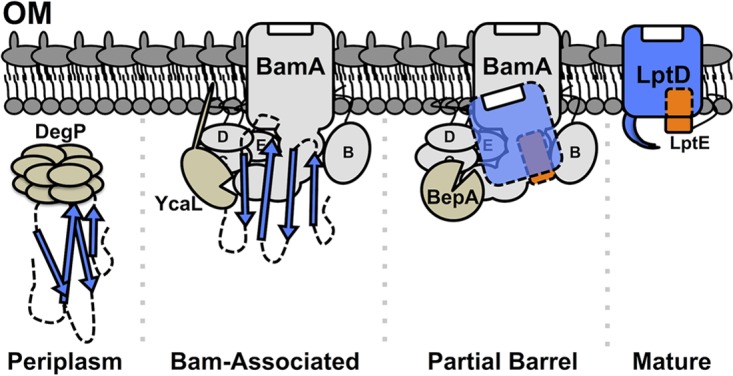
Model for the functional separation of DegP, YcaL, and BepA. DegP degrades LptD that misfolds in the periplasm, while YcaL degrades substrate that is engaged with the Bam complex but has not yet formed a partial barrel (Bam-associated). BepA degrades LptD substrate that has formed a partial barrel around LptE and is still bound to the Bam complex (partial barrel). D, E, and B, BamD, BamE, and BamB, respectively.
It is noteworthy that DegP and BepA appear to be unable to interact with substrates outside their functional spectra. For example, we see no evidence that DegP accesses and degrades LptD4213 or that BepA is able to access LptDY721D. The results from our investigation of LptE6 are consistent with this view. Wild-type LptE resides within and is protected by the LptD barrel (22). The lptE6 mutation interferes with LptD/E assembly, leaving a fraction of the mutant LptE accessible to DegP and Tsp, but this mutant protein is not a substrate for BepA or YcaL. We suggest that protease access to stalled substrates may be limited by association of the substrate or protease with the Bam complex or the OM. We favor a model in which DegP recognizes mostly unfolded, periplasmic substrates, while BepA and YcaL recognize distinct intermediates that are bound to the Bam complex (Fig. 4). This model is corroborated by the finding that BepA has genetic and physical interactions with the Bam complex (15) and YcaL is a predicted OM lipoprotein (16, 30).
How BepA accesses OMP substrates is not clear. BepA is a soluble periplasmic protein reported to have associations with the OM lipoprotein LoiP and the Bam complex (15, 31). During our characterization of BepA, we observed no restoration of LptD4213 levels in a ΔloiP mutant as we did with a ΔbepA mutant, indicating that BepA activity is not mediated through its association with LoiP (Table 1). However, it is possible that BepA reversibly associates with the Bam complex depending on the folding state and identity of the substrate being assembled. We do note that our finding that a soluble periplasmic protein (BepA) accesses a substrate that is an open barrel (LptD4213) supports the OMP assembly model, in which folding begins in the periplasm at the membrane interface before insertion (29).
Here, we report the first known function for the protease YcaL. Specifically, a ycaL deletion, like the suppressor mutation bamAF494L, rescues the unique synthetic lethality caused by BamD overexpression in a lptDY721D strain. However, a degP deletion, despite increasing the levels of reduced LptD significantly, does not rescue this BamD-mediated lethality. If excess BamD sequesters LptDY721D, then it stands to reason that YcaL must be degrading a subpopulation of LptDY721D that is present on a productive Bam complex and, given enough time, can be assembled properly.
The predicted lipoprotein YcaL is a member of the same M48 metalloprotease family that contains LoiP and BepA (16, 30). Like BepA, YcaL contains a well-defined zinc metalloprotease active site motif. YcaL has been reported to be essential in Salmonella enterica but does not contribute to the viability or membrane integrity of an otherwise wild-type E. coli strain (16) (Table 1). We propose that YcaL, like BepA and DegP, monitors OMP assembly and degrades substrates that exhibit impaired assembly. However, it is noteworthy that ycaL deletion uniquely suppresses many lptDY721D mutant phenotypes. Although we have observed this suppression of lptD mutant phenotypes only in this one mutant background, it is possible that YcaL also recognizes other substrates that are stalled at an early step in assembly and are still competent to fold into mature β-barrels. In this way, YcaL may act to increase the efficiency of OMP assembly by “cleaning” delayed OMPs from the Bam complex so that other substrates can be assembled properly.
Our work here contributes to an expanded integrated description of the multifaceted process of OMP assembly. It does not rule out further layers of complexity currently obfuscated by the multiple redundant features of the OMP quality control network (16, 23). While it is possible that the distinct activities of the three proteases described here are required only for OMPs such as LptD, the largest β-barrel and likely the most complex OMP in the E. coli OM, we are currently exploring whether this paradigm is applicable to the assembly of other OMPs.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Strains and plasmids used in this work are described in Table S1 in the supplemental material. All strains were constructed using standard microbiological techniques (32). When necessary, media were supplemented with 125 mg/liter ampicillin, 25 mg/liter kanamycin, 20 mg/liter chloramphenicol, or 25 mg/liter tetracycline. Unless otherwise noted, all bacterial cultures were grown under aerobic conditions at 37°C.
Quantification of small molecule and antibiotic sensitivity.
Efficiency-of-plating (EOP) assays were performed by growing overnight cultures in the appropriate medium and serially diluting the overnight cultures 10-fold in 200-μl volumes in a 96-well plate. The volumes were then spotted onto LB agar plates containing the indicated antibiotics, using a 48-pin replicator. Disc diffusion assays were conducted as follows. A 100-μl sample of an overnight culture was mixed with 3 ml of cooled molten LB “top” agar and poured over an LB agar plate. Once the agar solidified, sterile 6-mm filter discs (BBL) infused with antibiotic were placed, evenly spaced, on the top agar. Plates were incubated at the required temperature overnight, and the diameter of each zone of inhibition was measured (in millimeters) at multiple angles. All results presented are representative of at least three independent experiments.
Western blot analysis.
Culture samples (250 μl) were pelleted (13,000 × g for 1 min) and resuspended in sample buffer at a volume equal to the optical density at 600 nm (OD600)/60. Oxidized samples were resuspended in the same volume of sample buffer without β-ME. Samples were boiled for 10 min and subjected to electrophoresis through 10% SDS-PAGE gels. Immunoblotting was performed using rabbit polyclonal antisera that recognized LamB/OmpA (1:30,000 dilution), LptD (1:5,000 dilution), BamA (1:30,000 dilution), DegP (1:30,000 dilution), or SurA (1:8,000 dilution). Donkey anti-rabbit IgG secondary antibody conjugated to horseradish peroxidase (GE Healthcare) was used at a 1:8,000 dilution for all immunoblots. Immunoblots were visualized using the ECL antibody detection kit (Amersham) and HyBlot CL film (Denville Scientific). All results presented are representative of at least three independent experiments.
BamD overexpression assay.
Overnight cultures of the strains used were inoculated 1:500 in fresh LB medium (normalized by their OD600 values) and grown at the indicated temperature until the OD600 was ∼1.0. Samples (500 μl) from each culture were taken and normalized again by their OD600 values before being mixed with ice-cold 2× transformation and storage solution (TSS) (33) and then incubated on ice for 15 min. Samples of each strain (100 μl) were then transformed with the appropriate vector (normalized by absorbance at 260 nm) and supplemented with 1 ml of LB medium before recovery at the indicated temperature. Samples (100 μl) from the recovered culture and a 1:10 dilution of the recovered culture were plated on LB agar plates containing 25 mg/liter kanamycin to select for the bamD plasmids. After overnight growth at the indicated temperature, CFU were counted on each plate and compared to values for control-plasmid-transformed parent strains.
Supplementary Material
ACKNOWLEDGMENTS
We thank the Silhavy laboratory for critical reading of the manuscript.
This work was supported by National Institute of General Medical Sciences grants GM034821 and GM118024 (awarded to T.J.S.) and the National Science Foundation Graduate Research Fellowship Program under grant DGE1148900 (to H.A.S.).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00418-17.
REFERENCES
- 1.Silhavy TJ, Kahne D, Walker S. 2010. The bacterial cell envelope. Cold Spring Harb Perspect Biol 2:a000414. doi: 10.1101/cshperspect.a000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nikaido H. 2003. Molecular basis of bacterial outer membrane permeability revisited. Microbiol Mol Biol Rev 67:593–656. doi: 10.1128/MMBR.67.4.593-656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ruiz N, Kahne D, Silhavy TJ. 2009. Transport of lipopolysaccharide across the cell envelope: the long road of discovery. Nat Rev Microbiol 7:677–683. doi: 10.1038/nrmicro2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ricci DP, Silhavy TJ. 2012. The Bam machine: a molecular cooper. Biochim Biophys Acta 1818:1067–1084. doi: 10.1016/j.bbamem.2011.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Merdanovic M, Clausen T, Kaiser M, Huber R, Ehrmann M. 2011. Protein quality control in the bacterial periplasm. Annu Rev Microbiol 65:149–168. doi: 10.1146/annurev-micro-090110-102925. [DOI] [PubMed] [Google Scholar]
- 6.Mecsas J, Rouviere PE, Erickson JW, Donohue TJ, Gross CA. 1993. The activity of sigma E, an Escherichia coli heat-inducible sigma-factor, is modulated by expression of outer membrane proteins. Genes Dev 7:2618–2628. doi: 10.1101/gad.7.12b.2618. [DOI] [PubMed] [Google Scholar]
- 7.Sklar JG, Wu T, Kahne D, Silhavy TJ. 2007. Defining the roles of the periplasmic chaperones SurA, Skp, and DegP in Escherichia coli. Genes Dev 21:2473–2484. doi: 10.1101/gad.1581007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lazar SW, Kolter R. 1996. SurA assists the folding of Escherichia coli outer membrane proteins. J Bacteriol 178:1770–1773. doi: 10.1128/jb.178.6.1770-1773.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rizzitello AE, Harper JR, Silhavy TJ. 2001. Genetic evidence for parallel pathways of chaperone activity in the periplasm of Escherichia coli. J Bacteriol 183:6794–6800. doi: 10.1128/JB.183.23.6794-6800.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Strauch KL, Johnson K, Beckwith J. 1989. Characterization of depP, a gene required for proteolysis in the cell envelope and essential for growth at high temperature. J Bacteriol 171:2689–2696. doi: 10.1128/jb.171.5.2689-2696.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Costello SM, Plummer AM, Fleming PJ, Fleming KG. 2016. Dynamic periplasmic chaperone reservoir facilitates biogenesis of outer membrane proteins. Proc Natl Acad Sci U S A 113:E4794–E4800. doi: 10.1073/pnas.1601002113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rhodius VA, Suh WC, Nonaka G, West J, Gross CA. 2006. Conserved and variable functions of the σE stress response in related genomes. PLoS Biol 4:e2. doi: 10.1371/journal.pbio.0040002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krojer T, Garrido-Franco M, Huber R, Ehrmann M, Clausen T. 2002. Crystal structure of DegP (HtrA) reveals a new protease-chaperone machine. Nature 416:455–459. doi: 10.1038/416455a. [DOI] [PubMed] [Google Scholar]
- 14.Clausen T, Kaiser M, Huber R, Ehrmann M. 2011. HTRA proteases: regulated proteolysis in protein quality control. Nat Rev Mol Cell Biol 12:152–162. doi: 10.1038/nrm3065. [DOI] [PubMed] [Google Scholar]
- 15.Narita S, Masui C, Suzuki T, Dohmae N, Akiyama Y. 2013. Protease homolog BepA (YfgC) promotes assembly and degradation of β-barrel membrane proteins in Escherichia coli. Proc Natl Acad Sci U S A 110:E3612–E3621. doi: 10.1073/pnas.1312012110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weski J, Ehrmann M. 2012. Genetic analysis of 15 protein folding factors and proteases of the Escherichia coli cell envelope. J Bacteriol 194:3225–3233. doi: 10.1128/JB.00221-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu T, McCandlish AC, Gronenberg LS, Chng S-S, Silhavy TJ, Kahne D. 2006. Identification of a protein complex that assembles lipopolysaccharide in the outer membrane of Escherichia coli. Proc Natl Acad Sci U S A 103:11754–11759. doi: 10.1073/pnas.0604744103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Braun M, Silhavy TJ. 2002. Imp/OstA is required for cell envelope biogenesis in Escherichia coli. Mol Microbiol 45:1289–1302. doi: 10.1046/j.1365-2958.2002.03091.x. [DOI] [PubMed] [Google Scholar]
- 19.Sampson BA, Misra R, Benson SA. 1989. Identification and characterization of a new gene of Escherichia coli K-12 involved in outer membrane permeability. Genetics 122:491–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dong H, Xiang Q, Gu Y, Wang Z, Paterson NG, Stansfeld PJ, He C, Zhang Y, Wang W, Dong C. 2014. Structural basis for outer membrane lipopolysaccharide insertion. Nature 511:52–56. doi: 10.1038/nature13464. [DOI] [PubMed] [Google Scholar]
- 21.Qiao S, Luo Q, Zhao Y, Zhang XC, Huang Y. 2014. Structural basis for lipopolysaccharide insertion in the bacterial outer membrane. Nature 511:108–111. doi: 10.1038/nature13484. [DOI] [PubMed] [Google Scholar]
- 22.Chimalakonda G, Ruiz N, Chng S-S, Garner RA, Kahne D, Silhavy TJ. 2011. Lipoprotein LptE is required for the assembly of LptD by the β-barrel assembly machine in the outer membrane of Escherichia coli. Proc Natl Acad Sci U S A 108:2492–2497. doi: 10.1073/pnas.1019089108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schwalm J, Mahoney TF, Soltes GR, Silhavy TJ. 2013. A role for Skp in LptD assembly in Escherichia coli. J Bacteriol 195:3734–3742. doi: 10.1128/JB.00431-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ruiz N, Chng S-S, Hiniker A, Kahne D, Silhavy TJ. 2010. Nonconsecutive disulfide bond formation in an essential integral outer membrane protein. Proc Natl Acad Sci U S A 107:12245–12250. doi: 10.1073/pnas.1007319107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chng S-S, Xue M, Garner RA, Kadokura H, Boyd D, Beckwith J, Kahne D. 2012. Disulfide rearrangement triggered by translocon assembly controls lipopolysaccharide export. Science 337:1665–1668. doi: 10.1126/science.1227215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ehrmann M. 2007. The periplasm. ASM Press, Washington, DC. [Google Scholar]
- 27.Chng S-S, Ruiz N, Chimalakonda G, Silhavy TJ, Kahne D. 2010. Characterization of the two-protein complex in Escherichia coli responsible for lipopolysaccharide assembly at the outer membrane. Proc Natl Acad Sci U S A 107:5363–5368. doi: 10.1073/pnas.0912872107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ruiz N, Falcone B, Kahne D, Silhavy TJ. 2005. Chemical conditionality: a genetic strategy to probe organelle assembly. Cell 121:307–317. doi: 10.1016/j.cell.2005.02.014. [DOI] [PubMed] [Google Scholar]
- 29.Lee J, Xue M, Wzorek JS, Wu T, Grabowicz M, Gronenberg LS, Sutterlin HA, Davis RM, Ruiz N, Silhavy TJ, Kahne DE. 2016. Characterization of a stalled complex on the β-barrel assembly machine. Proc Natl Acad Sci U S A 113:8717–8722. doi: 10.1073/pnas.1604100113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Finn RD, Coggill P, Eberhardt RY, Eddy SR, Mistry J, Mitchell AL, Potter SC, Punta M, Qureshi M, Sangrador-Vegas A, Salazar GA, Tate J, Bateman A. 2016. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res 44:D279–D285. doi: 10.1093/nar/gkv1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lütticke C, Hauske P, Lewandrowski U, Sickmann A, Kaiser M, Ehrmann M. 2012. E. coli LoiP (YggG), a metalloprotease hydrolyzing Phe-Phe bonds. Mol Biosyst 8:1775–1782. doi: 10.1039/c2mb05506f. [DOI] [PubMed] [Google Scholar]
- 32.Silhavy TJ, Berman ML, Enquist L. 1984. Experiments with gene fusions. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. [Google Scholar]
- 33.Chung CT, Niemela SL, Miller RH. 1989. One-step preparation of competent Escherichia coli: transformation and storage of bacterial cells in the same solution. Proc Natl Acad Sci U S A 86:2172–2175. doi: 10.1073/pnas.86.7.2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.