Abstract
Systems metabolic engineering is a multidisciplinary area that integrates systems biology, synthetic biology and evolutionary engineering. It is an efficient approach for strain improvement and process optimization, and has been successfully applied in the microbial production of various chemicals including amino acids. In this review, systems metabolic engineering strategies including pathway-focused approaches, systems biology-based approaches, evolutionary approaches and their applications in two major amino acid producing microorganisms: Corynebacterium glutamicum and Escherichia coli, are summarized.
Keywords: Systems metabolic engineering, Amino acid, Corynebacterium glutamicum, Escherichia coli
1. Introduction
Systems metabolic engineering is an emerging discipline that combines the concepts of systems biology, synthetic biology and evolutionary engineering [1], as defined by Lee [2], it involves the application of omics data and the utilization of omics data for synthetic biology and evolutionary engineering for strain breeding and process improvement. With the development of high-throughput technologies, computational methods and simulation approaches, systems biology has become much more mature and applicable, and has already manifested its giant potential in providing genome-wide information and clues for synthetic biology and evolutionary engineering. The various combinations of systems biology, synthetic biology and evolutionary biology have been successfully applied for metabolic engineering of industrial strains [3], [4], [5]. Undoubtedly, systems metabolic engineering could dig out the maximum potentials of microbial cell factories.
The microbial production of amino acids is a large area where systems metabolic engineering strategies have been successfully applied, mainly in two important producing microorganisms: Corynebacterium glutamicum and Escherichia coli. Since the first discovery of the l-glutamate producing strain C. glutamicum in 1957, strain breeding has become a fierce competing spot of leading amino acid manufacturing enterprises with the expanding market demand for amino acids. l-Glutamate is the major bulk amino acid, which covers nearly two thirds of the amino acid market. The market demand of l-lysine ranks just next to l-glutamate, with an current annual production of over 2200000 tons [6]. Various strain breeding approaches have been developed, whilst genetically defined metabolic strategies have gradually taken the place of the conventional random mutagenesis-selection method and become the mainstream. While local metabolic engineering of microorganisms that focuses on the engineering of one or a few specific genes or metabolic pathways generally has the limitation of being not able to take the whole metabolic process into consideration. Systems metabolic engineering tries to overcome this limitation by combined approaches to obtain rationally designed strains.
In this review, systems metabolic engineering strategies and applications for amino acid producing strain improvement are summarized, mainly focusing on the two major industrial production microorganisms: C. glutamicum and E. coli.
2. Strategies for systems metabolic engineering of microorganisms for amino acids production
According to Lee et al. [7], strategies for systems metabolic engineering could be categorized into two groups, the rational intuitive approaches and the systematic and rational-random approaches. The former group covers the typical metabolic engineering process of the synthetic pathway of a certain product, that from the uptake of carbon source, elimination of byproducts, enrichment of precursors, to the reconstruction of related metabolic pathways, supply of cofactor, and so on [7], when the target genes to be engineered are obvious. The latter group mainly includes omics-based metabolic engineering techniques and various evolution approaches when no obvious target genes are known. Applications of systems metabolic engineering of microorganisms for amino acid production have been increasing, and representative examples are shown in Table 1. In this review, we classified the systems metabolic strategies for amino acid high-producing strains into three categories as illustrated in Fig. 1, which are summarized below.
Table 1.
Representative examples of the applications of systems metabolic engineering strategies for amino acids production.
| Strategy | Detailed method | Effect | microorganism | Product | Reference | |
|---|---|---|---|---|---|---|
| Pathway-focused approaches | Carbon source utilization engineering | Combined overexpression of iolT1 or iolT2 with ppgK | Non-PTS replacing the PTS for efficient PEP supply | C. glutamicum | l-lysine | [8] |
| Combined overexpression of heterogenous xylose isomerase and homogenous xylulokinase | Improved xylose utilization for accelerated production of amino acids | C. glutamicum |
l-lysine l-glutamate l-ornithine |
[17] | ||
| Precursor enrichment and byproduct elimination | ΔthrB, ΔmcbR/(pJYW-4-homm-lysCm) | Increased precursor supply | C. glutamicum | l-methionine | [20] | |
| Δddh, ΔlysE | Reduced l-lysine production with enhanced l-threonine production | C. glutamicum |
l-threonine l-isoleucine |
[23] | ||
| Transport engineering | Overexpression of brnFE, ΔbrnQ | Increased production of branched chain amino acids and l-methionine | C. glutamicum | Branched chain amino acids and l-methionine | [20], [31], [32], [33] | |
| Cofactor engineering | Mutation in gapA to alter the coenzyme specificity of a native NAD-dependent glyceraldehyde 3-phosphate dehydrogenase (GAPDH) to NADP | Improved production of l-lysine | C. glutamicum | l-lysine | [38] | |
| Systems biology-based approaches | Omics-based approach | Combined analysis of transcriptome, metabolome, and fluxome | Providing important information on the different phases of cell growth and lysine production | C. glutamicum | l-lysine | [41] |
| Metabolic engineering based on transcriptome analysis | Find the transporter system as the engineering target | E. coli | l-valine | [39] | ||
| In silico simulation | Flux response analysis, Δacs | Reduced acetic acid production | E. coli | l-threonine | [40] | |
| Evolutionary approaches | Biosensor-based evolution | The use of an l-valine responsive sensor based on Lrp | Increased l-valine titers (25%) and a 3–4-fold reduction of by-product formation | C. glutamicum | l-valine | [54] |
Fig. 1.
The constitution and strategies of systems metabolic engineering.
2.1. Pathway-focused approaches
Pathway-focused approaches usually aim to increase the production ability of certain products by combining local metabolic engineering methods, such as enhancing carbon source utilization and key enzyme expression, removing feedback inhibition and transcriptional attenuation, and blocking bypass pathway etc. A lot of endeavors have been made in the pathway-focused engineering of microorganisms for amino acid production.
2.1.1. Carbon source utilization engineering
The carbon source uptake and utilization process is the first crucial step for the production of amino acids. By enhancing the uptake and utilization of carbon sources, more carbon flux could be provided for the synthesis of amino acids. Generally, there are two types of carbon source transport systems, the phosphotransferase system (PTS) and non-phosphotransferase system. The PTS requires phosphoenolpyruvate (PEP) for the phosphorylation of carbon sources, which is usually an important intermediate for the synthesis of certain amino acids. In that situation, the replacement of the PTS with non-PTS could save more PEP for the following step of amino acid synthesis [8], [9]. Other ways of increasing the intracellular PEP have been tried. For example, Tatarko et al. [10] disrupted a global regulatory gene csrA encoding Csr (carbon storage regulator) to increase the gluconeogenesis and decrease the glycolysis, which elevated intracellular PEP for the synthesis of phenylalanine.
The enhancement of the expression of ptsG, encoding the glucose-specific EII permease of the PTS, could increase the utilization of glucose in C. glutamicum and E. coli. Except for the direct gene manipulation, the expression of ptsG could also be affected by the existence of other carbon sources. For example, the existence of acetate could reduce the expression of ptsG to 45% by the SugR-mediated repression of ptsG, while, the addition of maltose could increase the ptsG expression by counteracting the SugR-mediated repression [11] in the presence of acetate. It has been reported that the addition of maltose increased the glucose utilization of a pyruvate dehydrogenase complex-deficient C. glutamicum strain, and thus, improved its l-valine productivity [12]. Recently, Henrich et al. [13] found out that maltose uptake by the novel ABC transporter system MusEFGK2I was the reason that caused increased expression of ptsG in C. glutamicum.
Besides, to cope with the food crisis all over the world, the utilization of cellulose and hemicellulose derived sugars, such as xylose and arabinose, for the production of amino acids, has become more and more urgent. E. coli can grow efficiently on a wide range of carbon substrates including various pentose such as xylose, mannose, arabinose etc [14]. Simultaneous uptake of lignocellulose-based monosaccharides in E. coli has been reported [15]. In nature, C. glutamicum is not capable of utilizing xylose as a carbon source, due to its lack of xylose isomerase gene for xylose metabolism [16]. Considering the importance of C. glutamicum in the industrial area, xylose utilization has drew many researchers' attentions. By introducing heterogenous xylose isomerase into C. glutamicum, amino acids and their derivatives could be produced with xylose as carbon source [17], [18], [19]. While the transport system in C. glutamicum for xylose uptake still needs intensive investigation, which is crucial for the further improvement of xylose utilization.
2.1.2. Precursor enrichment, byproduct elimination and product degradation blocking
The biosynthetic process of a certain amino acid usually comprises several enzymes that perform divided functions to fulfill the conversion of carbon source to the desired amino acid product. The most commonly used metabolic engineering strategy is to enhance the expression of the key enzymes to obtain the maximum precursor enrichment [20], whilst, eliminate unnecessary byproduct formation by blocking or attenuating the competing pathways [21], [22], [23], and cutting off the further degradation of the desired amino acid. Elimination of feedback inhibition of the key enzyme in a metabolic pathway is frequently the first and most important step for the development of a high-producing strain [24], [25]. Usually, feedback inhibition resistant enzymes could be obtained by introducing site-directed mutations [26]. Besides, DNA-binding transcriptional regulation is another important approach to make use of to achieve precursor enrichment. Transcriptional repression could be removed by deleting the transcriptional repressor [27] or by changing the DNA binding motif. As byproduct elimination is usually achieved by deleting competing genes or interfering relative gene expressions, which sometimes could cause adverse effects to the normal cell growth and metabolism, thus the major challenge in applying this strategy in strain breeding is how to balance the cell growth, metabolism and the synthesis of the desired product.
2.1.3. Transporter engineering
The final titer of a desired amino acid product not only relies on its intracellular synthesis, but is also determined by the efficiency of the transporter system. With the discovery of various transporters for amino acids [28], such as BrnFE for the export of branched chain amino acids and l-methionine [20], ThrE for the export of l-threonine [29], LysE for the export of l-lysine and l-arginine [30] etc., transporter engineering has been increasingly used to obtain higher titer, yield or productivity. Currently, transporter engineering is conducted mainly by enhancing the excretion of the desired amino acid, and simultaneously blocking the inverse import of the excreted product. For example, branched-chain amino acids production could be increased by overexpressing the global regulator Lrp and the two-component export system BrnFE [27], [31], [32], and deleting the import carrier BrnQ [33].
2.1.4. Cofactor engineering
Cofactors are required in many biochemical reactions inside microbial cells [34], of which, NADPH and NADH are the main reducing powers for microbes [35], [36]. Their recycle and respective equilibration are vital for cell growth and intracellular metabolism. Different NADPH or NADH generating methods have been developed [35], [36], [37]. Bommareddy et al. [38] constructed a de novo NADPH generation pathway by altering the coenzyme specificity of a native NAD-dependent glyceraldehyde 3-phosphate dehydrogenase (GAPDH) to NADP, as a result of which, additional NADPH supply through the glycolytic pathway was obtained. They systematically manipulated the coenzyme specificity of GAPDH by rational protein design and evaluated the resulting l-lysine production. The results suggested that sufficient NADPH supply led to increased l-lysine production, with the highest increase approaching 60%, highlighting the importance of cofactor engineering for the overproduction of desired amino acids.
2.2. Systems biology-based approaches
As our understanding of microbial metabolism is limited and sometimes biased, pathway-focused engineering doesn't always work well. Systems biology-based approaches and evolutionary approaches on the other hand could provide more comprehensive views, the conduct of which could efficiently target the crucial genes for further metabolic engineering, and overcome the hidden bottlenecks for strain improvement.
2.2.1. Omics-based approaches
Systems biology is a fast growing discipline that has enormously broaden our understanding of the overall living world from different dimensions of views. The application of systems biology, i.e. omics-based approaches, have greatly accelerated the metabolic engineering of important industrial strains, including amino acids overproducers [39], [40]. During the construction processes of amino acid high-producing strains, bottlenecks are often inevitably encountered after basic rational engineering. The bottlenecks are largely due to the complex metabolic pathways and the uncertain perturbation consequences caused by the artificial engineering. For example, the enhancement of the synthesis pathway of a certain product could probably result in the imbalance of reducing power, energy, or carbon flux distribution [39] etc., which could further lead to undesirable cell growth or product yield. Therefore, accurately evaluation of the consequences of basic pathway focused engineering from an overall view is crucial to the metabolic engineering of industrial strains. Omics-based approaches, including genomics, transcriptomics, proteomics, metabolomics, fluxomics, and especially their integration [41], [42], could help to solve the bottleneck problems by providing comprehensive information on the intracellular metabolic processes for the further metabolic engineering. For example, comparative genomics analysis of an engineered lysine-producing strain and its parental strain identified beneficial point mutations that were further used to construct a high lysine-producing strain [43].
2.2.2. In silico simulation
In silico simulation has been playing an increasingly important role in the systems metabolic engineering of microorganisms [44], the performing of which is usually based on genome-scale information, metabolic reactions, literature information and experimental data [45]. The aim of in silico simulation is to understand cellular metabolic networks and predict the metabolic capability of cells in a particular condition. So far, various in silico algorithms have been developed, such as flux balance analysis (FBA) [46] and regulatory on/off minimization (ROOM) [47]. Choosing an appropriate algorithm is crucial to the successful conduct of in silico simulation, and without doubt the algorithm should be selected according to the aim of the simulation. Examples of different algorithms for in silico simulation are shown in Table 2. For example, if the aim is to predict the metabolic status after a certain gene knockout, the algorithm of MOMA, ROOM, or Flux-sum etc., could be used.
Table 2.
| Purpose of simulation | Algorithm | Objective |
|---|---|---|
| To accurately describe cellular physiology | OMNI | Identifies a set of bottleneck reactions to be removed in the model, to minimize the disagreement between the model predictions and experimental data |
| SR-FBA | Predicts gene expression and metabolic fluxes | |
| TMFA | Predicts intracellular flux distribution with thermodynamic constraints | |
| To predict metabolic capability after genetic perturbation | MOMA | Minimizes the Euclidian distance from a wild type flux distribution under knock-out condition |
| ROOM | Minimizes the number of significant flux changes in the knock-out mutant compared to the wild type | |
| OptKnock | Predicts gene knock-out targets through bilevel optimization framework | |
| OptGene | Predicts gene knock-out targets using genetic algorithm and constraints-based flux analysis | |
| OptReg | Determines the activation/inhibition and elimination reaction set for biochemical production |
In silico simulation has successfully aided the rational engineering of amino acid overproducers. For example, in the systems metabolic engineering of the l-valine overproducing strain performed by Park et al. [39], through in silico gene knockout simulation three important genes were identified as the knockout targets, which further increased the yield of l-valine. In another case, to solve the acetic acid production problem, in silico flux response analysis was performed by Lee et al. [40] to examine the flux that can most effectively reduce the acetic acid production. Besides, the response of the acetic acid production rate to the varying individual flux of central metabolic pathway was evaluated. After these analyses, acetic acid production was finally reduced by amplifying the acs gene encoding acetyl-CoA synthetase. With the fast development of computational science, more accurate algorithms are under development, which will promote the advance of in silico simulation. Hopefully, it will exert more contributions for systems metabolic engineering.
2.3. Evolutionary approaches
With the fast development of synthetic biology, various cellular biosensors have been designed to monitor and control microbial behaviors. As defined by Jay Keasling [49], ‘cellular biosensors’ are made by host cells that produce signals and can be recognized by the host cells to control their behavior or the behavior of heterologous pathways.
Recently, fluorescence-activated cell sorting (FACS) technique has been developed and applied in strain development for various products including amino acids [50]. By using optical biosensors that respond to specific metabolites in single cell by emitting fluorescence, cell with desired performance, such as quick synthesis of precursors, efficient product accumulation etc., could be efficiently screened or selected from huge amounts of mutations, making the engineering and optimization of metabolic pathways more efficient [51]. This technique has manifested its giant potential for strain improvement and process optimization in industry, especially its combination with evolutionary metabolic engineering [52], [53], [54], which comprises enzyme evolution, metabolic evolution, and adaptive evolution. Thus, the combined application of FACS and biosensor could greatly facilitate the conduct of systems metabolic engineering for industrial strains and processes.
Transcriptional regulator-based biosensor is the major type that has been successfully used in screening or selection of strains with good performance [55]. Currently, two amino acid-responsive transcriptional regulators, Lrp and LysG, have been successfully utilized in C. glutamicum to develop genetically-encoded biosensors (Fig. 2). Mustafi et al. [56] developed a biosensor based on Lrp for the detection of intracellular l-methionine and branched-chain amino acids (l-leucine, l-isoleucine, and l-valine) in C. glutamicum. As shown in Fig. 2A, Lrp could activate the expression of the brnFE operon with increased levels of branched-chain amino acids or l-methionine as effectors. In this way, increased intracellular concentration of branched-chain amino acids or l-methionine could be translated into fluorescence signal outputs. Based on this biosensor device, Mahr et al. [53] performed an adaptive laboratory evolution of an l-valine producing C. glutamicum strain, which led to a 25% increase in l-valine titer and 3–4-fold reduction of byproduct formation. The l-lysine biosensor based on LysG (Fig. 2B) was used to screen variants of murE-encoded UDP-N-acetylmuramoy-L-alanyl-d-glutamate: meso-diaminopimelate ligase for enhanced lysine production [50].
Fig. 2.
Transcriptional regulator-based biosensor construction in C. glutamicum. (A) Lrp-based biosensor for l-methionine and branched-chain amino acid production [47]; (B) LysG-based biosensor for l-lysine production [41]. BrnFE and LysE are the exporter of l-methionine & branched chain amino acids, and l-lysine, respectively. Lrp could activate the expression of the brnFE operon in the presence of increased levels of l-methionine or branched chain amino acids; LysG could activate the expression of the lysE operon in the presence of increased level of l-lysine.
3. Case studies of systems metabolic engineering in C. glutamicum
Since its discovery in 1957, C. glutamicum, a GRAS (generally recognized as safe) organism, has been playing a critical role in the industrial production of amino acids, organic acids, nucleosides and related derivatives. It has evolved to be a workhorse for industrial biotechnology, and is qualified to be a good chasis microorganism in synthetic biology. C. glutamicum is notably well-known for its potent amino acid producing capability, and the products range from the bulk amino acids: l-glutamate, l-lysine, to the high value-added amino acids, such as branched chain amino acids, making C. glutamicum one of the best characterized microorganisms [57]. The strain improvement in titer, yield and productivity has become the competing focus of leading amino acids producing companies, and has accelerated the application of systems metabolic engineering in this area. Representative case studies of systems metabolic engineering in C. glutamicum for amino acids production will be reviewed below.
3.1. l-glutamate
l-glutamate is mainly produced by fermentation of C. glutamicum. It is interesting that the wild-type C. glutamicum secretes little l-glutamate, while, under certain treatments [58], such as the suboptimal supply of biotin, the addition of penicillin or detergents etc., the non-producing strains could become efficient cell factories for l-glutamate production. For several decades, researchers have been trying to explore the mechanism of l-glutamate secretion. Current findings are correlated with the change of intracellular metabolism [59] and the structural and functional variation of cell envelope [60], [61]. Without the triggering effects, almost all 2-oxoglutarate is converted to succinyl-CoA catalyzed by ODHC (2-oxoglutartate dehydrogenase), while, under triggering conditions, the flux could be diverted to the synthesis of l-glutamate by GDH (glutamate dehydrogenase). It has been reported that the suboptimal supply of biotin and the addition of detergents could affect the synthesis of fatty acids, and subsequently affecting the phospholipids synthesis in cell membrane. It was thus suggested that the increase in cell membrane fluidity [62] and variation in cell envelope structure [58], [63], [64] lead to increased secretion of l-glutamate. We believe that the secretion of l-glutamate in great amount is the combined effects of the changes in intracellular metabolism and cell envelope.
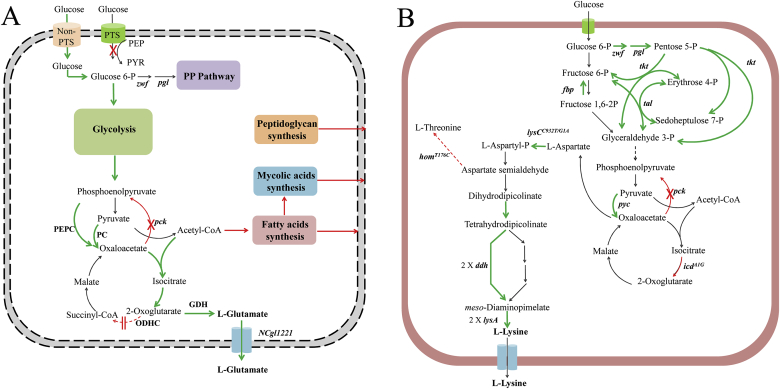
As there is still no clear conclusion for the mechanism of the l-glutamate secretion in C. glutamicum, the application of systems metabolic engineering strategies for l-glutamate high-producing strains has been hindered. Based on current knowledge, schematic systems metabolic engineering for l-glutamate production is proposed in Fig. 3A, which highlighted the combined engineering of intracellular metabolism, and the structure and function of cell envelope. As shown in the figure, the decrease in the ODHC activity and the simultaneous increase of the GDH activity is crucial and essential for the l-glutamate production [59]. Besides, the enhancement of the anaplerotic pathway of the PEP–pyruvate–oxaloacetate node has been proved to be an effective approach to increase the carbon flux for l-glutamate [65], [66], [67], [68]. As PEP is needed for the anaplerotic pathway, the replacement of the PEP dependent PTS with non-PTS could save more PEP for the following conversion [8]. Except for the above engineering, export of l-glutamate should also be enhanced.
Fig. 3.
Systems metabolic engineering of C. glutamicum for the production of l-glutamate (A) and l-lysine [69] (B). (A) The green colored arrows indicate the pathways that should be enhanced, and the red colored arrows indicate the pathways that should be attenuated or deleted. (B) The green colored arrows represent the amplification of relative genes; the red dotted lines and “X” represent attenuation or deletion of relative genes.
As for the engineering of structure and permeability of the cell envelope, fatty acid synthesis, mycolic acid synthesis and peptidoglycan synthesis etc. should be the potential targets. Systems biology-based approaches could provide valuable information for the coordination of the intracellular synthesis process and the export process across the cell envelope.
3.2. l-lysine
Systems metabolic engineering has been successfully applied in the breeding of l-lysine high-producing strains. For example, Becker et al. [69] obtained a genetically defined strain of l-lysine hyper-producing C. glutamicum from the wild-type C. glutamicum ATCC 13032 using systems metabolic engineering strategies (shown in Fig. 3B). Metabolic blueprint of the wild type was obtained through 13C flux and in silico analysis based on the genome information, according to which, systems-wide engineering was designed. The systematic design covered the increase in the flux of the l-lysine biosynthesis pathway, the anaplerotic carboxylation, and the pentose phosphate pathway. Meanwhile, the decrease in the flux of counteracting decarboxylating reactions, TCA cycle and the entire anabolism were also included. After which, the non-producing wild type was successfully turned into an efficient cell factory for l-lysine. The final obtained strain C. glutamicum LYS-12 is the first genetically defined strain that can compete with conventional producers optimized for more than 50 years, showing the giant potential of systems metabolic engineering.
4. Case studies of systems metabolic engineering in E. coli
E. coli is a typical model microorganism that has been most intensively studied, and it has become an efficient workhorse for the production of various bio-products including amino acids and their derivatives [70], [71].
4.1. l-threonine
Lee et al. [40] constructed the first genetically defined producing strain from the basal strain E. coli WL3110, a lacI-mutant strain of W3110. They rewired regulatory and metabolic circuits for the development of an initial threonine producer TH07 (pBRThrABC). As shown in Fig. 4A, the construction of TH07 was performed firstly by removing the feedback inhibition of asparate kinase I and III by bringing in site-directed mutation of thrAC1034T and lysCC1055T. As asparate kinase II encoded by metL was repressed by l-methionine, the deletion of metA not only block the synthesis of l-methionine, but also removed the feedback inhibition of asparate kinase II. By replacing the promoter of mutated thrAC1034TBC operon with a tac promoter, the carbon flux from l-asparate to l-threonine was increased, and at the same time, transcriptional attenuation was removed. Then, the competing pathway of l-threonine for l-lysine synthesis was blocked by deleting lysA (diaminopimelate decarboxylase) gene. The deletion of tdh gene encoding threonine dehydrogenase blocked the synthesis of l-glycine, and the mutated ilvAC290T decreased the synthesis of l-isoleucine. In this way, the degradation of l-threonine was minimized. In the second round of engineering, comparative transcriptomic analysis of TH07 and WL3110 was performed, based on the results of which, the ppc (phosphoenolpyruvate carboxylase) gene expression, l-threonine exporter system, and the glyoxylate shunt were enhanced. The l-threonine titer was stepwise increased by the above engineering approaches. At last, in silico flux response analysis was performed aiming to reduce the acetic acid production, and the acs gene encoding acetyl-CoA synthetase was amplified. The systems metabolic engineering approaches resulted in a final l-threonine producing strain TH28C with an l-threonine titer of 82.4 g/L.
Fig. 4.
Systems metabolic engineering of E. coli for the production of l-threonine [40] (A) and l-tryptophan [72] (B). (A) The green colored arrows, “X”, and dotted lines represent the strategies used in the first round of systems metabolic engineering, specifically the amplification of enzymes in the synthetic pathway, the deletion or decrease of competing and degradation pathway. The red colored arrows and “X” represent the strategies used in the second round of engineering based on transcriptome data and in silico flux response analysis, specifically the enhancement of the PPC flux, the glyoxylate shunt, and the export system of l-threonine, and the blocking of the import system of l-threonine. The blue colored arrow represents the strategy for the reduction of acetic acid in the third round of engineering based on in silico flux response analysis. (B) The green colored arrows represent the amplification of relative genes; the red dotted lines and “X” represent deletion of relative genes.
4.2. l-tryptophan
l-tryptophan is an important biosynthetic precursor of various bioactive components of great pharmaceutical interest, which makes it a high value-added amino acid. Over the past decades, various attempts have been made to achieve the bio-production of l-tryptophan, but due to the complexity of its synthetic pathway, it had been difficult to develop a genetically defined overproducing strain. Recently, Chen et al. [72] conducted systematic metabolic engineering of E. coli, the resulting strain E. coli S028 was able to produce 34–40 g/L of l-tryptophan, which was so far the highest amount for rationally designed l-tryptophan producers.
The systems metabolic engineering procedure is shown in Fig. 4B. They firstly deleted the degradation gene tnaA encoding tryptophanase, and thus blocked the degradation of l-tryptophan. Next, the l-tryptophan importer genes mtr and tnaB were deleted. As shown in Fig. 4B, three genes aroG, aroF, and aroH encoding 3-deoxy-D-arabino-heptulosonate-7-phosphate (DAHP) synthase are responsible for the first step of the synthesis of l-tryptophan. The problem is that they suffer from feedback inhibition by l-phenylalanine, l-tyrosine, and l-tryptophan, respectively. By deleting aroF and aroH, and bringing in site-directed mutated aroGS180F, the feedback inhibition of DAHP synthase was removed. Further enhancement of the expression of aroGS180F using a strong ribosome binding site under tac promoter increased the DAHP supply for l-tryptophan synthesis. In order to increase the l-serine supply, they integrated the feedback resistant gene serAH344A/N364A with a strong ribosome binding site under tac promoter into the genome. At the same time, the flux from chorismate to the synthesis of l-tryptophan was enhanced by replaced the native trpE gene with a feedback inhibition resistant gene trpES40F connected to a strong ribosome binding site. Following that, the native promoter of the L-trp operon was replaced with a strong trc promoter. The resulting strain E. coli S028 efficiently produced 34–40 g/L l-tryptophan with a yield of 0.15 g l-tryptophan/g glucose. The intracellular and extracellular concentrations of key metabolites for l-tryptophan synthesis were systematically measured, and the obtained information suggested that an increased availability of glutamine synthetase and overexpression of the l-tryptophan exporter could be the targets for further strain improvement.
5. Perspective
Systems metabolic engineering has become an efficient and necessary way to perform strain improvement and bio-production of chemicals including amino acids. With the development of advanced techniques of systems biology, synthetic biology, and evolutionary engineering, systems metabolic engineering will undoubtedly generate more excellent strains and valuable bio-products in the future. While, at the present time, the challenges of systems metabolic engineering for amino acid production lie in the following two aspects.
The integration of omics data has become a major challenge for systems biology. Different omics approaches could generate large quantities of data, which provides different levels of information. While, sometimes these information are apparently contradictive due to our analyzing technology and limited understanding of cellular metabolism. The integration of omics data is essential to the overall systems-level understanding of the microorganism, which largely depends on computational science and mathematic algorithms. To achieve the well integration of omics data, multidisciplinary knowledge is required.
Efforts should be made to realize the efficient genome editing in C. glutamicum. Besides, the complicated regulatory networks in amino acid producing strains still need to be further clarified for synthetic biology and evolutionary engineering applications. At present, the transcriptional regulators that could be used in evolutionary engineering still need to be explored. With all these efforts, systems metabolic engineering will contribute more to the industrial production of valuable chemicals including amino acids.
Acknowledgements
This research was supported by National High Technology Research and Development Program of China (2015AA021003), National Natural Science Foundation of China (31470211), Natural Science Foundation of Tianjin (17JCQNJC09500), and Foundation (No. 2016IM104) of Key Laboratory of Industrial Fermentation Microbiology of Ministry of Education and Tianjin Key Lab of Industrial Microbiology (Tianjin Universityof Science & Technology).
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Contributor Information
Xixian Xie, Email: xixianxie@tust.edu.cn.
Ning Chen, Email: ningch@tust.edu.cn.
References
- 1.Lee S.Y., Kim H.U. Systems strategies for developing industrial microbial strains. Nat Biotech. 2015;33(10):1061–1072. doi: 10.1038/nbt.3365. [DOI] [PubMed] [Google Scholar]
- 2.Lee J.W., Kim T.Y., Jang Y.S., Choi S., Lee S.Y. Systems metabolic engineering for chemicals and materials. Trends Biotechnol. 2011;29(8):370–378. doi: 10.1016/j.tibtech.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 3.Chubukov V., Mukhopadhyay A., Petzold C.J., Keasling J.D., Martín H.G. Synthetic and systems biology for microbial production of commodity chemicals. Npj Syst Biol Appl. 2016;2:16009. doi: 10.1038/npjsba.2016.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Soyer O.S., O'Malley M.A. Evolutionary systems biology: what it is and why it matters. BioEssays. 2013;35(8):696–705. doi: 10.1002/bies.201300029. [DOI] [PubMed] [Google Scholar]
- 5.Bassalo M.C., Liu R., Gill R.T. Directed evolution and synthetic biology applications to microbial systems. Curr Opin Biotech. 2016;39:126–133. doi: 10.1016/j.copbio.2016.03.016. [DOI] [PubMed] [Google Scholar]
- 6.Eggeling L., Bott M. A giant market and a powerful metabolism: l-lysine provided by Corynebacterium glutamicum. Appl Microbiol Biot. 2015;99(8):3387–3394. doi: 10.1007/s00253-015-6508-2. [DOI] [PubMed] [Google Scholar]
- 7.Lee J.W., Na D., Park J.M., Lee J., Choi S., Lee S.Y. Systems metabolic engineering of microorganisms for natural and non-natural chemicals. Nat Chem Biol. 2012;8(6):536–546. doi: 10.1038/nchembio.970. [DOI] [PubMed] [Google Scholar]
- 8.Lindner S.N., Seibold G.M., Henrich A., Krämer R., Wendisch V.F. Phosphotransferase system-independent glucose utilization in Corynebacterium glutamicum by inositol permeases and glucokinases. Appl Environ Microb. 2011;77(11):3571–3581. doi: 10.1128/AEM.02713-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen R., Yap W.M.G., Postma P.W., Bailey J.E. Comparative studies of Escherichia coli strains using different glucose uptake systems: metabolism and energetics. Biotechnol Bioeng. 1997;56:583–590. doi: 10.1002/(SICI)1097-0290(19971205)56:5<583::AID-BIT12>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 10.Tatarko M., Romeo M. Disruption of a global regulatory gene to enhance central carbon flux into phenylalanine biosynthesis in Escherichia coli. Curr Microbiol. 2001;43(1):26–32. doi: 10.1007/s002840010255. [DOI] [PubMed] [Google Scholar]
- 11.Engels V., Wendisch V.F. The DeoR-type regulator SugR represses expression of ptsG in Corynebacterium glutamicum. J Bacteriol. 2007;189(8):2955–2966. doi: 10.1128/JB.01596-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krause F.S., Henrich A., Blombach B., Krämer R., Eikmanns B.J., Seibold G.M. Increased glucose utilization in Corynebacterium glutamicum by use of maltose, and its application for the improvement of l-valine productivity. Appl Environ Microb. 2010;76(1):370–374. doi: 10.1128/AEM.01553-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Henrich A., Kuhlmann N., Eck A.W., Krämer R., Seibold G.M. Maltose uptake by the novel ABC transport system MusEFGK2I causes increased expression of ptsG in Corynebacterium glutamicum. J Bacteriol. 2013;195(11):2573–2584. doi: 10.1128/JB.01629-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aristidou A., Penttilä M. Metabolic engineering applications to renewable resource utilization. Curr Opin Biotech. 2000;11(2):187–198. doi: 10.1016/s0958-1669(00)00085-9. [DOI] [PubMed] [Google Scholar]
- 15.Jarmander J., Hallström B.M., Larsson G. Simultaneous uptake of lignocellulose-based monosaccharides by Escherichia coli. Biotechnol Bioeng. 2014;111(6):1108–1115. doi: 10.1002/bit.25182. [DOI] [PubMed] [Google Scholar]
- 16.Kawaguchi H., Vertes A.A., Okino S., Inui M., Yukawa H. Engineering of a xylose metabolic pathway in Corynebacterium glutamicum. Appl Environ Microb. 2006;72(5):3418–3428. doi: 10.1128/AEM.72.5.3418-3428.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meiswinkel T.M., Gopinath V., Lindner S.N., Nampoothiri K.M., Wendisch V.F. Accelerated pentose utilization by Corynebacterium glutamicum for accelerated production of lysine, glutamate, ornithine and putrescine. Microb Biotechnol. 2013;6(2):131–140. doi: 10.1111/1751-7915.12001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kang M.K., Lee J., Um Y., Lee T.S., Bott M., Park S.J. Synthetic biology platform of CoryneBrick vectors for gene expression in Corynebacterium glutamicum and its application to xylose utilization. Appl Microbiol Biot. 2014;98(13):5991–6002. doi: 10.1007/s00253-014-5714-7. [DOI] [PubMed] [Google Scholar]
- 19.Jo S., Yoon J., Lee S.M., Um Y., Han S.O., Woo H.M. Modular pathway engineering of Corynebacterium glutamicum to improve xylose utilization and succinate production. J Biotechnol. 2017 doi: 10.1016/j.jbiotec.2017.01.015. [DOI] [PubMed] [Google Scholar]
- 20.Qin T., Hu X., Hu J., Wang X. Metabolic engineering of Corynebacterium glutamicum strain ATCC13032 to produce l-methionine. Biotechnol Appl Biochem. 2015;62(4):563–573. doi: 10.1002/bab.1290. [DOI] [PubMed] [Google Scholar]
- 21.Hasegawa S., Suda M., Uematsu K., Natsuma Y., Hiraga K., Jojima T. Engineering of Corynebacterium glutamicum for high-yield l-valine production under oxygen deprivation conditions. Appl Environ Microb. 2013;79(4):1250–1257. doi: 10.1128/AEM.02806-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu Q., Zhang X., Luo Y., Guo W., Xu G., Shi J. l-Serine overproduction with minimization of by-product synthesis by engineered Corynebacterium glutamicum. Appl Microbiol Biot. 2015;99(4):1665–1673. doi: 10.1007/s00253-014-6243-0. [DOI] [PubMed] [Google Scholar]
- 23.Dong X., Zhao Y., Hu J., Li Y., Wang X. Attenuating l-lysine production by deletion of ddh and lysE and their effect on l-threonine and l-isoleucine production in Corynebacterium glutamicum. Enzyme Microb Tech. 2016;93–94:70–78. doi: 10.1016/j.enzmictec.2016.07.013. [DOI] [PubMed] [Google Scholar]
- 24.Vogt M., Krumbach K., Bang W.G., Ooyen J.V., Noack S., Klein B. The contest for precursors: channelling l-isoleucine synthesis in Corynebacterium glutamicum without byproduct formation. Appl Microbiol Biot. 2015;99(2):791–800. doi: 10.1007/s00253-014-6109-5. [DOI] [PubMed] [Google Scholar]
- 25.Guo Y., Han M., Xu J., Zhang W. Analysis of acetohydroxyacid synthase variants from branched-chain amino acids-producing strains and their effects on the synthesis of branched-chain amino acids in Corynebacterium glutamicum. Protein Expres Purif. 2015;109:106–112. doi: 10.1016/j.pep.2015.02.006. [DOI] [PubMed] [Google Scholar]
- 26.Chen Z., Bommareddy R.R., Frank D., Rappert S., Zeng A.P. Deregulation of feedback inhibition of phosphoenolpyruvate carboxylase for improved lysine production in Corynebacterium glutamicum. Appl Environ Microb. 2014;80(4):1388–1393. doi: 10.1128/AEM.03535-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vogt M., Haas S., Klaffl S., Polen T., Eggeling L., Ooyen J.V. Pushing product formation to its limit: metabolic engineering of Corynebacterium glutamicum for L-leucine overproduction. Metab Eng. 2014;22:40–52. doi: 10.1016/j.ymben.2013.12.001. [DOI] [PubMed] [Google Scholar]
- 28.Kalinowski J., Bathe B., Bartels D., Bischoff N., Bott M., Burkovski A. The complete Corynebacterium glutamicum ATCC 13032 genome sequence and its impact on the production of l-aspartate-derived amino acids and vitamins. J Biotechnol. 2003;104(1–3):5–25. doi: 10.1016/s0168-1656(03)00154-8. [DOI] [PubMed] [Google Scholar]
- 29.Simic P., Sahm H., Eggeling L. L-Threonine export: use of peptides to identify a new translocator from Corynebacterium glutamicum. J Biotechnol. 2001;183(18):5317–5324. doi: 10.1128/JB.183.18.5317-5324.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lubitz D., Jorge J.M.P., Pérez-García F., Taniguchi H., Wendischc V.F. Roles of export genes cgmA and lysE for the production of l-arginine and l-citrulline by Corynebacterium glutamicum. Appl Microbiol Biot. 2016;100(19):8465–8474. doi: 10.1007/s00253-016-7695-1. [DOI] [PubMed] [Google Scholar]
- 31.Yin L., Shi F., Hu X., Chen C., Wang X. Increasing l-isoleucine production in Corynebacterium glutamicum by overexpressing global regulator Lrp and two-component export system BrnFE. J Appl Microbiol. 2013;114(5):1369–1377. doi: 10.1111/jam.12141. [DOI] [PubMed] [Google Scholar]
- 32.Chen C., Li Y., Hu J., Dong X., Wang X. Metabolic engineering of Corynebacterium glutamicum ATCC13869 for l-valine production. Metab Eng. 2015;29:66–75. doi: 10.1016/j.ymben.2015.03.004. [DOI] [PubMed] [Google Scholar]
- 33.Xie X., Xu L., Shi J., Xu Q., Chen N. Effect of transport proteins on l-isoleucine production with the l-isoleucine-producing strain Corynebacterium glutamicum YILW. J Ind Microbiol Biot. 2012;39(10):1549–1556. doi: 10.1007/s10295-012-1155-4. [DOI] [PubMed] [Google Scholar]
- 34.Huang J., Wu Y., Wu W., Zhang Y., Liu D., Chen Z. Cofactor recycling for co-production of 1,3-propanediol and glutamate by metabolically engineered Corynebacterium glutamicum. Sci Rep. 2017;7:42246. doi: 10.1038/srep42246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li Y., Cong H., Liu B., Song J., Sun X., Zhang J. Metabolic engineering of Corynebacterium glutamicum for methionine production by removing feedback inhibition and increasing NADPH level. Ant Leeuw. 2016;109(9):1185–1197. doi: 10.1007/s10482-016-0719-0. [DOI] [PubMed] [Google Scholar]
- 36.Xu H., Zhou Z., Wang C., Chen Z., Cai H. Enhanced succinic acid production in Corynebacterium glutamicum with increasing the available NADH supply and glucose consumption rate by decreasing H+-ATPase activity. Biotechnol Lett. 2016;38(7):1181–1186. doi: 10.1007/s10529-016-2093-4. [DOI] [PubMed] [Google Scholar]
- 37.Wang Z., Chan S.H.J., Sudarsan S., Blank L.M., Jensene P.R., Solem C. Elucidation of the regulatory role of the fructose operon reveals a novel target for enhancing the NADPH supply in Corynebacterium glutamicum. Metab Eng. 2016;38:344–357. doi: 10.1016/j.ymben.2016.08.004. [DOI] [PubMed] [Google Scholar]
- 38.Bommareddy R.R., Chen Z., Rappert S., Zeng A.P. A de novo NADPH generation pathway for improving lysine production of Corynebacterium glutamicum by rational design of the coenzyme specificity of glyceraldehyde 3-phosphate dehydrogenase. Metab Eng. 2014;25:30–37. doi: 10.1016/j.ymben.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 39.Park J.H., Lee K.H., Kim T.Y., Lee S.Y. Metabolic engineering of Escherichia coli for the production of L-valine based on transcriptome analysis and in silico gene knockout simulation. P Natl Acad Sci. 2007;104(19):7797–7802. doi: 10.1073/pnas.0702609104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee K.H., Park J.H., Kim T.Y., Kim K.U., Lee S.Y. Systems metabolic engineering of Escherichia coli for L-threonine production. Mol Syst Biol. 2007;3(1):2025. doi: 10.1038/msb4100196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Krömer J.O., Sorgenfrei O., Klopprogge K., Heinzle E., Wittmannz C. In-depth profiling of lysine-producing Corynebacterium glutamicum by combined analysis of the transcriptome, metabolome, and fluxome. J Bacteriol. 2004;186(6):1769–1784. doi: 10.1128/JB.186.6.1769-1784.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Buchinger S., Strösser J., Rehm N., Hänßler E., Hans S., Bathe B. A combination of metabolome and transcriptome analyses reveals new targets of the Corynebacterium glutamicum nitrogen regulator AmtR. J Biotechnol. 2009;140(1–2):68–74. doi: 10.1016/j.jbiotec.2008.10.009. [DOI] [PubMed] [Google Scholar]
- 43.Ohnishi J., Mitsuhashi S., Hayashi M., Ando S., Yokoi H., Ochiai K. A novel methodology employing Corynebacterium glutamicum genome information to generate a new L-lysine-producing mutant. Appl Microbiol Biot. 2002;58(2):217. doi: 10.1007/s00253-001-0883-6. [DOI] [PubMed] [Google Scholar]
- 44.Toya Y., Shimizu H. Flux analysis and metabolomics for systematic metabolic engineering of microorganisms. Biotechnol Adv. 2013;31(6):818–826. doi: 10.1016/j.biotechadv.2013.05.002. [DOI] [PubMed] [Google Scholar]
- 45.Park J.M., Kim T.Y., Lee S.Y. Constraints-based genome-scale metabolic simulation for systems metabolic engineering. Biotechnol Adv. 2009;27(6):979–988. doi: 10.1016/j.biotechadv.2009.05.019. [DOI] [PubMed] [Google Scholar]
- 46.Nguyen A., Schneider J., Reddy G., Wendisch V.F. Fermentative production of the diamine putrescine: system metabolic engineering of Corynebacterium glutamicum. Metabolites. 2015;5(2):211. doi: 10.3390/metabo5020211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shlomi T., Berkman O., Ruppin E. Regulatory on/off minimization of metabolic flux changes after genetic perturbations. P Natl Acad Sci. 2005;102(21):7695–7700. doi: 10.1073/pnas.0406346102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim H.U., Kim T.Y., Lee S.Y. Metabolic flux analysis and metabolic engineering of microorganisms. Mol Biosyst. 2008;4(2):113–120. doi: 10.1039/b712395g. [DOI] [PubMed] [Google Scholar]
- 49.Zhang F., Keasling J. Biosensors and their applications in microbial metabolic engineering. Trends Microbiol. 2011;19(7):323–329. doi: 10.1016/j.tim.2011.05.003. [DOI] [PubMed] [Google Scholar]
- 50.Binder S., Siedler S., Marienhagen J., Bott M., Eggeling L. Recombineering in Corynebacterium glutamicum combined with optical nanosensors: a general strategy for fast producer strain generation. Nucleic Acids Res. 2013;41(12):6360. doi: 10.1093/nar/gkt312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu D., Evans T., Zhang F. Applications and advances of metabolite biosensors for metabolic engineering. Metab Eng. 2015;31:35–43. doi: 10.1016/j.ymben.2015.06.008. [DOI] [PubMed] [Google Scholar]
- 52.Williams T.C., Pretorius I.S., Paulsen I.T. Synthetic evolution of metabolic productivity using biosensors. Trends Biotechnol. 2016;34(5):371–381. doi: 10.1016/j.tibtech.2016.02.002. [DOI] [PubMed] [Google Scholar]
- 53.Mahr R., Gätgens C., Gätgens J., Polen T., Kalinowski J., Frunzke J. Biosensor-driven adaptive laboratory evolution of l-valine production in Corynebacterium glutamicum. Metab Eng. 2015;32:184–194. doi: 10.1016/j.ymben.2015.09.017. [DOI] [PubMed] [Google Scholar]
- 54.Binder S., Schendzielorz G., Stäbler N., Krumbach K., Hoffmann K., Bott M. A high-throughput approach to identify genomic variants of bacterial metabolite producers at the single-cell level. Genome Biol. 2012;13(5):R40. doi: 10.1186/gb-2012-13-5-r40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Eggeling L., Bott M., Marienhagen J. Novel screening methods—biosensors. Curr Opin Biotech. 2015;35:30–36. doi: 10.1016/j.copbio.2014.12.021. [DOI] [PubMed] [Google Scholar]
- 56.Mustafi N., Grünberger A., Kohlheyer D., Bott M., Frunzke J. The development and application of a single-cell biosensor for the detection of l -methionine and branched-chain amino acids. Metab Eng. 2012;14(4):449–457. doi: 10.1016/j.ymben.2012.02.002. [DOI] [PubMed] [Google Scholar]
- 57.Becker J, Gießelmann G, Hoffmann, SL, Wittmann C. Corynebacterium glutamicum for sustainable bioproduction: from metabolic physiology to systems metabolic engineering. Springer Berlin Heidelberg: Berlin, Heidelberg. 1–47. [DOI] [PubMed]
- 58.Nakamura J., Hirano S., Ito H., Wachi M. Mutations of the Corynebacterium glutamicum NCgl1221 gene, encoding a mechanosensitive channel homolog, induce l-glutamic acid production. Appl Environ Microb. 2007;73(14):4491–4498. doi: 10.1128/AEM.02446-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Asakura Y., Kimura E., Usuda Y., Kawahara Y., Matsui K., Osumi T. Altered metabolic flux due to deletion of odhA causes L-glutamate overproduction in Corynebacterium glutamicum. Appl Environ Microbiol. 2007;73:1308–1319. doi: 10.1128/AEM.01867-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Radmacher E., Stansen K.C., Besra G.S., Alderwick L.J., Maughan W.N., Hollweg G. Ethambutol, a cell wall inhibitor of Mycobacterium tuberculosis, elicits L-glutamate efflux of Corynebacterium glutamicum. Microbiology. 2005;151:1359–1368. doi: 10.1099/mic.0.27804-0. [DOI] [PubMed] [Google Scholar]
- 61.Hoischen C., Krämer R. Membrane alteration is necessary but not sufficient for effective glutamate secretion in Corynebacterium glutamicum. J Bacteriol. 1990;172(6):3409–3416. doi: 10.1128/jb.172.6.3409-3416.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bokas D., Uy D., Grattepanche F., Duportail G., Guedon E., Delaunay S. Cell envelope fluidity modification for an effective glutamate excretion in Corynebacterium glutamicum 2262. Appl Microbiol Biot. 2007;76(4):773. doi: 10.1007/s00253-007-1046-1. [DOI] [PubMed] [Google Scholar]
- 63.Becker M., Börngen K., Nomura T., Battle A.R., Marin K., Martinac B. Glutamate efflux mediated by Corynebacterium glutamicum MscCG, Escherichia coli MscS, and their derivatives. BBA-Biomembranes. 2013;1828(4):1230–1240. doi: 10.1016/j.bbamem.2013.01.001. [DOI] [PubMed] [Google Scholar]
- 64.Bayan N., Houssin C., Chami M., Leblon G. Mycomembrane and S-layer: two important structures of Corynebacterium glutamicum cell envelope with promising biotechnology applications. J Biotechnol. 2003;104(1–3):55–67. doi: 10.1016/s0168-1656(03)00163-9. [DOI] [PubMed] [Google Scholar]
- 65.Wada M., Sawada K., Ogura K., Shimono Y., Hagiwara T., Sugimoto M. Effects of phosphoenolpyruvate carboxylase desensitization on glutamic acid production in Corynebacterium glutamicum ATCC 13032. J Biosci Bioeng. 2016;121(2):172–177. doi: 10.1016/j.jbiosc.2015.06.008. [DOI] [PubMed] [Google Scholar]
- 66.Shirai T., Fujimura K., Furusawa C., Nagahisa K., Shioya S., Shimizu H. Study on roles of anaplerotic pathways in glutamate overproduction of Corynebacterium glutamicum by metabolic flux analysis. Microb Cell Fact. 2007;6(1):19. doi: 10.1186/1475-2859-6-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Guo X., Wang J., Xie X., Xu Q., Zhang C., Chen N. Enhancing the supply of oxaloacetate for l-glutamate production by pyc overexpression in different Corynebacterium glutamicum. Biotechnol Lett. 2013;35(6):943–950. doi: 10.1007/s10529-013-1241-3. [DOI] [PubMed] [Google Scholar]
- 68.Delaunay S., Uy D., Baucher M.F., Engasser J.M., Guyonvarch A., Goergen J.L. Importance of phosphoenolpyruvate carboxylase of Corynebacterium glutamicum during the temperature triggered glutamic acid fermentation. Metab Eng. 1999;1(4):334–343. doi: 10.1006/mben.1999.0131. [DOI] [PubMed] [Google Scholar]
- 69.Becker J., Zelder O., Häfner S., Schröder H., Wittmann C. From zero to hero—design-based systems metabolic engineering of Corynebacterium glutamicum for L-lysine production. Metab Eng. 2011;13(2):159–168. doi: 10.1016/j.ymben.2011.01.003. [DOI] [PubMed] [Google Scholar]
- 70.Ning Y., Wu X., Zhang C., Xu Q., Chen N., Xie X. Pathway construction and metabolic engineering for fermentative production of ectoine in Escherichia coli. Metab Eng. 2016;36:10–18. doi: 10.1016/j.ymben.2016.02.013. [DOI] [PubMed] [Google Scholar]
- 71.Zhang C., Qi J., Li Y., Fan X., Xu Q., Chen N. Production of α-ketobutyrate using engineered Escherichia coli via temperature shift. Biotechnol Bioeng. 2016;113(9):2054–2059. doi: 10.1002/bit.25959. [DOI] [PubMed] [Google Scholar]
- 72.Chen L., Zeng A.P. Rational design and metabolic analysis of Escherichia coli for effective production of L-tryptophan at high concentration. Appl Microbiol Biot. 2017;101(2):559–568. doi: 10.1007/s00253-016-7772-5. [DOI] [PubMed] [Google Scholar]