Abstract
Next-generation sequencing (NGS) provides unrestricted access to the genome, but it produces ‘big data’ exceeding in amount and complexity the classical analytical approaches. We introduce a bioinformatics-based classifying biomarker that uses emergent properties in genetics to separate pain patients requiring extremely high opioid doses from controls. Following precisely calculated selection of the 34 most informative markers in the OPRM1, OPRK1, OPRD1 and SIGMAR1 genes, pattern of genotypes belonging to either patient group could be derived using a k-nearest neighbor (kNN) classifier that provided a diagnostic accuracy of 80.6±4%. This outperformed alternative classifiers such as reportedly functional opioid receptor gene variants or complex biomarkers obtained via multiple regression or decision tree analysis. The accumulation of several genetic variants with only minor functional influences may result in a qualitative consequence affecting complex phenotypes, pointing at emergent properties in genetics.
Introduction
Genotyping-based drug therapy decisions are increasingly desired in clinical practice; however, their introduction is still limited. The so far published single functional genetic variants are increasingly perceived as insufficient in providing a genetic diagnosis of clinical phenotypes.1 However, exploitation of the whole genetic information becomes possible overcoming the restricted selection of known variants. Next-generation sequencing (NGS)2 provides unrestricted access to the subjects’ genome.
As the resulting ‘big data’ exceeds in its amount and complexity the classical approaches, the analysis of NGS derived data is an active research topic. It has already led to working solutions (for review, see Nielsen et al.3 and Pabinger et al.4) and new statistical methods for analyzing NGS data are continuously emerging.5, 6, 7, 8, 9 However, methods to convert NGS-derived big data into biomarkers are still sparse and solutions exploiting the whole genomic information content for patient classification are still needed. Novel types of classifying biomarkers derived from NGS information are needed, and we report a subsymbolic classifier that uses emergent properties in genetics10 with the potential of self-learned improvement from successively addable genetic and clinical information.
The biomarker was developed based on the clinical problem of extremely high opioid demands by some pain patients without any indication of addiction. These patients were subjected to our pharmacogenetic counseling for opioid receptor genetics as the primary candidates coding for the main targets of this class of drugs. The results show that the biomarker utilizing comprehensive DNA sequence information outperforms classical approaches at genetics-based patient classification and promises the utilization of complex information in NGS-derived genotypes for successful clinical diagnostics.
Materials and methods
Patients
The investigation followed the Declaration of Helsinki on Biomedical Research Involving Human Subjects and was approved by the ethics committee of the Medical Faculty of the Goethe-University, Frankfurt, Germany (ethics protocol number E 195/08). Patients were included for whom pharmacogenetic counseling, in particular opioid receptor genotyping, had been requested because of the perception of uncommonly high analgesic opioid dosing requirements without any obvious clinical reason and explicit denial of an addiction background. Using conversion to oral morphine equivalents (OMEs; for details, see Supplementary Table 1), these patients were divided into two groups with high (⩾400 mg day−1 OME; n=30; mean±s.d.: 3.04±0.34 log OME) and common (⩽100 mg day−1 OME; n=28; ‘controls’ mean±s.d.: 1.7±0.24 log OME) opioid dosing requirements for chronic pain therapy. Upon written informed consent, a venous blood sample was taken from each patient; the samples were anonymized and sent to our laboratory at the Institute of Clinical Pharmacology in Frankfurt.
Opioid receptor genotyping using NGS
DNA preparation and amplification
Genomic DNA was extracted from 200 μl venous blood on a BioRobot EZ1 workstation applying the blood and body fluid spin protocol provided in the EZ1 DNA Blood 200 μl Kit (Qiagen, Hilden, Germany). A multiplex amplification primer set for the exonic sequences of the opioid receptor genes (OPRM1, OPRK1, OPRD1 and SIGMAR1, located on chromosomes 6, 8, 1 and 9, respectively) was designed online using a web tool (Ion Ampliseq Designer; Life Technologies, Darmstadt, Germany) provided by the manufacturer of the NGS device at http://www.ampliseq.com. Sequencing gaps affected only noncoding regions (for details, see Supplementary Table 2). A total of 10 ng DNA per sample were used for the target enrichment by a multiplex PCR and each DNA pool was amplified with the Ion AmpliSeq Library Kit 2.0 in conjunction with the Ion AmpliSeq ‘custom Primer Pool’ protocols according to the manufacturer’s instructions (Life Technologies). After each pool had undergone 17 PCR cycles, amplicons were digested with FuPa Reagent partially removing the primer sequences followed by adaptors ligation. To enable multiplexing, sequencing adaptors with short lengths of index sequences (barcodes) were used (Ion Xpress Barcode Adapters Kit; Life Technologies). The adaptor-ligated amplicons were then purified using the Agentcourt AMPure XP beads (Beckman Coulter, Krefeld, Germany). After purification, fragment libraries were normalized to a final DNA concentration of 100 pM using the Ion Library Equalizer Kit (Life Technologies). Equalized barcoded libraries from 10 to 12 samples were pooled at a time. To clonally amplify the library DNA onto the Ion Sphere Particles (Life Technologies), a volume of 2 μl of the barcoded and pooled library was suspended to emulsion PCR using the Ion Personal Genome Machine (PGM) Template OT2 200 Kit on the Ion OneTouch 2 system (Life Technologies) according to the manufacturer’s recommended protocol.
DNA NGS and variant identification
The template-positive Ion Sphere Particles were enriched on the Ion OneTouch ES (Life Technologies) and loaded on Ion 316 v2 Chips (500 Mb of expected sequence data output). Sequencing was performed with the Ion (PGM) system using Ion PGM 200 Sequencing Kit v2 (Life Technologies) with the 200-bp single-end run configuration following the manufacturer’s instructions. Using the Torrent Suite software (version 4.4.2; Life Technologies), signal processing, base calling and the generation of unmapped and mapped BAM-files (hg19 reference genomic sequence) were performed. The variant calling (single-nucleotide polymorphisms, multi-nucleotide polymorphisms (MNPs), insertions (Ins) and deletions) across the hg19 reference genomic sequence was performed with the Torrent Variant Caller Plugin with following key parameters: minimum allele frequency=0.1, minimum quality=10, minimum coverage=6 and minimum coverage on either strand=0. The annotation of called variants was done with Ion Reporter software (version 4.2.2; Life Technologies) using VCF files from the Torrent Variant Caller as input. Data quality and coverage checks as well as variant identification were done using the single-nucleotide polymorphism and variation suite software (SVS version 8.3.3 for Linux 64-bit; Golden Helix, Bozeman, MT, USA). The correctness of the genotyping was verified using 10 amplifications of the coding parts of the genes that were completely conventionally sequenced by an independent commercial provider (LGC GmbH, Berlin, Germany).
Data analysis
Opioid receptor genotype differences between patients with high opioid dosage and controls were analyzed using the single-nucleotide polymorphism and variation suite software (SVS version 8.3.3 for Linux 64-bit; Golden Helix), the R software package (version 3.0.2 for Linux; http://CRAN.R-project.org/) and the Matlab numerical computing environment (version 8.3.0.532, MathWorks, Natick, MA, USA). The analyses are described briefly in the following; more detailed descriptions are provided in the Supplementary Materials.
Assessment of group differences in single opioid receptor variant or haplotype frequencies
A first analysis employed the classical approach to pharmacogenetic analyses consisting of χ2 statistics with an α-level of 0.05 corrected for multiple testing according to the conservative criterion of Bonferroni. The dominant hereditary model (DD, Dd versus dd, where D denotes the minor allele and d the wild-type allele) was applied. In addition, haplotypes identified in silico via the minimizing historical recombination algorithm11 were analogously assessed. To observe statistical power, only variants found at a frequency of >10%12 were included.
Assessment of combined genotypic group classifiers
Further analysis addressed combinations of genetic variants employing two classical methods comprising stepwise regression analysis and classification and regression tree (CART) analysis. In addition, advanced approaches at complex genetic group classifiers were used. As above approaches implicitly assumed homogeneity within the two patient cohorts, cluster analysis was used to reveal subgroups within each cohort. For this analysis, data were preprocessed to obtain the genotypes at each locus that could be expected by chance, which were obtained as the group size weighted means across all patients, separately for each variant. Subsequently, for each variant and patient the directed deviations were calculated as the difference between the actual observation and the above calculated expectation. These deviations were submitted to cluster analysis using the Ward algorithm and the Jaccard distance calculated as 1−Jaccard coefficient, the latter being identical to the percentage of nonzero values that differ. In a final step, these clusters served for the training of a k-nearest neighbor (kNN)13 classifier with k=3. Furthermore, the distance function was optimized by selection of the most informative variables using computed ABC analysis14 that identified those genetic variants that promised to provide the best distinction between the two patient groups based on the difference in absolute group means of the number of rare-type alleles.
Classification performance analyses
For all single and complex classifiers cross-validated prediction performances were assessed. Test data sets (sample size n=20) were drawn from the study cohort that always included (1) seven additional patients with high opioid dosage, and expanded by (2) further three patients randomly chosen from the already analyzed group with high opioid dosage to obtain a sufficiently large number for accuracy calculations and (3) 10 patients randomly chosen from the control group. Testing was 100 times repeated. The diagnostic accuracy, test sensitivity, specificity and positive predictive value were calculated using standard equations.
Results
Group differences in single opioid receptor variant frequencies
Nucleotide information was completely available and sequencing runs met the standard requirements. The average throughput was 540 mega-bases, using 316 sequencing chips, which is within the upper third of the expected sequencing output according to the manufacturer’s instructions, and the average chip loading was 71%, meeting the all expected assay quality parameters criteria.
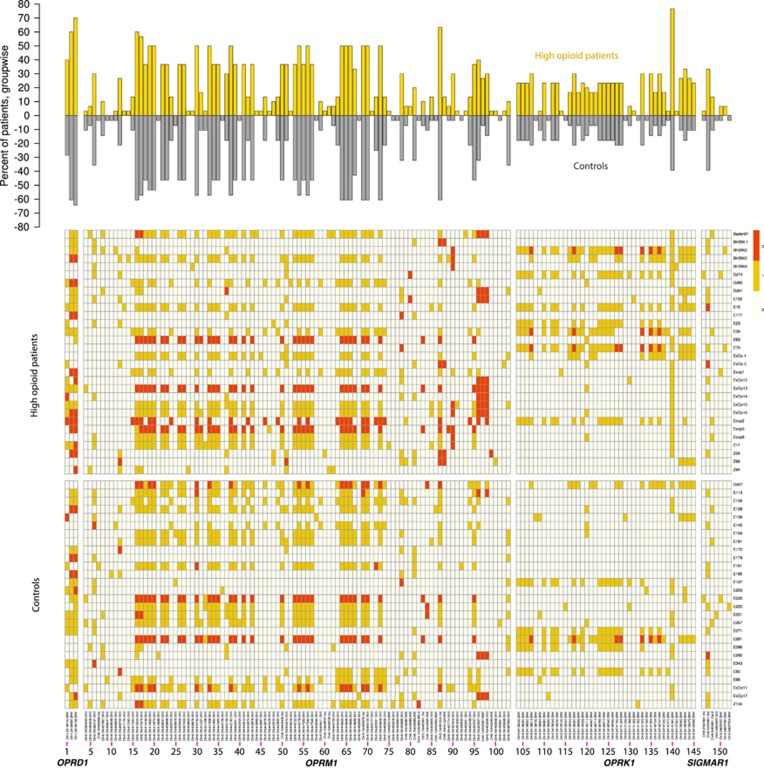
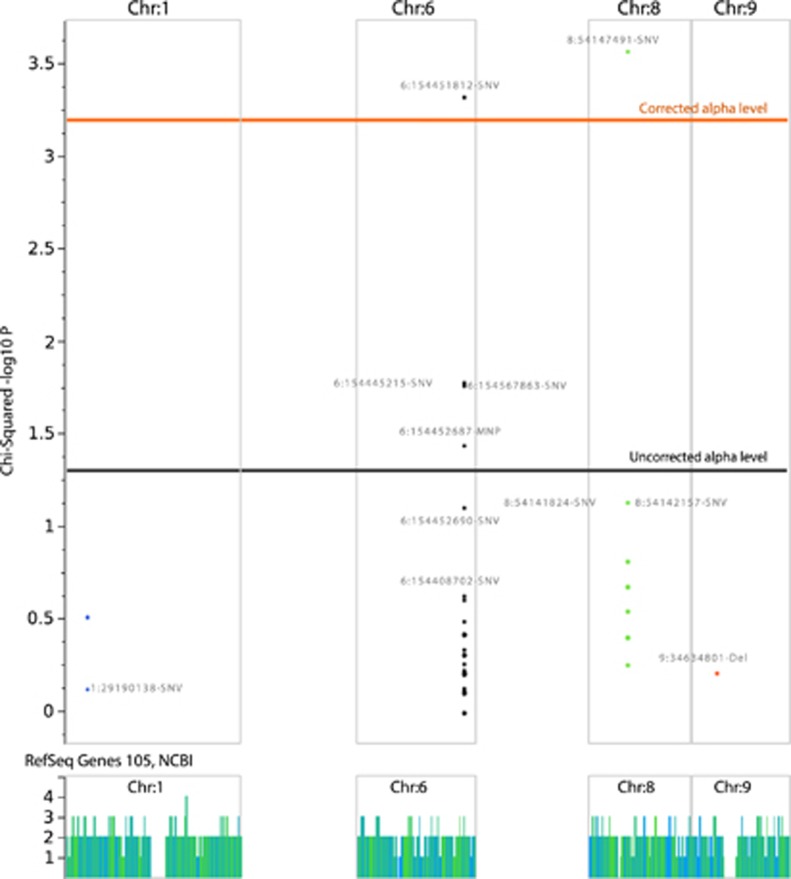
NGS identified 152 variants in opioid receptor genes in the whole study population (Figure 1). A number of 100, 42, 3 and 7 variants were located in the covered sequences of the OPRM1, OPRK1, OPRD1 and SIGMAR1 genes. Group differences in the allelic frequencies were analyzed for 77 gene loci where variant alleles were found in at least 10% of the patients. This resulted in a corrected α-level of 0.000649351 as the upper limit for acceptance of a significant difference (Figure 2). A P-value below this limit resulted for the group frequency comparisons of the Chr6:154451812-single-nucleotide variation (SNV) in the μ-opioid receptor gene OPRM1 (P=0.00049, α-corrected P=0.038) and for the Chr8:54147491-SNV in the κ-opioid receptor gene OPRK1 (P=0.000278, α-corrected P=0.021), whereas a few other variants in the μ-opioid receptor gene, that is, Chr6:154444436-Ins (uncorrected P=0.0169), Chr6:154452687-MNP (uncorrected P=0.036) and Chr6:154567863-SNV (uncorrected P=0.0174), merely displayed differences at an uncorrected significance level and were therefore rejected.
Figure 1.
Overview of the opioid receptor genetic pattern of the present patients (n=30 and 28 pain patients with high and average opioid dosage, respectively). The denominations of gene loci where variants have been detected is given at the bottom of the figure. (Top) Bar plot of the frequency of carriers of single-nucleotide polymorphisms (SNPs), with bar lengths indicating the percentage of patients carrying a variant allele (either heterozygously or homozygously); bars in the upper direction (yellow) show the high opioid patients (n=30) and bars directed toward the bottom (dark gray) show the controls (n=28). (Bottom) Matrix plot of the occurrence of variants (columns) per patient (lines; color coding is white: wild type, yellow: heterozygous, red: homozygous rare allele), separated by gaps for the two groups of patients and for the four opioid receptor genes (OPRD1, OPRM1, OPRK1 and SIGMAR1) from the left to the right; the codes at the right are the patient’s codes left in the figure for potential data validation purposes). For better visibility, the variants are numbered and the abscissa details are given in an enlarged version in the Supplementary Materials (DetailedAbscissa_Figure1.pdf).
Figure 2.
Manhattan plot showing the results of the genotype association test using the dominant hereditary model. Only chromosomes 1, 6, 8 and 9 are shown where the four opioid receptor genes are located. In addition, the α-levels before and after correction for multiple testing according to Bonferroni are indicated as horizontal lines.
All opioid receptor genetic variants previously proposed as modulating opioid requirements (Table 1) failed to display significant group differences with respect to their allelic frequencies. For example, the variant OPRM1 118 G allele was found in 9 patients requiring high opioid doses and in 13 controls (uncorrected P=0.589). However, none of the variants identified in this classical analysis provided an acceptable accuracy for the assignment of a patient to either group (51.5±2% and 51.4±5%) and the diagnostic sensitivities of these markers were also low (Table 3). Furthermore, a total of 24 different genetic variants carried only by pain patients who received very high opioid doses but not by any of the control patients provided an immediate biologically plausible cause for reduced or absent opioid receptor function (Table 2).
Table 1. Variants in opioid receptor genes for which functional consequences for opioid-based analgesic therapy had been reported previously, including example references for each variant.
| Gene | cDNA | Nucleotide position | Effect | dbSNP | Clinical consequence | Reference |
|---|---|---|---|---|---|---|
| OPRM1 | c.575G>T | chr6:154411245 | p.Cys192Phe | rs62638690 | Heroin and cocaine addiction | 26 |
| c.1206A>T | chr6:154414446 | p.Gln402His | rs540825 | Decreased effects in response to antidepressants | 27 | |
| c.118A>G | chr6:154360797 | p.Asn40Asp | rs1799971 | Decreased effects in response to opioids | 28 | |
| c.17C>T | chr6:154360696 | p.Ala6Val | rs1799972 | Heroin addiction; opioid dependence | 29 | |
| c.172C>T | chr6:154039373 | p.Gln5His | rs6912029 | Change-in-libido side effects; insomnia side effects | 30 | |
| c.1231C>T | chr6:154107531 | p.Glu411AMB | rs677830 | Decreased effects in response to opioids | 27 | |
| c.440C>G | chr6:154089975 | p.Ser47Cys | rs17174794 | Heroin and cocaine addiction | 26 | |
| c.1323A>G | chr6:154414563 | p.(=) | rs675026 | Increased risk in coronary heart disease | 31 | |
| c.1333C>T | chr6:154414573 | p.(=) | rs562859 | Decreased effects in response to antidepressants | 32 | |
| OPRD1 | c.921C>T | chr1:29189597 | p.(=) | rs2234918 | Increased effect sizes of pain | 33 |
| OPRK1 | c.36G>T | chr8:54163562 | p.(=) | rs1051660 | Heroin addiction; alcohol dependence | 34 |
| c.846C>T | chr8:54142154 | p.(=) | rs16918875 | Heroin addiction; alcohol dependence | 35 | |
| c.843A>G | chr8:54142157 | p.(=) | rs702764 | Heroin addiction; alcohol dependence | 35 | |
| SIGMAR1 | c.5A>C | chr9:34637690 | p.Gln2Pro | rs1800866 | Increased risk for developing Alzheimer’s disease; decreased effects in response to antidepressants | 36 |
Abbreviations: cDNA, complementary DNA; dbSNP, single-nucleotide polymorphism database; p.(=), synonymous variant.
Table 2. Genetic variants found only in patients receiving high opioid doses.
| Gene | Variant | DNA change | Molecular consequence | Potential functional effecta | dbSNP ID |
|---|---|---|---|---|---|
| OPRK1 | 54139145-SNV | c.*2712C>T | Noncoding | Reduced transcriptional efficiency | rs117602211 |
| 54141429-SNV | c.*428G>A | Noncoding | Reduced transcriptional efficiency | rs182444059 | |
| 54147531-SNV | c.398T>C | p.Ile133Thr | Missense mutation | rs146859342 | |
| OPRM1 | 154411110-SNV | c.440C>G | p.Ser147Cys | Missense mutation | rs17174794 |
| 154411245-SNV | c.575G>T | p.Cys192Phe | Missense mutation | rs62638690 | |
| 154439865-SNV | c.9C>T | — | — | rs11575858 | |
| 154439876-SNV | c.*20G>A | Noncoding | Reduced transcriptional efficiency | rs200778856 | |
| 154443060-SNV | — | — | — | — | |
| 154443067-SNV | — | — | — | — | |
| 154443459-SNV | — | — | — | — | |
| 154443510-SNV | c.*3689A>G | Noncoding | Reduced transcriptional efficiency | rs188792757 | |
| 154446218-SNV | — | — | — | rs190450820 | |
| 154448521-Ins | — | — | — | — | |
| 154449106-Ins | — | — | — | rs73022035 | |
| 154450157-SNV | — | — | — | — | |
| 154450988-MIX | — | — | — | — | |
| 154450973-MNP | — | — | — | — | |
| 154451224-MIX | — | — | — | — | |
| 154451843-SNV | c.*11987C>T | Noncoding | Reduced transcriptional efficiency | rs191957030 | |
| 154452251-SNV | c.*2395G>C | Noncoding | Reduced transcriptional efficiency | rs644261 | |
| 154453066-MIX | c.*13210A>G | Noncoding | Reduced transcriptional efficiency | rs184783311 | |
| 154453095-SNV | — | — | — | — | |
| SIGMAR1 | 34637690-SNV | c.5A>C | p.Gln2Pro | Missense mutation | rs1800866 |
Abbreviations: dbSNP, single-nucleotide polymorphism database; mutation variants are as follows: Ins, insertion; MIX, a mixture of variation types; MNP, multi-nucleotide polymorphism; SNV, single-nucleotide variation.
Potential consequences according to this review.21
Predictions using combined genetic markers obtained with classical approaches
An only moderate improvement of the opioid receptor genetics-based group classification performance was obtained using classical assessments of combined genotypic group classifiers. Stepwise regression analysis identified, based on the P<0.05 inclusion and P>0.1 rejection criteria, n=6 opioid receptor variants, that is, Chr6:154443510-SNV, Chr6:154449106-Ins, Chr6:154450991-MIX, Chr6:154451812-SNV, Chr8:54147491-SNV and Chr8:54155452-SNV, as suitable components of a combined genotypic classifier for the present group assignment. A linear regression model using these alleles performed with an average accuracy of 61.7±6% on the 100 randomly drawn test data sets (Table 3). A decision tree classifier constructed with the CART algorithm used n=12 opioid receptor variants, that is, Chr6:154360483-SNV, Chr6:154414666-SNV, Chr6:154439865-SNV, Chr6:154442128-SNV, Chr6:154443510-SNV, Chr6:154450065-SNV, Chr6:154450991-MIX, Chr6:154450996-MNP, Chr6:154451812-SNV, Chr6:154567863-SNV, Chr8:54147491-SNV and Chr8:54155452-SNV. The prediction of group assignment using this decision tree performed with an average accuracy of 74.4±4% on the 100 randomly drawn test data sets (Table 3).
Table 3. Performances of selected classifiers to predict a patient with high opioid dosing.
| Classifier | Group difference (P-uncorrected)* | No. of genes | Sensitivity (%) | Specificity (%) | PPV (%) | Accuracy (%) | No. of 7 independent samples correct |
|---|---|---|---|---|---|---|---|
| Chr6:154360797-SNV | P=0.589 | 1 | 13.9 | 87 | 51 | 50.6±5 | 0 |
| Chr6:154451812-SNV | 0.00049 | 1 | 2.9 | 100 | 29 | 51.5±2 | 0 |
| Chr8:54147491-SNV | 0.000278 | 1 | 10.5 | 92.2 | 52 | 51.4±5 | 4 |
| Regression | — | 6 | 67.7 | 55.7 | 60 | 61.7±6 | 4 |
| CART | — | 12 | 92.6 | 56.1 | 68 | 74.4±4 | 3 |
| Clustering+kNN classifier | — | 152 | 75.3 | 73.5 | 74 | 74.4±7 | 5 |
| ABC analysis+clustering+kNN classifier | — | 34 | 93.7 | 67.4 | 74 | 80.6±4 | 4 |
Abbreviations: CART, classification and regression tree; kNN, k-nearest neighbor; PPV, positive predictive value; SNV, single-nucleotide variation.
χ2 test: the α-corrected significance level was 0.000649351.
Predictions using advanced approaches at complex genetic group classifiers
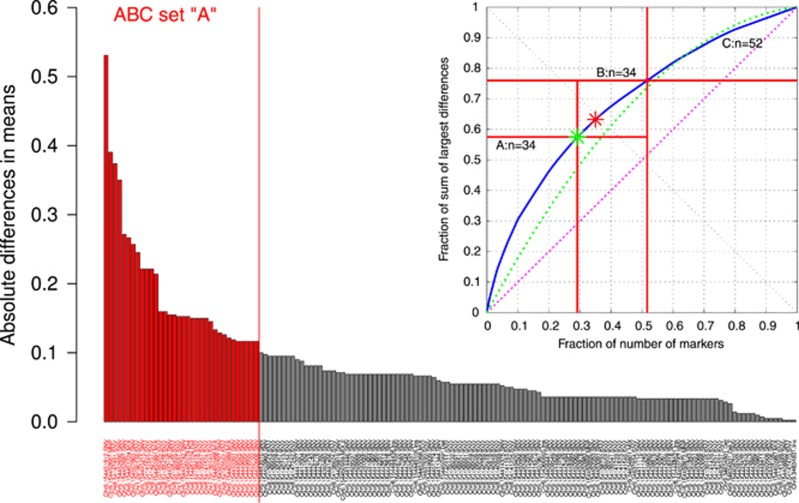
A Ward clustering using the Jaccard distance on all genetic markers led to clusters of n=5 each in both the high opioid patients and the controls. The kNN classifier was trained with the pattern of each cluster per group that resulted in a number of typical patterns given by the number of clusters per patient group. Each of the 100 randomly drawn test data sets was compared in the high-dimensional data space with the obtained complete genotypes according to the Jaccard distance, and the case was assigned to that cluster to which the majority of its three neighbors belonged. The performance of this classifier on the test data resulted in an accuracy of 74.4±7% matching that of the CART-derived classifier (Table 3). This result could be improved by eliminating those genetic markers that only introduced noise into the classifier. ABC analysis revealed that the genetic information contained at 34 loci in opioid receptor genes suffices for accurate group assignment to either patients with high opioid dosage or controls (Figure 3). With these markers, three clusters were identified for the control group, whereas four clusters appeared in the high opioid group. The performance of this kNN classifier on the test data, obtained as described in the previous paragraph, resulted in an accuracy of 80.6±4% (Table 3).
Figure 3.
Identification of those genetic variants that promised to provide the comparatively best distinction between the two patient groups. The bar plot shows the absolute group size standardized differences of the occurrences of each variant between the two patients groups. The 34 bars left of the solid vertical line indicate the genetic variants found by a computed ABC analysis (right upper part of the figure) to provide a statistically valid set of markers to be included in a complex biomarker. At the upper right corner is the ABC plot14 of the cumulative distribution function of the additional fraction of the absolute group differences in allelic numbers (solid curved line). In addition, ABC plots of the identity distribution, xi=constant (dotted line, that is, each genetic variant occurs at the same number in each group), and of the uniform distribution in the data range (dotted line, that is, each variant allele had the same chance to occur) are shown. Compared with the latter distributions, the solid curved line clearly indicates a highly unequal distribution of the group differences in the numbers of variant alleles. Further marks in this plot consist of a light grey and a darker grey star denoting the so-called Pareto and BreakEven points, respectively. The Pareto point A (Ax, Ay) is the point at the smallest distance (left oblique black line) to the ideal point at xy where the effort would be zero to obtain the whole yield. The BreakEven point B (Bx, By) marks the point on the ABC curve where its slope, dY/dE, equals 1, that is, the so-called profit gain dABC equals 1. Beyond that point, more information can only be gained with inadequately high efforts. The ABC analysis comes from economical informatics and, in the present context, aims at identifying the most informative genetic variant for group classification by dividing the 152 variants into 3 distinct subsets. Set A should contain the ‘critical few’, that is, those elements that allow obtaining a maximum of yield with a minimal effort.14 Set B comprises those elements where an increase in effort is proportional to the increase in yield. In contrast, set C contains the ‘trivial many’, that is, those elements that are not worth to be considered a biomarker. As a result, set A was used to establish a subsymbolic classifying biomarker.
Discussion
The present analysis showed that patterns of opioid receptor genotypes indeed provide a basis for the high opioid dose occasionally observed in pain patients. However, this was obtained using a ‘self-learning subsymbolic high-dimensional classifying biomarker’. That is, in machine learning a classifier is called symbolic if it can in principle answer the question of why a given data set has been assigned to a particular group; however, it is in principle not possible to get a reason for a particular classification. The presently used kNN classifier belongs to this type of algorithms. Its functioning is like an associative memory. For a given case the kNN classifier searches its data base of already learned correct classifications in order to find those cases that are most similar to the given data set.15 If a data set has been classified successfully to its proper class, the data base of the classifier can be enhanced with this data set and classification (‘machine learning’). In analogy to the subsymbolic/symbolic concept in machine learning we define a subsymbolic biomarker as a pattern of markers, here represented by the genetic changes, where (1) none of the markers needs to be directly related to any known biological function/process or a cellular component, although this is not excluded; and (2) the sequence of marker changes is not important, that is, the marker does not retrieve its classificatory value by its place in a sequence such as a nucleic or amino acid sequence in genes or proteins. In contrast, a symbolic biomarker would represent a genetic change that can be directly related to a biological function/process or a cellular component. However, if the pattern of the subsymbolic biomarkers is observed in an organism, it can be connected to biological function processes or a cellular component. This points at emergent properties in genetics.10
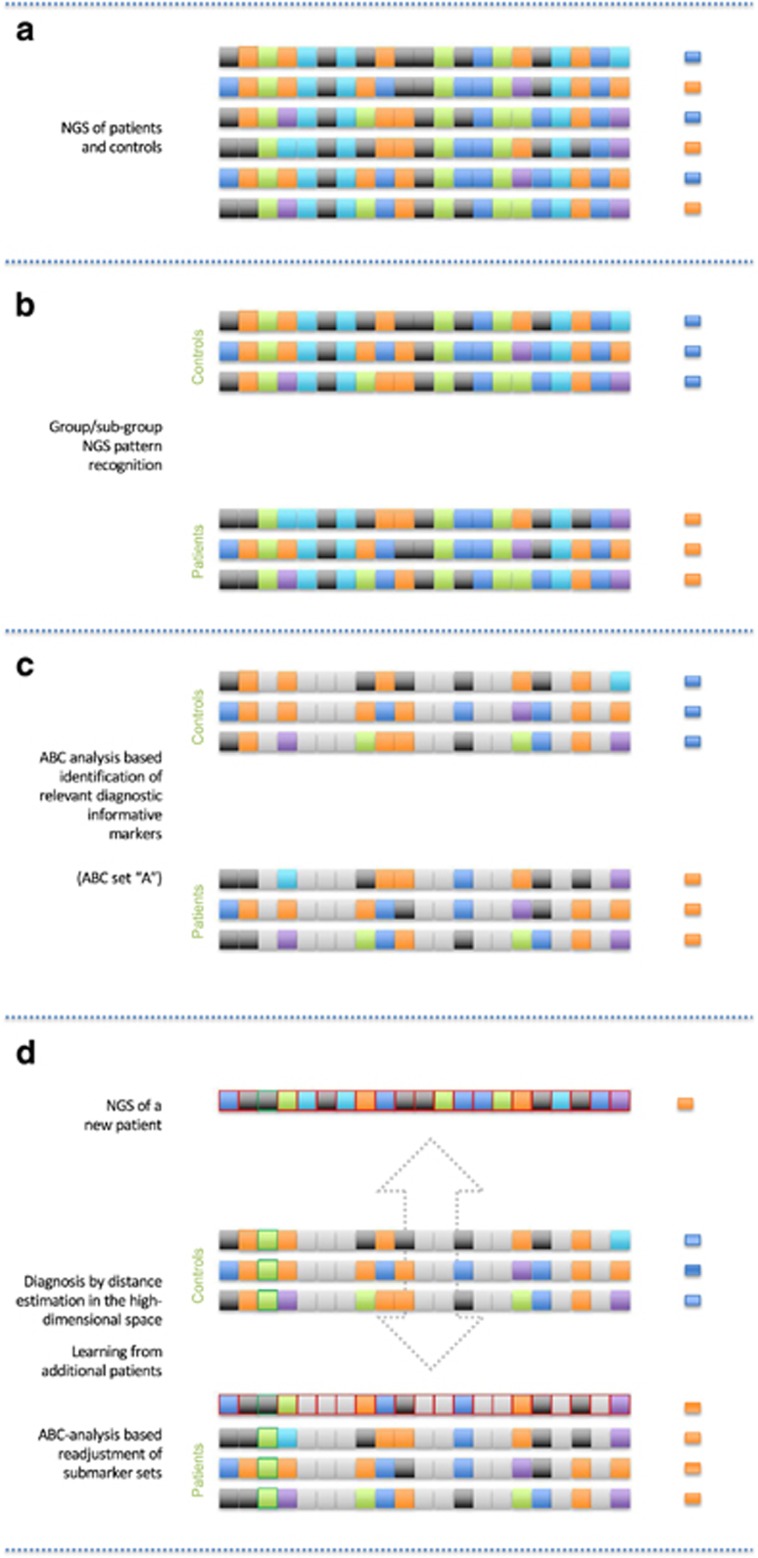
A subsymbolic high-dimensional classifying biomarker uses subsymbolic biomarkers and subsymbolic classifiers in order to decide between different biological conditions/classes/clusters. It is called self-learning, if successfully classified data are added to the knowledge base of the classifier. This can be called a ‘self-learning subsymbolic high-dimensional classifying biomarker’ (Figure 4). The present information from 30+28 patients can be successively enlarged by adding further patients. In the case that a new opioid receptor genotype pattern will be correctly identified by comparison with the present information from these 58 patients, the biomarker would be successful. In the opposite case, a wrongly classified genotype pattern can be implemented into the present data basis and new ABC marker sets. Jaccard distances can be computed to improve further diagnoses by a continuously developing system. Thus, the biomarker can ‘learn’.
Figure 4.
Schematic representation of main features of a prediction using the proposed ‘high-dimensional subsymbolic biomarker’ (from top to bottom). (a) The biomarker creation starts from the analysis of the relevant single markers, gene sequences, symbolized as arbitrarily colored squares (for example, let this be nucleotides and their heterozygous or homozygous presences). In the present analysis, let these squares denote the variants in the opioid receptor genes. (b) Based on the clinical background, the complex markers were grouped into either high-opioid doses demanding patients (orange small squares at the right) or controls (blue small squares at the right). Subgroups within the groups are possible and should be addressed by clustering (not shown). (c) From these composed makers, ABC analysis identifies the relevant submarkers that are most informative for the prediction (noninformative submarkers grayed out). (d) A new patient is analyzed (markers with red margins). Based on distance measuring in the high-dimensional space, this patient will be assigned to the most similar group, that is, to its nearest neighbors in the high-dimensional space based on the chosen distance measure (here, the Jaccard distance was used for genetic markers being either 0, 1 or 2). Concomitantly a learning process of the biomarker starts. In the present example, it proves useful, in a new ABC analysis, to include a further marker that, in a previous ABC analysis, had been found uninformative (third marker from the left, green margins). The process is repeated (dotted arrow) with each prediction where the disease background is known, that is, the biomarker ‘learns’ in the sense of Artificial Intelligence, where valid information about a patient’s background is used to improve the marker. This will improve its predictive accuracy in cases where the background is not known.
One of the keys to the success of the classification was the accounting for the heterogeneity of the patients. Beyond the main phenotype of high opioid dosage, the present patients were indeed a heterogeneous group. They had been submitted to our pharmacogenetic counseling mostly without further details of the clinical background except for a few cases such as those presented. Therefore, patients have different diseases underlying the pain. The high opioid doses may be accidental as most patients were sent from University tertiary care centers where the physicians were more inclined to raise the opioid doses, whereas in the periphery, the same patients might have been labeled as opioid resistant already at doses below 400 mg OME per day and therefore not included into this analysis. An opioid resistance might indeed apply to one of the presented patients as discussed below. Thus, the heterogeneity of opioid-treated pain patients is a clinical reality and has to be considered in a successful biomarker. Here, a self-learning biomarker provides an ideal basis.
However, although the subsymbolic biomarker successfully classified the patients, the question remained of how genetic changes of no singular identifiable major molecular consequence can nevertheless underlie the phenotype. A possible explanation may be the above-mentioned emergent properties in genetics10 to which the biomarker pointed. Specifically, we hypothesize that from the accumulation of several genetic variants that mostly provide only small quantitative modulations of gene transcription, a qualitative genetic change can emerge toward a substantially more or less efficiently transcribed gene that then gains the phenotypic consequences that were not produced by any single variant alone.
Indeed, molecular knowledge about members of the ‘A’ set of the ABC analysis (Figure 3) supports this hypothesis, and this will be illustrated by two examples. The Chr6:154451812-SNV, and various further variants that occurred only in patients receiving high analgesic opioid doses (not shown), is noncoding; yet, it is known that such variants can affect mRNA splicing, stability and structure, resulting in a reduced transcriptional efficiency.16, 17 Indeed, genome-wide association studies yielded that besides mutations in the coding regions of genes, even mutations in noncoding and intergenic regions can be associated with diseases.18 These changes can affect the function of proteins, change the cellular response to therapeutic targets and can explain the different responses of individual patients to medications.19 Furthermore, the Chr8:54147491-SNV variant (c.438G>T) changes the codon ATA, which translates to isoleucine, to codon ATC, which also translates to isoleucine. Although apparently this mutation does not lead to an alteration in the primary polypeptide sequence, that is, it is synonymous, this kind of mutation is now widely acknowledged to be able to cause changes in protein expression, conformation and function.20 Recent genetic and biomedical studies have identified multiple mechanisms that provide an indication of the means by which synonymous mutations can affect physiological changes and consequently influence disease.21
A synonymous substitution might produce changes in the phenotype by affecting splicing that in consequence can lead to a shortened mRNA and subsequently a nonfunctional protein.21, 22 Moreover, synonymous mutations might alter the mRNA stability, leading to faster degradation and reduced protein expression.23, 24 Another possibility of how synonymous mutations may influence protein levels is affecting translation elongation. The relative synonymous codon usage (RSCU) value represents the local translation elongation rates and is directly linked with protein expression, as the speed of translation is often important for accurate protein folding. For example, a negative ΔRSCU value (when a mutation introduces a rarer codon) might lead to a slower rate of translation elongation compared with the wild type.21 The c.438G>T variant causes, as a consequence of codon modification, a change in the RSCU value (the RSCU value changes from 1.41 to 0.51 with ΔRSCU=−0.9)25 as queried from the Codon Usage Database at www.kazusa.or.jp/codon/. A negative ΔRSCU value means that the mutation introduces a rarer codon and this might be associated with a slower rate of translation elongation compared with the wild type.21
Taking this idea one step further, multiple mutations that affect codon usage thus might increase or decrease translation rates and therefore appreciably bias protein expression. In its consequence, the opioidergic system might have been altered to a degree that translated into a clinical phenotype. Specifically, the present subsymbolic classifying biomarker included 21 OPRM1 variants, 9 OPRK1 variants and 3 OPRD1 variants. All coded receptors are involved in the endogenous nocifensive system. Hence, a pharmacological interpretation of the present results may involve the hypothesis of a general weakening of the endogenous opioid system, mainly but not exclusively μ-opioidergic, that was caused by the accumulation of genetic variants. This may have finally rendered the patient as nearly ‘opioid resistant’, directly because of a reduced activity of exogenous μ-opioid agonists and indirectly also owing to a disturbed opioidergic system. Alternatively, as the subsymbolic nature of the biomarker involves that its classification performance cannot be definitely assigned to one of its components, it could turn out that the main contributors to the successful classification are variants in the OPRM1 gene. This is also well within the possibility of a self-learning classifier that, as explained above, by successively re-evaluating and modifying its composition, may still evolve toward a OPRM1 variant-based classifier. In that case, the association with a reduced activity of mainly μ-opioid receptor agonists would become straightforward.
This analysis showed that opioid receptor genotyping, consistent with biological plausibility, has the potential to provide the desired predictively of particular (clinical) phenotypes as demonstrated with high opioid dose demands in pain patients. This may be obtained with a novel type of biomarker, called ‘self-learning subsymbolic high-dimensional classifying biomarker’. A self-learning biomarker offers an ideal basis to account for the heterogeneity of patients as it uses flexible patterns for the prediction. This is a novel concept of biomarkers and it is not limited to the present clinical diagnostic tasks. Related to this concept, results of the analysis can also be taken as evidence for emergent properties in genetics.10
Acknowledgments
This work has been funded by the European Union Seventh Framework Programme (FP7/2013) under Grant Agreement No. 602919 (JL, GLORIA). An additional contribution, in particular the funding of the next-generation genotyping equipment, has been received from the Landesoffensive zur Entwicklung wissenschaftlich-ökonomischer Exzellenz (LOEWE), Schwerpunkt: Anwendungsorientierte Arzneimittelforschung (JL). CS was supported by EU FP7-HEALTH-2013-INNOVATION-1 No. 602891-2.
Footnotes
Supplementary Information accompanies the paper on the The Pharmacogenomics Journal website (http://www.nature.com/tpj)
The authors declare no conflict of interest.
Supplementary Material
References
- Lötsch J, Geisslinger G. Relevance of frequent mu-opioid receptor polymorphisms for opioid activity in healthy volunteers. Pharmacogenomics J 2006; 6: 200–210. [DOI] [PubMed] [Google Scholar]
- Mardis ER. The impact of next-generation sequencing technology on genetics. Trends Genet 2008; 24: 133–141. [DOI] [PubMed] [Google Scholar]
- Nielsen R, Paul JS, Albrechtsen A, Song YS. Genotype and SNP calling from next-generation sequencing data. Nat Rev Genet 2011; 12: 443–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pabinger S, Dander A, Fischer M, Snajder R, Sperk M, Efremova M et al. A survey of tools for variant analysis of next-generation genome sequencing data. Brief Bioinform 2014; 15: 256–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarovich DS, Price EP. SPANDx: a genomics pipeline for comparative analysis of large haploid whole genome re-sequencing datasets. BMC Res Notes 2014; 7: 618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoddard JL, Niemela JE, Fleisher TA, Rosenzweig SD. Targeted NGS: a cost-effective approach to molecular diagnosis of PIDs. Front Immunol 2014; 5: 531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galindo-González L, Pinzón-Latorre D, Bergen EA, Jensen DC, Deyholos MK. Ion Torrent sequencing as a tool for mutation discovery in the flax (Linum usitatissimum L.) genome. Plant Methods 2015; 11: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glotov AS, Kazakov SV, Zhukova EA, Alexandrov AV, Glotov OS, Pakin VS et al. Targeted next-generation sequencing (NGS) of nine candidate genes with custom AmpliSeq in patients and a cardiomyopathy risk group. Clin Chim Acta 2015; 446: 132–140. [DOI] [PubMed] [Google Scholar]
- Tenedini E, Artuso L, Bernardis I, Artusi V, Percesepe A, De Rosa L et al. Amplicon-based NGS: an effective approach for the molecular diagnosis of Epidermolysis Bullosa. Br J Dermatol 2015; 173: 731–738. [DOI] [PubMed] [Google Scholar]
- Ultsch A (ed). Emergence in Self-Organizing Feature Maps In: International Workshop on Self-Organizing Maps (WSOM '07). Neuroinformatics Group: Bielefeld, Germany, 2007. [Google Scholar]
- Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B et al. The structure of haplotype blocks in the human genome. Science 2002; 296: 2225–2229. [DOI] [PubMed] [Google Scholar]
- Daly MJ, Rioux JD, Schaffner SF, Hudson TJ, Lander ES. High-resolution haplotype structure in the human genome. Nat Genet 2001; 29: 229–232. [DOI] [PubMed] [Google Scholar]
- Cover TM, Hart PE. Nearest neighbor pattern classification. IEEE Trans Inform Theory 1967; 13: 21–27. [Google Scholar]
- Ultsch A, Lotsch J. Computed ABC analysis for rational selection of most informative variables in multivariate data. PLoS One 2015; 10: e0129767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altman NS. An introduction to kernel and nearest-neighbor nonparametric regression. Am Stat 1992; 46: 175–185. [Google Scholar]
- Fung KL, Gottesman MM. A synonymous polymorphism in a common MDR1 (ABCB1) haplotype shapes protein function. Biochim Biophys Acta 2009; 1794: 860–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung KL, Pan J, Ohnuma S, Lund PE, Pixley JN, Kimchi-Sarfaty C et al. MDR1 synonymous polymorphisms alter transporter specificity and protein stability in a stable epithelial monolayer. Cancer Res 2014; 74: 598–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treutlein J, Cichon S, Ridinger M, Wodarz N, Soyka M, Zill P et al. Genome-wide association study of alcohol dependence. Arch Gen Psychiatry 2009; 66: 773–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glinskii AB, Ma J, Ma S, Grant D, Lim CU, Sell S et al. Identification of intergenic trans-regulatory RNAs containing a disease-linked SNP sequence and targeting cell cycle progression/differentiation pathways in multiple common human disorders. Cell Cycle 2009; 8: 3925–3942. [DOI] [PubMed] [Google Scholar]
- Hunt R, Sauna ZE, Ambudkar SV, Gottesman MM, Kimchi-Sarfaty C. Silent (synonymous) SNPs: should we care about them? Methods Mol Biol 2009; 578: 23–39. [DOI] [PubMed] [Google Scholar]
- Sauna ZE, Kimchi-Sarfaty C. Understanding the contribution of synonymous mutations to human disease. Nat Rev Genet 2011; 12: 683–691. [DOI] [PubMed] [Google Scholar]
- Wang GS, Cooper TA. Splicing in disease: disruption of the splicing code and the decoding machinery. Nat Rev Genet 2007; 8: 749–761. [DOI] [PubMed] [Google Scholar]
- Duan J, Wainwright MS, Comeron JM, Saitou N, Sanders AR, Gelernter J et al. Synonymous mutations in the human dopamine receptor D2 (DRD2) affect mRNA stability and synthesis of the receptor. Hum Mol Genet 2003; 12: 205–216. [DOI] [PubMed] [Google Scholar]
- Nackley AG, Shabalina SA, Tchivileva IE, Satterfield K, Korchynskyi O, Makarov SS et al. Human catechol-O-methyltransferase haplotypes modulate protein expression by altering mRNA secondary structure. Science 2006; 314: 1930–1933. [DOI] [PubMed] [Google Scholar]
- Sharp PM, Tuohy TM, Mosurski KR. Codon usage in yeast: cluster analysis clearly differentiates highly and lowly expressed genes. Nucleic Acids Res 1986; 14: 5125–5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke TK, Crist RC, Kampman KM, Dackis CA, Pettinati HM, O'Brien CP et al. Low frequency genetic variants in the mu-opioid receptor (OPRM1) affect risk for addiction to heroin and cocaine. Neurosci Lett 2013; 542: 71–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabalina SA, Zaykin DV, Gris P, Ogurtsov AY, Gauthier J, Shibata K et al. Expansion of the human mu-opioid receptor gene architecture: novel functional variants. Hum Mol Genet 2009; 18: 1037–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lötsch J, Skarke C, Grösch S, Darimont J, Schmidt H, Geisslinger G. The polymorphism A118G of the human mu-opioid receptor gene decreases the clinical activity of morphine-6-glucuronide but not that of morphine. Pharmacogenetics 2002; 12: 3–9. [DOI] [PubMed] [Google Scholar]
- Bond C, LaForge KS, Tian M, Melia D, Zhang S, Borg L et al. Single-nucleotide polymorphism in the human mu opioid receptor gene alters beta-endorphin binding and activity: possible implications for opiate addiction. Proc Natl Acad Sci USA 1998; 95: 9608–9613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S-C, Tsou H-H, Chen C-H, Chen Y-T, Ho I-K, Hsiao C-F et al. Genetic polymorphisms in the opioid receptor mu1 gene are associated with changes in libido and insomnia in methadone maintenance patients. Eur Neuropsychopharmacol 2012; 22: 695–703. [DOI] [PubMed] [Google Scholar]
- Lettre G, Palmer CD, Young T, Ejebe KG, Allayee H, Benjamin EJ et al. Genome-wide association study of coronary heart disease and its risk factors in 8,090 African Americans: the NHLBI CARe Project. PLoS Genet 2011; 7: e1001300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garriock HA, Tanowitz M, Kraft JB, Dang VC, Peters EJ, Jenkins GD et al. Association of mu-opioid receptor variants and response to citalopram treatment in major depressive disorder. Am J Psychiatry 2010; 167: 565–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doehring A, Küsener N, Fluhr K, Neddermeyer TJ, Schneider G, Lötsch J. Effect sizes in experimental pain produced by gender, genetic variants and sensitization procedures. PLoS One 2011; 6: e17724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levran O, Londono D, O'Hara K, Nielsen DA, Peles E, Rotrosen J et al. Genetic susceptibility to heroin addiction: a candidate gene association study. Genes Brain Behav 2008; 7: 720–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen S, Wang C, Berg A, Li Y, Chang MM, Fillingim RB et al. Modeling genetic imprinting effects of DNA sequences with multilocus polymorphism data. Algorithms Mol Biol 2009; 4: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishi T, Yoshimura R, Okochi T, Fukuo Y, Kitajima T, Okumura T et al. Association analysis of SIGMAR1 with major depressive disorder and SSRI response. Neuropharmacology 2010; 58: 1168–1173. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.