Abstract
Background
Protease-activated receptor-1 (PAR-1) potentiates diabetic nephropathy (DN) as evident from reduced kidney injury in diabetic PAR-1 deficient mice. Although thrombin is the prototypical PAR-1 agonist, anticoagulant treatment does not limit DN in experimental animal models suggesting that thrombin is not the endogenous PAR-1 agonist driving DN.
Objectives
To identify the endogenous PAR-1 agonist potentiating diabetes-induced nephropathy.
Methods
Unbiased protease expression profiling in glomeruli from human kidneys with DN was performed using publically available microarray data. The identified prime candidate PAR-1 agonist was subsequently analysed for PAR-1-dependent induction of fibrosis in vitro.
Results
Of the 553 proteases expressed in the human genome, 247 qualified as potential PAR-1 agonists of which 71 were significantly expressed above background in diabetic glomeruli. The recently identified PAR-1 agonist plasmin(ogen), together with its physiological activator tissue plasminogen activator, were among the highest expressed proteases. Plasmin did however not induce mesangial proliferation and/or fibronectin deposition in vitro. In a PAR-1 independent manner, plasmin even reduced fibronectin deposition.
Conclusion
Expression profiling identified plasmin as potential endogenous PAR-1 agonist driving DN. Instead of inducing fibronectin expression, plasmin however reduced mesangial fibronectin deposition in vitro. Therefore we conclude that plasmin may not be the endogenous PAR-1 agonist potentiating DN.
Keywords: PAR-1, Diabetic nephropathy, Plasmin, Protease profiling
Highlights
-
•
Plasmin is highly expressed in kidneys of diabetic nephropathy patients.
-
•
Plasmin limits fibronectin deposition by mesangial cells.
-
•
Plasmin-dependent PAR-1 activation does not drive diabetic nephropathy.
1. Introduction
The World Health Organization approximates that over 300 million people will suffer from diabetes in 2025 [1]. The health implications of this endemic disease are expected to be larger as diabetic patients frequently develop complications like (among others) diabetic nephropathy leading to end-stage renal disease (ESRD) [2]. Diabetic nephropathy actually emerged as the major causative pathology in patients entering ESRD worldwide and it is responsible for 30–40% of all ESRD cases. In individuals with diabetes, the presence and severity of nephropathy adversely affects their well-being, significantly contributes to disease morbidity and increases their risk of a premature death [3], [4]. Although the progression of diabetic nephropathy can be delayed by strict control of plasma glucose levels and/or by lowering blood pressure, the majority of patients eventually need renal replacement therapy. The large impact of this latter therapy, both on the social and economic level [5], [6], urges the need for alternative treatment options.
In the search for alternative targets to pursue in combatting diabetic nephropathy, we recently identified protease-activated receptor (PAR)−1 as an attractive candidate. Indeed, PAR-1 deficient mice showed reduced diabetes-induced albuminuria, plasma cystatin C levels, mesangial expansion and tubular atrophy as compared to wild type diabetic controls [7]. Subsequent mechanistic experiments showed that PAR-1 activation induces proliferation and fibronectin production by MES13 mesangial cells in vitro.
PAR-1 is a seven transmembrane domain receptor that is activated by proteolytic cleavage rather than by ligand binding [8], [9], [10]. Generally, PAR-1 is recognized as a blood coagulation factor receptor and thrombin is considered the prototypical PAR-1 agonist. Importantly however, anticoagulation with low-molecular-weight heparin did not protect against diabetic nephropathy in diabetic wild type mice despite the fact that it normalized markers of coagulation (i.e. thrombin-antithrombin, d-dimer and renal fibrin deposition). Indeed, albuminuria, kidney weight, histological PAS scores and glomerular size were similar in saline and low-molecular-weight heparin treated diabetic mice [11]. Similarly, the direct thrombin inhibitor hirudin had also no significant effect on key parameters of nephropathy in diabetic wild type mice [12]. In the setting of diabetic nephropathy, thrombin may therefore not be the endogenous PAR-1 agonist potentiating kidney injury.
Next to thrombin, activated protein C (APC) is a well-recognized PAR-1 agonist [13]. APC-dependent PAR-1 activation, however, prevents hyperglycemia-induced apoptosis of endothelial cells and podocytes in vitro, whereas APC overexpression reduces diabetes-induced kidney injury in vivo [11]. These data indicate that APC is not the endogenous PAR-1 agonist potentiating diabetic nephropathy but also imply that targeting the PAR-1 agonist driving diabetic nephropathy might be a more effective treatment strategy than targeting the receptor itself. Indeed, preventing PAR-1-dependent mesangial expansion and tubular atrophy without blocking the beneficial PAR-1 effects on endothelial cells and podocytes may even further decrease diabetes-induced nephropathy.
To fully appreciate the importance and potential clinical relevance of the PAR-1 pathway in diabetic nephropathy, it is important to identify the endogenous PAR-1 agonist that potentiates kidney injury during diabetes. In the current manuscript, we therefore aimed to pinpoint candidate proteases as endogenous PAR-1 agonists potentiating diabetic nephropathy. To this end, we employed an unbiased approach in which expression levels of all proteases expressed in the human genome were assessed in diabetic glomeruli.
2. Materials and methods
2.1. Mining of publically available RNA microarray dataset
The GSE10009 [14] dataset was downloaded from Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/gds). This dataset reports whole-genome gene expression of glomeruli isolated from patients with diabetes mellitus. Background expression was determined for individual chips using Affymetrix negative control probes and protease expression levels above the mean+2SD were considered significantly expressed and were used for further analysis. Subcellular localisation of proteases was assessed using the UniProtKB/Swiss-Prot and COMPARTMENTS databases [15].
2.2. Cell culture and stimulation
Mouse mesangial cells (SV40 MES13; CRL-1927 ATCC) were cultured according to the recommended protocol (https://www.lgcstandards-atcc.org/Products/Cells_and_Microorganisms/By_Tissue/Kidney/CRL-1927.aspx?geo_country=nl#culturemethod) using a 3:1 mixture of Dulbecco's Modified Eagle's Medium containing 1 g/L glucose with Ham's-F12 medium, supplemented with 5% heat inactivated fetal calf serum, 100 U/ml penicillin, 100 µg/ml streptomycin and 2 mM l-glutamine. Cells were cultured at 37 °C in an atmosphere of 5% CO2. Cells were serum starved overnight in low glucose (1 g/L) medium before stimulation with 100 μM PAR-1 agonist peptide (PAR-1-AP; H-SFLLRN-NH2; Biochem, Shanghai, China) or 2 or 8 μg/ml plasmin (Biopur, Switzerland) in high glucose (4 g/L) medium (concentrations based on [16], [17] for PAR-1-AP and plasmin, respectively). If indicated, cells were pretreated with 10 μM PAR-1 pepducin (P1pal12; palmitate-RCLSSSAVANRS-NH2; Biochem, Shanghai, China) or 100 nM vorapaxar (AdooQ BioScience, Irvine, CA; concentration based on [18], [19] for P1pal12 and vorapaxar, respectively).
2.3. MTT assay
Cells were seeded at a density of 5000 cells/well in 96 well plates. After stimulation with plasmin or PAR-1 agonist peptide for 24 h, MTT (St. Louis, MO, USA) was added to the culture medium. After 2 h incubation at 37 °C, cells were lysed with DMSO and OD570 was measured using a microplate reader (Synergy HT, BioTek).
2.4. Western Blot
Cells were seeded at a density of 50000 cells/well in 24 well plates. After stimulation for 24 h, cells were lysed with RIPA lysis buffer (50 mM Tris HCl (pH 7.4), 150 mM NaCl, 0.5% deoxycholate, 1.0% Triton X-100, 0.1% SDS and 1 mM EGTA) supplemented with 1x Halt Protease and Phosphatase Inhibitor Cocktail (Thermo Scientific, #78445). Cell lysates were subsequently diluted (1:1) in Laemmli buffer and separated on 10% SDS-PAGE gel and transferred onto Immobulin-PL membranes (Millipore) as described before [20]. Membranes were blocked for 1 h at room temperature in 5% bovine serum albumin (BSA) in TBS+0,1% tween-20 (TBS-T) and subsequently incubated with the following primary antibodies, diluted in TBS-T: mouse-anti-tubulin 1:2500 (Santa Cruz; sc-23948) or goat-anti-fibronectin 1:1000 (Santa Cruz; sc-6953). After overnight incubation, the membranes were washed 3 times with TBS-T and incubated 1 h at room temperature with horseradish peroxidase (HRP)-conjugated (1:1000, DakoCytomation, Glostrup, Denmark) secondary antibodies diluted 1:5000 in TBS-T. Membranes were washed 3 times with TBS-T and imaged using Luminata Forte western blot substrate (Merck Millipore, Billerica, Massachusetts, USA) on an ImageQuant LAS 4000 biomolecular imager (GE Healthcare, Zeist, the Netherlands).
2.5. RNA isolation and RT-qPCR
Cells were seeded at a density of 50000 cells/well in 24 well plates. After stimulation for the indicated time points, mRNA was isolated using TriReagent isolation reagent (#11667165001; Roche Diagnostics) according to the manufacturers recommendations. All mRNA samples were quantified by spectrophotometry and stored at −80 °C until further analysis. 0.75 μg of mRNA was treated with DNAse using the RQ1 DNAse kit (M6101, Promega, Madison, WI, USA) and subsequently converted to cDNA using M-MLV reverse transcriptase (M1705, Promega, Madison, WI, USA) and random hexamer primers (#SO142, Fisher scientific, Landsmeer, the Netherlands) according to the manufacturers recommendations. qPCR and subsequent analysis were performed using a Roche lightcycler with SYBR green PCR master mix (#04707516001; Roche, Almere, the Netherlands) on a Lightcycler 480 machine and corresponding software (Software release 1.5.0 (1.5.0.39), Roche, Almere, the Netherlands). Expression levels were normalized using the average expression levels of HPRT and TBP. The following primer sequences were used: Fibronectin forward 5′-CCATGTAGGAGAACAGTGGCA-3′ and reverse 5′-GAAGCACTCAATGGGGCA-3′; TBP forward: 5′-GGAGAATCATGGACCAGAACA-3′ and reverse: 5′-GATGGGAATTCCAGGAGTCA-3′; HPRT forward: 5′-TCCTCCTCAGACCGCTTTT-3′ and reverse: 5′-CCTGGTTCATCATCGCTAATC-3′.
2.6. Statistics
All values are expressed as mean ±SEM. Differences between groups were analysed using a Mann-Whitney U-test for non-parametric data. All analyses were performed using GraphPad Prism version 5.01.
3. Results
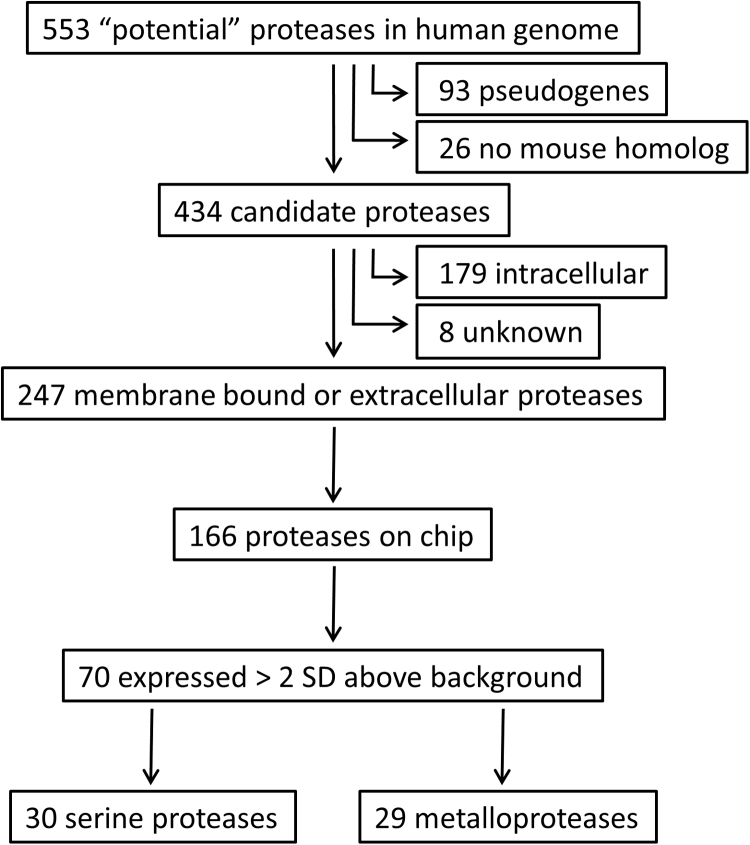
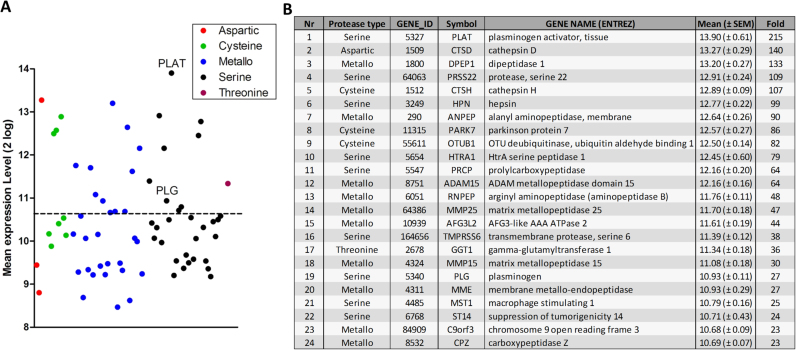
The proteolytic activation mechanism of PAR-1 dictates the endogenous PAR-1 agonist inducing glomerular expansion and subsequent diabetic nephropathy to be a protease. Consequently, we assessed protease mRNA expression levels in glomeruli obtained from patients with diabetic nephropathy using the publically available GSE1009 gene expression omnibus dataset [14]. From the total of 553 genes that have been annotated to encode proteases or protease homologues in the human genome [21], 93 genes were excluded as candidate PAR-1 agonist as they represent catalytically inactive pseudogenes and 26 were excluded as they do not have murine counterparts (Fig. 1). Based on subsequent subcellular location analysis another 187 intracellular proteases were excluded. From the remaining 247 secreted or outer cell membrane-bound proteases, expression data were available for 166 proteases of which 70 were significantly expressed above background levels (Supplemental Table 1; Fig. 2A). Serine and metalloproteases constitute the large majority of these 70 candidate PAR-1 agonists.
Fig. 1.
Schematic overview of data mining in the GS1009 database. The human genome contains 553 proteases of which 247 are catalytically active membrane bound or secreted proteases. Of these 166 were present on the Affymetrix chip and 70 were expressed above background.
Fig. 2.
Overview of proteases expressed in diabetic glomeruli. (A) Shown are all proteases with expression levels at least 2 standard deviations above the mean subdivided by protease type. The dotted line separates the upper tertile from the lower tertiles. (B) Details of the proteases in the upper tertile of expression.
Tissue plasminogen activator (tPA) is the highest expressed protease in diabetic glomeruli, whereas its physiological substrate plasminogen is also within the upper tertile of expressed proteases (Fig. 2B). This is particularly interesting as plasmin is a known PAR-1 agonist [17], [22], [23], [24], [25] and thus a likely candidate to drive PAR-1 activation during diabetic nephropathy. As shown in Fig. 3A, plasmin does not affect mesangial proliferation as opposed to PAR-1 agonist peptide that significantly induced proliferation of MES13 cells in vitro. PAR-1 activation on MES13 mesangial cells using PAR-1 agonist peptide also induced fibronectin expression, which was not observed after plasmin stimulation (Fig. 3B). In fact, plasmin even decreased fibronectin deposition by MES13 cells in a dose-dependent manner (Fig. 3C). Pretreatment of MES13 cells with the PAR-1 inhibitors P1pal12 or vorapaxar did not affect unstimulated fibronectin levels (Fig. 3D) but did also not prevent the plasmin-dependent reduction in fibronectin levels showing that this response is PAR-1 independent (Fig. 3E). The reduction in fibronectin deposition after plasmin stimulation was not accompanied by a decrease in fibronectin mRNA levels (Fig. 3F).
Fig. 3.
Plasmin limits fibronectin deposition by MES13 mesangial cells. (A) MES13 proliferation after stimulation with plasmin or PAR-1 agonist peptide (PAR-1-AP). Shown is the mean +/- SEM. (B) Western blot of fibronectin expression after plasmin (2 μg/ml) or PAR-1-AP (100 μM) simulation of mesangial cells for 24 h. (C) Fibronectin blot after stimulation with 2 or 8 μg/ml plasmin with tubulin as loading control. (D) Fibronectin expression of MES13 cells after pretreatment with PBS (control), P1pal12 or vorapaxar. (E) Fibronectin expression of MES13 cells after plasmin (2 μg/ml) or PBS stimulation in the presence of P1pal12 or vorapaxar. (F) Fibronectin mRNA levels of MES13 cells stimulated with plasmin for the indicated time intervals. Tubulin serves as loading control in all panels.
4. Discussion
PAR-1 activation potentiates diabetic nephropathy [7], but the endogenous agonist responsible for PAR-1 activation during diabetes remains elusive. In the current manuscript, we performed an unbiased protease profiling approach to identify candidate PAR-1 agonists driving diabetic nephropathy. This approach identified the tPA/plasmin axis as prime candidate but subsequent in vitro experiments seem to exclude plasmin as endogenous PAR-1 agonist aggravating diabetes-induced kidney injury.
In the upper tertile of expressed proteases, plasmin(ogen) is the only protease that has been implicated as potential PAR-1 agonist. Already in 1999, it was shown that plasmin cleaved a soluble N-terminal exodomain of PAR-1 as effectively as thrombin [23]. Plasmin-PAR-1 signaling was subsequently shown to, amongst others, induce Cyr61 expression -a growth factor-like gene involved in cell proliferation, adhesion, and migration [23]- and transforming growth factor beta production [24]. Moreover, plasmin stimulation of murine tubular epithelial cells induced their phenotypic transition to fibronectin-producing fibroblast-like cells which was inhibited by PAR-1 siRNA and by a specific PAR-1 antagonist [25]. Despite these data pinpointing plasmin-PAR-1 signaling as a key pathway in profibrotic responses, here we show that plasmin does not induce proliferation of mesangial cells whereas it even inhibits fibronectin deposition by these cells.
Although plasmin is probably best known as a key protease of the fibrinolytic pathway involved in clot lysis, it can also cleave/degrade extracellular matrix proteins like laminin and fibronectin [26]. Already in the early 1980's it was shown that plasmin efficiently cleaved purified fibronectin in vitro [27] whereas the proteolytic activity of plasmin towards fibronectin in vivo was confirmed more recently [28], [29]. As plasmin stimulation of MES13 cells did not affect fibronectin mRNA levels, reduced fibronectin deposition by MES13 cells after plasmin stimulation is most likely not the result of reduced fibrinogen synthesis but is actually caused by enhanced degradation.
Plasmin does not mimic the previously reported effect of thrombin-PAR-1 signaling on mesangial cells. Indeed, thrombin induced proliferation and extracellular matrix deposition in a PAR-1 dependent manner [7], whereas plasmin-PAR-1 signaling does not affect these processes. Such divergent responses of different PAR-1 agonists are well described, and the capacity of agonists to trigger distinct signaling pathways is referred to as biased signaling (excellently reviewed in [30]). The contrasting effects of thrombin and activated protein C (APC)-induced PAR-1 signaling on endothelial barrier function is a classic example hereof [31]. Interestingly however, PAR-1-induced biased signaling has also been described for podocytes as APC-PAR-1 signaling inhibits whereas thrombin-PAR-1 signaling induces apoptosis of podocytes [11]. Next to biased signaling, plasmin-cleaved PAR-1 may not induce profibrotic responses of mesangial cells because plasmin cleavage is not only described to activated PAR-1 signaling [17], [22], [23], [24], [25] but may actually also desensitise PAR-1 [32]. Irrespective the actual explanation, our data show that plasmin does not induce fibronectin production by mesangial cells.
The PAR-1 agonists described to date are either serine (i.e. APC [13], Factor X [33], thrombin [8], [9], [10], plasmin [22], [23], [24], [25], Granzyme K [34], kallikrein-1 [35] and −6 [36], kallikrein-related peptidase 4 [37], neutrophil elastase [38], PRSS3 [39], proteinase 3 [38]) or metalloproteases (MMP-1 [40] and MMP-13 [41]). Highly expressed proteases of these types may thus represent prime PAR-1 agonist candidates and actually all serine and metalloproteases in the upper tertile of expressed proteases in diabetic glomeruli (Fig. 2B) theoretically qualify as candidates. Except tPA/plasminogen, none of the proteases from the upper tertile have however been implicated in diabetic nephropathy and/or to modify PAR-1 signaling. Moreover, the endogenous PAR-1 agonist driving diabetic nephropathy may not be synthesized locally in the glomeruli at all, but may actually be produced at a distant site. Due to microvascular injury early on during diabetic nephropathy, the protease may reach PAR-1 expressing mesangial cells. Our approach, analysing mRNA expression levels in glomeruli, is unfortunately not qualified to identify such proteases.
Overall, unbiased protease profiling identified the tPA/plasmin axis as prime candidate for driving PAR-1-dependent diabetes-induced kidney injury but in vitro experiments do not support a role of plasmin in PAR-1-dependent fibronectin deposition. The endogenous PAR-1 agonist in the setting of diabetic nephropathy therefore remains elusive.
Acknowledgement
This work was supported by the Dutch Diabetes Foundation (2012.00.1471 and 2009.11.001). The authors declare that there is no conflict of interest associated with this manuscript.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.bbrep.2017.03.009.
Appendix A. Supplementary material
Supplementary material
.
References
- 1.Zimmet P., Alberti K.G., Shaw J. Global and societal implications of the diabetes epidemic. Nature. 2001;414:782–787. doi: 10.1038/414782a. [DOI] [PubMed] [Google Scholar]
- 2.Friedman E.A. Renal syndromes in diabetes. Endocrinol. Metab. Clin. North Am. 1996;25:293–324. doi: 10.1016/s0889-8529(05)70326-1. [DOI] [PubMed] [Google Scholar]
- 3.P.H. Groop, M.C. Thomas, J.L. Moran, et al., The presence and severity of chronic kidney disease predicts all-cause mortality in type 1 diabetes, Diabetes 58 (2209) 1651–1658. [DOI] [PMC free article] [PubMed]
- 4.Adler A.I., StevensJ R.J., Manley S.E. Development and progression of nephropathy in type 2 diabetes: the United Kingdom Prospective Diabetes Study (UKPDS 64) Kidney Int. 2003;63:225–232. doi: 10.1046/j.1523-1755.2003.00712.x. [DOI] [PubMed] [Google Scholar]
- 5.Foley R.N., Collins A.J. The growing economic burden of diabetic kidney disease. Curr. Diab. Rep. 2009;9:460–465. doi: 10.1007/s11892-009-0075-9. [DOI] [PubMed] [Google Scholar]
- 6.S.F. Niu, I.C. Li, Quality of life of patients having renal replacement therapy, J Adv Nurs. 51 (2205) 15-21. [DOI] [PubMed]
- 7.Waasdorp M., Duitman J.W., Florquin S., Spek C.A. Protease-activated receptor-1 deficiency protects against streptozotocin-induced diabetic nephropathy in mice. Sci. Rep. 2016;6:33030. doi: 10.1038/srep33030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coughlin S.R. Protease-activated receptors in vascular biology. Thromb. Haemost. 2001;86:298–307. [PubMed] [Google Scholar]
- 9.Gieseler F., Ungefroren H., Settmacher U. Proteinase-activated receptors (PARs) - focus on receptor-receptor-interactions and their physiological and pathophysiological impact. Cell Commun. Signal. 2013;11:86. doi: 10.1186/1478-811X-11-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hollenberg M.D., Mihara K., Polley D. Biased signalling and proteinase-activated receptors (PARs): targeting inflammatory disease. Br. J. Pharmacol. 2014;171:1180–1194. doi: 10.1111/bph.12544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Isermann B., Vinnikov I.A., Madhusudhan T. Activated protein C protects against diabetic nephropathy by inhibiting endothelial and podocyte apoptosis. Nat. Med. 2007;13:1349–1358. doi: 10.1038/nm1667. [DOI] [PubMed] [Google Scholar]
- 12.Wang H., Madhusudhan T., He T. Low but sustained coagulation activation ameliorates glucose-induced podocyte apoptosis: protective effect of factor V Leiden in diabetic nephropathy. Blood. 2011;117:5231–5242. doi: 10.1182/blood-2010-10-314773. [DOI] [PubMed] [Google Scholar]
- 13.Mosnier L.O., Zlokovic B.V., Griffin J.H. The cytoprotective protein C pathway. Blood. 2007;109:3161–3172. doi: 10.1182/blood-2006-09-003004. [DOI] [PubMed] [Google Scholar]
- 14.Baelde H.J., Eikmans M., Doran P.P. Gene expression profiling in glomeruli from human kidneys with diabetic nephropathy. Am. J. Kidney Dis. 2004;43:636–650. doi: 10.1053/j.ajkd.2003.12.028. [DOI] [PubMed] [Google Scholar]
- 15.J.X. Binder, S. Pletscher-Frankild, K. Tsafou K, et al., COMPARTMENTS: unification and visualization of protein subcellular localization evidence, Database (Oxford) bau012, 2014. [DOI] [PMC free article] [PubMed]
- 16.Lin C., Duitman J., Daalhuisen J. Targeting protease activated receptor-1 with P1pal-12 limits bleomycin-induced pulmonary fibrosis. Thorax. 2014;69:152–160. doi: 10.1136/thoraxjnl-2013-203877. [DOI] [PubMed] [Google Scholar]
- 17.Mandal S.K., Rao L.V., Tran T.T. A novel mechanism of plasmin-induced mitogenesis in fibroblasts. J. Thromb. Haemost. 2005;3:163–169. doi: 10.1111/j.1538-7836.2004.01054.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin C., Rezaee F., Waasdorp M. Protease activated receptor-1 regulates macrophage-mediated cellular senescence: a risk for idiopathic pulmonary fibrosis. Oncotarget. 2015;6:35304–35314. doi: 10.18632/oncotarget.6095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang C., Srinivasan Y., Arlow D.H. High-resolution crystal structure of human protease-activated receptor 1. Nature. 2012;492:387–392. doi: 10.1038/nature11701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Borensztajn K., Stiekema J., Nijmeijer S. Factor Xa stimulates proinflammatory and profibrotic responses in fibroblasts via protease-activated receptor-2 activation. Am. J. Pathol. 2008;172:309–320. doi: 10.2353/ajpath.2008.070347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Puente X.S., Sánchez L.M., Overall CM C.M. Human and mouse proteases: a comparative genomic approach. Nat. Rev. Genet. 2003;4:544–558. doi: 10.1038/nrg1111. [DOI] [PubMed] [Google Scholar]
- 22.Pendurthi U.R., Ngyuen M., Andrade-Gordon P. Plasmin induces Cyr61 gene expression in fibroblasts via protease-activated receptor-1 and p44/42 mitogen-activated protein kinase-dependent signaling pathway. Arterioscler. Thromb. Vasc. Biol. 2002;22:1421–1426. doi: 10.1161/01.atv.0000030200.59331.3f. [DOI] [PubMed] [Google Scholar]
- 23.Carmo A.A., Costa B.R., Vago J.P. Plasmin induces in vivo monocyte recruitment through protease-activated receptor-1-, MEK/ERK-, and CCR2-mediated signaling. J. Immunol. 2014;193:3654–3663. doi: 10.4049/jimmunol.1400334. [DOI] [PubMed] [Google Scholar]
- 24.Maeda S., Nakajima K., Tohyama Y. Characteristic response of astrocytes to plasminogen/plasmin to upregulate transforming growth factor beta 3 (TGFbeta3) production/secretion through proteinase-activated receptor-1 (PAR-1) and the downstream phosphatidylinositol 3-kinase (PI3K)-Akt/PKB signaling cascade. Brain Res. 2009;1305:1–13. doi: 10.1016/j.brainres.2009.09.025. [DOI] [PubMed] [Google Scholar]
- 25.Zhang G., Kernan K.A., Collins S.J. Plasmin(ogen) promotes renal interstitial fibrosis by promoting epithelial-to-mesenchymal transition: role of plasmin-activated signals. J. Am. Soc. Nephrol. 2007;18:846–859. doi: 10.1681/ASN.2006080886. [DOI] [PubMed] [Google Scholar]
- 26.Deryugina E.I., Quigley J.P. Cell surface remodeling by plasmin: a new function for an old enzyme. J. Biomed. Biotechnol. 2012;2012:564259. doi: 10.1155/2012/564259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liotta L.A., Goldfarb R.H., Brundage R. Effect of plasminogen activator (urokinase), plasmin, and thrombin on glycoprotein and collagenous components of basement membrane. Cancer Res. 1981;41:4629–4636. [PubMed] [Google Scholar]
- 28.Horowitz J.C., Rogers D.S., Simon R.H. Plasminogen activation-induced pericellular fibronectin proteolysis promotes fibroblast apoptosis. Am. J. Respir. Cell Mol. Biol. 2008;38:78–87. doi: 10.1165/rcmb.2007-0174OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tjwa M., Moura R., Moons L. Fibrinolysis-independent role of plasmin and its activators in the haematopoietic recovery after myeloablation. J. Cell Mol. Med. 2009;13:4587–4595. doi: 10.1111/j.1582-4934.2008.00521.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao P., Metcalf M., Bunnett N.W. Biased signaling of protease-activated receptors. Front. Endocrinol. 2014;5(9):67. doi: 10.3389/fendo.2014.00067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Feistritzer C., Riewald M. Endothelial barrier protection by activated protein C through PAR1-dependent sphingosine 1-phosphate receptor-1 crossactivation. Blood. 2005;105:3178–3184. doi: 10.1182/blood-2004-10-3985. [DOI] [PubMed] [Google Scholar]
- 32.Ruf W., Dorfleutner A., Riewald M. Specificity of coagulation factor signaling. J. Thromb. Haemost. 2003;1:1495–1503. doi: 10.1046/j.1538-7836.2003.00300.x. [DOI] [PubMed] [Google Scholar]
- 33.Kuliopulos A., Covic L., Seeley S.K. Plasmin desensitization of the PAR1 thrombin receptor: kinetics, sites of truncation, and implications for thrombolytic therapy. Biochemistry. 1999;38:4572–4585. doi: 10.1021/bi9824792. [DOI] [PubMed] [Google Scholar]
- 34.Cooper D.M., Pechkovsky D.V., Hackett T.L. Granzyme K activates protease-activated receptor-1. PLoS One. 2011;6:e21484. doi: 10.1371/journal.pone.0021484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gao L., Chao L., Chao J. A novel signaling pathway of tissue kallikrein in promoting keratinocyte migration: activation of proteinase-activated receptor 1 and epidermal growth factor receptor. Exp. Cell Res. 2010;316:376–389. doi: 10.1016/j.yexcr.2009.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yoon H., Radulovic M., Wu J. Kallikrein 6 signals through PAR1 and PAR2 to promote neuron injury and exacerbate glutamate neurotoxicity. J. Neurochem. 2013;127:283–298. doi: 10.1111/jnc.12293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ramsay A.J., Dong Y., Hunt M.L. Kallikrein-related peptidase 4 (KLK4) initiates intracellular signaling via protease-activated receptors (PARs). KLK4 and PAR-2 are co-expressed during prostate cancer progression. J. Biol. Chem. 2008;283:12293–12304. doi: 10.1074/jbc.M709493200. [DOI] [PubMed] [Google Scholar]
- 38.Mihara K., Ramachandran R., Renaux B. Neutrophil elastase and proteinase-3 trigger G protein-biased signaling through proteinase-activated receptor-1 (PAR1) J. Biol. Chem. 2013;288 doi: 10.1074/jbc.M113.483123. (32979-3290) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Knecht W., Cottrell G.S., Amadesi S S. Trypsin IV or mesotrypsin and p23 cleave protease-activated receptors 1 and 2 to induce inflammation and hyperalgesia. J. Biol. Chem. 2007;282:26089–26100. doi: 10.1074/jbc.M703840200. [DOI] [PubMed] [Google Scholar]
- 40.Goerge T., Barg A., Schnaeker E.M. Tumor-derived matrix metalloproteinase-1 targets endothelial proteinase-activated receptor 1 promoting endothelial cell activation. Cancer Res. 2006;66:7766–7774. doi: 10.1158/0008-5472.CAN-05-3897. [DOI] [PubMed] [Google Scholar]
- 41.Jaffré F., Friedman A.E., Hu Z. β-adrenergic receptor stimulation transactivates protease-activated receptor 1 via MMP13 in cardiac cells. Circulation. 2012;125:2993–3003. doi: 10.1161/CIRCULATIONAHA.111.066787. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material