Abstract
Ellagic acid (EA), a natural polyphenol evidence several pharmacological benefits. The binding profile of EA with human serum albumin (HSA) has been explored and investigated by Isothermal titration calorimetry (ITC), circular dichroism (CD) spectroscopy, time-correlated single-photon counting (TCSPC), absorbance spectroscopy, steady-state fluorescence spectroscopy, and modelling studies. The ITC data analysis revealed the binding Constant (Ka), ΔH, ΔS and ΔG values to be 15.5×104M−1, −116.2±18.1 Kcal mol−1, −366 cal mol−1K−1 and −7.13 Kcal mol−1 respectively with a unique binding site at HSA. EA effectively quenched the intrinsic fluorescence of HSA by static quenching, whereas TCSPC data also revealed association of dynamic quenching also. Thermodynamic analysis confirmed that hydrophobic and mainly hydrogen bonding interaction played important role in stabilizing the HSA-EA complex. It further dictates the binding reaction to be enthalpy driven. The secondary structure of HSA was altered upon binding with EA. CD spectroscopic data indicated the fraction of alpha helicity to be decreased from 52% to 40% upon binding to EA. This study will provide an insight on evaluation of this bioactive interaction during transport and releasing efficiency at the target site in human physiological system since HSA is the most important carrier protein in blood serum.
Abbreviation: EA, Ellagic acid; HSA, Human Serum albumin; CD, Circular dichroism; ITC, Isothermal calorimetry; TCSPC, Time resolved single photon counter
Chemical compounds used: Ellagic acid (Pub Chem CID: 5281855)
Keywords: Human serum albumin, Ellagic acid, Isothermal calorimetry, Time resolved single photon counter, Circular dichroism spectroscopy
Highlights
-
•
A single state binding mode of Human Serum Albumin with Ellagic Acid is proposed.
-
•
Fluoresence quenching of HSA with Ellagic acid is validated in this study.
-
•
Number of binding sites of HSA is characterized using Molecular Docking and ITC study.
1. Introduction
In biological system, intercommunication between biological macromolecule and small molecules constitute one of the most important parts of interconnected networks. Small molecules, both synthetic organic molecules and naturally occurring ones, are important agonists or antagonists to specific target proteins and nucleic acids. The characterization of the interactions between small ligands and various functional bio-macromolecules provide opportunities to design new drugs which can efficiently regulate biological processes and specific cellular pathways [1]. Ellagic acid (2,3,7,8-tetrahydroxy benzopyrano [5,4,3-cde] benzopyran-5,10-dione), a plant polyphenol, present in various fruits and nuts such as strawberry, raspberry and blackberry regularly consumed by humans [15], [2]. This compound can exist in free form, glycoside or linked as ellagitannins which are esterified with glucose [3], although the free form of this compound is rarely found. EA has also been found in several types of honey [4].
EA and different glycosidic derivatives are also present in these fruits including glucosides, arabinoside and the corresponding acetyl esters [5]. Recently dietary polyphenols are receiving increasing attention as potential protectors against variety of human diseases including cardiovascular diseases and cancer [[6], [7]]. EA has diverse properties such as anti-oxidant, anti-inflammatory, anti-fibrosis and anti-carcinogenic[[8], [9]]. EA has also been shown to stabilize the G-quadruplex DNA, formed in the promoter region of KRAS oncogene to prevent Cancer [10]. In our physiological system, drugs are mainly transported by proteins such as Serum albumin. To explore the molecular mechanism taking place between drugs (or ligands) and protein, it is of crucial importance for determining strategy regarding pharmacokinetics and pharmacodynamics. Human serum albumin (HSA) is one of the most studied protein for characterization of ligand-macromolecule interactions [[11], [12]]. HSA can bind drugs and helps in their absorption, distribution, metabolism and excretion. Drug binding and displacement reactions with serum albumins are important determinant of drug pharmacokinetics [[13], [14]]. For using EA as drug against diseases, it is of utmost importance to characterize the interaction of this small molecule with biological macromolecules, especially serum proteins.
But up to date there is no systemic study on HSA-EA interaction, except only a few fluorescence and docking studies by Jiang-Hong Tang et al. and RK Nanda et al. [[15], [2], [16]]. Our work explores the binding properties of HSA-EA in vitro using modern techniques. Moreover, we have also attempted to study if there are any changes in the secondary conformation of HSA due to this ligand interaction. To comply with our target we used various conventional as well as modern biophysical techniques including UV–vis absorption spectroscopy, Steady-state fluorescence spectroscopy, Circular dichroism spectroscopy (CD), Isothermal titration calorimetry (ITC), Time-correlated single-photon counting (TCSPC) supported by the results of molecular docking.
2. Materials and methods
2.1. Sample preparation
HSA and EA both were purchased from Sigma-Aldrich, HSA was dissolved in 100 mM sodium phosphate buffer (pH 7.4), whereas EA was dissolved in 0.1N NaOH solution to make 1654.54 µM stock solution. All chemicals were of analytical grade and used without further purification.
2.2. Measurement of absorption spectrum
The absorption measurements were recorded using a Cary UV–vis Spectrometer at a scan rate of 480 nm/min with 1 cm-path-length quartz cuvette in the visible wavelength range of 350-500nmat 28 °C. UV–vis absorption titrations were carried out by stepwise addition of HSA solutions to a cell containing 5 μM EA.
2.3. Measurement of steady state fluorescence spectrum
Fluorescence emission spectrum was recorded in the range of 300–500 nm using Hitachi F-7000 Spectro fluorimeter with excitation at 295 nm. Both the excitation and emission bandwidth were fixed at 5 nm. EA was added with an increasing concentration of 0–20 µM at 298–15 µM HSA.
2.4. Measurement of fluorescence lifetime
Fluorescence lifetime of both free HSA and HSA-EA were measured using TCSPC instrument (Fluoro Hub; Horiba JobinYvon, GB). The samples were excited at 295 nm using a Nano-LED pulsed laser. The emission decay data at 330 nm were analysed using the DAS6.4 software provided with the instrument [17].
2.5. Circular dichroism measurements
The far UV CD spectrum of HAS (5 µM) with increasing concentration of EA were recorded using a JASCO-J815 spectrometer equipped with a temperature controller at 25 °C (298 K) in a quartz cuvette of 2 mm path length. The scan rate was 100 nm/min, and each final spectrum was an average over three scans.
2.6. ITC measurement
The energetics of the binding of EA with HSA at 25 °C was estimated using a VM2: Cell - ITC 200 (micro cal) titration micro calorimeter. Calorimetric reference cell was loaded with 300 µl of HSA solution (7.5 µM) in 100 mM sodium phosphate buffer (pH 7.0). The protein and ligand concentration were adjusted at 7.5 µM and 300 µM, respectively, to maintain a high ligand: protein ratio. The titration curve for PBS-EA interaction was subtracted from the heat of binding reaction of HSA- EA to obtain the effective heat of binding. The resulting titration curves were fitted to two-binding-mode model using Micro Cal Origin software. Thus, the association constant (K), enthalpy change (H) and stoichiometry(N), were determined. The entropy variation (S) wascalculated according to the standard thermodynamic equation. Three independent experiments were averaged. The response signals were corrected for the small heat of dilution associated with titrating the EA solution into phosphate buffer in each case. The heat of dilution for titrating water into the peptide was negligible. A binding isotherm of heat released as a function of the molar ratio was constructed and the data fitted by nonlinear least square fitting analysis.
2.7. Molecular docking
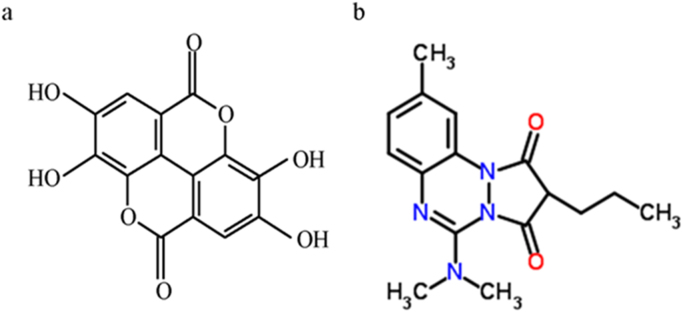
There are many co-crystal structures of HSA available in the protein data base that can be used for docking studies. But, it is important to choose the right PDB file for making the docking study more reliable. In this study, for the 3D structure of HSA we used the co-crystal structure (PDB id 2BX8, resolution 2.7 angstroms) where Azapropazone is the bound ligand. Comparison of the overall structural aspects with the EA indicates that these molecules are reasonably similar in nature (Fig. 1). Thus the binding site on HSA in this crystal structure may be the potent site for the binding of EA. The 3D structure of EA was generated and energy minimized by the Accelrys DS Visualizer2.0 and saved as PDB files [18]. We have used these 3D models for docking based binding studies using Autodock 4 [19], [20].
Fig. 1.
The structure of the compounds: a) EA, b) Azapropazone.
2.8. Setup and docking procedure
The docking procedure was done by using Autodock 4.0 according to the methods described by R. [10]. A cubic grid dimension 60 Å×60 Å ×60 Å with the grid points along the x, y and z axes and a grid spacing of 0.375 Å was used.
3. Results
3.1. UV–visible spectroscopy data analysis
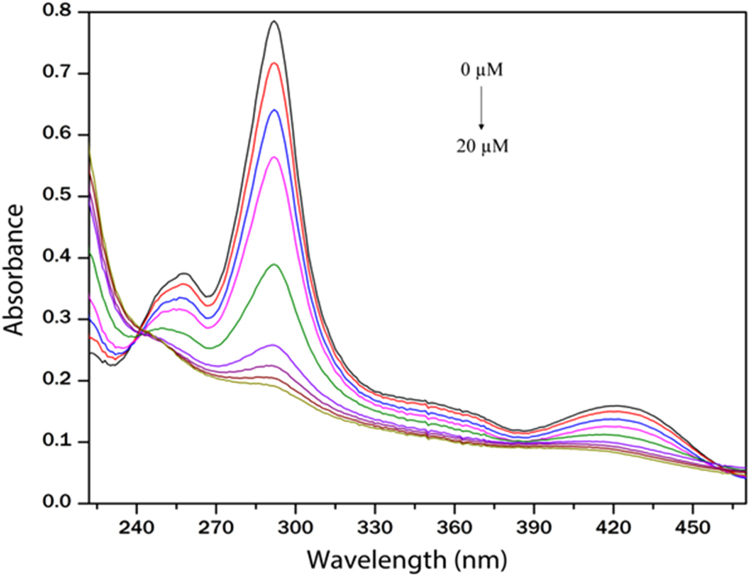
Fig. 2 shows the absorption spectra of EA in absence and presence of varying concentrations of HSA at 298 K. The UV–vis absorption spectrum of free EA had a strong absorption band at 291 nm and three weak absorption bands at 260, 349 and 422 nm.
Fig. 2.
UV–vis absorption spectra of EA when titrated with HSA. HSA concentration ranges from 0 to 20 μM.
In ligand-protein binding studies, we have utilized the Scatchard equation for calculation of binding constant and number of binding sites:
Where, r is the number of moles of EA bound to 1 mol of HSA, n is the number of equivalent binding sites, and K is the affinities of ligands for those sites [[21], [22]]. The concentrations of free EA (Cf) and bound EA (Cb) are calculated using and respectively, where C is the total EA concentration (15 μM). The fraction of bound EA (α) wascalculated using the equation,
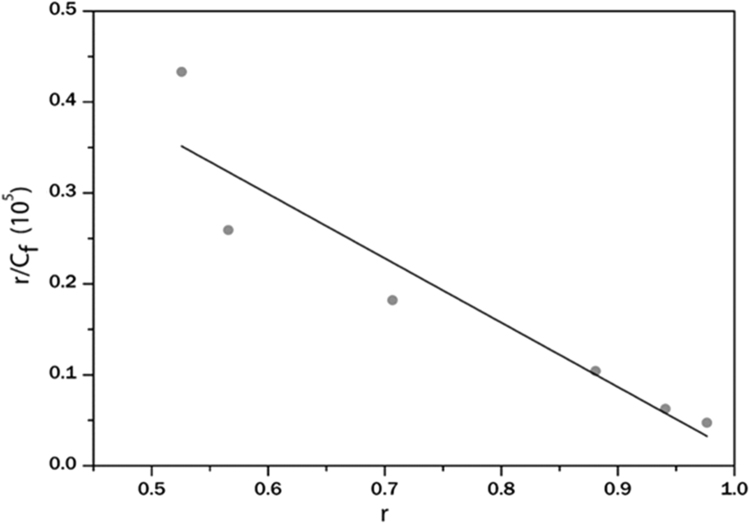
Where Af and Ab are the absorbance of the free and fully bound EA at 422 nm respectively, and A is the absorbance (291 nm) at any given point during the titration. A linearity relationship was seen between r/Cf and r (Fig. 3). Linear fitting generated an n value of 1.03. The binding constant K was determined to be 7.07×104 L mol−1.
Fig. 3.
Scatchard plot for EA with HSA in phosphate buffer (pH 7.4). Absorbance at 422 nm is used to construct the plot.
3.2. Steady state fluorescence measurements
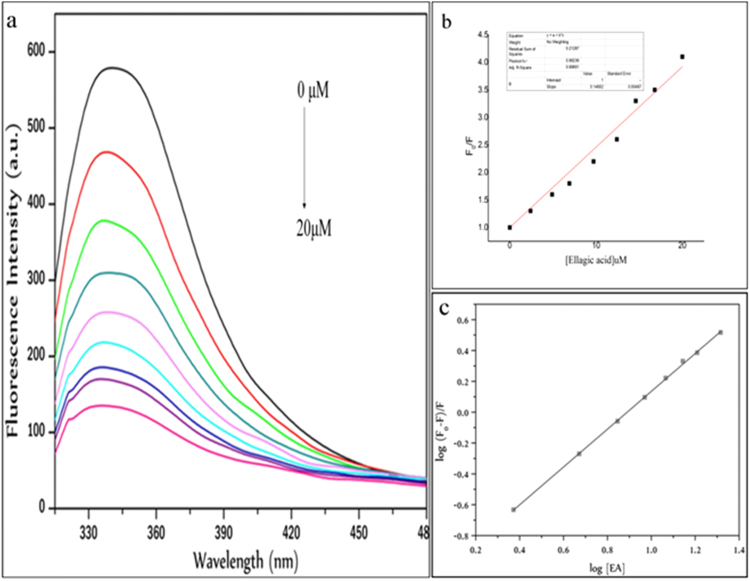
Upon excitation at 295 nm, strong emission from HSA at 337 nm and no emission from EA were registered in the emission wave length range of 300–500 nm. Fluorescence intensity of HSA was observed to decrease with gradual addition of EA in the range 0–25 µM as shown in Fig. 4. Stern-Volmer equation (Eq. (1)) was used to analyse the fluorescence data and the plot F0/F against [Q] was constructed.SV
| (1) |
Where F0 and F are the fluorescence intensities without and with ligand molecule respectively where Ksv is the Stern-Volmer constant and Q is the concentration of ligand. Binding affinity and number of binding sites were evaluated using modified Stern-Volmer equation (Eq. (2)).
| (2) |
Where, K and n are the binding constant and number of binding sites, respectively. The values of K and n for HSA –EA were calculated from the intercept and slope of the plot of log (F0–F)/F vs. log [Q] which revealed the values of Stern-Volmer constant to be 14.9×104M−1, modified Stern-Volmer constant to be 8.13×104M−1with 1 binding site.
Fig. 4.
(a) Normalized fluorescence emission spectra of HSA (15.04×10–6 mol L−1) in the presence of increasing concentrations of EA. EA/HSA corresponds to 0, 0.15, 0.31, 0.47, 0.62, 0.78, 0.94, 1.09 and 1.41 respectively. pH=7.4, T=298 K, λex=295 nm, λem=339 nm (b) Stern-Volmer plot for the quenching of HSA by EA, at 298 K. (c) Modified Stern-Volmer plot of HSA-EA system.
3.3. Excited-state fluorescence lifetime measurements
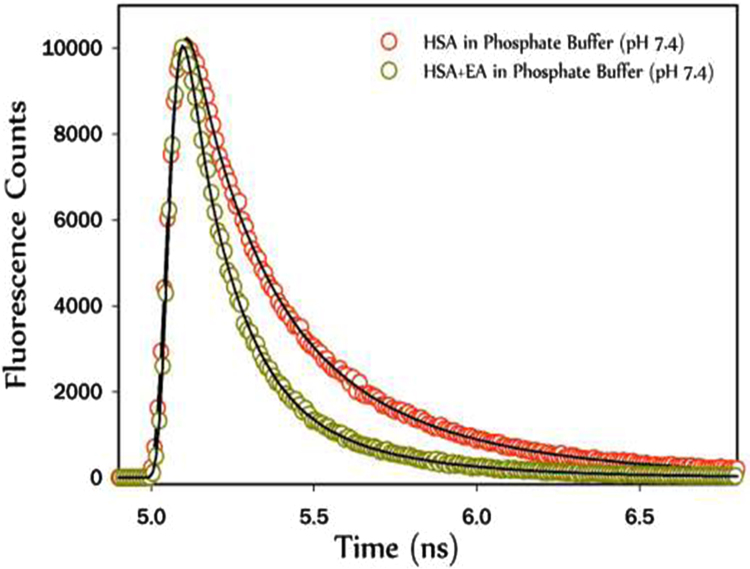
An excited state lifetime measurement is an important tool to investigate the micro-environment of a fluorophore. Depending upon polarity the life time value also changes. If the microenvironment is polar in nature, lifetime value decreases because the energy transfer from donor to acceptor is more favourable in that case. If lifetime value increases, the micro-environment becomes more non-polar [23]. Lifetime decay of Tryptophanresidue in HSA was monitored in both native and conjugated state with EA. The interaction of HSA with EA decreases the fluorescence life-times (Fig. 5).
Fig. 5.
Time-resolved fluorescence spectra at emission wavelength 435 nm of HSA in absence and presence of EA in sodium phosphate buffer (pH 7.4).
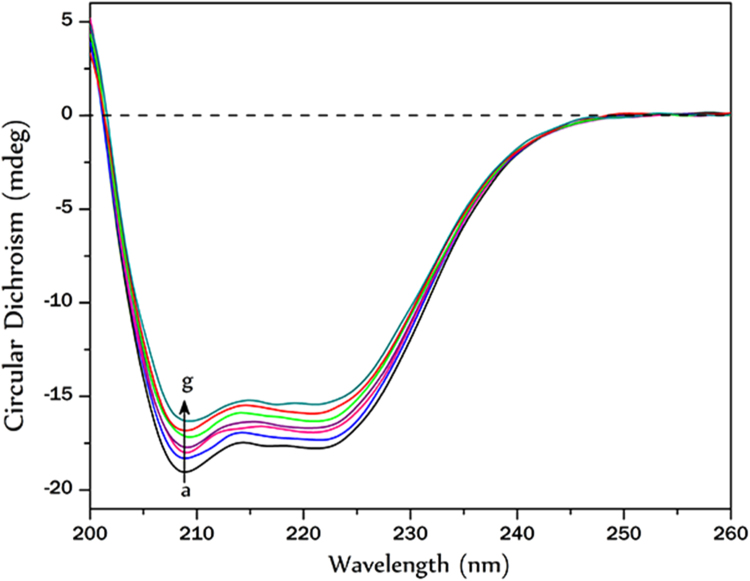
3.4. Alteration in secondary structure of HSA upon EA binding
The helical content of free and bound HSA was calculated from Fig. 6 using MRE (mean residue elipticity in deg cm2 dmol−1) values at 222 nm, which is defined as:
Where θ(mdeg) is the observed elipticity (CD) in mili-degrees, C is the concentration of HSA inmg/ml, l is the path length of the cell in centimetres (0.1 cm), M is the molecular mass of theprotein in Dalton (66,500 in case of HSA) and n is the number of peptide bonds (585 in case ofHSA). The mean residue molar elipticity [MRE] is expressed in deg cm2 dmol−1. Percentage α-helix is calculated using:
where MRE222 is the observed MRE value at 222 nm, 4000 is the MRE of the β- form and random coil conformation at 222 nm and 33,000 is the MRE value of a pure helix at 209 nm [23].
Fig. 6.
Far UV CD spectra of HSA in presence of increasing concentrations of EA; (a) 0 μM (b) 4.12 μM (c) 18.4 μM (d) 34.4 μM (e) 63.6 μM (f) 98.8 μM (g) 119.0 μM.
It was observed that in presence of increasing concentration of EA (0–120 µM) the α-helical content of HSA decreased from 52% to 40%.
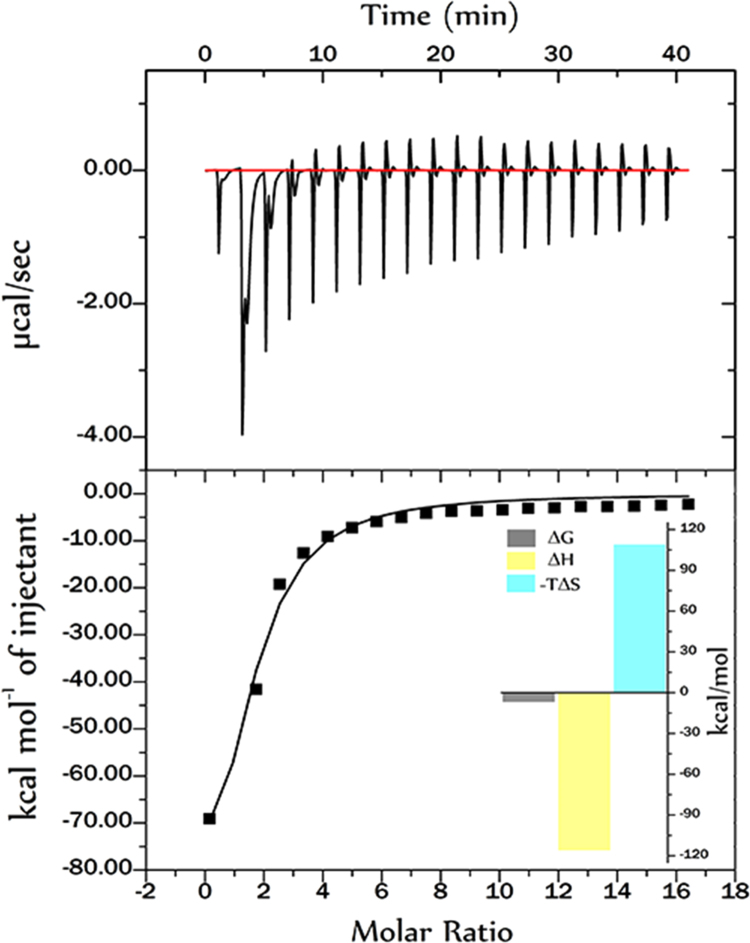
3.5. Thermodynamics of binding interaction
The enthalpic and entropic contributions to the Gibbs free energy of binding were used to understand the mechanism of binding. As seen in Fig. 7, the ITC titrations of 300 μM EA with 7.5 μM HSA yielded negative heat deflection, indicating the binding to be exothermic with a high affinity constant (15.5×104M−1) at 298 K.
Fig. 7.
Isothermal titration calorimetry of HSA and EA interaction at 298 K. Upper panel shows the isothermal titration plot of HSA (in cell) with EA (in syringe), whereas the lower panel shows the integrated heat profile of the calorimetric titration plot shown in upper panel. Inset shows graphic representation of different thermodynamic parameters analysed. The solid line represents the best nonlinear least-squares fit to a single binding site model.
The value of entropy, enthalpy and free energy during interaction between HSA and EA (Table 2) suggested the binding profile to beenthalpy driven.
Table 2.
Thermodynamic parameters and association constant for HSA-EA interaction as evaluated by isothermal titration calorimetry.
| Binding Constant (Ka) | N | ΔH | ΔS | -T ΔS | ΔG |
|---|---|---|---|---|---|
| (kcal mol−1) | (kcal mol−1 K−1) | (kcal mol−1) | (kcal mol−1) | ||
| (M−1) | |||||
| (15.5±0.39)×104 | 1.39±0.18 | −116.2±18.1 | −0.366 | 109.1 | −7.13 |
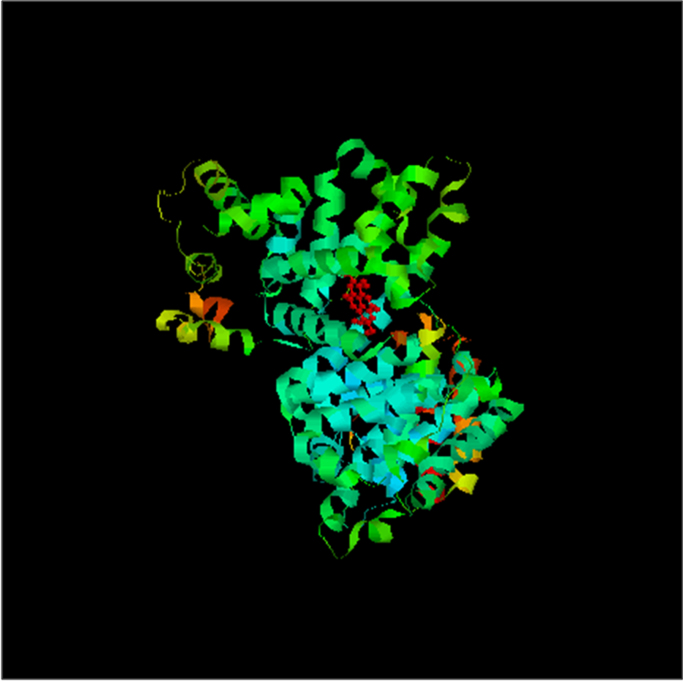
3.6. Molecular modelling and docking of EA to HSA
EA was successfully docked to HSA as seen as Fig. 8. The selected co-crystal structure has two binding sites of Azapropazone. We have docked EA separately to both the binding sites and have obtained estimated binding energies −7.18 kcal/mol and −6.50 kcal/mol. As the binding energy (−7.18 kcal/mol) is more close to our experimental data, we have selected that site as the potential binding site for EA. The estimated Free Energy of Binding of EA to HSA is −7.18 kcal/mol with binding Constant (Ki) of 5.48 µM and four H bonding interaction. Autodock estimates the binding energy considering the three interaction components, electrostatic, van der Waal and desolvation energies. Electrostatic interaction takes care of H-bonding interactions and desolvation energy partially takes care of hydrophobic interaction.
Fig. 8.
Docked picture of HSA with Ellagic acid. (HSA is shown in ribbon model and the ellagic acid is highlighted in ball and stick model).
4. Discussion
According to Fig. 2, it was found that the presence of HSA caused substantial hypochromicity (76% and 48% for 291 and 422 nm respectively for EA) of the bands, indicating strong interactions between EA and HSA. A very small red shift of ~2 nm of the absorption maxima was also observed during the absorption titration experiment. Most importantly an isosbestic point was observed at 241 nm confirming the binding between EA and HSA. A linearity relationship was noticed between r/Cf and r (Fig. 3). Linear fitting generated an n value of 1.03, indicating binding stoichiometry of one EA per HSA molecule. The binding constant K was determined to be 7.07×104M−1. This high value further indicates a strong interaction to exist between EA and HSA.
The shape of the SV plotindicates the quenching mechanisms to be involved. A linear plot suggests the presence ofeither static or dynamicquenching. The Stern-Volmer quenching constant Ksv was 14.9×104M−1 for EA - HSA complex. The value of the quenching rate constant kq was also computed, according to where is the excited state lifetime of the fluorophore. Taking Ksv∼105 M−1 and τ0 for HSA∼10−8 s, a value of kq∼1013 M−1 s−1 was obtained. The values of Kb and n obtained from the graph are 8.13×104 L mol−1 and 1.23. This is in good accordance with the value 7.07×104 M−1 obtained by UV–vis spectroscopy study described earlier (Fig. 2). As the n value approaches unity, we can consider that one molecule of EA combines with one molecule of HSA.
Quenching of fluorescence intensity of HSA by EA changed the micro-environmentof Trp-214 and causing a decrease in the fluorescence lifetime. These different conformational equilibrium existing both in the ground and in the excited states. The fluorescence decay profiles of the conjugated HSA under multi-exponential decay occurred due to such heterogeneous systems. The data was fitted by triple exponential function to carry out time resolved fluorescence study to explore the nature of binding interactions of EA with HSA. There were three lifetime components in native HSA protein τ1, τ2 and τ3, which contribute depending on the protein's multiple local configurations and changes in the extent of solvent accessibility. In our experiment, a decrease in lifetime value was observed from 2.74 to 1.85 ns (Table 1)which confirmed the presence of more polar environment around the tryptophan residue of HSA while interacting with EA.
Table 1.
Comparison of Fluorescence lifetimes (τ) of free HSA and HSA bound to EA.
| Sample | Lifetime (ns) |
Average lifetime (ns) | χ2 | ||
|---|---|---|---|---|---|
| τ 1 | τ 2 | τ 3 | |||
| HSA | 0.43 | 2.09 | 5.70 | 2.74 | 1.02 |
| HSA-EA | 0.16 | 1.36 | 4.04 | 1.85 | 0.96 |
ΔH and ΔS both being negative in this case signifies favourable non-covalent interactions, viz., electrostatic, H-bonding and Van der waals between the protein and the drug. The phenolic hydroxyl group of EA can interact with C, O, and NH of the main polypeptide chain of the protein with strong hydrogen bonding, resulting in the rearrangement of polypeptide carbonyl hydrogen bonding network [24] have demonstrated that the role of hydroxyl groups of phenolic compounds plays a significant factor in binding the interaction between phenolic compounds with serum albumin. Thus the present study suggested that both the hydrogen bonding and electrostatic forces are involved in such interactions at physiological pH. Alongside negative value of ΔG suggest that the formation of complex was spontaneous in nature. The value of ΔG obtained earlier by UV–vis spectroscopy is in good agreement with this value. The binding constant for binding of EA with HSA also indicates the involvement of strong binding affinity. Binding constant for the fluorescence study is due to the involvement of local tryptophan emission and for UV–vis absorption study, it is due to the involvement of global protein conformation [[25], [26]]. From the binding profile it can be concluded that binding of EA to HSA is strong with affinity constant being the range of 105 M−1.
CD spectroscopy is an important method to monitor the secondary structural change of a protein upon ligand binding. Strong binding nature of EA with HSA can alter the secondary structure of the protein. There was a decrease in alpha helical content from 52% to 40% for HSA upon EA binding.
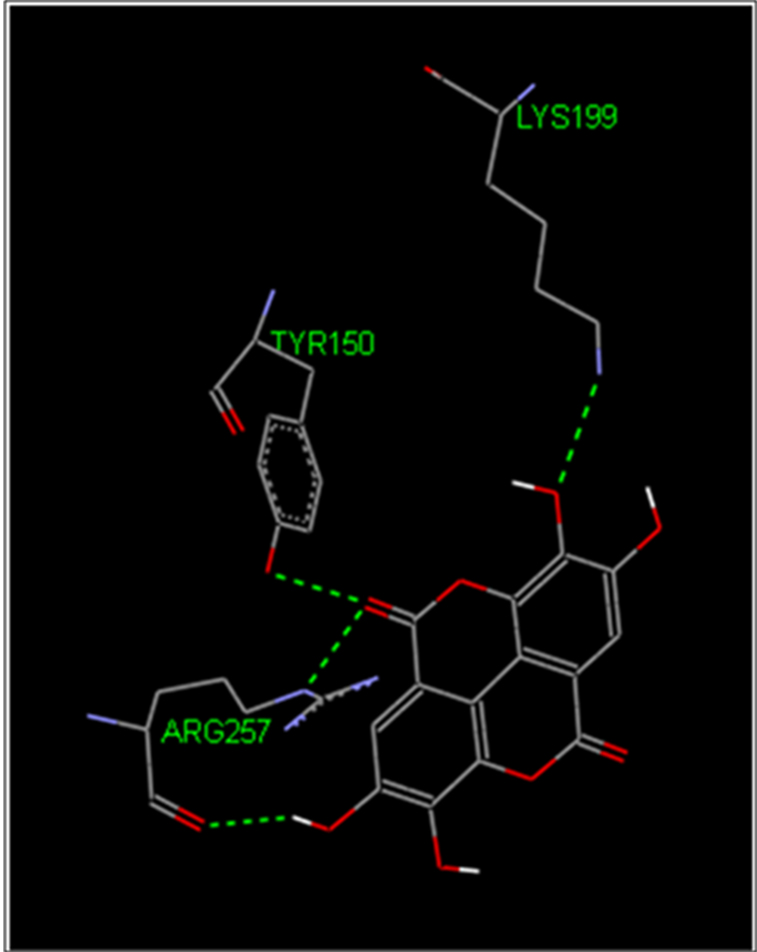
It appeared that EA formed four H-bonds with the amino acid residues Tyr-150, Lys-199 and Arg-257 of HSA with a binding constant of 5.48 µM (Fig. 9). The estimated binding free energy is −7.18 kcal/mol which is in good agreement with the ITC data. The spectroscopic studies provide detailed information on the binding property. However, spectroscopic studies in general cannot provide information of the binding at the atomic level. It cannot provide the direct evidence on the actual binding site as we all as the binding mode of the small molecule to the protein. On the other hand molecular modelling based studies essentially provide information about the binding site and the binding mode at the atomic level. Thus, spectroscopic studies and molecular modelling studies generally provide complementary information that provides a much complete story about the binding details. Docking of compounds to target biomolecule provides valuable information on the binding mode, binding energy which is very useful in designing improved compounds for binding to a specific target biomolecule.
Fig. 9.
H-bonding pattern of the interaction between HSA and Ellagic acid as obtained in the molecular docking exercise. The H-bonds are shown in broken green lines.
5. Conclusion
Steady state fluorescence and UV–vis absorption spectroscopy indicated ground state complex formation between EA and HSA which involved static quenching. Moreover, TCSPC study supplemented that dynamic quenching was also observed with the excited life time changing from 2.74 to 1.85 ns for Trp-214 of HSA molecule. Secondary structural alteration was noticed by Circular dichroism studies. ITC reveals the mode of binding to be enthalpy driven and spontaneous. Molecular modelling and docking indicated the binding constant of EA to HSA to be 5.48 µM. It is also evident, that mainly hydrogen bonding supported the stability of the HSA-EA complex. This study is very important to provide a better understanding about the prospect of transportation of EA through the Serum albumin.
Acknowledgements
We acknowledge World Bank for providing fellowship to R. Pattanayak. We are also grateful to DST (FIST), World Bank-ICZMP (54-ICZMP/3 P), UGC-CAS, UGC-UPE, and DBT-IPLS, Government of India for providing the instrumental facility in the Department of Biochemistry, Calcutta University.
References
- 1.Zian R., Xu S., Lei X., Jin W., Ye M., Zou H. Characterization of small-molecule biomacromolecule interactions: from simple to complex. Trends Anal. Chem. 2005;24(9):810–825. [Google Scholar]
- 2.Tang J.H., Liang G.B., Zheng C.Z., Lian N. Investigation on the binding behavior of ellagic acid to human serum albumin in aqueous solution. J. Solut. Chem. 2013;42(1):226–238. [Google Scholar]
- 3.Pinto M.D.S., Lajolo F.M., Genovese M.I. Bioactive compounds and quantification of total ellagic acid in strawberries (Fragaria×ananassaDuch) Food Chem. 2008;107(4):1629–1635. [Google Scholar]
- 4.Larrosa M., García-Conesa M.T., Espín J.C., Tomás-Barberán F.A. Ellagitannins, ellagic acid and vascular health. Mol. Asp. Med. 2010;31(6):513–539. doi: 10.1016/j.mam.2010.09.005. [DOI] [PubMed] [Google Scholar]
- 5.Zafrilla P., Ferreres F., Tomás-Barberán F.A. Effect of processing and storage on the antioxidant ellagic acid derivatives and flavonoids of red Raspberry (Rubusidaeus) Jams. J. Agric. Food Chem. 2001;49(8):3651–3655. doi: 10.1021/jf010192x. [DOI] [PubMed] [Google Scholar]
- 6.Loa J., Chow P., Zhang K. Studies of structure activity relationship on plant polyphenolinduced suppression of human liver cancer cells. Cancer ChemotherPharmacol.". 2009;63(6):1007–1016. doi: 10.1007/s00280-008-0802-y. [DOI] [PubMed] [Google Scholar]
- 7.Duthie G.G., Duthie S.J., Kyle J.A.M. Plant polyphenols in cancer and heart disease: implications as nutritional antioxidants. Nutr. Res. Rev. 2000;13(01):79–106. doi: 10.1079/095442200108729016. [DOI] [PubMed] [Google Scholar]
- 8.Meyer A.S., Heinonen M., Frankel E.N. Antioxidant interactions of catechin, cyanidin, caffeic acid, quercetin, and ellagic acid on human LDL oxidation. Food Chem. 1998;61(12):71–75. [Google Scholar]
- 9.Chao P.C., Hsu C.C., Yin M.C. Anti-inflammatory and anti-coagulatory activities of caffeic acid and ellagic acid in cardiac tissue of diabetic mice. NutrMetab (Lond.). 2009;6:33. doi: 10.1186/1743-7075-6-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pattanayak R., Basak P., Sen S., Bhattacharyya M. Interaction of KRAS G-quadruplex with natural polyphenols: Aspectroscopic analysis with molecular modeling. Int. J. Biol. Macromol. 2016;89:228–237. doi: 10.1016/j.ijbiomac.2016.04.074. [DOI] [PubMed] [Google Scholar]
- 11.Zaidi N., Ajmal M.R., Rabbani G., Ahmad E., Khan R.H. A comprehensive insight into binding of hippuric acid to human serum albumin: a study to uncover its Impaired elimination through hemodialysis. PLoS One. 2013;8(8):e71422. doi: 10.1371/journal.pone.0071422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matei I., Hillebrand M. Interaction of kaempferol with human serum albumin: AFluorescence and circular dichroism study. J. Pharm. Biomed. 2010;51(3):768–773. doi: 10.1016/j.jpba.2009.09.037. [DOI] [PubMed] [Google Scholar]
- 13.Sudlow G., Birkett D.J., Wade D.N. The characterization of two specific drug binding sites on human serum albumin. Mol. Pharmacol. 1975;11(6):824–832. [PubMed] [Google Scholar]
- 14.Fasano M., Curry S., Terreno E., Galliano M., Fanali G., Narciso P., Notari S., Ascenzi P. The extraordinary ligand binding properties of human serum albumin. IUBMB Life. 2005;57(12):787–796. doi: 10.1080/15216540500404093. [DOI] [PubMed] [Google Scholar]
- 15.Tang J.H., Liang G.B., Zheng C.Z., Lian N. Investigation on the binding behavior of ellagic acid to human serum albumin in aqueous solution. J. Solut. Chem. 2013;42(1):226–238. [Google Scholar]
- 16.Nanda R.K., Sarkar N., Banerjee R. Probing the interaction of ellagic acid with human serum albumin: a fluorescence spectroscopic study. J. Photochem. Photobiol. A:Chem. 2007;192(23):152–158. [Google Scholar]
- 17.Losytskyy M.Y., Kovalska V.B., Varzatskii O.A., Sergeev A.M., Yarmoluk S.M., Voloshin Y.Z. Interaction of the iron(II) cage complexes with proteins: protein fluorescence quenching study. J. Fluoresc. 2013;23:889–895. doi: 10.1007/s10895-013-1199-5. [DOI] [PubMed] [Google Scholar]
- 18.DassaultSystèmes BIOVIA, Discovery Studio Modeling Environment, Release 2017, San Diego: DassaultSystèmes, 2016. 〈http://softadvice.informer.com/Ds_Visualizer_2.0.html〉.
- 19.Goodsell D.S., Olson A.J. Automated docking of substrates to proteins by simulated annealing. Protein.: Str. Func. Genet. 1990;8:195–202. doi: 10.1002/prot.340080302. [DOI] [PubMed] [Google Scholar]
- 20.Morris G.M., Goodsell D.S., Halliday R.S., Huey R., Hart W.E., Belew R.K., Olson A.J. Automated docking using a Lama-rckian genetic algorithm and an empirical binding free energy function. J. Comput. Chem. 1998;19:1639–1662. [Google Scholar]
- 21.Anantha N.V., Azam M., Sheardy R.D. Porphyrin binding to quadruplexed T4G4. Biochemistry. 1998;37(9):2709–2714. doi: 10.1021/bi973009v. [DOI] [PubMed] [Google Scholar]
- 22.Chaires J.B. Analysis and interpretation of ligand-DNA binding isotherms. In: Jonathan B., Chaires M.J.W., editors. Vol. 340. Academic Press; 2001. pp. 3–22. (Methods in Enzymology.edn.). [DOI] [PubMed] [Google Scholar]
- 23.Basak P., Pattanayak R., Nag S., Bhattacharyya M. pH induced conformational isomerization of leghemoglobin from arachishypogea. Biochemistry. 2014;79(11):1255–1261. doi: 10.1134/S0006297914110133. [DOI] [PubMed] [Google Scholar]
- 24.He T., Liang Q., Luo T., Wang Y., Luo G. Study on interactions of phenolic acid-like drug candidates with bovine serum albumin by capillary electrophoresis and fluorescence spectroscopy. J. Solut. Chem. 2010;39(11):1653–1664. [Google Scholar]
- 25.Keswani N., Choudhary S., Kishore N. Interaction of weakly bound antibiotics neomycin and lincomycin with bovine and human serum albumin: biophysical approach. J. Biochem. 2010;148(1):71–84. doi: 10.1093/jb/mvq035. [DOI] [PubMed] [Google Scholar]
- 26.Nada T., Terazima M. A novel method for study of protein folding kinetics by monitoring diffusion coefficient in time domain. Biophys. J. 2003;85(3):1876–1881. doi: 10.1016/S0006-3495(03)74615-3. [DOI] [PMC free article] [PubMed] [Google Scholar]