Abstract
Starch, a very compact form of glucose units, is the most abundant form of storage polyglucan in nature. The starch synthesis pathway is among the central biochemical pathways, however, our understanding of this important pathway regarding genetic elements controlling this pathway, is still insufficient. Starch biosynthesis requires the action of several enzymes. Soluble starch synthases (SSs) are a group of key players in starch biosynthesis which have proven their impact on different aspects of the starch biosynthesis and functionalities. These enzymes have been studied in different plant species and organs in detail, however, there seem to be key differences among species regarding their contributions to the starch synthesis. In this review, we consider an update on various SSs with an emphasis on potato SSs as a model for storage organs. The genetics and regulatory mechanisms of potato starch synthases will be highlighted. Different aspects of various isoforms of SSs are also discussed.
Keywords: Bioinformatics, Glycosyltransferase, Potato, Starch biosynthesis, Starch synthases
Highlights
-
•
We have shown that transgenic plants with modified starches has been generated by manipulation of various SSs.
-
•
A detailed characterization of the biosynthetic machinery is required to understand the effects of inhibiting any particular SSs.
-
•
A comparison is made between the predicted 3D structure of potato SSs with those of rice, barley and glycogen synthases.
-
•
We have pointed out the similarities and differences between different potato SSs.
-
•
We have managed to summarize most of the finding regarding in planta modification of different SSs.
1. Introduction
Starch, a megadalton-size glucose polymer is a prominent storage carbohydrate in many higher plants. With many genes encoding starch biosynthesis enzymes known, starch has become very amenable for (bio) engineering in planta. Moreover, bacterial and other foreign genes involved in glycogen biosynthesis and other glucose polymers such as mutan, dextran, and alternan have also been applied to alter starch characteristics with varying success [1], [2], [3], [4].
Starch synthesis involves a number of enzymes, including soluble starch synthases (SSs; EC 2.4.1.21), starch branching enzymes (SBEs; EC 2.4.1.18), starch debranching enzymes (DBE; EC 3.2.1.68) and disproportionating enzymes (EC 2.4.1.25) [5]. Various SSs are involved in the elongation of the glucan chains by transferring glucose residues from ADP-glucose to the non-reducing end of the growing glucan chains. SBEs introduce the α−1,6 linkages by simultaneous cleavage of some short α−1,4 linked glucan chains and connecting them to other chains, thus providing amylopectin molecules as well as increasing the number of non-reducing ends for further elongation by various SSs isoforms. DBEs seem to trim the irregularly arranged glucan chains to maintain glucan branches in amylopectin molecules in a regular order, thus enabling formation of semi-crystalline structures. Disproportionating enzymes cleave short malto-oligosaccharides (MOS) producing glucose units which can either be used for the ADP-glucose synthesis or as an energy source for plant metabolism.
Higher plant SSs possess multiple isoforms which are grouped based on their amino acid sequence similarities [6]. All the SSs appear to share the same overall structure, consisting of a glass domain (substrate-binding site), a typical transit peptide [7] and different motifs [8]. SSs are further classified into three distinctly localised groups in the plastids, i.e., exclusively granule-bounded (Granular-Bound Starch synthase, GBSS) exclusive or nearly exclusive activity in the soluble phase; and those present in both the granule and soluble phase. Moreover, in potato SSs are further subdivided into four subclasses based upon cDNA and amino acid sequence similarities, i.e. GBSS (~60 kDa), SSI (~57 kDa), SSII (~77 kDa), and SSIII (~110–140 kDa). Nevertheless, pea GBSSI has been further subdivided into GBSSIa and GBSSIb isoforms [9]. Since different SSs contribute to starch biosynthesis, a better knowledge of the relationships among SSs enzymes involved will definitely provide guidelines for plant geneticists, biotechnologists, and breeders to modify starch properties as demanded by various sections. In this review, the function and contributions of different SSs isoforms with an emphasis on potato SSs are discussed.
2. Sucrose to starch conversion in storage organs
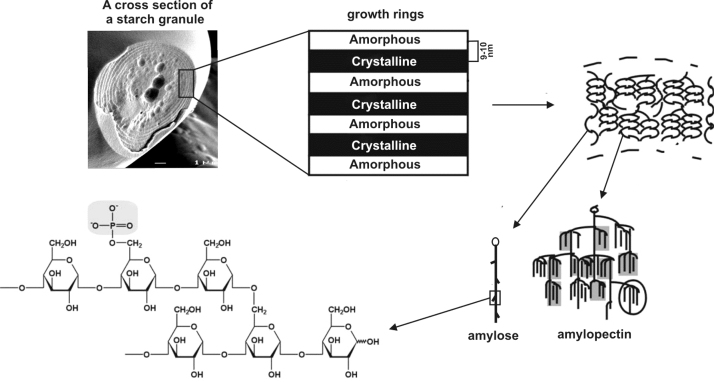
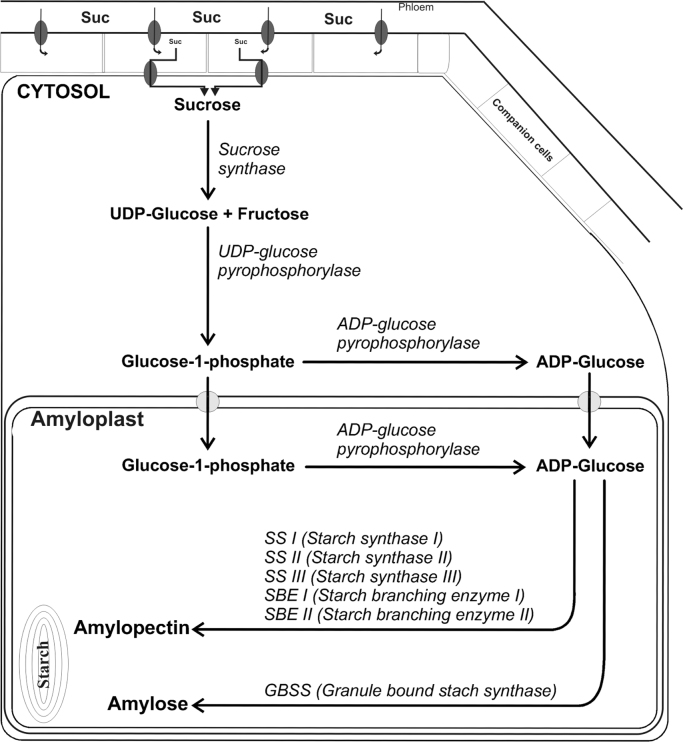
The polyglucan starch is made up of two glucose polymers, amylose and the more highly branched amylopectin. Amylose is a linear polymer of glucose units held together entirely by α−1,4 glucosidic bonds, whereas amylopectin is a highly-branched polysaccharide consisting of α−1,4 linked glucose with α−1.6 linkages at the branch points (Fig. 1). Sucrose to starch conversion is a relatively complicated pathway involving many known SSs as well as a number of sugar transporters (Fig. 2). Plasmalemma-bound transporters and/or diffusion not only transport hexose sugars but also translocate apoplastic sucrose directly to the cytosol [10]. In developing storage organs (e.g., potato tubers) sucrose present in the phloem is metabolised in different ways. Apoplastic or cytosolic invertases convert sucrose molecules to glucose and fructose. Alternatively, sucrose is converted to UDP-glucose (UDP-Glc) and fructose by a sucrose synthase (Susy, EC 2.4.1.13). A part of apoplastic sucrose, upon entry into the cytosol, is transported to vacuoles by endocytosis. The starch biosynthesis in higher plants takes place in a specialized compartment, plastids, which relies on translocation of precursors from the cytosol through the plastid envelop. Glucose-6-phosphate (Glc-6P) and ADP-glucose (ADP-Glc) transporters are actively involved in transferring these important nucleotide precursor molecules into plastids [11], [12]. In potato tubers and once inside the amyloplasts, Glc-6P is subsequently converted to glucose-1-phosphate (Glc-1P) and ADP-Glc by phosphoglucomutases (PGM, EC 2.7.5.1) and ADP-glucose pyrophosphorylase (AGPase, EC 2.7.7.27), respectively. In plants, at least five independent-conserved classes of genes encode SSs [13]. These SSs use ADP-Glc produced by AGPase as a substrate to catalyse the formation of new glycosidic linkages by transferring glucose moieties of ADP-Glc to the non-reducing end of an existing α−1,4 glucan chain.
Fig. 1.
Schematic representation of the structure of a starch granule, with alternating amorphous and semi-crystalline regions constituting the growth rings. The glucose residues are connected through α−1,4 and α−1,6 linkages. In potato starch one out of 200–300 glucose units of amylopectin is phosphorylated. Phosphate groups can be attached to the C-3 or the C-6 of a glucose residue. The position of the phosphate group with respect to the α−1,6 branch point is arbitrary (Figure reproduced with minor modifications from [98]).
Fig. 2.
Illustration of the starch biosynthesis pathway in potato tubers.  and
and  represent putative transporters.
represent putative transporters.
The semi-crystalline structure of amylose-free starches in potato (amf) and cereal crops (waxy) suggest that amylose is synthesized downstream of amylopectin, using either amylopectin or small soluble glucans as a primer [14], [15]. In principle, two mechanisms proposed for the amylose synthesis can occur side-by-side. Within the crystalline matrix, GBSSI is responsible for the amylose biosynthesis. It seems that once MOS molecules have been elongated to a certain extent, they may become too large to escape from the growing starch granules. After further extension, these moderate molecular weight (105–106) chains form amylose. It has also been shown that this mechanism appears to be at work in the starches extracted from higher plants [16]. Moreover, the GBSSI activity seems to be stimulated by MOS molecules in the course of starch synthesis [17]. In the small glucan-primed amylose biosynthesis, DBEs could also play an important role in generating small molecules which diffuse into the granule to serve as a preferred substrate for GBSSI. Apart from being exclusive granule-bounded, GBSSI elongates a growing α−1,4 linkage processively, suggesting that the enzyme does not dissociate from the growing granule right after each glucose unit is added. Very recently, a plastidial protein named Protein targeting to starch (PTST) has been identified which seems to be specifically required for amylose synthesis and targeting GBSSI to starch granules in Arabidopsis [18]. Arabidopsis ptst mutants did not produce amylose and they were phenotypically similar to GBSSI mutants.
3. Reaction catalyzed by SSs
Potato contains four different SSs isoforms (SSI, SSII, SSIII and GBSSI). Contrary to other SSs, SSI does not have multiple isoforms in plants, suggesting a presumably unique and important role in starch biosynthesis [19]. However, its precise role is not yet clear. For instance, although the activity of SSI enzyme was repressed to non-detectable levels in potato transgenic plants, neither amylopectin structure nor starch granule morphology was changed. The reason for no measurable changes might be due to the fact that SSI is predominantly expressed in the potato leaf tissue and mainly involved in the synthesis of transitory starch in the leaves [20]. SSI prefers the shortest amylopectin chains as substrate, and it is particularly responsible for synthesizing amylopectin short chains [21], [22]. Different analyses revealed that the ratio of DP 8–12/DP 17–20 branched chains in Arabidopsis [22] and the ratio of DP 6–7/DP 16–19 branched chains in rice [23] mutants were decreased. Furthermore, SSI-RNAi suppressed wheat lines showed an increased frequency of very short chains (DP6 and DP7), a lower proportion of short chains (DP8-DP12), and more intermediate chains (DP13-DP20), suggesting that very short glucose chains of DP6-DP7 residues are presumably the substrates for SSI enzyme, which SSI then extends to a length of 8–12 glucose residues within the amylopectin clusters [24]. Despite these findings, antisense down regulation of SSI in potato tubers did not change the starch structure, suggesting other SSs may partly compensate for the lack of SSI in tubers. SSIII is the major SSs in potato tubers and accounts for almost 80% of soluble starch synthase activity although a small proportion of the SSIII activity is found bound to the starch granules [25], [26]. Antisense down regulation of SSIII in potato tubers led to almost 80% loss of SSIII activity, and consequently alteration of starch granule morphology [27]. The suppression of the SSIII activity resulted in overall reduced amylopectin synthesis and a decrease in the amylopectin long chains with degree of polymerization between 25 and 35 (DP25-DP35), suggesting that SSIII preferentially synthesizes long B1(Dp13-DP24) and B2 (DP25-DP36) chains [27]. In other crop species, similar isoforms to those found in potato are present. However, the relevance of the individual SS isoforms and their distribution between stroma and starch granules within the plastids is very species-dependent [6], and these differences presumably contribute to variations observed in the structure of starches produced from different plant species [28]. The effects of SSII repression in many plants studied so far, show fairly similar phenotypes with respect to the amylopectin structure in such a way that the abundance of chains with DP6-DP10 has been increased in comparison to longer chains (DP12-DP28) [27], [29], [30], [31], [32]. Interestingly, the apparent redundant function of SSII and SSIII in amylopectin biosynthesis has recently been revealed [33].
Although Arabidopsis mutants lacking SSIV show abnormalities in their granule initiation [34], the precise role of SSIV is not yet known. Studies have revealed that two distinct isoforms of SSIV i.e. SSIVa and SSIVb, are present in cereal endosperms and leaves, respectively [13], [35]. To the best of our knowledge, an enzyme similar to any known SSIV has not been reported/or characterized in potato. Blast information using potato genome sequence database (www.potatogenome.net) has revealed a putative 3 kb open reading frame on chromosome number 2 of potato close to a tomato genetic marker (C2_At1g29950). Sequence based comparison showed that this putative enzyme belongs to the same family as other SSs, the GT5 glycosyltransferase family. Interestingly, ChloroP program (http://www.cbs.dtu.dk/services/ChloroP/) has revealed a transit peptide with a split site between 46 and 47 amino acids (unpublished data), meaning that this putatively SSIV may enter the plastids and potentially contribute to starch synthesis.
4. Domain structure of SSs
All SSs and glycogen synthases (GSs) belong to a very important superfamily of enzymes, glycosyltransferase (GTs, EC 2.4.x.y). Up to now, GTs members seem to possess two different folds (i.e. GT-A and GT-B) [36] and a new type of fold with similarities and differences with the GT-A fold found in bacterial sialyltransferase (CstII) belonging to family GT42 [37]. The GT-B fold (Pfam clan CL0113, CAZy) consists of two similar Rossmann fold domains (Pfam database: Glyco_transf_1(GT1; PF00534); and Glyco_transf_5 (GT5; PF08323). The catalytic activity of all SSs and GSs is conducted by a highly conserved catalytic [13]. This region in all SSs, include the conserved synthase catalytic domain (GT5) and a glycosyltransferase 1 domain (GT1), which is encompassed by almost the whole ~60 kDa protein of bacterial GSs. This 60 kDa “core” region is in the C-terminus of SSs from plant and algae [38](Fig. 3). With almost no exception, the GT1 domain is common among various GT-B members. This may suggest that this part of the catalytic domain is the nucleotide binding domain. The overall sequence similarity between bacterial GSs and plant SSs are about 30–36%, suggesting a similar structure and catalytic activity as both catalyse the same chemical reaction [39]. Prediction of structural relationship between the crystal structure of Agrobacterium tumafacians GS (AgtGS, PDB entry 1rzu; [40], the catalytic domain of Oryza sativa GBSSI (OsGBSSI-CD, PDB entries 3vue and 3vuf; [41]) and the catalytic domain of Hordeum vulgare SSI (HvSSI, PDB entry 4hln; [42]) with potato SSs, revealed strong similarities in terms of overall structural geometry (Fig. 4A). Similar to the 3D structure of AgtGS, the core of the N-terminal domain of potato SSs possess a number of α-helices and β-sheets, characteristic of GT-B members (Fig. 4A). The overall predicted structure of potato SSs reveal striking structural and topological similarities with the solved 3D structures.
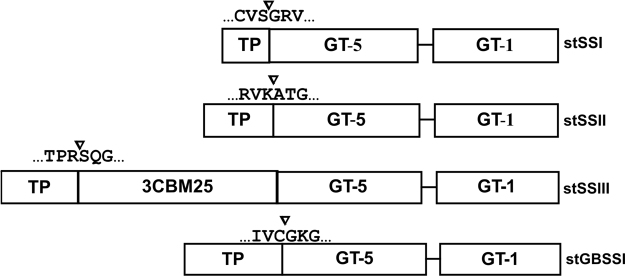
Fig. 3.
Domain structure and comparison of various SSs. Conserved domains were identified through a Genbank conserved domain database search service. The putative transit peptide cleavage sites are identified using the ChloroP neural network analysis at the N-terminal of each particular SS. Glycos_transf_1 (PF00534; GT1) and Glyco_transf_5 (PF08323; GT5). GT; Glycosyltransferase, TP: Transit peptide, CBM: Carbohydrate binding module.
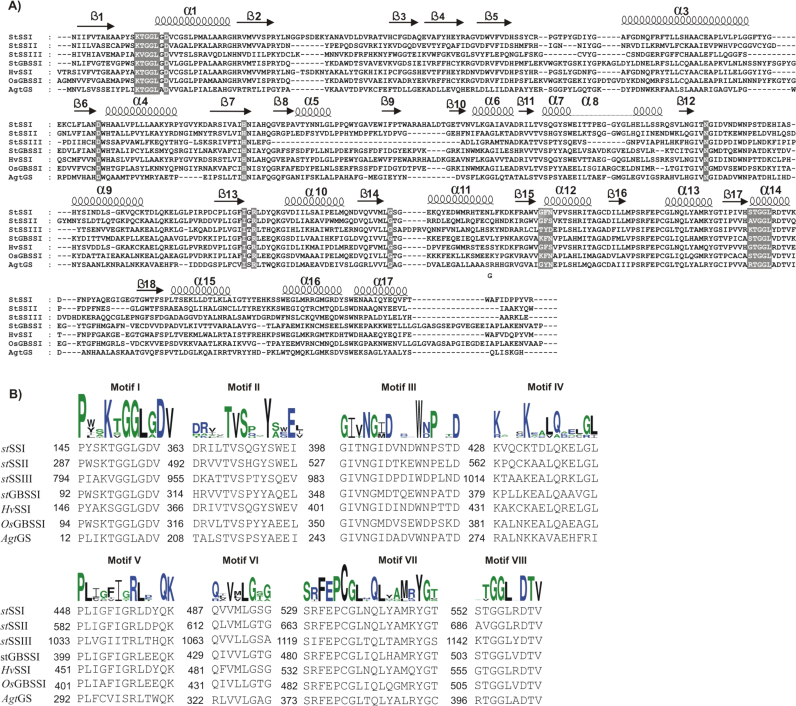
Fig. 4.
A) The sequence alignment of different potato SSs together with AgtGS, OsGBSSI and HvSSI. The ClustalW program (Larkin et al., 2007) was used to align the protein sequences. B) A partial alignment of different potato SSs together with AgtGS, OsGBSSI and HvSSI. The logo of conserved motifs was generated by WebLogo (http://weblogo.berkeley.edu/logo.cgi). All the sequences were obtained from EMBL/DDBJ database. stSSI(P93568.1), StSSII(CAA61241.2), StSSIII(Q43846.1), StGBSSI(Q00775.1), OsGBSSI (AK070431) and HvSSI (AAF37876.1).
In both GSs and SSs, the N-terminal regions of the GT5 domain possesses a highly conserved KXGGL motif (motif I) (Fig. 4B). This motif is very well conserved among different SSs as well as GSs and seems to play a pivotal role in substrate binding [41], [43], [44], [45]. One KTGGL motif has been shown to bind ADP-Glc/ADP in bacterial GS [39], [46]. In E.coli GS (EcGS, PDB entry 2qzs; [45]) and AgtGS closure of C and N-terminal domains, a phenomenon which may also be involved in SSs facilitates the simultaneous acceptor and donor substrates binding. However, in contrast to EcGS, the substitution of Lysine-193 in the so-called KTGGL motif did not change maize SSIIa capability towards ADP-Glc. Remarkably, the substitution changed the overall SSIIa activity towards different primers [47]. Moreover, all SSs contain a second “KTGGL look-alike” motif towards their C-termini (Motif VIII) (Fig. 4A and B). The look-alike motif is less well conserved between different SSs; STGGL (SSI and GBSSI), AVGGL (SSII) and KTGGL (SSIII). The significance of these sequence similarities is only partly understood although it seems likely that this second motif is also involved in substrate binding [48]. Nonetheless, this motif (STGGL/AVGGL) lacks the lysine (K) residue which is thought to interact with the ADP-Glc in AgtGS [44], [49], [50]. In contrast to OsGBSSI, Cystein (C337) has been replaced with valine (V335), meaning that the so-called disulfide bridge is not present in StGBSSI (Fig. 4A). It is thought that this disulfide bridge is present in Poaceae family and might be functionally important for efficient starch biosynthesis [41].
Although the exact roles of other recognized motifs are yet to be resolved, these motifs seem likely to be associated with SSs three-dimensional structure and may contribute to the ADP-binding and the stability of domains [51]. The GT1 domain accommodate a number of these conserved motifs that are found in a functionally heterogeneous group of glycosyl transferases, mediating an inverting mode of glycosyl transfer [52], [53].
Bioinformatic comparative analysis revealed that the N-terminal extensions of potato starch synthases classes are different. Experimental data shows that N-terminal extensions in SSIII and SSIV enzymes, are involved in protein–protein interactions. It seems that the conserved coiled-coil motifs covering almost one-third of the enzymes facilitate the interaction between SSs and other interacting proteins. Very recently, it has been shown that both localization and interaction with fibrillins, gylycoproteins essential for the formation of elastic fibers in connective tissues, are mediated by the N-terminal part of Arabidopsis thaliana SSIV [54].
All SSs except GBSSI contain additional sequence extensions located N-terminal to their catalytic region [55]. The differences in the molecular mass of the soluble SSs are primarily caused by differences in the lengths of N-terminal extensions to the catalytic domain: 85 amino acids for SSI, 275 amino acids for SSII, and 780 amino acids for SSIII. The function of these N-terminal extensions is not fully known [48], [56]. It has been suggested that these additional extensions play a role in the interaction with other enzymes involved in starch biosynthesis [42]. Another possibility is that they determine the partitioning of the synthases between the granule and soluble phase (stroma). However, it seems that this region determines the chain length specificity of enzymes or is partly involved in enzyme-protein interactions [57]. It has been shown that the N-terminal part of Arabidopsis thaliana SSIII has a sequence similarity to carbohydrate binding modules (CBM) [58], [59]. Based on different truncations made on three in-tandem Carbohydrate Binding Domains (CBDs) repeats of Arabidopsis thaliana SSIII, Wayllace et al. (2010) concluded that the presence of these domains is crucial for full activity of the enzyme [60]. Due to the presence of two binding sites for the polysaccharides, the binding ability of CBDs (i.e. SBDs) enable them to breakdown their substrate [61]. Moreover, the N-terminal CBDs of SSIII seem to have a regulatory role and enhance the capacity of the enzyme to bind starch [59]. There appears to be a correlation between the number of CBD modules and the capacity of enzyme to bind starch.
All GBSSI proteins have a highly-conserved C-terminal extension of their catalytic domain consisting of about 20 extra amino acids. This region is believed to confer some specific properties to GBSSI such as amylopectin biosynthesis [48].
5. Processive versus distributive mode of action
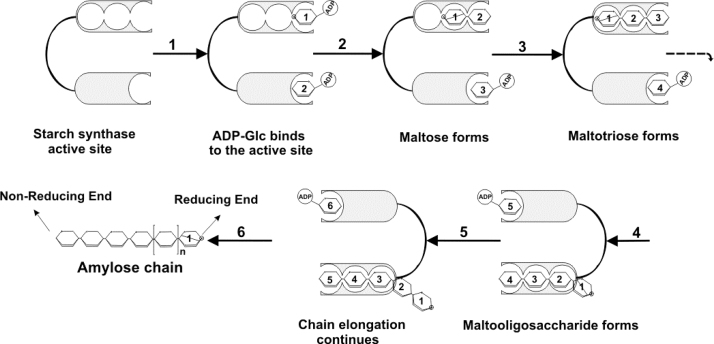
It is now well established that SSs cleave the donor substrate (ADP-Glc) and attach the glucosyl residue to the non-reducing end of a maltodextrin (the acceptor molecule) [28]. In the donor substrate, the ADP group is attached to glucose through an “α-linkage”. The product of the reaction is also in the α-configuration, meaning that the GT5 members are retaining enzymes [62]. At the beginning, it is possible that SSs enzymes accommodate two ADP-Glc molecules in their corresponding catalytic sites (Fig. 5). This mechanism is called two catalytic-site insertions which seem to take place in vivo [63]. According to this mechanism, switching and transferring D-glucose from ADP-Glc substrate between two catalytic sites results in elongating oligosaccharides during which SSs remain attached to growing glucose polymers. Since one of the catalytic sites seems to be able to accommodate maltotriose, it is possible that the two catalytic-site insertion mechanism does not involve switching growing oligos between two sites (Fig. 5). The actual transfer of a glucosyl residue to the acceptor substrate can take place in two different ways: processive or distributive. Processive enzymes remain attached to the acceptor molecule after addition of the glucosyl residue. As a consequence of this, these synthases add multiple residues to one nascent glucan chain, which can grow relatively fast. The distributive synthases are released from the acceptor molecule after each glucosyl addition. Thus, many different chains grow at a relatively slow rate. Only a few SSs have been investigated for processivity. For this, starch granules from mutant pea lines (lam containing SSII but no GBSSI; rug5 containing GBSSI but no SSII) were incubated with ADP-Glc and maltotriose. Subsequently, the elongation of the maltotriose was monitored [14]. It appeared that incubations with the lam granules yielded only maltotetraose, whereas incubations with the rug5 granules resulted in the formation of many products, having a DP ranging from 4 to more than 10. From this finding, it was concluded that GBSSI adds more than one glucose residue to the acceptor substrate for each enzyme-glucan encounter, whereas SSII adds only one glucose residue. Thus, pea GBSSI may have a processive mode of action, whereas SSII seem to have a distributive mode of action. To facilitate further biochemical analysis, several isoforms of SSs were expressed in Escherichia coli. Potato GBSSI and potato SSII were produced in this way. Comparison of the biochemical properties of these two enzymes showed the same activity as compared to the pea orthologs [14]. However, these experiments revealed an interesting new feature of GBSSI; maltotriose elongation by GBSSI shifted from a distributive to a processive mode when amylopectin was included in the reaction mixture. In addition, the rate of maltotriose elongation with GBSSI increased significantly in the presence of amylopectin [14], [27], particularly as amylopectin concentration increased to the critical point where crystallites start to form as a result of side chain interactions.
Fig. 5.
Proposed reactions catalyzed by SSs. Two catalytic-site insertion mechanism for starch polymer biosynthesis. The synthesis of amylose or α−1,4 glucans by the action of SSs which have been reported to occur by the addition of glucose moieties to the non-reducing end of a growing α−1,4-glucan. Glucose number one with a slanted line inside designates the reducing end of α−1,4 glucan chain.  : represent a glucose moiety with the reducing end.
: represent a glucose moiety with the reducing end.
6. Acceptor, substrate binding of SSs
From the proceeding discussion, it is clear that the various SSs isoforms can have different biochemical properties (processivity, granule-boundness, affinity for donor and acceptor substrates and activation by amylopectin), suggesting that each of them fulfils a particular role in starch biosynthesis. SS isoforms also vary in their affinity for donor and acceptor substrates. GBSSI has a 10-fold lower affinity (higher Km, Km=0.96 mM) for ADP-Glc than SSII [27]. This means that amylose synthesis by GBSSI will be more sensitive to limiting concentrations of ADP-Glc. When present in the granule, potato GBSSI has a much higher affinity for maltotriose than SSII [14]. For the acceptor substrate, amylopectin, the opposite is true [48]. The various isoforms play distinct roles in the elongation of glucan chains of a length.
Table 1 shows the conserved equivalent important residues among different potato SSs in comparison with those of AgtGS, OsGBSSI and HvSSI. These amino acids take part in substrate/acceptor binding and are crucial for SSs activity. Based on the crystal structure of a number of GSs, it is quite possible that the overall architecture of the ADP-Glc binding pocket of SSs is basically the same as that in EcGS, AgtGS, OsGBSSI and HvSSI. In AgtGS, the ADP-Glc molecule bind to a pocket located on the C-terminal of the enzyme (Fig. 5). During ADP-Glc binding, a number of amino acids stabilize the interaction between the substrate and the enzyme. The so-called KTGGL conserved motif plays a direct role in accommodating the nucleotide-diphosphate-glucose as has been reported in E.coli and Agrobacterium tumafaciance [40]. Therefore, such close conformation is formed in SSs. In KTGGL conserved motif, both glycine (G) residues are essential for enzyme activity (Furukawa et al., 1993). In AgtGS, Tyr354, which occupy the centre of a loop connecting β15 and α12 barrels, interact with the adenine ring of ADP-Glc [40]. It must be mentioned that this position is symmetrically present in other bacterial glycogen synthases and in SSs. Furthermore, the Lys (K) and Gly(G) of KTGGL conserved motif in collaboration with next door Asp (D) interact with the ribose of ADP.
Table 1.
List of important amino acids that are directly involve in substrate (ADP-Glc) binding in potato SS based on 3D structure of AgtGS, OsGBSSI and HvSSI. These amino acid residues were identified both base on site directed mutagenesis experiments and/ or sequence similarities with AgtGS, OsGBSSI and HvSSI 3D structures.
| AgtGS | OsGBSSI | HvSSI | StGBSSI | StSSI | StSSII | StSSIII |
|---|---|---|---|---|---|---|
| Lys(K)15 | Lys(K)97 | Lys(K)149 | Lys(K)95 | Lys(K)145 | Lys(K)290 | Lys(K)794 |
| Gly(G)18 | Gly(G)100 | Gly(G)152 | Gly(G)98 | Gly(G)148 | Gly(G)293 | Gly(G)797 |
| Asp(D)138 | Asp(D)234 | Asp(D)280 | Asp(D)232 | Asp(D)277 | Asp(D)414 | Asp(D)909 |
| His(H)163 | His(H)264 | His(H)310 | His(H)262 | His(H)307 | His(H)444 | His(H)938 |
| Asn(N)246 | Asn(N)353 | Asn(N)404 | Asn(N)351 | Asn(N)401 | Asn(N)530 | Asn(N)986 |
| Ile(I)297 | Ile(I)406 | Ile(I)456 | Ile(I)404 | Ile(I)453 | Ile(I)587 | Ile(I)1038 |
| Arg(R)299 | Arg(R)408 | Arg(R)458 | Arg(R)406 | Arg(R)455 | Arg(R)589 | Arg(R)1040 |
| Gly(G)327 | Gly(G)436 | Gly(G)486 | Gly(G)434 | Gly(G)483 | Gly(G)617 | Gly(G)1068 |
| Tye(Y)354 | Phe(F)463 | Phe(F)513 | Phe(F)461 | Phe(F)510 | Phe(F)644 | Tye(Y)1100 |
| Asn(N)355 | Asn(N)464 | Ser(S)514 | Asn(N)462 | Asn(N)511 | Ser(S)645 | Asp(D)1101 |
Considerable effort has been made to determine which structural elements of SSs underlie properties. A large number of truncations and chimeric proteins have been made between potato GBSSI and SSII [48]. Many of the chimeras showed little or no activity, suggesting that the swapped parts do not interact appropriately with more distant parts of the sequence. As a consequence, the proteins may be incorrectly folded and become inactive. From those chimeras that are active, the N-terminal extension of SSII does not seem to influence the catalytic activity of the core catalytic domain. A non-catalytic role in keeping the synthases of the granule, or a role in binding particular acceptor substrates is suggested by these experiments. The experiments showed that many of the specific properties of GBSSI (although not processivity) seem to reside in the C-terminal part of the protein that includes the KTGGL look-alike motif and the C-terminal extension unique to GBSSI isoforms. They hypothesized that amino acids with basic side chains (arginine [R], lysine [K], histidine [H]) are positively charged at neutral pH, and are well equipped to bind anionic substrates such as ADP-Glc which has been demonstrated for the lysine residue within the KTGGL motif of AgtGS, OsGBSSI and HvSSI [40], [45]. Although potato SSs show some differences in primer preferences, primer affinity and chain length specificities, they all possess a very high sequence similarity in their catalytic domains, as can be seen in Fig. 4. Such conserved motifs can also be seen in all other SSs from different sources. Arginine within an active site of a SS has been found far more reactive to arginine-specific reagent phenylglyoxal (PG) than one existing outside of the enzyme active site. Presumably, the guanidine group of the arginine (Agr211) residue in maize is interacting with the anionic pyrophosphate moiety of ADP-Glc. The treatment of maize SSIIa with the arginine-specific reagent phenylglyoxal inactivated the synthase in a time- and concentration-dependent manner [64]. Addition of ADP-Glc completely protected SSIIa from inactivation. This suggested that arginine residues of SSs are involved in ADP-glucose-binding. Single arginine residues were modified in SSIIa at 8 different locations, two of which had a profound effect on the enzyme's activity. In addition, the highly-conserved histidine (H213) residue was altered (H213K, H213W and H213N). The Km for ADP-Glc was not affected by the mutations, indicating that these residues are not involved in the binding of ADP-Glc. In another study the importance of acidic amino acids (aspartate [D21 and D139], glutamate [E391]) in catalysis by SSs was investigated. For this purpose, maize SSIIb was treated with the specific reagent 1-ethyl-3-(3-dimethyl-aminopropyl) carbodiimide (EDAC) [65]. Enzyme activity was abolished in a time and concentration dependent manner whereas addition of ADP-Glc completely protected the enzyme from inactivation by EDAC. Twelve conserved acidic amino acid residues were mutagenized. The mutant enzymes displayed activity ranging from 0% to 110% of that of the wild-type enzyme. These studies suggest that the starch synthase reaction proceeds via general acid/base catalysis, in which the aspartate and glutamate residues are necessary for the activity of the enzyme. Indeed, it is now generally accepted that the catalytic machinery of glycosyltransferase enzymes is likely to involve aspartate or glutamate residues. The side chains of these amino acids possess the appropriate reactivity to act as acceptor activation/nucleophile for the formation of a glycosyl-enzyme intermediate [66]. This idea has been further proven by the site-directed mutagenesis analysis of GTs. The aspartate residue may be involved in ADP-Glc-binding, presumably interacting with the adenosine ring. Since the closest E to the PCGL conserved box is absolutely present in all SSs (Fig. 4B), it is definitely involved in retaining the activity of the enzymes. Any substitution of this amino acid in EcGS which shares the same conserved sequence in this region, has resulted in a dramatic decrease of the enzyme activity [45].
7. Impact of various SSs isoforms on starch granules
The ultimate aim of starch manipulation in planta, is production of starches with improved or new functionalities to overcome major shortcomings demanded by various industries. As the activity of these SSs influence different aspects of starch properties and functionalities, genetic modification can be applied to uncover the role of these enzymes. Among many desired targets, modification of amylose/amylopectin ratio, chain length distribution (CLD), phosphate content and diversification of linkages are more common. Various starch synthase activities have been down-regulated in potato plants using antisense technology, either alone (all 4 isoforms) or in combination (e.g., SSII and SSIII). The rationale of these experiments was twofold; (i) these studies attempted to determine the contribution of each isoform to tuber starch biosynthesis and (ii) uncovering the specificity of different SSs isoforms towards different primers. Therefore, these studies have explored the possibilities of engineering novel starch types in planta with improved functional properties for certain industrial applications.
Inhibition of the major soluble isoform of SSs (SSIII) in potatoes in particular had a considerable impact on granule morphology. The granule surface contains deep grooves, and it seems as if each granule is composed of multiple subunits. This effect was even more pronounced when both SSIII and SSII were antisensed [27], [67]. Inhibition of SSII alone did not have significant consequences on granule morphology in potatoes. In contrast, inhibition of SSII in pea embryos through knock out mutations of the rug5 locus notably affected the granule morphology, making the granules very convoluted, which was in accordance with the fact that SSII is the major starch synthase in pea [29]. In cases where the level of the GBSSI activity was down-regulated, and the starch granules were subsequently treated with iodine then the granules were stained red instead of blue. It is important to note that the antisense effect can vary from 0% to 100%, meaning that potato starches with an amylose content between 0% and 20% can be produced in this way. As a result, the granules are only partially filled with amylose, and characteristic “blue cores” are obtained.
8. Modulation of starch properties
Genetic modification of SSs expression levels can also have an impact on the yield, fine structure and physical properties of the starch. Table 2 summarizes the characteristics of all transgenic potato starches (with respect to SSs) that have been obtained so far. Typically, down-regulation of synthase activity does not have much impact on the starch yield possibly because reduced activity in one SSs may result in compensatory increases in the activity of others. In case of antisense SSII+SSIII, in particular, this is surprising because the potato tuber's ability to polymerize α-glucans from ADP-Glc was largely impaired. Down-regulation of GBSSI activity led to a reduction of the amylose content. As a result, the physical properties of the starch were also altered, the melting temperature of the granule (Tonset) was increased by approximately 5 °C, and the stability of the starch solution after gelatinization was improved due to lack of retrogradation. Although no significant effect was found on the CLD, it should be noted that the methodology employed might not have been conclusive for the detection of very long (DP>50) side chains.
Table 2.
Overview of the consequences for starch yield and starch characteristics upon inhibition of various potato SSs using a reverse genetics approach, and upon heterologous expression of synthases in potato. Cassava GBSSI and E.coli glgA are mentioned for comparison.
| Starch type | Starch yield | Amylose content | CLD | Phosphorylation | Tonset | Peak viscosity | Granule morphology | References |
|---|---|---|---|---|---|---|---|---|
| WT | n.a | 20% | n.a | C6 and C3 | 63 °C | Yes | Normal | [90], [91], [92] |
| as-GBSSI | = | ↓ | = | = | ↑ | = | Normal | [90], [91], [93] |
| as-SSI | = | = | = | = | n.d | n.d | n.d | [20] |
| as-SSII | = | = | DP7,8,9↑ | C6↓, C3= | ↓ | ↓↓ | Fissured | [20], [27], [94] |
| as-SSIII | = | = | DP6↑ | C6↑, C3? | ↓ | ↑ | Fissured and multilobed | [25], [26], [27], [67] |
| as-(SSII+SSIII) | = | = | DP7,8↑; DP12,13↑ DP15–30↓, extra long chains↑ |
C6↓, C3? | ↓↓ | ↓↓ | Fissured and cracked | [27], [67] |
| as-(SSIII+GBSSI) | = | ↓ | DP6↑ | n.d | n.d | n.d | Normal | [95] |
| Cassava GBSSI (amf) | n.d | ↓ | = | n.d | ↑ | = | Normal | [96] |
| E.coli glgA | ↓ | ↓ | low DP↑; high DP↓ | ↓ | ↓ | ↓ | [97] |
When SSII was down-regulated in potato, small changes were observed in the CLD. This lends some support to the hypothesis that each synthase may play a distinct role in elongating side chains of a specific length. However, inhibition of SSII in rug5 mutants of pea caused an increase in very long chains within the amylopectin, and also in very short chains and a loss of intermediate length chains [29]. The physical properties of the potato starch down-regulated in the SSII activity were also altered, both the Tonset and the peak viscosity were decreased. Furthermore, the phosphate content of the antisense SSII starch was also decreased [20]. This is presumably related to the severely lowered peak viscosity. This is consistent with the finding that inhibition of an enzyme involved in starch phosphorylation (R1) may also lead to a collapse of the peak viscosity [68]. When SSII plays a more predominant role in assembling the starch granule in potato (for example when SSIII expression was inhibited), then an increase in the phosphate content, as well as in peak viscosity is observed. Thus, there seems to be a relationship between SSII and starch phosphorylation. It is tempting to speculate that the N-terminal extension of SSII may interact with a starch-phosphorylating enzyme. Another possibility is that SSII can introduce phosphorylated glucose residues into nascent glucan chains or preferentially synthesizes branch lengths more suitable for phosphorylation.
Table 2 also contains two examples of heterologous expression of SSs in potato tubers. The cassava GBSSI was expressed in the amylose-free (amf) potato mutant. Although the GBSSI activity in these starch granules was comparable to that of wild-type granules, the amylose content was only restored to 60% of that of wild-type ones. This study has provided evidence that the potato and cassava GBSSIs differ in their intrinsic properties, and that the cassava enzyme is not fully adapted to the conditions governing starch synthesis in potato. This study illustrated the importance of having certain mutant backgrounds for unravelling the structure-function relationships of particular enzymes.
Glycogen and amylopectin are both α−1,4 and α−1,6 glucans, which differ only in the amount of branching; glycogen is more heavily branched than amylopectin. Table 2 shows that both the fine structure and the physical properties of potato starch can be altered by introduction of a glycogen synthase A (glgA, EC 2.4.1.21). It is worth noting that both the degree of phosphorylation and the peak viscosity were lowered, which was in agreement with the data for antisense SSII starch.
9. Contributions of individual SSs in starch granule formation
Although the mechanism of starch biosynthesis in storage organs has been extensively studied, the exact process (s) involved in the initiation and starch granule formation have not yet been understood and remain to be resolved [28]. Despite the fact that there are protein sequences with homology to glycogenin encoded by the Arabidopsis genome [69], [70], the role and mechanism of such proteins in the initiation of starch biosynthesis still remains to be demonstrated. Our search for a potato homologous sequence for Arabidopsis amylogenin in the potato genome sequence database (www.potatogenome.net) resulted in a putative gene on chromosome 4 close to a known potato SSR marker (SSR450). Further bioinformatic analysis showed that this gene is about 1.2 kb long. Surprisingly, the ChloroP prediction server did not recognize a transit peptide like motif at N-terminal of this putative enzyme (unpublished data).
Starch and glycogen biosynthesis seem to have similarities with respect to linkages and side chains. However, in mammals and yeast, glycogen synthesis seems to be initiated by glycogenin, a self-glucosylating protein where glycoginin adds 10 glucose units onto one of its tyrosine (Y) residues, providing priming molecules for GSs to extend and branch the glycogen [71]. This prototype has led to the suggestion that starch synthesis may be initiated in a similar fashion and a self-glycosylating protein, termed amylogenin, has been reported from maize [70]. However, there has been no association of the activity of this amylogenin with plastidial starch biosynthesis and its activity may be more closely associated with polysaccharide biosynthesis in plant cell walls [72]. Antisense down regulation of a glycogenin-like starch initiation protein (PGSIP) in Arabidopsis leaves resulted in reduction of leaves starch contents, suggesting such a class of proteins may be involved in granule formation [69]. Interestingly, this protein has since been shown to function in the synthesis of the hemicellulose xylan by catalyzing the addition of glucuronic acid onto xylan during plant cell wall formation. These proteins are thus now referred to as GUX (for GlcA substitution of xylan) proteins [73].
In addition to their role as enzymes for elongation of glucose units during starch synthesis, there is mounting evidence that different SSs may directly or indirectly be involved in starch granule formation. Although GBSSI was one of the major enzymes identified, efforts to link granule formation with its activity provide no clue. However, its likely involvement in amylopectin synthesis, particularly in the formation of the extra-long unit chain fraction, may be considered as a member of a still unidentified multiprotien complex needed for the process of starch granule formation. Arabidopsis starch synthase IV mutant analysis has shown that this isoform of SSs may play a selective role in priming of starch granule formation [34], [74]. The number of starch granules per chloroplast was significantly reduced in Arabidopsis thaliana SSIV mutants, suggesting that SSIV is necessary for the production of starch granules [75]. Detailed analysis of a SSIV from wheat endosperms has revealed that this enzyme possesses an N-terminal extension which may play a role in binding. It is anticipated that this extra N-terminal may enable SSIV to interact with other proteins, hence priming the granule initiation [13], [28]. Despite such interesting findings about the role of SSIV in granule formation, starch granule formation in mutant plants lacking SSIV was not completely abolished, suggesting a substitution pathway or involvement of still undetermined isoforms. Taking these results into account, and since an orthologue of SSIV has not been identified in potato plants as yet, expression of SSIV may increase the amount of starch (by controlling the number of starch granule formed in the plastids) in potato tuber amyloplasts [76].
10. Genetics and regulation of potato starch synthase genes
Since various SSs isoforms are expressed in an organ/and or developmental specific manner, it was generally believed that in some plant species, including wheat and maize, multiple genes may encode each SSs isoform [7], [8]. Interestingly, Blast analyses and bioinformatics data have placed all four isoforms of potato SSs on different chromosomes (Table 3).
Table 3.
An overview of various isoforms of potato SSs together with some other biosynthesis enzymes. All potato SSs enzymes catalyse the same reaction known as ADP-glucose+(1,4-alpha-D-glucosyl)(n)=ADP+(1,4-alpha-D-glucosyl)(n+1).
| Enzyme | Chromosome locationa | Isoforms | Closest Molecular Markerb | PGS number |
Genome super scaffold number |
|---|---|---|---|---|---|
| Starch synthase | 3 2 2 8 |
SSI SSII SSIII GBSSI |
cLPT−5-E3 C2_At3g01180 T0562 C2_At1g32900 |
PGSC0003DMG200018552 PGSC0003DMG200001328 PGSC0003DMG200016481 PGSC0003DMG200012111 |
126 4 99 48 |
| Branching enzyme | 4 9 |
SBEI SBEII |
T1203 cLET−7-O3 |
PGSC0003DMG200009981 PGSC0003DMG200002712 |
32 21 |
| Debranching enzyme | 8 9 6 |
ISA1 ISA2 ISA3 |
C2_At1g31410 C2_At2g48120 C2_At4g01900 |
PGSC0003DMG200020699 PGSC0003DMG200000954 PGSC0003DMG200017932 |
361 207 208 |
| Starch phosphorylase | unknown | SP_H, SP_L | PGSC0003DMG200009711 | 644 |
The chromosome location was deduced from the potato genome sequence consortium database (http://www.potatogenome.net/index.php/Main_Page).
Based on tomato genetic map.
Starch synthesis in higher plants is being controlled in different ways. There is increasing evidence of transcriptional control, circadian and redox control, and regulation by phosphorylation. Although enzymatic key steps involved in starch syntheses were regarded as unregulated, a number of recently published papers have shown that starch biosynthesis and degradation enzymes are being modulated by various effector proteins. With respect to starch biosynthesis enzymes, AGPase is the only enzyme whose allosteric regulation has been studied in great detail [77], [78], [79], [80], [81]. It is estimated that 3000–4000 different proteins are present inside plant chloroplasts [82], of which only a small number are being made by the chloroplast genome and the rest are encoded by nuclear genes. Therefore, there must be mechanism(s) to coordinate the expression and activity of such enzymes far from the plant main genome. It is now evident that multi-enzyme complexes of SSs, SBEs and other enzymes may play a significant role in regulation of the pathway [83]. The role of Redox gene signalling to control the level of gene expression in various plant organs is currently a hot area of focus. It is now clear that reduction of oxygen plays a major role with respect to regulation of gene expression in many SSs [84], [85], [86]. It has been shown that when potato tubers were supplied with dithiothreitol, a strong reducing agent, due to redox inactivation of AGPase, starch biosynthesis was hampered. This phenomenon presumably indicates that starch content in the potato tubers is controlled by redox inactivation. The role of protein-protein interactions has recently been uncovered in Arabidopsis and potato plants. Mutant and/or simultaneous down regulation of some of the starch biosynthesis enzymes abolished assembling these proteins into a complex, indicating that high molecular weight complexes may play a regulatory role in starch biosynthesis. Interestingly, it has been shown that at least some small subunits of these enzymes are being phosphorylated [5], [57], [87]. For instance, it has been documented that many pairs of starch biosynthetic enzymes in maize endosperms are capable of associating with each other in multi-subunit complexes [57]. Also, a protein named At5g39790 has recently been identified in Arabidopsis which seems to follow a diurnal rhythm at both mRNA and protein level. This protein seems to have a kind of carbohydrate binding domain and plays a regulatory scaffold protein when interacting with the coil-coiled structure domain of SSIII [88]. The exact mechanism by which SSIII may modulate the activity of other SSs is not fully understood [57]. However, and worth noting, if formation of such complexes regulate the starch biosynthesis rate, why did simultaneous down regulation of SSII and SSIII in potato plants not lead to starch yield penalty? [27], [67]. Molecular analyses by promoter-reporter constructs of several starch synthesis genes in Arabidopsis have indicated that SSs genes were co-regulated spatially by transcriptional activity of the promoters, suggesting a common transcriptional co-regulation mechanism for such genes [89].
Concluding remarks
The examples given above show that an extensive repertoire of transgenic plants with novel starches have already been generated by modification of various starch synthases activities. Although molecular and biochemical analyses of starch synthases have revealed their role in starch biosynthesis, it is often difficult to predict the properties of the transgenic starches in advance. There are contradicting results not only with regard to different plants but also in the same plant species. One interesting finding is that a decrease in the ADP-Glc pool size caused by antisense inhibition of AGPase influences the amount of amylose that is produced. GBSSI has a lower affinity (higher Km) for ADP-Glc compared to other starch synthases. Thus, when the availability of ADP-Glc is limited, GBSSI is the first synthase to become limited by this situation, and consequently relatively less amylose is produced. This example shows that a detailed characterization of the biosynthetic machinery is required to understand the effects of inhibiting any particular enzyme activity during granule assembly. Currently, our understanding of the precise properties of the individual enzymes is still incomplete. An attractive strategy for unravelling the biochemical properties of starch synthases is the introduction of “optimized” enzymes into (their corresponding) mutant background(s). The example with the cassava GBSSI illustrates that the amf potato plants provide a very suitable model system to perform such experiments (especially because potato plants are relatively easy to transform). However, one may challenge the idea by saying that GBSSI is unique with no isoform while other starch synthases mostly possess various isoforms and can compensate the activity in one way or the other.
Acknowledgment
This work was supported by the Iranian ministry of Science and technology and Wageningen University, the Netherlands. The authors would like to thank Dr. Jean-Paul Vincken for helpful discussions.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.bbrep.2017.02.004.
Appendix A. Transparency document
Supplementary material
.
References
- 1.Kok-Jacon G.A., Vincken J.Pl, Suurs L.C.J.M., Visser R.G.F. Mutan produced in potato amyloplasts adheres to starch granules. Plant Biotechnol. J. 2005;3(3):341–351. doi: 10.1111/j.1467-7652.2005.00128.x. [DOI] [PubMed] [Google Scholar]
- 2.Kok-Jacon G.A., Vincken J.Pl, Suurs L.C.J.M., Wang D., Liu S., Visser R.G.F. Production of dextran in transgenic potato plants. Transgenic Res. 2005;14(4):385–395. doi: 10.1007/s11248-005-0439-0. [DOI] [PubMed] [Google Scholar]
- 3.Kok-Jacon G.A., Vincken J.Pl, Suurs L.C.J.M., Wang D., Liu S., Visser R.G.F. Expression of alternansucrase in potato plants. Biotechnol. Lett. 2007;29(7):1135–1142. doi: 10.1007/s10529-007-9348-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nazarian-Firouzabadi F., Kok-Jacon G., Vincken J.P., Ji Q., Suurs L.C.J.M., Visser R.G.F. Fusion proteins comprising the catalytic domain of mutansucrase and a starch-binding domain can alter the morphology of amylose-free potato starch granules during biosynthesis. Transgenic Res. 2007;16(5):645–656. doi: 10.1007/s11248-006-9053-z. [DOI] [PubMed] [Google Scholar]
- 5.Tetlow I.J., Wait R., Lu Z., Akkasaeng R., Bowsher C.G., Esposito S., Kosar-Hashemi B., Morell M.K., Emes M.J. Protein phosphorylation in amyloplasts regulates starch branching enzyme activity and protein-protein interactions. Plant Cell Online. 2004;16(3):694–708. doi: 10.1105/tpc.017400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ball S.G., Morell M.K. From bacterial glycogen to starch: understanding the biogenesis of the plant starch granule. Annu. Rev. Plant Biol. 2003;54(1):207–233. doi: 10.1146/annurev.arplant.54.031902.134927. [DOI] [PubMed] [Google Scholar]
- 7.Harn C., Knight M., Ramakrishnan A., Guan H., Keeling P.L., Wasserman B.P. Isolation and characterization of the zSSIIa and zSSIIb starch synthase cDNA clones from maize endosperm. Plant Mol. Biol. 1998;37(4):639–649. doi: 10.1023/a:1006079009072. [DOI] [PubMed] [Google Scholar]
- 8.Vrinten P.L., Nakamura T. Wheat granule-bound starch synthase I and II are encoded by separate genes that are expressed in different tissues. Plant Physiol. 2000;122(1):255–264. doi: 10.1104/pp.122.1.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Edwards A., Vincken J.P., Suurs L.C.J.M., Visser R.G.F., Zeeman S., Smith A., Martin C. Discrete forms of amylose are synthesized by isoforms of GBSSI in pea. Plant Cell Online. 2002;14(8):1767–1785. doi: 10.1105/tpc.002907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Etxeberria E., Gonzalez P. Evidence for a tonoplast-associated form of sucrose synthase and its potential involvement in sucrose mobilization from the vacuole. J. Exp. Bot. 2003;54(386):1407–1414. doi: 10.1093/jxb/erg148. [DOI] [PubMed] [Google Scholar]
- 11.Bowsher C.G., Scrase-Field E., Esposito S., Emes M.J., Tetlow I.J. Characterization of ADP-glucose transport across the cereal endosperm amyloplast envelope. J. Exp. Bot. 2007;58(6):1321–1332. doi: 10.1093/jxb/erl297. [DOI] [PubMed] [Google Scholar]
- 12.Kirchberger S., Leroch M., Huynen M.A., Wahl M., Neuhaus H.E., Tjaden J. Molecular and biochemical analysis of the plastidic ADP-glucose transporter (ZmBT1) from Zea mays. J. Biol. Chem. 2007;282(31):22481–22491. doi: 10.1074/jbc.M702484200. [DOI] [PubMed] [Google Scholar]
- 13.Leterrier M., Holappa L.D., Broglie K.E., Beckles D.M. Cloning, characterisation and comparative analysis of a starch synthase IV gene in wheat: functional and evolutionary implications. BMC Plant Biol. 2008;8(1):98. doi: 10.1186/1471-2229-8-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Denyer K., Waite D., Motawia S., Møller B.L., Smith A.M. Granule-bound starch synthase I in isolated starch granules elongates malto-oligosaccharides processively. Biochem. J. 1999;340(1):183–191. [PMC free article] [PubMed] [Google Scholar]
- 15.van de Wal M., D'Hulst C., Vincken J.P., Buleon A., Visser R., Ball S. Amylose is synthesized in vitro by extension of and cleavage from amylopectin. J. Biol. Chem. 1998;273(35):22232–22240. doi: 10.1074/jbc.273.35.22232. [DOI] [PubMed] [Google Scholar]
- 16.van de Wal M. Landbouwuniversiteit Wageningen (Wageningen Agricultural University); Wageningen: 2000. Amylose Biosynthesis in Potato: Interaction Between Substrate Availability and GBSSI Activity, Regulated at the Allelic Level. [Google Scholar]
- 17.Denyer K., Clarke B., Hylton C., Tatge H., Smith A.M. The elongation of amylose and amylopectin chains in isolated starch granules. Plant J. 1996;10(6):1135–1143. [Google Scholar]
- 18.Seung D., Soyk S., Coiro M., Maier B.A., Eicke S., Zeeman S.C. Protein targeting to starch is required for localising granule-bound starch synthase to starch granules and for normal amylose synthesis in arabidopsis. PLoS Biol. 2015;13(2) doi: 10.1371/journal.pbio.1002080. (e1002080-e1002080) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jeon J.-S., Ryoo N., Hahn T.-R., Walia H., Nakamura Y. Starch biosynthesis in cereal endosperm. Plant Physiol. Biochem. 2010;48(6):383–392. doi: 10.1016/j.plaphy.2010.03.006. [DOI] [PubMed] [Google Scholar]
- 20.Kossmann J., Abel G.J.W., Springer F., Lloyd J.R., Willmitzer L. Cloning and functional analysis of a cDNA encoding a starch synthase from potato (Solanum tuberosum L.) that is predominantly expressed in leaf tissue. Planta. 1999;208(4):503–511. doi: 10.1007/s004250050587. [DOI] [PubMed] [Google Scholar]
- 21.Commuri P.D., Keeling P.L. Chain-length specificities of maize starch synthase I enzyme: studies of glucan affinity and catalytic properties. Plant J. 2001;25(5):475–486. doi: 10.1046/j.1365-313x.2001.00955.x. [DOI] [PubMed] [Google Scholar]
- 22.Delvallé D., Dumez S., Wattebled F., Roldán I., Planchot V., Berbezy P., Colonna P., Vyas D., Chatterjee M., Ball S. Soluble starch synthase I: a major determinant for the synthesis of amylopectin in Arabidopsis thaliana leaves. Plant J. 2005;43(3):398–412. doi: 10.1111/j.1365-313X.2005.02462.x. [DOI] [PubMed] [Google Scholar]
- 23.Fujita N., Yoshida M., Asakura N., Ohdan T., Miyao A., Hirochika H., Nakamura Y. Function and characterization of starch synthase I using mutants in rice. Plant Physiol. 2006;140(3):1070–1084. doi: 10.1104/pp.105.071845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McMaugh S.J., Thistleton J.L., Anschaw E., Luo J., Konik-Rose C., Wang H., Huang M., Larroque O., Regina A., Jobling S.A. Suppression of starch synthase I expression affects the granule morphology and granule size and fine structure of starch in wheat endosperm. J. Exp. Bot. 2014:eru095. doi: 10.1093/jxb/eru095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abel G.J.W., Springer F., Willmitzer L., Kossmann J. Cloning and functional analysis of a cDNA encoding a novel 139 kDa starch synthase from potato (Solanum tuberosum L.) Plant J. 1996;10(6):981–991. doi: 10.1046/j.1365-313x.1996.10060981.x. [DOI] [PubMed] [Google Scholar]
- 26.Marshall J., Sidebottom C., Debet M., Martin C., Smith A.M., Edwards A. Identification of the major starch synthase in the soluble fraction of potato tubers. Plant Cell Online. 1996;8(7):1121–1135. doi: 10.1105/tpc.8.7.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Edwards A., Fulton D.C., Hylton C.M., Jobling S.A., Gidley M., Rössner U., Martin C., Smith A.M. A combined reduction in activity of starch synthases II and III of potato has novel effects on the starch of tubers. Plant J. 1999;17(3):251–261. [Google Scholar]
- 28.Zeeman S.C., Kossmann J., Smith A.M. Starch: its metabolism, evolution, and biotechnological modification in plants. Annu. Rev. Plant Biol. 2010;61:209–234. doi: 10.1146/annurev-arplant-042809-112301. [DOI] [PubMed] [Google Scholar]
- 29.Craig J., Lloyd J.R., Tomlinson K., Barber L., Edwards A., Wang T.L., Martin C., Hedley C.L., Smith A.M. Mutations in the gene encoding starch synthase II profoundly alter amylopectin structure in pea embryos. Plant Cell Online. 1998;10(3):413–426. doi: 10.1105/tpc.10.3.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morell M.K., Kosar-Hashemi B., Cmiel M., Samuel M.S., Chandler P., Rahman S., Buleon A., Batey I.L., Li Z. Barley sex6 mutants lack starch synthase IIa activity and contain a starch with novel properties. Plant J. 2003;34(2):173–185. doi: 10.1046/j.1365-313x.2003.01712.x. [DOI] [PubMed] [Google Scholar]
- 31.Nakamura Y., Francisco P.B., Hosaka Y., Sato A., Sawada T., Kubo A., Fujita N. Essential amino acids of starch synthase IIa differentiate amylopectin structure and starch quality between japonica and indica rice varieties. Plant Mol. Biol. 2005;58(2):213–227. doi: 10.1007/s11103-005-6507-2. [DOI] [PubMed] [Google Scholar]
- 32.Takahata Y., Tanaka M., Otani M., Katayama K., Kitahara K., Nakayachi O., Nakayama H., Yoshinaga M. Inhibition of the expression of the starch synthase II gene leads to lower pasting temperature in sweetpotato starch. Plant Cell Rep. 2010;29(6):535–543. doi: 10.1007/s00299-010-0842-8. [DOI] [PubMed] [Google Scholar]
- 33.Zhang X., Szydlowski N., Delvallé D., D'Hulst C., James M.G., Myers A.M. Overlapping functions of the starch synthases SSII and SSIII in amylopectin biosynthesis in Arabidopsis. BMC Plant Biol. 2008;8(1):96. doi: 10.1186/1471-2229-8-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roldán I., Wattebled F., Lucas M.M., Delvallé D., Planchot V., Jiménez S., Pérez R., Ball S., D'Hulst C., Mérida Á. The phenotype of soluble starch synthase IV defective mutants of Arabidopsis thaliana suggests a novel function of elongation enzymes in the control of starch granule formation. Plant J. 2007;49(3):492–504. doi: 10.1111/j.1365-313X.2006.02968.x. [DOI] [PubMed] [Google Scholar]
- 35.Dian W., Jiang H., Wu P. Evolution and expression analysis of starch synthase III and IV in rice. J. Exp. Bot. 2005;56(412):623–632. doi: 10.1093/jxb/eri065. [DOI] [PubMed] [Google Scholar]
- 36.Charnock S.J., Davies G.J. Structure of the nucleotide-diphospho-sugar transferase, SpsA from Bacillus subtilis, in native and nucleotide-complexed forms. Biochemistry. 1999;38(20):6380–6385. doi: 10.1021/bi990270y. [DOI] [PubMed] [Google Scholar]
- 37.Chiu C.P.C., Watts A.G., Lairson L.L., Gilbert M., Lim D., Wakarchuk W.W., Withers S.G., Strynadka N.C.J. Structural analysis of the sialyltransferase CstII from Campylobacter jejuni in complex with a substrate analog. Nat. Struct. Mol. Biol. 2004;11(2):163–170. doi: 10.1038/nsmb720. [DOI] [PubMed] [Google Scholar]
- 38.Liu H., Yu G., Wei B., Wang Y., Zhang J., Hu Y., Liu Y., Yu G., Zhang H., Huang Y. Identification and phylogenetic analysis of a novel starch synthase in maize. Front. Plant Sci. 2015;6:1013. doi: 10.3389/fpls.2015.01013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yep A., Ballicora M.A., Sivak M.N., Preiss J. Identification and characterization of a critical region in the glycogen synthase from Escherichia coli. J. Biol. Chem. 2004;279(9):83598367. doi: 10.1074/jbc.M312686200. [DOI] [PubMed] [Google Scholar]
- 40.Buschiazzo A., Ugalde J.E., Guerin M.E., Shepard W., Ugalde R.A., Alzari P.M. Crystal structure of glycogen synthase: homologous enzymes catalyse glycogen synthesis and degradation. EMBO J. 2004;23:3196–3205. doi: 10.1038/sj.emboj.7600324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Momma M., Fujimoto Z. Interdomain disulfide bridge in the rice granule bound starch synthase I catalytic domain as elucidated by X-ray structure analysis. Biosci. Biotechnol. Biochem. 2012;76(8):1591–1595. doi: 10.1271/bbb.120305. [DOI] [PubMed] [Google Scholar]
- 42.Cuesta-Seijo J.A., Nielsen M.M., Marri L., Tanaka H., Beeren S.R., Palcic M.M. Structure of starch synthase I from barley: insight into regulatory mechanisms of starch synthase activity. Acta Crystallogr. Sect. D. Biol. Crystallogr. 2013;69(6):1013–1025. doi: 10.1107/S090744491300440X. [DOI] [PubMed] [Google Scholar]
- 43.Furukawa K., Tagaya M., Inouye M., Preiss J., Fukui T. Identification of lysine 15 at the active site in Escherichia coli glycogen synthase. Conservation of Lys-X-Gly-Gly sequence in the bacterial and mammalian enzymes. J. Biol. Chem. 1990;265(4):2086–2090. [PubMed] [Google Scholar]
- 44.Furukawa K., Tagaya M., Tanizawa K., Fukui T. Role of the conserved Lys-X-Gly-Gly sequence at the ADP-glucose-binding site in Escherichia coli glycogen synthase. J. Biol. Chem. 1993;268(32):23837–23842. [PubMed] [Google Scholar]
- 45.Sheng F., Jia X., Yep A., Preiss J., Geiger J.H. The crystal structures of the open and catalytically competent closed conformation of Escherichia coli glycogen synthase. J. Biol. Chem. 2009;284(26):17796–17807. doi: 10.1074/jbc.M809804200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yep A., Ballicora M.A., Preiss J. The ADP-glucose binding site of the Escherichia coli glycogen synthase. Arch. Biochem. Biophys. 2006;453(2):188–196. doi: 10.1016/j.abb.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 47.Gao Z., Keeling P., Shibles R., Guan H. Involvement of lysine-193 of the conserved. Arch. Biochem. Biophys. 2004;427(1):1–7. doi: 10.1016/j.abb.2004.01.010. [DOI] [PubMed] [Google Scholar]
- 48.Edwards A., Borthakur A., Bornemann S., Venail J., Denyer K., Waite D., Fulton D., Smith A., Martin C. Specificity of starch synthase isoforms from potato. Eur. J. Biochem. 1999;266(3):724–736. doi: 10.1046/j.1432-1327.1999.00861.x. [DOI] [PubMed] [Google Scholar]
- 49.Asare E.K., Baìšga M., Rossnagel B.G., Chibbar R.N. Polymorphism in the barley granule bound starch synthase 1 (gbss1) gene associated with grain starch variant amylose concentration. J. Agric. Food Chem. 2012;60(40):10082–10092. doi: 10.1021/jf302291t. [DOI] [PubMed] [Google Scholar]
- 50.Sparla F., Falini G., Botticella E., Pirone C., Talamè V., Bovina R., Salvi S., Tuberosa R., Sestili F., Trost P. New starch phenotypes produced by TILLING in barley. PLoS One. 2014;9(10):e107779. doi: 10.1371/journal.pone.0107779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang X., Feng B., Xu Z., Sestili F., Zhao G., Xiang C., Lafiandra D., Wang T. Identification and characterization of granule bound starch synthase I (GBSSI) gene of tartary buckwheat (Fagopyrum tataricum Gaertn.) Gene. 2014;534(2):229–235. doi: 10.1016/j.gene.2013.10.053. [DOI] [PubMed] [Google Scholar]
- 52.Brust H., Orzechowski S., Fettke J., Steup M. Starch synthesizing reactions and paths: in vitro and in vivo studies. J. Appl. Glycosci. 2013;60(1):3–20. [Google Scholar]
- 53.Ross J., Li Y., Lim E.-K., Bowles D.J. Higher plant glycosyltransferases. Genome Biol. 2001;2(2) doi: 10.1186/gb-2001-2-2-reviews3004. (reviews3004. 1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Raynaud S., Ragel P., Rojas T., Mérida Á. The N-terminal part of Arabidopsis thaliana starch synthase 4 determines the localization and activity of the enzyme. J. Biol. Chem. 2016;291(20):10759–10771. doi: 10.1074/jbc.M115.698332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Keeling P.L., Myers A.M. Biochemistry and genetics of starch synthesis. Annu. Rev. Food Sci. Technol. 2010;1:271–303. doi: 10.1146/annurev.food.102308.124214. [DOI] [PubMed] [Google Scholar]
- 56.Imparl-Radosevich J.M., Nichols D.J., Li P., McKean A.L., Keeling P.L., Guan H. Analysis of purified maize starch synthases iia and iib: ss isoforms can be distinguished based on their kinetic properties. Arch. Biochem. Biophys. 1999;362(1):131–138. doi: 10.1006/abbi.1998.1028. [DOI] [PubMed] [Google Scholar]
- 57.Hennen-Bierwagen T.A., Liu F., Marsh R.S., Kim S., Gan Q., Tetlow I.J., Emes M.J., James M.G., Myers A.M. Starch biosynthetic enzymes from developing maize endosperm associate in multisubunit complexes. Plant Physiol. 2008;146(4):1892–1908. doi: 10.1104/pp.108.116285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Senoura T., Asao A., Takashima Y., Isono N., Hamada S., Ito H., Matsui H. Enzymatic characterization of starch synthase III from kidney bean (Phaseolus vulgaris L.) FEBS J. 2007;274(17):4550–4560. doi: 10.1111/j.1742-4658.2007.05984.x. [DOI] [PubMed] [Google Scholar]
- 59.Valdez H.A., Busi M.V., Wayllace N.Z., Parisi G., Ugalde R.A., Gomez-Casati D.F. Role of the N-terminal starch-binding domains in the kinetic properties of starch synthase III from Arabidopsis thaliana. Biochemistry. 2008;47(9):3026–3032. doi: 10.1021/bi702418h. [DOI] [PubMed] [Google Scholar]
- 60.Wayllace N.Z., Valdez H.A., Ugalde R.A., Busi M.V., Gomez-Casati D.F. The starch†binding capacity of the noncatalytic SBD2 region and the interaction between the N†and C†terminal domains are involved in the modulation of the activity of starch synthase III from Arabidopsis thaliana. FEBS J. 2010;277(2):428–440. doi: 10.1111/j.1742-4658.2009.07495.x. [DOI] [PubMed] [Google Scholar]
- 61.Southall S.M., Simpson P.J., Gilbert H.J., Williamson G., Williamson M.P. The starch-binding domain from glucoamylase disrupts the structure of starch. FEBS Lett. 1999;447(1):58–60. doi: 10.1016/s0014-5793(99)00263-x. [DOI] [PubMed] [Google Scholar]
- 62.Lim E.-K. Plant glycosyltransferases: their potential as novel biocatalysts. Chem.-A Eur. J. 2005;11(19):5486–5494. doi: 10.1002/chem.200500115. [DOI] [PubMed] [Google Scholar]
- 63.Mukerjea R., Robyt J.F. De novo biosynthesis of starch chains without a primer and the mechanism for its biosynthesis by potato starch-synthase. Carbohydr. Res. 2012;352:137–142. doi: 10.1016/j.carres.2011.11.024. [DOI] [PubMed] [Google Scholar]
- 64.Imparl-Radosevich J.M., Keeling P.L., Guan H. Essential arginine residues in maize starch synthase IIa are involved in both ADP-glucose and primer binding. FEBS Lett. 1999;457(3):357–362. doi: 10.1016/s0014-5793(99)01066-2. [DOI] [PubMed] [Google Scholar]
- 65.Nichols D.J., Keeling P.L., Spalding M., Guan H. Involvement of conserved aspartate and glutamate residues in the catalysis and substrate binding of maize starch synthase. Biochemistry. 2000;39(26):7820–7825. doi: 10.1021/bi000407g. [DOI] [PubMed] [Google Scholar]
- 66.Campbell J.A., Davies G.J., Bulone V., Henrissat B. A classification of nucleotide-diphospho-sugar glycosyltransferases based on amino acid sequence similarities. Biochem. J. 1997;326(3):929–939. doi: 10.1042/bj3260929u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lloyd J.R., Landschütze V., Kossmann J. Simultaneous antisense inhibition of two starch-synthase isoforms in potato tubers leads to accumulation of grossly modified amylopectin. Biochem. J. 1999;338(2):515–521. [PMC free article] [PubMed] [Google Scholar]
- 68.Lorberth R., Ritte G., Willmitzer L., Kossmann J. Inhibition of a starch-granule–bound protein leads to modified starch and repression of cold sweetening. Nat. Biotechnol. 1998;16(5):473–477. doi: 10.1038/nbt0598-473. [DOI] [PubMed] [Google Scholar]
- 69.Chatterjee M., Berbezy P., Vyas D., Coates S., Barsby T. Reduced expression of a protein homologous to glycogenin leads to reduction of starch content in Arabidopsis leaves. Plant Sci. 2005;168(2):501–509. [Google Scholar]
- 70.Singh D.G., Lomako J., Lomako W.M., Whelan W.J., Meyer H.E., Serwe M., Metzger J.W. [beta]-Glucosylarginine: a new glucose-protein bond in a self-glucosylating protein from sweet corn. FEBS Lett. 1995;376(1–2):61–64. doi: 10.1016/0014-5793(95)01247-6. [DOI] [PubMed] [Google Scholar]
- 71.Roach P.J., Skurat A.V. Self-glucosylating initiator proteins and their role in glycogen biosynthesis. Prog. Nucleic Acid Res. Mol. Biol. 1997;57:289–316. doi: 10.1016/s0079-6603(08)60284-6. [DOI] [PubMed] [Google Scholar]
- 72.Nakamura Y. Towards a better understanding of the metabolic system for amylopectin biosynthesis in plants: rice endosperm as a model tissue. Plant Cell Physiol. 2002;43(7):718–725. doi: 10.1093/pcp/pcf091. [DOI] [PubMed] [Google Scholar]
- 73.Rennie E.A., Hansen S.F., Baidoo E.E., Hadi M.Z., Keasling J.D., Scheller H.V. Three members of the Arabidopsis glycosyltransferase family 8 are xylan glucuronosyltransferases. Plant Physiol. 2012;159(4):1408–1417. doi: 10.1104/pp.112.200964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Szydlowski N., Ragel P., Raynaud S., Lucas M.M., Roldan I., Montero M., Munoz F.J., Ovecka M., Bahaji A., Planchot V. Starch granule initiation in Arabidopsis requires the presence of either class IV or class III starch synthases. Plant Cell Online. 2009;21(8):2443–2457. doi: 10.1105/tpc.109.066522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Crumpton-Taylor M., Pike M., Lu K.J., Hylton C.M., Feil R., Eicke S., Lunn J.E., Zeeman S.C., Smith A.M. Starch synthase 4 is essential for coordination of starch granule formation with chloroplast division during Arabidopsis leaf expansion. New Phytol. 2013;200(4):1064–1075. doi: 10.1111/nph.12455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.D'Hulst C., Mérida Á. The priming of storage glucan synthesis from bacteria to plants: current knowledge and new developments. New Phytol. 2010;188(1):13–21. doi: 10.1111/j.1469-8137.2010.03361.x. [DOI] [PubMed] [Google Scholar]
- 77.Ballicora M.A., Erben E.D., Yazaki T., Bertolo A.L., Demonte A.M., Schmidt J.R., Aleanzi M., Bejar C.M., Figueroa C.M., Fusari C.M. Identification of regions critically affecting kinetics and allosteric regulation of the Escherichia coli ADP-glucose pyrophosphorylase by modeling and pentapeptide-scanning mutagenesis. J. Bacteriol. 2007;189(14):5325–5333. doi: 10.1128/JB.00481-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ghosh H.P., Preiss J. Adenosine diphosphate glucose pyrophosphorylase: a regulatory enzyme in the biosynthesis of starch in spinach leaf chloroplasts. J. Biol. Chem. 1966;241(19):4491–4504. [PubMed] [Google Scholar]
- 79.Gómez-Casati D.F., Iglesias A.A. ADP-glucose pyrophosphorylase from wheat endosperm. Purification and characterization of an enzyme with novel regulatory properties. Planta. 2002;214(3):428–434. doi: 10.1007/s004250100634. [DOI] [PubMed] [Google Scholar]
- 80.Kleczkowski L.A., Villand P., Luthi E., Olsen O.A., Preiss J. Insensitivity of barley endosperm ADP-glucose pyrophosphorylase to 3-phosphoglycerate and orthophosphate regulation. Plant Physiol. 1993;101(1):179–186. doi: 10.1104/pp.101.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nagata T., Saitou K. Regulation of expression of D3-type cyclins and ADP-glucose pyrophosphorylase genes by sugar, cytokinin and ABA in sweet potato (Ipomoea batatas Lam.) Plant Prod. Sci. 2009;12(4):434–442. [Google Scholar]
- 82.Fey V., Wagner R., Brautigam K., Pfannschmidt T. Photosynthetic redox control of nuclear gene expression. J. Exp. Bot. 2005;56(416):1491–1498. doi: 10.1093/jxb/eri180. [DOI] [PubMed] [Google Scholar]
- 83.Kötting O., Kossmann J., Zeeman S.C., Lloyd J.R. Regulation of starch metabolism: the age of enlightenment? Curr. Opin. Plant Biol. 2010;13(3):320–328. doi: 10.1016/j.pbi.2010.01.003. [DOI] [PubMed] [Google Scholar]
- 84.Fu Y., Ballicora M.A., Leykam J.F., Preiss J. Mechanism of reductive activation of potato tuber ADP-glucose pyrophosphorylase. J. Biol. Chem. 1998;273(39):25045–25052. doi: 10.1074/jbc.273.39.25045. [DOI] [PubMed] [Google Scholar]
- 85.Michalska J., Zauber H., Buchanan B.B., Cejudo F.J., Geigenberger P. NTRC links built-in thioredoxin to light and sucrose in regulating starch synthesis in chloroplasts and amyloplasts. Proc. Natl. Acad. Sci. USA. 2009;106(24):9908–9913. doi: 10.1073/pnas.0903559106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sokolov L.N., Dominguez-Solis J.R., Allary A.L., Buchanan B.B., Luan S. A redox-regulated chloroplast protein phosphatase binds to starch diurnally and functions in its accumulation. Proc. Natl. Acad. Sci. USA. 2006;103(25) doi: 10.1073/pnas.0603329103. (973297-37) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tetlow I.J., Beisel K.G., Cameron S., Makhmoudova A., Liu F., Bresolin N.S., Wait R., Morell M.K., Emes M.J. Analysis of protein complexes in wheat amyloplasts reveals functional interactions among starch biosynthetic enzymes. Plant Physiol. 2008;146(4):1878–1891. doi: 10.1104/pp.108.116244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lohmeier-Vogel E.M., Kerk D., Nimick M., Wrobel S., Vickerman L., Muench D.G., Moorhead G.B.G. Arabidopsis At5g39790 encodes a chloroplast-localized, carbohydrate-binding, coiled-coil domain-containing putative scaffold protein. BMC Plant Biol. 2008;8(1):120. doi: 10.1186/1471-2229-8-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tsai H.-L., Lue W.-L., Lu K.-J., Hsieh M.-H., Wang S.-M., Chen J. Starch synthesis in Arabidopsis is achieved by spatial cotranscription of core starch metabolism genes. Plant Physiol. 2009;151(3):1582–1595. doi: 10.1104/pp.109.144196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Visser R.G.F., Suurs L., Bruinenberg P.M., Bleeker I., Jacobsen E. Comparison between amylose-free and amylose containing potato starches. Starch-Stärke. 1997;49(11):438–443. [Google Scholar]
- 91.Visser R.G.F., Suurs L., Steeneken P.A.M., Jacobsen E. Some physicochemical properties of amylose-free potato starch. Starch-Stärke. 1997;49(11):443–448. [Google Scholar]
- 92.Muhrbeck P., Tellier C. Determination of the posphorylation of starch from native potato varieties by 31P NMR. Starch‐Stärke. 1991;43(1):25–27. [Google Scholar]
- 93.Kozlov S.S., Blennow A., Krivandin A.V., Yuryev V.P. Structural and thermodynamic properties of starches extracted from GBSS and GWD suppressed potato lines. Int. J. Biol. Macromol. 2007;40(5):449–460. doi: 10.1016/j.ijbiomac.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 94.Edwards A., Marshall J., Sidebottom C., Visser R.G.F., Smith A.M., Martin C. Biochemical and molecular characterization of a novel starch synthase from potato tubers. Plant J. 1995;8(2):283–294. doi: 10.1046/j.1365-313x.1995.08020283.x. [DOI] [PubMed] [Google Scholar]
- 95.Fulton D.C., Edwards A., Pilling E., Robinson H.L., Fahy B., Seale R., Kato L., Donald A.M., Geigenberger P., Martin C. Role of granule-bound starch synthase in determination of amylopectin structure and starch granule morphology in potato. J. Biol. Chem. 2002;277(13):10834–10841. doi: 10.1074/jbc.M111579200. [DOI] [PubMed] [Google Scholar]
- 96.Salehuzzaman S.N.I.M., Jacobsen E., Visser R.G.F. Expression patterns of two starch biosynthetic genes in in vitro cultured cassava plants and their induction by sugars. Plant Sci. 1994;98(1):53–62. [Google Scholar]
- 97.Shewmaker C.K., Boyer C.D., Wiesenborn D.P., Thompson D.B., Boersig M.R., Oakes J.V., Stalker D.M. Expression of Escherichia coli glycogen synthase in the tubers of transgenic potatoes (Solanum tuberosum) results in a highly branched starch. Plant Physiol. 1994;104(4):1159–1166. doi: 10.1104/pp.104.4.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Buléon A., Colonna P., Planchot V., Ball S. Starch granules: structure and biosynthesis. Int. J. Biol. Macromolec. 1998;23(2):85–112. doi: 10.1016/s0141-8130(98)00040-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material