Abstract
Background
Genomic DNA extracted from species of Cactaceae is often contaminated with significant amounts of mucilage and pectin. Pectin is one of the main components of cellular walls, whereas mucilage is a complex polysaccharide with a ramified structure. Thus, pectin- and mucilage-free extraction of DNA is a key step for further downstream PCR-based analyses.
Results
We tested our DNA extraction method on cladode tissue (juvenile, adult, and herbaria exemplars) of 17 species of Opuntia Mill., which are characterized by a large quantity of pectin and mucilage.
Conclusion
We developed a method for the extraction of gDNA free of inhibitory compounds common in species of Opuntia Mill., such as pectin and mucilage. Compared to previously extraction protocols, our method produced higher yields of high-quality genomic DNA.
Keywords: DNA quality, DNA quantity, Genomic DNA, Mucilage, Opuntia, Pectin
Background
Present-day DNA-based molecular studies are useful tools with a wide-range of applications in different biological disciplines. Molecular studies, especially in species with similar morphologies, can be used to characterize and differentiate species [1, 2]. Such studies have used molecular techniques involving PCR amplification of DNA [3, 4] to successfully solve taxonomic and phylogenetic controversies [5]. More specifically, DNA analyses have been used at different taxonomic levels, from communities of bacteria, fungi, yeast, plants and animals, to the cloning of specific genes [6]. High-quality DNA extraction is a necessary first step to conduct molecular studies. This can be performed using conventional methods or commercial kits specifically designed for particular types of samples. Most commercial kits efficiently capture DNA using extraction columns and resins, but the cost of these kits limits their application to large numbers of samples [7].
Conventional methods of DNA extraction involve three basic steps: (1) lysis of cellular walls and membranes; (2) removal of cell debris and other molecular compounds (e.g., polysaccharides, secondary metabolites, proteins, tannins, alkaloids, and polyphenols); (3) DNA precipitation and purification [8]. Currently, fast and cost-efficient DNA extraction protocols yielding large quantities of high-quality DNA are key to the study of species’ molecular genetics [9]. For example, DNA extracted from species of cacti (Cactaceae) are often contaminated with high quantities of mucilage and pectin [10–15].
In these species, pectin is the main component of the cellular wall and its composition often varies among species (e.g., Opuntia), location and environments. The main molecular components of pectin are α-(1 → 4) chains linked to d-galacturonic acid interspersed by the insertion of (1 → 2) residues linked to adjacent or alternate residues of l-rhamnopyranosyl. The lineal segments are predominantly composed of homogalacturone [16].
Mucilage is an organic component present in large cells (idioblasts) in the chlorenchyma and adjacent water-retaining parenchymal cells [17, 18]. Mucilage is composed of complex polysaccharides with ramified structures [16] containing varying proportions of different sugars (e.g., l-arabinose, pyranose, furanose, d-galactose, l-rhamnose and d-xylose) and galacturonic acid. The primary structure of the molecule consists of lineal repetitive chains of 1,4-β-d-galacturonic acid and α-1,2-l-rhamnose with a trisaccharide of β-1,6-d-glucose with a lateral chain joined to O-4-l-residues of rhamnose [19, 20]. Mucilage is found throughout all body parts, including flowers [11]. In most species of cacti, mucilage is secreted in response to wounds and during the DNA extraction process. More specifically, during the DNA extraction process mucilage appears as soon as the tissue is pulverized, which significantly hinders the efficiency of the extraction and purification [21].
Generally, extraction and purification of high-quality genomic DNA (gDNA) is hindered by the presence of pectin that precipitates alongside DNA [22], thus reducing the quality and yield of the extraction process [23]. Although efficient DNA extraction is crucial for downstream PCR-based analyses, there are relatively few studies focusing on gDNA extraction efficiency in species of cacti [11, 13, 22, 24–27]. In this context, the aim of the present study was to develop a simple and cost-effective method to obtain large yields of high-quality gDNA from cladode tissue of Opuntia species.
Methods
We obtained tissues samples from the national Opuntia collection of the Botanical Garden at Instituto de Biología, Universidad Nacional Autónoma de México.
Protocol
CTAB 2X buffer
Prepare CTAB 2X buffer solution (Tris 10 mM pH8.0; EDTA 20 mM, pH 8.0; CTAB 2; NaCl 1.4 M) and preheat to 80 °C for 5 min.
Pulverize 2–3 mg of tissue using liquid nitrogen.
Mix the pulverized tissue with 700 µl of CTAB 2X in a 2 mL eppendorf tube. Mix vigorously for 20 s.
Heat to 85 °C for 2 h and mix vigorously for 20 s.
Add 750 µl of chloroform: isoamyl alcohol (24: 1) and mix vigorously for 20 s.
Centrifuge for 60 min at 12,000 g (4 °C).
Transfer the aqueous phase to a 1.5 mL eppendorf tube.
Add 400 µl of isopropyl alcohol previously cooled to − 20 °C. Mix gently for 1 min.
Centrifuge for 25 min at 10,000 g. Discard the supernatant.
Add 500 µl HPLC-grade water to the DNA pellet to dissolve the pectin (evident as a gelatinous substance). Do not mix and discard the disolved pectin with a micropipette.
Resuspend the pellet in 1 mL of ethanol (70) previously cooled to − 20 °C.
Centrifuge for 5 min at 10,000 g. Discard the supernatant.
Air-dry pellet at room temperature for 40 min.
Resuspend the pellet in 50 µl of HPLC-grade water.
Heat to 60 °C for 15 min.
Integrity of the extracted DNA
We analyzed the integrity of extracted gDNA from 17 species of Opuntia by electrophoresis (1 h with a 87 V cm−3 current) using 1.5 agarose gels prepared with TAE buffer (Tris Acetate-EDTA) and stained with Gel red (Biotium, USA). DNA bands were visualized under UV light with an Infinity 3000 transilluminator (Vilber Lourmat, Germany), which confirmed the presence of intact high quality gDNA without conspicuous contamination by proteins or other compounds (Fig. 1).
Fig. 1.
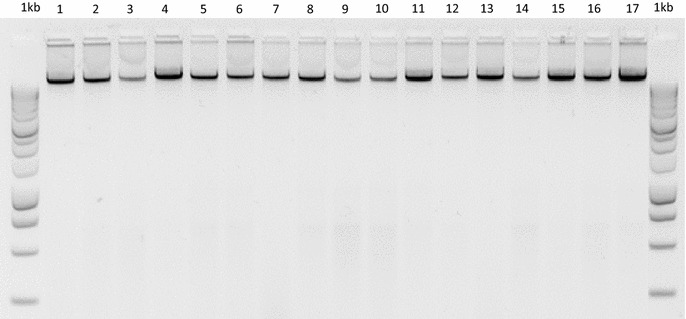
Image of the agarose gel of genomic DNA (gDNA) ran by electrophoresis extracted from 17 tissue samples of Opuntia Mill., using the improved extraction method (Promega™ 1 kb DNA Ladder Molecular Weight Marker)
Evaluation of gDNA concentration
We determined gDNA concentration with a spectrophotometry analysis using a NanoDrop 8000 (Thermo, USA) and with a fluorometry analysis using Quant-iT™ PicoGreen® dsDNA Assay Kit (Invitrogen™) according to the manufacturer’s instructions.
Evaluation of the quality of gDNA
We assessed the purity of all the gDNA samples by spectrophotometry with a Nanodrop 8000 (Thermo, USA) (Table 1).
Table 1.
Genomic DNA (gDNA) concentration and quality extracted from 17 tissue samples of Opuntia Mill. using the improved extraction method
| Species | PicoGreen ng/µl | NanoDrop ng/µl | C B ratio PicoGreen concentration/Nanodrop concentration | A260/A280 NanoDrop | A260/A230 NanoDrop |
|---|---|---|---|---|---|
| 1. Opuntia auberi Pfeiff. | 1250 | 1500 | 0.83 | 1.9 | 2.1 |
| 2. Opuntia decumbens Salm-Dyck | 3199 | 3642 | 0.87 | 1.9 | 2.2 |
| 3. Opuntia delafuentiana Martínez-González et al. | 8021 | 8126 | 0.98 | 1.9 | 2.2 |
| 4. Opuntia depressa Britton and Rose | 2191 | 2588 | 0.84 | 1.9 | 2.0 |
| 5. Opuntia durangensis Britton and Rose | 8220 | 8853 | 0.92 | 1.8 | 2.1 |
| 6. Opuntia ficus-indica Mill. | 5898 | 6196 | 0.95 | 1.9 | 2.1 |
| 7. Opuntia heliabravoana Scheinvar | 8341 | 9147 | 0.91 | 2.0 | 1.9 |
| 8. Opuntia huajuapensis Bravo | 3624 | 4497 | 0.80 | 1.9 | 2.1 |
| 9. Opuntia joconostle F. A. C. Weber | 1091 | 1304 | 0.83 | 1.8 | 2.2 |
| 10. Opuntia lasiacantha Pfeiff. | 1892 | 2088 | 0.90 | 1.9 | 2.1 |
| 11. Opuntia leiascheinvariana Martínez-González | 4799 | 5407 | 0.88 | 1.9 | 2.2 |
| 12. Opuntia leucotricha DC. | 6258 | 7000 | 0.89 | 1.9 | 2.2 |
| 13. Opuntia matudae Scheinvar | 2354 | 2802 | 0.84 | 1.9 | 2.2 |
| 14. Opuntia megacantha Salm-Dyck | 6895 | 7861 | 0.87 | 1.8 | 2.1 |
| 15. Opuntia microdasys Pfeiff. | 7526 | 8592 | 0.87 | 1.9 | 2.1 |
| 16. Opuntia oligacantha Förster | 1548 | 1897 | 0.81 | 1.8 | 2.2 |
| 17. Opuntia olmeca Joel Pérez et al. | 2112 | 2568 | 0.82 | 1.9 | 2.1 |
PCR amplifications
The purity of gDNA was confirmed through PCR of three different molecular markers: (1) nDNA internal transcribed spacer (ITS, 600 bp) [28–32]; (2) cpDNA RuBisCO gene (rbcL, 500 pb) [33, 34]; (3) mtDNA cytrochrome oxidase subunit 3 (cox3, 1000pb) [35]. We used a negative control (without target gDNA) to confirm no contamination with extraneous DNA before the PCR. PCRs were performed on a final volume 25 µL containing 1 × buffer, 0.8 mM dNTPs mix, 20 pmol of each primer, 2 units of GoTaq DNA (Promega, USA) and 100 ng of template DNA. For each gene, PCRs consisted of an initial denaturation step at 96 °C for 2 min, followed by 35 cycles at 94 °C for 1 min, annealing temperature differing according to the primer for 1 min (Table 2), 72 °C elongation temperature for different time durations, depending on the length of the product. PCRs were performed using a Peltier Thermal Cycler PTC-200 (BIORAD, México). Amplification products were subjected to electrophoresis (1 h with a 87 V cm−3 current) using 1.5 agarose gels prepared with TAE buffer (Tris Acetate-EDTA), stained with Gel red (Biotium, USA) and visualized with an Infinity 3000 transilluminator (Vilber Lourmat, Germany). PCR products were purified with the ExoSAP Purification kit (Affymetrix, USA) and sequenced using the Bigdye terminator v.3.1 Cycle Sequencing kit (Applied Biosystem) and an Applied Biosystems 3730 × L automated sequencer (Applied BioSystems, USA).
Table 2.
Primers used in the amplification and sequencing of the DNA fragments
| Locus/segment | Name | Sequence 5′–3′ | Tm (°C) |
|---|---|---|---|
| ITS | ITS5 | GGAAGTAAAAGTCGTAACAAGG | 57 |
| ITS4 | TCCTCCGCTTATTGATATGC | 57 | |
| rbcL | 1f | ATGTCACCACAAACAGAAAC | 56 |
| 724r | TCGCATGTACCTGCAGTAGC | 56 | |
| cox3 | Cox3f | CCGTAGGAGGTGTGATGT | 51 |
| Cox3r | CTCCCCACCAATAGATAGAG | 51 |
Sequence assembly
DNA sequences were visualized, edited and assembled using BioEdit vers. 7.0.5 [36]. For each gene, consensus sequences were compared with those deposited in GenBank using the BLASTN 2.2.19 search algorithm [37].
Comparison with previous methods
Our protocol was compared with two previous methods [11, 13] using 17 species of Opuntia.
Only one species (Opuntia ficus-indica) was shared with the protocol of Mondragón et al. [11].
Results
The list of the 17 species of Opuntia studied is shown in Table 1.
Our new extraction method allowed us to obtain high quality gDNA from young and mature cladodes using standard protocols using CTAB (Cetyl Trimethyl Ammonium Bromide), which efficiently extracts polysaccharides from leaf tissue. The Agarose gel electrophoresis showed the presence of large quantities of gDNA free of contaminants (Fig. 1). Accordingly, the large amount of gDNA was confirmed with two different methods (i.e., spectrophotometry and fluorimetry). These analyses yielded a mean gDNA ratio (PicoGreen concentration/Nanodrop concentration) of 0.80–0.98 ng/µl for all of the samples tested (Table 1). We obtained reliable absorbance readings from the spectrophotometric analysis.
The estimation of the A260/A280 absorbance ratio is a common way to measure DNA purity. Nucleic acids have a maximum absorbance at a wavelength of 260 nm, thus absorbance at this wavelength is directly proportional to DNA concentration. On the other hand, proteins show a maximum absorbance at 280 nm wavelength (mainly resulting from tryptophan residues), thus absorbance readings at 280 nm measure the concentration of proteins in the sample. Depending on the base composition of DNA, reading for the A260/A280 ratio between 1.6 and 1.9 are indicative of high-quality DNA. In addition, absorbance readings at 230 nm wavelength measure the concentration of salts, carbohydrates and other contaminants, so the A260/A230 absorbance ratio should also be considered. Both A260/A280 and A260/A230 absorbance ratios are typically used to determine the purity of DNA samples that were extracted using biological, organic and inorganic compounds. Sambrook et al. [8] suggested that when measuring pure double-stranded DNA, the A260/A280 and A260/A230 absorbance ratios should ideally be in the range of 1.6–1.9 and 2.0–2.2, respectively. Accordingly, our absorbance analysis for all samples yielded values for A260/A280 and A260/A230 within the ideal range (Table 1), which is indicative of high quality of the extracted gDNA.
PCRs of rbcL, cox3 and ITS regions were successful for all samples (Fig. 2). DNA sequencing for all three regions was successful (Fig. 3), which allowed us to construct high-quality consensus sequences for all three regions.
Fig. 2.
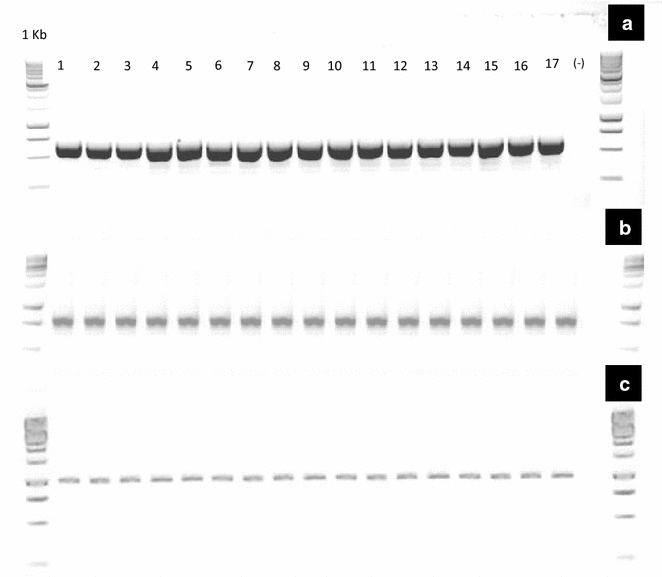
Image of the agarose gel of the PCR products (gDNA) ran by electrophoresis obtained from genomic DNA (gDNA) extracted from 17 tissue samples of Opuntia Mill., using the improved extraction method. a nDNA internal transcribed spacer (ITS), b cpDNA RuBisCO gene (rbcL), c mtDNA cytochrome oxidase subunit 3 (cox3) (Promega™ 1 kb DNA Ladder Molecular Weight Marker)
Fig. 3.
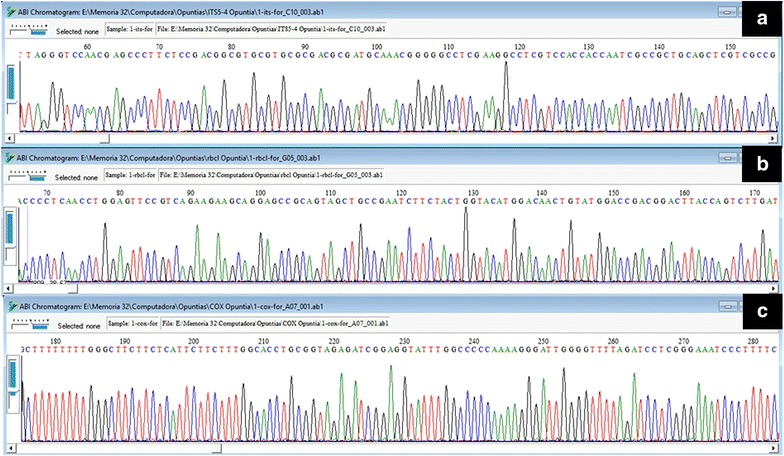
DNA sequence chromatograms for PCR products obtained from genomic DNA (gDNA) samples extracted from 17 tissue samples of Opuntia Mill using the improved extraction method. a nDNA internal transcribed spacer (ITS), b cpDNA RuBisCO gene (rbcL), c mtDNA cytochrome oxidase subunit 3 (cox3). Sequences were visualized using BioEdit v 7.0.5
In order to complement sequence quality assessment, we decided to assess the identity of sequences, at least preliminary, with a basic BLAST search. It has been documented that BLAST is not the proper mean for taxonomical identification, but it provides an easy way to broadly verify if the sequence belongs to the sample (e.g., verifying a potential contamination).
We conducted a BLAST search for each of the 17 sequences and the first hit on each search was recorded (Table 3). All the searches hit in sequences of Opuntia, but only five ITS sequences matched with the corresponding species. The other loci (rbcL and cox3) matched on Opuntia as well, but with non-corresponding species.
Table 3.
Blast search for the three markers
| Species number | Description | Max score | Total score | Query cover (%) | E value | Ident (%) | Accession |
|---|---|---|---|---|---|---|---|
| ITS | |||||||
| 1 | Opuntia sp. | 865 | 865 | 100 | 0.0 | 100 | JF787077.1 |
| 2 | Opuntia bravoana | 929 | 929 | 100 | 0.0 | 100 | JF87044.1 |
| 3 | Opuntia delafuentiana | 968 | 968 | 100 | 0.0 | 100 | KM67822.1 |
| 4 | Opuntia depressa | 822 | 822 | 100 | 0.0 | 99 | JF787089.1 |
| 5 | Opuntia martiniana | 963 | 963 | 100 | 0.0 | 100 | JF787066.1 |
| 6 | Opuntia ficus-indica | 1059 | 1059 | 100 | 0.0 | 100 | JF78710.1 |
| 7 | Opuntia robusta | 1048 | 1048 | 100 | 0.0 | 99 | JF787122.1 |
| 8 | Opuntia velutina | 850 | 850 | 100 | 0.0 | 100 | HQ872589.1 |
| 9 | Opuntia martiniana | 1094 | 1094 | 100 | 0.0 | 100 | JF787066.1 |
| 10 | Opuntia pittieri | 1109 | 1109 | 100 | 0.0 | 100 | JF787105.1 |
| 11 | Opuntia leiascheinvariana | 970 | 970 | 100 | 0.0 | 100 | KM507353.1 |
| 12 | Opuntia cubensis | 1027 | 1027 | 100 | 0.0 | 100 | JF787058.1 |
| 13 | Opuntia martiniana | 1003 | 1003 | 100 | 0.0 | 100 | JF787066.1 |
| 14 | Opuntia pittieri | 1120 | 1120 | 100 | 0.0 | 100 | JF787105.1 |
| 15 | Opuntia carstenii | 992 | 992 | 100 | 0.0 | 100 | JF787112.1 |
| 16 | Opuntia oligacantha | 953 | 953 | 100 | 0.0 | 100 | KX247005.1 |
| 17 | Opuntia bakeri | 1059 | 1059 | 100 | 0.0 | 99 | JF787101.1 |
| rbcL | |||||||
| 1 | Opuntia maxima | 1245 | 1245 | 100 | 0.0 | 100 | HM850212.1 |
| 2 | Opuntia dillenii | 1262 | 1262 | 99 | 0.0 | 100 | HM850211.1 |
| 3 | Opuntia maxima | 1254 | 1254 | 100 | 0.0 | 100 | HM850212.1 |
| 4 | Opuntia maxima | 1262 | 1262 | 99 | 0.0 | 100 | HM850212.1 |
| 5 | Opuntia maxima | 1258 | 1258 | 99 | 0.0 | 100 | HM850212.1 |
| 6 | Opuntia dillenii | 1258 | 1258 | 99 | 0.0 | 100 | HM850211.1 |
| 7 | Opuntia dillenii | 1262 | 1262 | 99 | 0.0 | 100 | HM850211.1 |
| 8 | Opuntia maxima | 1260 | 1260 | 99 | 0.0 | 100 | HM850212.1 |
| 9 | Opuntia maxima | 1262 | 1262 | 99 | 0.0 | 100 | HM850212.1 |
| 10 | Opuntia maxima | 1260 | 1260 | 99 | 0.0 | 100 | HM850212.1 |
| 11 | Opuntia maxima | 1260 | 1260 | 99 | 0.0 | 100 | HM850211.1 |
| 12 | Opuntia maxima | 1260 | 1260 | 99 | 0.0 | 100 | HM850212.1 |
| 13 | Opuntia maxima | 1253 | 1253 | 99 | 0.0 | 100 | HM850212.1 |
| 14 | Opuntia maxima | 1090 | 1090 | 100 | 0.0 | 100 | HM850212.1 |
| 15 | Opuntia dillenii | 1262 | 1262 | 99 | 0.0 | 100 | HM850211.1 |
| 16 | Opuntia maxima | 1085 | 1085 | 100 | 0.0 | 100 | HM850212.1 |
| 17 | Opuntia maxima | 1254 | 1254 | 99 | 0.0 | 100 | HM850212.1 |
| cox3 | |||||||
| 1 | Opuntia megacantha | 1117 | 1117 | 100 | 0.0 | 100 | EU930402.1 |
| 2 | Opuntia megacantha | 1033 | 1033 | 100 | 0.0 | 100 | EU930402.1 |
| 3 | Opuntia megacantha | 1125 | 1125 | 100 | 0.0 | 100 | EU930402.1 |
| 4 | Opuntia megacantha | 900 | 900 | 100 | 0.0 | 100 | EU930402.1 |
| 5 | Opuntia megacantha | 1212 | 1212 | 100 | 0.0 | 100 | EU930402.1 |
| 6 | Opuntia albicarpa | 1179 | 1179 | 100 | 0.0 | 100 | EU930396.1 |
| 7 | Opuntia megacantha | 1249 | 1249 | 100 | 0.0 | 100 | EU930402.1 |
| 8 | Opuntia megacantha | 1175 | 1175 | 100 | 0.0 | 100 | EU930402.1 |
| 9 | Opuntia megacantha | 1236 | 1236 | 100 | 0.0 | 100 | EU930402.1 |
| 10 | Opuntia megacantha | 1234 | 1234 | 100 | 0.0 | 100 | EU930402.1 |
| 11 | Opuntia megacantha | 1201 | 1201 | 100 | 0.0 | 100 | EU930402.1 |
| 12 | Opuntia megacantha | 1223 | 1223 | 100 | 0.0 | 100 | EU930402.1 |
| 13 | Opuntia matudae | 1225 | 1225 | 100 | 0.0 | 100 | EU930401.1 |
| 14 | Opuntia megacantha | 1238 | 1238 | 100 | 0.0 | 100 | EU930402.1 |
| 15 | Opuntia megacantha | 1171 | 1171 | 100 | 0.0 | 100 | EU930388.1 |
| 16 | Opuntia megacantha | 1173 | 1173 | 100 | 0.0 | 100 | EU930402.1 |
| 17 | Opuntia megacantha | 985 | 985 | 100 | 0.0 | 100 | EU930402.1 |
In this table is only recorded the first hit on each search
BLAST results on rbcL and cox3 are due to the fact that those loci have very low variability at species level. Sequence variability was not enough for proper species identity, but sufficient for genera identity.
On the other hand, ITS is a loci with larger variability at species level. We found five searches that matched with the corresponding species. At four searches, the corresponding species were not available in GenBank, and no correct match was possible, but the search hit in Opuntia. The remaining searches on the ITS sequences did not match on the correct species, but did match in Opuntia. This result is due to two main reasons: 1) the BLAST search is not designed for species match, even if the species are available in the database, and in consequence it is not a suitable tool for specimens identification; and 2) because in most cases our sequences are longer (including ITS1 and 2 as well as 5.8S region) than those available in GenBank; this extra length may induce some errors.
Comparison with previous methods
We replicated the protocols of Mondragón-Jacobo et al. [11] and Griffith and Porter [13] using the same 17 species of Opuntia (Table 4). We confirmed that our method got better performance (quality and quantity of gDNA), and that it has some advantages over other protocols (Table 5). In addition, our protocol is the cheapest one and considered as a micro-method due to the amounts of reagents and tissue involved.
Table 4.
Comparison among three different protocols to obtain total genomic DNA using NanoDrop
| Species | Mondragón-Jacobo et al. [11] | Griffith and Porter [13] | This protocol | |||
|---|---|---|---|---|---|---|
| DNA yield (ng/µl) | OD ratio 260.280 | DNA yield (ng/µl) | OD ratio 260.280 | DNA yield (ng/µl) | OD ratio 260.280 | |
| 1. Opuntia auberi Pfeiff. | 256 | 1.4 | 423 | 1.7 | 1600 | 1.9 |
| 2. Opuntia decumbens Salm-Dyck | 35 | 1.7 | 30 | 1.9 | 2930 | 1.9 |
| 3. Opuntia delafuentiana Martínez-González et al. | 75 | 1.6 | 56 | 1.9 | 4937 | 1.8 |
| 4. Opuntia depressa Britton and Tose | 95 | 1.9 | 73 | 1.8 | 8755 | 1.9 |
| 5. Opuntia durangensis Britton and Tose | 134 | 1.5 | 123 | 1.7 | 5835 | 1.9 |
| 6. Opuntia ficus-indica Mill. | 34 | 1.8 | 258 | 1.8 | 3829 | 2.0 |
| 7. Opuntia heliabravoana Scheinvar | 198 | 1.6 | 43 | 1.8 | 8743 | 1.8 |
| 8. Opuntia huajuapensis Bravo | 57 | 1.5 | 78 | 1.7 | 1573 | 1.9 |
| 9. Opuntia joconostle F.A.C. Weber | 86 | 1.9 | 196 | 1.0 | 8375 | 1.8 |
| 10. Opuntia lasiacantha Pfeiff. | 67 | 1.7 | 356 | 1.7 | 2943 | 1.9 |
| 11. Opuntia leiascheinvariana Martínez-González | 110 | 1.8 | 98 | 1.9 | 3980 | 1.9 |
| 12. Opuntia leucotricha DC. | 248 | 1.5 | 34 | 1.8 | 3789 | 1.9 |
| 13. Opuntia matudae Scheinvar | 93 | 1.7 | 63 | 1.8 | 7947 | 1.9 |
| 14. Opuntia megacantha Salm-Dyck | 117 | 1.6 | 78 | 1.8 | 7000 | 1.8 |
| 15. Opuntia microdasys Pfeiff. | 44 | 1.5 | 39 | 1.7 | 6578 | 1.9 |
| 16. Opuntia oligacantha Förster | 87 | 1.8 | 70 | 1.9 | 2395 | 1.8 |
| 17. Opuntia olmeca Joel Pérez et al. | 94 | 1.5 | 57 | 1.7 | 9200 | 1.9 |
Table 5.
Advantages of our protocol
| Mondragón-Jacobo et al. [11] | Griffith and Porter [13] | This protocol |
|---|---|---|
| They tried to use young tissues, avoiding older ones because their higher content of fiber and cuticular wax | They tried to use epidermal tissue free of waxes | We can use tissue from any part of the plant |
| They used β-mercaptoethanol | They used β-mercaptoethanol | We did not use β-mercaptoethanol |
| 8000 mg of cactus pear tissue | 30–50 mg of epidermal tissue | 2–3 mg of tissue from every part of the plant |
| They used more CTAB (25 ml) | They used more CTAB (15 ml) | We used few CTAB (0.7 ml) |
| They used more chloroform-isoamyl alcohol (10 ml) | They used more chloroform-isoamyl alcohol (5 ml) | We used few chloroform-isoamyl alcohol (0.75 ml) |
| They used ethanol (8.7 ml) | They used more isopropanol (5 ml) | We used few isopropanol (0.4 ml) |
| They used bigger and expensive tubes (15 ml) | They used bigger and expensive tubes (15 ml) | We used smaller tubes (2 ml) |
| They used RNAse to eliminate RNA | They did not use RNAse | We did not use RNAse |
Discussion
Several gDNA extraction protocols were developed recently, but few of these have been focused on the elimination of pectin and polysaccharides. These two compounds are among the most difficult contaminants to separate from the DNA [38] and significantly interfere with the activity of DNA polymerases. Therefore, the elimination of these compounds during the extraction of gDNA favors the efficiency of PCR amplification [39]. Pectin and mucilage (polysaccharides) are two of the main tissue components tissue in Opuntia. More specifically, pectin is the main component of the middle layer of cell walls and mucilage is one of the principal components of the parenchyma.
Mondragón-Jacobo et al. [11] developed a DNA extraction method for several cacti species (e.g., Cleistocactus spp., Echinocereus spp., Nopalea spp., Opuntia spp., Stenocereus spp.). The amount of tissue used in this extraction protocol is species-dependent due to varying mucilage content among species. Griffith and Porter [13] extracted DNA from epidermal cells from several species of Austrocylindropuntia, Brasilopuntia, Consolea, Cumulopuntia, Cylindropuntia, Grusonia, Maihueniopsis, Miqueliopuntia, Nopalea, Opuntia, Pereskiopsis, Pterocactus, Tephrocactus and Tunilla. In recent years, Mihalte et al. [25] showed that the protocol of Pop et al. [30] yielded sufficient amounts of DNA from small amounts of tissue for species of Rebutia, Mediolobivia, Sulcorebutia and Aylostera. Accordingly, Yu et al. [26] introduced a protocol, similar to that of Pop et al. [30], for reliable DNA extraction from Hylocereus spp. Montiel et al. [27] used root tissue from Opuntia to extract DNA due to the difficulties encountered during extraction from cladode tissue. Wong et al. [22] developed a method to extract DNA from Hylocereus spp. Out of these studies, only those of De la Cruz et al. [10], Mondragón-Jacobo et al. [11], Griffith and Porter [13], Montiel et al. [27] and Fehlberg et al. [40] tested extraction efficiency on species of Opuntia.
Our improved gDNA extraction method is based on the protocols of Mondragón-Jacobo et al. [11] and Griffith and Porter [13]. We developed this method for the extraction of DNA from Opuntia cladodes, which contain large quantities of mucilage and pectin [20]. More specifically, improvements in the method involved changes to centrifugation and incubation steps (e.g., increased times and temperatures), the addition of water to remove pectin and the elimination of various reactive agents, such as polyvinylpyrrolidone (PVP), β-mercaptoethanol and protein and RNA degrading enzymes.
The increased centrifugation times allowed for a better separation of gDNA from fiber cells and non-soluble cellular components, such as proteins. As pectin is water-soluble, the addition of water permitted the extraction of this compound, forming a gelatinous substance over the precipitated gDNA [41, 42].
Generally, polyvinylpyrrolidone (PVP) is used to suppress polyphenolic oxidation during the extraction process [43]. However, PVP was not used because the main issue associated with DNA extraction from Opuntia samples is the presence of pectin and mucilage, and not of phenolic compounds.
The longer time of incubation at higher temperatures results in a more efficient denaturation of the proteins and enzymes found in tissue samples of Opuntia. Therefore, the extra step of incubation with proteinases is not needed.
The Β-mercaptoethanol inhibits the activity of DNAs and RNAs and thus protects gDNA from degradation. However, we do not use this compound in our extraction protocol because EDTA (contained in CTAB) forms a molecular complex with Mg2 + ions that prevents the functioning of DNAs [8]. In turn, we do not use RNAse because we included a final drying step for 40 min, followed by 15 min at 60 °C, that allows for the efficient degradation of RNA.
Ribonucleases (RNAses) are abundant in all biological and most of these are fairly stable and difficult to inactivate even when extraction reagents and materials have been autoclaved. Thus, when extracting RNA from biological samples RNAses should be eliminated rapidly with denaturing compounds [8]. The presence of RNA in the samples is controlled with the fluorimetry analysis using the Quant-iT ™ PicoGreen® Kit (Invitrogen™), which is an ultra-sensitive method for quantifying double-stranded DNA. The determination of absorbance at 260 nm wavelength is the commonly used technique for measuring the overall concentration of nucleic acids. However, absorbance measures have the main disadvantage of confounding the absorbance contribution of single-stranded nucleic acids, thus being unable to distinguish between DNA and RNA.
The purity of the extracted gDNA was confirmed by spectrophotometry. Generally, a higher A260/A280 value is indicative of RNA contamination, whereas lower values are indicative of protein contamination. On the other hand, lower A260/A230 values indicate the presence of phenolic compounds and carbohydrates, whereas higher values are usually associated with calibration errors [44]. The A260/A280 and A260/A230 ratios for dsDNA ideally range from 1.6 to 1.9 and from 2.0 to 2.2, respectively [8]. Our analyses showed A260/A280 and A260/A230 within these ideal ranges (Table 1), which confirm the purity of the gDNA samples. Through the improvement of DNA extraction protocols, we were able to improve the overall yield and purity of gDNA (1500–9147 ng/μl, Table 1) extracted from different species of Opuntia. In addition, with these changes, the extraction protocol becomes cheaper and the use of toxic reagents is diminished.
When we compared our method with other two previous protocols [11, 13], we observed that the necessary amount of tissue in these two protocols is huge. Also, both methods need a great amount of expensive chemical reagents, making them impractical. With our new protocol, we obtained a higher DNA performance with high molecular weight (1500 ng/μl), and an average of the ratio A260/A280 of 1.8.
Our protocol is a good alternative to these methods, since it requires milligrams of tissue and small volumes of reagents, facilitating the handling of a large number of samples. In short, our method is cheaper, quick and simple, and it does not need to carry out additional purification.
Conclusion
In this study, we developed a method of DNA extraction that yields high-quality gDNA free of inhibitory organic compounds common in species of Opuntia, such as pectin and mucilage. This improved method allowed us to obtained higher yields of gDNA of excellent quality. Our method works in other species of cacti (e.g., Nopalxochia [45]); it will be interesting to test it in other Cactaceae and succulent plants such as Crassulaceae. Finally, we are demonstrating that the addition of RNAses is not necessary to remove RNA from the genomic DNA samples. The use of RNAse is replaced by a heat treatment to remove the RNA making the protocol cheaper.
Authors’ contributions
CRMG performed the protocol methodology, standardization, as well the coordination and integration of laboratory results, RRM provided technical support in the laboratory for standardization of the protocol and protocol replicate the species of interest and conducting PCR´s, JJR performed field identification and collection of the species distributed in the central region of Mexico, CGV performed field identification and collection of the species distributed in the North central region of Mexico and ILV performed protocol integration, results and coordinated the development of the final work. All authors read and approved the final manuscript.
Acknowledgements
The editor of this journal, Ricardo García-Sandoval and three anonymous referees did great suggestions to our manuscript.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
Not applicable.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Funding
This research was financially supported by the Red del Nopal belonging to the Sistema Nacional de Recursos Fitogenéticos para la alimentación y la agricultura del Servicio Nacional de Inspección y certificación de semillas and by the Laboratorio de Biogeografía of the Departamento de Biología Evolutiva, Universidad Nacional Autónoma de México.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
César Ramiro Martínez-González, Email: ramiro_mg.unam@ciencias.unam.mx.
Rosario Ramírez-Mendoza, Email: rosaricuqui@yahoo.com.mx.
Jaime Jiménez-Ramírez, Email: jjimenez_ramirez@yahoo.com.
Clemente Gallegos-Vázquez, Email: cgallegosvazquez@gmail.com.
Isolda Luna-Vega, Email: luna.isolda@gmail.com.
References
- 1.Scheinvar L. 2004. Caracteres macroscópicos, microscópicos y moleculares valiosos para la identificación de los recursos genéticos del nopal. In: Memorias X Congreso Nacional y VIII Congreso Internacional sobre el conocimiento y aprovechamiento del nopal. Agosto. UACh. Texcoco. Mexico City.
- 2.Reyes-Agüero JA, Aguirre-Rivera JR, Hernández HM. Nota sistemática y una descripción detallada de Opuntia ficus-indica (L.) Mill. (Cactaceae) Agrociencia. 2005;39:395–408. [Google Scholar]
- 3.Wang X, Falher P, Burrow MD, Peterson AH. Comparison of RAPD marker patterns to morphological and physiological data in the classification of Opuntia accessions. J Prof Assoc Cactus. 1998;3:3–14. [Google Scholar]
- 4.Srikanth K, Whang S. Phylogeny of Korean Opuntia spp. based on multiple DNA regions. Turk J Bot. 2015;39:635–641. doi: 10.3906/bot-1405-10. [DOI] [Google Scholar]
- 5.Eguiarte L, Souza V, Aguirre X. Ecología Molecular. Secretaría de Medio Ambiente y Recursos Naturales. Instituto Nacional de Ecología. Universidad Nacional Autónoma de México. Mexico City; 2007. 594 p.
- 6.Plaza GA, Upchurch R, Brigmon RL, Whitman WB, Ulfi K. Rapid DNA extraction for screening soil filamentous fungi using PCR amplification. Polish J Environ Stud. 2003;13:315–318. [Google Scholar]
- 7.Park D. Genomic DNA isolation from different biological materials. Methods Mol Biol. 2007;353:3–13. doi: 10.1385/1-59745-229-7:3. [DOI] [PubMed] [Google Scholar]
- 8.Sambrook J, Fritsch EF, Maniatis T. Molecular cloning A laboratory manual. 2. New York: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 9.Boiteux LS, Fonseca M, Simon PW. Effects of plant tissue and DNA purification method randomly amplified polymorphic DNA-based genetic fingerprinting analysis in carrot. J Am Soc Hortic Sci. 1999;124:32–38. [Google Scholar]
- 10.De la Cruz M, Ramírez F, Hernández H. DNA isolation and amplification from cacti. Plant Mol Biol Rep. 1997;1:19–21. [Google Scholar]
- 11.Mondragón-Jacobo C, Doudareva N, Bordeleon BP. DNA extraction from several cacti. HortScience. 2000;35:1124–1126. [Google Scholar]
- 12.Nyffeler R. Phylogenetic relationships in the cactus family (Cactaceae) based on evidence from trnK/matK and trnL-trnF sequences. Am J Bot. 2002;89:312–326. doi: 10.3732/ajb.89.2.312. [DOI] [PubMed] [Google Scholar]
- 13.Griffith MP, Porter JM. Back to the basics: a simple method of DNA extraction for mucilaginous cacti. Bradleya. 2003;21:126–128. [Google Scholar]
- 14.Edwards E, Niffeler R, Donoghue J. Basal cactus phylogeny: implications of Pereskia (Cactaceae) paraphyly for the transition to the cactus life form. Am Naturalist. 2005;92:1177–1188. doi: 10.3732/ajb.92.7.1177. [DOI] [PubMed] [Google Scholar]
- 15.Korotkova N, Borsch T, Quandt D, Taylor P, Muller F, Barthlott W. What does it take to resolve relationships and to identify species with molecular markers? An example from the epiphytic Rhipsalideae (Cactaceae) Am J Bot. 2011;99:847–864. doi: 10.3732/ajb.1000502. [DOI] [PubMed] [Google Scholar]
- 16.Goycoolea F, Cárdenas A. Pectins from Opuntia spp.: a short review. J Prof Assoc Cactus. 2003;5:17–23. [Google Scholar]
- 17.Nobel P, Cavalier J. Mucilage in cacti: its apoplastic capacitance, associated solutes, and influence on tissue water relations. J Exp Bot. 1992;43:641–648. doi: 10.1093/jxb/43.5.641. [DOI] [Google Scholar]
- 18.Sepúlveda E, Sáenz C, Aliaga E, Aceituno C. Extraction and characterization of mucilage in Opuntia spp. J Arid Environ. 2007;68:534–545. doi: 10.1016/j.jaridenv.2006.08.001. [DOI] [Google Scholar]
- 19.Matsuhiro B, Lillo L, Sáenz C, Urzúa C, Zárate O. Chemical characterization of the mucilage from fruits of Opuntia ficus-indica. Carbohyd Polym. 2006;63:263–267. doi: 10.1016/j.carbpol.2005.08.062. [DOI] [Google Scholar]
- 20.Guevara-Arauza J, Ornelas-Paz J, Pimentel-González D, Rosales S, Soria-Guerra R, Paz-Maldonado T. Prebiotic effect of mucilage and pectin-derived oligosaccharides from nopal (Opuntia ficus-indica) Food Sci Biotechnol. 2012;21:997–1003. doi: 10.1007/s10068-012-0130-1. [DOI] [Google Scholar]
- 21.Aljanabi SM, Forget L, Dookun A. An improved and rapid protocol for the isolation of polysaccharide-and polyphenol-free sugarcane DNA. Plant Mol Biol Rep. 1999;17:1–8. doi: 10.1023/A:1007692929505. [DOI] [Google Scholar]
- 22.Wong LM, Silvaraj S, Phoon LQ. An optimised high-salt CTAB protocol for both DNA and RNA isolation from succulent stems of Hylocereus sp. J Med Biol Eng. 2014;3:236–240. [Google Scholar]
- 23.Katterman F, Shattuck VL. An effective method of DNA isolation from the mature leaves of Gossypium species that contain large amounts of phenolic terpenoids and tannins. Prep Biochem. 1983;13:347–359. doi: 10.1080/00327488308068177. [DOI] [PubMed] [Google Scholar]
- 24.Tel-Zur N, Abbo S, Myslabodski D, Mizrahi Y. Modified CTAB procedure for DNA isolation from epiphytic cacti of the genera Hylocereus and Selenicereus (Cactaceae) Plant Mol Biol Rep. 1999;17:249–254. doi: 10.1023/A:1007656315275. [DOI] [Google Scholar]
- 25.Mihalte L, Sestras R, Feszt G. Assessing genetic variability at different genotypes of cacti plants by means of RAPD analysis. Bull UASVM Hortic. 2008;65:110–115. [Google Scholar]
- 26.Yu ZX, Ou GZ, Chen QX, Yuan YF. Study on comparison of methods for dragon fruit total DNA extraction. Chin Agric Sci Bull. 2010;26:300–303. [Google Scholar]
- 27.Montiel D, Valadez-Moctezuma E, Palomino G, Bermúdez M, Fernández F. DNA extraction from roots of xoconostle. J Prof Assoc Cactus. 2012;14:35–40. [Google Scholar]
- 28.Kim SR, Yang J, An G, Jena KK. A simple DNA preparation method for high quality polymerase chain reaction in rice. Plant Breed Biotechnol. 2016;4:99–106. doi: 10.9787/PBB.2016.4.1.099. [DOI] [Google Scholar]
- 29.Pop IF, Pamfil D, Raica PA, Petricele IV, Botu I, Vicol AC, Harta M, Sisea CR. Evaluation of the genetic diversity of several Corylus avellana accessions from the Romanian National Hazelnut Collection. Not Bot Horti Agrobo. 2010;38:61–67. [Google Scholar]
- 30.Rogers SO, Bendich AJ. Ribosomal genes in plants: variability in copy number and in the intergenic spacer. Plant Mol Biol. 1987;9:509–520. doi: 10.1007/BF00015882. [DOI] [PubMed] [Google Scholar]
- 31.Hamby RK, Zimmer EA. Ribosomal RNA as a phylogenetic tool in plant systematics. In: Soltis PS, editor. Molecular systematics of plants. New York: Springer, Chapman and Hall; 1992. pp. 50–91. [Google Scholar]
- 32.Weider L, Elser J, Crease T, Mateos M, Cotner J, Markow T. The functional significance of ribosomal (r)DNA variation: impacts on the evolutionary ecology of organisms. Annu Rev Ecol Evol Syst. 2005;36:219–242. doi: 10.1146/annurev.ecolsys.36.102003.152620. [DOI] [Google Scholar]
- 33.Berg S, Krause K, Krupinska K. The rbcL genes of two Cuscuta species, C. gronovii and C. subinclusa, are transcribed by the nuclear-encoded plastid RNA polymerase (NEP) Planta. 2004;219:541–546. doi: 10.1007/s00425-004-1260-3. [DOI] [PubMed] [Google Scholar]
- 34.Galmes J, Flexas J, Keys AJ, Cifre J, Mitchell RAC. Rubisco specificity factor tends to be larger in plant species from drier habitats and in species with persistent leaves. Plant Cell Environ. 2005;28:571–579. doi: 10.1111/j.1365-3040.2005.01300.x. [DOI] [Google Scholar]
- 35.Duminil J, Pemonge H, Petit M. A set of 35 consensus primer pairs amplifying genes and introns of plant mitochondrial DNA. Mol Ecol Notes. 2002;2:428–430. doi: 10.1046/j.1471-8286.2002.00263.x. [DOI] [Google Scholar]
- 36.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- 37.Zhang Z, Schwartz S, Wagner L, Miller W. A greedy algorithm for aligning DNA sequences. J Comput Biol. 2000;7:203–214. doi: 10.1089/10665270050081478. [DOI] [PubMed] [Google Scholar]
- 38.Murray MG, Thompson WF. Rapid isolation of high molecular weight DNA. Nucleic Acids Res. 1980;8:4321–4325. doi: 10.1093/nar/8.19.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lodhi MA, Ye GN, Weeden NF, Reisch BI. A simple and efficient method for DNA lysis from grapevine cultivars and Vitis species. Plant Mol Biol Rep. 1994;12:6–13. doi: 10.1007/BF02668658. [DOI] [Google Scholar]
- 40.Fehlberg SD, Allen JM, Church K. A novel method of genomic DNA extraction for Cactaceae. Appl Plant Sci. 2013;1:1–4. doi: 10.3732/apps.1200013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cárdenas A, Higuera-Ciapara I, Goicoolea FM. Rheology and aggregation of cactus (Opuntia ficus-indica) mucilage in solution. J Prof Assoc Cactus. 1997;2:152–159. [Google Scholar]
- 42.Majdoub H, Roudesli S, Picton L, Le Cerf D, Muller G, Grisel M. Prickly pear nopals pectin from Opuntia ficus-indica physicochemical study in dilute and semi-dilute solutions. Carbohyd Polym. 2001;46:69–79. doi: 10.1016/S0144-8617(00)00284-8. [DOI] [Google Scholar]
- 43.Porebski S, Bailey LG, Baum BR. Modification of a CTAB DNA extraction protocol for plants containing high polysaccharide and polyphenol components. Plant Mol Biol Rep. 1997;15:8–15. doi: 10.1007/BF02772108. [DOI] [Google Scholar]
- 44.Meyer R. Detection methods for genetically modified crops. In: Heller KJ, editor. Genetically engineered food: methods and detection. KGaA. Hoboken: Wiley; 2003. pp. 188–200. [Google Scholar]
- 45.Martínez-González CR, Alcántara-Ayala O, Luna-Vega I, García-Sandoval R. Phylogenetic placement and new data on macro and micro morphology of Nopalxochia phyllanthoides (Cactaceae), an endangered species from Mexico. Phytotaxa. 2015;222(4):241–250. doi: 10.11646/phytotaxa.222.4.1. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.


