Abstract
What adaptive changes in brain structure and function underpin the evolution of increased cognitive performance in humans and our close relatives? Identifying the genetic basis of brain evolution has become a major tool in answering this question. Numerous cases of positive selection, altered gene expression or gene duplication have been identified that may contribute to the evolution of the neocortex, which is widely assumed to play a predominant role in cognitive evolution. However, the components of the neocortex co-evolve with other functionally interdependent regions of the brain, most notably in the cerebellum. The cerebellum is linked to a range of cognitive tasks and expanded rapidly during hominoid evolution. Here we present data that suggest that, across anthropoid primates, protein-coding genes with known roles in cerebellum development were just as likely to be targeted by selection as genes linked to cortical development. Indeed, based on currently available gene ontology data, protein-coding genes with known roles in cerebellum development are more likely to have evolved adaptively during hominoid evolution. This is consistent with phenotypic data suggesting an accelerated rate of cerebellar expansion in apes that is beyond that predicted from scaling with the neocortex in other primates. Finally, we present evidence that the strength of selection on specific genes is associated with variation in the volume of either the neocortex or the cerebellum, but not both. This result provides preliminary evidence that co-variation between these brain components during anthropoid evolution may be at least partly regulated by selection on independent loci, a conclusion that is consistent with recent intraspecific genetic analyses and a mosaic model of brain evolution that predicts adaptive evolution of brain structure.
Keywords: Brain evolution, Cerebellum, Neocortex, Molecular evolution, Primates
Introduction
The proximate basis of primate brain expansion and the evolution of increased cognitive performance lies in changes in gene function and regulation. Identifying the genetic basis of phenotypic change can provide insights into how developmental mechanisms evolve, how they are constrained, and how changes at a cellular level contribute to broad-scale anatomical evolution [Rausher and Delph, 2015]. The potential to dissect the biological basis of brain and behavioural evolution motivates many genomic comparisons across primates [Enard, 2014]. These have identified numerous genes associated with brain development with high rates of evolution [Enard et al., 2002a; Pollard et al., 2006; Montgomery et al., 2011; Montgomery and Mundy, 2012a, b; Kamm et al., 2013; Boyd et al., 2015; Boddy et al., 2017], divergent expression profiles [Enard et al., 2002b; Khaitovich et al., 2004; Brawand et al., 2011; Bauernfeind et al., 2015] or duplicated sequences [Burki and Kaessmann, 2004; Keeney et al., 2014; Florio et al., 2015; Zimmer and Montgomery, 2015] either across primates or during recent human evolution. In several of these cases the genetic changes have been demonstrated to have functional effects on neuronal proliferation or maturation [Enard et al., 2009; Pulvers et al., 2010; Kamm et al., 2013; Boyd et al., 2015; Florio et al., 2015]. These results highlight the potential cellular adaptations that drive changes in brain size, and they provide a powerful means to investigate human-specific adaptations.
The majority of these examples investigate genes linked to neocortical evolution, reflecting the widely held view that the neocortex has a predominant role in “higher” cognition. This view has been critiqued as “corticocentric myopia” by Parvizi [2009] for ignoring the importance of subcortical brain regions in supporting neocortical function. Indeed, the neocortex co-evolves with other brain components with which it is functionally connected, suggesting that a complete understanding of primate brain expansion will not be found by focusing solely on neocortex development [Barton and Harvey, 2000; Whiting and Barton, 2003; Barton, 2012]. Of particular importance is the relationship between the neocortex and the cerebellum, which share extensive functional and anatomical connections [Ramnami, 2006]. Indeed, cortico-cerebellar co-evolution pervades biological levels, occurring at a coarse volumetric scale, at the level of individual nuclei, and at the cellular level [Barton and Harvey, 2000; Whiting and Barton, 2003; Balsters et al., 2010; Herculano-Houzel and Sherwood, 2010]. Across primates, patterns of co-evolution between components of the cortical-cerebellar complex closely mirror their anatomical connectivity, with neocortex volume correlating strongly with cerebellar cortex volume, whilst the size of the cerebellar nuclei correlates with their downstream projection target, the thalamus, which in turn projects back to the neocortex [Whiting and Barton, 2003].
In humans, the cerebellum's role in the cortico-cerebellar complex is increasingly recognised to be important for both motor and “higher cognitive” function, including the capacity to plan and execute complex behavioural sequences [Ramnani, 2006; Parvizi, 2009; Barton, 2012]. Across primates, variation in cerebellum volume is linked to extractive foraging, after controlling for variation in neocortex volume [Barton, 2012]. The importance of the cerebellum in primate brain evolution is further bolstered by comparative analyses that demonstrate the cerebellum is expanded in hominoids [Rilling and Insel, 1998; MacLeod et al., 2003] due to a non-allometric expansion of the cerebellum relative to neocortex size that was caused by a faster-than-expected increase in cerebellum volume and neuron number that persisted throughout hominoid evolution [Barton and Venditti, 2014]. Although data at increasingly fine grained levels are more limited, the volumetric expansion of the whole cerebellum appears to be driven by a specific increase in the cerebellar lateral hemispheres [MacLeod et al., 2003] and possibly specific areas which receive direct functional connections with the prefrontal cortex [Balsters et al., 2010], an area of the neocortex long thought to be selectively expanded in hominoids [e.g., Semendeferi et al., 2001; Schoenemann et al., 2005; Sherwood et al., 2005; Smaers et al., 2011, but see Barton and Venditti, 2013; Gabi et al., 2016]. These data are indicative of an adaptive specialisation in the cortico-cerebellar functional relationship during hominoid evolution. Hominin evolution may also be characterised by reciprocal expansion of the neocortex and the cerebellum, with recent modern humans reportedly being distinguished from early modern humans by an increase in cerebellum volume beyond that observed for the neocortex [Weaver, 2005]. The cerebellum's role in computing complex sequences of motor behaviour suggests a possible link between cerebellar expansion and an enhanced capacity to plan and execute the kind of complex behavioural sequences necessary for both tool use and language development [Ackermann, 2008; Sultan et al., 2010; Leggio et al., 2011; Barton, 2012; Stout and Chaminade, 2012].
The importance of a coordinated expansion of components of the cortico-cerebellum suggests that we must look beyond neocortical evolution to obtain a full picture of the genetic architecture of primate brain and cognitive evolution. The likely action of positive selection on genes involved in cerebellum development is further suggested by the accelerated rate of evolution of AHI1, a gene associated with developmental disorders of the cerebellum, during human evolution [Ferland et al., 2004]. Phylogenetic approaches to studying gene-phenotype associations may be useful in addressing long-standing debates about the constraints governing co-evolving brain networks. For example, do the neocortex and the cerebellum co-evolve through genetically independent developmental changes maintained by selection [sensu Barton and Harvey, 2000]? Or is their co-evolution the result of common developmental control or pleiotropic genetic effects shared across anatomical boundaries [sensu Finlay and Darlington, 1995]? If the latter is true, do these pleiotropic effects reflect an evolutionary constraint, or have they evolved to maintain the functional relationship between components during brain expansion? These questions will be central to providing a full understanding of brain evolution, and to addressing questions of long-standing interest in evolutionary biology centred on the trade-offs between adaptation and constraint in the genetic basis of composite traits [Montgomery et al., 2016].
Here, we ask three questions that provide an initial assessment of the role of protein-coding genes contributing to the development of the neocortex and the cerebellum in anthropoid brain evolution. First, we ask whether protein-coding genes with known roles in the development of the cerebral cortex, which is predominantly composed of the neocortex, are more likely to be targets of positive selection than those affecting cerebellum development. Second, we ask whether patterns of molecular evolution mirror non-allometric changes in whole component volume, with greater evidence of an accelerated rate of evolution in hominoids for genes implicated in the development of the cerebellum than for those genes with roles in cerebral cortex development. Finally, to explore whether cortico-cerebellar co-evolution at the volumetric level is maintained by selection acting on a common or independent set of genes, we test whether each gene evolves in a manner suggesting a specific evolutionary association with either or both brain components.
Materials and Methods
Data Collection
We used Gene Ontology data to build lists of candidate genes with known roles in cerebral cortex or cerebellum development. These lists are inevitably incomplete but provide an initial assessment of patterns of molecular evolution across genes with known roles in different aspects of brain development, based on currently available data. We obtained a list of human genes with known roles in “cerebral cortex development” or “cerebellum development” from two ontology databases: Amigo [Carbon et al., 2009] (GO:0021987 and GO:0021549) and QuickGO of EBI [Binns et al., 2009] (GO:0021987 and GO:0021549). These GO terms have the best-matched definitions among those relevant for each component. In anthropoids, the neocortex comprises the vast majority of the cerebral cortex (∼90% [Stephan et al., 1981]). This GO search resulted in 198 Amigo and 300 QuickGO genes associated with cerebral cortex development, and 144 Amigo and 222 QuickGO genes associated with cerebellum development, which are not mutually exclusive, that were then combined to form the starting gene set. This starting human gene set was then used to obtain 1:1 orthologs from 11 anthropoid genomes (Fig. 1a) using a reciprocal best hit BLASTn [Altschup et al., 1990] approach between each species and the human coding sequence with an e-value cut-off of 1e−10 and a minimum percentage identify of 30. Eleven-way 1:1 ortholog sets were aligned using codon aware PRANK v 140615 [Löytynoja and Goldman, 2010], converted to phylip format in preparation for PAML analyses [Yang, 2007] and filtered using the conservative alignment filtering program SWAMP v 1.0 [Harrison et al., 2014]. SWAMP identifies short stretches in a gene alignment with unusually high numbers of non-synonymous mutations on individual branches of a tree that are typically caused by sequence or alignment errors and that produce false signatures of positive selection. The program removes these regions of poorly aligned or error-rich sequences on a branch-specific basis, reducing their effect on site-specific analyses of selection pressures [Harrison et al., 2014]. SWAMP was run twice, first using a threshold of 5 and a window size of 15, and then a second run with a threshold of 2 with a window size of 3, with a minimum sequence length of 300 bases and interscan masking for both runs. The final, strict and conservatively filtered 11-species dataset consisted of three non-overlapping groups: 53 genes with known roles in cerebral cortex development, 47 genes with known roles in cerebellum development and 10 genes with known roles in both (online suppl. File 1; online suppl. Table S1; see www.karger.com/doi/10.1159/000477432 for all online suppl. material). Additional analyses were conducted with broader GO terms to assess the potential for biases produced by GO selection; these are discussed in online supplementary File 2.
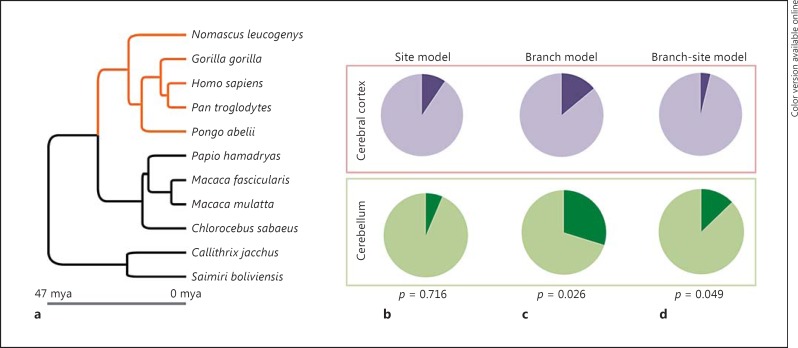
Fig. 1.
Selection on genes with annotated roles in cerebral cortex and cerebellum development in anthropoids. a Phylogeny of the 11 anthropoids included in this study; hominoid lineages are shown in a contrasting shade. b-d Proportion of genes significant (darker) under each test for cerebral cortex (top) and cerebellum genes (bottom). p values from a Fisher's exact test comparing the groups also shown.
Selection Analyses
Estimation of dN/dS ratios (ω), a common measure of the strength of selection acting on a protein coding gene, was carried out using a codon-based maximum likelihood method (PAML v.4) [Yang, 2007]. The dN/dS ratio estimates the rate at which non-synonymous substitutions are fixed (dN) relative to the rate at which synonymous substitutions are fixed (dS). Non-synonymous substitutions are assumed to affect protein function and they are subject to positive or negative selection, whilst dS is assumed to reflect the neutral rate of evolution [Yang, 2007]. Nested models were compared using the likelihood ratio test statistic (-2[log likelihood 1 - log likelihood 2]) to critical values of the χ2 distribution and degrees of freedom as the difference in the number of parameters estimated in each model. We compared the frequency of positive selection acting on genes associated with cerebral cortex and cerebellum development in two ways. First, we used the site model tests of positive selection (“model 8/8a”) to identify genes evolving under positive selection across anthropoids. The site models allow ω to vary across sites, but not across branches. Second, we used the branch-site models (“new model A”) to identify genes under positive selection specifically in hominoids (Fig. 1a). The branch-site models allow ω to vary across both sites and branch categories defined a priori. We repeated this test for accelerated rates of evolution in hominoids using a branch model test, where ω is fixed across sites but varies between branch categories. For each analysis, the percentage of each category of gene that experienced selection was compared using a Z test and the more conservative Fisher's exact test.
Gene-Phenotype Co-Evolution
We sought to test the link between the molecular evolution of our gene set and the evolution of neocortex and cerebellum volume using a phylogenetic, comparative approach [Montgomery et al., 2011]. Data on neocortex, cerebellum and total brain volume were taken from Stephan et al. [1981], with additional data for Pongo from Zilles and Rehkemper [1988]. Rest-of-brain volume was calculated as total brain volume minus neocortex and cerebellum volume. Branch models in PAML were used to calculate the root-to-tip dN/dS for each species. These were then regressed against the phenotypic trait of interest using a one-tailed phylogenetic generalised least squares regression implemented in BayesTraits [Pagel, 1999], which corrects for the non-independence of interspecific data. Each gene was regressed against absolute neocortex volume and absolute cerebellum volume as predictor variables simultaneously: log10(dN/dS) ∼ log10(neocortex) + log10(cerebellum). This permits the identification of genes with a specific, positive co-evolutionary association with either neocortex or cerebellum volume, or genes that co-vary with the volume of both traits. We also repeated the analyses including absolute rest-of-brain volume as an additional predictor variable in the regression model: log10(dN/dS) ∼ log10(neocortex) + log10(cerebellum) + log10(rest-of-brain). This tests whether the results are robust to correction for a measure of overall brain volume. This analysis leads to results similar to those presented in the main text, identifying similar distributions of significant results and highlighting the same genes (online suppl. File 1; online suppl. Table S6). For the sole purpose of data visualisation we calculated residuals from a neocortex∼cerebellum volume regression to display the relationships for selected loci in Figure 2. Residuals were not used in any statistical analyses contributing to the main text.
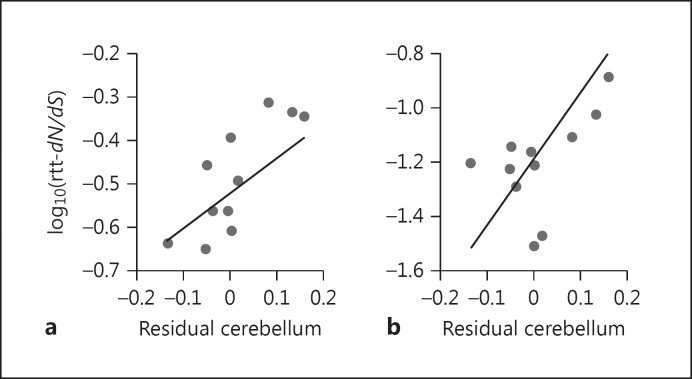
Fig. 2.
Phenotypic associations for two genes, i.e., RGRIP1L (a) and ATRN (b). The log10-transformed root-to-tip (rtt) dN/dS is plotted against residual cerebellum volume, calculated from a phylogenetic generalised least squares regression of cerebellum against neocortex volume. The line shows the result of a phylogenetically controlled regression between rtt-dN/dS and residual cerebellum volume. This figure is for illustrative purposes; in the main analyses the two brain components were included as variables in a multiple regression.
Results
What Proportion of Genes with Known Roles in Cerebral Cortex or Cerebellum Development Are Targeted by Pervasive Positive Selection?
Across anthropoids, the average rate of evolution of protein-coding genes with known roles in the development of the cerebellum did not differ from those with known functions in the development of the cerebral cortex, which is predominantly comprised of neocortex (Mann-Whitney U[98] = 1,167.5, Z = 0.539, p = 0.590) (online suppl. File 1; online suppl. Table S2). Genes with annotated functions in the development of both brain components did not differ in their average rate of evolution from those specifically linked to either cerebral cortex (U[61] = 296.5, Z = 0.592, p = 0.554) or cerebellum development (U[55] = 241.5, Z = 0.136, p = 0.892). Site model tests for positive selection acting at a subset of codons also identified a similar proportion of genes associated with the cerebral cortex or cerebellum development genes with evidence of positive selection across anthropoids. Three of forty-seven cerebellum genes (6.4%) were significant at a nominal α of 0.05, compared to 5/53 cerebral cortex genes (9.4%). These proportions were not significantly different (Fisher's exact test, p = 0.716, Z test: z = 0.5613, p = 0.575) (Table 1; Fig. 1b; online suppl. File 1; online suppl. Table S3). The same conclusion was reached after correcting for multiple testing using the Benjamini-Hochberg method to control the false discovery rate [Benjamini and Hochberg, 1995], after which the site model test was significant for two genes linked to cerebellum development (i.e., RPGRIP1L and PCNT) and one gene linked to cerebral cortex development (TACC2). None of the 10 genes with annotated functions in the development of both brain components showed evidence of positive selection. Given current GO data, these results suggest that protein-coding genes affecting cerebral cortex development are no more likely to be targeted by positive selection than those affecting cerebellum development.
Table 1.
Summary of the results of dN/dS analyses and tests of positive selection
| Test | Inference | Significant results (proportion) |
Difference in proportion |
||
|---|---|---|---|---|---|
| cerebral cortex (n = 47) | cerebellum (n = 53) | Z test | Fisher's exact test | ||
| Site model test | pervasive positive selection | 5 (9.4) | 3 (6.4) | z = 0.561, p = 0.575 | p = 0.716 |
| Branch model test | accelerated evolution in apes | 6 (11.3) | 14 (29.8) | z = 2.304, p = 0.021 | p = 0.026 |
| Branch-site model test | |||||
| All | episodic positive selection in apes | 2 (3.8) | 6 (12.8) | z = 1.654, p = 0.099 | p = 0.143 |
| Restricted1 | episodic positive selection in apes | 0 (0.0) | 4 (10.0) | z = 2.136, p = 0.049 | p = 0.032 |
Values are presented as numbers (%) unless otherwise stated.
Excludes genes with significant site-model results.
Do Rates of Molecular Evolution Reflect Rates of Brain Component Expansion?
A significantly greater proportion of genes annotated in cerebellum development (14/47; 29.8%) than those annotated in cerebral cortex development (6/53; 11.3%) experienced an accelerated rate of evolution in hominoids, taking a nominal significance threshold of 0.05 (Fisher's exact test, p = 0.026, Z test: z = 2.304, p = 0.021) (Table 1; Fig. 1c; online suppl. File 1; online suppl. Table S4). This result was also reflected in the branch-site test, where the proportion of our cerebellum genes (6/47; 12.8%) with evidence of episodic positive selection in hominoids again exceeded the proportion of our cerebral cortex genes (2/53, 3.8%). In this case the trend did not reach significance (Fisher's exact test, p = 0.143, Z test: z = 1.654, p = 0.099) (online suppl. File 1; online suppl. Table S5). However, one assumption of the branch-site test is the absence of positive selection in the background (i.e., non-hominoid) branches [Anisimova and Yang, 2007]. After excluding genes with evidence of positive selection across anthropoids under the site model test (online suppl. File 1; online suppl. Table S3), the proportions were 0/48 for the cerebral cortex and 4/44 for the cerebellum, and the trend observed in the branch-site test was narrowly significant at p < 0.05 (Fisher's exact test, p = 0.049, Z test: z = 2.136, p = 0.032) (Table 1; Fig. 1d). Again, given available GO data, these contrasting proportions suggest that the strength of positive selection acting on genes controlling cerebellum development may have increased during hominoid evolution, a clade in which the rate of cerebellar expansion significantly accelerated [Barton and Venditti, 2014] beyond the rate predicted based on scaling with the neocortex in other primates [Barton and Harvey, 2000; Barton and Venditti, 2013].
Is Selection Associated with Interspecific Variation in the Size of Specific Brain Components?
We identified 11/47 genes annotated in cerebellum development (23.4%) that co-evolved with cerebellum volume, independently of neocortex volume; 4 of these were significant at p < 0.001 and 2 (RPGRIP1L and ATRN) remained significant after FDR correction (Table 1; Fig. 2; online suppl. File 1; online suppl. Table S6). We re-analysed the top 4 genes associated with cerebellum volume, separating dN and dS whilst accounting for co-variation in neocortex volume, to test if the association is driven by variation in dN. For 3/4 genes we found a significant partial regression with dN (ATRN t5 = 3.789, p = 0.006; EZH2 t5 = 3.990, p = 0.005; KNDC1 t5 = 3.586, p = 0.008), the remaining locus showed a non-significant trend (RGRIP1L t5 = 1.582, p = 0.087). Only one cerebellum development gene (2.1%) showed an association with neocortex volume, which is a significantly lower proportion (Fisher's exact test, p = 0.004, Z test: z = 3.091, p = 0.002). One of fifty-three genes annotated as functioning in cerebral cortex development (DICER1) showed evidence of an association with neocortex volume (1.9%), whilst 6 (11.3%) showed evidence of an association with cerebellum size (online suppl. File 1; online suppl. Table S6). This was not a significant difference in proportion (Fisher's exact test, p = 0.113, Z test: z = 1.955, p = 0.050), only one of these associations was significant at p < 0.001, and none survived FDR correction. One of the ten genes annotated as functioning in both neocortex and cerebellum development (GART) showed an association with cerebellum, but not neocortex, volume (online suppl. File 1; online suppl. Table S6). Similar results were obtained when rest-of-brain was included in the regression model with the same region-specific pattern identified and a high degree of overlap between the genes highlighted as showing positive phenotypic associations (online suppl. File 1; online suppl. Table S6).
Our regression analyses, which included the absolute volumes of both brain regions as predictor variables, therefore failed to identify any gene that showed a co-evolutionary association with variation in both neocortex and cerebellum volume, regardless of the gene category or the inclusion of rest-of-brain as a third variable. These results are consistent with the action of selection on genes associated with phenotypic variation specific to each brain component. To test whether this pattern is robust to the choice of GO term we performed a series of additional analyses using three further GO terms higher in the GO hierarchy: “brain development,” “forebrain development” and “hindbrain development.” The results were consistent with our main analyses, with an absence of genes which co-evolve with both cerebellum and neocortex volume (online suppl. File 2).
Discussion
Primate brain expansion reflects increases in the volume and neuron number of multiple co-evolving structures [Barton and Harvey, 2000]. This pattern of distributed adaptation must be reflected in the molecular evolution of genes controlling brain development. Our results provide two contributions to understanding the genetic basis of brain evolution. First, on the basis of currently available GO annotation data, we found no evidence that protein-coding genes associated with cerebral cortex development are more likely to have evolved adaptively across anthropoids than protein-coding genes linked to cerebellum development. Indeed, significantly more of these cerebellum genes experienced an increased rate of evolution in hominoids, consistent with evidence for a non-allometric expansion of the cerebellum in apes [Rilling and Insel, 1998; MacLeod et al., 2003; Barton and Venditti, 2014]. Second, we provide evidence that the selection pressures acting on some of these genes may be linked to evolutionary changes in the development of particular brain components, independently of others.
Our results suggest that a significant proportion of the genetic changes that underpin the adaptive evolution of primate brain size, structure and therefore cognition may affect aspects of non-cortical development. This conclusion is consistent with evidence of adaptive expansion in cerebellum volume during hominoid evolution [Rilling and Insel, 1998; MacLeod et al., 2003; Barton and Venditti, 2014]. Our results are, of course, wholly dependent on the quality of gene ontology annotation. Our incomplete knowledge of gene function makes it is inevitable that the genes included in this study reflect a minority of those that influence cerebral cortex and cerebellum development. Gene ontology datasets are routinely used in post hoc tests for functional enrichment where variable quality of GO annotation may also affect the power to detect enrichment among different gene classes. Here, we have used the available data to facilitate initial tests of specific hypotheses to determine whether, given existing data, there is any justification for a restricted focus on the genetics of neocortical expansion. In doing so, we adopted a conservative approach, analysing only genes with strict 11-way 1:1 orthologs in published anthropoid genomes and removing short and/or poorly aligned sequence. This further reduces the gene set, but produces more reliable estimates of selection regimes [Harrison et al., 2014]. Although we cannot formally rule out the possibility that our gene sets may be biased in a way that would produce contrasting patterns of results between gene categories, our supplementary analyses suggest that the patterns we observed are robust among ontology terms higher in the GO hierarchy.
Changes in protein-coding genes offer one route for selection to alter development, but many evolutionary changes in brain development may be caused by altered gene regulation. Whether or not regulatory evolution is biased towards changes affecting particular brain structures has not been robustly tested. However a phylogenetic analysis of lineage-specific shifts in gene expression across mammals also found an over-abundance of hominoid-specific expression shifts in genes expressed in the cerebellum compared to those expressed in the rest of the brain [Brawand et al., 2011]. A comparison of the expression profiles of microRNA in humans, chimpanzees and macaques similarly showed high rates of evolution among cerebellar genes, although in this case the rate of change was higher for microRNA expressed in the prefrontal cortex [Somel et al., 2011]. These results at least suggest that future comparative studies of primate gene expression should not be limited to samples derived from cortical tissue, and functional tests of candidate genes identified as targets of positive selection in genome-wide scans should consider the phenotypic relevance of changes in gene function in non-cortical structures.
We also note that our analyses were designed to detect patterns of molecular evolution associated with changes in whole component volume. We do not rule out the possibility that the internal structure or physiology of either the neocortex or the cerebellum has, in part, evolved independently of their total volumes. Indeed, available data suggest that the internal structure of both the neocortex [Barton, 2007] and the cerebellum [Whiting and Barton, 2003] evolves in a mosaic manner. Across primates neocortical expansion is particularly associated with increases in the size of visual cortices [Barton, 2007] and cortical association areas [Sherwood et al., 2012], whilst cerebellar expansion is particularly due to increases in the size of the lateral hemisphere [MacLeod et al., 2003] and those areas within the lateral hemisphere that receive projections from prefrontal areas the neocortex [Balsters et al., 2010]. This internal remodelling, which extends to fine grained morphology [see Verendeev and Sherwood, 2017] and has been suggested by some analyses of postnatal gene expression [e.g., Somel et al., 2009, 2011], may show a distinct phylogenetic signal and involve genetic changes that are at least partially independent of component volume (and by proxy neuron number), potentially by altering patterns of regional differentiation of proliferating progenitor cells [e.g., Sylvester et al., 2010]. Future work combining more fine grained neuroanatomical data with larger genetic datasets may provide more precise resolution of the genetic basis of the size and structure of brain components.
Nevertheless, our analyses highlight several genes with patterns of molecular evolution that link them to interspecific differences in cerebellum volume. These include two genes, i.e., RGRIP1L and ATRN, with a particularly strong signal of co-evolution between the strength of selection acting on their coding sequence and cerebellum volume, independently of variation in neocortex or rest-of-brain volume, in our gene-phenotype association tests (Fig. 2). RGRIP1L, which also shows evidence of positive selection hominoids, is one of a small number of loci linked to Joubert syndrome, a rare genetic disorder associated with severe hypoplasia of the mid-hindbrain and cerebellar vermis [Joubert et al., 1969; Doherty, 2009]. The cellular role of RGRIP1L appears to be in the correct function of the cilia and basal bodies [Arts et al., 2007; Delous et al., 2007] which are necessary for expansion of the cerebellar neural progenitor pool [Spassky et al., 2008]. Disruption of ATRN, an E3 ubiquitin ligase, in Mus causes vacuolization and degeneration of the cerebellum [Bronson et al., 2001; He et al., 2003]. Another ubiquitin ligase, UBE3A, has been implicated in brain and cognitive development [Wilkinson et al., 2007] and interacts with ASPM, a key regulator of brain size [Singhmar and Kumar, 2011]. A third gene, EZH2, also shows evidence of a phenotypic association with cerebellum volume but is narrowly nonsignificant after FDR correction. EZH2 functions to regulate neuronal migration of precerebellar neurons during the development of the cortico-cerebellar connectivity [Meglio et al., 2013].
We also found multiple genes with significantly accelerated rates of evolution in hominoids, coincident with an accelerated rate of cerebellar expansion [Barton and Venditti, 2014]. Several of these genes also showed an association with cerebellum volume in our gene-phenotype association tests at a nominal significance threshold of 0.05. These include AGTPBP1, the disruption of which causes cerebellar Purkinje cell degeneration [Lalonde et al., 2006], PCNT, which causes primordial dwarfism with microcephaly [Rauch et al., 2008], TH, a gene linked to two disorders that affect motor control, Segawa syndrome and Parkinson's [Ludecke et al., 1995; Haavik and Toska, 1998], and MYO16 and GART which have been putatively linked to autistic spectrum disorders and Down syndrome, respectively [Brodsky et al., 1997; Liu et al., 2015].
Finally, we identified patterns of molecular evolution that may implicate a separate group of genes in the evolution of neocortex size. Although our GO category “cerebral cortex” may include genes specifically involved in the development of cortical structures outside the neocortex, that are likely to co-vary with absolute neocortex volume, all of the genes highlighted by our analyses have known functions in the development of the neocortex. The sole gene to show a phenotypic association with neocortex volume, DICER1 regulates neurogenesis during the formation of the neocortex and the hippocampus in a time-dependent manner [Davis et al., 2008; Kawase-koga et al., 2009]. Notably, although neither showed a phenotypic association in our gene-phenotype regression analyses, the two genes with the strongest evidence for episodic positive selection in hominoids belong to the same gene family. TACC1 and TACC2 are both associated with regulation of nuclear migration and they are required for normal patterns of self-renewal in neural progenitors. TACC interact with a centrosomal protein, CEP120, to regulate nuclear migration and the self-renewal of cortical neural progenitor cells by controlling microtubule growth [Xie et al., 2007]. A similar function is thought to mediate the influence of microcephaly genes on brain development [Thornton and Woods, 2009; Pulvers et al., 2010].
Beyond the functional effects of individual genes, our approach has the potential to tackle fundamental questions about how composite or modular tissues, such as the brain, evolve. For example, two principle models of brain evolution dominate debates surrounding the adaptive significance of variation in brain structure. One model proposes that a conserved developmental program drives a “concerted” pattern of brain evolution, with selection shaping the overall size of the system rather than its individual components through a coordinated lengthening or contracting of a conserved developmental schedule governing the development of the size of brain components [Finlay and Darlington, 1995; Finlay et al., 2001]. An alternative model instead argues that different brain regions evolve independently of overall brain size to meet species-specific behavioural needs, resulting in a “mosaic” pattern of brain evolution, but stresses that brain regions may also co-evolve due to their functional interdependence [Barton and Harvey, 2000; de Winter and Oxnard, 2001]. These models are potentially non-mutually exclusive and reconcilable at a phenotypic level [Herculano-Houzel et al., 2014], and there has been movement to accommodate some taxon-specific shifts in brain structure within the concerted framework [Clancey et al., 2000, 2001; Workman et al., 2013]. However, focusing on the “polarised” interpretation of these hypotheses allows us to recognise that these two models implicitly make contrasting predictions about the genetic architecture and proximate basis of variation in brain structure [Airey and Williams, 2001; Montgomery et al., 2016]. The concerted model predicts that variation in the size of different brain regions will be determined by genetic correlations between those structures, i.e., variation in a common set of genes [Montgomery et al., 2016]. Although cerebellar and cortical cells are produced by distinct progenitor pools [Carletti and Rossi, 2007], concerted expansion could be driven by changes in the global developmental schedules that define the order of neurogenesis across brain structures [Finlay and Darlington, 1995; Finlay et al., 2001], extrinsic morphogens that may act on multiple developing brain regions [Menuet et al., 2007; Sylvester et al., 2010], or, in theory, pleiotropic effects of mutations in genes controlling cell division in multiple progenitor pools. The mosaic model instead predicts that the development of different brain regions must be at least partially distinct in order to facilitate independent evolution. In this case, both co-evolution between brain structures and non-allometric changes in component size may be explained by selection acting of distinct sets of genes [Montgomery et al., 2016].
In recent years these predictions have been tested using quantitative genetics within a range of vertebrates, using wild pedigrees [Rogers et al., 2007, 2010; Fears et al., 2009], inbred mouse strains [Hager et al., 2012] or divergent populations/domestic breeds [Henriksen et al., 2016; Noreikiene et al., 2015]. In support of the mosaic brain hypothesis, these have found little evidence for widespread genetic co-variation between major brain components. Similarly, large genome-wide association studies within humans have identified independent genetic bases associated with brain regions [Toro et al., 2013; Hibar et al., 2015].
Quantitative genetics assesses the phenotypic associations of standing genetic variation within populations. Their relevance to macroevolution therefore depends on the relative frequency at which selection acts on de novo mutations that may cause different patterns of genetic correlation. Our phylogenetic approach complements these intraspecific studies, providing the first interspecific test designed to identify genes associated with neocortex and/or cerebellum evolution. Even after accounting for variation in total brain size, the size of these two structures shows a consistent pattern of co-evolution across primates, both at a volumetric and at a cellular level [Barton and Harvey, 2000; Whiting and Barton, 2003; Herculano-Houzel and Sherwood, 2010], reflecting their functional interdependence [Ramnani, 2006; Barton, 2012]. However, our analyses did not identify any gene that shows a pattern of molecular evolution that co-varies with the volume of both structures across anthropoids, whereas it does identify multiple genes with a specific evolutionary association with either cerebellum or neocortex volume. This suggests that the co-evolution of these structures is unlikely to be solely due to genetic integration, common developmental control or pleiotropy, although such mechanisms may still play a role. However, if more broadly true, this conclusion bolsters the interpretation of cortico-cerebellar co-evolution as indicative of an adaptive relationship maintained by selection acting on distinct developmental pathways. We have focused here on two brain components that have clearly expanded during primate evolution, whereas the concerted and mosaic arguments extend across all brain regions. It remains possible, and indeed likely, that other brain regions show greater evidence of genetic integration. Understanding how and when such genetic and developmental links shape and constrain brain evolution remains central to interpreting the adaptive significance of brain size and structure [Montgomery et al., 2016].
Comparative functional analysis of the genes highlighted by our analyses will be necessary to confirm and extend these conclusions. These will also be needed to address key questions beyond functional effects. For example, when disrupted, several of the genes highlighted by our analyses affect the development of multiple organs. For example, both AGTPBP1 and PAFAH1B have known roles in spermatogenesis [Yan et al., 2003; Kim et al., 2011], whilst disruption of PCNT can affect global somatic growth [Rauch et al., 2008]. If these genes do have an evolutionary role in cerebellar development, how are these pleiotropic effects avoided? Similar questions have been raised regarding previous candidate genes [Pulvers et al., 2010; Montgomery et al., 2011], further emphasising the importance of coupling comparative and functional data. Finally, volumetric data can mask a hidden diversity of neuron number. In particular, neuron density in the cerebellum is substantially higher than in the cerebral cortex [Herculano-Houzel et al., 2007]. Given that our primary interest is in the regulation of neurogenesis, future gene-phenotype analyses may benefit from an increase in the taxonomic sampling of studies which estimate neuron number across brain regions [Herculano-Houzel et al., 2015]. Currently the overlap between genomic datasets and estimates of neuron number in primates is insufficient for meaningful comparisons.
In summary, we have presented an analysis aimed at providing an initial assessment of the strength of selection targeting genes with development roles in distinct brain regions. Although our understanding of gene ontology is far from complete, we have illustrated how this information can be used to test hypotheses in a phylogenetic comparative setting, in addition to post-hoc enrichment analyses. Our data suggests that, among protein-coding genes, there is currently no evidence that selection is limited or biased towards genes affecting cerebral cortex development, and encourage evolutionary geneticists to adopt a cohesive view of brain evolution that encompasses the recognised importance or non-allometric expansion of non-cortical regions and to tackle the central question of co-evolution and the relative genetic independence of brain components. Finally, we further illustrate the potential of hypothesis driven comparative genetics in dissecting the genetic basis of phenotypic evolution. The ever-increasing numbers of sequenced genomes will permit increasingly powerful analyses of the targets of selection and their phenotypic relevance.
Disclosure Statement
The authors declare that they have no competing interests.
Acknowledgements
S.H.M. is grateful for an Early Career Research Fellowship from the Leverhulme Trust and an Independent Research Fellowship from NERC for funding. We thank Amy Boddy, Robert Barton, Nicholas Mundy, and Judith Mank and her lab at UCL for comments on previous drafts of this paper.
References
- Ackermann H. Cerebellar contributions to speech production and speech perception: psycholinguistic and neurobiological perspectives. Trends Neurosci. 2008;31:265–272. doi: 10.1016/j.tins.2008.02.011. [DOI] [PubMed] [Google Scholar]
- Airey DC, Williams RW. Quantitative neurogenetic perspectives. Behav Brain Sci. 2001;24:279–280. [Google Scholar]
- Altschup SF, Gish W, Miller W, Myers E, Lipman D. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Anisimova M, Yang Z. Multiple hypothesis testing to detect lineages under positive selection that affects only a few sites. Mol Biol Evol. 2007;24:1219–1228. doi: 10.1093/molbev/msm042. [DOI] [PubMed] [Google Scholar]
- Arts HH, Doherty D, van Beersum SE, Parisi MA, Letteboer SJF, Voesenek K, et al. Mutations in the gene encoding the basal body protein RPGRIP1L, a nephrocystin-4 interactor, cause Joubert syndrome. Nat Genet. 2007;39:882–888. doi: 10.1038/ng2069. [DOI] [PubMed] [Google Scholar]
- Balsters JH, Cussans E, Diedrichsen J, Phillips KA, Preuss TM, Rilling JK, et al. Evolution of the cerebellar cortex: the selective expansion of prefrontal-projecting cerebellar lobules. Neuroimage. 2010;49:2045–2052. doi: 10.1016/j.neuroimage.2009.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton RA. Evolutionary specialization in mammalian cortical structure. J Evol Biol. 2007;20:1504–1511. doi: 10.1111/j.1420-9101.2007.01330.x. [DOI] [PubMed] [Google Scholar]
- Barton RA. Embodied cognitive evolution and the cerebellum. Phil Trans R Soc Lond B Biol Sci. 2012;367:2097–2107. doi: 10.1098/rstb.2012.0112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton RA, Harvey PH. Mosaic evolution of brain structure in mammals. Nature. 2000;405:1055–1058. doi: 10.1038/35016580. [DOI] [PubMed] [Google Scholar]
- Barton RA, Venditti C. Human frontal lobes are not relatively large. Proc Natl Acad Sci USA. 2013;110:9001–9006. doi: 10.1073/pnas.1215723110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton RA, Venditti C. Rapid evolution of the cerebellum in humans and other great apes. Curr Biol. 2014;24:2440–2444. doi: 10.1016/j.cub.2014.08.056. [DOI] [PubMed] [Google Scholar]
- Bauernfeind AL, Soderblom EJ, Turner ME, Moseley MA, Ely JJ, Hof PR, et al. Evolutionary divergence of gene and protein expression in the brains of humans and chimpanzees. Genome Biol Evol. 2015;7:2276–2288. doi: 10.1093/gbe/evv132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple yesting. J Stat Soc B. 1995;57:289–300. [Google Scholar]
- Binns D, Dimmer E, Huntley R, Barrell D, O'Donovan C, Apweiler R. QuickGO: a web-based tool for Gene Ontology searching. Bioinformatics. 2009;25:3045–2046. doi: 10.1093/bioinformatics/btp536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boddy AM, Harrison PW, Montgomery SH, Caravas JA, Raghanti MA, Phillips KA, Mundy NI, Wildman DE. Evidence of a conserved molecular response to selection for increased brain size in primates. Genome Biol Evol. 2017;9:700–713. doi: 10.1093/gbe/evx028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd JL, Skove SL, Rouanet JP, Pilaz L-J, Bepler T, Gordân R, et al. Human-chimpanzee differences in a FZD8 enhancer alter cell-cycle dynamics in the developing neocortex. Curr Biol. 2015;25:772–779. doi: 10.1016/j.cub.2015.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brawand D, Soumillon M, Necsulea A, Julien P, Csárdi G, Harrigan P, et al. The evolution of gene expression levels in mammalian organs. Nature. 2011;478:343–348. doi: 10.1038/nature10532. [DOI] [PubMed] [Google Scholar]
- Brodsky G, Barnes T, Bleskan J, Becker L, Cox M, Patterson D. The human GARS-AIRS-GART gene encodes two proteins which are differentially expressed during human brain development and temporally overexpressed in cerebellum of individuals with Down syndrome. Hum Mol Genet. 1997;6:2043–2050. doi: 10.1093/hmg/6.12.2043. [DOI] [PubMed] [Google Scholar]
- Bronson RT, Donahue LR, Samples R, Kim JH, Naggert JK. Mice with mutations in the mahogany gene Atrn have cerebral spongiform changes. J Neuropathol Exp Neurol. 2001;60:724–730. doi: 10.1093/jnen/60.7.724. [DOI] [PubMed] [Google Scholar]
- Burki F, Kaessmann H. Birth and adaptive evolution of a hominoid gene that supports high neurotransmitter flux. Nat Genet. 2004;36:1061–1063. doi: 10.1038/ng1431. [DOI] [PubMed] [Google Scholar]
- Carbon S, Ireland A, Mungall CJ, Shu S, Marshall B, Lewis S, et al. AmiGO: online access to ontology and annotation data. Bioinformatics. 2009;25:288–289. doi: 10.1093/bioinformatics/btn615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carletti B, Rossi F. Neurogenesis in the cerebellum. Neurosci. 2007;14:91–100. doi: 10.1177/1073858407304629. [DOI] [PubMed] [Google Scholar]
- Clancy B, Darlington RB, Finlay BL. The course of human events: predicting the timing of primate neural development. Dev Sci. 2000;3:57–66. [Google Scholar]
- Clancy B, Darlington RB, Finlay BL. Translating developmental time across mammalian species. Neuroscience. 2001;105:7–17. doi: 10.1016/s0306-4522(01)00171-3. [DOI] [PubMed] [Google Scholar]
- Davis TH, Cuellar TL, Koch SM, Barker AJ, Harfe BD, McManus MT, et al. Conditional loss of Dicer disrupts cellular and tissue morphogenesis in the cortex and hippocampus. J Neurosci. 2008;17:4322–4330. doi: 10.1523/JNEUROSCI.4815-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delous M, Baala L, Saloman R, Laclef C, Vierkotten J, Golzio C, et al. The ciliary gene RPGRIP1L is mutated in cerebello-oculo-renal syndrome (Joubert syndrome type B) and Meckel syndrome. Nat Genet. 2007;39:875–881. doi: 10.1038/ng2039. [DOI] [PubMed] [Google Scholar]
- de Winter W, Oxnard CE. Evolutionary radiations and convergences in the structural organization of mammalian brains. Nature. 2001;409:710–714. doi: 10.1038/35055547. [DOI] [PubMed] [Google Scholar]
- Doherty D. Joubert syndrome: insights into brain. Semin Pediatr Neurol. 2009;16:143–154. doi: 10.1016/j.spen.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enard W. Comparative genomics of brain size evolution. Front Hum Neurosci. 2014;8:345. doi: 10.3389/fnhum.2014.00345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enard W, Gehre S, Hammerschmidt K, Hölter SM, Blass T, Somel M, et al. A humanized version of Foxp2 affects cortico-basal ganglia circuits in mice. Cell. 2009;137:961–971. doi: 10.1016/j.cell.2009.03.041. [DOI] [PubMed] [Google Scholar]
- Enard W, Przeworski M, Fisher SE, Lai CSL, Wiebe V, Kitano T, et al. Molecular evolution of FOXP2, a gene involved in speech and language. Nature. 2002a;418:869–872. doi: 10.1038/nature01025. [DOI] [PubMed] [Google Scholar]
- Enard W, Khaitovich P, Klose J, Zöllner S, Heissig F, Giavalisco P, et al. Intra- and interspecific variation in primate gene expression patterns. Science. 2002b;296:340–343. doi: 10.1126/science.1068996. [DOI] [PubMed] [Google Scholar]
- Fears SC, Melega WP, Service SK, Lee C, Chen K, Tu Z, et al. Identifying heritable brain phenotypes in an extended pedigree of vervet monkeys. J Neurosci. 2009;29:2867–2875. doi: 10.1523/JNEUROSCI.5153-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferland RJ, Eyaid W, Collura RV, Tully LD, Hill RS, Al-nouri D, et al. Abnormal cerebellar development and axonal decussation due to mutations in AHI1 in Joubert syndrome. Nat Genet. 2004;36:1008–1013. doi: 10.1038/ng1419. [DOI] [PubMed] [Google Scholar]
- Finlay B, Darlington R. Linked regularities in the development and evolution of mammalian brains. Science. 1995;268:1578–1584. doi: 10.1126/science.7777856. [DOI] [PubMed] [Google Scholar]
- Finlay BL, Darlington RB, Nicastro N. Developmental structure in brain evolution. Behav Brain Sci. 2001;24:263–278. discussion 278–308. [PubMed] [Google Scholar]
- Florio M, Albert M, Taverna E, Namba T, Brandl H, Lewitus E, et al. Human-specific gene ARHGAP11B promotes basal progenitor amplification and neocortex expansion. Science. 2015;347:1465–1470. doi: 10.1126/science.aaa1975. [DOI] [PubMed] [Google Scholar]
- Gabi M, Neves K, Masseron C, Ribeiro PF, Ventura-Antunes L, Torres L, Mota B, Kaas JH, Herculano-Houzel S. No relative expansion of the number of prefrontal neurons in primate and human evolution. Proc Natl Acad Sci USA. 2016;113:9617–9622. doi: 10.1073/pnas.1610178113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haavik J, Toska K. Tyrosine hydroxylase and Parkinson's disease. Mol Neurobiol. 1999;16:285–309. doi: 10.1007/BF02741387. [DOI] [PubMed] [Google Scholar]
- Hager R, Lu L, Rosen GD, Williams RW. Genetic architecture supports mosaic brain evolution and independent brain-body size regulation. Nat Commun. 2012;3:1079. doi: 10.1038/ncomms2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison PW, Jordan GE, Montgomery SH. SWAMP: Sliding Window Alignment Masker for PAML. Evol Bioinform Online. 2014;10:197–204. doi: 10.4137/EBO.S18193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, Lu X, Jolly AF, Eldridge AG, Watson SJ, Jackson PK, et al. Spongiform degeneration in mahoganoid mutant mice. Science. 2003;299:710–712. doi: 10.1126/science.1079694. [DOI] [PubMed] [Google Scholar]
- Henriksen R, Johnsson M, Andersson L, Jensen P, Wright D. The domesticated brain: genetics of brain mass and brain structure in an avian species. Sci Rep. 2016;30:34031. doi: 10.1038/srep34031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herculano-Houzel S, Catania K, Manger PR, Kaas JH. Mammalian brains are made of these: a dataset of the numbers and densities of neuronal and nonneuronal cells in the brain of glires, primates, scandentia, eulipotyphlans, afrotherians and artiodactyls, and their relationship with body mass. Brain Behav Evol. 2015;2015:145–163. doi: 10.1159/000437413. [DOI] [PubMed] [Google Scholar]
- Herculano-Houzel S, Collins CE, Wong P, Kaas JH. Cellular scaling rules for primate brains. Proc Natl Acad Sci USA. 2007;104:3562–3567. doi: 10.1073/pnas.0611396104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herculano-Houzel S, Manger PR, Kaas JH. Brain scaling in mammalian evolution as a consequence of concerted and mosaic changes in numbers of neurons and average neuronal cell size. Front Neuroanat. 2014;8:77. doi: 10.3389/fnana.2014.00077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herculano-Houzel S, Sherwood CC. Coordinated scaling of cortical and cerebellar numbers of neurons. Front Neuroanat. 2010;4:1–8. doi: 10.3389/fnana.2010.00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibar DP, Stein LP, Renteria ME, Arias-Vasquez A, Desrivières S, Jahanshad N, et al. Common genetic variants influence human subcortical brain structures. Nature. 2015;520:224–229. doi: 10.1038/nature14101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joubert M, Eisenring J, Robb JP, Andermann F. Familial agenesis of the cerebellar vermis A syndrome of episodic hyperpnea, abnormal eye movements, ataxia, and retardation. Neurology. 1969;19:813. doi: 10.1212/wnl.19.9.813. [DOI] [PubMed] [Google Scholar]
- Kamm GB, López-Leal R, Lorenzo JR, Franchini LF. A fast-evolving human NPAS3 enhancer gained reporter expression in the developing forebrain of transgenic mice. Philos Trans R Soc Lond B Biol Sci. 2013;368 doi: 10.1098/rstb.2013.0019. 20130019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawase-koga Y, Otaegi G, Sun T. Different timings of dicer deletion affect neurogenesis and gliogenesis in the developing mouse central nervous system. Dev Dyn. 2009;238:2800–2812. doi: 10.1002/dvdy.22109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeney J, Dumas L, Sikela J. The case for DUF1220 domain dosage as a primary contributor to anthropoid brain expansion. Front Hum Neurosci. 2014;8:1–11. doi: 10.3389/fnhum.2014.00427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaitovich P, Muetzel B, She X, Lachmann M, Hellmann I, Dietzsch J, et al. Regional patterns of gene expression in human and chimpanzee brains. Genome Res. 2004;14:1462–1473. doi: 10.1101/gr.2538704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim N, Xiao R, Choi H, Jo H, Kim J, Uhm S, et al. Abnormal sperm development in pcd 3J-/- mice: the importance of Agtpbp1 in spermatogenesis. Mol Cell. 2011;2005:39–48. doi: 10.1007/s10059-011-0002-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalonde R, Strazielle C, Inserm U, Electronique SDM, De Médecine F. Spontaneous and induced mouse mutations with cerebellar dysfunctions: behavior and neurochemistry. Brain Res. 2006;1140:51–74. doi: 10.1016/j.brainres.2006.01.031. [DOI] [PubMed] [Google Scholar]
- Leggio MG, Chiricozzi FR, Clausi S, Tedesco AM, Molinari M. The neuropsychological profile of cerebellar damage: the sequencing hypothesis. Cortex. 2011;47:137–144. doi: 10.1016/j.cortex.2009.08.011. [DOI] [PubMed] [Google Scholar]
- Liu YF, Sowell SM, Luo Y, Chaubey A, Cameron RS, Kim H, et al. Autism and intellectual disability-associated KIRREL3 interacts with neuronal proteins MAP1B and MYO16 with potential roles in neurodevelopment. PLoS One. 2015;10 doi: 10.1371/journal.pone.0123106. e0123106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löytynoja A, Goldman N. webPRANK: a phylogeny-aware multiple sequence aligner with interactive alignment browser. BMC Bioinformatics. 2010;11:579. doi: 10.1186/1471-2105-11-579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludecke B, Dworniczak B, Bartolome K. A point mutation in the tyrosine hydroxylase gene associated with Segawa's syndrome. Hum Genet. 1995;6:123–125. doi: 10.1007/BF00225091. [DOI] [PubMed] [Google Scholar]
- MacLeod CE, Zilles K, Schleicher A, Rilling JK, Gibson KR. Expansion of the neocerebellum in Hominoidea. J Hum Evol. 2003;44:401–429. doi: 10.1016/s0047-2484(03)00028-9. [DOI] [PubMed] [Google Scholar]
- Meglio T Di, Kratochwil CF, Vilain N, Loche A, Vitobello A, Yonehara K, et al. Ezh2 orchestrates topographic migration and connectivity of mouse precerebellar neurons. Science. 2013;339:204–207. doi: 10.1126/science.1229326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menuet A, Alunni A, Joly J-S, Jeffery WR, Rétaux S. Expanded expression of Sonic Hedgehog in Astyanax cavefish: multiple consequences on forebrain development and evolution. Development. 2007;134:845–855. doi: 10.1242/dev.02780. [DOI] [PubMed] [Google Scholar]
- Montgomery SH, Capellini I, Venditti C, Barton RA, Mundy NI. Adaptive evolution of four microcephaly genes and the evolution of brain size in anthropoid primates. Mol Biol Evol. 2011;28:625–638. doi: 10.1093/molbev/msq237. [DOI] [PubMed] [Google Scholar]
- Montgomery SH, Mundy NI. Evolution of ASPM is associated with both increases and decreases in brain size in primates. Evolution. 2012a;66:927–932. doi: 10.1111/j.1558-5646.2011.01487.x. [DOI] [PubMed] [Google Scholar]
- Montgomery SH, Mundy NI. Positive selection on NIN, a gene involved in neurogenesis, and primate brain evolution. Genes Brain Behav. 2012b;11:903–910. doi: 10.1111/j.1601-183X.2012.00844.x. [DOI] [PubMed] [Google Scholar]
- Montgomery SH, Mundy NI, Barton RA, Montgomery SH. Brain evolution and development: adaptation, allometry and constraint. Proc Biol Sci. 2016;283 doi: 10.1098/rspb.2016.0433. 20160433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noreikiene K, Herczeg G, Gonda A, Balazs G, Husby A, Merilä J. Quantitative genetic analysis of brain size variation in sticklebacks: support for the mosaic model of brain evolution. Proc Biol Sci. 2015;282 doi: 10.1098/rspb.2015.1008. 20151008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagel M. Inferring the historical patterns of biological evolution. Nature. 1999;401:877–884. doi: 10.1038/44766. [DOI] [PubMed] [Google Scholar]
- Parvizi J. Corticocentric myopia: old bias in new cognitive sciences. Trends Cogn Sci. 2009;13:354–359. doi: 10.1016/j.tics.2009.04.008. [DOI] [PubMed] [Google Scholar]
- Pollard KS, Salama SR, King B, Kern AD, Dreszer T, Katzman S, et al. Forces shaping the fastest evolving regions in the human genome. PLoS Genet. 2006;2:1599–1611. doi: 10.1371/journal.pgen.0020168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulvers JN, Bryk J, Fish JL, Wilsch-Bräuninger M, Arai Y, Schreier D, et al. Mutations in mouse Aspm (abnormal spindle-like microcephaly associated) cause not only microcephaly but also major defects in the germline. Proc Natl Acad Sci USA. 2010;107:16595–16600. doi: 10.1073/pnas.1010494107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramnani N. The primate cortico-cerebellar system: anatomy and function. Nat Rev Neurosci. 2006;7:511–522. doi: 10.1038/nrn1953. [DOI] [PubMed] [Google Scholar]
- Rauch A, Thiel CT, Schindler D, Wick U, Crow YJ, Ekici AB, et al. Mutations in the Pericentrin (PCNT) gene cause primordial dwarfism. Science. 2008;319:816–819. doi: 10.1126/science.1151174. [DOI] [PubMed] [Google Scholar]
- Rausher MD, Delph LF. When does understanding phenotypic evolution require identification of the underlying genes? Evolution. 2015;69:1655–1664. doi: 10.1111/evo.12687. [DOI] [PubMed] [Google Scholar]
- Rilling J, Insel T. Evolution of the cerebellum in primates: differences in relative volume among monkeys, apes and humans. Brain Behav Evol. 1998;52:308–314. doi: 10.1159/000006575. [DOI] [PubMed] [Google Scholar]
- Rogers J, Kochunov P, Lancaster J, Shelledy W, Glahn D, Blangero J, et al. Heritability of brain volume, surface area and shape: an MRI study in an extended pedigree of baboons. Hum Brain Mapp. 2007;28:576–583. doi: 10.1002/hbm.20407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers J, Kochunov P, Zilles K, Shelledy W, Lancaster J, Thompson P, et al. On the genetic architecture of cortical folding and brain volume in primates. Neuroimage. 2010;53:1103–1108. doi: 10.1016/j.neuroimage.2010.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenemann PT, Sheehan MJ, Glotzer LD. Prefrontal white matter volume is disproportionately larger in humans than in other primates. Nat Neurosci. 2005;8:242–252. doi: 10.1038/nn1394. [DOI] [PubMed] [Google Scholar]
- Semendeferi K, Armstrong E, Schleicher A, Zilles K, Van Hoesen GW. Prefrontal cortex in humans and apes: a comparative study of area 10. Am J Phys Anthropol. 2001;114:224–241. doi: 10.1002/1096-8644(200103)114:3<224::AID-AJPA1022>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- Sherwood CC, Bauernfeind AL, Bianchi S, Raghanti MA, Hof PR. Human brain evolution writ large and small. Prog Brain Res. 2012;195:237–254. doi: 10.1016/B978-0-444-53860-4.00011-8. [DOI] [PubMed] [Google Scholar]
- Sherwood CC, Holloway RL, Semendeferi K, Hof PR. Is prefrontal white matter enlargement a human evolutionary specialization? Nature Neurosci. 2005;8:537–538. doi: 10.1038/nn0505-537. [DOI] [PubMed] [Google Scholar]
- Singhmar P, Kumar A. Angelman syndrome protein UBE3A interacts with primary microcephaly protein ASPM, localizes to centrosomes and regulates chromosome segregation. PLoS One. 2011;6:e20397. doi: 10.1371/journal.pone.0020397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smaers JB, Steele J, Case CR, Cowper A, Amunts K, Zilles K. Primate prefrontal cortex evolution: human brains are the extreme of a lateralized ape trend. Brain Behav Evol. 2011;77:67–78. doi: 10.1159/000323671. [DOI] [PubMed] [Google Scholar]
- Somel M, Franz H, Yan Z, Lorenc A, Guo S, Giger T, Kelso J, Nickel B, Dannemann M, Bahn S, Webster MJ. Transcriptional neoteny in the human brain. Proc Natl Acad Sci USA. 2009;106:5743–5748. doi: 10.1073/pnas.0900544106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somel M, Liu X, Tang L, Yan Z, Hu H, Guo S, Jiang X, Zhang X, Xu G, Xie G, Li N. MicroRNA-driven developmental remodeling in the brain distinguishes humans from other primates. PLoS Biol. 2011;9 doi: 10.1371/journal.pbio.1001214. e1001214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spassky N, Han Y, Aguilar A, Strehl L, Besse L, Laclef C, et al. Primary cilia are required for cerebellar development and Shh-dependent expansion of progenitor pool. Dev Biol. 2008;317:246–259. doi: 10.1016/j.ydbio.2008.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephan H, Frahm H, Baron G. New and revised data on volumes of brain structures in Insectivores and Primates. Folia Primatol. 1981;35:1–29. doi: 10.1159/000155963. [DOI] [PubMed] [Google Scholar]
- Stout D, Chaminade T. Stone tools, language and the brain in human evolution. Philos Trans R Soc B Biol Sci. 2012;367:75–87. doi: 10.1098/rstb.2011.0099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sultan F, Hamodeh S, Baizer JS. The human dentate nucleus: a complex shape untangled. Neuroscience. 2010;167:965–968. doi: 10.1016/j.neuroscience.2010.03.007. [DOI] [PubMed] [Google Scholar]
- Sylvester JB, Rich CA, Loh Y-HE, van Staaden MJ, Fraser GJ, Streelman JT. Brain diversity evolves via differences in patterning. Proc Natl Acad Sci USA. 2010;107:9718–9723. doi: 10.1073/pnas.1000395107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton GK, Woods CG. Primary microcephaly: do all roads lead to Rome? Trends Genet. 2009;25:501–510. doi: 10.1016/j.tig.2009.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toro R, Poline J-B, Huguet G, Loth E, Frouin V, Banaschewski T, et al. Genomic architecture of human neuroanatomical diversity. Mol Psychiatry. 2015;20:1011–1016. doi: 10.1038/mp.2014.99. [DOI] [PubMed] [Google Scholar]
- Verendeev A, Sherwood CC. Human brain evolution. Curr Opin Behav Sci. 2017;16:41–45. doi: 10.1016/j.cobeha.2017.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver AH. Reciprocal evolution of the cerebellum and neocortex in fossil humans. Proc Natl Acad Sci USA. 2005;102:3576–3580. doi: 10.1073/pnas.0500692102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiting BA, Barton RA. The evolution of the cortico-cerebellar complex in primates: anatomical connections predict patterns of correlated evolution. J Hum Evol. 2003;44:3–10. doi: 10.1016/s0047-2484(02)00162-8. [DOI] [PubMed] [Google Scholar]
- Wilkinson LS, Davies W, Isles AR. Genomic imprinting effects on brain development and function. Nat Rev Neurosci. 2007;8:832–843. doi: 10.1038/nrn2235. [DOI] [PubMed] [Google Scholar]
- Workman AD, Charvet CJ, Clancy B, Darlington RB, Finlay BL. Modelling transformations of neurodevelopmental sequences across mammalian species. J Neurosci. 2013;33:7368–7383. doi: 10.1523/JNEUROSCI.5746-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Moy LY, Sanada K, Zhou Y, Buchman JJ, Tsai L. Cep120 and TACCs control interkinetic nuclear migration and the neural progenitor pool. Neuron. 2007;56:79–93. doi: 10.1016/j.neuron.2007.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan W, Assadi AH, Wynshaw-boris A, Eichele G, Matzuk MM, Clark GD. Previously uncharacterized roles of platelet-activating factor acetylhydrolase 1b complex in mouse spermatogenesis. Proc Natl Acad Sci USA. 2003;100:7189–7194. doi: 10.1073/pnas.1236145100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Zilles K, Rehkemper G. The brain with special reference to the telencephalon. In: Schwartz JH, editor. Orang-utan Biology. Oxford: Oxford University Press; 1988. pp. 157–176. [Google Scholar]
- Zimmer F, Montgomery SH. Phylogenetic analysis supports a link between DUF1220 domain number and primate brain expansion. Genome Biol Evol. 2015;7:2083–2088. doi: 10.1093/gbe/evv122. [DOI] [PMC free article] [PubMed] [Google Scholar]