Abstract
Bacteria sometimes cooperate with co-inhabiting cells. Pathogenic bacteria, for example, often produce and excrete virulence factors, eventually benefitting both producer and non-producer cells. The role of social interactions involving antibiotic resistance, however, has been more elusive. Enzymes that inactivate β-lactam antibiotics such as ampicillin or penicillin (β-lactamases) are good candidates as public goods. Nonetheless, it has been claimed that bacteria harbouring plasmids of natural origin coding for β-lactamase almost do not protect sensitive bacteria. This does not fit with the fact that ampicillin-sensitive bacteria can be isolated from subjects undergoing ampicillin treatment. We hypothesised that there are two non-exclusive explanations for the discrepancy between previous works: (1) the range of values of demographic conditions (such as initial strain frequency, initial total cell density or habitat structure) has not been broad enough to include most scenarios, or (2) there are interactions between some of these factors. We performed experiments with Escherichia coli bacterial cells to measure the degree of protection of sensitive cells when co-cultured with cells harbouring RP4, R16a or the R1 plasmids, all of natural origin and coding for β-lactamases, and in presence of ampicillin. In these co-cultures, performed in structured and non-structured environments, both the initial total cell density and the initial frequency of sensitive cells spanned four orders of magnitude. We found protection of sensitive cells in 63% of tested conditions. All factors (plasmid, structure, frequency and density) significantly affect levels of protection. Moreover, all factors interact, with interactions revealing large or very large effect sizes.
Introduction
Cooperative behaviour has been observed in the bacterial world, notably in processes involving pathogenicity (West et al., 2006, 2007). A common form of cooperation in bacterial populations is the production of public goods, which are metabolically costly molecules produced by one individual that become available to neighbouring individuals (West et al., 2006, 2007). Different molecules may act as public goods, ranging from products responsible for scavenging, motility and communication to substances involved in allelopathy or virulence (West et al., 2007).
There are three main ways of antibiotic resistance: pumping out the antibiotic from the cell, inhibiting the interaction of the drug with the target or by modifying or degrading the antibiotic molecule (Wright, 2005; Nicoloff and Andersson, 2016; Sorg et al., 2016). Enzymes that modify the antibiotic may be considered public goods because, by decreasing antibiotic concentration in the local environment, they may protect nearby bacteria. In this study, we focus on the case of β-lactamase enzymes, which degrade antibiotics such as ampicillin or penicillin.
Degrading enzymes may detoxify the surrounding environment even if they remain within the cell; for example, in Gram-negative bacterial cells, β-lactamase locates primarily at the periplasmic space (Livermore, 1995), a space located between the inner and outer membranes. This phenomenon will possibly be stronger than it seems because molecules of β-lactamase may escape the periplasm, albeit by leakage rather than by secretion (Livermore, 1995).
The ability of β-lactam-sensitive cells to survive when located in the vicinity of β-lactamase producer cells has been noticed several times by molecular biologists in the process of gene cloning, by observing the presence of the so-called ‘satellite colonies’ (Sambrook and Russell, 2001). These small colonies, composed by ampicillin-sensitive bacteria, are able to grow in ampicillin-enriched agar plates because of their proximity to β-lactamase-producing cells. This phenomenon has also been hypothesised to occur in polymicrobial infections, as ampicillin-sensitive bacteria were isolated from subjects undergoing ampicillin treatment (Brook, 2004). More recently, Dugatkin et al. (2005) showed that a β-lactamase enzyme, encoded on an artificial plasmid, more specifically on a gene that was modified to inactivate ampicillin outside the cell (and not in the periplasmic space), could indeed protect nearby cells from death (Clark et al., 2009; Perlin et al., 2009). Furthermore, Gore and colleagues (Yurtsev et al., 2013) showed that Escherichia coli strains harbouring artificial plasmids coding for non-modified β-lactamases could detoxify the environment, hence protecting otherwise ampicillin-sensitive cells from β-lactam antibiotics.
More recently, Medaney et al. (2016) also tested for protection mediated by a β-lactamase enzyme, this time with a natural isolated plasmid (called pCT). Using a natural plasmid is important because it elucidates whether the protection of sensitive cells previously observed using artificial plasmids (Yurtsev et al., 2013), sometimes even artificially increasing the secretion of β-lactamase (Dugatkin et al., 2005), can occur in more natural settings. Using a plasmid of natural origin, however, Medaney et al. (2016) achieved a different conclusion, observing that sensitive cells do not typically benefit from the presence of resistant cells. Rather, the authors showed that the few sensitive cells allowed to form colonies are those undergoing a state of persistence, in which bacteria stop dividing (Balaban et al., 2004; Lewis, 2010). Such delay in cell division saves those few sensitive cells from death if it provides the time required for antibiotic degradation or direct detoxification by β-lactamase-producing cells (Medaney et al., 2016).
On one hand, studies involving artificial plasmids consistently demonstrate the ability of β-lactamase producer cells to protect sensitive cells, unlike the study involving a plasmid of natural origin. A possible explanation for the lack of protection with the pCT plasmid could be that many relevant demographic conditions were not examined, as their experiments focused on very narrow intervals of initial frequencies of plasmid-bearing and plasmid-free cells and of initial total cell densities of both cell strains (Medaney et al., 2016).
Indeed, Ross-Gillespie et al. (2007, 2009) have shown the importance of demographic conditions in the exploitation of public goods production among bacteria. They used the production of siderophores (molecules that scavenge iron from the environment) as a model of cooperative trait. In a structured environment, non-producers of siderophores, or cheaters, are better able to exploit the cooperative individuals at higher initial total population density—positive density dependence (Ross-Gillespie et al., 2009). In an unstructured environment, there is negative frequency dependence—that is, non-producers of siderophores have an advantage when rare (Ross-Gillespie et al., 2007).
To our knowledge, there is no experimental study of the conjoint effect of different combinations of demographic conditions (such as habitat structure, total cell density and cell frequencies) on the cooperative production of β-lactamase. Through the testing of several combinations one may test for interactions between factors—for example, between initial total cell density and initial relative strain frequency, a complication expected to occur (Ross-Gillespie et al., 2009). For example, in a liquid environment, if the initial total cell density is low, the number of β-lactamase producers may be insufficient to detoxify the environment, eventually leading to the death of all sensitive cells irrespectively of the initial frequencies of sensitive and producer cells; in this case, we would not observe frequency dependence. On the other hand, if density is above a certain value, the fitness of the sensitive population is expected to depend on the frequency of producers and sensitive cells.
Our work aims to study the ability of ampicillin-sensitive cells to thrive in environments with lethal doses of ampicillin, focusing on the antibiotic protection conferred by resistant bacteria harbouring three plasmids of natural origin. Using three different plasmids, we perform a systematic study of the levels of protection under different combinations of demographic conditions—population structures, initial strain frequencies and initial total cell densities—as well as the interactions between these factors.
Materials and methods
Bacterial Strains
In this work we used two isogenic strains of E. coli:the wild-type E. coli K12 MG1655 and E. coli K12 MG1655 Δara ValR. The latter strain harbours two chromosomal markers: Δara—deletion of the arabinose operon, which disables the metabolism of arabinose monosaccharide; and ValR—a phenotype resulting from a mutation that enables the growth in the presence of valine (40 μg l−1) when isoleucine is absent.
Conjugative plasmids R1, R16a and RP4, of natural origin, were inserted into E. coli K12 MG1655 Δara ValR through bacterial conjugation, resulting in the three ampicillin-resistant strains: E. coli K12 MG1655 Δara ValR (R1), E. coli K12 MG1655 Δara ValR (R16a) and E. coli K12 MG1655 Δara ValR (RP4). All three β-lactamase-producing strains are identical (except for the different plasmids) and will therefore be mentioned solely according to the harboured plasmid. The wild-type strain will be mentioned henceforth as the ampicillin-sensitive strain.
Experimental conditions of co-culture
The sensitive strain was co-cultured with an ampicillin-resistant strain in a set of four different conditions: (i) ampicillin-resistant strain containing one of the three β-lactamase-producing plasmids; (ii) two different habitat structures—structured (solid medium) and unstructured (liquid medium with constant agitation); (iii) three initial total cell densities—high (107 cells ml−1), intermediate (105 cells ml−1) and low (103 cells ml−1); and (iv) three initial frequencies of sensitive cells—high frequency (99S:1R), intermediate frequency (50S:50R) and low frequency of sensitive cells (1S:99R), where S stands for sensitive and R for resistant.
Each condition of co-culture was performed in parallel, both in medium containing ampicillin (100 μg ml−1) and in ampicillin-free media. All co-cultures were performed in triplicate.
Additionally, sensitive and resistant strains were cultured in isolation in media containing ampicillin to confirm the ability of the resistant strains to grow and to confirm that the sensitive strain does not. This control was performed both in structured and unstructured media, and at all initial total cell densities. After a 24-h period of incubation, cultures were plated on selective media and incubated for 48 h; the next steps are described in the following two sections for unstructured and structured media.
Co-cultures in unstructured medium
The sensitive and ampicillin-resistant strains were inoculated in each of the appropriate combinations of initial total cell density and strain frequency in 10 ml of Luria broth (LB) and incubated at 37 °C with constant agitation (170 r.p.m.). We allowed the bacteria to grow in co-culture for 24 h (reaching the stationary phase).
After incubation, samples of the co-cultures were retrieved, appropriately diluted and plated on selective media for 48 h: M9 Minimal Medium with 3 mm MgSO4, supplemented with 4 g l−1 of arabinose to select the sensitive strain and supplemented with 4 g l−1 glucose, 40 μg ml−1 valine and 100 μg ml−1 of both ampicillin and kanamycin to select ampicillin-resistant strains. Additionally, all samples were plated on M9 Minimal Medium with 3 mm MgSO4, supplemented with 4 g l−1 of arabinose, 40 μg ml−1 valine and 100 μg ml−1 of both ampicillin and kanamycin to select transconjugant cells (which result from the transfer of plasmids, via conjugation, between ampicillin-resistant (donors) and sensitive (recipient) strains).
Co-cultures in structured medium
We inoculated the sensitive and ampicillin-resistant strains of each demographic combination on Petri dishes containing solid LB medium (supplemented with 1.5% agar) and incubated at 37 °C for 24 h.
After the incubation period, the whole resulting bacterial culture was retrieved and suspended in 2 ml of MgSO4 (0.01 m). Samples of the co-cultures were then retrieved, appropriately diluted and plated on selective medium, as described for the unstructured medium.
Data analysis
We calculated the relative fitness (WS) of the sensitive strain in each combination of conditions, both for ampicillin and ampicillin-free co-cultures, using the following equation (Lenski et al., 1991):
where S is the population of ampicillin-sensitive strain and R is the population of the ampicillin-resistant strain, calculated in CFU ml−1, at the beginning and end of the experiments.
As transconjugant cells also produce β-lactamase, those present in the final population were subtracted from the original sensitive population (whence they were obtained, having the same chromosomal marker) and added to the resistant population.
The relative fitness of sensitive cells in the presence of ampicillin was normalised with the relative fitness of sensitive cells in the absence of ampicillin:
In this equation, the numerator and the denominator stand for fitness values of sensitive cells if ampicillin is present or absent, respectively. By dividing these two fitness values, we automatically discount the fitness effect of the Δara ValR chromosomal marker in the producer cells.
The resulting value was designated relative fitness of susceptible cells (rFS), which represents the ability or the inability of sensitive cells to survive and/or grow under the protection of β-lactamase-producing strains in an ampicillin-enriched environment. The calculation of this ratio allowed us to discount potential pre-existing interference detected in the relative fitness of sensitive cells resulting from the co-cultures without ampicillin. A negative value of rFS means that sensitive cells survived but their numbers diminished over time. If bacterial replication is faster than bacterial death, rFS becomes positive, implying that the number of sensitive cells is higher at the end than initially.
To understand the impact of transconjugants, we also calculated a second quantity, rFSb, which assumes that transconjugants do not produce β-lactamase and, as such, remain sensitive to ampicillin. In other words, contrary to the method used to calculate rFS, there is no subtraction of transconjugants from the sensitive population or their addition to the resistant population. Considering that transconjugants do not produce β-lactamase is a non-realistic assumption, but a comparison between rFS and rFSb is useful to assess the impact of conjugation on the protection levels.
Statistical tests and graphical analysis were performed in IBM SPSS Statistics version 23 for Windows (Armonk, NY, USA) and R version 3.2.0, available at http://www.rstudio.com/.
Results
β-lactamase producers protect sensitive cells from ampicillin in most conditions
We started by confirming that after being in the presence of the clinically relevant concentration of ampicillin (Foulds, 1986) for 24 h, sensitive cells do not form colonies when plated in selective medium and incubated for 48 h. This result was obtained regardless of the initial conditions: three total initial cell densities (circa 107 cells ml−1, 105 cells ml−1 and 103 cells ml−1) and two habitat structures (in liquid medium with agitation and on a Petri dish). In the same conditions, and contrary to what happened with sensitive cells, β-lactamase-producing cells (cells harbouring each of the three plasmids) are able to grow and form colonies.
We then tested the ability of different β-lactamase-producing strains to protect the sensitive strain in ampicillin-enriched medium. To do so, the sensitive strain was co-cultured with β-lactamase-producing strains in 54 different conditions.
In the presence of ampicillin and when co-cultured with β-lactamase-producing strains, sensitive cells can either be (i) protected if the environment is indeed detoxified, leading to the survival of some sensitive cells, or (ii) eliminated if producers are unable to effectively detoxify the environment, letting all sensitive cells die from the action of ampicillin. When protection of sensitive cells was observed, we further distinguish two possible outcomes: (i) survival of sensitive cells—when, after incubation, the number of sensitive cells decreased, but not all cells disappear—or (ii) growth of sensitive cells—when the numbers of sensitive cells increased over the incubation period. We observed protection in 34 out of the 54 tested conditions, among which there were 13 cases of growth of sensitive cells and 21 cases of survival.
Number of protection cases increases with initial total cell density
Supplementary Figure S1A shows that the number of protection cases increases with initial total cell density (henceforth referred to as ‘density’): two cases with the lower density, 14 cases with intermediate density and all the 18 cases when density is high. The number of cases where we observed growth of sensitive cells also revealed positive density dependence: two cases when density is low, four cases for intermediate density and seven cases at high density.
Structured habitat has more cases of protection
In the structured habitat, we observed protection of sensitive cells in 20 out of 27 conditions (Supplementary Figure S1A or S1B) and growth of sensitive cells in 11 of those conditions. In the unstructured habitat, sensitive cells were protected in 14 out of 27 conditions (Supplementary Figure S1A or S1B) and only had a net increase in numbers in two of those.
Bacteria harbouring natural plasmids efficiently detoxify ampicillin-enriched environments
All three plasmids used in this work detoxify the environment in several conditions (Supplementary Figure S1A or S1B). Even for plasmid R1, the lowest protective plasmid of the three, we observed nine cases of protection (out of 18 conditions), which correspond to 50% of the cases. Furthermore, we observed 13 protection cases (out of 18) with the RP4 plasmid (72%) and 12 protection cases (out of 18) with the R16a plasmid (67%). The difference between plasmids is even stronger if one looks to the cases of growth of sensitive cells: plasmid RP4 enabled the growth of sensitive cells in 7 out of 18 cases (39%), whereas with plasmid R16a we observed only in 4 cases (22%) and only in 2 cases with R1 (11%).
Effect of initial cell frequency in the number of protection cases
Cases of protection increase with the initial frequency of sensitive cells (henceforth referred to as ‘Frequency’ ‘frequency’) but only slightly: we observed 10 protection cases with low frequency, 11 cases at intermediate frequency and 13 cases at the highest frequency (Supplementary Figure S1B). A single trend is absent if one looks at the cases of growth of sensitive cells: four, three and six cases when frequency was high, intermediate and low, respectively.
ANOVA shows that plasmids, the three demographic conditions and all interactions significantly affect the levels of protection
We quantified protection levels by measuring the fitness of sensitive cells in relation to producer cells, rFS (Supplementary Table S1). A four-way analysis of variance was conducted to compare the effects of the conditions on protection. All the conditions, as well as all their interactions, significantly affect the levels of protection (P<0.001 for all cases, Table 1).
Table 1. Four-way ANOVA with P-values and effect size (partial η 2) for each factor and for each interaction between factors.
| Variable | df | F | P-value | Partialη2 |
|---|---|---|---|---|
| Plasmid | 2 | 546.9 | <0.001 | 0.910 |
| Structure | 1 | 1189.1 | <0.001 | 0.917 |
| Density | 2 | 1852.0 | <0.001 | 0.972 |
| Frequency | 2 | 81.2 | <0.001 | 0.601 |
| Plasmid × Structure | 2 | 56.2 | <0.001 | 0.510 |
| Plasmid × Density | 4 | 71.4 | <0.001 | 0.726 |
| Plasmid × Frequency | 4 | 22.4 | <0.001 | 0.454 |
| Structure × Density | 2 | 53.8 | <0.001 | 0.499 |
| Structure × Frequency | 2 | 30.4 | <0.001 | 0.360 |
| Density × Frequency | 4 | 318.1 | <0.001 | 0.922 |
| Plasmid × Structure × Density | 4 | 115.9 | <0.001 | 0.811 |
| Plasmid × Structure × Frequency | 4 | 73.6 | <0.001 | 0.732 |
| Plasmid × Density × Frequency | 8 | 116.1 | <0.001 | 0.896 |
| Structure × Density × Frequency | 4 | 120.6 | <0.001 | 0.817 |
| Plasmid × Structure × Density × Frequency | 8 | 47.4 | <0.001 | 0.778 |
Abbreviation: ANOVA, analysis of variance.
We also quantified the effect size of each factor using the partial η2, an index that describes the extent of an effect after controlling for the impact of other variables in a model (Levine and Hullett, 2002), and found that all factors and their interactions have high or very high effect sizes.
Effect of initial total cell density on the level of protection
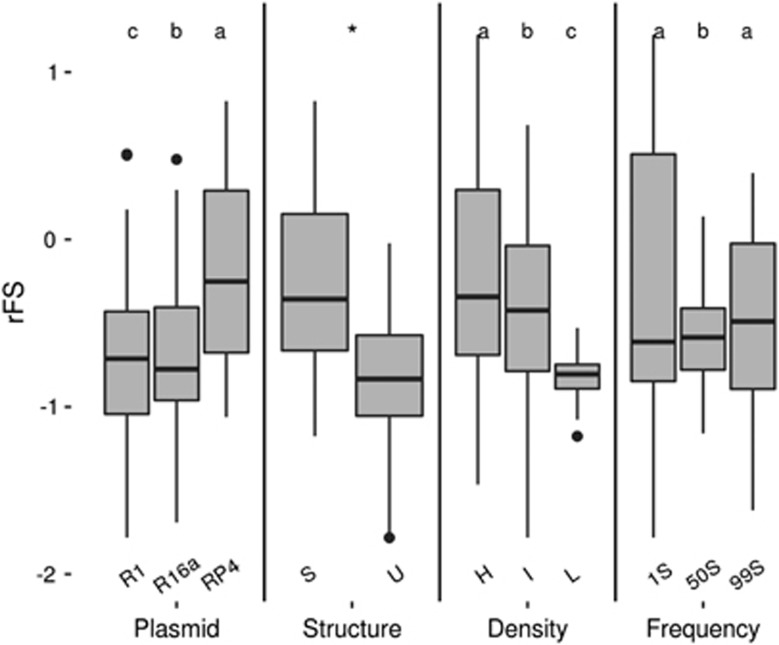
Initial total cell density (density) is the condition with stronger effect size on the protection of sensitive cells (rFS) (F(2,108)=1851.98, P<0.001, partial η2=0.972). Post hoc comparisons using the Tukey HSD test (Figure 1) indicated that the mean score (M) for high density (M=−0.229, s.d.=0.013) is significantly higher than that of intermediate density (M=−0.511, s.d.=0.013). Additionally, lower density is where the lowest mean score of rFS is observed (M=−1.305, s.d.=0.013). Overall, these results reveal that rFS increases with density.
Figure 1.
Level of protection of the sensitive bacteria (rFS) for each subgroup of conditions. rFS values not sharing the same letter are significantly different (Tukey HSD, P<0.05). *rFS values are significantly different. ‘R1’, ‘R16a’ and ‘RP4’ stand for Escherichia coli cells harbouring plasmids R1, R16a or RP4, respectively. ‘S’ stands for structured and ‘U’ for unstructured habitats. ‘H’, ‘I’ and ‘L’ stand for high, intermediate and low initial total densities, respectively. ‘1S’, ‘50S’ and ‘99S’ stand for the initial strain frequencies 1S:99R, 50S:50R and 99S:1R, respectively, where ‘S’ stands for sensitive and ‘R’ stands for resistant (producers of beta-lactamase). Outlier values are not represented. For the categories ‘Plasmid’, ‘Density’ and ‘Frequency’ n=18 for each group, whereas for the category ‘Structure’ n=27.
Effect of habitat structure on the level of protection
Habitat structure also significantly affects the protection levels (F(1,108)=1189.07, P<0.001, partial η2=0.917). The observed mean scores of rFS, for each structure, showed higher levels of protection in the structured habitat (M=−0.423, s.d.=0.011) compared with that in the unstructured habitat (M=−0.940, s.d.=0.011) (Figure 1).
Effect of plasmids on the level of protection
The plasmid harboured by the β-lactamase-producing cells also has a significant effect on the levels of protection (F(2,108)=546.88, P<0.001, partial η2=0.910). We further conducted a post hoc comparison using the Tukey HSD test to understand the differences of protection between plasmids. We observed that the mean score (M) of rFS for plasmid RP4 (M=−0.368, s.d.=0.013) is significantly higher than that of the other two plasmids. Furthermore, the mean score for the R16a plasmid (M=−0.703, s.d.=0.013) is significantly higher than that of the R1 plasmid (M=−0.974, s.d.=0.013).
Effect of initial frequency on the level of protection
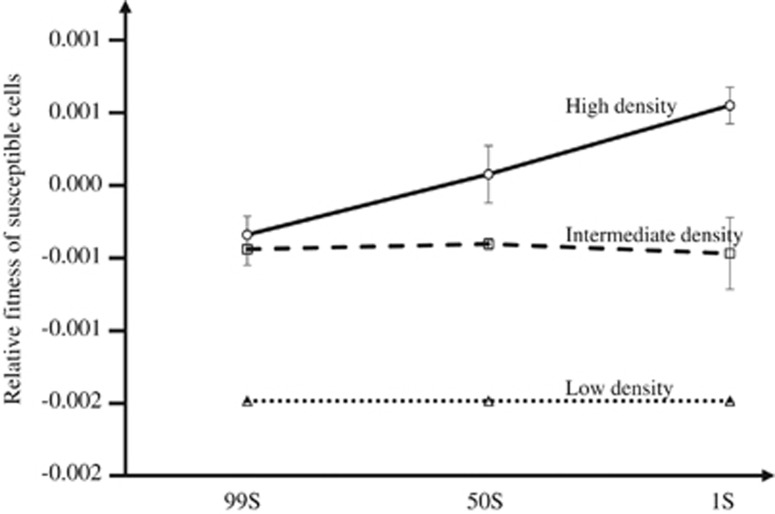
The initial frequency of sensitive cells (frequency) also significantly affects the levels of protection. This demographic factor also has a strong effect size on the protection of sensitive cells, but not so strong as the other isolated conditions (F(2,108)=81.22, P<0.001, partial η2=0.601). The effect size resulting from the interaction between density and frequency was more pronounced (F(4,108)=318.10, P⩽0.001, partial η2=0.922), being superior to that of the structure, plasmid or frequency alone or other interactions (for example, the interaction ‘Structure × Density’ has partial η2=0.499 only). For example, in the case involving producers of β-lactamase harbouring the R1 plasmid, at structured habitat, Figure 2 illustrates the effect of the interaction between density and frequency: the line obtained at high density is not parallel to the lines obtained at low and intermediate densities. In other words, for this case, protection levels are higher at higher densities, but even higher if frequency is intermediate or low (Figure 2).
Figure 2.
Interaction plot of fitted rFS, showing how the protection levels depend on frequency and density. For this specific case (involving producers of β-lactamase harbouring the R1 plasmid at structured habitat), one can see that protection levels are higher at higher densities, but even higher if frequency is intermediate (50S:50R) or low (1S:99R). Error bars show twice the s.e. around the means.
Additionally, a post hoc Tukey HSD test that was used to assess the differences between the three studied initial frequencies of sensitive cells revealed that the intermediate frequency (50S:50R) has a mean score significantly lower than the other two frequencies (M=−0.816, s.d.=0.013) (Figure 1).
Initial total density and structure are demographic predictors of protection
To measure the weight of each condition on the level of protection (rFS), we also performed a forward stepwise regression analysis of the effects of these conditions on the level of protection (Table 2). Results were in agreement with the conclusions withdrawn from the analysis of variance analysis. The factor density alone is the demographic condition that has a more pronounced effect on rFS values. Indeed, density alone explains 26% of the observed protection of sensitive cells (R2=0.260, P<0.001), followed by structure (+18%, R2=0.440, P<0.001), and the presence of RP4 plasmid in the β-lactamase producer cells (+5.9%, R2=0.499, P<0.001).
Table 2. Stepwise regression for rFS values.
| Variable | IndividualR2 | CumulativeR2 | s.e. |
|---|---|---|---|
| Density | 0.260 | 0.260 | 0.669 |
| Structure | 0.180 | 0.440 | 0.582 |
| RP4 Plasmid | 0.059 | 0.499 | 0.551 |
These results show that about 50% of the variance of the protection (rFS) values is explained by these three conditions: density, habitat structure and harbouring the RP4 plasmid.
The remaining conditions, such as frequency or harbouring the R16a or R1 plasmid, have no further significant contribution to the stepwise regression.
Plasmid transfer does not influence the impact of demographic conditions on the levels of protection
β-lactamase genes of producer cells were all encoded on plasmids that are able to transfer to ampicillin-sensitive cells, during the co-culture experiments, originating from transconjugant cells (Supplementary Table S2). Transconjugant cells are also β-lactamase producers; hence, rFS values were always calculated assuming that transconjugant cells are also producers (see Materials and methods section).
Therefore, we compared rFS values with a second set of values (rFSb) assuming that transconjugants do not produce β-lactamase and as such remain sensitive to ampicillin. Despite being a non-realistic assumption, rFSb was calculated to assess the impact of conjugation on the protection levels. In 10 cases, the rFSb values were significantly higher than rFS values (Supplementary Table S1). Despite this difference, the analysis of variance using rFSb values yielded the same qualitative conclusions as with rFS (Supplementary Table S3). Furthermore, the conditions in which there is protection are the same.
Discussion
With this work we had two main objectives. First, to confirm that there is protection of sensitive cells by producers of β-lactamase. As the answer could depend on the plasmid, we use three plasmids of natural origin, and because the answer could also depend on specific conditions, we checked different demographic conditions of habitat structure, initial strain frequency and initial total cell density, in a total of 18 combinations for each of the plasmids. Contrary to what was done in previous studies, here we used initial strain frequencies (frequency) and total initial cell densities (density) spanning four orders of magnitude each. We used this broader range of initial cell frequencies and densities as it was important to include most contrasting scenarios. Given that we tested all combinations of these conditions, we could also achieve our second goal: to understand how these demographic conditions interact and affect the levels of protection.
We found that in 34 of the 54 studied conditions bacteria expressing β-lactamase protect ampicillin-sensitive cells from otherwise lethal doses of ampicillin. The best condition for β-lactamase producers to protect sensitive cells is the combination of the highest density when initial sensitive cells are rare (low frequency). In 13 conditions, the population of sensitive bacteria was even able to increase in numbers.
Although there are more cases of protection and growth of sensitive cells in structured habitat (20 cases), it is interesting that so many cases of protection were observed in the unstructured environment (14 cases). As in liquid medium the effect of β-lactamase is diluted across the whole environment (and not localised around a producer cell as in structured environment); this indicates that producers are highly effective at detoxifying the environment.
In most cases, the relative fitness of sensitive cells increases with initial total cell density, as predicted by Ross-Gillespie et al. (2009). The effect of initial strain frequency, however, is not so easy to predict. As pointed out by Ross-Gillespie et al. (2007) the fitness of cheaters (here sensitive cells) should not depend on cheaters’ frequency in the population if selection is weak (Hamilton, 1964). However, with some degree of population structuring or in cases in which higher frequency of cooperators would lead to greater population growth, the fitness of sensitive cells should depend on strain frequency (Ross-Gillespie et al., 2007).
Our results show that relative fitness of sensitive cells indeed depends on the initial strain frequency. However, the effect of frequency is markedly dependent on other conditions, mainly initial total cell density, as shown by the very low P-values of their interactions (Table 1). This is clearer by looking at effect sizes (Table 1, partial η2 values): the effect size of the interaction ‘Frequency × Density’ is the second highest value appearing in Table 1 (partial η2=0.922), whereas that of ‘Frequency’ condition isolated is partial η2=0.601, one of the lowest values appearing in Table 1. For example, in structured habitat and using the RP4 or R16a plasmids, the level of protection is higher when sensitive cells are rare with high density, whereas at low density the level of protection is higher when the initial proportion of sensitive cells is higher (Supplementary Figure S1B).
E. coli’s main habitats are believed to be the lower intestine of warm-blooded animals (Savageau, 1983). Given that these habitats are structured, probably most E. coli cells close to a given E. coli cell belong to the same ‘colony’ that is, probably most of them harbour the β-lactamase gene. Protection may be useful in this case if a certain proportion of kin cells, despite coding for β-lactamase, is transiently not producing the enzyme—the so-called phenotypic heterogeneity (Ackermann, 2015). Therefore, by protecting neighbouring cells, producers of β-lactamase are protecting their kin, thus maximising their inclusive fitness (Hamilton, 1964). Moreover, given that cells are in a structured environment, there is accumulation of β-lactamase near producer cells, hence allowing for higher (local) concentration of the enzyme.
However, other (different) bacterial strains of E. coli may be present in the same habitat, for example, mammalian gut (Caugant et al., 1981; Faith et al., 2013), so producers of β-lactamase may be protecting other strains as well. In this case, one should expect β-lactamase producer E. coli cells to be more refractory to exploitation by other cells not producing public goods. At least in many other systems, mechanisms to avoid exploitation by cheaters have evolved (West et al., 2006).
The theoretical prediction is that selection for such mechanisms is expected to occur when relatedness is low (if relatedness is high, control mechanisms are not necessary) (Frank, 1995; Dionisio and Gordo, 2007). This theoretical prediction, together with our observations that there is protection in so many conditions (hence not avoiding helping other cells), is an indication that, even if different E. coli cells co-inhabit, the two populations are not mingled. Therefore, within the surrounding of each E. coli cell, genetic variance is much lower than between patches, which implicates that relatedness is high (Frank, 1998).
Mammalian hosts harbour many bacterial species (Qin et al., 2010; Faith et al., 2013; Martinez et al., 2013), implying that some of the protected bacterial cells may belong to other species. The ability of E. coli to protect sensitive bacterial cells from other species has already been experimentally demonstrated (Perlin et al., 2009). This may be advantageous to transferable plasmids. Indeed, as they are able to transfer to many bacterial species (Dionisio et al., 2002), protected cells are, potentially, new hosts for the plasmid. From this point of view, the production of high amounts of β-lactamase to save bacteria not carrying the plasmid become advantageous to the plasmid itself.
An important point of this work was the use of plasmids of natural origin, contrary to previous works in which protection was also observed (Dugatkin et al., 2005; Clark et al., 2009; Perlin et al., 2009; Yurtsev et al., 2013). None of the three plasmids were engineered to increase or optimised for β-lactamase production. In fact, all three plasmids used in this work (R1, R16a and RP4) were obtained from natural isolates. RP4 was isolated from Pseudomonas aeruginosa (Datta et al., 1971), R1 from Salmonella enterica serovar paratyphi (Anderson and Datta, 1965) and R16a from Providencia stuartii (Chabbert et al., 1972). The three plasmids are low copy number (Grinsted et al., 1972; Uhlin and Nordstrom, 1975; Szabo et al., 2016) and encode TEM β-lactamases (Matthew and Hedges, 1976; Szabo et al., 2016), so the three enzymes are very similar. It has been reported that the level of transcription of RP4 β-lactamase is higher than that of R1 plasmid (Crowlesmith and Howe, 1980), which explains the higher levels of protection displayed by RP4 in our results and the predominance of RP4 in our stepwise analysis (Table 2).
It could be argued that a weakness of our study is that we used a domesticated bacterial strain to house the plasmids, rather than wild-type isolates. However, our strain was not specifically engineered to increase or optimise β-lactamase production. Moreover, the fact that we used a domesticated strain revealed serendipitous because it happened to be the same E. coli strain used in the study in which protection was not observed (Medaney et al., 2016).
The interaction between demographic conditions explored in this study explains why previous works did not agree on the existence of protection. As contrasting results are obtained with different demographic conditions, it is possible that some previous works explored propitious conditions (hence observing protection of sensitive cells), whereas others did not.
Our results also suggest an explanation for how is it possible that ampicillin-sensitive bacteria are isolated from subjects undergoing ampicillin treatment (Brook, 2004). Despite the presence of the antibiotic in these patients, probably some sensitive cells never encountered it, because of the β-lactamase produced by neighbouring resistant cells.
The experiments described here were performed in a laboratory environment; in the future, however, this should be tested in a natural environment (for example, in mice gut, as recently done with the antibiotic chloramphenicol (Sorg et al., 2016)).
Acknowledgments
We thank editors and reviewers for their corrections and suggestions, which substantially ameliorated the manuscript. ILD (SFRH/BD/82375/2011) and JAG (SFRH/BD/86103/2012) received PhD fellowships and LMC (SFRH/BPD/33633/2009) received a post-doc fellowship, all from Fundação para a Ciência e a Tecnologia, Portugal http://www.fct.pt.
Footnotes
Supplementary Information accompanies this paper on Heredity website (http://www.nature.com/hdy)
The authors declare no conflict of interest.
Supplementary Material
References
- Ackermann M (2015). A functional perspective on phenotypic heterogeneity in microorganisms. Nat Rev Microbiol 13: 497–508. [DOI] [PubMed] [Google Scholar]
- Anderson ES, Datta N (1965). Resistance to penicillins and its transfer in Enterobacteriaceae. Lancet 1: 407–409. [DOI] [PubMed] [Google Scholar]
- Balaban NQ, Merrin J, Chait R, Kowalik L, Leibler S (2004). Bacterial persistence as a phenotypic switch. Science 305: 1622–1625. [DOI] [PubMed] [Google Scholar]
- Brook I (2004). Beta-lactamase-producing bacteria in mixed infections. Clin Microbiol Infect 10: 777–784. [DOI] [PubMed] [Google Scholar]
- Caugant DA, Levin BR, Selander RK (1981). Genetic diversity and temporal variation in the Escherichia-coli population of a human host. Genetics 98: 467–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabbert YA, Scavizzi MR, Witchitz JL, Gerbaud GR, Bouanchaud DH (1972). Incompatibility groups and the classification of fi - resistance factors. J Bacteriol 112: 666–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark DR, Alton TM, Bajorek A, Holden P, Dugatkin LA, Atlas RM et al (2009). Evolution of altruists and cheaters in near-isogenic populations of Escherichia coli. Front Biosci 14: 4815–4824. [DOI] [PubMed] [Google Scholar]
- Crowlesmith I, Howe TG (1980). Quantitative correlation between penicillin resistance and beta-lactamase activity specified by the R plasmids R1, R1 bla-45, and RP1 in Escherichia coli K-12. Antimicrob Agents Chemother 18: 675–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta N, Hedges RW, Shaw EJ, Sykes RB, Richmond MH (1971). Properties of an R factor from Pseudomonas aeruginosa. J Bacteriol 108: 1244–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dionisio F, Gordo I (2007). Controlling excludability in the evolution of cooperation. Evol Ecol Res 9: 365–373. [Google Scholar]
- Dionisio F, Matic I, Radman M, Rodrigues OR, Taddei F (2002). Plasmids spread very fast in heterogeneous bacterial communities. Genetics 162: 1525–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugatkin LA, Perlin M, Lucas JS, Atlas R (2005). Group-beneficial traits, frequency-dependent selection and genotypic diversity: an antibiotic resistance paradigm. Proc Biol Sci 272: 79–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faith JJ, Guruge JL, Charbonneau M, Subramanian S, Seedorf H, Goodman AL et al (2013). The long-term stability of the human gut microbiota. Science 341: 1237439 44-+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foulds G (1986). Pharmacokinetics of sulbactam ampicillin in humans - a review. Rev Infect Dis 8: S503–S511. [DOI] [PubMed] [Google Scholar]
- Frank SA (1995). Mutual policing and repression of competition in the evolution of cooperative groups. Nature 377: 520–522. [DOI] [PubMed] [Google Scholar]
- Frank SA (1998)Foundations of Social Evolution. Princeton University Press: Princeton, NJ, USA. [Google Scholar]
- Grinsted J, Saunders JR, Ingram LC, Sykes RB, Richmond MH (1972). Properties of a R factor which originated in Pseudomonas aeruginosa 1822. J Bacteriol 110: 529–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton WD (1964). Genetical evolution of social behaviour I. J Theor Biol 7: 1–16. [DOI] [PubMed] [Google Scholar]
- Lenski RE, Rose MR, Simpson SC, Tadler SC (1991). Long-term experimental evolution in Escherichia-coli.1. Adaptation and divergence during 2,000 generations. Am Nat 138: 1315–1341. [Google Scholar]
- Levine TR, Hullett CR (2002). Eta squared, partial eta squared, and misreporting of effect size in communication research. Hum Commun Res 28: 612–625. [Google Scholar]
- Lewis K (2010). Persister cells. Annu Rev Microbiol 64: 357–372. [DOI] [PubMed] [Google Scholar]
- Livermore DM (1995). Beta-lactamases in laboratory and clinical resistance. Clin Microbiol Rev 8: 557–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez I, Muller CE, Walter J (2013). Long-term temporal analysis of the human fecal microbiota revealed a stable core of dominant bacterial species. PLos ONE 8: e69621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthew M, Hedges RW (1976). Analytical isoelectric focusing of R factor-determined beta-lactamases: correlation with plasmid compatibility. J Bacteriol 125: 713–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medaney F, Dimitriu T, Ellis RJ, Raymond B (2016). Live to cheat another day: bacterial dormancy facilitates the social exploitation of beta-lactamases. ISME J 10: 778–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicoloff H, Andersson DI (2016). Indirect resistance to several classes of antibiotics in cocultures with resistant bacteria expressing antibiotic-modifying or -degrading enzymes. J Antimicrob Chemother 71: 100–110. [DOI] [PubMed] [Google Scholar]
- Perlin MH, Clark DR, McKenzie C, Patel H, Jackson N, Kormanik C et al (2009). Protection of Salmonella by ampicillin-resistant Escherichia coli in the presence of otherwise lethal drug concentrations. Proc Biol Sci 276: 3759–3768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin JJ, Li RQ, Raes J, Arumugam M, Burgdorf KS, Manichanh C et al (2010). A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464: 59–U70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross-Gillespie A, Gardner A, Buckling A, West SA, Griffin AS (2009). Density dependence and cooperation: theory and a test with bacteria. Evolution 63: 2315–2325. [DOI] [PubMed] [Google Scholar]
- Ross-Gillespie A, Gardner A, West SA, Griffin AS (2007). Frequency dependence and cooperation: theory and a test with bacteria. Am Nat 170: 331–342. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell DW (2001). Molecular Cloning—A Laboratory Manual Vol 1, 3rd Edition. Cold Spring Harbour Laboratory Press: Cold Spring Harbour, NY, USA.
- Savageau MA (1983). Escherichia-Coli habitats, cell-types, and molecular mechanisms of gene-control. Am Nat 122: 732–744. [Google Scholar]
- Sorg RA, Lin L, van Doorn GS, Sorg M, Olson J, Nizet V et al (2016). Collective resistance in microbial communities by intracellular antibiotic deactivation. PLos Biol 14: e2000631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo M, Nagy T, Wilk T, Farkas T, Hegyi A, Olasz F et al (2016). Characterization of two multidrug-resistant IncA/C plasmids from the 1960s by using the MinION sequencer device. Antimicrob Agents Chemother 60: 6780–6786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlin BE, Nordstrom K (1975). Plasmid incompatibility and control of replication: copy mutants of the R-factor R1 in Escherichia coliK-12. J Bacteriol 124: 641–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West SA, Diggle SP, Buckling A, Gardner A, Griffins AS (2007). The social lives of microbes. Annu Rev Ecol Evol Syst 38: 53–77. [Google Scholar]
- West SA, Griffin AS, Gardner A, Diggle SP (2006). Social evolution theory for microorganisms. Nat Rev Microbiol 4: 597–607. [DOI] [PubMed] [Google Scholar]
- Wright GD (2005). Bacterial resistance to antibiotics: enzymatic degradation and modification. Adv Drug Deliver Rev 57: 1451–1470. [DOI] [PubMed] [Google Scholar]
- Yurtsev EA, Chao HX, Datta MS, Artemova T, Gore J (2013). Bacterial cheating drives the population dynamics of cooperative antibiotic resistance plasmids. Mol Syst Biol 9: 683. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.