Abstract
Alternative polyadenylation (APA) is increasingly recognized to regulate gene expression across different cell-types, but obtaining APA maps from individual cell-types typically requires prior purification, a stressful procedure that can itself alter cellular states. Here, we describe a new platform, cTag-PAPERCLIP, that generates APA profiles from single cell populations in intact tissues; cTag-PAPERCLIP requires no tissue dissociation and preserves transcripts in native states. Applying cTag-PAPERCLIP to profile four major cell-types in the mouse brain revealed common APA preferences between excitatory and inhibitory neurons distinct from astrocytes and microglia, regulated in part by neuron-specific RNA-binding proteins NOVA2 and PTBP2. We further identified a role of APA in switching Araf protein isoforms during microglia activation, impacting production of downstream inflammatory cytokines. Our results demonstrate the broad applicability of cTag-PAPERCLIP and a previously undiscovered role of APA in contributing to protein diversity between different cell-types and cellular states within the brain.
ETOC BLURB
Hwang et al. develop cTag-PAPERCLIP to profile mRNA 3′ ends in individual cell-types from intact tissues in vivo. They use cTag-PAPERCLIP to show that mRNA alternative polyadenylation contributes to protein diversity between different cell-types and cellular states within the brain.
INTRODUCTION
Alternative polyadenylation (APA), which generates diverse mRNA species with distinct 3′ termini, has been increasingly recognized as a mechanism to regulate gene expression across different tissues (reviewed in (Tian and Manley, 2017)). Tissue-specific APA preferences were documented in early studies of transcriptome (EST) diversity and subsequently in recent high-throughput sequencing analyses in animals (Lianoglou et al., 2013; Smibert et al., 2012; H. Zhang et al., 2005). Studies of specific genes revealed that they could have distinct APA patterns between different cell-types within a living tissue (Boutet et al., 2012; L. Wang et al., 2013). These studies have suggested that APA profiles of individual cell-type level would be of great value for developing a thorough understanding of the relationships between diverse biological functions and APA in individual cell-types within a tissue.
A common requirement for RNA-seq or existing high-throughput APA mapping methods (Derti et al., 2012; Hoque et al., 2013; Jan et al., 2011; Jenal et al., 2012; Lianoglou et al., 2013; Shepard et al., 2011) to profile individual cell-types from an intact tissue is to harvest pure cell populations in order to generate the input RNAs. However, the process of tissue dissociation and cell-type purification, which commonly involves enzymatic digestion and fluorescent activated cell sorting (FACS), in some cases have been shown to result in cellular stress and alter the gene expression profile (Cardona et al., 2006; Okaty et al., 2011), particularly for transcripts that respond to acute stress.
Furthermore, standard RNA-seq has limited abilities in precisely pinpointing poly(A) sites and therefore depends on existing poly(A) site annotations to identify potential APA shifts, while oligo(dT) primer-based APA mapping methods suffer from low specificity and commonly misidentify internal adenine-rich regions as the poly(A) sites (Miura et al., 2014; Shi, 2012). We recently developed a new mRNA 3′ end mapping method, PAPERCLIP (Hwang et al., 2016), based on the popular CLIP (Crosslinking Immunoprecipitation) technique. CLIP was originally developed for studying the RNA interactome of the neuron-specific RNA-binding protein Nova in the living brain (Ule et al., 2003) and has since been applied to additional organs such as the heart and muscle to study cell-type-enriched RNA-binding proteins (Maatz et al., 2014; E. T. Wang et al., 2015). When combined with high-throughput sequencing, CLIP generates high-resolution RNA-protein interaction maps from intact tissues (i.e. whole brain) (Licatalosi et al., 2008). Therefore, a key advantage of PAPERCLIP over other APA mapping methods, in addition to its high specificity for the mRNA 3′ ends, is that it can potentially be adapted to profile individual cell-types from whole tissues without cell purification.
In this study, we used the PAPERCLIP technique to profile selected cell populations from intact tissues. We generated a new knock-in mouse platform technology, termed “cTag-PABP” (“conditionally”-tagged PABP), in which GFP-tagged PABP is conditionally expressed in vivo in selected cell populations using the Cre/Lox system. We termed the resulting combination of cTag-PABP mouse model and the new GFP-PAPERCLIP protocol “cTag-PAPERCLIP” to distinguish it from the original PAPERCLIP technique. cTag-PAPERCLIP replaces the antibody purification of PABP with high-affinity GFP antibody mixtures to allow purification of PABP-RNA complexes from Cre-expressing cells.
As a proof-of-principle analysis, we applied cTag-PAPERCLIP to globally map and compare the poly(A) site usage in four major cell-types in the mouse brain (excitatory neurons, inhibitory neurons, astrocytes and microglia). We found that the two types of neurons as a group have distinct APA preferences compared with astrocytes and microglia, and we demonstrated a contributing role of neuron-specific RNA-binding proteins to this differential regulation. Furthermore, cTag-PAPERCLIP identified a role of APA during microglia activation following lipopolysaccharide (LPS) challenge in mice. This in turn allowed us to discover an APA-mediated switch in Araf protein isoforms that affects their ability to produce inflammatory cytokines. Our results reveal biological roles of APA in contributing to transcriptional and protein diversity between different cell-types and distinct cellular states within a single cell-type that were undetectable with current approaches based on cell-purification.
RESULTS
The cTag-PABP mouse and GFP-PAPERCLIP provide a universal solution to individual cell-type APA profiling from intact tissues in vivo
The original PAPERCLIP protocol generates an all-inclusive profile of any cell-type in a tissue because of the ubiquitous expression of poly(A)-binding protein (PABP) (Fig. 1A). To confer cell-type specificity on PAPERCLIP and expand its application to profile distinct cell populations from a tissue, we generated a new mouse model that conditionally and selectively expresses GFP-tagged poly(A)-binding protein (PABP-GFP) in a Cre-dependent fashion (Fig. 1B, Pabpc1fl & Pabpc1Δneo). We named the mouse model “cTag-PABP” (“conditionally”-tagged PABP). We employed a knock-in strategy targeting the endogenous Pabpc1 locus to maintain the PABP-RNA stoichiometry, a critical factor in protein-RNA interaction (Darnell, 2010; Ince-Dunn et al., 2012; Licatalosi and Darnell, 2010). The cTag-PABP mice were born at Mendelian ratios, grew into adulthood and bred without observable abnormalities (data not shown). To examine the proper induction of PABP-GFP, we bred the cTag-PABP mice to CMV-Cre mice, a near-ubiquitous Cre driver. Genotyping PCR and immunoblot experiments on the resulting CMV-Cre; Pabpc1cTag mice demonstrated successful Cre-mediated recombination and Cre-dependent expression of PABP-GFP in multiple organs (Fig. 1C&D). We next modified the original PAPERCLIP protocol by replacing the PABP antibody pulldown with one in which we used a mixture of two high-affinity monoclonal GFP antibodies (Heiman et al., 2014) to pulldown PABP-GFP from CMV-Cre; Pabpc1cTag mice (Fig. 1E), and performed a side-by-side comparison of the resulting transcript profiles. The results from the two versions of PAPERCLIP are highly similar (R2 = 0.83; Fig. 1F), indicating that our tagging strategy does not alter the binding profile of PABP. Overall, these results demonstrate the success of our knock-in strategy and the feasibility of using GFP-PAPERCLIP for in vivo RNA profiling.
Figure 1.

Creation of the cTag-PABP mouse to restrict PAPERCLIP profiling to specific cell populations in intact tissues. (A) Schematics illustrating the strategy for selective PAPERCLIP profiling by expressing “conditionally-tagged” (cTag)-PABP (PABP-GFP) in the targeted cell population (“Tagged” PAPERCLIP, right) and comparing it to the conventional PAPERCLIP (left). (B) Schematics illustrating the targeting strategy for generating the knock-in cTag-PABP mouse (right) and the corresponding Pabpc1 allele nomenclature (left). FNF: Flox-Neo-Flox selection cassette. Black arrowheads denote loxP sites. pA: a synthetic poly(A) site. (C) Genotyping PCR experiments demonstrating successful Cre-mediated excision. Lanes: wildtype (left); Pabpc1fl (middle); CMV-Cre;Pabpc1cTag (right). Rapsn serves as a loading control. (D) Fluorescent immunoblots showing Cre-dependent expression of PABP-GFP. Asterisk denotes a non-specific band that was also observed in non-transgenic wildtype mice. (E) Autoradiogram from a cTag-PAPERCLIP experiment. The red arrow denotes the PABP-GFP-RNA complex. The red box shows the area of the nitrocellulose membrane used for RNA extraction and subsequent sequencing library construction. (F) Left, Schematic illustrating the experimental design to compare conventional PAPERCLIP to GFP-PAPERCLIP. Right, a scatter plot showing the correlation of read counts between conventional PAPERCLIP and GFP-PAPERCLIP. Each dot represents a poly(A) site. R2, the coefficient of determination.
We next applied GFP-PAPERCLIP and the cTag-PABP mice (referred to henceforth as cTag-PAPERCLIP) to profile four major cell-types in the adult mouse brain: excitatory neurons, inhibitory neurons, astrocytes and microglia. These were generated by breeding cTag-PABP mice to four established Cre mouse lines to generate the following mouse lines for cTag-PAPERCLIP profiling (Fig. 2A): Camk2a-Cre; Pabpc1cTag (excitatory neurons), Gad2-Cre; Pabpc1cTag (inhibitory neurons), mGfap-Cre; Pabpc1cTag (astrocytes) and Cx3cr1-Cre; Pabpc1cTag (microglia). We performed immunohistochemistry on brain sections from the four mouse lines to examine the expression patterns of PABP-GFP (Fig. 2B–E) and compared them to those of established cell-type markers (Fig. 2F–I). Overall, PABP-GFP showed highly restricted and anticipated expression patterns for each cell-type, correlating well with the corresponding cell-type markers. We proceeded to carry out cTag-PAPERCLIP profiling in different cell-types within brain cortex (Camk2a-Cre; Pabpc1cTag, Gad2-Cre; Pabpc1cTag and mGfap-Cre; Pabpc1cTag) or whole brain (Cx3cr1-Cre; Pabpc1cTag) from adult mouse (Table S1). We examined the abundance of eight prototypic cell-type specific marker genes (Butovsky et al., 2014; Spiegel et al., 2014; Vong et al., 2011) in cTag-PAPERCLIP profiles from the four mouse lines. We found that all markers showed high expression only in the expected mouse lines (Fig. 2J–M), consistent with the highly restricted expression patterns of PABP-GFP in immunohistochemistry experiments and provides strong support for specificity of the cTag-PAPERCLIP technique. To further substantiate the analysis, we assembled an 80-gene panel that differentiates individual cell-type RNA-seq data for the four cell-types (Mo et al., 2015; Y. Zhang et al., 2014) (Fig. 2N, columns). Examining the expression level of the 80 genes in both RNA-seq and cTag-PAPERCLIP profiles revealed a high correlation between cTag-PAPERCLIP and RNA-seq from the same cell-type (Fig. 2N, columns vs. rows, R>0.8 for all four cell-types). The results were especially notable given that cTag-PAPERCLIP and RNA-seq datasets were generated independently from the brains of distinct mouse crosses using different techniques. In summary, we provide multiple lines of evidence to demonstrate that the cTag-PAPERCLIP is able to identify distinct gene expression profiles from different cell populations from within intact tissues.
Figure 2.
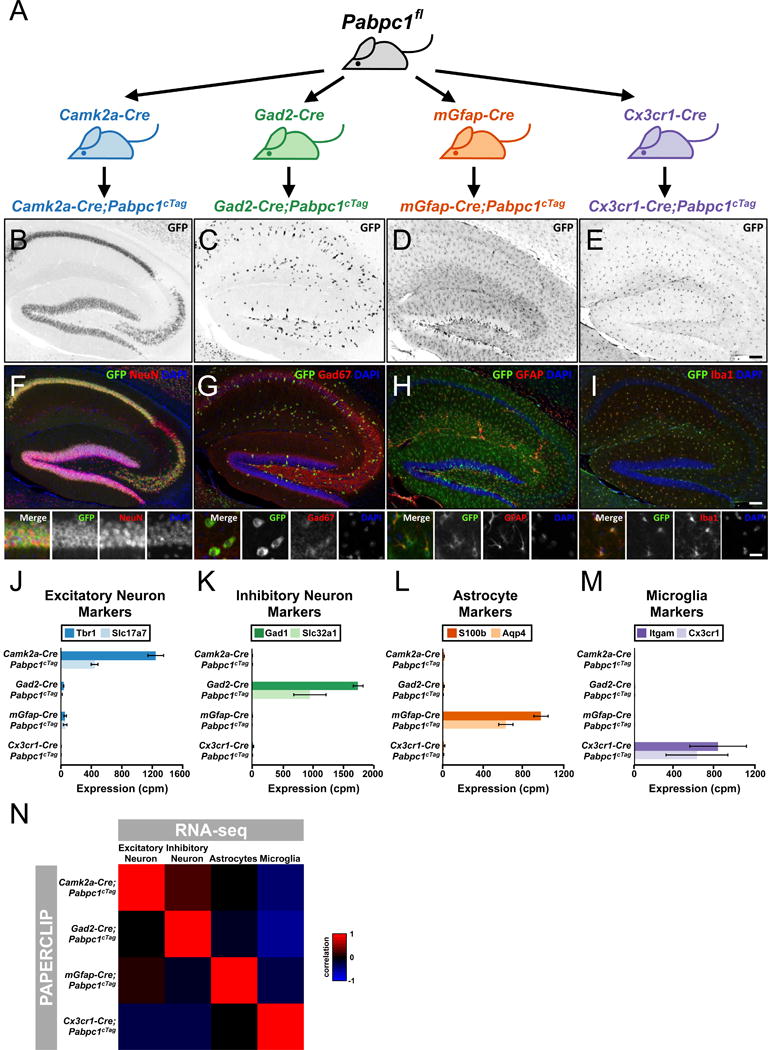
The cTag-PABP mouse and GFP-PAPERCLIP provide a universal solution to individual cell-type profiling in vivo. (A) Schematics illustrating the breeding strategy and experimental design for PAPERCLIP profiling in excitatory neurons (Camk2a-Cre), inhibitory neurons (Gad2-Cre), astrocytes (mGfap-Cre) and microglia (Cx3cr1-Cre). (B–I) Immunofluorescent staining showing the expression of PABP-GFP alone (B-E) or in combination with cell-type markers (F–I) in the hippocampus of Camk2a; Pabpc1cTag (B&F), Gad2-Cre; Pabpc1cTag (C&G), mGfap-Cre; Pabpc1cTag (D&H), and Cx3cr1-Cre; Pabpc1cTag (E&I) mice. (Insets, F–I): high-power images taken from the pyramidal layer (F) or the molecular layer (G–I) of the hippocampus to show individual cells. NeuN: neuronal marker. Gad67: inhibitory neuron marker. GFAP: astrocyte marker. Iba1: microglia marker. Scale Bar: 100 μM. (J–M) Bar graphs showing the expression level of established cell-type markers in the GFP-PAPERCLIP profiles from brain cortex of Camk2a; Pabpc1cTag, Gad2-Cre; Pabpc1cTag, mGfap-Cre; Pabpc1cTag, and the whole brain of Cx3cr1-Cre; Pabpc1cTag mice. Error bars: standard error. cpm: counts per million. (N) A heatmap showing the correlation of gene expression profiles between GFP-PAPERCLIP and publicly available cell-type-specific RNA-seq datasets (Mo et al., 2015; Y. Zhang et al., 2014) from an 80-gene panel.
Distinct APA preference between neurons and glia results in diversity in protein expression
PAPERCLIP offers transcriptome-wide assessment of APA patterns and accurately detects APA shifts (Hwang et al., 2016). Next, we investigated whether cTag-PAPERCLIP profiles can also identify distinct patterns of poly(A) site usage in different brain cell-types. We annotated 22,799 poly(A) sites in 15,603 genes from the various cell-specific cTag-PAPERCLIP profiles. Among the 15,603 genes, 10,234 genes (66%) have a single poly(A) site while 5,369 genes (34%) have multiple poly(A) sites. We next selected 2,606 dual-poly(A) site genes that have sufficient sequencing coverage to compare their APA patterns in different cell-types through permutation, in which all the cell-types are designated into two groups for comparison without a priori hypothesis (Table S2). The only positive hit from this permutation analysis was identified when neurons (excitatory and inhibitory as a group) were compared against the rest of the cell-types as a second group, from which we identified 27 dual-poly(A) site genes that have different APA patterns between neurons and glia (FDR<0.05). No genes reached the statistical significance threshold in all the other permutations. To provide an independent verification for the permutation results, we analyzed the aforementioned individual cell-type RNA-seq data (Y. Zhang et al., 2014) with input from our cTag-PAPERCLIP data, which provided exact poly(A) site annotations and pairs of differentially expressed APA isoforms. The RNA-seq analysis was performed by counting mapped reads for genes with tandem poly(A) sites or by performing RNA isoform analysis when the poly(A) sites are associated with distinct RNA isoforms. Analyzing this stringently defined dataset, we found 12 statistically significant differences in poly(A) site usage between neurons and both non-neuronal cell-types from among 14 genes (Table S3). Thirteen additional genes for which gene models can not be unequivocally established based on the cTag-PAPERCLIP annotated poly(A) sites and the standard annotations (Ensembl and RefSeq), or for which there was insufficient sequence coverage, were not included in the RNA-seq analysis. Taken together, these results provide strong support for the ability of cTag-PAPERCLIP and permutation analysis results to distinguish cell-specific APA profiles.
Next, we selected six genes (Itsn1, Map4, Atp2a2, Rnf130, Klc1 and Ptpn2) for further analysis based on the following criteria: first, the difference in poly(A) site usage results in a shift in the predominant poly(A) site between neurons and glia (Fig. 3A–B, Fig. S1A–D); second, the APA shift results in a switch in mRNA transcript isoforms between neurons and glia that are predicted to generate distinct protein isoforms based on Ensembl or RefSeq annotations (Fig. S2A–F). We also included Cdc42 in the analysis, which in neurons clearly gains a new isoform that is absent in glia (Fig. 3C) and is predicted to produce different proteins from the two isoforms (Fig. S2G). Again, RNA isoform analysis using an independent RNA-seq dataset demonstrated a statistically significant difference in neuronal/glial APA isoform expression between neurons and astrocyte and/or microglia for all seven genes (Fig. 3D–F, Fig. S1E–H and Table S3), corroborating the cTag-PAPERCLIP results. We further extended the neuronal/glial isoform RNA-seq analysis to five additional mouse tissues (liver, spleen, kidney, heart and lung) for the seven genes. Interestingly, the glial isoform is always preferred in the five non-neuronal tissues for these genes except for Atp2a2 (Fig. 3D–F and Fig. S1E–H), suggesting that the glial isoform might be the default isoform for the six genes (Itsn1, Map4, Cdc42, Rnf130, Klc1 and Ptpn2) and a shift to the neuronal isoform might be specifically regulated.
Figure 3.
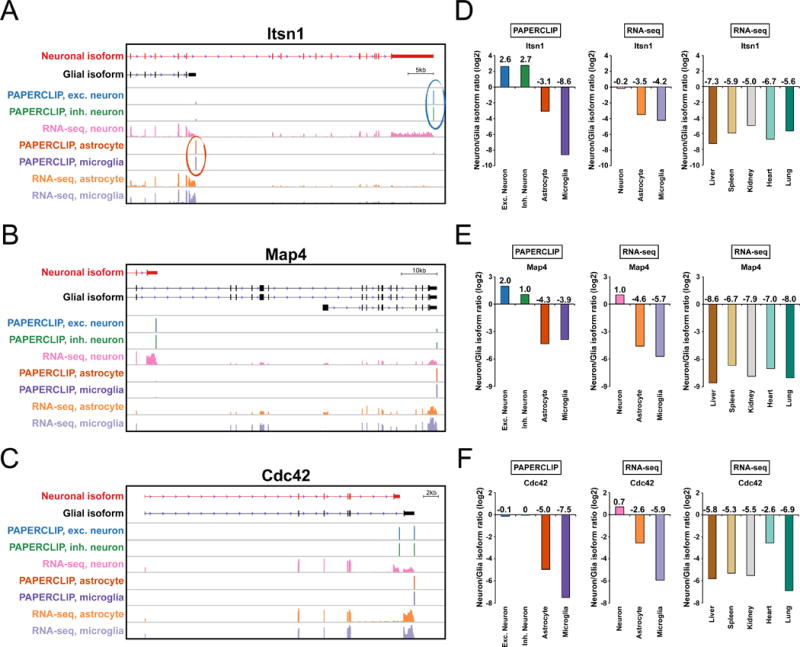
cTag-PAPERCLIP identifies distinct APA preferences in neurons. (A, B and C) Diagrams demonstrating differential APA patterns between neurons and glia at Itsn1 (A), Map4 (B) and Cdc42 (C) loci. X-axis: position of PAPERCLIP or RNA-seq reads across RNA transcripts, as indicated in the top two tracks. Exc.: excitatory. Inh.: inhibitory. Y-axis: normalized read depth (scaled in each individual track). RNA-seq data is from (Y. Zhang et al., 2014). (D, E and F) Bar graphs showing the relative ratio of neuronal and glial APA isoforms of Itsn1 (D), Map4 (E) and Cdc42 (F) in different cell-types (indicated by colors) as determined by PAPERCLIP (left), RNA-seq in mouse brain (middle), or RNA-seq in five different mouse tissues (right). Exc.: excitatory. Inh.: inhibitory.
NOVA2 and PTBP2 directly regulate the neuronal APA isoform expression of Itsn1, Map4 and Cdc42
Neuron-specific RNA-binding proteins (RBPs) such as NOVA1/2 and PTBP2 are key players in shaping the neuronal transcriptome, and both proteins have been shown to regulate APA (Castelo-Branco et al., 2004; Licatalosi et al., 2008). As NOVA2 is the predominant NOVA protein in mouse brain cortex (Saito et al., 2016), we hypothesized that NOVA2 and PTBP2 might regulate the neuronal APA isoform expression for some of the seven genes that we identified. To test the hypothesis, we newly generated RNA-seq data from E18.5 brain cortex of Ptbp2−/− mice (Ptbp2-KO) (Licatalosi et al., 2012) and re-analyzed our previously published RNA-seq data from E18.5 brain cortex of wildtype and Nova2−/− mice (Nova2-KO) (Saito et al., 2016) to compare the relative abundance of APA isoforms of the seven genes in Nova2-KO and Ptbp2-KO mice to the wildtype mice. Interestingly, the relative abundance of Itsn1 and Klc1 neuronal APA isoforms is decreased in Nova2-KO mice (Fig. 4A) while the relative abundance of Map4 and Cdc42 neuronal APA isoforms is increased in Ptbp2-KO mice (Fig. 4B). The observed isoform changes were unlikely reflecting alterations of neuron-to-glia cell number ratio as the neuronal/glial marker ratio is maintained in Nova2-KO mice and decreased in Ptbp2-KO mice compared with the wildtype mice (Fig. S2H). A similar neuronal/glial marker ratio between Nova2-KO and the wildtype mice is consistent with our previous reports that loss of Nova2 does not affect neuronal differentiation (Leggere et al., 2016; Saito et al., 2016). In contrast, a decrease in both the pan-neuron/astrocyte marker ratio and the excitatory neuron/astrocyte marker ratio suggests a possible decrease in the number of excitatory neurons in Ptbp2-KO mice, which is consistent with a known role of Ptbp2 in neuronal maturation (Li et al., 2014; Licatalosi et al., 2012). To further support the RNA-seq results, we performed qRT-PCR to specifically measure the expression of neuronal and glial isoforms for all four genes using RNAs from E18.5 brain cortex from Nova2-KO, Ptbp2-KO and the wildtype mice. In all cases, the results confirmed the RNA-seq observations, and we also found a reciprocal change in the other isoform with the exception of Klc1 (Fig. 4C&D). These data demonstrate that cTag-PAPERCLIP is able to detect APA isoforms regulated by NOVA2 and PTBP2. We conclude that these proteins are regulating the balance between neuronal and glial APA isoforms of the four genes; NOVA2 promotes while PTBP2 suppresses usage of the neuronal APA isoform.
Figure 4.
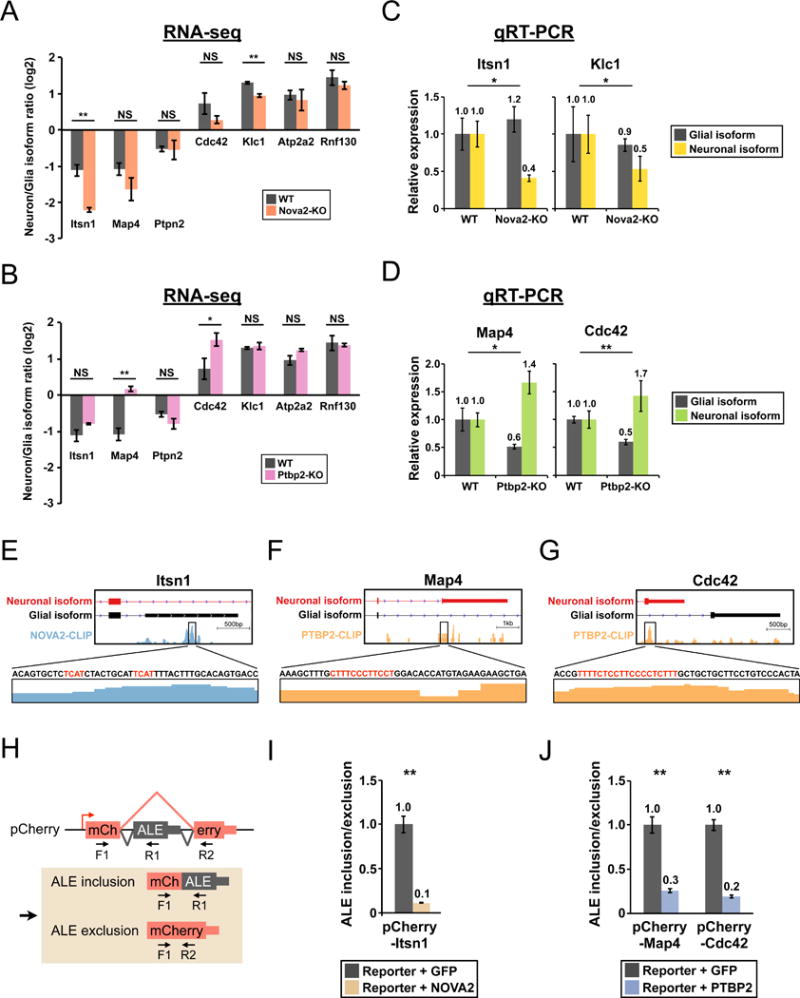
NOVA2 and PTBP2 directly regulate the neuronal APA isoform expression of Itsn1, Map4 and Cdc42. (A and B) Bar graphs comparing the neuron/glia isoform ratio in E18.5 brain cortex of (A) wildtype (WT) and Nova2−/− (Nova2-KO) mice or (B) wildtype (WT) and Ptbp2−/− (Ptbp2-KO) mice as determined by RNA-seq. Error bars: standard deviation. *: p<0.05. **: p<0.01. (C) Bar graphs showing quantitation of the neuronal and glial isoforms at the Itsn1 and Klc1 loci in E18.5 brain cortex of Nova2−/− mice (Nova2-KO) by qRT-PCR. WT: wildtype. Error bars: standard error. *: p<0.05. (D) Bar graphs showing an increase in abundance of the neuronal isoform and a decrease in abundance of the glial isoform at the Map4 and Cdc42 loci in E18.5 brain cortex of Ptbp2−/− mice (Ptbp2-KO) by qRT-PCR. WT: wildtype. Error bars: standard error. *: p<0.05. **: p<0.01. (E, F and G) Diagrams showing NOVA2 and PTBP2 HITS-CLIP footprints in the alternative last exons of Itsn1, Map4 and Cdc42. The genomic sequence is shown in the 5′−3′ direction. NOVA and PTBP binding motifs (YCAY and poly-pyrimidines) are highlighted in red. Due to space limitation, additional NOVA and PTB binding motifs in Itsn1 and Map4 are not shown. (H) Schematics showing the experimental design and possible outcomes for the mini-gene assay for exon inclusion/exclusion. Top: Illustration of the pCherry vector. Thick box: coding sequence. Thin box: untranslated region. ALE: alternative last exon. Red arrow: the transcription start site. Black arrows (F1, R1 and R2): primers for qRT-PCR assays. Bottom: Two possible mRNA products from pCherry and the respective qRT-PCR amplicons. (I and J) Bar graphs showing qRT-PCR results from the mini-gene assays. Y-axis denotes the ratio of (ALE inclusion product)/(ALE exclusion product). Co-transfection of a GFP-expressing plasmid serves as the control. Error bars: standard error. **: p<0.01.
Next, we examined NOVA2 and PTBP2 HITS-CLIP datasets (Licatalosi et al., 2012; Saito et al., 2016; C. Zhang et al., 2010) for all four genes. No NOVA or PTBP2 footprints can be identified at or near the alternative last exon (ALE) of Klc1 (data not shown). In contrast, we identified NOVA2 and PTBP2 footprints at the ALEs of Itsn1, Map4 and Cdc42 (Fig. 4E–G), indicating that NOVA2 and PTBP2 directly bind these regions and, as previously demonstrated for both proteins (Licatalosi et al., 2012; 2008) such binding can regulate processing of the alternative exon. To demonstrate a direct regulatory relationship experimentally, we respectively cloned the ALEs and the flanking intronic sequence of Itsn1, Map4 and Cdc42 into a split-exon expression plasmid, pCherry, to generate reporter constructs that allow the examination of ALE inclusion/exclusion by qRT-PCR assays in HEK293 cells (Fig. 4H). Co-transfection of a NOVA2-expression plasmid suppressed the inclusion of Itsn1 glial ALE (Fig. 4I), consistent with a role of NOVA2 in promoting generation of the Itsn1 neuronal APA isoform. In contrast, co-transfection of a PTBP2-expression plasmid repressed the inclusion of Map4 and Cdc42 neuronal ALEs (Fig. 4J), providing further support that PTBP2 suppresses the neuronal APA isoforms of Map4 and Cdc42. To demonstrate that the pCherry reporter can positively respond to NOVA2 co-expression, we created another pCherry reporter by inserting Aplp2 exon 13, the usage of which was promoted by direct NOVA binding (Saito et al., 2016), along with the entire flanking intronic sequence. As expected, co-transfection of NOVA2 increased its inclusion (Fig. S2I), providing evidence that our mini-gene assay utilizing the pCherry construct recapitulates the full range of alternative processing events regulated by RBPs. Taken together, these results reveal a role of neuron-specific RBPs in regulating APA to generate distinct protein isoform expression in neurons. Furthermore, our results illustrate how cTag-PAPERCLIP guides biological discoveries through generating individual cell-type APA maps in vivo.
cTag-PAPERCLIP profiles microglia with minimal perturbation and better preserves their naïve states
Current protocols for microglia gene expression profiling by microarray or RNA-seq require prior purification that needs at least a few hours to complete and includes enzymatic digestion to dissociate cells (Bennett et al., 2016; Cardona et al., 2006; Nikodemova and Watters, 2012). Such purification process often results in various degrees of cellular stress and immunological activation of microglia, which is known to be very sensitive to experimental manipulations (Cardona et al., 2006). In contrast, the cTag-PAPERCLIP protocol “freezes” PABP to its binding sites by UV-irradiation of intact brain, and therefore does not require any enzymatic digestion, allowing sampling of the entire brain microglia population in a few minutes (Fig. 5A), which should lead to better preservation of microglia at the naïve state. To assess the technical advantages of cTag-PAPERCLIP, we compared our data from Cx3cr1-Cre; Pabpc1cTag mouse, in which microglia RNAs are purified from acutely dissected brain snap-frozen in liquid nitrogen (Fig. 5A) to eight publicly available microglia RNA-seq datasets generated through ex vivo microglia purification. As most of the studies in the eight RNA-seq datasets utilized FACS to isolate microglia, we also performed RNA-seq to evaluate column-based microglia isolation (Nikodemova and Watters, 2012), which might activate microglia less because of its shorter duration (< 2 hours) and more gentle tissue dissociation compared with FACS. Strikingly, in what are expected to be resting microglia isolated from the mouse brain, the expression levels of microglia activation markers and immediate early genes were significantly lower in the cTag-PAPERCLIP profile compared with those in both RNA-seq profiles, which were themselves indistinguishable (Fig. 5B). In contrast, microglia cell-type markers are highly enriched in all three profiles without statistically significant differences (Fig. 5B). We attribute these differences to the ability in the cTag-PAPERCLIP protocol to immediately preserve acutely dissected brain in liquid nitrogen, with subsequent UV cross-linking of frozen tissue, minimizing activation artifacts introduced by either existing cell purification method. Taken together, these analyses demonstrate that cTag-PAPERCLIP better preserves native resting cell-specific transcriptome states and therefore is suitable for accurate in vivo RNA profiling, even for difficult to purify cell-types such as microglia.
Figure 5.
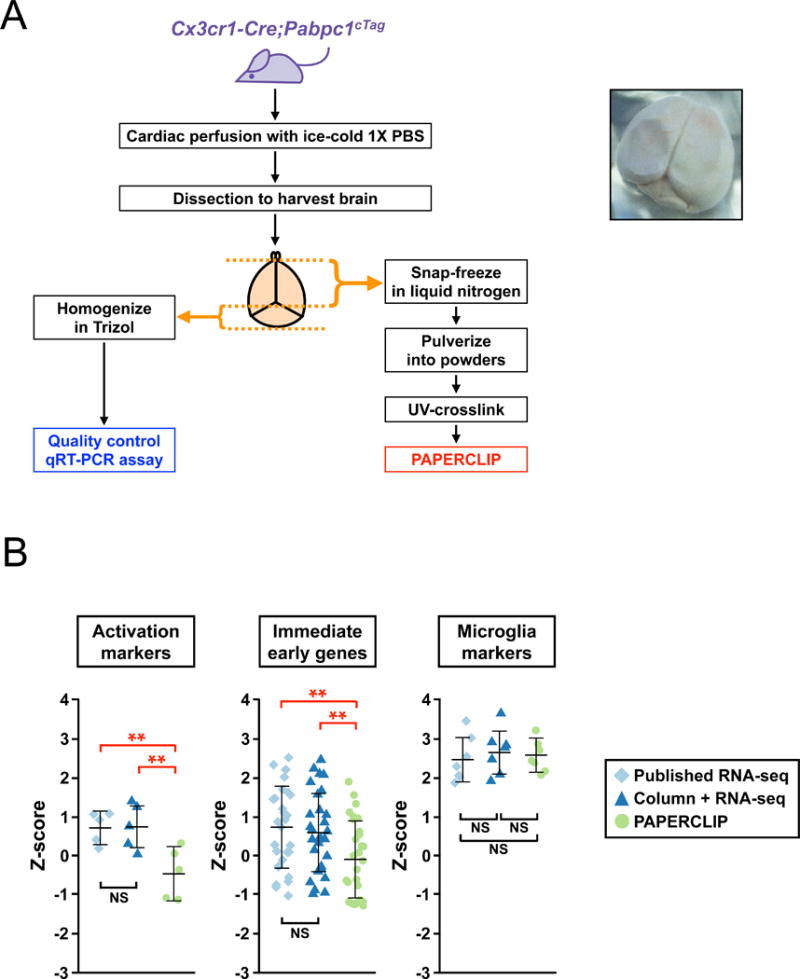
cTag-PAPERCLIP profiles brain microglia with minimal perturbation and better preserves their naïve states. (A) (Left) A diagram showing the procedure to harvest Cx3cr1-Cre; Pabpc1cTag mouse brain for microglia profiling by cTag-PAPERCLIP. A quality-control qPCR assay was performed for every sample before cTag-PAPERCLIP profiling. (Right) An example image of a snap-frozen mouse brain from a cTag-PAPERCLIP experiment. (B) Dot plots comparing the expression level of microglia activation markers (left), immediate early genes (middle) and microglia cell-type markers (right) identified by cTag-PAPERCLIP to those identified by RNA-seq through ex vivo microglia purification. **: p<0.01; NS: not significant.
Activated brain microglia prefers proximal poly(A) site and upregulates full-length ARAF through an APA switch
Microglia activation is a common event in neurological disease (Prinz and Priller, 2014) that is yet to be fully characterized in vivo due to difficulties in purifying microglia in their resting state. We took advantage of the technical advances of cTag-PAPERCLIP to characterize microglia activation in vivo by comparing their RNA profile in a true resting state in vivo to a fully activated state. Blood monocytic cells, which have overlapping gene expression program with microglia but are believed to perform distinct functions in the brain, are potential contaminants in microglia molecular profiling (Cardona et al., 2006; Yamasaki et al., 2014; Yona et al., 2013) and they enter the brain parenchyma when the blood-brain barrier (BBB) is disrupted. To globally activate all brain microglia in vivo and at the same time minimize BBB disruption, we adapted a previously described protocol (Chen et al., 2012) by performing daily low-dose (1mg/Kg) LPS injection in Cx3cr1-Cre; Pabpc1cTag mice for four consecutive days before cTag-PAPERCLIP profiling (Fig. 6A). We first performed flow cytometry analysis on brain microglia harvested from PBS- and LPS-injected Cx3cr1-Cre; Pabpc1cTag mice to examine the efficiency of the LPS induction. Consistent with the previous report (Chen et al., 2012), the LPS injection protocol resulted in a universal activation of brain microglia (Fig. 6B) without overt heavy infiltration of blood monocytic cells at the time of harvest (Fig. 6C).
Figure 6.
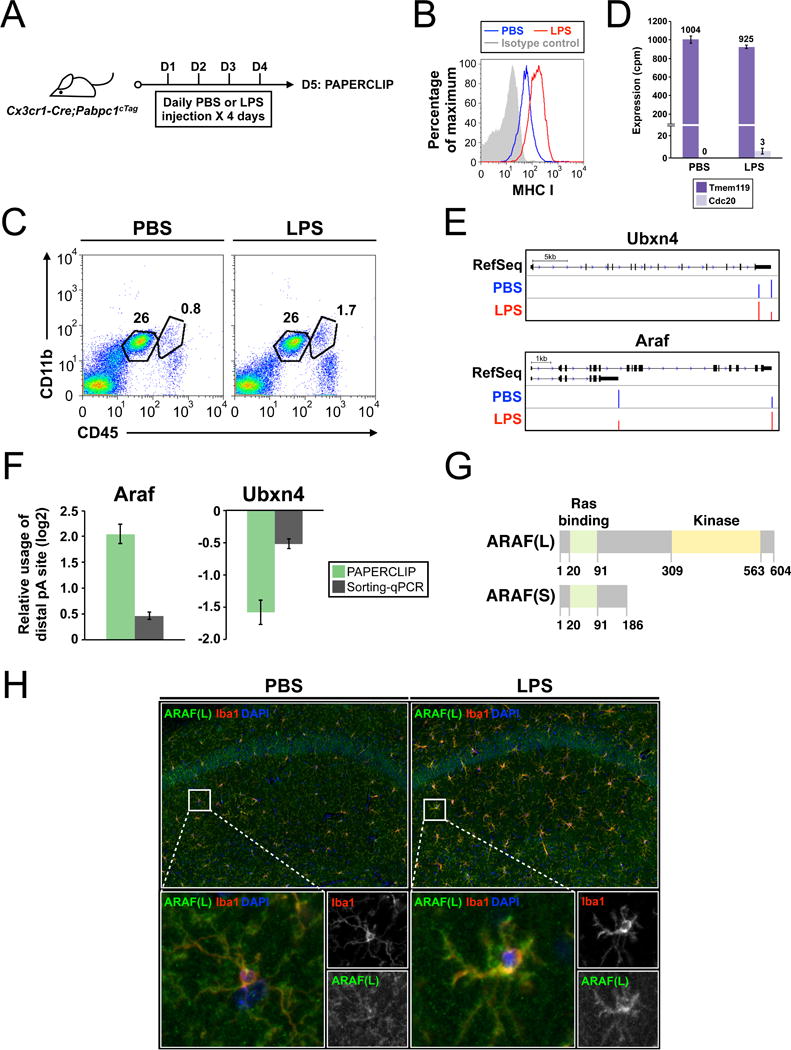
cTag-PAPERCLIP reveals that activated microglia upregulates full-length ARAF through APA switch. (A) A schema showing the experimental design to identify APA events at microglia activation by cTag-PAPERCLIP. (B) Flow cytometry results showing a full activation of brain microglia by the LPS injection protocol. (C) Representative flow cytometry results from PBS- or LPS-treated Cx3cr1-Cre; Pabpc1cTag mouse brain showing that the LPS injection protocol does not result in heavy infiltration of blood monocytic cells [the (Cd11b+, Cd45high) population on the right]. The (Cd11b+, Cd45intermediate) population on the left is microglia. (D) Bar graphs showing the cell-type-specific marker expression in cTag-PAPERCLIP data from PBS- or LPS-injected Cx3cr1-Cre; Pabpc1cTag mice. Tmem119, a microglia-specific marker. Cdc20, a monocyte-specific marker. Error bars: standard error. cpm: counts per million. (E) Diagrams showing switches in the major poly(A) sites at the Ubxn4 and Araf loci (top row, ‘RefSeq’) in activated microglia as identified by cTag-PAPERCLIP, shown as the sum of the three biological replciates (middle and bottom rows; ‘PBS’ and ‘LPS’). Y-axis: normalized read depth (scaled in each individual track). (F) Bar graphs showing the relative usage of the distal poly(A) site (LPS/PBS) at the Ubxn4 and Araf loci as determined by cTag-PAPERCLIP in Cx3cr1-Cre; Pabpc1cTag mice or qRT-PCR on sorted microglia from wildtype B6 mice. Error bars: standard error. (G) Schematics showing the lengths and the annotated domains in ARAF proteins generated by the two Araf APA isoforms. Numbers denote amino acid residues. (H) Immunofluoresence microscopy of PBS-treated (left) or LPS-treated (right) wildtype B6 mouse brain demonstrates an increase in full-length Araf protein [ARAF(L)] expression in activated microglia. Images from the two groups were taken under the same conditions. The results were reproduced in an independent experiment and representative images are shown. Iba1: a microglia marker.
More detailed flow cytometry analysis revealed that GFP-positive microglia strongly outnumber GFP-positive monocytes (~50:1 in PBS-injected brain and ~25:1 in LPS-injected brain; data not shown). Furthermore, the expression of Cdc20 RNA, which is exclusively expressed in monocytes but not in microglia (Goldmann et al., 2016) was negligible compared with that of microglia-specific Tmem119 (Bennett et al., 2016) in the cTag-PAPERCLIP expression profiles (Fig. 6D). Altogether, these results demonstrate that we were able to globally activate brain microglia and at the same time minimize the infiltration of blood monocytic cells into the brain parenchyma and their contamination of the microglia cTag-PAPERCLIP profiles.
Next, we performed three biologically independent experiments to profile brain microglia from Cx3cr1-Cre; Pabpc1cTag mice injected with PBS (resting microglia) or with LPS (activated microglia) by cTag-PAPERCLIP. We observed that the unique read counts for the PBS group were constantly much lower than the LPS group with the same amount of input (Fig. S3A), which is consistent with the quiescent nature of the resting microglia (Ponomarev et al., 2011). Nevertheless, the overall correlation between replicates remains high for both groups (R≥0.86, Fig. S3B), demonstrating the robustness of our LPS-activation and cTag-PAPERCLIP protocols. Previous reports suggest that activated cells have a tendency to use the proximal poly(A) sites (Flavell et al., 2008; Sandberg et al., 2008). Indeed, we found that activated microglia are more likely to use the proximal poly(A) sites for dual-poly(A) site genes compared with resting microglia (Fisher’s exact test, p=0.0076).
The relative low unique read counts for each poly(A) site in resting microglia (~300,000 unique reads for ~15,000 poly(A) sites) pose a challenge for APA shift identification. Therefore, we modified our analysis pipeline to stringently identify 27 genes that showed highly significant APA shifts (8: distal; 19: proximal; Table S4). Four of the 27 genes switched the major poly(A) site between resting and activated microglia. Among those four genes, we selected two, Ubxn4 (proximal shift) and Araf (distal shift) (Fig. 6E), for further validation, as these consistently switched their predominant poly(A) sites upon activation in all three individual experiments. To provide an entirely independent measure for APA shift, we repeated the same LPS injection protocol in wildtype B6 mice to activate microglia and harvested them by cell sorting based on surface markers. Despite the aforementioned caveat of activating PBS-treated microglia during the lengthy purification process (Cardona et al., 2006), we observed the same trend of APA shift for both Ubxn4 and Araf (Fig. 6F) by qRT-PCR experiments on purified microglia, although the magnitude of the changes after activation were much greater in cTag-PAPERCLIP data. These results underscore the advantages of cTag-PAPERCLIP for in vivo profiling.
Interestingly, while both poly(A) sites of Ubxn4 are located in the 3′ UTR, the APA shift of Araf is accompanied by a change in coding capacity of the resulting transcripts, which is predicted to generate different protein isoforms [referred to henceforth as ARAF short (S) and ARAF long (L), respectively] (Fig. 6G). ARAF(S) is a truncated, kinase-null protein previously shown to act as a dominant-negative antagonist of the Ras-ERK pathway during myogenic differentiation (Yokoyama et al., 2007). Switches in the usage of these ARAF isoforms in microglia have not previously been reported. Importantly, however, the microglial-specific APA switch detected by cTag-PAPERCLIP is predicted to cause a corresponding switch in microglial Araf protein isoform usage in vivo.
To examine the abundance of ARAF(L) in vivo, we used an isoform-specific antibody (Fig. S3C) to perform immunohistochemistry on brain sections obtained from PBS- or LPS-injected wildtype B6 mice. The lack of commercially available ARAF(S)-specific antibody precluded us from examining its abundance in the same experiment. Consistent with an increased expression of the full-length Araf mRNA isoform from the APA switch, we observed stronger signals of the ARAF(L) protein in brain microglia from LPS-injected mice compared with PBS-injected mice (Fig. 6H). Altogether, we present multiple lines of evidence suggesting that activated brain microglia increases full-length Araf mRNA and protein isoform production through an APA switch.
The Araf APA switch is a common event for microglia/macrophage in response to multiple TLR ligands and plays a role in their inflammatory cytokine production
Multiple toll-like receptor (TLR) pathways are present in microglia and macrophages with distinct downstream transcriptional programs, and the LPS response is mediated by TLR4 (Fig. S4A) (Chevrier et al., 2011). We sought to address whether the Araf APA switch is specific to the TLR4 pathway or if it can be induced through other TLR pathways. We studied immortalized BV2 microglia and RAW264.7 macrophages, two established mouse cell-lines that are more accommodative to experimental manipulations than primary microglia and are commonly used for signaling pathway studies (Häcker et al., 2005; Henn et al., 2009). We first tested whether the LPS-induced switch of Araf APA isoforms can be observed in these two cell-lines in culture. We treated BV2 and RAW264.7 cells with or without LPS in culture and measured the mRNA levels of Araf(S) and Araf(L). Indeed, both cell-lines exhibited down-regulation of Araf(S) upon LPS treatment, a response similar to brain microglia (Fig. 7A), and RAW264.7 cells replicated the Araf isoform switch response of brain microglia (Fig. 6E and Fig. 7A).
Figure 7.
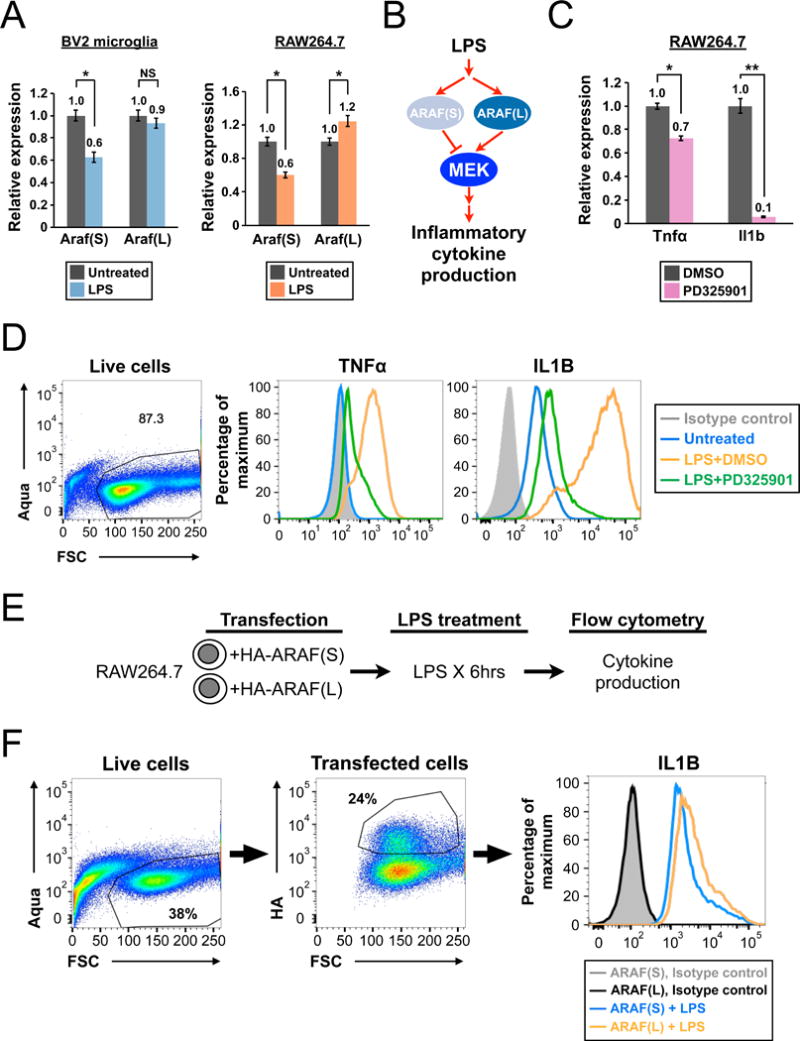
The Araf APA switch is a common event for microglia/macrophage in response to LPS and plays a role in their inflammatory cytokine production. (A) Bar graphs showing qRT-PCR results comparing the abundance of Araf mRNA isoforms between untreated and LPS-treated BV2 microglia (left) or RAW264.7 macrophages (right). Error bars: standard error. *: p<0.05. (B) A diagram showing the hypothesized effects of Araf isoforms in LPS-mediated inflammatory cytokine production. (C) Bar graphs showing the qRT-PCR results comparing Tnfα or Il1b mRNA expression levels between DMSO- or PD325901-treated RAW264.7 macrophages after LPS induction. PD, PD325901, a specific MEK inhibitor. Error bars: standard error. *: p<0.05; **< p<0.01. (D) Flow cytometry results showing the suppression of TNFα and IL1B production by MEK inhibition in LPS-treated RAW264.7 macrophages. (E) Schematics showing the experimental design to assay the effects of Araf isoforms on LPS-induced cytokine production in RAW 264.7 cells. (F) Flow cytometry results comparing the effects on LPS-induced IL1B production from ARAF(S) overexpression to ARAF(L) overexpression in RAW264.7 cells.
To begin to assay for functional consequences of the Araf APA switch in stimulated cells, we next treated both cell-lines with TLR2, TLR3 or TLR4 ligands separately and measured the abundance of representative downstream target genes, Il1b and Cxcl10, at the end of treatment to establish the TLR-ligand response profiles for each cell-line. We found that RAW264.7 cells overall had a stronger response to the three TLR ligands compared with BV2, which was almost unresponsive to Poly(I:C) (Polyinosinic:polycytidylic acid, a TLR3 agonist) (Fig. S4B). Therefore, we chose RAW264.7 cells to address whether the Araf isoform switch can also be induced by other TLR ligands. qRT-PCR experiments demonstrated that Poly(I:C) induced an APA switch similar to that of LPS while the same trend can be observed in PAM (palmitoyl(3)-cysteine-serine-lysine(4), a TLR2 agonist) treatment (Fig. S4C). Taken together, these results demonstrate that the Araf APA switch is a common event in both brain microglia and microglia/macrophage cell-lines in culture to multiple TLR ligands.
The Ras-ERK pathway is known to participate in LPS-induced inflammatory cytokine production (Chevrier et al., 2011). We hypothesized that the balance between Araf protein isoforms, which are predicted to have opposing effects on downstream targets such as MEK, affects LPS-induced inflammatory cytokine production (Fig. 7B). To further examine the functional effects of Araf isoforms on LPS-induced cytokine production, we assayed the effects of MEK inhibition on LPS-induced Araf APA switching and production of inflammatory cytokines TNFα and IL1B. We inhibited MEK in RAW264.7 cells with a chemical inhibitor, PD325901, and examined Tnfα and Il1b expression by qRT-PCR (Fig. 7C) and flow cytometry (Fig. 7D). The induction of both cytokines by LPS was suppressed by MEK inhibition. Notably, Il1b is especially sensitive to MEK inhibition in RAW264.7 cells as its induction is suppressed by 10-fold at both mRNA and protein levels (Fig. 7C & D). Having established that MEK inhibition does have a suppressive effect on Tnfα and Il1b induction by LPS in RAW264.7 cells, we proceeded to transfect cells with either ARAF(S)- or ARAF(L)-expressing plasmid to compare the effects of expressing either ARAF protein on Tnfα and Il1b induction by LPS (Fig. 7E). Indeed, we consistently observed a lower IL1B production in Araf(S)-transfected cells compared with Araf(L)-transfected cells upon LPS induction (Fig. 7F), providing evidence that the balance between the Araf APA isoforms in microglia and macrophages mediates functional outcomes of protein activity that in turn are able to affect changes in LPS-induced inflammatory cytokine production.
DISCUSSION
In this study, we describe cTag-PAPERCLIP, a new method that meets the technical challenge of generating APA profiles for selected cell populations without tissue dissociation, providing a universal solution to individual cell-type APA profiling from intact tissues in vivo. We believe that two key innovations separate cTag-PAPERCLIP from existing technologies: in vivo cell-type specificity and preservation of tissue integrity. The importance of achieving cell-type specificity in studying RNA processing is highlighted from the findings of a recent study of alternative splicing (AS) in mouse liver development (Bhate et al., 2015): 1) two-thirds of identified AS events (87 of 132) were cell-type specific; 2) importantly, about a quarter of all AS events (30 of 132) were only revealed by assaying different cell-types separately and were not detectable by assaying the whole liver. The abilities of cTag-PAPERCLIP to focus in vivo profiling on restricted cell populations, exemplified by a more than 190-fold enrichment of microglia cell-type markers in the cTag-PAPERCLIP profile (Fig. S5A) compared with the whole cortex PAPERCLIP profile from our original PAPERCLIP study (Hwang et al., 2016), are crucial in the discovery of APA shifts between different cell-types or between distinct cellular states within a single cell-type in current study.
The full advantage of cTag-PAPERCLIP from preserving tissue integrity is best illustrated in our microglia profiling studies: cTag-PAPERCLIP allows transcript analysis from a single cell-type within a complex tissue, minimizes sample processing time while freezing cellular transcripts in their native states (Fig. 5). This in turn leads to otherwise unattainable sensitivity in detecting molecular changes between activated and naïve states in single cell populations in the brain (Fig. 6E), which is especially critical for less abundant genes such as Araf. An alternative way to generate cell-type specific expression profiles is to isolate the cell-type of interest ex vivo followed by RNA-seq analysis. Therefore, to provide additional evidence for the technical advantage of cTag-PAPERCLIP, we performed an RNA-seq experiment to assess whether ex vivo microglia isolation by magnetic column, a rapid and high-yield alternative to FACS, is able to detect the Araf APA switch in activated microglia (Fig. S5B). Although the LPS treatment fully activated brain microglia as expected (Fig. S5C), RNA-seq failed to identify a statistically significant difference in Araf APA isoform usage between the two treatment groups (Fig. S5D). Importantly, in contrast to cTag-PAPERLCIP, which consistently showed that Araf(S) is the major Araf isoform in resting microglia (Fig. 6E), RNA-seq demonstrated that Araf(L) expression was higher (~1.4-fold) than Araf(S) in column-isolated microglia from PBS-injected mice (Fig. S5D). As we showed in the same experiment that column isolation of microglia resulted in partial activation (Fig. 5B), it is therefore possible that the isolation process altered the expression of the two Araf isoforms and diminished the difference in Araf APA preference between the PBS and LPS groups. These results fully demonstrate the benefits of skipping cell purification and the distinctive advantage of cTag-PAPERCLIP in profiling sensitive cell-types compared with existing technologies.
Our study presents the first in-depth analysis of APA patterns at the individual cell-type level in adult brain in vivo, which detects a distinct APA preference shared by either excitatory or inhibitory neurons compared with non-neuronal cells. We note that many of the identified APA events are not covered by standard annotations and therefore would not be detectable by RNA-seq analysis alone. Further analysis focused on the extreme cases identify 7 genes in which distinct APA isoforms are preferred by neurons over glia, such as the discovery of a mixture of long and short Cdc42 APA isoforms in neurons, one of which is undetectable in astrocytes or microglia (Fig. 3 and Fig. S1). Recent studies demonstrated distinct gene expression patterns and biological behaviors in microglia from different brain regions (Grabert et al., 2016; Tay et al., 2017). While we use the whole brain to generate a representative microglia APA profile and to minimize dissection time in current study, it is an intriguing possibility that microglia might also have different APA preferences by anatomical regions. Characterization of additional cell-types and/or same cell-types from distinct regions by cTag-PAPERCLIP in the future might reveal more genes with distinct APA patterns across different cell populations in the brain.
Combining HITS-CLIP and RNA-seq analysis, we demonstrated a role of PTBP2 in suppressing two of the neuronal isoforms (Map4 and Cdc42) at E18.5 in mouse brain (Fig. 4). MAP4 is known to be a “non-neuronal” member of the MAP2/Tau family of microtubule-associated proteins: MAP4 protein is widely expressed in different tissues but absent from neurons (Dehmelt and Halpain, 2004). While PTBP2 is expressed in mature neurons (Darnell, 2013; Licatalosi et al., 2012), the PTB paralog PTBP1 is highly expressed in glia, non-neural cells and neural progenitor cells (NPCs) (Boutz et al., 2007), and the two isoforms have antagonistic actions (Boutz et al., 2007; Makeyev et al., 2007; Spellman et al., 2007; Zheng et al., 2012). Our analysis of a recent mouse E14.5 RNA-seq data (X. Zhang et al., 2016) showed a concurrent decrease in PTBP1 expression and an increase in Map4 and Cdc42 neuronal isoforms when NPCs differentiate into immature neurons (Fig. S6A–B). Furthermore, an examination of ENCODE eCLIP data identified a strong PTBP1 footprint at the MAP4 neuronal isoform ALE in HepG2 cells (Fig. S6C). Taken together, these results suggest that PTBP1 contributes to the repression of the neuronal ALE usage to produce the full-length MAP4 protein in NPC during development and in glia/non-neural cells at the adult stage (Fig. S6D).
Consistent with our results, the expression of full-length ITSN1 protein (generated by Itsn1 neuronal APA isoform) is restricted to neurons while the truncated ITSN1 protein (generated by Itsn1 glial APA isoform) is expressed in glia and other tissues (Hussain et al., JBC 1999). Our data suggest that Nova2 contributes to the restricted expression of Itsn1 neuronal APA isoform expression (Fig. S6E). Recently, Makeyev and colleagues have independently observed the differential Cdc42 APA isoform usage during neuron development and between neuron and non-neuronal cells (Yap et al., 2016). They identified a role of PTB in suppressing Cdc42 neuronal APA isoform expression in non-neuronal cells and demonstrated an adequate balance between two Cdc42 APA isoforms is critical for normal neuronal function. The full-length ITSN1 protein functions as a guanine nucleotide exchange factor for Cdc42, an activity that is lacking in the truncated ITSN1 (Fig. S2) (Hunter et al., 2013). Therefore, it is likely that the Itsn1 neuronal APA isoform plays an important role in neuronal function, which is supported by the abnormal axon development and deficits in learning and memory observed in the Itsn1 knockout mice (Sengar et al., 2013). Overall, our results and two aforementioned studies (Yap et al., 2016; X. Zhang et al., 2016) together suggest that neurons utilize post-transcriptional mechanisms to regulate and diversify their gene expression program to suit the specialized functions not present in other cell-types.
Our in vivo microglia studies revealed a previously unknown role of Araf APA isoform switching in microglia activation. A role of Araf(S), the Araf short APA isoform, in antagonizing the RAS-ERK signaling and promoting differentiation was recently described in myogenic differentiation and zebrafish embryo development (Xiong et al., 2015; Yokoyama et al., 2007). Our data also suggest that dysregulation of the balance of Araf APA isoforms may result in abnormal microglia activation and aberrant production of inflammatory cytokines, a common feature for many neurological diseases. Future genetic experiments altering the ratio of Araf APA isoforms specifically in microglia should provide further insights into the contribution of their balance in microglia quiescence and activation. We also demonstrate that the Araf APA isoform switch is a common response for microglia/macrophage to multiple TLR ligands. Our results suggest that a detailed dissection of the molecular mechanism responsible for the Araf APA isoform switch will be of interest; we note that inhibition of TAK1, a MAP3K kinase controlling major inflammatory response pathways in microglia and macrophage (Goldmann et al., 2013; Levin et al., 2016), does not prevent the downregulation of Araf(S) in RAW264.7 cells (Fig. S7), suggesting that the p38, JNK and NF-kappaB pathways are not major contributors to the Araf isoform switch in this cell-type.
A major challenge in biomedical studies is to understand the biological functions of individual cell-types. In the past few years, several new tools utilizing mouse transgenic technology for molecular profiling at individual cell-type level have been described (Reviewed in (Handley et al., 2015)). We show the cTag-PABP mouse provides a robust platform for assessing the normal mouse transcriptome, as evidenced by the consistency between our cTag-PAPERCLIP data and data independently obtained through analysis of endogenous PABP using the original PAPERCLIP (Fig. 1F), as well as comparison with transcripts identified with a variety of different methods from mouse brain (Fig. 3, Fig. S1 and Fig. 6F). Furthermore, its compatibility with the popular Cre/Lox technology makes it versatile and directly applicable for studying different biological questions in multiple cell-types. We demonstrate the sensitivity of cTag-PAPERCLIP in identifying APA from individual cell-types that are not evident from analysis of whole brain (Fig. 3, Fig. S1 and Fig. 6E); importantly, cTag-PAPERCLIP can also detect dynamic changes within a single cell-type, as demonstrated in analysis of quiescent versus activated microglia, cells which produce about one-tenth of RNA that neurons do (Srinivasan et al., 2016). Future development of the cTag-PAPERCLIP method by incorporating new CLIP techniques should further enhance its sensitivity (Van Nostrand et al., 2016; Zarnegar et al., 2016). Establishing cTag-PAPERCLIP as an important in vivo molecular profiling tool with application to individual cell-types and/or cell-states offers a new means of gaining insights into cell-specific biology within intact living tissues.
STAR METHODS
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| HtzGFP19C8 (Mouse monoclonal anti-GFP) | Heiman et al. Nat Protocols. 2014 | N/A |
| HtzGFP19F7 (Mouse monoclonal anti-GFP) | Heiman et al. Nat Protocols. 2014 | N/A |
| Mouse monoclonal anti-GFP | Santa Cruz | Cat#sc-9996; RRID: AB_627695 |
| Mouse monoclonal anti-HA | Biolegend | Cat#901501; RRID: AB_2565006 |
| Mouse monoclonal anti-Araf | Santa Cruz | Cat#sc-166771; RRID: AB_2060508 |
| Rat anti-GFP | Nacalai Tesque | Cat#GF090R; RRID: AB_2314545 |
| Rabbit anti-NeuN | Millipore | Cat#ABN78; RRID: AB_10807945 |
| Mouse anti-Gad67 | Millipore | Cat#MAB5406; RRID: AB_2278725 |
| Mouse anti-GFAP | Progen | Cat#GF12.24 |
| Rabbit anti-Iba1 | Wako | Cat#019-197411 |
| FITC Mouse Anti-Mouse H-2Kb, Clone AF6-88.5 | BD Biosciences | Cat#562002; RRID: AB_10924590 |
| PE Rat Anti-Mouse CD11b, Clone M1/70 | BD Biosciences | Cat#553311; RRID: AB_394775 |
| PerCP Rat Anti-Mouse CD45, Clone 30-F11 | BD Biosciences | Cat#557235; RRID: AB_396609 |
| APC Hamster Anti-Mouse CD11c, Clone HL3 | BD Biosciences | Cat#561119; RRID: AB_10562405 |
| APC Rat Anti-Mouse TNF, Clone MP6-XT22 | BD Biosciences | Cat#554420; RRID: AB_398553 |
| IL-1 beta (Pro-form) Monoclonal Antibody (NJTEN3), PE-Cyanine7 | eBioscience | Cat#25-7114-82; RRID: AB_2573526 |
| Custom rabbit anti-serum against PABP | This Paper | N/A |
| Bacterial and Virus Strains | ||
| None | ||
| Biological Samples | ||
| None | ||
| Chemicals, Peptides, and Recombinant Proteins | ||
| Lipopolysaccharide | Sigma | Cat#L4524 |
| Lipopolysaccharide | InvivoGen | Cat#LPS-EK |
| Pam3CSK4 | InvivoGen | Cat#Pam3CSK4 |
| Polyinosinic-polycytidylic acid | Enzo Life Sciences | Cat#ALX-746-021 |
| PD325901 | Millipore | Cat#444968 |
| (5Z)-7-OxoZeaenol | Millipore | Cat#499610 |
| Critical Commercial Assays | ||
| Neural Tissue Dissociation Kit (P) | Miltenyi Biotec | Cat#130-092-628 |
| Debris Removal Solution | Miltenyi Biotec | Cat#130-109-398 |
| CD11b (Microglia) MicroBeads | Miltenyi Biotec | Cat#130-093-636 |
| Dynabeads mRNA Purification Kit | Ambion | Cat#61006 |
| Ribo-Zero rRNA Removal Kit (Human/Mouse/Rat) | Illumina | Cat#MRZH116 |
| TruSeq RNA Sample Preparation Kit v2 | Illumina | Cat#RSS-122-2001 |
| Deposited Data | ||
| cTag-PAPERCLIP | This Paper | GEO: GSE94054 |
| RNA-seq | This Paper | GEO: GSE94054 |
| See Table S1 for additional GEO/SRA datasets. | ||
| Experimental Models: Cell Lines | ||
| Mouse: Neuro2a | ATCC | Cat#CCL-131 |
| Mouse: RAW264.7 | ATCC | Cat#TIB-71 |
| Mouse: BV2 | Jellinck et al. J Steroid Biochem Mol Biol 2006. | N/A |
| Human: HEK293 | ATCC | Cat#CRL-1573 |
| Experimental Models: Organisms/Strains | ||
| cTag-PABP | This Paper | N/A |
| CMV-Cre | The Jackson Lab | Cat#006054 |
| Camk2a-Cre | The Jackson Lab | Cat#005359 |
| Gad2-Cre | The Jackson Lab | Cat#010802 |
| mGfap-Cre | The Jackson Lab | Cat#012886 |
| Cx3cr1-Cre | MMRRC | Cat#036395-UCD |
| Ptbp2 knockout | Licatalosi et al., Genes Dev. 2012 | N/A |
| Oligonucleotides | ||
| See Table S5 for the list of primers. | ||
| Recombinant DNA | ||
| Flag/HA-pcDNA3.1 | Addgene | #52535 |
| Flag/HA-ARAF(L) | This Paper | N/A |
| Flag/HA-ARAF(S) | This Paper | N/A |
| pCherry | This Paper | N/A |
| pCherry-Itsn1 | This Paper | N/A |
| pCherry-Map4 | This Paper | N/A |
| pCherry-Cdc42 | This Paper | N/A |
| pCherry-Aplp2 | This Paper | N/A |
| Software and Algorithms | ||
| CIMS package | Hwang et al. Cell Reports. 2016. | https://github.com/chaolinzhanglab/ctk/tree/v1.0.3 |
| STAR 2.5.2 | Dobin et al. Bioinformatics. 2013. | https://github.com/alexdobin/STAR |
| Kallisto 0.43.0 | Bray et al. Nat Biotechnology. 2016. | https://github.com/pachterlab/kallisto |
| Sleuth | Pimentel et al. Nat Methods. 2017. | https://github.com/pachterlab/sleuth |
| Other | ||
| cTag-PAPERCLIP protocol | This Paper | N/A |
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents generated in this study should be directed to and will be fulfilled by the Lead Contact, Robert Darnell (darnelr@rockefeller.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Generation of the cTag-PABP mouse line
The targeting vector, generated through standard restriction cloning, contains the endogenous Pabpc1 exons 14–15 sequence, a synthetic poly(A) site preceding a FRT-Neo-FRT cassette and the last coding exon of Pabpc1 fused to AcGFP sequence (Fig. 1B). The construct was electroporated into Bruce 4 ES cells. Correctly targeted ES cells were then injected into C57BL/6J blastocysts to screen for chimeras. Chimeric males were bred to C57BL/6J females to generate Pabpc1fl. Removal of the FRT-Neo-FRT cassette was done by breeding Pabpc1fl to ACTB-FLPe mice (Stock No. 005703, JAX) to generate Pabpc1Δneo. No phenotypical difference was observed between Pabpc1fl and Pabpc1Δneo. Genotyping of cTag-PABP mouse was performed at Transnetyx with standard assays for AcGFP and Neomycin.
Animal experiments
All procedures were conducted according to the Institutional Animal Care and Use Committee (IACUC) guidelines at the Rockefeller University. CMV-Cre (Stock No. 006054), Camk2a-Cre (Stock No. 005359), Gad2-Cre (Stock No. 010802), mGfap-Cre (Stock No. 012886) and wildtype C57BL/6J mice were obtained from the Jackson Lab. Cx3cr1-Cre mouse was generated by the GENSAT project (Gong et al., 2007) and obtained from MMRRC. Mice of both sexes were pooled for the study. Sex-specific effects were not tested. To activate microglia, 8–14 week-old adult Cx3cr1-Cre; Pabpc1cTag or wildtype B6 mice received daily intraperitoneally injection of 1mg/Kg lipopolysaccharide (L4524, Sigma) or phosphate-buffered saline for 4 consecutive days before sacrifice. For genotyping PCR experiment in Fig. 1C, mouse genomic DNA was extracted from a tail snip using Gentra Puregene kit (Qiagen); the primer sequences are listed in Table S5. For cTag-PAPERCLIP microglia profiling, Cx3cr1-Cre; Pabpc1cTag mice were perfused manually with ice-cold phosphate-buffered saline before brain removal. The dissected out brain was immediately snap-frozen in liquid nitrogen and pulverized in the Cellcrusher tissue pulverizer (pre-chilled in liquid nitrogen). Pulverized brain powder was transferred to petri dishes on dry ice for UV-crosslinking. The crosslinked brain powder was stored at −80°C until usage. Dissection and processing of mouse brain cortex for other cTag-PAPERCLIP profiling was performed following standard procedures (Hwang et al., 2016). We observed occasionally leakiness of GFP expression in neuronal lineages in Cx3cr1-Cre; Pabpc1cTag mice (<10% of tested animals). A quality-control qPCR assay was performed for every sample before cTag-PAPERCLIP profiling (Fig. S3A). The initial cTag-PAPERCLIP experiments were performed in double heterozygous progeny from breeding cross between Pabpc1fl and Cre drivers. To maintain consistency, all subsequent cTag-PAPERCLIP experiments were performed using Pabpc1fl as the breeding partner to Cre drivers.
Cell culture
HEK293, Neuro-2a, RAW264.7 and BV2 (Jellinck et al., 2006) cells (gender: not known) were grown in Dulbecco’s modified Eagle’s medium supplemented with 10% FBS. Transfection of HA/FLAG-tagged ARAF APA isoform expression constructs were transfected into Neuro-2a cells using Lipofectmine 2000 or into RAW264.7 cells using Lipofectamine LTX following manufacturer’s instructions. For TLR activation, BV2 and RAW264.7 cells were treated by the following ligands for 6 hours: 10μg/mL LPS (InvivoGen, LPS-EK), 250ng/mL PAM (InvivoGen, Pam3CSK4), 10μg/mL Poly(I:C) (Enzo Life Sciences). For MEK inhibition, RAW264.7 cells were pre-treated with DMSO or 10μM PD325901 (Millipore, 444968) for an hour before being treated with 10μg/mL LPS with DMSO or 10μg/mL LPS with 1μM PD325901 for 6 hours. For TAK inhibition, (5Z)-7-Oxozeaenol (Millipore, 499610) was used at a final concentration of 100nM. To assay Araf APA isoform function in inflammatory cytokine production, RAW 264.7 cells were first transfected with Araf APA isoform expression constructs overnight followed by LPS treatment for 6 hours before fixation for flow cytometry analysis. For mini-gene assay, 0.8 μg of pCherry reporter and 1.5 μg of GFP, NOVA2 or PTBP2-expressing constructs per well were transfected into HEK293 cells in 6-well plates with Lipofectamine 2000 transfection reagent (Thermo Fisher Scientific, Cat. #11668027). Cells were harvested 24 hrs after transfection to assay exon inclusion/exclustion by qRT-PCR.
METHD DETAILS
cTag-PAPERCLIP and analysis
The procedures for sample preparation and library construction were previously described (Hwang et al., 2016). Mouse monoclonal anti-GFP clones 19F7 and 19C8 (Heiman et al., 2014) were used for immunoprecipitation. The complete protocol is detailed in Methods S1. To minimize batch effects, the entire process was performed independently for replicate experiments, sometimes using primers with different indices. Individual cTag-PAPERCLIP libraries were multiplexed and sequenced by HiSeq 2500 or MiSeq (Illumina) to obtain 100-nt (HiSeq) or 75-nt (MiSeq) single-end reads. The procedures for raw read processing, mapping and poly(A) site annotation were previously described (Hwang et al., 2016).
RNA-seq and analysis
For Ptbp2-KO brain cortex RNA-seq, E18.5 mouse cortex RNA (three biological replicates) from Ptbp2-KO mice (Licatalosi et al., 2012) was prepared using Trizol (Ambion); ribosomal RNA was removed from 1 μg RNA using Ribo-Zero rRNA removal Kit (Illumina). RNA-seq libraries were prepared using TruSeq RNA Sample Preparation Kit v2 (Illumina) following manufacturer’s instructions. High-throughput sequencing was performed on HiSeq 2500 (Illumina) to obtain 125 nucleotide paired-end reads at New York Genome Center. RNA-seq mapping to mouse genome (mm10) was performed using STAR aligner (Dobin et al., 2013). APA isoform abundance in RNA-seq datasets was estimated using Kallisto and Sleuth (Bray et al., 2016; Pimentel et al., 2017) with customized aggregation index. Additional validation of cTag-PAPERCLIP results was performed using bedtools to count mapped reads (v2.26.0). The GEO/SRA datasets used for analysis are listed in Table S1.
Immunohistochemistry
Immunohistochemistry was performed as previously described (Saito et al., 2016). Briefly, adult mice brains were perfused with PBS and fixed with 4% praraformaldehyde (PFA)/PBS at 4 degrees overnight, and sequentially replaced to 15% sucrose/PBS and 30% sucrose/PBS at 4 degrees for cryo-protection, then embedded in OCT compound. Frozen brains were sliced into 30 μm thick sections on a cryostat (CM3050S, LEICA). Brain sections were subjected to immunohistochemistry. These samples were washed three times with PBS at room temperature (RT), incubated with 0.2% Triton X-100/PBS at RT, blocked with 1.5% normal donkey serum (NDS)/PBS at RT, and then incubated overnight at 4 degrees with primary antibodies in 1.5% NDS/PBS followed by incubation with Alexa 488, 555 or 647 conjugated donkey secondary antibodies (1:1000) in 1.5% NDS/PBS. Images of immunostained specimens were collected by BZ-X700 (KEYENCE) microscope.
Primary antibodies used for immunohistochemistory were: rat anti-GFP (nacalai tesque, GF090R, 1/1,000), rabbit anti-NeuN (Millipore, ANB78, 1/1,000), mouse anti-Gad67 (Millipore, MAB5406, 1/1,000), mouse anti-GFAP (Progen, GF12.24, 1/1,000), rabbit anti-Iba1 (Wako, 019-197411/1,000), mouse anti-Araf (Santa Cruz, sc-166771, 1/500).
Column-based microglia isolation and RNA-seq analysis
All procedures were performed per manufacturer’s instructions. Mice were perfused with 30ml of heparinized PBS. Brains were excised, minced and two brains were placed into C tube and processed as per instructions for the Neural Tissue Dissociation Kit P (Miltenyi Biotec) using the gentleMACS™ Octo Dissociator with Heaters (Miltenyi Biotec, protocol 37_ABDK). Once digested, the debris and myelin was removed using the Debris Removal Solution (Miltenyi Biotec) followed by CD11b positive selection using Microglia CD11b beads (Miltenyi). Cells were counted, washed in PBS and pelleted prior to isolation of RNA. Total RNA from isolated microglia was prepared using Trizol (Ambion) and High Pure RNA Isolation Kit (Roche). The Total RNA was then further purified for polyadenylated RNA by using 150ng Total RNA in Dynabeads mRNA Purification Kit (Ambion). The libraries were prepared by TruSeq RNA Sample Preparation Kit v2 (Illumina) following manufacturer’s instructions. High-throughput sequencing was performed on MiSeq (Illumina) to obtain 67 nucleotide single-end reads.
Flow cytometry analysis
For monitoring mouse brain microglia activation and peripheral monocytic blood cell infiltration, microglia were isolated from each individual brain as previously described (Cardona et al., 2006); for microglia sorting, we pooled 18 mice per group. Briefly, brains were removed from mice perfused with 20 ml cold heparinized saline and mechanically dissociated using a dounce homogenizer (VWR) in HBSS with Ca2+ and Mg2+ (Gibco) containing 500ug/ml Collagenase D (Roche), 5mg/ml Dispase II (Roche), 20U/ml DNase I (Roche) and 10mM HEPES (Gibco). The resulting cell suspension was fractionated on a 30/37/70% percoll gradient. Cells were collected from the 37/70% interface and FcR blocked (CD16/CD32, clone 2.4G2) prior to staining with anti-MHC 1 (H-2Kb, clone AF6-88.5)-FITC, anti-CD45 Ab (clone 30-F11)-PerCP, anti-CD11b (clone M1/70)-PE anti-CD11c Ab (clone HL3)-APC. All antibodies were obtained from Becton Dickenson Biosciences. Samples were analyzed with BD FACSCalibur™ and FlowJo10.6. For cell sorting, live microglia (DAPI excluding, CD45int and CD11b+) were isolated using the FACS AriaII cell sorter (BD Biosciences).
Inflammatory cytokine production was assayed by first blocking secretion using GolgiStop (BD Biosciences) at the same time of LPS treatment. Cells were harvested, and FcR blocked (CD16/CD32, clone 2.4G2), and where indicated surface stained with anti-HA.11 Epitope Tag (clone16B12, BioLegend) followed by Goat anti-mouse FITC (Jackson ImmunoResearch). Cells were stained using LIVE/DEAD® Fixable Aqua (Invitrogen), prior to fixing, permeabilization (BD Fix Permeabilization kit, BD Biosciences) and intracellular staining with IL1-Beta Pro-form (clone NJTEN3, eBioscience)-PE-Cy7 or TNF-α (clone MP6-XT22, BD Biosciences)-APC. Samples were analyzed on MACSQuant® (Miltenyi Biotec) and FlowJo10.6.
SDS-PAGE and western blots
30~50μg total protein per lane was separated on 10% Novex NuPAGE Bis-Tris gels (Invitrogen) and transferred to nitrocellulose membrane following standard procedures. The following antibodies are used for western blotting: mouse monoclonal anti-GFP (sc-9996), mouse monoclonal anti-HA (Biolegend, 901501), mouse monoclonal anti-Araf (sc-166771) and custom rabbit anti-serum (Covance) that is able to recognize both endogenous mouse PABP and conditionally expressed PABP-GFP.
Quantitative PCR
Reverse transcription was performed using an anchored primer and SuperScript III (Invitrogen) following manufacturer’s instructions on 1μg DNase I (Invitrogen)-digested total RNA or 20 ng poly(A)-selected RNA. qRT-PCR was performed using FastStart SYBR Green Master mix (Roche) in triplicates. All primer sequences are listed in Table S5. The cycling parameters were: 95°C for 10 min. followed by 40 cycles of 95°C for 15 sec., 58°C for 30 sec., 72°C for 20 sec. Quantification was calculated using the ΔΔCt method with Rpl13a as the endogenous control.
Molecular Cloning
Mouse Araf short and long APA isoforms were PCR-amplified from wildtype B6 mouse cortex cDNAs and then inserted into Flag/HA-pcDNA3.1 (#52535, Addgene) vector between XbaI and EcoRI sites. All insert sequences were verified by Sanger sequencing. All primer sequences are listed in Table S5, and plasmids are listed in Table S6.
QUANTIFICATION AND STATISTICAL ANALYSIS
Permutation test to compare the cTag-PAPERCLIP APA profiles from different cell-types was performed by the R package “perm” (Fay and Shaw, 2010) followed by multiple test correction of the derived p-values using the local FDR method (Storey and Tibshirani, 2003) (Table S2). Statistical analyses to confirm differential APA pattern using individual cell-type RNA-seq data were performed using Sleuth to compute q-values or by Fisher’s exact test in R (Table S3). Analysis of APA shift during microglia activation in Cx3cr1-Cre; Pabpc1fl mice was performed as described (Hwang et al., 2016), with the following modification of pooling the PAPERCLIP biological replicate reads for each condition and conducting Fisher’s exact test in R to statistically assess the shift in poly(A) site usage given the library complexity (Table S4). Expression of APA isoforms in RNA-seq data was estimated using Kallisto (Figs. 3, 4 and S1). Statistical significance for differential gene/isoform expression was calculated using Sleuth or Student’s t-test (Figs. 4, 6, S2, and S6–9). For the analysis shown in Fig. S3, the gene expression levels were first estimated by read counting (cTag-PAPERCLIP) or using Kallisto (RNA-seq); for each library, the estimated gene expression value was then converted to a Z-score in R before assessing the statistical significance by Wilcoxon rank-sum test.
DATA AND SOFTWARE AVAILABILITY
The GEO accession number for the high-throughput sequencing data (cTag-PAPERCLIP and RNA-seq) originated from this study is GSE94054.
ADDITIONAL RESOURCES
The complete cTag-PAPERCLIP protocol is detailed in Method S1.
Supplementary Material
Table S1. Information of cTag-PAPERCLIP libraries, gene lists, and publicly available datasets used in the study, related to Figure 1.
Table S2. Permutation analysis results, related to Figure 2.
Table S3. Summary of RNA-seq analysis for cTag-PAPERCLIP validation, related to Figure 2.
Table S4. List of genes identified by cTag-PAPERCLIP to exhibit APA shift in LPS-activated brain microglia, related to Figure 6.
Table S5. List of primers, related to the STAR Methods.
Highlights.
cTag-PAPERCLIP generates in vivo profiles of cell-type-specific polyadenylated mRNAs.
cTag-PAPERCLIP preserves tissue integrity, capturing mRNAs in the cell’s native state.
Distinct APA preference contributes to protein diversity between brain cell-types.
Activated brain microglia upregulate full-length kinase-active ARAF through APA.
Acknowledgments
We thank Chingwen Yang (the Rockefeller University Gene Targeting Resource Center) and Sarah Van Driesche (the Darnell Lab) for their help in generation of the cTag-PABP mouse. We thank Connie Zhao and Christine Lai at the Rockefeller University Genomics Resource Center for their support in high-throughput sequencing. We thank Aldo Mele for guidance during the development of cTag-PAPERCLIP protocol and Salina Parveen for help with flow cytometry experiments. H-W.H. is an HHMI Fellow of the Life Sciences Research Foundation. Y.S. is a JSPS Postdoctoral Fellow for Research Abroad. This work was also in part supported by grants from the National Institutes of Health (NS034389, NS081706 and NS097404) and Simons Foundation (SFARI 240432) to R.B.D. R.B.D. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions: H-W.H. and R.B.D. conceived the project. H-W.H. designed experiments. H-W.H. designed and generated the cTag-PABP mouse. H-W.H. developed the cTag-PAPERCLIP protocol. Y.S. and I.Z-S. performed immunohistochemistry experiments. N.E.B. performed flow cytometry and microglia purification experiments. Y.S. and Y.T. performed NOVA2 and PTBP2 reporter experiments. Y.T. performed qRT-PCR experiments from wildtype/Nova2-KO/Ptbp2-KO E18.5 mouse cortex. J.J.F. performed RNA-seq experiments. H-W.H. performed all other experiments. H-W.H. and C.Y.P. performed informatics analyses. H-W.H. and R.B.D wrote the manuscript.
Conflict of interest statement: None declared.
References
- Bennett ML, Bennett FC, Liddelow SA, Ajami B, Zamanian JL, Fernhoff NB, Mulinyawe SB, Bohlen CJ, Adil A, Tucker A, et al. New tools for studying microglia in the mouse and human CNS. Proc Natl Acad Sci USA. 2016;113:E1738–E1746. doi: 10.1073/pnas.1525528113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhate A, Parker DJ, Bebee TW, Ahn J, Arif W, Rashan EH, Chorghade S, Chau A, Lee JH, Anakk S, et al. ESRP2 controls an adult splicing programme in hepatocytes to support postnatal liver maturation. Nat Commun. 2015;6:8768. doi: 10.1038/ncomms9768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutet SC, Cheung TH, Quach NL, Liu L, Prescott SL, Edalati A, Iori K, Rando TA. Alternative polyadenylation mediates microRNA regulation of muscle stem cell function. Cell Stem Cell. 2012;10:327–336. doi: 10.1016/j.stem.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutz PL, Stoilov P, Li Q, Lin CH, Chawla G, Ostrow K, Shiue L, Ares M, Black DL. A post-transcriptional regulatory switch in polypyrimidine tract-binding proteins reprograms alternative splicing in developing neurons. Genes Dev. 2007;21:1636–1652. doi: 10.1101/gad.1558107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray NL, Pimentel H, Melsted P, Pachter L. Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol. 2016;34:525–527. doi: 10.1038/nbt.3519. [DOI] [PubMed] [Google Scholar]
- Butovsky O, Jedrychowski MP, Moore CS, Cialic R, Lanser AJ, Gabriely G, Koeglsperger T, Dake B, Wu PM, Doykan CE, et al. Identification of a unique TGF-β–dependent molecular and functional signature in microglia. Nat Neurosci. 2014;17:131–143. doi: 10.1038/nn.3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardona AE, Huang D, Sasse ME, Ransohoff RM. Isolation of murine microglial cells for RNA analysis or flow cytometry. Nat Protoc. 2006;1:1947–1951. doi: 10.1038/nprot.2006.327. [DOI] [PubMed] [Google Scholar]
- Castelo-Branco P, Furger A, Wollerton M, Smith C, Moreira A, Proudfoot NJ. Polypyrimidine tract binding protein modulates efficiency of polyadenylation. Mol Cell Biol. 2004;24:4174–4183. doi: 10.1128/MCB.24.10.4174-4183.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Jalabi W, Shpargel KB, Farabaugh KT, Dutta R, Yin X, Kidd GJ, Bergmann CC, Stohlman SA, Trapp BD. Lipopolysaccharide-induced microglial activation and neuroprotection against experimental brain injury is independent of hematogenous TLR4. J Neurosci. 2012;32:11706–11715. doi: 10.1523/JNEUROSCI.0730-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevrier N, Mertins P, Artyomov MN, Shalek AK, Iannacone M, Ciaccio MF, Gat-Viks I, Tonti E, DeGrace MM, Clauser KR, et al. Systematic Discovery of TLR Signaling Components Delineates Viral-Sensing Circuits. Cell. 2011;147:853–867. doi: 10.1016/j.cell.2011.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell RB. RNA protein interaction in neurons. Annu Rev Neurosci. 2013;36:243–270. doi: 10.1146/annurev-neuro-062912-114322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell RB. HITS-CLIP: panoramic views of protein-RNA regulation in living cells. WIREs RNA. 2010;1:266–286. doi: 10.1002/wrna.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehmelt L, Halpain S. The MAP2/Tau family of microtubule-associated proteins. Genome Biol. 2004;6:204. doi: 10.1186/gb-2004-6-1-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derti A, Garrett-Engele P, MacIsaac KD, Stevens RC, Sriram S, Chen R, Rohl CA, Johnson JM, Babak T. A quantitative atlas of polyadenylation in five mammals. Genome Res. 2012;22:1173–1183. doi: 10.1101/gr.132563.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay MP, Shaw PA. Exact and Asymptotic Weighted Logrank Tests for Interval Censored Data: The interval R package. J Stat Softw. 2010;36:i02. doi: 10.18637/jss.v036.i02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavell SW, Kim TK, Gray JM, Harmin DA, Hemberg M, Hong EJ, Markenscoff-Papadimitriou E, Bear DM, Greenberg ME. Genome-Wide Analysis of MEF2 Transcriptional Program Reveals Synaptic Target Genes and Neuronal Activity-Dependent Polyadenylation Site Selection. Neuron. 2008;60:1022–1038. doi: 10.1016/j.neuron.2008.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldmann T, Wieghofer P, Jordão MJC, Prutek F, Hagemeyer N, Frenzel K, Amann L, Staszewski O, Kierdorf K, Krueger M, et al. Origin, fate and dynamics of macrophages at central nervous system interfaces. Nat Immunol. 2016;17:797–805. doi: 10.1038/ni.3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldmann T, Wieghofer P, Müller PF, Wolf Y, Varol D, Yona S, Brendecke SM, Kierdorf K, Staszewski O, Datta M, et al. A new type of microglia gene targeting shows TAK1 to be pivotal in CNS autoimmune inflammation. Nat Neurosci. 2013;16:1618–1626. doi: 10.1038/nn.3531. [DOI] [PubMed] [Google Scholar]
- Gong S, Doughty M, Harbaugh CR, Cummins A, Hatten ME, Heintz N, Gerfen CR. Targeting Cre recombinase to specific neuron populations with bacterial artificial chromosome constructs. J Neurosci. 2007;27:9817–9823. doi: 10.1523/JNEUROSCI.2707-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabert K, Michoel T, Karavolos MH, Clohisey S, Baillie JK, Stevens MP, Freeman TC, Summers KM, McColl BW. Microglial brain region–dependent diversity and selective regional sensitivities to aging. Nat Neurosci. 2016;19:504–516. doi: 10.1038/nn.4222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handley A, Schauer T, Ladurner AG, Margulies CE. Designing Cell-Type-Specific Genome-wide Experiments. Mol Cell. 2015;58:621–631. doi: 10.1016/j.molcel.2015.04.024. [DOI] [PubMed] [Google Scholar]
- Häcker H, Redecke V, Blagoev B, Kratchmarova I, Hsu LC, Wang GG, Kamps MP, Raz E, Wagner H, Häcker G, et al. Specificity in Toll-like receptor signalling through distinct effector functions of TRAF3 and TRAF6. Nature. 2005;439:204–207. doi: 10.1038/nature04369. [DOI] [PubMed] [Google Scholar]
- Heiman M, Kulicke R, Fenster RJ, Greengard P, Heintz N. Cell type–specific mRNA purification by translating ribosome affinity purification (TRAP) Nat Protoc. 2014;9:1282–1291. doi: 10.1038/nprot.2014.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henn A, Lund S, Hedtjärn M, Schrattenholz A, Pörzgen P, Leist M. The suitability of BV2 cells as alternative model system for primary microglia cultures or for animal experiments examining brain inflammation. ALTEX. 2009;26:83–94. doi: 10.14573/altex.2009.2.83. [DOI] [PubMed] [Google Scholar]
- Hoque M, Ji Z, Zheng D, Luo W, Li W, You B, Park JY, Yehia G, Tian B. Analysis of alternative cleavage and polyadenylation by 3′ region extraction and deep sequencing. Nat Methods. 2013;10:133–139. doi: 10.1038/nmeth.2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter M, Russo A, O’Bryan J. Emerging Roles for Intersectin (ITSN) in Regulating Signaling and Disease Pathways. Int J Mol Sci. 2013;14:7829–7852. doi: 10.3390/ijms14047829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang HW, Park CY, Goodarzi H, Fak JJ, Mele A, Moore MJ, Saito Y, Darnell RB. PAPERCLIP Identifies MicroRNA Targets and a Role of CstF64/64tau in Promoting Non-canonical poly(A) Site Usage. Cell Rep. 2016;15:423–435. doi: 10.1016/j.celrep.2016.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ince-Dunn G, Okano HJ, Jensen KB, Park WY, Zhong R, Ule J, Mele A, Fak JJ, Yang C, Zhang C, et al. Neuronal Elav-like (Hu) Proteins Regulate RNA Splicing and Abundance to Control Glutamate Levels and Neuronal Excitability. Neuron. 2012;75:1067–1080. doi: 10.1016/j.neuron.2012.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan CH, Friedman RC, Ruby JG, Bartel DP. Formation, regulation and evolution of Caenorhabditis elegans 3′UTRs. Nature. 2011;469:97–101. doi: 10.1038/nature09616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jellinck PH, Kaufmann M, Gottfried-Blackmore A, Croft G, Byford V, McEwen BS, Jones G, Bulloch K. Dehydroepiandrosterone (DHEA) metabolism in the brain: Identification by liquid chromatography/mass spectrometry of the delta-4-isomer of DHEA and related steroids formed from androstenedione by mouse BV2 microglia. J Steroid Biochem Mol Biol. 2006;98:41–47. doi: 10.1016/j.jsbmb.2005.07.006. [DOI] [PubMed] [Google Scholar]
- Jenal M, Elkon R, Loayza-Puch F, van Haaften G, Kühn U, Menzies FM, Oude Vrielink JAF, Bos AJ, Drost J, Rooijers K, et al. The poly(A)-binding protein nuclear 1 suppresses alternative cleavage and polyadenylation sites. Cell. 2012;149:538–553. doi: 10.1016/j.cell.2012.03.022. [DOI] [PubMed] [Google Scholar]
- Leggere JC, Saito Y, Darnell RB, Tessier-Lavigne M, Junge HJ, Chen Z, Zoghbi HY. NOVA regulates Dcc alternative splicing during neuronal migration and axon guidance in the spinal cord. Elife. 2016;5:e14264. doi: 10.7554/eLife.14264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin RS, Hertz NT, Burlingame AL, Shokat KM, Mukherjee S. Innate immunity kinase TAK1 phosphorylates Rab1 on a hotspot for posttranslational modifications by host and pathogen. Proc Natl Acad Sci USA. 2016;113:E4776–E4783. doi: 10.1073/pnas.1608355113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Zheng S, Han A, Lin CH, Stoilov P, Fu XD, Black DL, Blencowe BJ. The splicing regulator PTBP2 controls a program of embryonic splicing required for neuronal maturation. Elife. 2014;3:e01201. doi: 10.7554/eLife.01201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lianoglou S, Garg V, Yang JL, Leslie CS, Mayr C. Ubiquitously transcribed genes use alternative polyadenylation to achieve tissue-specific expression. Genes Dev. 2013;27:2380–2396. doi: 10.1101/gad.229328.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licatalosi DD, Darnell RB. RNA processing and its regulation: global insights into biological networks. Nat Rev Genet. 2010;11:75–87. doi: 10.1038/nrg2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licatalosi DD, Mele A, Fak JJ, Ule J, Kayikci M, Chi SW, Clark TA, Schweitzer AC, Blume JE, Wang X, et al. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature. 2008;456:464–469. doi: 10.1038/nature07488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licatalosi DD, Yano M, Fak JJ, Mele A, Grabinski SE, Zhang C, Darnell RB. Ptbp2 represses adult-specific splicing to regulate the generation of neuronal precursors in the embryonic brain. Genes Dev. 2012;26:1626–1642. doi: 10.1101/gad.191338.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maatz H, Jens M, Liss M, Schafer S, Heinig M, Kirchner M, Adami E, Rintisch C, Dauksaite V, Radke MH, et al. RNA-binding protein RBM20 represses splicing to orchestrate cardiac pre-mRNA processing. J Clin Invest. 2014;124:3419–3430. doi: 10.1172/JCI74523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makeyev EV, Zhang J, Carrasco MA, Maniatis T. The MicroRNA miR-124 Promotes Neuronal Differentiation by Triggering Brain-Specific Alternative Pre-mRNA Splicing. Mol Cell. 2007;27:435–448. doi: 10.1016/j.molcel.2007.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura P, Sanfilippo P, Shenker S, Lai EC. Alternative polyadenylation in the nervous system: to what lengths will 3′ UTR extensions take us? Bioessays. 2014;36:766–777. doi: 10.1002/bies.201300174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo A, Mukamel EA, Davis FP, Luo C, Henry GL, Picard S, Urich MA, Nery JR, Sejnowski TJ, Lister R, et al. Epigenomic Signatures of Neuronal Diversity in the Mammalian Brain. Neuron. 2015;86:1369–1384. doi: 10.1016/j.neuron.2015.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikodemova M, Watters JJ. Efficient isolation of live microglia with preserved phenotypes from adult mouse brain. J Neuroinflammation. 2012;9:147. doi: 10.1186/1742-2094-9-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okaty BW, Sugino K, Nelson SB. A Quantitative Comparison of Cell-Type-Specific Microarray Gene Expression Profiling Methods in the Mouse Brain. PLoS ONE. 2011;6:e16493. doi: 10.1371/journal.pone.0016493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimentel H, Bray NL, Puente S, Melsted P, Pachter L. Differential analysis of RNA-seq incorporating quantification uncertainty. Nat Methods. 2017;14:687–690. doi: 10.1038/nmeth.4324. [DOI] [PubMed] [Google Scholar]
- Ponomarev ED, Veremeyko T, Barteneva N, Krichevsky AM, Weiner HL. MicroRNA-124 promotes microglia quiescence and suppresses EAE by deactivating macrophages via the C/EBP-α-PU. 1 pathway. Nat Med. 2011;17:64–70. doi: 10.1038/nm.2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prinz M, Priller J. Microglia and brain macrophages in the molecular age: from origin to neuropsychiatric disease. Nat Rev Neurosci. 2014;15:300–312. doi: 10.1038/nrn3722. [DOI] [PubMed] [Google Scholar]
- Saito Y, Miranda-Rottmann S, Ruggiu M, Park CY, Fak JJ, Zhong R, Duncan JS, Fabella BA, Junge HJ, Chen Z, et al. NOVA2-mediated RNA regulation is required for axonal pathfinding during development. Elife. 2016;5:e14371. doi: 10.7554/eLife.14371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandberg R, Neilson JR, Sarma A, Sharp PA, Burge CB. Proliferating cells express mRNAs with shortened 3′ untranslated regions and fewer microRNA target sites. Science. 2008;320:1643–1647. doi: 10.1126/science.1155390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengar AS, Ellegood J, Yiu AP, Wang H, Wang W, Juneja SC, Lerch JP, Josselyn SA, Henkelman RM, Salter MW, et al. Vertebrate Intersectin1 Is Repurposed to Facilitate Cortical Midline Connectivity and Higher Order Cognition. J Neurosci. 2013;33:4055–4065. doi: 10.1523/JNEUROSCI.4428-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepard PJ, Choi EA, Lu J, Flanagan LA, Hertel KJ, Shi Y. Complex and dynamic landscape of RNA polyadenylation revealed by PAS-Seq. RNA. 2011;17:761–772. doi: 10.1261/rna.2581711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y. Alternative polyadenylation: New insights from global analyses. RNA. 2012;18:2105–2117. doi: 10.1261/rna.035899.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smibert P, Miura P, Westholm JO, Shenker S, May G, Duff MO, Zhang D, Eads BD, Carlson J, Brown JB, et al. Global Patterns of Tissue-Specific Alternative Polyadenylation in Drosophila. Cell Rep. 2012;1:277–289. doi: 10.1016/j.celrep.2012.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spellman R, Llorian M, Smith CWJ. Crossregulation and functional redundancy between the splicing regulator PTB and its paralogs nPTB and ROD1. Mol Cell. 2007;27:420–434. doi: 10.1016/j.molcel.2007.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiegel I, Mardinly AR, Gabel HW, Bazinet JE, Couch CH, Tzeng CP, Harmin DA, Greenberg ME. Npas4 regulates excitatory-inhibitory balance within neural circuits through cell-type-specific gene programs. Cell. 2014;157:1216–1229. doi: 10.1016/j.cell.2014.03.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan K, Friedman BA, Larson JL, Lauffer BE, Goldstein LD, Appling LL, Borneo J, Poon C, Ho T, Cai F, et al. Untangling the brain’s neuroinflammatory and neurodegenerative transcriptional responses. Nat Commun. 2016;7:11295. doi: 10.1038/ncomms11295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay TL, Mai D, Dautzenberg J, Fernández-Klett F, Lin G, Sagar Datta M, Drougard A, Stempfl T, Ardura-Fabregat A, et al. A new fate mapping system reveals context-dependent random or clonal expansion of microglia. Nat Neurosci. 2017;10:793–803. doi: 10.1038/nn.4547. [DOI] [PubMed] [Google Scholar]
- Tian B, Manley JL. Alternative polyadenylation of mRNA precursors. Nat Rev Mol Cell Biol. 2017;18:18–30. doi: 10.1038/nrm.2016.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ule J, Jensen KB, Ruggiu M, Mele A, Ule A, Darnell RB. CLIP Identifies Nova-Regulated RNA Networks in the Brain. Science. 2003;302:1212–1215. doi: 10.1126/science.1090095. [DOI] [PubMed] [Google Scholar]
- Van Nostrand EL, Pratt GA, Shishkin AA, Gelboin-Burkhart C, Fang MY, Sundararaman B, Blue SM, Nguyen TB, Surka C, Elkins K, et al. Robust transcriptome-wide discovery of RNA-binding protein binding sites with enhanced CLIP (eCLIP) Nat Methods. 2016;13:508–514. doi: 10.1038/nmeth.3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vong L, Ye C, Yang Z, Choi B, Chua S, Jr, Lowell BB. Leptin Action on GABAergic Neurons Prevents Obesity and Reduces Inhibitory Tone to POMC Neurons. Neuron. 2011;71:142–154. doi: 10.1016/j.neuron.2011.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ET, Ward AJ, Cherone JM, Giudice J, Wang TT, Treacy DJ, Lambert NJ, Freese P, Saxena T, Cooper TA, et al. Antagonistic regulation of mRNA expression and splicing by CELF and MBNL proteins. Genome Res. 2015;25:858–871. doi: 10.1101/gr.184390.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Dowell RD, Yi R. Genome-wide maps of polyadenylation reveal dynamic mRNA 3′-end formation in mammalian cell lineages. RNA. 2013;19:413–425. doi: 10.1261/rna.035360.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong C, Liu X, Meng A. The Kinase Activity-deficient Isoform of the Protein Araf Antagonizes Ras/Mitogen-activated Protein Kinase (Ras/MAPK) Signaling in the Zebrafish Embryo. J Biol Chem. 2015;290:25512–25521. doi: 10.1074/jbc.M115.676726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki R, Lu H, Butovsky O, Ohno N, Rietsch AM, Cialic R, Wu PM, Doykan CE, Lin J, Cotleur AC, et al. Differential roles of microglia and monocytes in the inflamed central nervous system. J Exp Med. 2014;211:1533–1549. doi: 10.1084/jem.20132477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap K, Xiao Y, Friedman BA, Je HS, Makeyev EV. Polarizing the Neuron through Sustained Co-expression of Alternatively Spliced Isoforms. Cell Rep. 2016;15:1316–1328. doi: 10.1016/j.celrep.2016.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama T, Takano K, Yoshida A, Katada F, Sun P, Takenawa T, Andoh T, Endo T. DA-Raf1, a competent intrinsic dominant-negative antagonist of the Ras–ERK pathway, is required for myogenic differentiation. J Cell Biol. 2007;177:781–793. doi: 10.1083/jcb.200703195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yona S, Kim KW, Wolf Y, Mildner A, Varol D, Breker M, Strauss-Ayali D, Viukov S, Guilliams M, Misharin A, et al. Fate Mapping Reveals Origins and Dynamics of Monocytes and Tissue Macrophages under Homeostasis. Immunity. 2013;38:79–91. doi: 10.1016/j.immuni.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarnegar BJ, Flynn RA, Shen Y, Do BT, Chang HY, Khavari PA. irCLIP platform for efficient characterization of protein–RNA interactions. Nat Methods. 2016;13:489–492. doi: 10.1038/nmeth.3840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Frias MA, Mele A, Ruggiu M, Eom T, Marney CB, Wang H, Licatalosi DD, Fak JJ, Darnell RB. Integrative Modeling Defines the Nova Splicing-Regulatory Network and Its Combinatorial Controls. Science. 2010;329:439–443. doi: 10.1126/science.1191150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Lee JY, Tian B. Biased alternative polyadenylation in human tissues. Genome Biol. 2005;6:R100. doi: 10.1186/gb-2005-6-12-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Chen MH, Wu X, Kodani A, Fan J, Doan R, Ozawa M, Ma J, Yoshida N, Reiter JF, et al. Cell-Type-Specific Alternative Splicing Governs Cell Fate in the Developing Cerebral Cortex. Cell. 2016;166:1147–1162.e15. doi: 10.1016/j.cell.2016.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Chen K, Sloan SA, Bennett ML, Scholze AR, O’Keeffe S, Phatnani HP, Guarnieri P, Caneda C, Ruderisch N, et al. An RNA-Sequencing Transcriptome and Splicing Database of Glia, Neurons, and Vascular Cells of the Cerebral Cortex. J Neurosci. 2014;34:11929–11947. doi: 10.1523/JNEUROSCI.1860-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S, Gray EE, Chawla G, Porse BT, O’Dell TJ, Black DL. PSD-95 is post-transcriptionally repressed during early neural development by PTBP1 and PTBP2. Nat Neurosci. 2012;15:381–388. doi: 10.1038/nn.3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Information of cTag-PAPERCLIP libraries, gene lists, and publicly available datasets used in the study, related to Figure 1.
Table S2. Permutation analysis results, related to Figure 2.
Table S3. Summary of RNA-seq analysis for cTag-PAPERCLIP validation, related to Figure 2.
Table S4. List of genes identified by cTag-PAPERCLIP to exhibit APA shift in LPS-activated brain microglia, related to Figure 6.
Table S5. List of primers, related to the STAR Methods.


