Abstract.
Several species of Plasmodium are responsible for causing malaria in humans. Proper diagnoses are crucial to case management, because severity and treatment varies between species. Diagnoses can be made using rapid diagnostic tests (RDTs), which detect Plasmodium proteins. Plasmodium falciparum causes the most virulent cases of malaria, and P. falciparum histidine-rich protein 2 (PfHRP2) is a common target of falciparum malaria RDTs. Here we report a case in which a falciparum malaria patient in Bangladesh tested negative on PfHRP2-based RDTs. The negative results can be attributed to a deletion of part of the pfhrp2 gene and frameshift mutations in both pfhrp2 and pfhrp3 gene. This finding may have implications for malaria diagnostics and case management in Bangladesh and other regions of South Asia.
More than 90% of symptomatic malaria cases in Bangladesh are caused by Plasmodium falciparum.1 Species-specific diagnoses are crucial to proper case management because falciparum malaria cases require different treatments and can rapidly lead to severe complications and death within days. Rapid diagnostic tests (RDTs) are extensively used for fast and proper diagnosis, and they are accessible even in areas without electricity or trained microscopists.2
Commonly used RDTs that are capable of specifically detecting P. falciparum target either P. falciparum histidine-rich protein 2 (PfHRP2) or P. falciparum lactate dehydrogenase (PfLDH);3 PfHRP2 has several repeats of the antibody-binding epitopes,4 which makes it ideal for immunodetection by RDT.5 PfHRP3 can also be detected by PfHRP2 RDTs because of the sequence homology of pfhrp2 and pfhrp3 gene although the expression is much lower than PfHRP2.5,6 PfHRP2 RDTs also have limitations. The pfhrp2 gene has greater variability than the pfldh gene,7 and pfhrp2 polymorphism can affect its detection by RDTs.5 In addition, strains with partial or total pfhrp2 deletions have been reported in South America, Africa, and India.3,8–10 A recent study in India reported 2.4% and 1.8% prevalence of pfhrp2 and pfhrp3 gene deletion, respectively; including an area in Tripura state, which shares its border with our study area.11 Here we report a case in which a falciparum malaria blood sample tested negative on a PfHRP2-based RDT in Bangladesh. A molecular basis for the negative result is provided, and its implications are discussed.
In August 2013, a blood sample was obtained from a 24-year-old male with high fever, chills, nausea, and headache at the Kamalganj Upazilla Health Complex in Sylhet, Bangladesh, located at 91°50′60″ E longitude and 24°21′36″ N latitude. The patient had no records of traveling abroad. The Falcivax Pv/Pf Combo RDT (Zephyr Biomedicals, Verna, Goa, India) was used, and the sample tested negative for both PfHRP2 and Plasmodium vivax lactate dehydrogenase (PvLDH). However, because the patient’s symptoms reflected falciparum malaria, a thin blood film was examined under a microscope. Falciparum malaria was diagnosed and, as per national guideline, the patient was treated with artemisinin-based combination therapy (ACT) for 3 days. A written informed consent was obtained from the patient, and the original study was approved by the institutional ethics review committee of the International Center for Diarrheal Disease Research, Bangladesh (icddr,b). Venous blood was obtained from the patient in ethylenediaminetetraacetic acid (EDTA) tubes and sent to icddr,b for further testing.
Presence of P. falciparum was confirmed via reexamination by two expert microscopists, and a parasitemia of 5,120 parasites/µL was counted in a thin blood film, corresponding to 0.1024% of erythrocytes infected. The blood sample was tested again, using three brands of RDTs: InTec® Pan/Pf Combo and Pf/Pv Combo (InTec Products Inc., Xiamen, China), SD Bioline Pf/Pv Combo (Standard Diagnostics, Yongin, Republic of Korea), and Falcivax Pv/Pf Combo (Falcivax; Zephyr Biomedicals, India). The sample tested negative for PfHRP2 on each RDT and positive for pLDH, shown by the “PAN” line on the InTec test. Diagnostic polymerase chain reaction (PCR) reconfirmed P. falciparum infection following a protocol described previously.12 In addition, msp2 marker (3D7 of central domain) region was amplified to confirm the validity of the DNA preparation.13,14
Given the high parasitemia of the sample (KHC 225) and PCR-confirmed diagnosis, variation or deletion in the hrp2 and hrp3 gene was the suspected cause for negative RDT results. The sample was subjected to PCR targeting the hrp2 and hrp3 gene. Two pairs of primers targeting the untranslated region (UTR), exon 1 and intronic region (PCR reaction 1), and exon 2 region (PCR reaction 2) of pfhrp2 were derived from a previous study.15 The other two sets of primers targeting the same sites (PCR reaction 3 and 4) of pfhrp3 gene were derived from other studies.16,17 The reverse primer for PCR reaction 4 is the reverse compliment of the forward primer for PCR reaction 3. PCR cycle conditions were thus modified; for 35 cycles, the annealing temperature for PCR reactions 1 and 3 was set to 55°C for 35 seconds and extension temperature was set to 68°C for 40 seconds followed by final extension at 68°C for 10 minutes. For PCR reaction 2 and 4, the conditions and cycles were the same, except the annealing stage was modified to 30 seconds and extension stage was 1 minute and 20 seconds. Both reactions included DNA extracted from a P. falciparum culture as a positive control (MRA-156, MR4, ATCC, Manassas, VA). Water was used as a common negative control along with genomic DNA of strain D10 and HB3 (MRA-201G and MRA-155G, MR4, ATCC, Manassas, VA) as negative control for hrp2 and hrp3 gene amplification, respectively. Amplification products were visualized by gel electrophoresis.
In the hrp2 gene amplification cycle, PCR reaction 1 yielded no visible amplification product for the experimental sample. PCR reaction 2 provided the expected DNA fragment approximately 1,021 bp long. For the hrp3 amplification, PCR reactions 3 and 4 yielded fragments of approximately 460 and 483 bp. The amplified products were sequenced using ABI3500 Genetic Analyzer (Life Technologies, Foster City, CA,) after purification by ExoSAP-IT (Affymetrix). ClustalW was used to align the sequences with that of P. falciparum strain 3D7 (PF3D7_0831800 and PF3D7_1372200, PlasmoDB Release 28).
In pfhrp2 exon 2, several point mutations and insertions were found, as well as a frameshift deletion very early in exon 2. The aligned sequence of KHC 225 hrp2 shares 93% identity with PF3D7_0831800. In KHC 225 pfhrp3, the region spanning UTR and exon 1 was conserved with some single-nucleotide polymorphisms (SNPs) in the intronic region. The exon 2 portion had various SNPs along with a 3 bp insertion and a frameshift deletion. KHC 225 hrp3 and PF3D7_1372200 share 81% identity. PCR and sequencing was performed at least three times for each portion of both the genes to confirm the mutations. The partial exon 2 sequences of KHC 225 hrp2 and hrp3 reported in this article has been deposited in the GenBank database (accession number KX388531 and MF176231).
Low parasitemia is one possible explanation for false negatives using RDTs, but this was ruled out as the parasitemia of 5,120 parasites/µL, which is approximately 50 times than that normally required for RDTs.5 Interestingly, PfHRP3 can also bind to PfHRP2-based RDTs. It has been found, however, that when PfHRP2 is missing, PfHRP3 expression is reduced.6
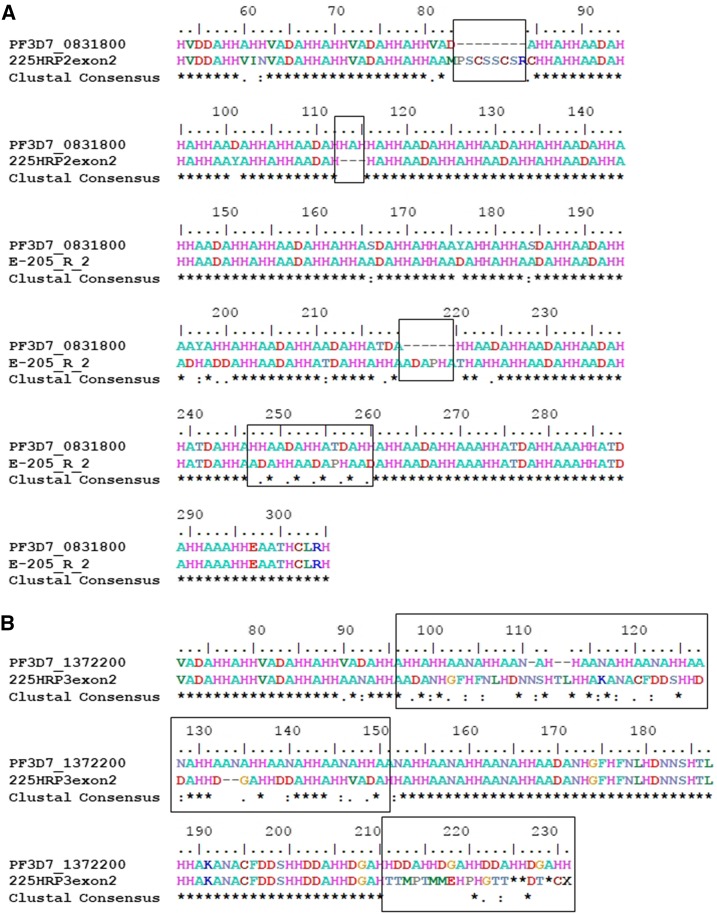
The lack of amplification in PCR reaction 1 suggests a deletion upstream of the forward primer for reaction 2, which is consistent with other strains found to have partial pfhrp2 deletions.9 Chromosome breakage in this unstable subtelomeric region is often accompanied by addition of new telomeric sequences.18 The gene cannot be transcribed because part of it, as well as the entire upstream region of the chromosome, has been deleted. This deletion accounts for the negative results on PfHRP2-based RDTs. If the upstream regulatory region somehow were present, the frameshift deletion in exon 2 would result in an unrecognizable amino acid sequence after translation. Furthermore, several mutations were found in pfhrp3. The variation of the pfhrp3 sequence resulted in an amino acid that is not recognizable to the RDT binding sites (Figure 1).
Figure 1.
(A) HRP2 alignment of KHC 225 with PF3D7_0831800 (3D7). Major changes in the HRP2 amino acid sequence of KHC 225 include insertion of eight amino acids after the 83 position, deletion of positions 113–115, insertion of six amino acids after the 219th and mismatches in several positions. (B) HRP3 alignment of KHC 225 with PF3D7_1372200 (3D7). Several major mismatches between KHC 225 HRP3 amino acid sequence and PF3D7_1372200 were found from position 96 to 152 and 221 to 231. This figure appears in color at www.ajtmh.org.
Because failure to detect PfHRP2 has not previously been reported in Bangladesh, the findings of this study may have several implications. According to the National Malaria Control Program, PfHRP2 and pan- Plasmodium-specific (pLDH) combo RDTs have been recently introduced to diagnose both P. falciparum and any other type of malaria; but they are still not implemented widely. The microscopy centers are still not enough to cater to the need for early detection and verification of the malarial species. In areas where pan RDTs are not used, these cases might be misdiagnosed as nonmalaria, increasing the likelihood that this strain will survive and spread in the endemic community. Conversely, in areas where pan RDTs are used, the case may be diagnosed as P. vivax infection. Because 80% of P. falciparum isolates in Bangladesh are resistant to chloroquine,14 which is still administered to patients with P. vivax infections, misdiagnosis of P. falciparum malaria as P. vivax malaria can result in improper treatment. Thus, misdiagnosis can hinder patient outcomes and increase the spread of chloroquine-resistant P. falciparum in the endemic population.
We recommend that the healthcare staff working in endemic areas should be informed about the various reasons, such as pfhrp2 mutation or deletion, for false-negative results. As there have been reports of such cases in other side of the border in India, it may be wise to increase surveillance for similar cases as many may go unreported or misclassified as P. vivax. We also advise that, whenever practical, nonfalciparum diagnoses by HRP2 RDT should be verified by either microscopy, P. falciparum–specific LDH-based RDT, or PCR.
Acknowledgments:
This study was supported by the Swiss Academy of Medical Science (SAMS) and the Velux Foundation. icddr,b acknowledges with gratitude the commitment of SAMS and Velux Foundation to its research efforts. icddr,b is also grateful to the Governments of Bangladesh, Canada, Sweden, and the United Kingdom for providing core/unrestricted support. We are grateful to Rubayet Elahi and Hans-Peter Führer for their review of this manuscript. We thank MR4 for kindly providing us Plamodium falciparum reference strain and genomic DNA.
REFERENCES
- 1.WHO , 2015. World Malaria Report 2014. Available at: http://www.who.int/malaria/publications/world_malaria_report_2014/en/. Accessed October 24, 2016.
- 2.Wongsrichanalai C, Barcus MJ, Muth S, Sutamihardja A, Wernsdorfer WH, 2007. A review of malaria diagnostic tools: microscopy and rapid diagnostic test (RDT). Am J Trop Med Hyg 77: 119–127. [PubMed] [Google Scholar]
- 3.Houzé S, Boly MD, Le Bras J, Deloron P, Faucher J-F, 2009. Pf HRP2 and Pf LDH antigen detection for monitoring the efficacy of artemisinin-based combination therapy (ACT) in the treatment of uncomplicated falciparum malaria. Malar J 8: 211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wellems TE, Howard RJ, 1986. Homologous genes encode two distinct histidine-rich proteins in a cloned isolate of Plasmodium falciparum. Proc Natl Acad Sci USA 83: 6065–6069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee N, Baker J, Andrews KT, Gatton ML, Bell D, Cheng Q, McCarthy J, 2006. Effect of sequence variation in Plasmodium falciparum histidine-rich protein 2 on binding of specific monoclonal antibodies: implications for rapid diagnostic tests for malaria. J Clin Microbiol 44: 2773–2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baker J, Gatton ML, Peters J, Ho M-F, McCarthy JS, Cheng Q, 2011. Transcription and expression of Plasmodium falciparum histidine-rich proteins in different stages and strains: implications for rapid diagnostic tests. PLoS One 6: e22593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mariette N, Barnadas C, Bouchier C, Tichit M, Ménard D, 2008. Country-wide assessment of the genetic polymorphism in Plasmodium falciparum and Plasmodium vivax antigens detected with rapid diagnostic tests for malaria. Malar J 7: 219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deme AB, Park DJ, Bei AK, Sarr O, Badiane AS, Gueye PEHO, Ahouidi A, Ndir O, Mboup S, Wirth DF, 2014. Analysis of pfhrp2 genetic diversity in Senegal and implications for use of rapid diagnostic tests. Malar J 13: 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gamboa D, Ho M-F, Bendezu J, Torres K, Chiodini PL, Barnwell JW, Incardona S, Perkins M, Bell D, McCarthy J, 2010. A large proportion of P. falciparum isolates in the Amazon region of Peru lack pfhrp2 and pfhrp3: implications for malaria rapid diagnostic tests. PLoS One 5: e8091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kumar N, Pande V, Bhatt R, Shah NK, Mishra N, Srivastava B, Valecha N, Anvikar AR, 2013. Genetic deletion of HRP2 and HRP3 in Indian Plasmodium falciparum population and false negative malaria rapid diagnostic test. Acta Trop 125: 119–121. [DOI] [PubMed] [Google Scholar]
- 11.Bharti PK, Chandel HS, Ahmad A, Krishna S, Udhayakumar V, Singh N, 2016. Prevalence of pfhrp2 and/or pfhrp3 gene deletion in Plasmodium falciparum population in eight highly endemic states in India. PLoS One 11: e0157949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Snounou G, Viriyakosol S, Zhu XP, Jarra W, Pinheiro L, do Rosario VE, Thaithong S, Brown KN, 1993. High sensitivity of detection of human malaria parasites by the use of nested polymerase chain reaction. Mol Biochem Parasitol 61: 315–320. [DOI] [PubMed] [Google Scholar]
- 13.Alam MS, Elahi R, Mohon AN, Al-Amin HM, Kibria MG, Khan WA, Khanum H, Haque R, 2016. Plasmodium falciparum genetic diversity in Bangladesh does not suggest a hypoendemic population structure. Am J Trop Med Hyg 94: 1245–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Snounou G, Zhu X, Siripoon N, Jarra W, Thaithong S, Brown KN, Viriyakosol S, 1999. Biased distribution of msp1 and msp2 allelic variants in Plasmodium falciparum populations in Thailand. Trans R Soc Trop Med Hyg 93: 369–374. [DOI] [PubMed] [Google Scholar]
- 15.Trouvay M, Palazon G, Berger F, Volney B, Blanchet D, Faway E, Donato D, Legrand E, Carme B, Musset L, 2013. High performance of histidine-rich protein 2 based rapid diagnostic tests in French Guiana are explained by the absence of pfhrp2 gene deletion in P. falciparum. PLoS One 8: e74269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baker J, McCarthy J, Gatton M, Kyle DE, Belizario V, Luchavez J, Bell D, Cheng Q, 2005. Genetic diversity of Plasmodium falciparum histidine-rich protein 2 (PfHRP2) and its effect on the performance of PfHRP2-based rapid diagnostic tests. J Infect Dis 192: 870–877. [DOI] [PubMed] [Google Scholar]
- 17.Sullivan DJ, Ayala YM, Goldberg DE, 1996. An unexpected 5′ untranslated intron in the P. falciparum genes for histidine-rich proteins II and III. Mol Biochem Parasitol 83: 247–251. [DOI] [PubMed] [Google Scholar]
- 18.Scherf A, Mattei D, 1992. Cloning and characterization of chromosome breakpoints of Plasmodium falciparum: breakage and new telomere formation occurs frequently and randomly in subtelomeric genes. Nucleic Acids Res 20: 1491–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]