Abstract.
Dengue virus (DENV) circulates in tropical and subtropical areas around the world, where it causes high morbidity and mortality. There is no effective treatment of infection, with supportive care being the only option. Furthermore, early detection and diagnosis are important to facilitate clinical decisions. In this study, seven monoclonal antibodies (mAbs) recognizing nonstructural protein 1 (NS1) of DENV were generated by hybridoma techniques. These antibodies can be divided into two groups: serotype-specific (DB6-1, DB12-3, and DB38-1) and nonspecific (consisting of antibodies DB16-1, DB20-6, DB29-1, and DB41-2). The B-cell epitopes of DB20-6 and DB29-1 were identified by phage display and site-directed mutagenesis, and its binding motif, WXXWGK, was revealed to correspond to amino acid residues 115–120 of the DENV-2 NS1 protein. A diagnostic platform, consisting of a serotype-specific capture antibody and a complex detection antibody, exhibited a detection limit of about 1 ng/mL, which is sufficient to detect NS1 in clinical serum samples from dengue patients. This diagnostic platform displayed better specificity and sensitivity than two examined commercial NS1 diagnostic platforms. In summary, our results indicate that these newly generated mAbs are suitable for detection of NS1 protein of DENV-2 in clinical samples.
INTRODUCTION
Dengue fever (DF) and its more severe manifestations, dengue hemorrhagic fever (DHF) and dengue shock syndrome (DSS), are the most critical arboviral diseases in tropical and subtropical areas. Up to 390 million infections are reported each year, including 500,000 cases of life-threatening DHF and DSS, which cause 25,000 deaths annually.1–3 These diseases are classified into mild febrile illness, classical DF, and severe or fatal hemorrhagic disease, according to the severity of the clinical manifestations. Symptoms include fever, frontal headache, stomachache, and myalgias, and frequently arthralgias, joint pain, retro-orbital eye pain, nausea, vomiting, rash, taste aberrations, and lymphadenopathy. In severe states, that is, DHF and/or DSS, patients present with thrombocytopenia, coagulopathy, vascular leakage, and hypotension.1,4 At the time of writing, there is no antiviral drug or suitably well-developed vaccine (although the first licensed dengue vaccine, Dengvaxia® [CYD-TDV] developed by Sanofi Pasteur [Lyon, France], was approved in Mexico in December 2015, the efficacy of the vaccine varies based on different serotypes and the age of the patient at the time of vaccination) that can treat or prevent the disease, leaving supportive interventions as the only clinical option.1,5 Moreover, our understanding of the disease mechanisms is limited due to the lack of appropriate animal models.1,6,7
The causative agent of this global issue is the dengue virus (DENV). The spread of this pathogen is through its primary vector, the Aedes aegypti mosquito, and secondary vectors, Aedes albopictus and Aedes polynesiensis.5,8 DENV belongs to the genus Flavivirus of the family flaviviridae, which is composed of several other important viruses, such as Japanese encephalitis virus (JEV), West Nile virus (WNV), yellow fever virus (YFV), St. Louis encephalitis (SLE), tick-borne encephalitis virus, and Zika virus, which was recently circulating in Brazil.9,10 There are four closely linked, but serologically distinct, serotypes of DENV: DENV-1, DENV-2, DENV-3, and DENV-4. Infection with any single serotype will provide lifelong immunity against the same serotype.
Nonstructural protein 1 (NS1) is a highly conserved glycoprotein expressed in all DENVs, with a molecular weight of about 46- to 50-kilodaltons.11,12 During the viral life cycle, NS1 is translated in the endoplasmic reticulum, and then transported through the Golgi apparatus, where it is glycosylated. Finally, the modified NS1 protein inserts into the cell membrane, or is secreted from the cell.13 NS1 is displayed as monomer, dimer, or hexamer, and the hexamer is the major form secreted into the blood stream.11,14 Although the role of NS1 protein is not clear, it is known to be important in DENV replication and assembly.13,15,16 Some experimental data suggest that the NS1 protein is modified by glycosylphosphatidylinositol, and associates with lipid rafts at the plasma membrane, where it mediates signaling.11,17 Other data suggest that the hexamer may play a role in DHF pathogenesis.11 Since NS1 is secreted into the blood stream of infected individuals, the presence of NS1 in sera may serve as a useful diagnostic marker, especially during acute phase.18
Current methods used to detect DENV infection include virus isolation, RNA detection, antigen detection, and serological tests. For virus isolation or RNA detection, the duration of virus persistence in the blood stream is important. However, viral isolation and RNA detection both have short windows for detecting the disease due to their short half-life in blood circulation and unstable molecular structure, which means they peak between day 1 and 3 (days postonset of symptoms), and decline thereafter. For developing countries where dengue is endemic, this is a heavy burden. Alternatively, IgM antibody capture enzyme-linked immunosorbent assay (ELISA) is commonly used for serological diagnosis; however, IgM appears later than viral RNA genome or NS1 protein in the blood stream of infected patients. Moreover, dengue antibodies may cross-react with other flaviviruses, such as YFV, WNV, JEV, SLE, or Zika, which may lead to misdiagnosis by clinicians.10
At present, there are various diagnostic platforms for DENV NS1 detection such as Platelia dengue NS1 Ag test (Bio-Rad Laboratories, Marnes La Coquette, France),15,16,19,20 Platelia dengue IgA capture assay (Bio-Rad Laboratories).21 Pan-E dengue early ELISA test (Panbio Diagnostics, Brisbane, Australia),19 dot blot immunoassay,22 DEN antigen detection kit (denKEY Blue kit; Globio Co., Beverly, MA),22 InBios DENV Detect NS1 ELISA kit23 Dengue early ELISA (MyBioSource, Vancouver, Canada), DENV NS1 ELISA test kit (EUROIMMUN, Luebeck, Germany),15,19,24–26 dengue NS1 Ag STRIP (Bio-Rad Laboratories),27 and NS1 lateral flow rapid test (LFRT).15,28 Most of these platforms are based on the NS1 capture ELISA; other platforms, such as paper-based ELISA29 for NS1 detection, apply the LFRT concept, enabling rapid detection of the NS1 antigen. Compared with the NS1 protein assay, detection of dengue infection through isolation of virus particles in sera, through reverse transcription polymerase chain reaction (PCR) or other serological approaches, is time consuming and requires well-trained personnel and expensive equipment. Hence, there is an urgent requirement for the development of inexpensive, convenient, sensitive, and specific diagnostic platforms for pathogen detection, especially in the developing countries.
In this study, we generated and characterized monoclonal antibodies (mAbs) against the NS1 protein of DENV-2. We then proceeded to test various combinations of mAbs for detection of NS1, to develop a diagnostic platform based on conventional capture ELISA principles. The resulting platform was used to examine immunoaffinity-purified DENV-2 NS1 protein and clinical sera, establishing the diagnostic potential of this system. Furthermore, the B-cell epitopes of mAbs were further identified from a random phage-displayed peptide library. These mAbs and epitope-based peptides can be used to develop a convenient, efficient serologic test that identifies DENV serotypes.
MATERIALS AND METHODS
Cells and viruses.
Baby hamster kidney (BHK)-21 cells were grown at 37°C with 5% CO2 in minimal essential medium (Gibco, Grand Island, NY) supplemented with 10% heat-inactivated fetal bovine serum (FBS, Gibco) and 100 U/mL penicillin, 100 μg/mL streptomycin, and 0.25 μg/mL amphotericin B (Antibiotic-antimycotic, Gibco). Aedes albopictus C6/36 cells were grown at 28°C in 1:1 Mitsuhashi and Maramorosch insect medium (Sigma-Aldrich, St. Louis, MO)/Dulbecco’s modified Eagle’s medium (DMEM, Gibco) containing 10% FBS and 100 U/mL penicillin, 100 μg/mL streptomycin, and 0.25 μg/mL amphotericin B (Antibiotic-Antimycotic, Gibco). Human umbilical vein endothelial cells (HUVECs) were grown in endothelial cell basal medium-2 (Lonza, Basel, Switzerland) containing essential supplements (FBS, hydrocortisone, human fibroblast growth factor-basic, vascular endothelial growth factor, R3-IGF-1, ascorbic acid, human epidermal growth factor, GA-1000, heparin) at 37°C with 5% CO2. The four DENVs (DENV-1 Hawaii, DENV-2 16681, DENV-3 H87, DENV-4 H241) were provided by Duane J. Gubler of the Centers for Disease Control and Prevention (CDC), Fort Collins, CO. These viruses were passaged in C6/36 cells.
Human serum samples.
Sera from patients infected with DENV serotype 2 were provided by Chwan-Chuen King, Day-Yu Chao, and Yen-Hsu Chen (12 serum samples). Control serum samples were provided by healthy donors (10 serum samples). Two types of “fever control” samples were provided by Chwan-Chuen King: 1) 20 serum samples from patients who visited Emergency Department at the National Taiwan University Hospital which is located in Taipei, northern Taiwan; and 2) 30 serum samples from the measles outbreak studies in Taipei County (renamed as New Taipei City since 2010, also located in northern Taiwan). The influenza virus-infected patients’ samples were provided by Yen-Hsu Chen (10 serum samples); the JEV-infected patients’ samples were provided by Day-Yu Chao (seven serum samples); the malaria-infected patients’ samples (Plasmodium falciparum and Plasmodium vivax) were provided by Ming-Chu Kuo (Taiwan CDC; 10 serum samples). The study protocols were approved by the National Taiwan University Institutional Review Board (NTUH-REC No. 200903086R) and Kaohsiung Medical University Hospital Institutional Review Board (KMUHIRB-2011-04-02(I)). DENV2-infected patients’ sera were confirmed by PCR with DENV2-specific primers and NS1 Ag rapid test (Bio-Rad or Standard Diagnostic, Inc.) by CDC, Taiwan. For the determination of primary/secondary infection, two different kits, anti- DENV ELISA (IgG) and anti-DENV ELISA (IgM) (EUROIMMUN), were used. All experimental procedures were performed in accordance with the instructions provided by the kits (Supplemental Table 1).
Virus infection.
C6/36 cells were seeded in T75 flasks at a density of 1 × 107 cells/flask. The culture media contained 10% FBS, and the cells were cultured at 28°C overnight. Once the cell density reached 80–90% confluence, the culture media was replaced with 3-mL fresh media/flask, containing 2% FBS. Titrated DENV-2 virus was added to flasks at a multiplicity of infection (MOI) of 0.1, and the flasks were then cultured at 28°C for 2 hours, with shaking once every 30 minutes. After 2 hours, fresh culture media containing 2% FBS was added to the flasks to a final volume of 8 mL/flask. To monitor the cytopathic effect of infection, cells were harvested at various time points. Media were centrifuged (800 × g, 5 minutes, 4°C), and the supernatant was stored at −80°C for future use.
DENV antigen preparation.
C6/36 cells were infected with one of the four serotypes of DENV. Infected cells were lysed in lysis buffer (25 mM Tris-HCl, pH 7.4, 150 mM NaCl, and 1% Nonidet P-40) in the presence of protease inhibitors (Roche, Basel, Switzerland). Cell debris was removed by centrifugation at 3,000 × g for 10 minutes at 4°C, and protein concentration was determined by measuring absorbance at 280 nm using a ultraviolet (UV) spectrophotometer.
Purification of DENV-2 NS1 protein.
Normal human serum (NHS)-activated Sepharose 4 Fast Flow beads (Amersham Bioscience Cat no. 17-0906-01) were loaded onto a column, and washed with ddH2O. Freshly prepared 1 mM HCl solution was chilled to 4°C, and then used to activate the beads. The beads were subsequently washed with phosphate-buffered saline (PBS), and the PBS was removed. Anti-NS1 mAb (DB16-1) was added to the beads (the volume ratio of antibody solution to beads was between 0.5 and 1.0), and the column top and bottom were sealed. The column was inverted several times, and then placed on a shaker at room temperature for 2–4 hours or 4°C overnight. After the coupling reaction, the antibody solution was drained, and the beads were washed several times with PBS. Subsequently, a volume of 0.5 M Tris buffer (pH 8) equal to 2× bead volume was added to block the unreacted sites on beads, and the column was left to stand at room temperature for 1–2 hours or 4°C overnight. The beads were cleaned by washing in three different buffers, in the following order: PBS, 0.1 M Tris (pH 9)/0.5 M NaCl, PBS, 0.1 M glycine (pH 2.3)/1 M NaCl. The cycle was repeated three times, and then the beads were equilibrated in PBS prior to use. Harvested DENV-2-infected C6/36 cell supernatant was added into the prepared column; after collecting the flow through, the column was washed with PBS. Captured NS1 protein was eluted with 0.1 M glycine (pH 2.3), and the fractions were harvested. Next, each fraction was neutralized with 1 M Tris (pH 9), and a UV spectrophotometer was used to measure protein concentration as described earlier. DENV-1 NS1 protein was purified by following the same procedures.
Generation of mAbs against DENV-2 NS1 protein.
The procedure for producing anti-NS1 protein was modified from an earlier method.30 Briefly, female BALB/c mice were intraperitoneally immunized with immunoaffinity-purified DENV-2 NS1 protein (described earlier under “Purification of DENV-2 NS1 protein”) four times at 3-week intervals. On day 4 after the final boost, lymphocytes were harvested from the spleen of the immunized mouse, and then fused with NSI/1-Ag4-1 myeloma cells using 50% polyethylene glycol (PEG) (Invitrogen, Carlsbad, CA). The fused cells were resuspended in DMEM containing 20% FBS, hypoxanthine–aminopterin–thymidine, and hybridoma cloning factor (ICN Biomedicals, Aurora, OH). All animal experiments were performed in accordance with the guidelines of the National Laboratory Animal Center. The protocol was approved by the Committee on the Ethics of Animal Experiments of Academia Sinica. To confirm the specificity of antibodies, cultured hybridoma supernatant was incubated with DENV-2-infected C6/36 cells (as described under “Cellular ELISA”). Hybridoma cell lines were grown in DMEM with 10% FBS. Ascites were generated in pristine-primed BALB/c mice, and mAbs purified with protein G-Sepharose 4 Fast Flow gel.
Immunofluorescence assay.
BHK21 cells were seeded on coverslips (Deckglaser Cover Glasses, 12 mm in diameter) in 24-well plates, 1 × 105 cells/well, and then incubated at 37°C overnight. Cells were infected with DENV-2 virus (strain 16681) at a MOI of 0.5; the plates were subsequently incubated at 37°C for 2 hours, with shaking once every 30 minutes. Fresh 2% FBS-containing medium was added to each well, and the plates were cultured at 37°C for 2 days. After the infection period, cells were washed twice with PBS. Methanol/acetone (1:1) was used to fix and permeabilize the cells, and the plates were then incubated at −20°C for 20 minutes. Methanol/acetone solution was removed, and the cells were washed with PBS. Blocking buffer (1% bovine serum albumin (BSA)/PBS) was added to the cells, which were then incubated at room temperature for 30 minutes. Each mAb (DB6-1, DB12-3, DB16-1, DB20-6, DB29-1, DB38-1, and DB41-2) was separately diluted in blocking buffer (5 μg/mL), and the dilutions were incubated with infected cells at room temperature for 1 hour. After treatment, cells were washed with PBS. Secondary antibody (fluorescein isothiocyanate-conjugated goat antimouse immunoglobulin G (IgG); Jackson ImmunoResearch Laboratories, West Grove, PA) was diluted in blocking buffer (1:250) and supplemented with 4',6-diamidino-2-phenylindole (Invitrogen) at a 1 to 2,000 ratio (DAPI: antibody). The secondary antibody and DAPI mixtures were incubated with the cells at room temperature for 1 hour. After the incubation period, cells were washed three times with PBS, 5 minutes/wash. Finally, cells were mounted on glass slides, covered with coverslips, and fixed using polish oil. A Zeiss microscope imaging system was used to visualize the cells. For HUVEC cells, the permeabilized cells were incubated with DB16-1, DB20-6, DB29-1, or control antibody (normal mouse [NM] IgG). The bound antibodies were detected by fluorochrome (Hylite-488)-conjugated secondary antibodies and the images were visualized and recorded by Zeiss microscope imaging system (Oberkochen, Germany).
Cellular ELISA.
C6/36 cells were seeded in 96-well plates at 2 × 104 cells/well, and incubated at 37°C overnight. Cells were infected with each DENV serotype 1–4 at an MOI of 0.1, and further incubated at 28°C for 5–7 days. Following the infection period, cells were washed twice with PBS, fixed and permeabilized with methanol/acetone (1:1), and incubated at −20°C for 20 minutes. Next, the cells were washed three times with PBS, blocked with 5% skimmed milk, and incubated at 4°C overnight. The cells were given a final wash with PBS prior to ELISA. Individual mAbs (DB6-1, DB12-3, DB16-1, DB20-6, DB29-1, DB38-1, DB41-2, or NM IgG) were added to each well, and the plates were then incubated at room temperature for 1 hour. The cells were then washed three times with PBS. The secondary antibody, horseradish peroxidase (HRP)-conjugated goat antimouse IgG (Jackson ImmunoResearch Laboratories), was added to the wells (1:2,000 dilution), and the plates were incubated at room temperature for a further 1 hour. Cells were washed four times with PBS containing 0.1% Tween 20. Signals were developed using the peroxidase substrate, o-phenylenediamine dihydrochloride (OPD; Sigma, St. Louis, MO). The reaction was stopped with 3 N HCl, and the plates were read using the Spectra Max M5 (Molecular Devices, Sunnyvale, CA) microplate reader at an absorbance of 490 nm.
Western blot analysis.
DENV-infected C6/36 cells lysates were added to the wells of a reducing gel. After electrophoresis, proteins within the gel were transferred to nitrocellulose (NC) paper. The NC paper was blocked with 5% skimmed milk at 4°C overnight, washed three times with PBS containing 0.1% Tween 20 (PBST0.1), and hybridized with the specified mAb (DB6-1, DB12-3, DB16-1, DB20-6, DB29-1, DB38-1, or DB41-2) at room temperature for 1 hour. The NC paper was then washed three times with PBST0.1, and hybridized with the secondary antibody, HRP-conjugated goat antimouse IgG, at room temperature for 1 hour. After washing three times with PBST0.1, the NC paper was incubated with enhanced chemiluminescence substrate (Millipore, Billerica, MA). Signals were recorded using a BioSpectra 600 Imaging System (UVP, Upland, CA).
Phage display biopanning.
Biopanning was performed as described previously.30 Briefly, an 8-well module was coated with 100 μg/mL of DB20-6 or DB29-1, and blocked at 4°C overnight. A phage displayed peptide library (New England Biolabs, Inc., Ipswich, MA) was diluted to 4 × 1010 phages, and incubated with the DB20-6- or DB29-1-coated wells for 50 minutes at room temperature. After washing with PBS containing 0.5% Tween 20 (PBST0.5), the bound phages were eluted with 0.2 M glycine, pH 2.2. The eluates were neutralized with 1 M Tris-HCl, pH 9.1. Next, the eluted phages were amplified in an ER2738 (New England Biolabs, Inc., Ipswich, MA) overnight culture, which was vigorously shaken for 4.5 hours at 37°C. The amplified phages were precipitated with 20% PEG 8000 in 2.5 M NaCl (PEG/NaCl) at 4°C overnight, and then centrifuged (8,000 × g, 20 minutes, 4°C), before being suspended in PBS. The phages were subsequently reprecipitated with PEG/NaCl, centrifuged at 4°C for 10 minutes, and resuspended in PBS. The amplified phages were titered on LB/isopropyl-β-D-thiogalactoside (IPTG)/X-gal plates. The second round was identical to the first, except for the addition of 2 × 1011 plaque-forming units (pfu) of the previously amplified phages. The third round of biopanning was performed with 2 × 1011 pfu of phages amplified from the second round. Phages generated from the third round were titered on LB/IPTG/X-gal plates, and selected for ELISA.
Identification and sequencing of immunopositive phage clones.
Immunopositive phage clones were identified and sequenced as previously described.30 Briefly, the ELISA plate was coated with 50 μg/mL of DB20-6, DB29-1, or NM IgG in coating buffer for 2 hours at room temperature, and then blocked with blocking buffer at 4°C overnight. The diluted phages were incubated with coated plates for 1 hour at room temperature. After washing, the bound phages were probed with HRP-conjugated mouse anti-M13 mAb (GE Healthcare Biosciences, Little Chalfont, England) following the same procedures described under “Cellular ELISA.” The immunopositive phage clones were further sequenced with the −96 primer 5′-CCCTCATAGTTAGCGTAACG-3′, which corresponds to the pIII gene sequence of the M13 phage. The phage-displayed peptide sequences were translated with the ExPASy Proteomics Server.
Phage binding assay.
ELISA plates were coated with DB20-6 or DB29-1 at a concentration of 10 μg/mL, and then blocked with blocking buffer at 4°C overnight. The plates were incubated with PC20-3 or PC29-2 and control phage, Ab4-C4, which were serially diluted from 109 to 104 pfu and 0 pfu. After washing with PBST0.5, the plate was incubated with HRP-conjugated anti-M13 mAb (GE Healthcare) for 1 hour at room temperature, and subjected to the procedures described under “Cellular ELISA.”
Peptide binding assay.
P29M and arbitrary control peptide P7M (this control peptide binds to serotype-specific epitope of DENV-2 and does not interact with dengue viral antigen)31 were coated onto a 96-well plate at a concentration of 5 μg/mL. DB20-6 and DB29-1 were serially diluted (2-fold) from 20–0 μg/mL, and the various dilutions (20, 10, 5, 2.5, 1.25, 0.625, 0.31, and 0 μg/mL) were added to the plate. After incubation for 1 hour at room temperature, the plate was washed three times with PBS. Next, HRP-conjugated goat antimouse IgG (1:2000) was added to the plates, and incubated at room temperature for 1 hour. The plates were washed four times with PBST0.1, and signals were developed with the peroxidase substrate, OPD (Sigma). The reaction was stopped by the addition of 3 N HCl, and the plates were read using the Spectra Max M5 (Molecular Devices) microplate reader at an absorbance of 490 nm.
Peptide competitive inhibition assay.
DENV-2-infected C6/36 cell lysate was separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), and then transferred to NC paper. After blocking with 5% skimmed milk, the NC paper was probed with a mixture of DB20-6 or DB29-1 (1 μg/mL) with or without P29M and P7M (100 μg/mL). The NC paper was then washed, before being hybridized with HRP-conjugated goat antimouse IgG. Following a final wash, the NC paper was incubated with enhanced chemiluminescent substrate. Signals were recorded using a BioSpectra 600 Imaging System (UVP, Upland, CA).
Immunohistochemical staining.
HUVEC cells were grown on coverslips at 37°C overnight. The cells were then washed with PBS and fixed with 2% paraformaldehyde. Individual antibodies (DB16-1, DB20-6, DB29-1, CD31, or 4G2) at a concentration of 10 μg/mL were incubated with HUVEC cells for 1 hour at room temperature. Next, the HUVEC cells were washed with PBS containing 0.1% Tween 20 (PBS0.1), before being analyzed with the polymer-based Super Sensitive IHC detection system (Biogenex, San Ramon, CA). Briefly, the coverslips were treated with super enhancer reagent for 20 minutes at room temperature, and then washed with PBS0.1. Following the addition of Poly HRP, the plates were incubated at room temperature for 30 minutes, and then washed with PBS0.1. Signals were developed with diaminobenzidine hydrochloride (0.02%) containing 0.03% H2O2, and cells were counterstained with hematoxylin. The coverslips were coated on the glass slides and fixed with polish oil. The images were examined under light microscopy.
Screening of binding activities of anti-NS1 mAbs by direct ELISA.
Serial dilutions (1,000, 333, 111, 37, 12, 4, 1, and 0.4 ng/mL) of immunoaffinity-purified DENV-2 NS1 protein were made in coating buffer (0.1 M NaHCO3), and used to coat 96-well plates (50 μL/well). The coated plate was incubated at room temperature for 2 hours. After incubation, the plate was washed with PBS, and blocked by incubation with 1% BSA/PBS at 4°C overnight. On the second day, the plate was washed three times with PBS. Individual mAbs (DB6-1, DB16-1, DB20-6, DB29-1, or NM IgG) were added to the wells (1 μg/mL and 50 μL/well), and incubated at room temperature for 1 hour. After washing with PBS, HRP-conjugated goat antimouse IgG was added to the wells, and the plate was incubated at room temperature for a further 1 hour before being washed four times with PBS containing 0.1% (v/v) Tween 20 (PBST0.1). Signals were developed by addition of the peroxidase substrate, OPD (Sigma), to each well, and the signals were read using a Spectra Max M5 (Molecular Devices) microplate reader at an absorbance of 490 nm.
Biotinylation of mAb.
Biotinylation of DB20-6 was performed according to the instructions provided with EZ-Link NHS-Biotin Reagents (ThermoFisher Scientific, Waltham, MA). Briefly, the biotin powder was dissolved in dimethyl sulfoxide solution to a final concentration of 10 mM, and then the appropriate volume (calculated using the formula provided by the manufacturer) was added to 1 mg DB20-6 dissolved in 200 μL of PBS. The mixture was incubated on ice for 2 hours or at room temperature for 30 minutes. The mixture was resuspended in PBS following Amicon (Millipore) centrifugation, and protein concentration was measured using a UV spectrophotometer as described earlier.
Establishment of NS1 standard curves.
A 96-well plate was coated with 50 μg/mL of capture antibody (DB6-1), and incubated at 4°C overnight. Next, the plate was washed with PBS and blocked with 1% BSA/PBS at room temperature for 2 hours. Immunoaffinity-purified DENV-2 NS1 protein was serially diluted using different buffer systems: PBS, 1% BSA/PBS, 1% NHS, or 10% NHS. After removal of the blocking buffer and three washes with PBS, serially diluted NS1 protein was added to the wells and incubated at room temperature for 1 hour. Following three washes with PBS, biotin-labeled DB20-6 (2 μg/mL) was added to the wells (50 μL/well) and incubated at room temperature for 1 hour. After a further three washes with PBS, HRP-conjugated streptavidin (1:1,000) was added to the wells and incubated at room temperature for 1 hour. Finally, the plates were washed four times with PBST0.1, and signals were developed with the peroxidase substrate, OPD (Sigma). The reaction was stopped by the addition of 3 N HCl, and the plates were read using the Spectra Max M5 (Molecular Devices) microplate reader at an absorbance of 490 nm.
Detection of NS1 protein in clinical samples.
An ELISA plate was coated with 50 μg/mL of DB6-1 in 0.1 M sodium bicarbonate buffer (NaHCO3, pH 8.6) at room temperature for 2 hour. After washing with PBS, the plate was blocked with blocking buffer (1% BSA in PBS) at 4°C overnight. Clinical samples were diluted with Thermo IgG elution buffer (pH 2.8, Product No. 21009) for 20 minutes, and then neutralized with 1 M Tris (pH 9.0) at a final ratio of 1 to 100. The plate was washed with PBS, and then incubated with diluted clinical samples (50 μL/well) at room temperature for 1 hour. Following another wash with PBS, the plate was incubated with biotin-labeled DB20-6 (2 μg/mL) at room temperature for 1 hour. The plate was then washed with PBS, and hybridized with HRP-conjugated streptavidin (1:1,000) at room temperature for 1 hour. After a further four washes with PBST0.1, signals were developed by the addition of the peroxidase substrate, OPD (Sigma). The reaction was stopped by the addition of 3 N HCl, and the plates were read using a Spectra Max M5 (Molecular Devices) microplate reader at an absorbance of 490 nm.
Detection of NS1 protein in clinical samples by commercial kits.
Two commercial kits, dengue early ELISA (MyBioSource) and DENV NS1 ELISA (EUROIMMUN), were purchased for comparison. All experimental procedures were performed in accordance with the instructions provided with the kits.
RESULTS
Generation and identification of mAbs against DENV-2 NS1 protein.
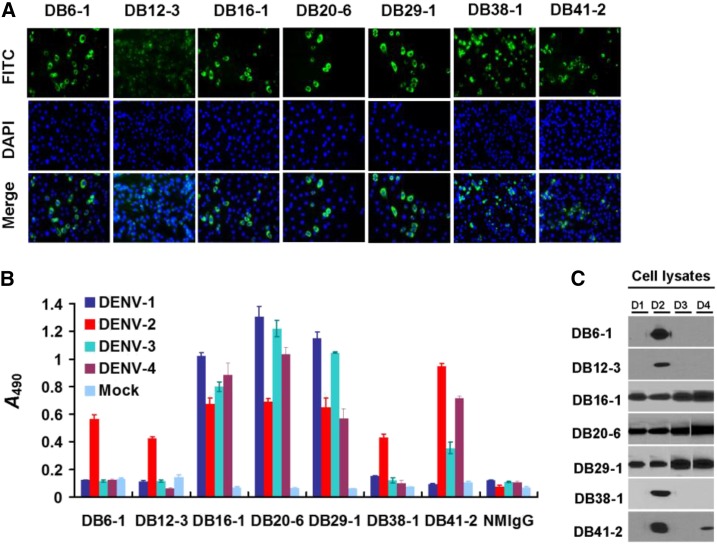
In this study, we generated a series of mAbs against immunoaffinity-purified DENV-2 NS1 protein. We characterized and analyzed the specificity of these mAbs to develop diagnostic tools for the clinical detection of NS1 protein. To acquire basic information regarding these mAbs, we performed immunofluorescence assay (IFA) (Figure 1A), cellular ELISA (Figure 1B), and Western blot analysis (Figure 1C). We report that these mAbs exhibit specific (DB6-1, DB12-3, and DB38-1), and nonspecific (DB16-1, DB20-6, DB29-1, and DB41-2) recognition of NS1 proteins (Figure 1B and C, Table 1). Of the four mAbs that recognize more than one serotype, DB16-1, DB20-6, and DB29-1 recognize NS1 protein from all serotypes, whereas DB41-2 recognizes the NS1 protein from DENV serotypes 2 and 4 (Figure 1B and C). All of the integrated information is shown in Table 1.
Figure 1.
Identification of monoclonal antibodies (mAbs) against nonstructural protein 1 (NS1) by immunofluorescence assay, cellular enzyme-linked immunosorbent assay (ELISA), and Western blot analysis. (A) The indicated mAbs (DB6-1, DB12-3, DB16-1, DB20-6, DB29-1, DB38-1, and DB41-2) were individually incubated with BHK21 cells infected by dengue virus type 2 (DENV-2) (strain 16681). After secondary antibody and DAPI staining, the signals were detected by immunofluorescence microscopy. (B) C6/36 cells infected with the indicated serotype were incubated with the indicated mAbs for analysis by cellular ELISA. (C) Lysates from C6/36 cells infected with the indicated serotypes of DENV were separated on a reducing gel. Antigens of four DENV serotypes were recognized by DB16-1, DB20-6, and DB29-1. DENV2-specific NS1 protein was recognized by DB6-1, DB12-3, and DB38-1. DB41-2 can recognize NS1 proteins of DENV-2 and DENV-4. This figure appears in color at www.ajtmh.org.
Table 1.
Characterization of DENV-2 mAbs by IFA, ELISA, WB
| mAbs | Isotype | Specificity | IFA | ELISA | WB | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Light chain | D2 | D1 | D2 | D3 | D4 | D1 | D2 | D3 | D4 | ||
| DB6-1 | IgG2a, κ | NS1 | + | − | + | − | − | − | + | − | − |
| DB12-3 | IgG1, κ | NS1 | + | − | + | − | − | − | + | − | − |
| DB16-1 | IgG2a, κ | NS1 | + | + | + | + | + | + | + | + | + |
| DB20-6 | IgG1, κ | NS1 | + | + | + | + | + | + | + | + | + |
| DB29-1 | IgG1, κ | NS1 | + | + | + | + | + | + | + | + | + |
| DB38-1 | IgG1, κ | NS1 | + | − | + | − | − | − | − | − | − |
| DB41-2 | IgG1, κ | NS1 | + | − | + | + | + | − | + | − | + |
DENV = dengue virus; ELISA = enzyme-linked immunosorbent assay; IFA = immunofluorescence assay; mAbs = monoclonal antibodies; NS1 = nonstructural protein 1; WB = Western blot analysis; D1, D2, D3, and D4, DENV-1 to -4; Ig, immunoglobulin;. (+) positive result to DENV, A490 > 0.2; (−) negative result to DENV, A490 < 0.2.
Identification of epitopes recognized by anti-NS1 mAbs.
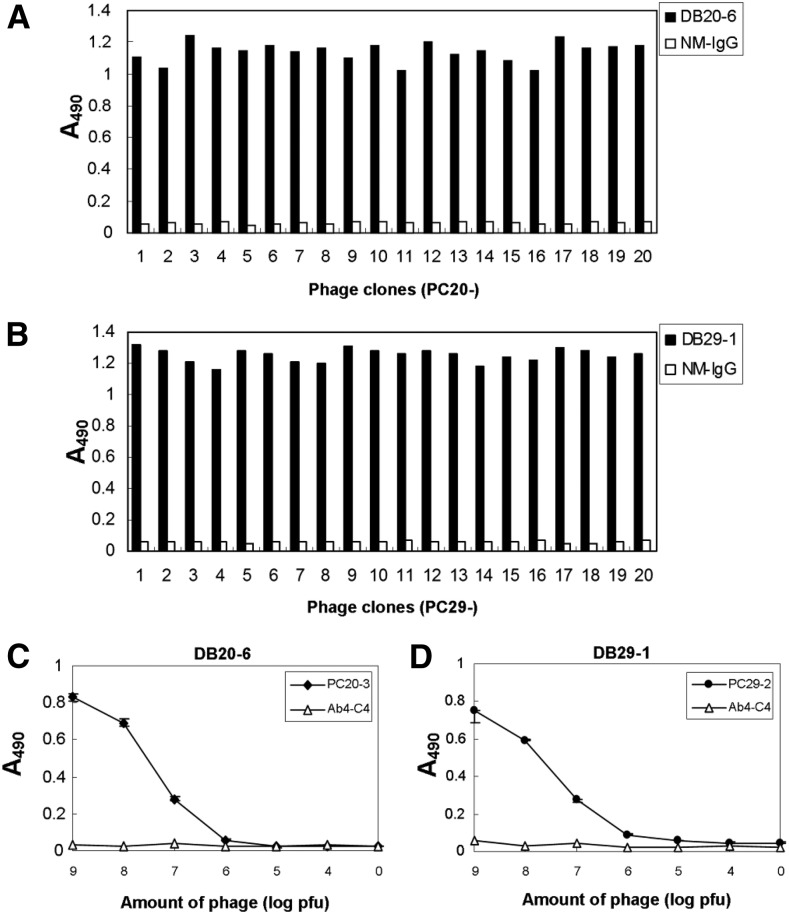
We proceeded to determine which epitopes are recognized by these mAbs. DB16-1, DB20-6, and DB29-1, recognize the NS1 proteins from all serotypes of DENV (Figure 1B and C), suggesting that there may be a common epitope on the NS1 protein recognized by these three mAbs. We previously used a phage-displayed random peptide library to identify the epitope recognized by DB16-1.30 Here, we used phage display and site-directed mutagenesis to identify the epitopes recognized by DB20-6 and DB29-1. These two mAbs were used as bait to capture candidate phage clones from the phage-displayed random peptide library. After three rounds of biopanning, the binding specificity and avidity of the bound phage clones were confirmed by ELISA (Figure 2A and B). The immunopositive phage clones selected from DB20-6 (prefixed with PC20 and numbered from 1 to 20) or DB29-1 (prefixed with PC29 and numbered from 1 to 20) all exhibited high binding activities to the respective mAbs, whereas exhibiting little or no binding ability to the negative control, NM IgG. Phage clones PC20-3 and PC29-2, which exhibited high affinity to their respective mAbs, were used to examine the dose-dependent effects. PC20-3 and PC29-2 bound to DB20-6 and DB29-1 mAbs in a dose-dependent manner. However, the control peptide, Ab4-C4,31 did not bind to either mAb (Figure 2C and D).
Figure 2.
Identification of DB20-6- or DB29-1-selected phage clones by enzyme-linked immunosorbent assay (ELISA) and characterization of the specificity of ELISA-positive phage clones. ELISA was performed to screen for phage clones with high binding affinity to DB20-6 and DB29-1. After three rounds of biopanning, all (A) 20 phage clones from DB20-6 and (B) 20 phage clones from DB29-1 showed high reactivity to DB20-6 or DB29-1, but not to normal mouse IgG (NM IgG). (C) The DB20-6-selected positive phage clone (PC20-3) bound to DB20-6 in a dose-dependent manner with high specificity, but control phage clone (Ab4-C4) did not. (D) The DB29-1-selected positive phage clone (PC29-2) bound to DB29-1 in a dose-dependent manner with high specificity, whereas Ab4-C4 did not.
To identify the binding motif of DB20-6 and DB29-1, 20 immunopositive phage clones (PC20-1 to PC-10 for DB20-6; PC29-1 to PC-10 for DB29-1) were amplified, and phage DNAs were extracted and sequenced. These 20 immunopositive phage clones possessed different amino acid sequences (Table 2A and B); however, alignment of the sequences revealed the presence of conserved motifs. For the DB20-6 epitope, a conserved tryptophan (W)-glycine (G)-lysine (K) motif was observed, which corresponds to amino acid residues 118–120 of DENV NS1. For the DB29-1 epitope, a tryptophan (W)-X-X-tryptophan (W)-glycine (G) motif was identified, which corresponds to amino acid residues 115-119 of DENV NS1 (Supplemental Figure 2 and Table 2).
Table 2.
Alignment of phage-displayed peptide sequences selected by DB20-6 and DB29-1
| Clone | Insert* | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DB20-6 | |||||||||||||||||||
| PC20-1 | G | W | S | Y | T | F | A | P | W | G | W | G | |||||||
| PC20-2 | L | E | H | Q | W | S | T | G | W | R | H | P | |||||||
| PC20-3 | S | S | H | R | I | W | T | E | L | W | G | K | |||||||
| PC20-4 | E | V | S | P | H | E | W | R | K | W | G | K | |||||||
| PC20-5 | D | H | N | M | W | G | K | R | L | P | P | S | |||||||
| PC20-6 | T | G | P | T | M | H | W | R | Q | W | G | K | |||||||
| PC20-7 | A | P | G | L | W | Q | T | W | G | W | S | P | |||||||
| PC20-8 | I | P | S | W | S | S | W | G | K | N | G | A | |||||||
| PC20-9 | D | V | K | H | Q | T | W | Y | N | W | G | K | |||||||
| PC20-10 | N | D | L | E | K | L | K | Y | W | G | K | Q | |||||||
| DEN-2† | E | L | R | Y | S | W | K | T | W | G | K | A | K | ||||||
| DEN-1† | E | H | K | Y | S | W | K | T | W | G | K | A | K | ||||||
| DEN-3† | E | L | K | Y | S | W | K | T | W | G | L | A | K | ||||||
| DEN-4† | D | L | K | Y | S | W | K | T | W | G | K | A | K | ||||||
| DB29-1 | |||||||||||||||||||
| PC29-1 | M | P | S | Y | S | D | S | S | W | L | M | W | |||||||
| PC29-2 | W | Q | K | P | W | N | S | W | G | V | S | G | |||||||
| PC29-3 | E | R | S | L | S | T | P | S | W | R | H | W | |||||||
| PC29-4 | Y | V | G | R | A | Y | A | P | W | L | E | W | |||||||
| PC29-5 | Q | P | A | I | P | L | G | H | W | S | N | W | |||||||
| PC29-6 | A | P | Y | H | T | W | G | W | P | S | L | Q | |||||||
| PC29-7 | V | V | W | D | S | W | G | S | L | M | T | A | |||||||
| PC29-8 | L | N | T | D | W | Y | Q | W | G | L | E | P | |||||||
| PC29-9 | K | T | Q | P | N | I | H | S | W | V | L | W | |||||||
| PC29-10 | Q | Y | P | Q | W | D | Y | W | G | H | S | H | |||||||
| DEN-2† | E | L | R | Y | S | W | K | T | W | G | K | A | K | ||||||
| DEN-1† | E | H | K | Y | S | W | K | T | W | G | K | A | K | ||||||
| DEN-3† | E | L | K | Y | S | W | K | T | W | G | L | A | K | ||||||
| DEN-4† | D | L | K | Y | S | W | K | T | W | G | K | A | K | ||||||
Phage-displayed consensus amino acids are shown in boldface.
Amino acid sequence of residues 110-122 of the NS1 protein of dengue virus.
Epitope validation by binding and competition assays.
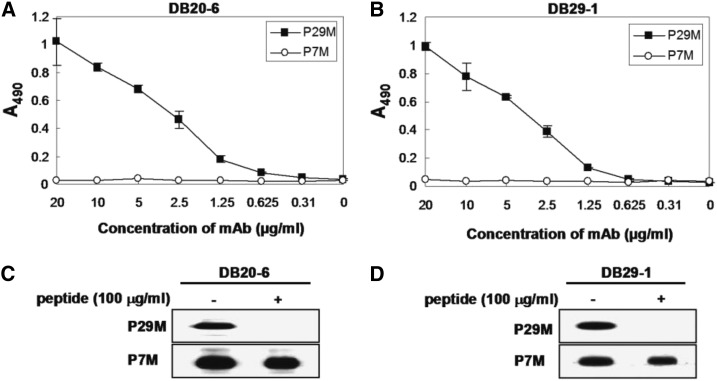
To validate the epitopes of these mAbs, we synthesized the P29M peptide (LKYSWKTWGKAK), which corresponds to amino acids 111 to 122 of DENV-4 NS1. The synthetic peptide P29M was used as bait for serial dilutions of DB20-6 and DB29-1. We observed that both mAbs bound to the synthetic peptide in a dose-dependent manner (Figure 3A and B); the P7M control peptide,31 however, did not bind. Peptide competitive inhibition assays were performed to confirm the specificity of binding; synthetic peptide P29M, but not control peptide P7M, inhibited the binding of DB20-6 and DB29-1 to NS1 (Figure 3C and D).
Figure 3.
Characterization of the synthetic peptide, P29M, which corresponds to amino acid residues 111-122 of dengue virus type 2 (DENV2-) nonstructural protein 1 (NS1). (A) and (B) Synthetic peptide P29M was used as an antigen for the detection of DB20-6 and DB29-1. Both DB20-6 and DB29-1 bound to P29M in a dose-dependent manner, but not to control peptide P7M. (C) and (D) Synthetic peptide P29M, but not control peptide P7M, inhibited the binding of DB20-6 and DB29-1 to DENV2- NS1 protein.
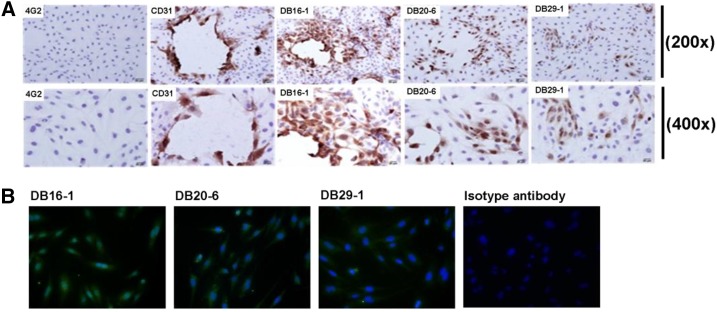
Epitope validation on endothelial cells.
As mentioned earlier, the epitopes of DB20-6 and DB29-1 possess similar amino acid sequences to the epitope of DB16-1.30 As DB16-1 recognizes both NS1 and the LYRIC protein on endothelial cells through the molecular mimicry effect,30 we compared the binding abilities of DB16-1, DB20-6, and DB29-1 to endothelial cells through immunohistochemistry and IFA. We report that both DB20-6 and DB29-1 could recognize endothelial cells, but their binding activities are not as strong as that of DB16-1 (Figure 4A and B).
Figure 4.
DB20-6 and DB29-1 cross-reacts with endothelial cells. (A) Human umbilical vein endothelial cells (HUVECs) were incubated with DB16-1, DB20-6, DB29-1, CD31, or 4G2, and the bound antibodies were visualized by light microscopy. CD31 antibody was used as the positive control and 4G2 antibody was used as the negative control. (B) HUVEC cells were incubated with DB16-1, DB20-6, DB29-1, or control antibody, and the image were visualized by fluorescence microscopy. This figure appears in color at www.ajtmh.org.
Establishment of a standard curve for detection of NS1 protein.
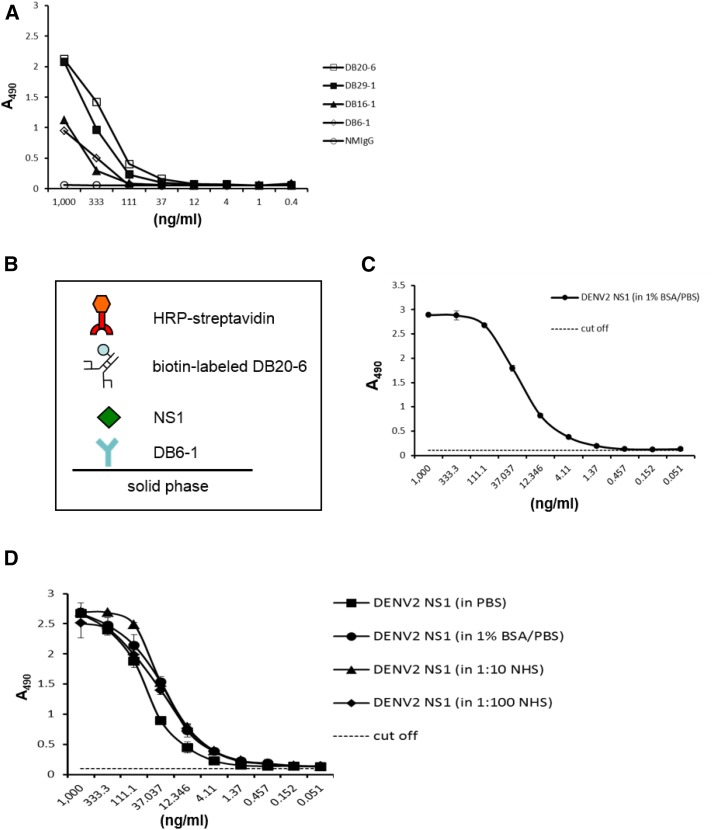
Based on the information obtained from the above experiments, we established a platform for detection of NS1 protein. DENV-2 (strain 16681)-infected C6/36 cell supernatant was collected, and NS1 protein was purified using anti-NS1 antibody (DB16-1)-conjugated Sepharose 4 Fast Flow beads (Supplemental Figure1). The presence of purified NS1 protein was confirmed by SDS-PAGE (Supplemental Figure 1B) and Western blot analysis (Supplemental Figure 1C). The purified NS1 protein was used as an antigen to test the binding abilities and sensitivities of different mAbs. Of the four screened mAbs (DB6-1, DB16-1, DB20-6, and DB29-1), DB20-6 showed the strongest binding to NS1, and this mAb was therefore used for further experiments (Figure 5A).
Figure 5.
Development of a diagnostic platform for dengue virus type 2 (DENV-2). (A) Direct enzyme-linked immunosorbent assay (ELISA) was used to compare a panel of monoclonal antibodies (mAbs) against serial dilutions of immunoaffinity-purified DENV-2 nonstructural protein 1 (NS1) protein. The ELISA plate was coated with a 3-fold dilution of purified DENV-2 NS1 protein. After washing, the coated NS1 protein was detected with cross-reactive mAbs (DB16-1, DB20-6, and DB29-1) or type-specific mAb (DB6-1) at a concentration of 1 μg/mL. Normal mouse immunoglobulin G was used as the negative control. (B) Schematic describing the diagnostic platform. (C) Standard curve of DENV-2 NS1. DENV-2 NS1-specific mAb, DB6-1, was used to coat an ELISA plate at a concentration of 50 μg/mL. Immunoaffinity-purified DENV-2 NS1 protein was diluted 3-fold in 1% bovine serum albumin/phosphate-buffered saline solution and incubated with the capture mAb. NS1 protein was detected through the scheme shown in panel (B). Data points represent the mean ± standard deviation (SD) for three replicates. The dashed line represents the cutoff value. (D) Effect of human serum constituents on the detection of NS1. Immunoaffinity-purified DENV-2 NS1 protein was dissolved in the indicated buffers, and subsequently incubated with the capture mAb, DB6-1. After washing, the bound NS1 protein was probed with biotin-labeled mAb, DB20-6, and detected as shown in panel (B). Data points represent the mean ±SD for three replicates. The dashed line represents the cutoff value. This figure appears in color at www.ajtmh.org.
As the platform needs to be used for serotyping, we used a serotype-specific mAb (DB6-1) as the capture antibody. In the platform, captured NS1 is detected by biotinylated DB20-6, and the signal is detected using HRP-streptavidin, as shown in Figure 5B. ELISA was used to demonstrate that the detection limit for DENV-2 NS1 protein in our platform is about 1 ng/mL (Figure 5C).
We dissolved immunoaffinity-purified NS1 protein in PBS, 1% BSA/PBS, or NHS at different dilutions (1:10 or 1:100) to compare the resulting standard curves (Supplemental Figure 4B) and found that the platform can detect NS1 protein in different buffer systems with high sensitivity and specificity.
Detection of NS1 protein in sera from patients infected with DENV-2.
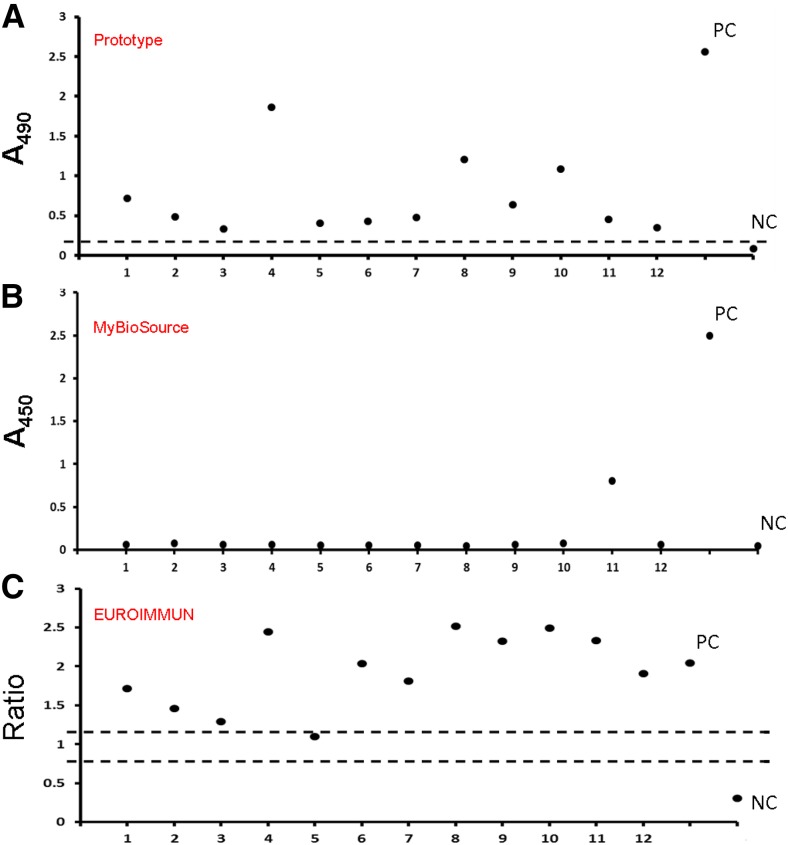
A literature review revealed that NS1 protein is present during the early stage of dengue infection, especially the acute phase.10,18,32 This phenomenon is related to the underlying pathogenesis of severe dengue diseases, such as DHF. We proceeded to use serum samples from dengue patients to evaluate the reliability of our platform. Sera were diluted in Thermo IgG elution buffer (pH 2.8, Product No. 21009) at a ratio of 1 to 100, as described in the section entitled “Detection of NS1 protein in clinical samples” of the “Materials and methods.” As shown in Figure 6A, our platform was able to detect various levels of NS1 protein concentrations in the serum samples obtained from 12 dengue patients, whereas no NS1 protein was detected in the sera obtained from 12 healthy volunteers. Two other commercial NS1 diagnostic platforms (MyBioSource and EUROIMMUN) were used to detect the target protein in the same serum samples, with their respective data shown in Figure 6B and C. As the data suggested, our platform displayed better performance than the commercial kits (Table 3).
Figure 6.
Detection of nonstructural protein 1 (NS1) in sera from patients infected with dengue virus type 2. (A) Sera were collected and analyzed with the capture enzyme-linked immunosorbent assay platform. All analyzed sera were treated by Thermo IgG elution buffer (pH 2.8) at room temperature for 20 minutes, and then neutralized with 1 M Tris (pH 9) at a final dilution of 1–100. Acid-treated sera were incubated with the capture mAb, DB6-1. After sequential recognition by biotin-DB20-6 and horseradish peroxidase-streptavidin, signals were developed, and the optical density at 490 nm was read. The dashed line represents the cutoff value, which is the mean plus three times the standard derivation from 12 normal human serum samples. (B) The same batch of serum samples was analyzed using a commercial NS1 diagnostic kit (MyBioSource); PC = positive control; NC = negative control. (C) The same batch of serum samples was analyzed using a commercial NS1 diagnostic kit (EUROIMMUN). The y axis is the ratio of each sample or controls (ratio = extinction of the control or patient sample/extinction of calibrator 2). Ratio ≥ 1.1: positive; ratio ≥ 0.8 to < 1.1: borderline; ratio < 0.8: negative.This figure appears in color at www.ajtmh.org.
Table 3.
Sensitivity and specificity of three different NS1 ELISA platforms
| DENV-2 NS1 ELISA kit | |||
|---|---|---|---|
| Prototype platform | MvBioSource (dengue early ELISA) | EUROIMMUN (dengue virus NS1 ELISA) | |
| Sensitivity | 100% (12/12) | 8.3% (1/12) | 91.6% (11/12) |
| Specificity | 100% (12/12) | N/A | N/A |
DENV-2 = dengue virus type 2; ELISA = enzyme-linked immunosorbent assay; N/A = not assayed; NS1 = nonstructural protein 1.
Serotyping specificity of the NS1 diagnostic platform.
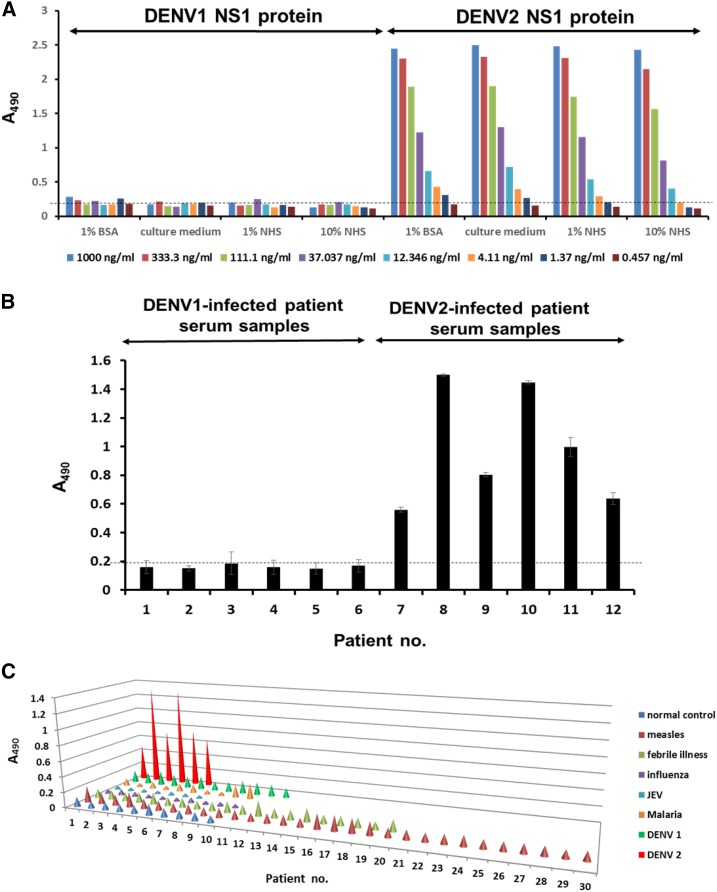
To confirm the serotyping specificity of our platform, we used it to detect NS1 protein in human serum. We found that our platform could detect the NS1 protein of DENV-2 with high specificity, but not the NS1 protein of DENV-1 (Figure 7A). Furthermore, we used purified NS1 proteins from four serotypes of DENV and from Zika virus as the antigens and performed the ELISA using our platform. The result indicated that only DENV-2 NS1 protein could be detected by DB6-1 (Supplemental Figure 4A). We further demonstrated that our platform can detect the NS1 protein in DENV-2-infected patient sera, but not in DENV-1-infected patient sera (Figure 7B). Our results demonstrate that our platform can detect the NS1 protein of DENV-2 with high specificity, without showing any cross-reactivity to the NS1 protein of DENV-1. To further confirm the specificity of the platform, we used the sera from other patients who displayed the symptom of fever from other infectious diseases, including measles, fever with unknown causes, influenza virus, JEV, and malaria. The data implicated that our platform could specifically bind to the NS1 protein from DENV-2-infected patient serum samples without showing any false positive bindings (Figure 7C).
Figure 7.
Serotyping specificity of the nonstructural protein 1 (NS1) diagnostic platform. (A) DB6-1 was used to coat a 96-well plate, and incubated with serially diluted immunoaffinity-purified dengue virus type 1 (DENV-1) and DENV-2 NS1 proteins dissolved in different buffer systems. After binding to biotin-DB20-6 and horseradish peroxidase (HRP)-streptavidin, the signals were detected using a spectrophotometer at 490 nm after processing of the substrate with HRP. Dashed lines represented the cutoff value. (B) The NS1 protein of serum samples from patients infected with DENV-1 or DENV-2 were detected using DENV-2 NS1 diagnostic platforms. Dashed lines represented the cutoff value. (C) Clinical serum samples from patients infected with DENV-1, DENV-2, measles, febrile illnesses, influenza, Japanese encephalitis virus, malaria were detected by DENV-2 NS1 diagnostic platforms, normal control sera were used as the negative control. This figure appears in color at www.ajtmh.org.
DISCUSSION
The goal of this study was to establish a diagnostic kit for early detection of the DENV using our newly generated mAbs for providing an alternative in clinical diagnosis. The target antigen in this platform is the secreted form of NS1, which is present during the early acute phase of DENV infection. The lack of effective antiviral drugs and vaccines for DENV has necessitated the development of diagnostic tools for early detection. Such tools could enable clinical physicians to make more accurate diagnoses for this prevalent arboviral disease that causes almost 390 million infections annually.
In this study, we newly generated mAbs against the NS1 protein of DENV serotype 2 (strain 16681). Through our pilot experiments, we characterized the binding specificity and affinity of these mAbs (Figure 1, Table 1). To fully characterize mAbs for the subsequent development of epitope-based diagnosis and elucidation of viral antigen–antibody interactions, it is important to first identify the epitope recognized by the mAbs. To identify the epitope, we used a phage displayed random peptide library composed of 12-mer peptide-displayed M13 phage clones. These 12-mer peptides were fused with the structural protein of M13 phage, pIII, and exposed to the surface of the phage. This enabled the exposed peptides to mimic linear or conformational epitopes, including tertiary or quaternary structure, and fit into the pockets of the antigen binding sites. Subsequent sequencing and alignment of the bound phage clones revealed the presence of possible epitopes.
From the phage display study, we found the WXXWGK motif to be a common motif among the four serotypes of DENV (corresponding to amino acid residues 115-120 of NS1 protein) (Table 2A and B, Figure 4A). This motif is similar to that of DB16-1, KXWG (detected in our previous study), emphasizing the importance of this motif.30 NS1 protein was previously reported to play a role in inducing autoimmunity and enhancing the severity of disease, through leakage of the plasma.30,33,34 In this study, we found that the B-cell epitopes of DB20-6 and DB29-1 are very similar to that of DB16-1, and these three epitopes exhibit certain overlaps in their motifs. As previously reported, DB16-1 is an autoantibody that can cross-react with endothelial cells (Liu and others).30 Much like DB16-1, DB20-6, and DB29-1 can also cross-react with endothelial cells, but their binding activities were not as great as that of DB16-1 (Figure 4B). B-cell epitope mapping of these three different mAbs revealed that autoantibodies may be induced by this epitope. After peptide sequences alignment, we found there are the similar epitopes in different flavivirus strains, including JEV, WNV, and SLE (data not shown). Recently, a paper revealed the structure of flavivirus NS1, which showed that the epitope recognized by our mAbs is located in the “wing domain” of NS1 protein.35 The epitope is a linear epitope and is exposed on the outer most surface of the NS1 protein. As such, this epitope can be readily detected by our mAbs in different conformations (monomer, dimer, and hexamer). The DB16-1, DB20-6, and DB29-1 have the potential to cross react the closely related flavivirus, nevertheless, the specificity of capture antibody (DB6-1) should avoid the cross reactivity in the detection.
ELISA was used to compare the binding abilities of mAbs that could recognize NS1. The antibody with the strongest binding activity was combined with a serotype-specific antibody to form the basis of the diagnostic platform. The detection limit of our platform is about 1 ng/mL, which is more sensitive than earlier systems.12,29 Using our platform, we can detect purified NS1 protein dissolved in various buffer systems, including PBS, 1% BSA/PBS, and NHS at different dilutions (1:10 and 1:100). Of particular note, NHS mimics the condition of clinical samples, suggesting our platform is suitable for use in a clinical setting.
We confirmed the platform’s clinical suitability by using it to detect NS1 protein in the sera of patients infected with DENV. NS1 protein was previously reported to be present from the onset of disease to about 2 weeks later.10 However, NS1 usually peaks from day 1 to 3. Moreover, NS1 protein is more abundant in the sera of patients with DHF than that in sera of patients with DF.18 The data generated using our platform are consistent with earlier reports that sera from DHF patients contain more NS1 protein, but more clinical samples are needed to confirm this finding. In the present study, we found that not every serum sample contained high levels of NS1, even for samples from the acute phase, possibly due to the formation of an immune complex: when anti-NS1 antibodies bind to NS1, these antibodies may shield or obstruct access to the epitope, preventing detection using our platform. Such a phenomenon may depend on the immune conditions of the patient.
False negatives or reductions in target protein readout due to immune complexes have also been observed in other cases, such as detection of the HIV p24 antigen.36 It has been reported that such immune complexes can be dissociated by various methods, including treatment with acid, alkali, or heat. Although acidification or alkali treatment may cause the immune complex to dissociate, the release of p24 protein was reported to be limited, causing false-negative readouts; this was particularly true when the antibody against p24 protein was used at very high titers. Alternatively, boiling diluted samples may be a possible solution for detecting linear epitopes, as immune complexes are unlikely to reform after heat treatment. It was previously reported that heat-mediated immune complex dissociation could enhance the ELISA signal of HIV p24 protein.37,38 The serum samples used in this study were treated with Thermo IgG elution buffer (pH 2.8); however, the untreated samples could still be detected by our platform (Supplemental Figure 3).
In this study, we demonstrated the high sensitivity (100%) and specificity (100%) of our platform. In the future, we intend to examine more clinical patients’ sera samples including both DENV-2-infected serum and sera from other febrile illnesses to further demonstrate the clinical benefits of our diagnostic platform. Serum samples from non-DENV infected patients may lead to false-positive results, which need to be resolved to ensure the accuracy of the diagnostic outcome of the platform developed. We expect this platform could be another choice or improve the detection limit of some commercially available diagnostic platforms on DENV-2 NS1 detection. At present, the threat from arboviral diseases is increasing, a problem compounded by the lack of effective therapies and preventive measures against DENV infection. A rapid, specific, and sensitive detection system is urgently needed, especially in developing countries where health-care resources are limited. The current treatment of dengue infection focuses primarily on solution infusions, which is only prophylactic, and does not require the physicians to know the specific serotype of dengue infection. However, the clinical benefit of serotyping NS1 ELISA is that, by providing more precise information on the serotype of dengue infection, it will help physicians to provide more accurate diagnosis, and hence lead to more appropriate treatment. So far, there is no drug or vaccine for DENV infection. We expect that serotype-specific therapeutic antibodies will be able to neutralize the virus after early serotyping diagnosis, thereby eliminating possible antibody-dependent enhancement effects in the future. For epidemiological analyses, information on the serotype(s) involved in a given outbreak of DENV infection is important, especially in areas in which four serotypes circulated. The serotype-specific diagnostic platform may make up for the shortcomings of current commercial platforms.
Here, we describe a possible platform for DENV detection, which may also serve as a basis for the development of detection platforms for other infectious diseases. Based on the model, we also need to develop the same diagnostic platforms to detect the other DENV NS1 proteins, such as DENV-1, DENV-3, and DENV-4 for future clinical diagnosis. On the other hand, the idea could also be applied to other diagnostic platforms, such as strip, paper, or disk.28,29,39 The detection procedures using these materials are similar to those of our platform in terms of detection mechanism; however, these diagnostic platforms require less time due to the properties of the different materials used. The low cost and ease of use make the device well-suited as a point-of-care diagnostic system. A complete network combining basic point of care and medical supportive systems should aid in the successful containment of the disease. All these efforts are made to provide an alternative choice for the monitoring, control, or prevention of dengue virus infection worldwide.
Supplementary Material
Acknowledgments:
We thank the Core Facility of the Institute of Cellular and Organismic Biology for their technical assistance. We are particularly grateful to Ms. Ming-Chu Kuo for the malaria patients’ samples.
Note: Supplemental table and figures appear at www.ajtmh.org.
REFERENCES
- 1.Back AT, Lundkvist A, 2013. Dengue viruses: an overview. Infect Ecol Epidemiol 3: 10.3402/iee.v3i0.19839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li PC, Liao MY, Cheng PC, Liang JJ, Liu IJ, Chiu CY, Lin YL, Chang GJ, Wu HC, 2012. Development of a humanized antibody with high therapeutic potential against dengue virus type 2. PLoS Negl Trop Dis 6: e1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Malavige GN, Ogg GS, 2013. T cell responses in dengue viral infections. J Clin Virol 58: 605–611. [DOI] [PubMed] [Google Scholar]
- 4.Chuang YC, Lin YS, Liu CC, Liu HS, Liao SH, Shi MD, Lei HY, Yeh TM, 2013. Factors contributing to the disturbance of coagulation and fibrinolysis in dengue virus infection. J Formos Med Assoc 112: 12–17. [DOI] [PubMed] [Google Scholar]
- 5.Tang KF, Ooi EE, 2012. Diagnosis of dengue: an update. Expert Rev Anti Infect Ther 10: 895–907. [DOI] [PubMed] [Google Scholar]
- 6.Prestwood TR, May MM, Plummer EM, Morar MM, Yauch LE, Shresta S, 2012. Trafficking and replication patterns reveal splenic macrophages as major targets of dengue virus in mice. J Virol 86: 12138–12147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prestwood TR, Prigozhin DM, Sharar KL, Zellweger RM, Shresta S, 2008. A mouse-passaged dengue virus strain with reduced affinity for heparan sulfate causes severe disease in mice by establishing increased systemic viral loads. J Virol 82: 8411–8421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jupatanakul N, Sim S, Dimopoulos G, 2014. Aedes aegypti ML and Niemann-Pick type C family members are agonists of dengue virus infection. Dev Comp Immunol 43: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou Y, Austin SK, Fremont DH, Yount BL, Huynh JP, de Silva AM, Baric RS, Messer WB, 2013. The mechanism of differential neutralization of dengue serotype 3 strains by monoclonal antibody 8A1. Virology 439: 57–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hu D, Di B, Ding X, Wang Y, Chen Y, Pan Y, Wen K, Wang M, Che X, 2011. Kinetics of non-structural protein 1, IgM and IgG antibodies in dengue type 1 primary infection. Virol J 8: 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gutsche I, et al., 2011. Secreted dengue virus nonstructural protein NS1 is an atypical barrel-shaped high-density lipoprotein. Proc Natl Acad Sci USA 108: 8003–8008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Young PR, Hilditch PA, Bletchly C, Halloran W, 2000. An antigen capture enzyme-linked immunosorbent assay reveals high levels of the dengue virus protein NS1 in the sera of infected patients. J Clin Microbiol 38: 1053–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Winkler G, Maxwell SE, Ruemmler C, Stollar V, 1989. Newly synthesized dengue-2 virus nonstructural protein NS1 is a soluble protein but becomes partially hydrophobic and membrane-associated after dimerization. Virology 171: 302–305. [DOI] [PubMed] [Google Scholar]
- 14.Flamand M, Megret F, Mathieu M, Lepault J, Rey FA, Deubel V, 1999. Dengue virus type 1 nonstructural glycoprotein NS1 is secreted from mammalian cells as a soluble hexamer in a glycosylation-dependent fashion. J Virol 73: 6104–6110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hang VT, et al., 2009. Diagnostic accuracy of NS1 ELISA and lateral flow rapid tests for dengue sensitivity, specificity and relationship to viraemia and antibody responses. PLoS Negl Trop Dis 3: e360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sylvestre G, Gandini M, de Araujo JM, Kubelka CF, Lourenco-de-Oliveira R, Maciel-de-Freitas R, 2014. Preliminary evaluation on the efficiency of the kit Platelia Dengue NS1 Ag-ELISA to detect dengue virus in dried Aedes aegypti: a potential tool to improve dengue surveillance. Parasit Vectors 7: 155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jacobs MG, Robinson PJ, Bletchly C, Mackenzie JM, Young PR, 2000. Dengue virus nonstructural protein 1 is expressed in a glycosyl-phosphatidylinositol-linked form that is capable of signal transduction. FASEB J 14: 1603–1610. [DOI] [PubMed] [Google Scholar]
- 18.Libraty DH, et al. , 2002. High circulating levels of the dengue virus nonstructural protein NS1 early in dengue illness correlate with the development of dengue hemorrhagic fever. J Infect Dis 186: 1165–1168. [DOI] [PubMed] [Google Scholar]
- 19.Bessoff K, Delorey M, Sun W, Hunsperger E, 2008. Comparison of two commercially available dengue virus (DENV) NS1 capture enzyme-linked immunosorbent assays using a single clinical sample for diagnosis of acute DENV infection. Clin Vaccine Immunol 15: 1513–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lapphra K, Sangcharaswichai A, Chokephaibulkit K, Tiengrim S, Piriyakarnsakul W, Chakorn T, Yoksan S, Wattanamongkolsil L, Thamlikitkul V, 2008. Evaluation of an NS1 antigen detection for diagnosis of acute dengue infection in patients with acute febrile illness. Diagn Microbiol Infect Dis 60: 387–391. [DOI] [PubMed] [Google Scholar]
- 21.De Decker S, Vray M, Sistek V, Labeau B, Enfissi A, Rousset D, Matheus S, 2015. Evaluation of the diagnostic accuracy of a new dengue IgA capture assay (platelia dengue IgA capture, Bio-Rad) for dengue infection detection. PLoS Negl Trop Dis 9: e0003596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koraka P, Burghoorn-Maas CP, Falconar A, Setiati TE, Djamiatun K, Groen J, Osterhaus AD, 2003. Detection of immune-complex-dissociated nonstructural-1 antigen in patients with acute dengue virus infections. J Clin Microbiol 41: 4154–4159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hermann LL, Thaisomboonsuk B, Poolpanichupatam Y, Jarman RG, Kalayanarooj S, Nisalak A, Yoon IK, Fernandez S, 2014. Evaluation of a dengue NS1 antigen detection assay sensitivity and specificity for the diagnosis of acute dengue virus infection. PLoS Negl Trop Dis 8: e3193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barniol J, et al., 2011. Usefulness and applicability of the revised dengue case classification by disease: multi-centre study in 18 countries. BMC Infect Dis 11: 106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Castleberry JS, Mahon CR, 2003. Dengue fever in the Western Hemisphere. Clin Lab Sci 16: 34–38. [PubMed] [Google Scholar]
- 26.Dussart P, Petit L, Labeau B, Bremand L, Leduc A, Moua D, Matheus S, Baril L, 2008. Evaluation of two new commercial tests for the diagnosis of acute dengue virus infection using NS1 antigen detection in human serum. PLoS Negl Trop Dis 2: e280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lima Mda R, Nogueira RM, Schatzmayr HG, dos Santos FB, 2010. Comparison of three commercially available dengue NS1 antigen capture assays for acute diagnosis of dengue in Brazil. PLoS Negl Trop Dis 4: e738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Y, Bai J, Ying JY, 2015. A stacking flow immunoassay for the detection of dengue-specific immunoglobulins in salivary fluid. Lab Chip 15: 1465–1471. [DOI] [PubMed] [Google Scholar]
- 29.Wang HK, Tsai CH, Chen KH, Tang CT, Leou JS, Li PC, Tang YL, Hsieh HJ, Wu HC, Cheng CM, 2014. Cellulose-based diagnostic devices for diagnosing serotype-2 dengue fever in human serum. Adv Healthc Mater 3: 187–196. [DOI] [PubMed] [Google Scholar]
- 30.Liu IJ, Chiu CY, Chen YC, Wu HC, 2011. Molecular mimicry of human endothelial cell antigen by autoantibodies to nonstructural protein 1 of dengue virus. J Biol Chem 286: 9726–9736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu HC, Jung MY, Chiu CY, Chao TT, Lai SC, Jan JT, Shaio MF, 2003. Identification of a dengue virus type 2 (DEN-2) serotype-specific B-cell epitope and detection of DEN-2-immunized animal serum samples using an epitope-based peptide antigen. J Gen Virol 84: 2771–2779. [DOI] [PubMed] [Google Scholar]
- 32.Alcon S, Talarmin A, Debruyne M, Falconar A, Deubel V, Flamand M, 2002. Enzyme-linked immunosorbent assay specific to dengue virus type 1 nonstructural protein NS1 reveals circulation of the antigen in the blood during the acute phase of disease in patients experiencing primary or secondary infections. J Clin Microbiol 40: 376–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Avirutnan P, et al. , 2006. Vascular leakage in severe dengue virus infections: a potential role for the nonstructural viral protein NS1 and complement. J Infect Dis 193: 1078–1088. [DOI] [PubMed] [Google Scholar]
- 34.Srikiatkhachorn A, Kelley JF, 2014. Endothelial cells in dengue hemorrhagic fever. Antiviral Res 109: 160–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Akey DL, et al. , 2014. Flavivirus NS1 structures reveal surfaces for associations with membranes and the immune system. Science 343: 881–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schupbach J, Flepp M, Pontelli D, Tomasik Z, Luthy R, Boni J, 1996. Heat-mediated immune complex dissociation and enzyme-linked immunosorbent assay signal amplification render p24 antigen detection in plasma as sensitive as HIV-1 RNA detection by polymerase chain reaction. AIDS 10: 1085–1090. [PubMed] [Google Scholar]
- 37.Nadal D, Boni J, Kind C, Varnier OE, Steiner F, Tomasik Z, Schupbach J, 1999. Prospective evaluation of amplification-boosted ELISA for heat-denatured p24 antigen for diagnosis and monitoring of pediatric human immunodeficiency virus type 1 infection. J Infect Dis 180: 1089–1095. [DOI] [PubMed] [Google Scholar]
- 38.Ledergerber B, Flepp M, Boni J, Tomasik Z, Cone RW, Luthy R, Schupbach J, 2000. Human immunodeficiency virus type 1 p24 concentration measured by boosted ELISA of heat-denatured plasma correlates with decline in CD4 cells, progression to AIDS, and survival: comparison with viral RNA measurement. J Infect Dis 181: 1280–1288. [DOI] [PubMed] [Google Scholar]
- 39.Zhang B, Salieb-Beugelaar GB, Nigo MM, Weidmann M, Hunziker P, 2015. Diagnosing dengue virus infection: rapid tests and the role of micro/nanotechnologies. Nanomedicine (Lond) 11: 1745–1761. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.