Abstract.
Malaria control in West Africa is impeded by the large reservoir of chronic asymptomatic Plasmodium falciparum infections in the human population. This study aimed to assess the extent of diversity in the P. falciparum reservoir in Bongo District (BD), Ghana, at the end of the dry season, the lowest point in malaria transmission over the course of the year. Analysis of the variation in 12 microsatellite loci was completed for 200 P. falciparum isolates collected from a cross-sectional survey of residents of all ages from two catchment areas in BD. Analysis of the multilocus haplotypes showed high levels of genetic diversity (He = 0.74), no population differentiation yet significant linkage disequilibrium (LD) (ISA = 0.0127, P = 0.006) in BD. Multilocus LD was significant between and within catchment areas even though every haplotype in the population was unique and the majority of individuals (84.0%) harbored multiple-clone infections. The linkage structure among multilocus haplotypes was not associated with sampling location. These data provide the first study with deep sampling of the P. falciparum reservoir in an area of seasonal malaria transmission in West Africa. The co-occurrence of high multiplicity of infection (multiple-clone infections) with significant multilocus LD is surprising given the likelihood of high recombination rates in BD. The results suggest that the linkage structure among multilocus haplotypes has not been shaped by geographic separation of parasite populations. Furthermore, the observed LD levels provide a baseline population genetic metric with putatively neutral markers to evaluate the effects of seasonality and malaria control efforts in BD.
INTRODUCTION
The majority of Plasmodium falciparum infections in areas of high malaria transmission in West Africa are asymptomatic with few infections developing into uncomplicated or severe clinical disease. These chronic, asymptomatic infections pose a major challenge to elimination efforts in high-transmission areas across sub-Saharan Africa (SSA) as they continuously fuel transmission. A limited number of contemporary community surveys in this region have reported that ≥ 75% of individuals of all ages harbor asymptomatic infections in malaria-endemic areas.1 These infections comprise the reservoir of infection and are often undetected by conventional screening programs; thus, they remain untreated and can persist for over a year in an individual.2,3 In areas of highly seasonal malaria transmission in West Africa, there is a marked rainy season when transmission peaks and a subsequent dry season when transmission drops. Asymptomatic infections persist through the dry season and can act as a reservoir that can initiate transmission when the mosquito population reemerges during the following wet season. Therefore, understanding parasite population structure in this reservoir, in areas with seasonal transmission, could inform local control and elimination strategies by providing insight into isolated parasite populations that could be targeted and/or patterns of gene flow in the context of seasonality.
Multilocus microsatellite genotyping has allowed us to map the worldwide P. falciparum population genetic structure and detect ancestral migration patterns; it has also proven to be a valuable molecular tool to characterize parasite population structure on a global, national, and local level.4–6 In the West African region, multilocus microsatellite genotyping has revealed high levels of genetic diversity and multiple-clone infections across a malaria endemicity gradient spanning from Senegal to the Republic of Guinea.7 Additionally, no multilocus linkage disequilibrium (LD) and limited evidence of population genetic differentiation were reported. In contrast, other studies in the region have reported high levels of genetic diversity with significant LD and evidence of genetic structure in P. falciparum populations from Zimbabwe,4 Senegal,8 and the Republic of Congo9 due to a Wahlund effect, whereby geographical separation of parasite populations results in genetic heterogeneities. These contrasting results in the region may be due to differences in sampling method with respect to the epidemiology of the isolates collected, geographical space, as well as malaria transmission intensity. Indeed, individual studies have tended to focus on genotyping clinical isolates, often over broad catchment areas, with insufficient sampling. Population genetic studies in West Africa that characterize parasite population structure on a local scale, especially with deep sampling, are limited. Within this context, it is critical to perform community-level genetic studies of the local reservoir of P. falciparum infections and consider “all infections,” not just clinical episodes in children to accurately inform control and elimination efforts.
This study describes the population genetics of P. falciparum populations sampled from individuals of all ages who resided in two proximal catchment areas in Bongo District (BD), Ghana, during June 2012. We determined the patterns of parasite diversity at the end of the dry season, when diversity is predicted to be at its lowest point over the course of the year, to describe the genetic structure of the P. falciparum reservoir in an area of highly seasonal malaria transmission. Using a panel of 12 microsatellite markers to define multilocus infection haplotypes, we investigated the extent of genetic diversity, LD, and local population structure in these proximal P. falciparum populations. To our knowledge, this is the first study to use multilocus microsatellite genotyping to investigate the population structure of the P. falciparum reservoir in an area of highly seasonal malaria transmission in Ghana.
MATERIALS AND METHODS
Human subjects ethical approval.
The study was reviewed and approved by the ethics committees at Navrongo Health Research Centre (Ghana), Noguchi Memorial Institute for Medical Research (Ghana), New York University (United States), University of Melbourne (Australia), University of Michigan (United States), and University of Chicago (United States). Individual informed consent was obtained in the local language from each enrolled participant by signature/thumbprint accompanied by the signature of an independent witness. A parent or guardian provided consent for children under the age of 18 years, and all children between the ages of 12 and 17 years also provided assent. Pregnant women, individuals with disabilities, and individuals presenting with a serious or acute disease (including symptomatic/clinical malaria) on the day the survey was conducted were not eligible for enrollment and were excluded. All individuals requiring treatment were referred to the Vea or Soe Health Center and/or the Bongo District Hospital for appropriate care.
Study area.
This age-stratified cross-sectional survey was designed to evaluate the reservoir of Plasmodium spp. in BD, located in the Upper East Region of Ghana (a more detailed description of the study area is published elsewhere10). Malaria in BD is hyperendemic (stable malaria) and is characterized by marked seasonal transmission of P. falciparum (minority: Plasmodium malariae and Plasmodium ovale) with a short rainy season (May–September) and a prolonged dry season (October–April). The annual entomological inoculation rate in BD is estimated to be > 25 (Dadzie, S. and Appawu, M., personal communication). Using demographic enumeration data collected prior to the start of the survey, two broad “catchment areas” in BD were selected (Vea/Gowrie and Soe) as they were considered to be similar in population size and ethnic composition (Figure 1). The two catchment areas were hypothesized to differ in terms of malaria transmission intensity. Vea/Gowrie is proximal to an irrigation dam and was expected to have higher and less seasonal transmission compared with Soe, which is not near a dam. In addition, the enumeration data were also used to further classify the “villages” (Vea, Gowrie, Soe Boko, Soe Sanabisi) in each of the catchment areas. Over a 2-week period in June 2012 at the end of the dry season, blood samples (thick/thin blood films and dried blood spots) and malaria-related questionnaires were collected after obtaining informed consent from 698 participants between the ages of 1 and 85 years during this cross-sectional survey (Supplemental Information).
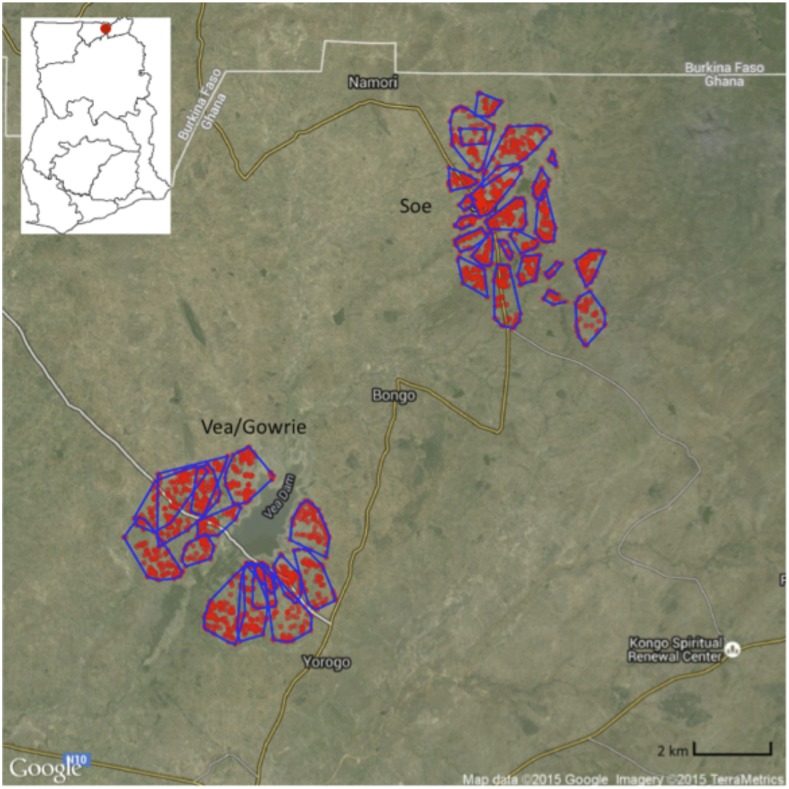
Figure 1.
The location of the study area in Bongo District within Ghana (insert map, upper left) and the distribution of compounds included in the study (red dots) within each catchment area: Vea/Gowrie catchment area (lower left) and Soe catchment area (upper right). Reproduced with permission from Tiedje and others.10 This figure appears in color at www.ajtmh.org.
Parasitological measurements.
Parasite densities were counted against white blood cells (WBCs) on 10% Giemsa-stained thick film blood smears and examined under oil immersion of a 100-fold magnification. Two experienced technicians independently read all the study slides and the results were reported into separate structured laboratory books. Parasite densities were counted per 200 WBCs and were calculated by averaging the two independent readings. When the slide readings were non-concordant by more than 50%, the slide was reread by a third independent technician, recorded into a separate laboratory book, and used as the tiebreaker. The parasite densities were further quantified as parasites per microliter of blood assuming the average WBC count was about 8,000 per μL of blood.11 Parasite species were also identified using a 100-fold magnification of the thin film smears and categorized based on morphology.
DNA extraction.
From each DBS sample two 5 mm × 5 mm sections were cut and placed in a 1.5-mL centrifuge tube. Genomic DNA was extracted from the DBS using the QIAamp DNA blood mini kit extraction procedure as described by the manufacturer (Qiagen, Valencia, CA). The DNA samples were eluted into 50 μL of buffer AE and stored at −20°C for short-term use and −80°C for long-term storage.
Molecular estimation of P. falciparum multiplicity of infection.
For all Plasmodium spp. microscopy-positive samples, a nested polymerase chain reaction (PCR) was performed to determine P. falciparum positivity and the number of P. falciparum merozoite surface protein 2 (msp2) (IC/3D7 and FC27) gene clones. The PCR was completed as described by Snounou and others with modifications.12 The first round was carried out in a 20 μL reaction mixture consisting of 10 μL Taq PCR Master Mix (Qiagen), 0.25 μL of each primer (OF and OR, 10 μM), 1 μL of sample DNA, and 8.5 μL of ddH20. The second round of the nested PCR was carried out in two separate tubes, each containing a single msp2 primer pair as previously described in the literature.12 The 20 μL reaction mixture contained 10 μL Taq PCR Master Mix (Qiagen), 0.25 μL of each primer (FC1/FC2 or IC1/IC2, 10μM), 1 μL of PCR product from round 1, and 8.5 μL of ddH20. Modifications were made to the cycling conditions for both the first and second rounds: Round 1 (95°C for 15 minutes, 25 cycles of 94°C for 1 minute, 58°C for 2 minutes, and 72°C for 2 minutes, followed by 72°C for 10 minutes) and Round 2 (95°C for 15 minutes, 30 cycles of 94°C for 1 minute, 58°C for 2 minutes, and 72°C for 2 minutes, followed by 72°C for 10 minutes). For each PCR reaction, positive and negative controls were included for quality control and all PCRs were carried out on an Eppendorf Mastercycler Nexus and visualized on a 1.5% agarose gel stained with EZ VisionTM DNA Dye/Loading Buffer (Amresco, Solon, OH) to determine the number of infecting IC/3D7 and FC27 clones, and to calculate the multiplicity of infection (MOI).
Microsatellite genotyping.
Each P. falciparum isolate was genotyped using 12 neutral microsatellite markers previously developed by Anderson and colleagues: TA1, 2490, TA81, TA109, TA60, TA87, POLYα, TA42, ARA2, PfG377, PfPK2, and TA40.13 Each field isolate was genotyped using a previously described hemi-nested PCR strategy with modifications.13 Briefly, first-round PCR reactions were multiplexed so that each PCR reaction was performed for pairs of primers (six reactions). In each first-round reaction, a total of 2μL of isolate genomic DNA was amplified with 1.5μL of 25 mM MgCl2, 1.5μL of 2 mM dNTPs, 3μL of 5× buffer, 1μL of each primer at a starting concentration of 5 pmol, 0.2μL of GoTaq Hot Start polymerase (Promega, Madison, WI), and water up to 15μL total reaction volume. The cycling conditions for the first round were as follows: 2 minutes at 95°C, 25 cycles of (30 seconds at 94°C, 30 seconds at 42°C, 30 seconds at 60°C, and 40 seconds at 65°C), and 2 minutes at 65°C. The second-round reactions contained fluorescent-labeled primers as described in Anderson and others,13 with the same reaction conditions as the first round except for the addition of 1μL of PCR product. The cycling conditions were as follows: 2 minutes at 95°C, 25 cycles of (20 seconds at 94°C, 20 seconds at 45°C, and 30 seconds at 65°C), and 2 minutes at 65°C.
Fluorescent-labeled PCR products were sent to the Biotechnology Resource Center at Cornell Institute of Biotechnology for fragment analysis. Capillary electrophoresis was performed on an Applied Biosystems 3730xl DNA analyzer (ThermoFisher Scientific, Waltham, MA). After electrophoresis, raw data files were imported and scored using GeneMarker (SoftGenetics LLC, State College, PA). Raw data files were analyzed using customized panels and normalized automatically based on the size standard LIZ500 by the scoring software. All major peaks for a sample were considered true alleles if they were spaced at intervals corresponding to trinucleotide repeats and within the expected marker base pair range. Any peaks less than 33% of the primary peak (local max) for a locus were considered stutter peaks and not interpreted as true alleles. Background noise was defined as any peak less than 200 fluorescent units. All microsatellites amplified are single-copy genes; therefore, the presence of two or more alleles at one or more loci was interpreted as a multiple infection4. “True” single-clone infections were defined as any isolate that had only one peak at all microsatellite loci. Multiple-clone infections were defined as any isolate that had more than two peaks at one or more loci. For multiple-clone infections, infection haplotypes were reconstructed from the predominant peaks at each locus.4 Results from each panel analysis were merged and exported for further analysis.
Population genetic analysis.
The exported data from GeneMarker were processed using TANDEM v1.09,14 which automatically bins the observed microsatellite alleles and is optimal for use with microsatellite loci that have tri-nucleotide repeats. Binned data files were then processed using Convert v1.3115 to calculate allele frequencies and to generate input files for various population genetics software. We used the software LOSITAN v1.016 to confirm that all loci were neutral and not under positive or balancing selection. We ran 100,000 simulations under both an infinite alleles and stepwise model. Patterns of genetic diversity were analyzed using ARLEQUIN v3.5,17 which calculates the number of haplotypes, number of alleles per locus, expected heterozygosity, and pairwise LD between microsatellite marker pairs. To minimize the effects of small sample sizes on allelic measures, allelic richness was also calculated using FSTAT v2.9,18 which takes into account the smallest sample size and normalizes the number of alleles per locus accordingly. We examined the genetic relatedness between complete infection haplotypes (i.e., no missing data) by calculating pairwise allele sharing (PAS). Specifically, PAS was calculated as the number of alleles shared between two infection haplotypes divided by the number of microsatellite markers (i.e., 12). Multilocus LD was investigated using LIAN v3.719 to calculate the standardized index of association (ISA) among loci using a Monte Carlo simulation method and 10,000 resamplings, where alleles are reshuffled at random among haplotypes. The LIAN software cannot process datasets with missing data; therefore, the standardized index of association was calculated only for isolates with complete haplotypes. Jost’s differentiation indices D were calculated using the R package DEMEtics v0.8-7.20 The D differentiation index is a robust estimator of population differentiation when within-population diversity (i.e., allelic diversity within a locus) is high and when the number of observed alleles per locus is greater than two.20 A minimum spanning tree was constructed using Phyloviz v1.121 to display the relationship among haplotypes and possible associations to geographic origin and sampling location, where haplotypes matched at least four out of 12 loci.
Statistical analysis.
Statistical analyses were carried out using IBM SPSS v22 Statistics software (IBM, Armonk, NY). For all analyses, study participants were categorized into defined age groups (1–5, 6–10, 11–20, 21–39, and ≥ 40 years), catchment areas, and villages as described in the study design. Continuous variables are presented as medians with inter quartile ranges (IQRs) and discrete variables are presented using the calculated/observed prevalence values. Fisher exact or χ2 tests were used for univariate analyses of discrete variables to compare proportions; non-parametric Mann–Whitney U (comparing distributions across two groups) and Kruskal–Wallis (comparing distributions across k groups) tests were used for comparing distributions across groups of continuous variables. A test was deemed to be statistically significant if the P value was less than 0.05.
RESULTS
For this cross-sectional study, 698 participants across all ages were recruited in BD from two broad catchment areas (Vea/Gowrie and Soe; Figure 1) that can be further classified into four villages (Vea, Gowrie, Soe Sanabisi, and Soe Boko; Figure 1). Using 12 microsatellite markers, a subset of 200 participants from the 267 participants identified to have a Plasmodium spp. infection (including mixed P. falciparum/P. malariae infection) (Supplemental Table 1) was used to define the multilocus infection haplotypes and evaluate the population structure of the P. falciparum reservoir in BD (Table 1). On microscopic analysis, one participant was found to have P. malariae infection; this participant was also harboring P. falciparum (determined by msp2 PCR), therefore they were included in the analyses as a mixed P. falciparum/P. malariae infection. Two hundred participants were chosen randomly and there were no statistically significant differences between the 200 included and the 67 excluded individuals in the microsatellite analyses for any of the key variables (P > 0.05) (Supplemental Table 1) except for catchment area (P = 0.036) and village (P < 0.001). The demographic and parasitological characteristics of the 200 P. falciparum infections analyzed by multilocus microsatellite genotyping are presented in Table 1.
Table 1.
Demographics and parasitological characteristics of the 200 participants analyzed with P. falciparum infections (including mixed P. falciparum/P. malariae infections)
| Characteristic | Total | Vea/Gowrie | Soe |
|---|---|---|---|
| Age groups* | |||
| All | 200 | 116 (58.0) | 84 (42.0) |
| 1–5 years | 41 (20.5) | 23 (19.8) | 18 (21.4) |
| 6–10 years | 63 (31.5) | 35 (30.2) | 28 (33.3) |
| 11–20 years | 58 (29.0) | 38 (32.8) | 20 (23.8) |
| 21–39 years | 18 (8.0) | 8 (6.9) | 8 (9.5) |
| ≥ 40 years | 22 (11.0) | 12 (10.3) | 10 (11.9) |
| Gender* | |||
| Female | 100 (50.0) | 53 (45.7) | 47 (56.0) |
| Male | 100 (50.0) | 63 (54.3) | 37 (44.0) |
| Microscopic Plasmodium spp. prevalence (N = 200) | |||
| P. falciparum | 196 (98.0) | 112 (96.5) | 84 (100.0) |
| P. falciparum/P. malariae | 3 (1.5) | 3 (2.6) | 0 (0.0) |
| P. malariae | 1 (0.5) | 1 (0.9) | 0 (0.0) |
| Plasmodium spp. median density (N = 200)† | |||
| All | 280 [130–870] | 280 [120–680] | 340 [160–990] |
| 1–5 years | 880 [500–1,800] | 800 [520–1,280] | 1,040 [480–3,080] |
| 6–10 years | 240 [120–920] | 200 [120–600] | 400 [200–1,000] |
| 11–20 years | 240 [120–480] | 240 [110–450] | 240 [130–510] |
| 21–39 years | 240 [90–390] | 240 [130–350] | 240 [80–730] |
| ≥ 40 years | 140 [80–250] | 120 [50–280] | 160 [80–210] |
| P. falciparum median MOI (N = 187)‡ | |||
| All | 2 [1–3] | 2 [1–3] | 2 [1–3] |
| 1–5 years | 2 [1–3] | 1.5 [1–3] | 2 [1–4] |
| 6–10 years | 2.5 [1–3.25] | 2 [1–3] | 3 [2–4] |
| 11–20 years | 2 [1–3] | 2 [1.25–3] | 2 [1–3] |
| 21–39 years | 1 [1–2] | 1 [1–3] | 1 [1–2] |
| ≥ 40 years | 1 [1–2] | 1 [1–2] | 1 [1–2] |
IQR = inter quartile range; MOI = multiplicity of infection; PCR = polymerase chain reaction.
Data reflect No. (% (n/N)) of participants sampled that were positive for P. falciparum (including mixed P. falciparum/P. malariae infections) by microscopy and msp2 PCR.
Median parasite density for microscopically positive P. falciparum (including mixed P. falciparum/P. malariae infections) (value/µL, [IQR]) samples.
Data reflect the median MOI for the participants sampled that were positive for P. falciparum (including mixed P. falciparum/P. malariae infections) by msp2 PCR (median, [IQR]).
For the 200 participants included in these analyses, there were no significant differences observed in the proportion of participants analyzed across all age categories and genders for both the complete study population and within/between the catchment areas or villages (Table 1). During the survey, 82.0% (95% confidence interval [CI] = 79.7–84.4%) individuals reported sleeping under a bed net the previous night and there were no significant differences in bed net usage between age groups, gender, catchment area, or village. Overall 6.0% (95% CI = 4.5–7.4%) of participants reported taking an antimalarial medication in the previous 2 weeks prior to being surveyed, with no significant differences between age groups, gender, catchment area, or village. Utilizing msp2 genotyping, MOI was successfully genotyped for 187 of the 200 (93.5%) P. falciparum (including mixed P. falciparum/P. malariae infections)-positive samples. MOI ranged from 1 to 6 (median = 2 [IQR = 1–3]) with 114 (61.0%) of infections having an MOI > 1. Median MOI significantly varied across the age groups surveyed (P = 0.001; Table 1) with younger age groups (1–5, 6–10, and 11–20 years) having median MOIs that were greater compared with the older participants surveyed (21–39, and ≥ 40 years). There were no significant differences in the median MOI between the catchment areas (P = 0.543; Table 1) or villages (P = 0.500).
Plasmodium spp. parasitemia.
This study was focused on describing the reservoir of Plasmodium spp. infections in BD. As anticipated, we observed a high proportion of participants—97.5% (N = 195)—with low-to-moderate parasitemias (40–9,999 parasites/µL), and only 2.5% (N = 5) having parasitemias ≥ 10,000 parasites/μL. For those participants with more than 10,000 parasites/μL, generally described as the fever threshold for malaria,22,23 only one participant was febrile (temperature ≥ 37.5°C) on the day the survey was conducted. For the 200 participants analyzed, median Plasmodium spp. density was significantly higher for 1–5 year olds compared with all other age groups (P < 0.001; Table 1), with Plasmodium spp. density gradually decreasing with age (Table 1). In this survey, the median Plasmodium spp. density for the total study population was 280 parasites/μL (Table 1) with no significant difference in median density between the Vea/Gowrie and Soe catchment areas (280 parasites/μL versus 340 parasites/µL, respectively, P = 0.117; Table 1).
Microsatellite genetic diversity of P. falciparum in BD.
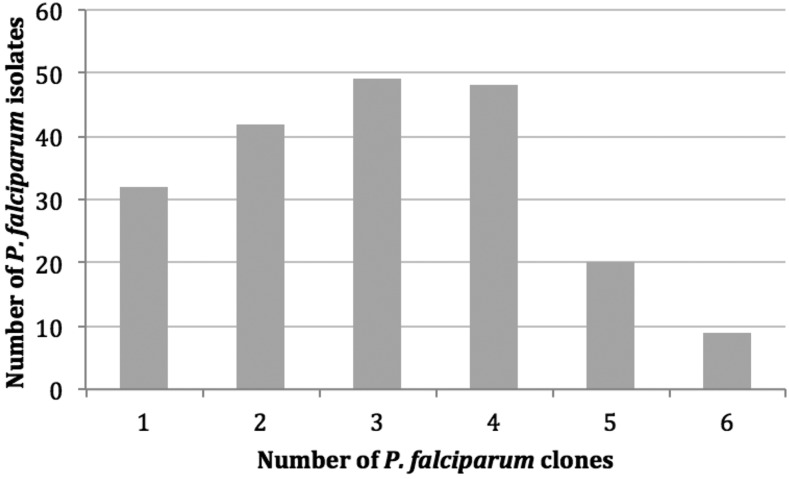
Microsatellite genotyping success ranged from 83.0% to 99.0% for each of the 12 microsatellite loci (Supplemental Table 2). All the loci genotyped were polymorphic and ranged from 5 to 26 alleles per locus (Supplemental Table 2). There was no indication that any of the microsatellite loci were non-neutral (i.e., under positive or balancing selection) using an infinite alleles and stepwise model,16 thus, all microsatellite markers were included for further analysis. A high number of isolates harbored multiple-clone infections indicative of high transmission intensity in the study area. There were 168 isolates (84.0%) with multiple-clone infections and only 32 isolates (16.0%) with true single-clone infections (Figure 2). In BD, there were high levels of genetic diversity with similar patterns observed within each catchment area (Vea/Gowrie and Soe) and within each of the four villages (Vea, Gowrie, Soe Sanabisi and Soe Boko) (Table 2). We observed the same number of haplotypes as isolates, indicating that every haplotype in the population was unique (Table 2). There was considerable allelic variation as defined by number of alleles per locus (A) and allelic richness estimates (Rs), which are normalized based on the smallest sample size.18 In BD, there were 13.25 alleles per locus (standard deviation [SD] = 6.36) and a comparable estimated allelic richness of 13.20 (SD = 6.32) (Table 2). Similarly, the A and Rs within each catchment area were high (Table 2). Within each of the four villages, the number of alleles per locus ranged from 7.17 to 9.75 alleles and the estimated allelic richness ranged from 7.07 to 8.60 alleles. Because the Soe Sanabisi village had fewer participants included in the microsatellite analyses (29 isolates), it also had a lower number of alleles per locus. However, the normalized Rs was comparable to the Rs estimates for the other villages. There was limited variability of the expected heterozygosity (He) among the villages, with all showing high He values. In BD, He was 0.74 (SD = 0.19) and ranged from 0.73 to 0.75 within the catchment areas and villages, indicative of high levels of heterozygosity in all P. falciparum populations (Table 2). The allele frequencies for each locus are presented in Supplemental Table 3.
Figure 2.
Number of P. falciparum clones in each P. falciparum isolate sampled in Bongo District based on microsatellite genotyping.
Table 2.
Patterns of genetic diversity for P. falciparum populations from Bongo District
| Population | n | h | A ± SE | Rs ± SE | He ± SE |
|---|---|---|---|---|---|
| Vea/Gowrie catchment area | 116 | 116 | 11.92 ± 5.30 | 11.37 ± 5.05 | 0.75 ± 0.18 |
| Vea | 60 | 60 | 9.75 ± 4.09 | 8.60 ± 3.46 | 0.75 ± 0.18 |
| Gowrie | 56 | 56 | 9.03 ± 3.80 | 8.29 ± 3.33 | 0.74 ± 0.20 |
| Soe catchment area | 84 | 84 | 10.42 ± 5.02 | 10.00 ± 5.04 | 0.73 ± 0.20 |
| Soe Sanabisi | 29 | 29 | 7.17 ± 3.41 | 7.07 ± 3.54 | 0.73 ± 0.19 |
| Soe Boko | 55 | 55 | 9.25 ± 4.03 | 8.02 ± 3.48 | 0.73 ± 0.20 |
| Total | 200 | 200 | 13.25 ± 6.36 | 13.20 ± 6.32 | 0.74 ± 0.19 |
A = number of alleles per locus; h = number of haplotypes; He = expected heterozygosity; n = number of isolates; Rs = allelic richness estimate.
This analysis was repeated for single-clone infections and here any isolate with one or two alleles at a given locus was included to maximize the analysis sample size. In the 42 isolates with two alleles at any locus, the dominant peak is easily identified based on fluorescence intensity when constructing the infection haplotypes.4 We thus considered these to be “dominant” constructed haplotypes and included them in this analysis. The dominant infection dataset comprised 74 isolates and the patterns of genetic diversity were comparable to the multiple-clone infections with high levels of expected He and allelic variation and only unique haplotypes (i.e., same number of haplotypes as isolates) observed (Supplemental Table 4). The allele frequencies for the 12 loci are presented in Supplemental Table 3.
Genetic relatedness of P. falciparum infection haplotypes.
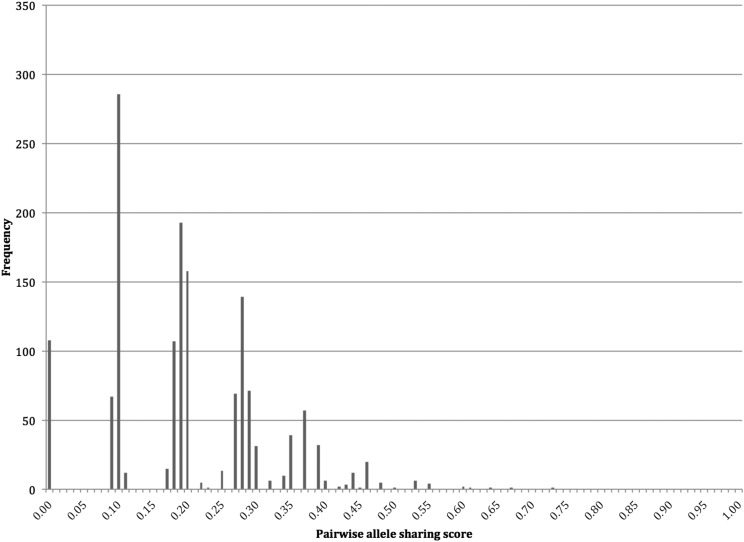
It is possible that although all infection haplotypes are unique, they only differ at one or two loci and are in fact highly related parasites.24 To investigate whether there were any highly related infection haplotypes in our population, we calculated the genetic relatedness of P. falciparum infections through PAS comparisons between haplotypes. For this, we used only the more robust dominant haplotype dataset. The dominant infection dataset comprised 74 isolates, of which 55 isolates had complete haplotypes, or no missing data (N haplotypes = 55, N pairwise comparisons = 1,485). The PAS comparisons showed that the majority of infection haplotype pairs shared on average ≤ 0.2 of their alleles (i.e., identical at only three loci) (Figure 3). There was one highly related pair of isolates with haplotypes that were identical at nine out of 12 loci (PAS = 0.75).
Figure 3.
Frequency distribution of the pairwise allele sharing scores between the dominant P. falciparum infection haplotypes.
Multilocus LD.
ISA was used to assess multilocus LD in the more robust dataset of 55 dominant infections with complete haplotypes. Similar patterns of significant multilocus LD in BD, within each catchment area and also within each of the four villages, were found (Table 3). To investigate whether the inclusion of isolates with two clones (i.e., a maximum of two alleles at any locus) in the dominant infection dataset biased the LD values, only the 32 “true” single-clone infections (i.e., only one allele at every locus) were considered and ISA was recalculated for the 27 isolates with complete haplotypes. Multilocus LD was no longer significant in BD, Vea/Gowrie, or Soe catchment areas (Table 3), however, it is likely that the small sample sizes did not enable enough power to detect multilocus LD.
Table 3.
Multilocus linkage disequilibrium in P. falciparum populations from Bongo District.
| Dominant infections | True single-clone infections | |||
|---|---|---|---|---|
| Population | n | ISA (P value) | n | ISA (P value) |
| Vea/Gowrie catchment area | 27 | 0.0227 (0.005)* | 12 | 0.0288 (0.058) |
| Vea | 19 | 0.0438 (< 0.001)* | 7 | 0.0694 (0.020)* |
| Gowrie | 8 | 0.0640 (0.011)* | 5 | 0.0926 (0.025)* |
| Soe catchment area | 28 | 0.0216 (0.008)* | 15 | 0.0233 (0.066) |
| Soe Sanabisi | 10 | 0.0780 (< 0.001)* | 6 | 0.0900 (0.009)* |
| Soe Boko | 18 | 0.0351 (0.003)* | 9 | 0.0443 (0.037)* |
| Total | 55 | 0.0127 (0.006)* | 27 | 0.0288 (0.051) |
n = number of isolates.
P < 0.05.
Exploration of significant multilocus LD.
To further explore our observation of significant multilocus LD, we completed additional analyses. To examine the possible clonal propagation of particular clones, ISA can be recalculated by only including unique haplotypes and treating any repeated haplotype as an individual.25 In this study, however, this step was not necessary since all the haplotypes in the population were unique (Supplemental Table 4). Nonetheless, it is still possible that although every haplotype is unique, highly related parasites may still propagate in the population, thus ISA can also be recalculated after excluding these parasite pairs.4,7 There was one highly related pair of isolates with haplotypes that were identical at nine out of 12 loci (Figure 3); however, multilocus LD was still significant after removing this isolate pair (ISA = 0.0131, P = 0.005).
Next we examined whether physical linkage between microsatellite markers found on the same chromosome may have confounded our results. Two of the markers used (TA42 and TA81) localize to chr5, three markers (TA109, TA87, and TA1) localize to chr6, two markers (TA40, 2490) localize to chr10, and two markers (Pfg377 and PfPk2) localize to chr12. The overall ISA for BD was recalculated with only one marker per chromosome (i.e., with seven markers located on seven different chromosomes) and multilocus LD was no longer significant except for two combinations (TA42–TA1–TA40‐Pfg377 and TA42–TA1–TA40‐PfPk2) (Supplemental Table 5).
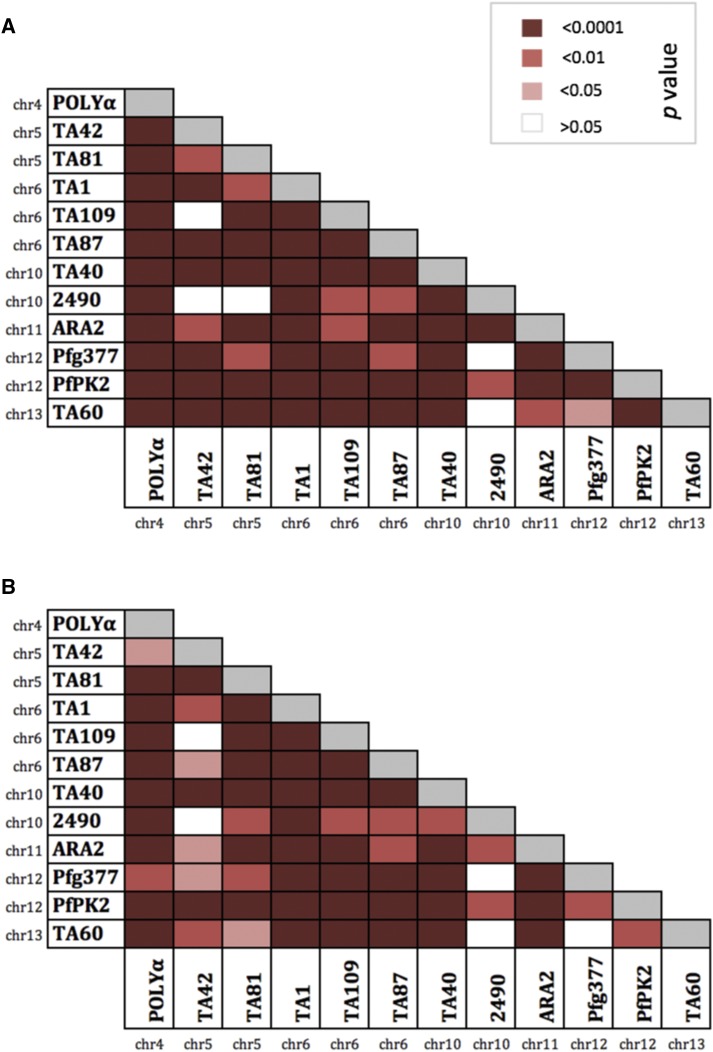
We cannot rule out the possibility that some of these microsatellite markers are physically linked to a gene under strong selection (e.g., a drug resistance gene or antigen). Because two known drug-resistant genes (Pfdhfr on chr4 and Pfmdr1 on chr5) also localize to the same chromosomes as three of the microsatellite markers used (POLYα on chr4 and TA42–TA81 on chr5), multilocus LD was recalculated after removing the markers on chr4 and chr5 separately. LD was no longer significant when the markers on chr5 (TA42 and TA81) were excluded (data not shown). Together with the loss of significant LD when only one marker per chromosome was considered (Supplemental Table 5), this suggests that the microsatellite markers that localize to chr5 may be physically linked to the Pfmdr1 gene, which may be under strong selection. Although it is possible that the significant LD may be influenced by Pfmdr1 antimalarial drug pressures, we interpreted this observation with care since pairwise LD for all 66 microsatellite marker pairs was significant for 61/66 (92.4%) pairs in all infections (Figure 4A) and 61/66 (92.4%) pairs in the dominant infections (Figure 4B), including pairs of markers located on different chromosomes. This may not simply reflect physical linkage between markers but may actually occur when multilocus haplotypes are transmitted together as a genomic unit.26
Figure 4.
Pairwise linkage disequilibrium for (A) 144 Plasmodium falciparum isolates with complete haplotypes and (B) the 55 dominant infections with complete haplotypes. The color key provided corresponds to the P value for each pairwise comparison where white indicates a nonsignificant P value (> 0.05) and the reds represent significant P values. This figure appears in color at www.ajtmh.org.
Geographic population structure between P. falciparum populations.
To measure levels of population differentiation between the villages that were hypothesized to differ in terms of transmission intensity, Jost’s pairwise index of differentiation (D) was calculated. Pairwise Jost’s D indices were compared among the four villages and resulted in relatively low pairwise D values that ranged from 0.0127 to 0.0491 and no significant pairwise values were identified (Table 4, lower diagonal). To assess whether patterns of population differentiation were detectable in the absence of possible false recombinant haplotypes from isolates with multiple-clone infections, Jost’s D values were also measured for the dominant infections. Overall, higher pairwise D values were found ranging from 0.0870 to 0.2026 with significant pairwise comparisons between Soe Boko and Vea (D = 0.1528, P < 0.05) as well as between Soe Boko and Soe Sanabisi (D = 0.2026, P < 0.05) (Table 4, upper diagonal). Jost’s D values can be interpreted as the mean proportion of pairwise private alleles between populations, therefore < 5% of alleles in each village were private when considering all infections, whereas 9–20% of alleles were private in the dominant infections. Although the significant D values in the dominant infections are indicative of possible geographic structure, these results must be interpreted with caution since the sample sizes in each village were small, which may also explain the large number of private alleles.
Table 4.
Genetic differentiation between local P. falciparum populations from Bongo District
| Vea | Gowrie | Soe Boko | Soe Sanabisi | |
|---|---|---|---|---|
| Vea | – | 0.0871 | 0.1528* | 0.0870 |
| Gowrie | 0.0404 | – | 0.1257 | 0.1540 |
| Soe Boko | 0.0400 | 0.0446 | – | 0.2026* |
| Soe Sanabisi | 0.0127 | 0.0491 | 0.0425 | – |
Lower diagonal: matrix of pairwise Jost’s D indices between villages for all infections (N = 200 isolates); upper diagonal: matrix of pairwise Jost’s D indices values between villages for dominant infections (N = 74 isolates).
P < 0.05.
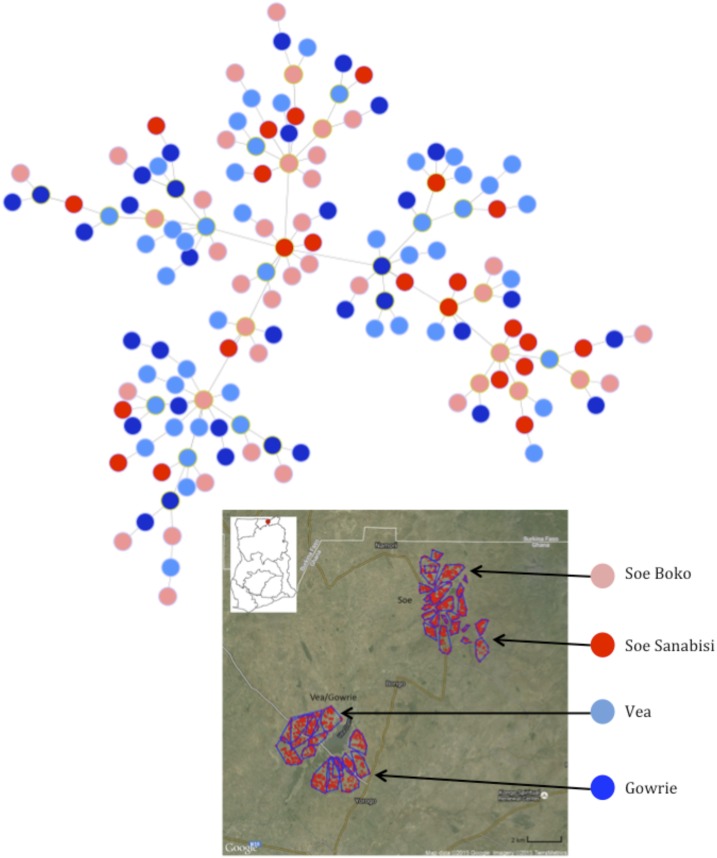
To explore possible patterns of geospatial population structure further, a minimum spanning tree was used to display the phylogenetic relationship among P. falciparum haplotypes (Figure 5). There was evidence of substructuring among haplotypes, although it was not associated with sampling location by catchment area or village (Figure 5). We conclude that the linkage structure and signal of relatedness among haplotypes has not been shaped by spatiotemporal forces, such as geographical separation among parasite populations.
Figure 5.
Network analysis displaying the genetic relatedness among the multilocus Plasmodium falciparum haplotypes, where haplotypes matched at at least 4 out of 12 loci. The haplotypes are colored based on geographical location as indicated in the map: light and dark blue correspond to Vea and Gowrie, respectively, and together the blues correspond to the Vea/Gowrie catchment area. The light and dark red corresponds to Soe Boko and Soe Sanabisi, respectively, and together the reds correspond to the Soe catchment area. Map reproduced with permission from Tiedje and others.10 This figure appears in color at www.ajtmh.org.
DISCUSSION
The goal of malaria elimination strategies is to reduce the local reservoir of malaria infection to zero to achieve any real and sustainable progress. Here we analyze the genetic structure of the P. falciparum reservoir of infection at the community level and across all ages in an area of highly seasonal malaria transmission in Ghana, using multilocus microsatellite genotyping. Our data set, representing extensive sampling of a local population, is the first to show the following features of population structure and diversity in P. falciparum from a West African malaria-endemic area. These include a high proportion of multiple-clone infections with high levels of genetic diversity and, surprisingly, significant multilocus LD with no detectable geographic population differentiation. In fact, our results contradict the expectation that a high proportion of multiple-clone P. falciparum infections in our population should translate into linkage equilibrium, as frequent outcrossing would be expected.
There are a few possible explanations for our finding of significant LD with no geographic population differentiation: 1) Increased levels of inbreeding (i.e., reduced levels of recombination) due to selfing or transmission of clonal or highly related parasite clones4; 2) The rate of recombination may be low relative to mutation, leading to increased mutation or strand slippage events that generate new haplotypes but are not sufficient to disrupt overall association patterns among loci27,28; (3) Vector behavior, whereby a reduction in transmission events at the end of the dry season leads to limited dispersion of the vector and may result in mosquitoes biting locally on small spatial scales. However, in the asymptomatic parasite reservoir in BD, the isolates shared on average three alleles. This lack of any closely related parasite clones across small distances (as little as ∼5 km apart) provides evidence against these explanations (i.e., 1–3) and is consistent with free gene flow and no apparent barriers that may impact allele frequencies based on geography. Additionally, the multilocus LD in BD cannot not be explained by “epidemics” of highly resistance parasite clones, as was observed in clinical isolates from Guinea, the Gambia, and Senegal7; 4) Selection at drug-resistance genes seems the most plausible, whereby the maintenance of resistance mutations in linked loci may lead to hitchhiking or selective sweeps29 of otherwise neutral microsatellite markers on the same chromosomes.
In fact, the loss of significant multilocus LD when excluding microsatellite markers that localize to chr5 suggests that the detectable LD may be driven by Pfmdr1 selection. A previous study in 2011 by Alam and colleagues demonstrated that Pfmdr1 is under selection in Ghanaian parasites, likely due to chloroquine use in the past since mutations in Pfmdr1 may confer or modulate chloroquine resistance.30 Indeed, the absence of any evidence of geospatial clustering of parasite populations (i.e., as defined by multilocus microsatellite haplotypes; Figure 5) suggests that other selective forces such as drug or host immune pressures may shape the parasite population such that there is an underlying linkage structure that cannot be explained by geographical separation. This warrants further investigation to characterize drug resistance to a succession of antimalarials and immune-selected markers in this population to describe the combined effects of population history and natural selection on parasite genetic structure.
The two catchment areas examined, Vea/Gowrie and Soe, were hypothesized to differ in terms of their malaria transmission intensity since Vea/Gowrie is proximal to an irrigation dam compared with Soe (the study area is described in more detail in10). Contrary to our expectations, the genetic diversity estimates were similar within and among all catchment areas and villages, consistent with little or no geographic population structure. It is possible that the small spatial scales examined in our study may have limited our ability to detect significant geographic differentiation between catchment areas and/or villages. Sampling all asymptomatic infections at the compound level may in fact reveal spatial heterogeneities as has been observed in a spatially dense dataset of P. vivax infections from Solomon Islands.31 Nevertheless, the lack of population structure we observed may be advantageous to maintain beneficial traits such as drug resistance genes.32
Our cross-sectional survey of this population revealed a large proportion of asymptomatic infections (99.5%; N = 199), consistent with a large parasite reservoir contributing to transmission. Interestingly, our findings are comparable to those reported for P. vivax, where strong multilocus LD has been shown to co-occur with high genetic diversity and a high proportion of multiple-clone infections in P. vivax isolates from a community-based study in Brazil.33 Additionally, P. vivax parasite populations exhibit high genetic diversity and large effective population sizes compared with sympatric P. falciparum populations in high-transmission areas of Papua New Guinea,32 as well as in areas of Sri Lanka28 and Colombia24 that have experienced significant reductions in malaria prevalence. Although cross-species comparisons must be made with caution, it is interesting to note that the dormant hypnozoite stage in P. vivax provides a possible mechanism for maintaining high levels of genetic diversity and chronicity, perhaps analogous to asymptomatic infections in P. falciparum. Indeed, the high proportion of asymptomatic infections maintained through the dry season in the P. falciparum reservoir could facilitate superinfections and/or coinfections with genetically distinct parasite clones, which may in turn have major implications for the success of future control strategies. It is, therefore, crucial to continuously monitor both diversity and LD levels in the asymptomatic parasite reservoir over time in response to interventions, since only sustained interruption of transmission is likely to disrupt a highly diverse parasite population with little genetic differentiation.32,34 Depth of sampling must also be considered.
Importantly, the significant though relatively low LD (i.e., index of association measures) reported here in the P. falciparum reservoir of infection can be used as a baseline population genetic metric to examine the effects of seasonality (i.e., during the wet season) and control interventions being implemented by National Malaria Control Programmes. Monitoring the parasite reservoir in response to malaria control measures is crucial if we are going to understand within-host competition among genetically distinct parasite clones that are under selection by antimalarial drug and/or host immune pressures.
Supplementary Material
Acknowledgments:
We thank the participants, communities, and the Ghana Health Service in Bongo District, Ghana for their willingness to participate in this study. We would like to thank the field teams for their technical assistance in the field and the laboratory personnel at the Navrongo Health Research Centre for sample collection and parasitological assessment/expertise. Additionally, we would like to thank laboratory personnel at New York University and The University of Melbourne for their assistance with laboratory experiments. Finally, we would like to acknowledge Aleksandra Leliwa-Sytek for her laboratory expertise and Mercedes Pascual for valuable discussions and her input related to this work.
Note: Supplemental information and tables appear at www.ajtmh.org.
REFERENCES
- 1.Bousema T, Okell L, Felger I, Drakeley C, 2014. Asymptomatic malaria infections: detectability, transmissibility and public health relevance. Nat Rev Microbiol 12: 833–840. [DOI] [PubMed] [Google Scholar]
- 2.Bruce MC, Donnelly CA, Packer M, Lagog M, Gibson N, Narara A, Walliker D, Alpers MP, Dar KP, 2000. Age- and species-specific duration of infection in asymptomatic malaria infections in Papua New Guinea. Parasitology 121: 247–256. [DOI] [PubMed] [Google Scholar]
- 3.Felger I, Maire M, Bretscher MT, Falk N, Tiaden A, Sama W, Beck H-P, Owusu-Agyei S, Smith TA, 2012. The dynamics of natural Plasmodium falciparum infections. PLoS One 7: e45542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anderson TJ, et al. , 2000. Microsatellite markers reveal a spectrum of population structures in the malaria parasite Plasmodium falciparum. Mol Biol Evol 17: 1467–1482. [DOI] [PubMed] [Google Scholar]
- 5.Yalcindag E, et al. , 2012. Multiple independent introductions of Plasmodium falciparum in South America. Proc Natl Acad Sci USA 109: 511–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schultz L, et al. , 2010. Multilocus haplotypes reveal variable levels of diversity and population structure of Plasmodium falciparum in Papua New Guinea, a region of intense perennial transmission. Malar J 9: 336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mobegi VA, Loua KM, Ahouidi AD, Satoguina J, Nwakanma DC, Amambua-Ngwa A, Conway DJ, 2012. Population genetic structure of Plasmodium falciparum across a region of diverse endemicity in West Africa. Malar J 11: 223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leclerc MC, Durand P, De Meeûs T, Robert V, Renaud F, 2002. Genetic diversity and population structure of Plasmodium falciparum isolates from Dakar, Senegal, investigated from microsatellite and antigen determinant loci. Microbes Infect 4: 685–692. [DOI] [PubMed] [Google Scholar]
- 9.Durand P, Michalakis Y, Cestier S, Oury B, Leclerc MC, Tibayrenc M, Renaud F, 2003. Significant linkage disequilibrium and high genetic diversity in a population of Plasmodium falciparum from an area (Republic of the Congo) highly endemic for malaria. Am J Trop Med Hyg 68: 345–349. [PubMed] [Google Scholar]
- 10.Tiedje KE, Oduro A, Agongo G, Anyorigiya T, Azongo D, Awine T, Ghansah A, Pascual M, Koram K, Day KP, 2017. Seasonal variation in the epidemiology of asymptomatic Plasmodium falciparum infections across two catchment areas in Bongo District, Ghana. Am J Trop Med Hyg. 97: 199–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.WHO, 2010.. Basic Malaria Microscopy. Geneva, Switzerland: World Health Organization. [Google Scholar]
- 12.Snounou G, Zhu X, 1999.. Biased distribution of msp1 and msp2 allelic variants in Plasmodium falciparum populations in Thailand. Trans R Soc Trop Med Hyg 93: 369–374. [DOI] [PubMed] [Google Scholar]
- 13.Anderson TJ, Su XZ, Bockarie M, Lagog M, Day KP, 1999. Twelve microsatellite markers for characterization of Plasmodium falciparum from finger-prick blood samples. Parasitology 119: 113–125. [DOI] [PubMed] [Google Scholar]
- 14.Matschiner M, Salzburger W, 2009. TANDEM: integrating automated allele binning into genetics and genomics workflows. Bioinformatics 25: 1982–1983. [DOI] [PubMed] [Google Scholar]
- 15.Glaubitz JC, 2004. CONVERT: a user-friendly program to reformat diploid genotypic data for commonly used population genetic software packages. Mol Ecol Notes 4: 309–310. [Google Scholar]
- 16.Antao T, Lopes A, Lopes RJ, Beja-Pereira A, Luikart G, 2008. LOSITAN: a workbench to detect molecular adaptation based on a F st -outlier method. BMC Bioinformatics 9: 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Excoffier L, Lischer HEL, 2010. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour 10: 564–567. [DOI] [PubMed] [Google Scholar]
- 18.Goudet J, 1995.. FSTAT (Version 1.2): a computer program to calculate F-statistics. J Hered 86: 485–486.
- 19.Haubold B, Hudson RR, 2000. LIAN 3.0: detecting linkage disequilibrium in multilocus data. Linkage Analysis. Bioinformatics 16: 847–848. [DOI] [PubMed] [Google Scholar]
- 20.Gerlach G, Jueterbock A, Kraemer P, Deppermann J, Harmand P, 2010. Calculations of population differentiation based on GST and D: forget GST but not all of statistics. Mol Ecol 19: 3845–3852. [DOI] [PubMed] [Google Scholar]
- 21.Francisco AP, Vaz C, Monteiro PT, Melo-Cristino J, Ramirez M, Carriço JA, 2012. PHYLOViZ: phylogenetic inference and data visualization for sequence based typing methods. BMC Bioinformatics 13: 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rogier C, Commences D, Trape JF, 1996. Evidence for an age-dependent pyrogenic threshold of Plasmodium falciparum parasitemia in highly endemic populations. Am J Trop Med Hyg 54: 613–619. [DOI] [PubMed] [Google Scholar]
- 23.Rogier C, 2000. Natural history of Plasmodium falciparum malaria and determining factors of the acquisition of antimalaria immunity in two endemic areas, Dielmo and Ndiop (Senegal). Bull Mem Acad R Med Belg 155: 218–226. [PubMed] [Google Scholar]
- 24.Chenet SM, Schneider KA, Villegas L, Escalante AA, 2012. Local population structure of Plasmodium : impact on malaria control and elimination. Malar J 11: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smith JM, Smith NH, O’Rourke M, Spratt BG, 1993. How clonal are bacteria? Proc Natl Acad Sci USA 90: 4384–4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Razakandrainibe FG, Durand P, Koella JC, De Meeüs T, Rousset F, Ayala FJ, Renaud F, 2005. “Clonal” population structure of the malaria agent Plasmodium falciparum in high-infection regions. Proc Natl Acad Sci USA 102: 17388–17393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Anderson TJ, Day KP, 2000. Geographical structure and sequence evolution as inferred from the Plasmodium falciparum S-antigen locus. Mol Biochem Parasitol 106: 321–326. [DOI] [PubMed] [Google Scholar]
- 28.Gunawardena S, Ferreira MU, Kapilananda GMG, Wirth DF, Karunaweera ND, 2014. The Sri Lankan paradox: high genetic diversity in Plasmodium vivax populations despite decreasing levels of malaria transmission. Parasitology 2014: 880–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hedrick PW, 1980. Hitchhiking: a comparison of linkage and partial selfing. Genetics 94: 791–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alam MT, et al. , 2011. Selective sweeps and genetic lineages of Plasmodium falciparum drug -resistant alleles in Ghana. J Infect Dis 203: 220–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Waltmann A, et al. , 2017. Increasingly inbred and fragmented populations of Plasmodium vivax with declining transmission. BioRxiv, 10.1101/100610. [Google Scholar]
- 32.Jennison C, et al. , 2015. Plasmodium vivax populations are more genetically diverse and less structured than sympatric Plasmodium falciparum populations. PLoS Negl Trop Dis 9: 1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Batista CL, Barbosa S, Silva Bastos M, Viana SAS, Ferreira MU, 2015. Genetic diversity of Plasmodium vivax over time and space: a community-based study in rural Amazonia. Parasitology 2014: 1–11. [DOI] [PubMed] [Google Scholar]
- 34.Escalante AA et al. , 2015. Malaria molecular epidemiology: lessons from the International Centers of Excellence for Malaria Research Network. Am J Trop Med Hyg 93 (Suppl): 79–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.