Abstract
Based on the data of the NIH-funded Human Connectome Project, we have computed structural connectomes of 426 human subjects in five different resolutions of 83, 129, 234, 463 and 1015 nodes and several edge weights. The graphs are given in anatomically annotated GraphML format that facilitates better further processing and visualization. For 96 subjects, the anatomically classified sub-graphs can also be accessed, formed from the vertices corresponding to distinct lobes or even smaller regions of interests of the brain. For example, one can easily download and study the connectomes, restricted to the frontal lobes or just to the left precuneus of 96 subjects using the data. Partially directed connectomes of 423 subjects are also available for download. We also present a GitHub-deposited set of tools, called the Brain Graph Tools, for several processing tasks of the connectomes on the site http://braingraph.org.
Keywords: Connectome, Brain connections, Braingraph, Human Connectome Project
Introduction
Mapping all the inter-neuronal connections of the human brain with more than 80 billion neurons is not possible today. The discovery of connections between much larger areas of the gray matter of the human brain is feasible by applying diffusion weighted imaging data acquisition and a subsequent data processing workflow.
The NIH-funded large Human Connectome Project (HCP) (McNab et al. 2013) regularly releases its high-quality functional- and high-angular resolution diffusion imaging (HARDI) MRI datasets of hundreds of healthy human subjects. One of the most interesting applications of the published data is the mapping of the connections of the human brain on a macroscopic level: State-of-the-art computational methods make possible of discovering neural fiber connections between 1015 anatomically identified gray matter areas (also called Regions of Interests, ROIs) of the brain (Daducci et al. 2012; Fischl 2012; Desikan et al. 2006; Tournier et al. 2012). If we have HARDI data from several human subjects, then these 1015 anatomically labeled cerebral areas can be corresponded to 1015 nodes, shared by all subjects.
We will get braingraphs or connectomes from this workflow if we identify the nodes (or vertices) with the 1015 ROIs, and we connect two such vertices by an edge if the workflow finds neural fibers, connecting the ROIs, corresponded to the two vertices. Therefore, by studying braingraphs we ignore the spatial trajectories of the neural fibers in the white matter that connect the gray matter areas and can focus on the presence and the absence of connections between those ROIs. The edges of the graphs can be labeled by physical properties of the neural fibers connecting the corresponding ROIs.
Since the nodes of these graphs are corresponded to the very same set of 1015 anatomical areas, one can make comparisons between the braingraphs of individual subjects or groups of subjects in several ways (e.g., Szalkai et al. 2015a, b, 2016a, b, c, d, 2017; Kerepesi et al. 2015, 2016a, b).
Here we present the http://braingraph.org repository of connectomes, computed from the high-angular resolution diffusion imaging data of the Human Connectome Project (McNab et al. 2013), and some related software tools for the analysis and the visualization of braingraphs at the GitHub depository https://github.com/kerepesi/Brain-Graph-Tools.
Results
The human braingraphs can be downloaded from the site http://braingraph.org/download-pit-group-connectomes/.
The Brain Graph Tools is a GitHub-based repository of software programs for the easy processing of the http://braingraph.org-deposited data. The depository can be accessed at https://github.com/kerepesi/Brain-Graph-Tools.
Discussion
The following repositories are available:
Full set
The set contains the connectomes of 426 subjects from the Human Connectome Project’s public data release (McNab et al. 2013). For each subject, we have prepared five graphs, with 83, 129, 234, 463 and 1015 nodes. Each graph is available as a separate GraphML file of name nnnnnn_connectome_scale_xxx.graphml. Here the first 6 digits refer to the subject ID from the Human Connectome Project’s public release; and the last 2 or 3 digits to the vertex number in the graph. Scale 33 corresponds to 83 vertices, scale 60–129 vertices, scale 125–234 vertices, scale 251–463 vertices and scale 500–1015 vertices.
In each file (i.e., in each graph) the following weights are given for each edge:
FA_std: the standard deviation of the fractional anisotropies (Basser and Pierpaoli 1996) of the fiber(s);
fiber_length_mean: The mean of the fiber lengths, defining the graph edge, in millimeters.
fiber_length_std: The standard deviation of the fiber lengths, defining the graph edge;
FA_mean: The mean of the fractional anisotropies (Basser and Pierpaoli 1996) of the fibers;
number_of_fibers: the count of the fibers, defining the edge in question.
Directed graphs
The set contains 423 braingraphs with the 1015 nodes resolution. The edges of the graphs are directed by the Consensus Connectome Dynamics-based (Kerepesi et al. 2016a, b; Szalkai et al. 2016d) method, detailed in Szalkai et al. (2016d). Every edge description field in GraphML format contains the directed status of the edge: if it is directed, then the source and target nodes are given, if the edge is not directed then it is noted as: <edge directed="false" source= u target= v > , where u and v stand for vertex numbers, specified with anatomical information in the vertex-description field of the file. The edge weights are also noted as separate attributes.
Partial set
Contains only the graphs of 96 subjects, otherwise the format is the same as the entries of the full set. This smaller set formed the basis for the studies (Szalkai et al. 2015a, b).
Per-lobe connectomes
Contain the subgraphs of the braingraphs of 96 subjects that are induced by the different lobes of the brain. That is, for each lobe, only those edges are listed that have both endpoints in the lobe. The edges carry the five weights, specified above.
Per-ROI connectomes
Contain the subgraphs of the braingraphs of 96 subjects that are induced by the different ROIs of the brain. That is, for each ROI, only those edges are listed that have both endpoints in the ROI in question. All the edges carry the five weights, specified above. Small ROIs, even with just one vertex (e.g., the left amygdala) and large ROIs, with dozens of vertices (e.g., the right inferior-parietal lobule with 26 nodes) are also present in the set.
The brain graph tools
Is a GitHub-based repository of some software programs for the easy processing of the http://braingraph.org-deposited data. The depository can be accessed at https://github.com/kerepesi/Brain-Graph-Tools. There are three main set of tools on the site:
The Budapest Reference Connectome workflow (with RefBrainGraph.pl), contains the tools of preparing consensus connectomes from a set of braingraphs, as in Szalkai et al. (2015a, 2016a). Graphs, called k-consensus connectomes, contain the edges of n connectomes that are present in k or more braingraphs ().
The Brain Diversity workflow contains the tools GenPreFile.pl and BrainDiversity.pl, and is capable of performing a related task: from n connectomes, the individual variability of the edges of the distinct lobes or ROIs are calculated as in Kerepesi et al. (2015). The output contains interactive Google Charts visualizing the variabilities.
-
The Brain Evolution Workflow, containing GenPreFile.pl and BrainEvolution.pl, is capable of comparing the random evolution of graph edges with the phenomenon, described as the “Consensus Connectome Dynamics” in Kerepesi et al. (2016a, b). The generated figures are also given as interactive Google Charts.
Further information is given in a README file at the site https://github.com/kerepesi/Brain-Graph-Tools/blob/master/README.
Materials and methods
The data source used was the Human Connectome Project’s website: http://www.humanconnectome.org/documentation/S500 (McNab et al. 2013).
The connectomes were computed by using the Connectome Mapper Toolkit (Daducci et al. 2012) http://cmtk.org for segmentation and partitioning. For tractography, we used the MRtrix processing tool (Tournier et al. 2012) applying randomized seeding and the deterministic streamline method with a maximum of 20,000 fibers.
The braingraphs are deposited in compressed form (by either 7-zip or zip) and are labeled by the HCP subject IDs. Some misconfigured systems contain 7-zip decompressing tools that do not decode properly the end-of-line characters; in this case, we suggest using a Linux system for decompressing the files (Fig. 1).
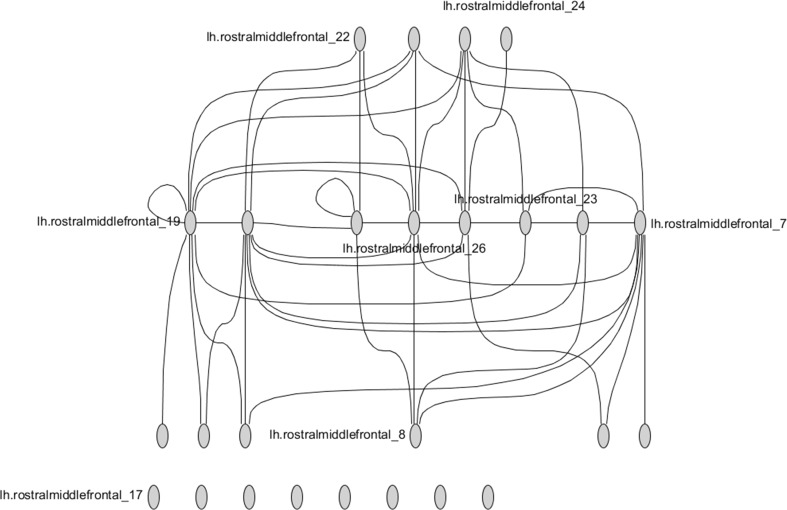
Fig. 1.
One example for the ROI subgraphs of the http://braingraph.org/download-pit-group-connectomes/, downloaded from the Repository Per-ROI connectomes. The graph describes the connections within the left rostral-middle-frontal region-of-interest (ROI) of subject ID 127933, with the 1015-vertex resolution (counted for the whole graph), the original filename is lh.rostralmiddlefrontal-127933_connectome_scale500.graphml. The graph contains 26 nodes and 44 edges, running between these nodes; some nodes are isolated. We show only some of the vertex-labels for clarity. The figure was prepared by the yED GraphML editor and viewer https://www.yworks.com/products/yed
Data availability
The Human Connectome Project’s MRI data is accessible at: http://www.humanconnectome.org/documentation/S500 (McNab et al. 2013).
The graphs (both undirected and directed) that were prepared by us from the HCP data can be downloaded at the site http://braingraph.org/download-pit-group-connectomes/. The Brain Graph Tools are available at https://github.com/kerepesi/Brain-Graph-Tools.
Acknowledgements
Data were provided in part by the Human Connectome Project, WU-Minn Consortium (Principal Investigators: David Van Essen and Kamil Ugurbil; 1U54MH091657) funded by the 16 NIH Institutes and Centers that support the NIH Blueprint for Neuroscience Research; and by the McDonnell Center for Systems Neuroscience at Washington University. CK was supported by Momentum Grant of the Hungarian Academy of Sciences (LP2012-19/2012).
Compliance with ethical standards
Conflict of interest
The authors declare no conflict of interest.
Footnotes
Csaba Kerepesi, Balázs Szalkai, and Bálint Varga: joint first authors.
Contributor Information
Csaba Kerepesi, Email: kerepesi@pitgroup.org.
Balázs Szalkai, Email: szalkai@pitgroup.org.
Bálint Varga, Email: balorkany@pitgroup.org.
Vince Grolmusz, Email: grolmusz@pitgroup.org.
References
- Basser PJ, Pierpaoli C. Microstructural and physiological features of tissues elucidated by quantitative-diffusion-tensor MRI. J Magn Reson. 1996;213(2):560–570. doi: 10.1016/j.jmr.2011.09.022. [DOI] [PubMed] [Google Scholar]
- Daducci A, Gerhard S, Griffa A, Lemkaddem A, Cammoun L, Gigandet X, Meuli R, Hagmann P, Thiran J-P. The connectome mapper: an open-source processing pipeline to map connectomes with MRI. PLoS ONE. 2012;7(12):e48121. doi: 10.1371/journal.pone.0048121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desikan RS, Ségonne F, Fischl B, Quinn BT, Dickerson BC, Blacker D, Buckner RL, Dale AM, Maguire RP, Hyman BT, Albert MS, Killiany RJ. An automated labeling system for subdividing the human cerebral cortex on mri scans into gyral based regions of interest. Neuroimage. 2006;31(3):968–980. doi: 10.1016/j.neuroimage.2006.01.021. [DOI] [PubMed] [Google Scholar]
- Fischl B. Freesurfer. Neuroimage. 2012;62(2):774–781. doi: 10.1016/j.neuroimage.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerepesi C, Szalkai B, Varga B, Grolmusz V (2015) Comparative connectomics: mapping the inter-individual variability of connections within the regions of the human brain. arXiv preprint arXiv:1507.00327 [DOI] [PubMed]
- Kerepesi C, Szalkai B, Varga B, Grolmusz V. How to direct the edges of the connectomes: dynamics of the consensus connectomes and the development of the connections in the human brain. PLoS ONE. 2016;11(6):e0158680. doi: 10.1371/journal.pone.0158680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerepesi C, Varga B, Szalkai B, Grolmusz V (2016b) The dorsal striatum and the dynamics of the consensus connectomes in the frontal lobe of the human brain. arXiv:1605.01441 [DOI] [PubMed]
- McNab JA, Edlow BL, Witzel T, Huang SY, Bhat H, Heberlein K, Feiweier T, Liu K, Keil B, Cohen-Adad J, Tisdall MD, Folkerth RD, Kinney HC, Wald LL. The human connectome project and beyond: initial applications of 300 mT/m gradients. Neuroimage. 2013;80:234–245. doi: 10.1016/j.neuroimage.2013.05.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szalkai B, Kerepesi C, Varga B, Grolmusz V. The Budapest reference connectome server v2. 0. Neurosci Lett. 2015;595:60–62. doi: 10.1016/j.neulet.2015.03.071. [DOI] [PubMed] [Google Scholar]
- Szalkai B, Varga B, Grolmusz V. Graph theoretical analysis reveals: women’s brains are better connected than men’s. PLoS ONE. 2015;10(7):e0130045. doi: 10.1371/journal.pone.0130045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szalkai B, Kerepesi C, Varga B, Grolmusz V (2016a) Parameterizable consensus connectomes from the human connectome project: the Budapest reference connectome server v3.0. Cogn Neurodyn. doi:10.1007/s11571-016-9407-z [DOI] [PMC free article] [PubMed]
- Szalkai B, Varga B, Grolmusz V (2016b) The graph of our mind. arXiv preprint arXiv:1603.00904 [DOI] [PMC free article] [PubMed]
- Szalkai B, Varga B, Grolmusz V (2016c) Mapping correlations of psychological and connectomical properties of the dataset of the human connectome project with the maximum spanning tree method. arXiv:1602.04776 [DOI] [PubMed]
- Szalkai B, Kerepesi C, Varga B, Grolmusz V (2016d) High-resolution directed human connectomes and the consensus connectome dynamics. arXiv:1609:09036 [DOI] [PMC free article] [PubMed]
- Szalkai B, Varga B, Grolmusz V (2017) Brain size bias-compensated graph-theoretical parameters are also better in women’s connectomes. Brain Imaging Behav. doi:10.1007/s11682-017-9720-0 [DOI] [PubMed]
- Tournier J, Calamante F, Connelly A, et al. Mrtrix: diffusion tractography in crossing fiber regions. Int J Imaging Syst Technol. 2012;22(1):53–66. doi: 10.1002/ima.22005. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The Human Connectome Project’s MRI data is accessible at: http://www.humanconnectome.org/documentation/S500 (McNab et al. 2013).
The graphs (both undirected and directed) that were prepared by us from the HCP data can be downloaded at the site http://braingraph.org/download-pit-group-connectomes/. The Brain Graph Tools are available at https://github.com/kerepesi/Brain-Graph-Tools.