Abstract
Chicken egg yolk immunoglobulin (IgY) is a functional substitute for mammalian IgG for antigen detection. Traditional IgY purification methods involve multi‐step procedures resulting in low purity and recovery of IgY. In this study, we developed a simple IgY purification system using IgY‐specific peptides identified by T7 phage display technology. From disulfide‐constrained random peptide libraries constructed on a T7 phage, we identified three specific binding clones (Y4‐4, Y5‐14, and Y5‐55) through repeated biopanning. The synthetic peptides showed high binding specificity to IgY‐Fc and moderate affinity for IgY‐Fc (Kd: Y4‐4 = 7.3 ± 0.2 μM and Y5‐55 = 4.4 ± 0.1 μM) by surface plasmon resonance analysis. To evaluate the ability to purify IgY, we performed immunoprecipitation and affinity high‐performance liquid chromatography using IgY‐binding peptides; the result indicated that these peptides can be used as affinity ligands for IgY purification. We then used a peptide‐conjugated column to purify IgY from egg yolks pre‐treated using an optimized delipidation technique. Here, we report the construction of a cost‐effective, one‐step IgY purification system, with high purity and recovery. © 2017 The Authors. Journal of Peptide Science published by European Peptide Society and John Wiley & Sons Ltd.
Keywords: T7 phage display, peptide library, IgY purification, chicken egg yolk
Abbreviations
- BSA
bovine serum albumin
- ELISA
enzyme‐linked immunosorbent assay
- Fmoc
9‐fluorenymethoxycarbonyl
- hIg
human immunoglobulin
- HPLC
high‐performance liquid chromatography
- HRP
horse‐radish peroxidase
- IgY
immunoglobulin Y
- Kd
equilibrium dissociation constant
- KLH
keyhole limpet haemocyanin
- mIg
mouse immunoglobulin
- PBS
phosphate‐buffered saline
- SA
streptavidin
- SPR
surface plasmon resonance
Introduction
Mammalian monoclonal and polyclonal antibodies are frequently used as research and diagnostic reagents. Recently, IgY (a category of immunoglobulins occurring in avians) was used as an alternative for mammalian antibodies 1. IgY antibodies are a predominant class of serum immunoglobulins found in oviparous animals; mammals contain an equivalent, IgG. As homologs of mammalian IgG, IgY antibodies are transferred to the egg yolk via a specific receptor expressed on the membrane of the ovarian follicles to confirm passive immunity to the developing offspring 2, 3. The phylogenetic and genetic properties of IgY that differ from mammalian immunoglobulin can be used to develop highly specific antibodies against conserved mammalian proteins. IgY does not cross‐react with class IgG, rheumatoid factor, human anti‐mice antibodies, and heterogeneity glycoproteins. Therefore, IgY has distinctive features useful for various immunology and diagnostic identification methods, reducing the risk of false‐positive results 4. Additionally, IgY technology reduces ethical concerns because it is isolated from egg yolks and does not require animal trauma 4. Furthermore, because 100 mg of IgY can be obtained from one egg yolk and a low quantity of antigen is required to induce a humoral response, IgY technology is both readily available and economical 3, 4, 5.
Several relatively easy, inexpensive, and highly efficacious procedures such as polyethylene glycol or ammonium sulfate precipitation, water dilution, ultrafiltration, gel filtration, thiophilic gel chromatography, and ion exchange chromatography have been developed for the isolating and purifying IgY from egg yolks 4, 5, 6. However, most traditional IgY extraction procedures involve changing temperature, ion strength, and pH, which are non‐specific and time‐consuming processes that achieve less than 50% yield and less than 80% purity 7. Because IgY‐Fc has an amino acid sequence deferent from IgG‐Fc, IgY structures lack protein A and G binding specificity, which are commonly used for mammalian antibody affinity purification 8. Recently, Grover et al. reported a transmembrane protein from human mycoplasma named as Protein M, which binds to all human and non‐human IgG with high specificity. Xuemei et al. recently reported a Protein M‐based affinity chromatography column with approximately 125‐fold greater activity in the IgY eluate compared with traditional IgY extraction methods 9. However, bacterial protein‐based antibody purification in the pharmaceutical industry requires extra attention to prevent bacterial endotoxin or bacterial protein contamination 10. Human mycoplasma is highly toxic and antigenic, and there is a high risk of contamination during large‐scale pharmaceutical methods of purification.
Phage display technology has been used as a screening method for the generation/identification of functional peptides, proteins, or monoclonal antibodies 10, 11, studying protein/DNA–protein interactions, screening cDNA expression, epitope mapping of antibodies, engineering human antibodies 12, optimizing antibody specificities, identifying peptides that home to specific organs or tissues, and generating immunogens for vaccine design 13, 14, 15, 16. The most common bacteriophages used for phage display are M13, fd filamentous phage, T4, T7, and λ phage. Among these, T7 phage display enables various peptides or proteins to be displayed on the surface of lytic T7 phage particles with a reduced bias of amino acids generated by the mixed nucleotides in the display peptides. By using T7 phage display, IgG 17 and IgA 18 purification columns were generated by isolating mammalian IgG‐specific and IgA‐specific peptides, respectively. Increased peptide diversity and rapid plaque formation properties (2–3 h) 19 of T7 phage‐based screening procedures are superior to those of filamentous phage‐based techniques 20. In addition, T7 phage is stable to detergents and denaturants including 1% sodium dodecyl sulfate (SDS) and urea (up to 4 M) 20.
In this study, we identified IgY binding peptides from a disulfide‐constrained random peptide library constructed on a T7 phage display system 18. The isolated peptide sequences indicated the essential residues for binding, which included two conserved tryptophan residues. Our novel peptides showed suitable binding affinity (Kd = 4–7 μM) and specificity for IgY, which will enable the development of an affinity column for IgY purification.
Experimental Procedures
Construction of T7 Phage Display Library and Biopanning
T7 phage libraries typically displaying X3CX7–10CX3 random peptides, where X represents randomized amino acid positions generated using mixed oligonucleotides on template DNA, were constructed using the T7Select vector 10‐3b from Merck (Kenilworth, NJ, USA), as previously 21. Next, 96‐well microplate's wells (Maxisorp; Nunc, Roskilde, Denmark) were coated with IgY (Jackson ImmunoResearch Laboratories, West Gove, PA, USA) solution (5.5 μg/200 μL/well) and blocked with 0.25% bovine serum albumin (BSA) in phosphate‐buffered saline (PBS; 50 mM phosphate butter containing 0.25 M NaCl, pH 7.0). T7 phage libraries (1.0 × 1010 pfu) of X3CX7–10CX3 were incubated for 1 h in wells coated with BSA to remove non‐specific phage and then added to IgY‐coated wells. After incubation for 1 h, the plate was washed 5–30 times with PBST. Escherichia coli BLT5615 cells (300 μL) (Novagen, Madison, WI, USA) in log‐phase growth (OD600 0.7–0.8) were added to the wells, infected with phage for 10 min, and propagated in 2TY medium at 37 °C. After bacteriolysis, 5 M NaCl (0.1‐fold the bacterial culture solution) was added, and the phages were recovered from the culture supernatant by centrifugation (15 000 rpm for 20 min). Next, 50% polyethylene glycol (0.2‐fold the bacterial culture solution) was added for precipitation. After centrifugation (15 000 rpm for 20 min), the supernatant was discarded, the remaining phage pellet was dissolved in PBS, and the recovered phage solution was used for the next round of biopanning (Figure 1).
Figure 1.
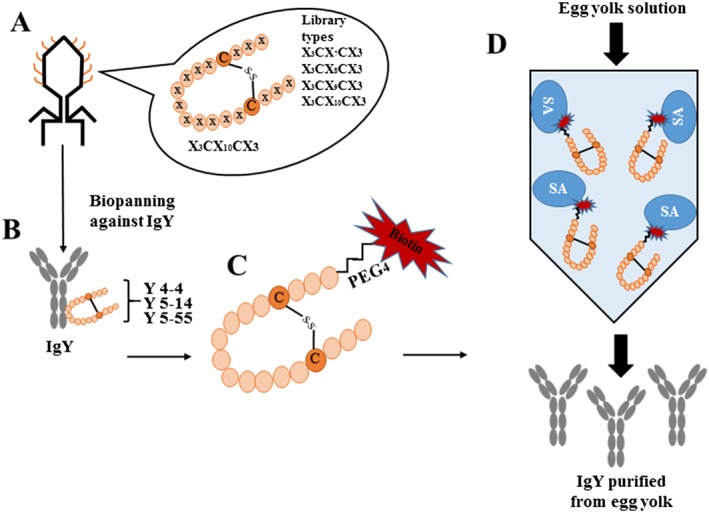
Schematic overview of this research. (A) Disulfide‐constrained random peptide libraries (X3CX7–10CX3 random peptides, where X represents the randomized amino acid positions) constructed on T7 phage using phage display technology. (B) Three IgY binding phage clones (Y4‐4, Y5‐14, and Y5‐55) identified thorough repeated biopanning against IgY from random peptide libraries. (C) Biotinylated IgY‐binding peptides derived from Y4‐4 and Y5‐55 phage clones. (D) Affinity column conjugated with biotinylated Y4‐4 peptide on a HiTrap™ SA HP column to purify IgY from egg yolk solution.
Preparation of Synthetic Peptides
C‐terminally amidated synthetic peptides Y4‐44 and Y5‐55 were synthesized by solid phase synthesis using Fmoc chemistry. The protected peptides were coupled on the resin with NHS‐PEG4‐biotin (ThermoFisher, Waltham, MA, USA). After removal of the protecting groups, the peptides were mildly oxidized to form intra‐molecular disulfide bonds in DMSO containing 1% pyridine. The generated disulfide‐constrained peptides were purified by reversed phase‐HPLC. Purity of the peptides were checked on Acquity SQD ultra‐performance liquid chromatography system (Waters Corp., Milford, MA, USA), and the disulfide bond formation of the peptides was confirmed by MALDI‐TOF mass spectrometry on Voyager System 6366 (Applied Biosystems, Foster City, CA, USA). The physicochemical properties of these peptides are summarized in Table S1. After lyophilization, the peptides were dissolved in the appropriate buffers and used for assay after centrifugation.
ELISA
The wells of a Microplate (Maxisorp; Nunc, Roskilde, Denmark) were coated with IgY, IgY‐Fc, hIgG (Chugai Pharmaceutical Co., Ltd., Tokyo, Japan), mIgG (PharMingen, San Diego, CA, USA), human serum albumin, and BSA (50 ng/50 μL/well) overnight at 4 °C, washed three times with PBST and blocked for 2 h with 0.5% BSA in PBS. Phage solution was added to each well and incubated for 2 h. After washing the plate, bound phages were detected with biotinylated anti‐T7 phage antibody (Novagen) and horseradish peroxidase (HRP)‐conjugated streptavidin (SA) (Vector Laboratories, Burlingame, CA, USA).
For peptide binding, biotinylated peptide (40 nM) was pre‐incubated with HRP‐conjugated SA (10 nM) to form a tetrameric peptide complex. The mixture was added to IgY‐coated wells of a plastic plate. After 1 h incubation, the wells were washed five times with PBST, and binding was detected with tetramethylbenzidine (Wako Pure Chemical, Osaka, Japan) reagent. Finally, 1 M hydrochloric acid (40 μL/well) was added to stop the reaction, and binding was measured by the absorbance at 450 nm in a microplate reader (680XR; Bio‐Rad, Hercules, CA, USA).
For anti‐KLH antibody measurements, KLH (50 ng/50 μL/well) and 0.5% BSA were coated overnight at 4 °C, washed three times with PBST, blocked with 0.5% BSA for 2 h, and washed again with PBST. Egg samples (before and after peptide‐conjugated column purification) were diluted by 1000‐fold using distilled water. Next, 50 μL/well of the solution was added, and binding was detected with anti‐IgY‐conjugated with HRP (diluted by 2000‐fold).
Surface Plasmon Resonance (SPR) Analysis
SPR analysis was performed on a BIAcore T200 (GE Healthcare, Little Chalfont, UK) at 25 °C. All reagents and sensor chips were purchased from GE Healthcare. IgY and IgY‐Fc were immobilized on a CM5 sensor chip according to the manufacturer's instructions. The amount of the immobilized IgY was adjusted to within 1300–4800 response units. The association reaction was monitored by injecting the peptides into the sensor chip at a flow rate of 50 μL/min for 180 s. The dissociation reaction was performed in HBS‐EP buffer (10 mM HEPES; pH 7.4 containing 150 mM NaCl, 3 mM EDTA, and 0.005% Tween 20).Binding kinetic parameters were calculated using BIAevalution Version 3.2 Software (GE HealthCare).
Immunoprecipitation by Magnetic Beads
Immunoprecipitation was performed on SA beads by washing with PBS. Biotinylated IgY‐binding peptides (100 mM) were added and shaken for 30 min. Next, IgY and human IgG (30 μg/100 μL) were added, shaken for 1 h, and centrifuged. The supernatant was removed, and dispersed precipitated beads were mixed with buffer and centrifuged. Finally, 30 μL of precipitated beads was used for SDS‐PAGE analysis.
For SDS‐PAGE analysis of the obtained fractions, the sample was mixed with SDS sample buffer and subjected to SDS‐PAGE on a 4–20% gradient gel (Mini‐PROTEIN TGX; Bio‐Rad).After electrophoresis, the gel was stained with Coomassie Brilliant blue R‐250 stain solution (Bio‐Rad).
HPLC Analysis
Y4‐4 and Y5‐55 peptides were biotinylated via PEG4 linker to the N‐terminus, and then 385 and 324 nmol/column were immobilized, respectively, on a HiTrap™ Streptavidin HP column (1 mL; GE Healthcare) according to the manufacturer's instructions. IgY, IgY‐Fc, and human IgG were used as standard proteins to identify binding against peptides. After injecting the standard proteins, the column was washed with PBS and peptide‐absorbed proteins were eluted with elution buffer (0.1 M glycine‐HCl, pH 2.5 and pH 3).
IgY Extraction by De‐Lipid Solution
Egg yolk was separated from egg white, and after washing the egg with distilled water, 5 mL of egg yolk was mixed with five volumes of (25 mL) of delipidation solution containing 0.072% κ‐Carrageenan (Sigma, St. Louis, USA), 0.12% low‐methoxyl pectin (Cp Kelco APS, Atlanta, GA, USA), and 12 μM CaCl2 (Sigma). The solution was then incubated at 4 °C for 2 h with shaking and then centrifuged for 20 min at 12 000 ×g at 4 °C. After centrifugation, soluble IgY in the delipidated supernatant was collected and precipitated by (NH4)2SO4, which accounted for up to 35–40% of the total solution, followed by 1 h incubation with shaking at 4 °C and centrifugation for 20 min at 12 000 ×g. Finally, the supernatant was removed, and the pellet was suspended in 10 or 50 mM phosphate buffer.
IgY Extraction by Distilled Water
First, 5 mL of egg yolk separated from egg white was mixed with 10 volumes of distilled water and incubated at 4 °C with shaking overnight followed by centrifugation at 10 000 ×g for 25 min. After centrifugation, the supernatant was collected and precipitated using four different concentrations (30%, 35%, 40%, and 50% w/w) of (NH4)2SO4. Finally, all the samples were incubated with shaking at 4 °C for 1 h, followed by centrifugation for 20 min at 12 000 ×g, and the pellet was suspended in 10 mM phosphate buffer (pH 7.0) after removing the supernatant.
Chicken Immunization and Egg Collection
Two Boris Brown Chickens were injected intramuscularly with JWH‐KLH (JWH conjugated to Keyhole Limpet Hemocyanin) (ARC Resources, Calgary, Canada), and the first immunization was carried out in complete adjuvant. Four booster injections with incomplete adjuvant were given at 14, 28, 42, and 63 days after the first immunization. The eggs were collected each day starting on the second day after immunization and stored at 4 °C.
Purification of Chicken Egg Yolk IgY
Approximately 385 nmol of Y4‐4 peptide was immobilized on a HiTrap Streptavidin HP column (1 mL; GE Healthcare), according to the manufacturer's instructions. Egg yolk IgY was extracted from a commercial egg by injecting distilled water directly into the peptide‐immobilized affinity column (1 mL), which was connected to a Profinia purification system (Bio‐Rad). Washing buffer (PBS) was used to remove unbound materials. Binding IgY was eluted with 0.1 M glycine‐HCl/0.25 M NaCl (pH 2.5 and pH 3), and the eluate was immediately neutralized with neutralization buffer (1 M Tris–HCl, pH 8.5) and stored at 4 °C until use. The obtained fractions were evaluated for immunoreactivity by enzyme‐linked immunosorbent assay (ELISA) and purity by SDS‐PAGE analysis. From the SDS‐PAGE data, the purity of the obtained fractions was estimated using GelAnalyzer2010a software. Protein concentrations were estimated from the absorbance at 280 nm using an absorbance coefficient of 1.55 mL/mg for IgY and other proteins.
Results
Isolation of IgY‐Specific Phage Clones
IgY‐binding T7 phage clones were enriched by five rounds of biopanning against IgY from two random libraries, X3CX7–8CX and X3CX9–10CX3, where X represents random amino acid positions. The binding activities of the phage libraries after biopanning were examined by ELISA (Figure 2(A)). Phage binding increased after repeated biopanning in both libraries, but in the X3CX7–8CX3 library, binding to human serum albumin was observed indicating non‐specific binding. In contrast, the X3CX9–10CX3 library showed increased binding to IgY. The phage were randomly cloned after 4 and 5 rounds of biopanning of the X3CX9–10CX3 library and subjected to binding screening by ELISA. Among the 30 clones, 25 clones showed high binding to IgY (Figure 2(B)). Sequence analysis of 16 clones displaying peptides revealed three individual motifs (Y4‐4, Y5‐14, and Y5‐15), as shown in Table 1. These binding clones bound specifically to IgY but not to other immunoglobulins or proteins (Figure 3(A)).
Figure 2.
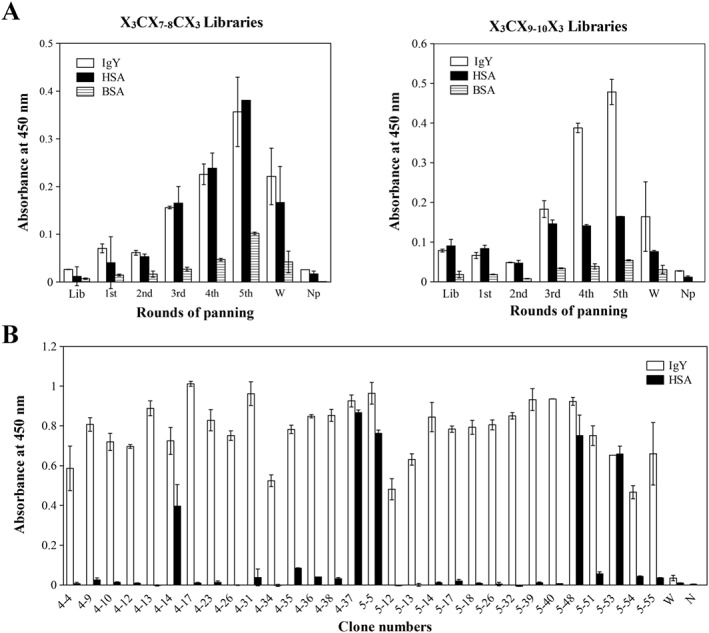
Isolation of IgY‐specific phage clones from T7 phage‐displayed random peptide libraries. (A) Isolation of IgY‐specific phage from the initial libraries (X3CX7‐8C3 or X3CX9‐10C3). After five rounds of biopanning, specific phage enrichment against IgY was examined by ELISA. Lib, phage libraries before biopanning; 1st–5th, phage after 1–5 rounds of biopanning; W, wild‐type phage; Np, measurement without phage. (B) Identification of IgY‐binding phage clones isolated from 4th and 5th phage pool by ELISA.
Table 1.
Comparison of amino acid sequences of IgY binding peptides
| Clone | Sequence | Frequency | Library source |
|---|---|---|---|
| — | 1 5 10 15 | — | — |
| 4–4 | GVKCTWSSIVDWVCVDM | 11/16 | X3CX9CX3 |
| 5–14 | GTRCDWSAAYGWLCYDY | 4/16 | X3CX9CX3 |
| 5–55 | RSVCVWTAVTGWDCRND | 1/16 | X3CX9CX3 |
Amino acid positions are numbered based on X3CX9CX3 sequence.
Figure 3.
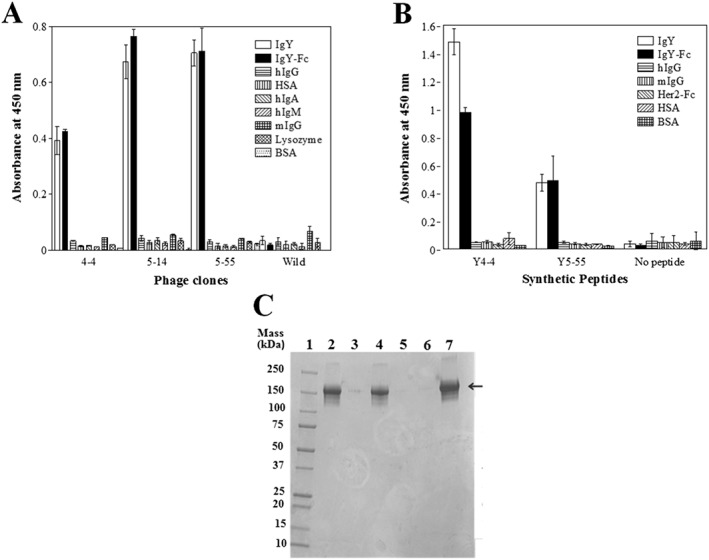
Binding specificity analysis of IgY‐binding peptides. The binding specificity of (A) identified phage clones and (B) synthetic peptide was examined by ELISA. No peptide indicates measurement without peptide. (C) Immunoprecipitation analysis of Y4‐4 and Y 5‐55 synthetic peptides. Lane 1, molecular weight marker (Precision Plus Protein™ All Blue Standards, Bio‐Rad). Lane 2, Y4‐4 peptide beads mixed with IgY. Lane 3, Y4‐4 peptide beads mixed with IgG. Lane 4, Y5‐55 peptide beads mixed with IgY. Lane 5, Y5‐55 peptide beads mixed with IgG. Lane 6, beads only. Lane 7, standard IgY (indicated by arrow).
Functional Evaluation of IgY Binding Synthetic Peptides
The peptide sequences displayed on the 16 phage clones shared a common peptide sequence pattern, X3CXWX5WXCX3 (Table 1), on three peptide‐displaying phage clones. We synthesized the most and least abundant peptide sequence derived from the Y4‐4 and Y5‐55 phage clones (Table 1) and analyzed their functionality by ELISA, SPR, and immunoprecipitation. Synthetic peptide binding affinity for IgY was analyzed by SPR. The equilibrium constants for the dissociation (Kd) between IgY immobilized on a CM5 sensor chip and Y4‐4 or Y5‐55 peptides were estimated to be 7.3 ± 0.2 and 4.4 ± 0.1 μM, respectively, using a pH 7 running buffer (Figure S1 ).
The binding specificities of the synthetic peptides Y4‐4 and Y5‐55 were compared with various proteins by ELISA (Figure 3(B)). Both peptides showed higher binding against IgY and IgY‐Fc, indicating that they were specific.
Finally, to confirm that the peptides had potential binding ability to recover IgY from the solution, SA‐agarose beads immobilized with biotinylated IgY binding peptides (Y4‐4 and Y5‐55) were mixed with IgY solution (2 μM) and, after washing the beads, subjected to SDS‐PAGE (Figure 3(C)). The result shows that IgY was specifically recovered by both peptides but hIgG was not.
Assessment of Y4‐4 and Y5‐55 Immobilized Affinity Column by HPLC
To investigate the absorption ability of Y4‐4 and Y5‐55 peptide‐conjugated affinity columns, an N‐terminal biotinylated peptide was immobilized on a HiTrap Streptavidin HP column. IgY, IgY‐Fc, or human IgG were injected into the column connected to HPLC. To elute the adsorbed protein fraction from the Y4‐4 and Y5‐55 peptide columns, two acidic elution buffers (0.1 M glycine‐HCl) were used with pH 2.5 and pH 3.0, respectively. Although human IgG passed through both peptide‐immobilized columns, IgY and IgY‐Fc proteins were absorbed onto both columns ( Figure S2 ). Furthermore, IgY and IgY‐Fc proteins were eluted from the column at pH 2.5 and 3.0, although the elution peaks at pH 2.5 were sharper than those at pH 3.0 for both peptides.
IgY Extraction from Eggs
We then attempted to develop an IgY purification procedure from egg yolk using an IgY binding peptide‐conjugated affinity column. We attempted to develop an IgY extraction method from egg yolk. Although several IgY extraction methods have been reported 4, 6, we evaluated IgY extraction using a de‐lipid solution containing κ‐Carrageenan 22. However, addition of the de‐lipid solution to the egg yolk considerably increased the viscosity of the samples subjected to ammonium sulfate precipitation, which would inhibit subsequent processes (date not shown).
Next, we attempted to extract IgY from the egg by diluting the egg yolk tenfold with distilled water with shaking at 4 °C overnight, followed by ammonium sulfate precipitation. The supernatant after centrifugation of the dialysate of the precipitate was clear and not viscous. SDS‐PAGE analysis of the precipitate (Figure 4(A)) by precipitation using different concentrations of ammonium sulfate indicated that IgY was sufficiently recovered by suppressing the contamination of low molecular weight proteins by water‐extraction coupled with 35% (w/w) ammonium sulfate precipitation (Figure 4(A), lane 3).
Figure 4.
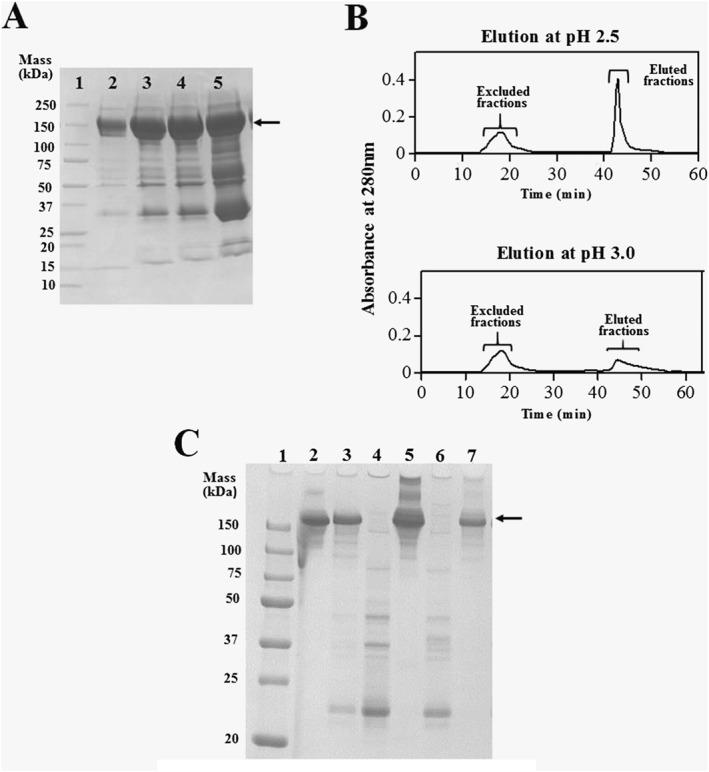
IgY purification from chicken egg yolk. (A) Optimization for extraction condition of IgY from delipitated egg yolk solution. Lane 1, IgY (standard control). Proteins (lanes 2–4) precipitated by 30, 35, 40, and 45% (w/w) (NH4)2SO4, respectively, were subjected to SDS‐PAGE. (B) IgY purification from egg yolk extract by peptide column at pH 2.5 and 3.0. (C) Confirmation of IgY purity by SDS‐PAGE. Lane 1, molecular weight marker (Precision Plus ProteinTM All Blue Standards, Bio‐Rad). Lane 2, IgY (standard control). Lane 3, egg yolk extract before peptide column purification. Lane 4, flow‐through solution. Lane 5, eluted fraction at pH 2.5. Lane 6, flow‐through fraction. Lane 7, eluted fraction at pH 3.0 in B. Arrows in panels A and C indicate the bands of IgY.
Purification of Egg Yolk IgY by Peptide‐Affinity Column
A peptide‐conjugated column prepared by immobilization of the biotinylated Y4‐4 peptide (385 nmol) in a HiTrap Streptavidin HP column was used for one‐step purification chromatography on Profinia (Bio‐Rad) to purify IgY from egg yolk extract precipitated with 35% (w/w) ammonium sulfate (Figure 4(B)). After applying the egg yolk solution to the peptide affinity column, the column was washed with 10 column volumes of PBS and eluted with 0.1 M glycine‐HCl (pH 2.5 and 3.0).The excluded and eluted fractions were subjected to SDS‐PAGE to evaluate the purity of IgY in each fraction (Figure 4(C)). IgY (150 kDa) was not detected in the flow‐through fractions but appeared as a single band in the eluted fractions at pH 3.0, indicating the successful isolation of IgY from egg yolk with high purity (93%). In contrast, proteins with higher molecular mass (>250 kDa) were observed by SDS‐PAGE following elution at pH 2.5, resulting in low purity (63%) of IgY. The recovery and purity of IgY after purification are summarized in Table 2.
Table 2.
Recovery and purity of IgY in peptide‐affinity column purification
| pH of elution buffer | Protein in egg yolk delipidation solution | Flow through fractions | Elution fractions | |||
|---|---|---|---|---|---|---|
| — | Amount (mg) | Purity (%) | Amount (mg) | Purity (%) | Amount (mg) | Purity (%) |
| 2.5 | 1.47 | 70 | 0.68 | 0 | 0.74 | 63 |
| 3.0 | 0.65 | 0 | 0.43 | 93 | ||
The protein amount (starting from 1 mL delipidation solution corresponding to 1 mL egg yolk) was indicated at each step of purification. The purity of IgY in each fraction was estimated from CBB‐gel image on SDS‐PAGE using GelAnalyzer2010a software.
Finally, we attempted to purify functional IgY from eggs collected from chickens 30–37 days after immunization with KLH antigen four times every week after initial immunization. IgY was purified from these eggs as described in Figure 4. The antigen binding ability of the purified IgY was tested by ELISA. As shown in Figure 5, purified IgY from eggs of immunized chickens showed KLH‐specific binding ability, which was also observed in the egg yolk extraction before column chromatography. This suggests that our method can be used for to purify functional IgY from the eggs of immunized chickens.
Figure 5.
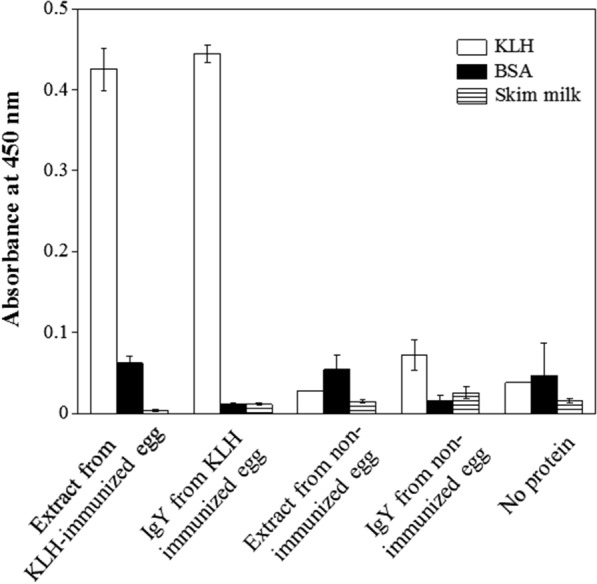
Function of purified IgY. The binding specificity of IgY from KLH‐immunized chicken egg was analyzed by ELISA. Extract from KLH‐immunized egg; delipidated solution from KLH‐immunized chicken egg yolk. IgY from KLH immunized egg; IgY purified by Y4‐4 peptide conjugated column from KLH‐immunized chicken egg. Extract from non‐immunized egg: delipidated solution from non‐immunized chicken egg yolk. IgY from non‐immunized egg; IgY purified by Y4‐4 peptide conjugated column from non‐immunized chicken egg. No protein; measurement without protein.
Discussion
In the field of biological research, chicken immunoglobulins (IgY) have recently gained attention for their biomedical potential compared with mammalian antibodies. However, the use of IgY is limited because of the lack of a simple and effective IgY purification technique. In this study, we identified unique IgY binding peptides from random peptide libraries constructed on a T7 phage display system and established an IgY‐purification technique from egg yolk using a peptide‐conjugated affinity column.
In this study, we used two groups of random peptide libraries (X3CX7–8CX3 and X3CX9–10CX3) constrained by an intramolecular disulfide bond 21 and successfully enriched IgY‐specific phage only from the X3CX9–10CX3library. The phage clones (4‐4, 5‐14, and 5‐55) isolated from the enriched phage pool showed clear binding specificity to IgY, but their sequences showed relatively low sequence identity (21–42%). Two characteristic conserved Trp residues were found at positions 6 and 12 in addition to two fixed Cys residues, indicating the importance of these Trp residues in binding. Other similar residues were found at several positions. For example, a Ser or Thr at position 7, Ser or Ala at position 8, and hydrophobic residues (Ile, Ala, and Val) at position 9 were observed. The common characteristics of these residues indicate that they are involved in binding, but further mutational studies are required to determine their roles.
Our peptides were very specific to IgY‐Fc but showed moderate binding affinity (Kd = 7.3 ± 0.2 μM and 4.4 ± 0.1 μM for Y4‐4 and Y5‐55) for IgY. Despite this moderate affinity, these peptides functioned as affinity ligands to isolate IgY from an IgY solution using peptide‐conjugated beads. Furthermore, the peptides conjugated to the column were tested as affinity ligands for IgY absorption on HPLC (Figure S2). As a result, the peptide‐conjugated column completely absorbed IgY and IgY‐Fc, but not human IgG. Dong et al. 7 reported a synthetic affinity ligand for IgY purification. However, their synthetic ligand bound to both IgY and human IgG. This is the first to report an IgY‐specific binding peptide with highly specific affinity for IgY without cross‐reactivity to human or mouse/rat IgG (data not shown).
The high lipid content of egg yolk interferes with affinity‐based IgY purification. To avoid this, a delipidation step is required to remove insoluble lipids and lipoproteins 22. We first investigated an IgY de‐lipid extraction procedure using vegetable gum (κ‐Carrageenan) followed by ammonium sulfate precipitation, as reported by Tan et al. 22. However, the κ‐Carrageenan de‐lipid solution‐based IgY extraction procedure cause the extract solution to become very viscous, which interrupted column chromatography. Therefore, we employed a simple dilution of egg yolk with distilled water followed by ammonium sulfate precipitation. This water‐based IgY extraction offers a clear and non‐viscous IgY extraction solution, removing lipids as a floating bubble layer. We confirmed that IgY extraction by distilled water and 35% (w/w) ammonium precipitation to remove insoluble lipids from the egg yolk was effective for extracting IgY.
Using the peptide‐conjugated column, we purified IgY from egg yolks of non‐immunized and KLH‐immunized chickens and successfully isolated highly pure IgY (>90%) in both cases with a high recovery yield (approximately 70%). However, the purified IgY contained several bands with high molecular weights larger than IgY (lane 5 of Figure 4(C)). Because these bands were not observed before chromatography, they were generated by the chromatographic procedures, likely by the formation of oligomers of IgY induced by acid treatment (pH 2.5) during elution. To avoid this, a mildly acidic condition of pH 3.0 was used for elution (lane 7 of Figure 4(C)), although the recovery yield from the column decreased from 0.74 to 0.43 mg (Table 2).
In summary, we successfully refined a highly specific and functional IgY binding peptide from the T7 phage library. This was achieved by identifying a specific IgY binding peptide‐displaying phage by biopanning from a random peptide‐displaying library. Our IgY‐binding novel peptide is compact and highly functional as an affinity ligand and may be an excellent reagent for low‐cost purification of IgY from chicken egg yolks with high yield and purity.
Supporting information
Figure S1. Interaction analysis of Y4–4 peptide (A, B) and Y5–55 peptide (C, D) with IgY by SPR: First IgY was immobilized on a sensor chip, and then different concentrations of peptides (0.156–10 μM) were injected on BIAcore T200 (GE Healthcare). The affinity was estimated from the equilibrium data using BIA evaluation software.
Figure S2. IgY absorption/elution profiles on IgY binding peptide immobilized column by HPLC. Biotinylated Y4–4 and Y5–55 peptides were immobilized into SA‐HiTrap columns. After equilibration with PBS, IgY, IgY‐Fc or IgG were injected. Elution was performed by 0.1 M glycine‐HCl buffers at pH 3.0 (A, C) or 2.5 (B, D) to monitor the absorption/elution properties. Arrows 1, 2 and 3 indicate starting point for sample injection, elution and regeneration with PBS, respectively.
Table S1. Physicochemical characterization of IgY binding peptides used in this study.
Khan, K. H. , Himeno, A. , Kosugi, S. , Nakashima, Y. , Rafique, A. , Imamura, A. , Hatanaka, T. , Kato, D.‐I. , and Ito, Y. (2017) IgY‐binding peptide screened from a random peptide library as a ligand for IgY purification. J. Pept. Sci., 23: 790–797. doi: 10.1002/psc.3027.
References
- 1. Spillner E, Braren I, Greunke K, Seismann H, Blank S, du Plessis D. Avian IgY antibodies and their recombinant equivalents in research, diagnostics and therapy. Biologicals 2012; 40: 313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ward ES. Acquiring maternal immunoglobulin; different receptors, similar functions. Immunity 2004; 20: 507. [DOI] [PubMed] [Google Scholar]
- 3. Zhang WW. The use of gene‐specific IgY antibodies for drug target discovery. Drug Discov. Today 2003; 8: 364. [DOI] [PubMed] [Google Scholar]
- 4. Schade R, Calzado EG, Sarmiento R, Chacana PA, Porankiewicz‐Asplund J, Terzolo JHR. Chicken egg yolk antibodies (IgY‐technology): a review of progress in production and use in research and human and veterinary medicine. Altern. Lab. Anim 2005; 33: 129. [DOI] [PubMed] [Google Scholar]
- 5. Larsson A, Sjoquist J. Chicken IgY: utilizing the evolutionary difference. Comp. Immunol. Microbiol. Infect. Dis. 1990; 13: 199. [DOI] [PubMed] [Google Scholar]
- 6. Ko KY, Ahn DU. Preparation of immunoglobulin Y from egg yolk using ammonium sulfate precipitation and ion exchange chromatography. Poult. Sci. 2007; 86: 400. [DOI] [PubMed] [Google Scholar]
- 7. Dong D, Liu H, Xiao Q, Li R. Affinity purification of egg yolk immunoglobulins (IgY) with a stable synthetic ligand. J Chromatogr B Analyt Technol Biomed Life Sci 2008; 870: 51–54. [DOI] [PubMed] [Google Scholar]
- 8. Grover RK, Zhu X, Nieusma T, Jones T, Boero I, MacLeod AS, Mark A, Niessen S, Kim HJ, Kong L, Assad‐Garcia N, Kwon K, Chesi M, Smider VV, Salomon DR, Jelinek DF, Kyle RA, Pyles RB, Glass JI, Ward AB, Wilson IA, Lerner RA. A structurally distinct human mycoplasma protein that generically blocks antigen‐antibody union. Science 2014; 343: 656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jiang X, Diraviyam T, Zhang X. Affinity purification of egg yolk immunoglobulins (IgY) using a human mycoplasma protein. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2016; 1012‐1013: 37. [DOI] [PubMed] [Google Scholar]
- 10. Food and Drug Administration, Control of microbiological contamination, Code of Federal Regulations 21, 211.113, US Government Printing Office, Washington, DC 2003.
- 11. Brissette R, Prendergast JK, Goldstein NI. Identification of cancer targets and therapeutics using phage display. Curr. Opin. Drug Discov. Devel. 2006; 9: 363. [PubMed] [Google Scholar]
- 12. Lu TK, Koeris MS. The next generation of bacteriophage therapy. Curr. Opin. Microbiol. 2011; 14: 524. [DOI] [PubMed] [Google Scholar]
- 13. Skerra A. Alternative binding proteins: anticalins – harnessing the structural plasticity of the lipocalin ligand pocket to engineer novel binding activities. FEBS J. 2008; 275: 2677. [DOI] [PubMed] [Google Scholar]
- 14. Smith GP, Petrenko VA. Phage display. Chem. Rev. 1997; 97: 391. [DOI] [PubMed] [Google Scholar]
- 15. Azzazy HM, Highsmith WE, Jr . Phage display technology: clinical applications and recent innovations. Clin. Biochem. 2002; 35: 425. [DOI] [PubMed] [Google Scholar]
- 16. Christensen DJ, Gottlin EB, Benson RE, Hamilton PT. Phage display for target‐based antibacterial drug discovery. Drug Discov. Today 2001; 6: 721. [DOI] [PubMed] [Google Scholar]
- 17. Sakamoto K, Ito Y, Hatanaka T, Soni PB, Mori T, Sugimura K. Discovery and characterization of a peptide motif that specifically recognizes a non‐native conformation of human IgG induced by acidic pH conditions. J. Biol. Chem. 2009; 284: 9986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hatanaka T, Ohzono S, Park M, Sakamoto K, Tsukamoto S, Sugita R, Ishitobi H, Mori T, Ito O, Sorajo K, Sugimura K, Ham S, Ito Y. Human IgA‐binding peptides selected from random peptide libraries: affinity maturation and application in IgA purification. J. Biol. Chem. 2012; 287: 43126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bratkovic T. Progress in phage display: evolution of the technique and its application. Cell. Mol. Life Sci. 2010; 67: 749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fukunaga K, Taki M. Practical tips for construction of custom peptide libraries and affinity selection by using commercially available phage display cloning systems. J. Nucleic Acids 2012; 2012: 295719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Krumpe LR, Atkinson AJ, Smythers GW, Kandel A, Schumacher KM, McMahon JB, Makowski L, Mori T. T7 lytic phage‐displayed peptide libraries exhibit less sequence bias than M13 filamentous phage‐displayed peptide libraries. Proteomics 2006; 6: 4210. [DOI] [PubMed] [Google Scholar]
- 22. Tan SH, Mohamedali A, Kapur A, Lukjanenko L, Baker MS. A novel, cost‐effective and efficient chicken egg IgY purification procedure. J. Immunol. Methods 2012; 380: 73. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Interaction analysis of Y4–4 peptide (A, B) and Y5–55 peptide (C, D) with IgY by SPR: First IgY was immobilized on a sensor chip, and then different concentrations of peptides (0.156–10 μM) were injected on BIAcore T200 (GE Healthcare). The affinity was estimated from the equilibrium data using BIA evaluation software.
Figure S2. IgY absorption/elution profiles on IgY binding peptide immobilized column by HPLC. Biotinylated Y4–4 and Y5–55 peptides were immobilized into SA‐HiTrap columns. After equilibration with PBS, IgY, IgY‐Fc or IgG were injected. Elution was performed by 0.1 M glycine‐HCl buffers at pH 3.0 (A, C) or 2.5 (B, D) to monitor the absorption/elution properties. Arrows 1, 2 and 3 indicate starting point for sample injection, elution and regeneration with PBS, respectively.
Table S1. Physicochemical characterization of IgY binding peptides used in this study.


