Summary
Class I KNOTTED‐LIKE HOMEOBOX (KNOX) proteins regulate development of the multicellular diploid sporophyte in both mosses and flowering plants; however, the morphological context in which they function differs.
In order to determine how Class I KNOX function was modified as land plants evolved, phylogenetic analyses and cross‐species complementation assays were performed.
Our data reveal that a duplication within the charophyte sister group to land plants led to distinct Class I and Class II KNOX gene families. Subsequently, Class I sequences diverged substantially in the nonvascular bryophyte groups (liverworts, mosses and hornworts), with moss sequences being most similar to those in vascular plants. Despite this similarity, moss mutants were not complemented by vascular plant KNOX genes. Conversely, the Arabidopsis brevipedicellus (bp‐9) mutant was complemented by the PpMKN2 gene from the moss Physcomitrella patens. Lycophyte KNOX genes also complemented bp‐9 whereas fern genes only partially complemented the mutant. This lycophyte/fern distinction is mirrored in the phylogeny of KNOX‐interacting BELL proteins, in that a gene duplication occurred after divergence of the two groups.
Together, our results imply that the moss MKN2 protein can function in a broader developmental context than vascular plant KNOX proteins, the narrower scope having evolved progressively as lycophytes, ferns and flowering plants diverged.
Keywords: Arabidopsis, Ceratopteris richardii, cross‐species complementation, KNOTTED homeobox genes, land plant evolution, phylogeny, Physcomitrella patens, Selaginella kraussiana
Introduction
Morphogenesis in all major eukaryotic lineages is regulated at least in part by homeodomain transcription factors (Holland, 2013). Based on phylogeny, two classes of homeodomain transcription factors are assumed to have been present in the eukaryote ancestor – the canonical homeodomain proteins which have a 60 amino acid DNA binding domain and the three amino acid loop extension (TALE) proteins which have a 63 amino acid DNA binding domain (Bharathan et al., 1997; Burglin, 1997; Derelle et al., 2007). In the green plant lineage, two families of TALE proteins have been distinguished: KNOTTED‐LIKE HOMEOBOX (KNOX) and BELL (Mukherjee et al., 2009). KNOX proteins are encoded by a single gene in the unicellular chlorophyte algae that are sister group to the streptophytes (charophyte algae and land plants) (Serikawa & Mandoli, 1999; Lee et al., 2008). In the chlorophyte alga Chlamydomanas reinhardtii, the KNOX protein GSM accumulates in the cytoplasm of ‘minus’ type gametes, and the BELL protein GSP similarly accumulates in ‘plus’ type gametes. When the two types of gamete fuse, KNOX/BELL dimerization leads to nuclear localization and formation of the diploid zygote (Lee et al., 2008). The KNOX protein in the unicellular green alga Acetabularia acetabulum similarly becomes nuclear‐localized during the vegetative to reproductive transition (Serikawa & Mandoli, 1999). KNOX/BELL interactions may therefore have evolved in the chlorophytes to regulate the transition from vegetative to reproductive development and/or to specifically facilitate formation of the diploid zygote.
In contrast to the single copy KNOX genes found in chlorophyte algae, several duplication events have led to multiple copies being present in all land plant genomes examined to date (Mukherjee et al., 2009). Similarly, BELL gene numbers have expanded from the one or two seen in algal genomes to the many that are present in flowering plants (Mukherjee et al., 2009). Members of the KNOX gene family are split into two classes, with representatives of both Class I and Class II genes present in the bryophyte moss Physcomitrella patens (Singer & Ashton, 2007; Sakakibara et al., 2008), the lycophyte genus Selaginella (Harrison et al., 2005; Kawai et al., 2010), the monilophyte fern Ceratopteris richardii (Sano et al., 2005) and in all flowering plants examined (Kerstetter et al., 1994; Mukherjee et al., 2009). This phylogenetic distribution suggests that the gene duplication that led to distinct Class I and Class II KNOX genes occurred in a common ancestor of the mosses and vascular plants.
In P. patens, although KNOX genes are expressed in the gametophyte, triple mutants that are null for the three Class I genes PpMKN2, PpMKN4 and PpMKN5 form normal gametophytes, and zygote formation is not disrupted (Sakakibara et al., 2008). However, mutants exhibit perturbed cell proliferation in the diploid generation of the life cycle, which in bryophytes is represented by a multicellular sporophyte as opposed to the unicellular zygotes of chlorophyte algae. Although Class I genes promote cell proliferation in the sporophyte, the Class II genes PpMKN1 and PpMKN6 promote tissue differentiation, with loss‐of‐function leading to diploid structures having gametophyte bodyplans (Sakakibara et al., 2013). Neofunctionalization following the duplication of KNOX genes thus enabled the development of a multicellular diploid sporophyte, which was a bryophyte innovation.
Opposing roles for Class I and Class II KNOX genes also are observed in the flowering plant Arabidopsis, where Class I genes promote cell proliferation in the shoot apical meristem (Long et al., 1996; Venglat et al., 2002; Belles‐Boix et al., 2006) and Class II genes promote tissue differentiation (Furumizu et al., 2015). Although the role of Class I genes in promoting cell proliferation in the multicellular sporophyte is conserved in mosses and flowering plants, the context in which the genes operate was modified during the course of land plant evolution. For example, bryophyte sporophytes are determinate, developing as a single unbranched stalk subtending a sporangium (Ligrone et al., 2012a), whereas vascular plant sporophytes have highly variable and complex shoot architecture that results from the sustained activity of indeterminate shoot meristems giving rise to determinate lateral organs (leaves) (Sussex & Kerk, 2001). The indeterminate nature of shoot growth in the three vascular plant lineages further varies in terms of the structure of the shoot apex, with lycophyte and monilophyte shoots developing from one or a few apical initials (Steeves & Sussex, 1989; Harrison et al., 2007; Jones & Drinnan, 2009; Harrison & Langdale, 2010; Sanders et al., 2011; Vasco et al., 2013), and seed plant shoots developing from multicellular meristems (Steeves & Sussex, 1989; Barton, 2010). There is currently very little understanding of how Class I KNOX gene function evolved as the developmental mechanisms underpinning shoot growth changed.
In Arabidopsis and other flowering plants with simple leaves, Class I KNOX genes function in the indeterminate shoot meristem but are switched off in determinate leaf primordia (Hay & Tsiantis, 2010). Gene function maintains indeterminacy in the meristem by promoting cytokinin (CK) signalling and inhibiting gibberellic acid (GA) signalling (Sakamoto et al., 2001; Hay et al., 2002; Jasinski et al., 2005). Repression of KNOX activity in the leaf therefore de‐represses GA and suppresses CK, leading to determinate leaf development. A failure to suppress KNOX gene expression in the leaf, through transgenic manipulation or dominant gain of function mutation, results in continued repression of GA, promotion of CK activity, ectopic cellular proliferation, and a resultant lobed or knotted leaf phenotype (Smith et al., 1992; Lincoln et al., 1994; Sakamoto et al., 2001, 2006; Hay et al., 2002). Notably, constitutive expression of Class I KNOX genes from both P. patens and C. richardii, also leads to a lobed leaf phenotype in Arabidopsis (Sano et al., 2005; Sakakibara et al., 2008). The similar phenotypes seen in lines with transgenes from mosses, ferns and flowering plants indicate that genes from the different lineages have equivalent potential to regulate GA and/or CK targets in the context of the Arabidopsis leaf. However, a true understanding of differences in KNOX gene activity between lineages cannot be obtained from constitutive overexpression studies; instead gene function must be analysed in endogenous expression domains.
In this report we demonstrate that the duplication leading to Class I and Class II KNOX genes occurred in the charophyte algae, before the evolution of a multicellular diploid sporophyte. Representatives of both classes are retained in all land plant lineages except hornworts, where Class I genes could not be identified in the species examined. Although Class I genes from the moss P. patens share sequence similarity with those from vascular plants, lycophyte, fern and flowering plant genes all failed to complement loss‐of‐function moss mutants. By contrast, MKN2 from P. patens fully complemented loss‐of‐function brevipedicellus (bp) mutants in Arabidopsis. Selaginella kraussiana Class I genes also complemented bp mutants, whereas genes from C. richardii only partially complemented them. These results suggest that genes in earlier diverging land plant lineages are able to function in a broader developmental context than those in later diverging lineages. Notably, the BELL gene phylogeny indicates that specialized function in ferns and flowering plants may have been achieved at least in part through modified KNOX‐BELL interactions.
Materials and Methods
Plant strains and growth conditions
The Gransden strain Physcomitrella patens subsp. patens (Engel, 1968) was grown and maintained under sterile conditions on BCD or BCDAT medium (Nishiyama et al., 2000) and grown at 24°C with a diurnal cycle of 16 h : 8 h, light (300 μmol m−2 s−1) : dark. For sporophyte induction, 1‐wk‐old protonemal tissue was transferred into Magenta pots containing 100 ml solidified BCD medium supplemented with 1 mM CaCl2, and grown at 25°C for 6 wk with a 16 h : 8 h, light (300 μmol m−2 s−1): dark cycle. Magenta pots containing tall gametophores were then transferred to 16°C, with an 8 h : 16 h, light (150 μmol m−2 s−1): dark cycle. Sterile water (5–6 ml) was added to each pot immediately after transfer to 16°C. Gametangia formation started within 2–3 wk and sporophytes were visible within 5–6 wk. The triple mkn2;mkn4;mkn5 mutant (Sakakibara et al., 2008) was obtained from Mitsuyasu Hasebe, Okazaki, Japan. Arabidopsis (Arabidopsis thaliana) ecotypes Columbia and Landsberg erecta were grown at 22°C in a glasshouse with a 16 h : 8 h, light (300 μmol m−2 s−1) : dark cycle. The bp‐9 (Col) mutant (Smith & Hake, 2003) was obtained from the Nottingham Arabidopsis Stock Centre, UK. Conditions for growth of Marchantia polymorpha, Spirogyra pratensis, Anthoceros agrestis, Selaginella kraussiana and Ceratopteris richardii are provided in Supporting Information Methods S1.
Phylogenetic analysis
KNOTTED‐LIKE HOMEOBOX (KNOX) and BELL homologous sequences were obtained by interrogating databases at NCBI (http://ncbi.nlm.nih.gov), Phytozome (http://phytozome.jgi.doe.gov) and OneKP (http://bioinfodata.org/Blast4OneKP) (Johnson et al., 2012; Wickett et al., 2014; Xie et al., 2014), plus rice (http://rice.plantbiology.msu.edu), Klebsormidium (http://www.plantmorphogenesis.bio.titech.ac.jp/~algae_genome_project/klebsormidium), Amborella (http://amborella.org) and spruce (http://congenie.org) genome projects, as well as in house genomic and transcriptomic resources for Anthoceros punctatus, A. agrestis, M. polymorpha, C. richardii and S. pratensis (Li et al., 2014; Szövényi et al., 2015; unpublished data), with a variety of KNOX or BELL homologues as Blast queries. Homology was assessed using reciprocal Blast and examination of alignments. Selaginella kraussiana BELL sequences were identified in this study (see Methods S1; Notes S1, S2).
Sequences were only included in the dataset if they comprised the conserved domains defined for each gene family: MEINOX, ELK and TALE‐HD domains for the KNOX family; SKY, PBC and TALE‐HD domains for the BELL family. Marchantia polymorpha, S. pratensis and A. agrestis KNOX sequences were verified by PCR amplification (see Methods S1; Notes S1) and dideoxy sequencing (Notes S2). In addition to species used in the KNOX dataset (Notes S3), sequences from Picea abies and Amborella trichopoda were added to the BELL dataset (Notes S4). A number of sequences from Arabidopsis, rice and P. patens could not be placed with confidence and consequently were excluded to improve overall tree robustness (listed in Notes S4). Putative homologues were identified in S. pratensis and other charophytes but these sequences appeared only remotely related to land plant BELL proteins, and when added to the dataset, showed very long branches and destabilized the resulting phylogenetic trees. As a consequence, charophyte sequences were not included in the final dataset. For the same reason, Chlorophyta sequences also were omitted. Because of these restrictions, the number of sequences for any particular species in the resulting phylogenies does not always reflect the total number of homologues in the genome of that species (Notes S3, S4).
KNOX protein alignment was achieved using Clustal Omega v.1.2.0 (Sievers et al., 2011) with 10 iterations and a HMM profile that was built using Pfam database alignments (http://pfam.xfam.org) of the MEINOX, ELK and TALE‐HD domains as a guide. The amino acids were then translated back to their corresponding codon sequences using a custom Perl script. BELL protein alignment was obtained using MergeAlign (Collingridge & Kelly, 2012) with default parameters. The aligned sequences were then edited and trimmed manually to conserve homologous sites only (Fig. S1; Notes S5, S6). Maximum‐likelihood trees were obtained with RAxML v.8.2.3 (Stamatakis, 2014) and Bayesian trees were obtained using MrBayes v.3.2.5 (Ronquist & Huelsenbeck, 2003) (full details in Methods S1).
Generation of constructs for complementation in Physcomitrella patens
MKN2 coding sequence was synthesized by Integrated DNA Technologies (Coralville, IA, USA). Remaining sequences were amplified from genomic DNA or by reverse transcription polymerase chain reaction (RT‐PCR) (Methods S1) and cloned into pJET1.2 before dideoxy sequencing. The p35S‐loxP‐BSD plasmid (NCBI no. AB537973) was used as a backbone for all P. patens transformation constructs. Constructs were designed to express PpMKN2, SkKNOX1, SkKNOX2, CrKNOX1, CrKNOX2, BP or STM genes under the control of the endogenous PpMKN2 gene promoter (MKN2pro:KNOX constructs) (Fig. S2). During the generation of the triple mkn2;mkn4;mkn5 mutant, the entire genomic sequence of PpMKN2 and a 208‐bp fragment of 5′ MKN2 flanking sequence was replaced by the G418 resistance cassette (Sakakibara et al., 2008). To reintroduce this 208‐bp fragment into the endogenous locus, it was included as part of the 5′ MKN2 flanking sequence that was used as homologous sequence for recombination. A total of 1.211 kb of 5′ flanking sequence was fused with the coding sequence of PpMKN2, SkKNOX1, SkKNOX2, CrKNOX1 CrKNOX2, BP or STM. The nos terminator was fused 3′ to the coding sequence followed by the blasticidin resistance cassette and then a 1.833‐kb fragment of 3′ MKN2 flanking sequence as homologous sequence for recombination. Expression of the blasticidin resistance gene was driven by the 35S CaMV promoter.
Physcomitrella patens transformation
Physcomitrella patens was transformed using the polyethylene glycol‐based transformation method (Schaefer et al., 1991). In five to seven independent transformation experiments, the MKN2pro:KNOX constructs were introduced into mkn2;mkn4;mkn5 triple mutant protoplasts. Protoplasts were regenerated for 7 d and then transferred to fresh media containing antibiotics for selection (blasticidin at 75 mg ml−1). After 2 wk, plants were transferred back to nonselective BCDAT medium for another week and then stable transformants were selected by plating on selective media. After generating stable transgenic moss lines, each one was analysed by PCR to check that the construct was integrated into the desired locus (using primers listed in Notes S1). To determine the transgene copy number in each line, DNA gel blot analysis was carried out using 6 μg of genomic P. patens DNA according to Langdale et al. (1988). A 510‐bp fragment of the blasticidin resistance gene was hybridized to NdeI restricted genomic DNA – if the construct was correctly targeted to the PpMKN2 locus, a single fragment was detected after hybridization. Genomic DNA from the mkn2;mkn4;mkn5 triple mutant lines was used as a negative control.
Expression analysis of Physcomitrella patens transformants by RT‐PCR
Total RNA was extracted from approximately 300 archegonia that contained either unfertilized eggs or early developing embryos. cDNA was synthesized using Superscript III reverse transcriptase (Invitrogen) and 500 ng of DNase treated (Turbo DNAse; Ambion) total RNA per sample. Primer pairs (Notes S1) designed to amplify a c.500‐bp fragment of the 5′ region of the Arabidopsis, S. kraussiana or C. richardii Class I KNOX genes, or to amplify PpMKN2, were used to check transcript accumulation. Amplification of the Glycolytic glyceraldehyde‐3‐phosphate dehydrogenase (GAPC1) gene (×72381) was used as an internal control (as used in Sakakibara et al., 2008). Each PCR was performed with both sets of primer pairs and adjusted so that the reactions were in the exponential phase. Optimization of RT‐PCR conditions to generate identical levels of GAPC1 amplification was not possible due to the difficulty of extracting sufficient amounts of RNA.
Arabidopsis transformation
Six constructs were designed to express P. patens, S. kraussiana, C. richardii and Arabidopsis Class I KNOX genes under the endogenous BP promoter. The final constructs (Fig. S3) consisted of the BP 5′ cis‐regulatory sequence (from −5474 bp to −1 bp relative to the ATG) fused to the coding sequence of PpMKN2, SkKNOX1, SkKNOX2, CrKNOX1, CrKNOX2 or BP. The octopine synthase (ocs3′) terminator sequence was fused to the 3′ end of the various KNOX gene coding sequences. The BPpro:KNOX:ocs3′ cassette was transferred as a NotI fragment into the binary vector pART27 (Gleave, 1992) and transformed into bp‐9 plants by floral dipping (Clough & Bent, 1998). Transformed plants were selected on the basis of kanamycin resistance and were self‐pollinated to generate T1, T2, and T3 populations that segregated the transgenes. Five T3 lines with a single transgenic copy were analysed for each combination of transgene/background, with the exception of BPpro:SkKNOX1 and BPpro:MKN2 for which 6 and 3 lines were analysed, respectively.
Expression analysis of Arabidopsis transformants by qPCR
Transgene expression was analysed in Arabidopsis by quantitative real‐time reverse transcription‐PCR (qPCR). Total RNA was extracted from 12 inflorescences and then cDNA was synthesized using Superscript III reverse transcriptase (Invitrogen) and 1 μg of DNase treated (Turbo DNAse, Ambion) total RNA. Primer pairs were designed to amplify a c. 100‐bp fragment of the 3′ end of specific KNOX genes, with the exception of PpMKN2 and BP which were designed to amplify the middle of the coding region (Notes S1). Amplification was detected using the SYBR Green master mix (Life Technologies, Carlsbad, CA, USA) according to manufacturer's instructions on a 7300 Real‐Time PCR System (Applied Biosystems, Foster City, CA, USA). Cycling conditions were: 95°C for 5 min, and 40 cycles of 95°C for 15 s and 60°C for 1 min. Three technical replicates were performed for three different biological samples of each independent line. The mean value between the two technical replicates with the closest values was calculated. Two housekeeping genes, EF1Balpha2 (At5g19510) and UBP6 (At1g51710) were used as constitutive controls. To test for genomic DNA contamination, primers that amplify only the genomic sequence of EF1Balpha2 (At5g19510) were used (Wang et al., 2007).
Data analysis was performed using the LinRegPCR program (Ramakers et al., 2003). LinRegPCR calculates a value called N 0, expressed in arbitrary fluorescence units, which reflects the initial amount of template in the reaction mix, using the following formula: N 0 = N q/E m Cq. N q is the fluorescence threshold, C q is the cycle number at which the PCR reaction reaches N q, and E m is the mean PCR efficiency for a particular primer pair amplicon. Samples were normalized using the geometric mean of the expression values of EF1BalphaA2 and UBP6 reference genes. Finally, N 0 values obtained from the reactions that amplify genomic sequence of EF1Balpha2, reflecting genomic contamination, were subtracted from N 0 value for each line.
Silique angle measurement
Measurements were performed on 7‐wk‐old plants. Silique angles on the main inflorescence were measured for 10 plants per line using a protractor. Statistical analyses were carried out as in Methods S1 and Notes S7.
Microscopy
Physcomitrella patens images were captured using a QImaging Micropublishing 5.0 RTV camera on a Leica M165C microscope. Photographs of 30 sporophytes were taken and then the surface area of the sporophyte was compared with the area occupied by spores. Area measurements were carried out using the ImageJ software (http://rsb.info.nih.gov/ij/). The mean of the sporophyte area divided by the area occupied by spores was calculated for each line.
Results
KNOX genes duplicated before the evolution of land plants
To date, liverwort and hornwort sequences have not been included in KNOX gene phylogenies, and in the one example where charophyte sequences have been included (Sakakibara, 2016) the topology contradicted a better‐supported tree that defined distinct Chlorophyta, KNOX1 and KNOX2 clades (Furumizu et al., 2015). To better define relationships in early diverging lineages, transcriptome and/or genome data from the charophyte alga S. pratensis, the liverwort M. polymorpha, and the hornworts A. punctatus (Li et al., 2014) and A. agrestis (Szövényi et al., 2015) were searched for KNOX‐homologous sequences using TBlastN and BlastP algorithms and a variety of KNOX homologues as queries. Retrieved sequences were classified as KNOX genes if they encoded both a MEINOX domain and a TALE‐HD, using careful sequence examination of alignments and reciprocal Blast (a number of MEINOX‐only sequences from M. polymorpha were discarded). A single KNOX gene sequence was identified in both A. punctatus and A. agrestis, whereas two gene sequences were identified in both S. pratensis and M. polymorpha. To validate gene models, coding sequences were confirmed by sequencing PCR‐amplified cDNA (Notes S2). Notably, the annotation model for the MpKNOX2 gene includes a long repetitive region between the MEINOX and ELK domains that could not be amplified by PCR (Notes S2). As this region could not be validated molecularly, it was excluded from subsequent alignments used for tree inference. In summary, a single KNOX gene was verified in each of the hornwort genomes, whereas two genes were verified in the charophyte S. pratensis and the liverwort M. polymorpha.
Phylogenetic analyses were carried out using the five newly identified sequences from S. pratensis, M. polymorpha and A. agrestis, all of the KNOX sequences identified in the model flowering plants Arabidopsis (eight genes) and rice (10 genes) (Mukherjee et al., 2009), and sequences that have previously been molecularly validated in the monilophyte fern C. richardii (Sano et al., 2005), the lycophyte S. kraussiana (Harrison et al., 2005), the moss P. patens (Sakakibara et al., 2008) and the chlorophyte algae A. acetabulum (Serikawa & Mandoli, 1999) and C. reinhardtii (Lee et al., 2008). To provide a more balanced species distribution, additional nonflowering plant and charophyte sequences were retrieved from the OneKP dataset (Matasci et al., 2014) and from the Klebsormidium genome project database (Hori et al., 2014). Further chlorophyte sequences were retrieved from the Phytozome database (Notes S3). After alignment of protein sequences and reverse translation back to nucleotide sequence (Fig. S1; Notes S5), both maximum‐likelihood and Bayesian trees were inferred (Figs 1, S4). The topology of both trees essentially reflected the species tree, with three statistically well‐supported KNOX gene clades – Chlorophyta, Class I and Class II. Notably, charophyte sequences are present in both Class I and Class II clades, whereas hornwort sequences are restricted to Class II. The duplication leading to Class I and Class II KNOX genes therefore occurred within the charophyte sister group to land plants, with subsequent events in the bryophytes leading to conserved Class II sequences but substantially different (or absent) Class I sequences.
Figure 1.
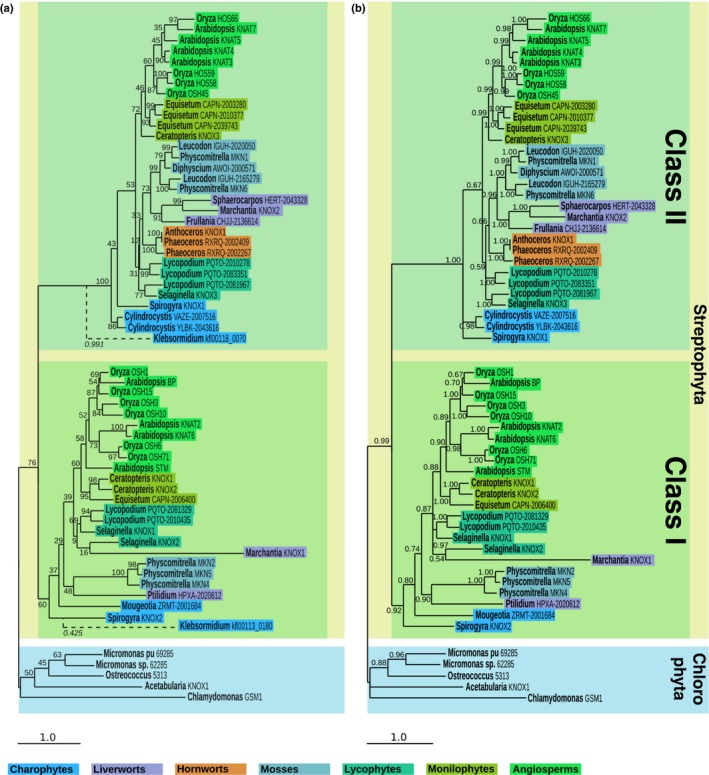
Maximum‐likelihood and Bayesian phylogenetic analysis of KNOTTED‐LIKE HOMEOBOX (KNOX) homologues. (a, b) Phylogenetic trees inferred with (a) RA x ML or (b) MrBayes, using a partitioned nucleotide dataset of 59 + 2 sequences (Supporting Information Fig. S1) evolving under the GTR + Γ + I model. Support values (a, bootstrap values; b, posterior probabilities) are indicated next to the corresponding branch. To avoid destabilization at the base of the Class I clade by Klebsormidium flaccidum sequences, kfl00113_0180 and kfl00118_0070 were added to the maximum‐likelihood tree after the phylogeny was inferred using the evolutionary placement algorithm of RAxML. As a consequence, K. flaccidum sequence branches are dashed and their associated likelihood weight ratios (Stamatakis, 2014) are indicated in italics. Each sequence was placed on its recipient tree branch with respect to the actual distance derived from a maximum likelihood tree inferred using all 61 sequences (Fig. S2). Colour‐coded boxes correspond to the species’ phyla. Both trees were rooted using the Chlorophyta clade as an outgroup.
Vascular plant Class I KNOX genes are unable to complement loss‐of‐function mutations in the moss Physcomitrella patens
In order to determine the extent to which Class I KNOX gene function diverged beyond the bryophytes, experiments were designed to test whether Class I KNOX genes from vascular plants can complement loss‐of‐gene‐function in P. patens. Sporophyte defects in triple loss‐of‐function mkn2;mkn4;mkn5 mutants are more severe than those in mkn4 and mkn5 single mutants but are very similar to those in single mkn2 mutants (Sakakibara et al., 2008). This suggests that PpMKN2 plays a more substantial role in sporophyte development than the other two Class I KNOX genes. To test this, a construct was designed to replace the G418 marker in the disrupted mkn2 locus of the triple mutant with a functional PpMKN2 gene and a blasticidin selectable marker (Fig. S2). Homologous recombination should lead to both the PpMKN2 coding sequence and the blasticidin resistance gene being driven by the endogenous PpMKN2 promoter, with both genes being flanked by the endogenous PpMKN2 3′ sequence. Three independent transgenic lines were selected on blasticidin and a single insertion of the expected size was confirmed in each line by DNA gel blot analysis (Fig. 2a). RT‐PCR analysis confirmed that the PpMKN2 coding sequence was expressed in each case (Fig. 2b) and a comparison between the phenotype of wild‐type (Fig. 3a), triple mutant (Fig. 3b) and triple mutant plus the MKN2pro:MKN2 transgene (Fig. 3c) lines demonstrated that sporophyte size and morphology was restored by the presence of the functional PpMKN2 gene (Fig. 3j,k), with the extent of complementation broadly correlated with transgene expression level (compare Figs 2b, 3k). Loss‐of‐function in all three P. patens Class I KNOX genes can thus be complemented by PpMKN2 alone.
Figure 2.
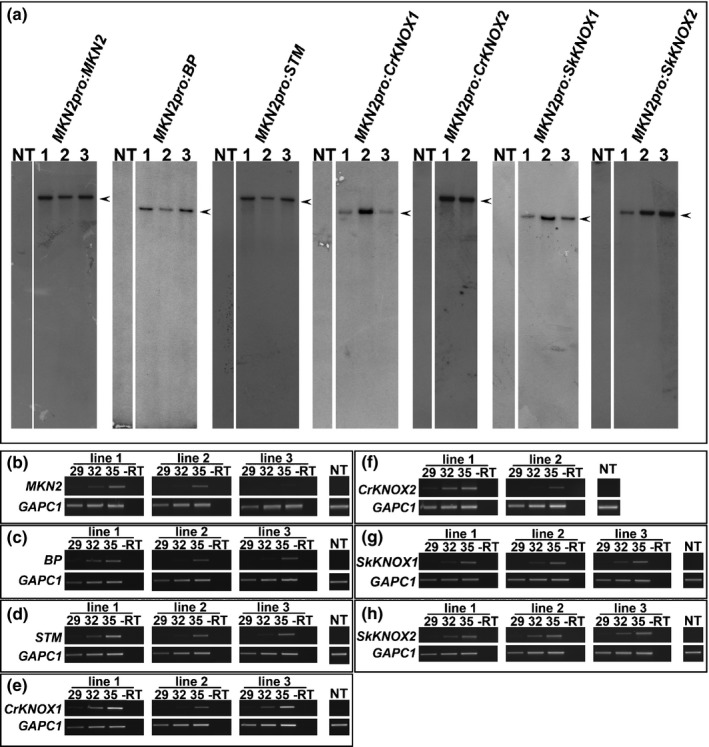
Transgene integration and expression in Physcomitrella patens mkn triple mutants. (a) DNA gel blot analysis of NdeI digested genomic DNA from mkn2;mkn4;mkn5 mutants transformed with MKN2pro:KNOX constructs. Blots were hybridized with a fragment of the blasticidin resistance gene present in the transgene construct (see Supporting Information Fig. S2). Hybridizing fragments were not detected in DNA samples from nontransformed mutants (NT), whereas 11.8‐kb (MKN2pro:MKN2), 10‐kb (MKN2pro:BP), 11.59‐kb (MKN2pro:STM), 9.9‐kb (MKN2pro:CrKNOX1), 11.78‐kb (MKN2pro:CrKNOX2), 9.42‐kb (MKN2pro:SkKNOX1) and 9.72‐kb (MKN2pro:SkKNOX2) fragments were detected in transgenic lines. (b–h) reverse transcription polymerase chain reaction (RT‐PCR) showing transgene transcript accumulation in mkn2;mkn4;mkn5 mutant plants transformed with (b) MKN2pro:MKN2, (c) MKN2pro:BP, (d) MKN2pro:STM, (e) MKN2pro:CrKNOX1, (f) MKN2pro:CrKNOX2, (g) MKN2pro:SkKNOX1 and (h) MKN2pro:SkKNOX2. GAPC1 was used as an amplification and loading control in each case. Amplifications were performed for 29, 32 and 35 cycles (as indicated) to ensure amplification in the exponential phase. Negative control reactions were carried out in the absence of reverse transcriptase (‐RT) and with cDNA isolated from nontransformed triple mutants (NT).
Figure 3.
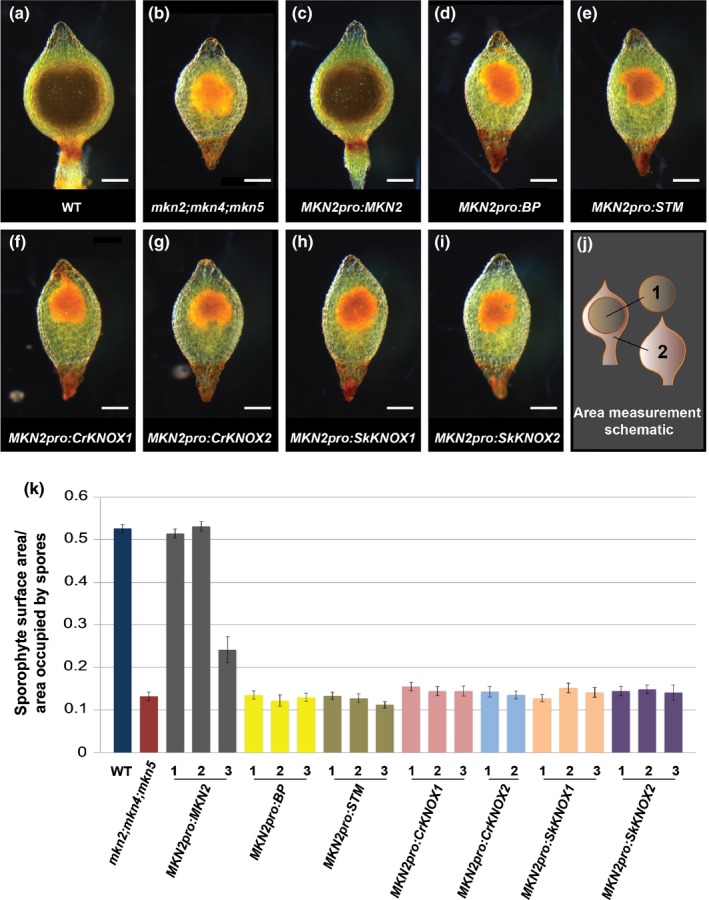
Cross‐species complementation tests with Physcomitrella patens mkn triple mutants. (a–i) Representative sporophyte phenotype of (a) wild‐type (WT), (b) mkn2;mkn4;mkn5 mutant, and (c–i) mkn2;mkn4;mkn5 mutants transformed with (c) MKN2pro:MKN2, (d) MKN2pro:BP, (e) MKN2pro:STM, (f) MKN2pro:CrKNOX1, (g) MKN2pro:CrKNOX2, (h) MKN2pro:SkKNOX1 and (i) MKN2pro:SkKNOX2 constructs. Bars, 200 μm. (j, k) Schematic depiction of measurements taken (j) and average ratios of sporophyte surface area to the area occupied by spores (k) for the lines exemplified in (a–i). Thirty sporophytes from each of three independent transformed lines were analysed for each construct, with the exception of MKN2pro:CrKNOX2 for which only two independent lines were recovered. Error bars represent ± SEM.
In order to determine whether Class I KNOX genes from vascular plant lineages can substitute for PpMKN2 function in P. patens, constructs analogous to that used for PpMKN2 were designed for homologous recombination with two of the four Class I KNOX genes in Arabidopsis (BP and SHOOTMERISTEMLESS–STM), the two genes in C. richardii (CrKNOX1 and CrKNOX2) and the two genes in S. kraussiana (SkKNOX1 and SkKNOX2) (Fig. S2b–g). As with MKN2pro:MKN2, three independent transgenic lines were selected for each construct (with the exception of MKN2pro:CrKNOX2 for which only two lines regenerated), all were confirmed by DNA gel blot analysis to have single transgene insertions of the expected size (Fig. 2a) and all showed transgene expression (Fig. 2c–h). Unlike MKN2pro:MKN2 lines, however, sporophyte phenotypes in lines expressing genes from the vascular plant lineages were the same as in the triple mutant (Fig. 3d–i). Quantification of the total sporophyte area and the area occupied by spores (Fig. 3j,k) confirmed the lack of complementation in each case. As such, Class I KNOX genes from vascular plants cannot substitute for PpMKN2 function in P. patens.
Class I KNOX genes from Physcomitrella patens and Selaginella kraussiana fully complement loss‐of‐function mutations in the Arabidopsis BP gene – but those from Ceratopteris richardii do not
Class I KNOX genes from vascular plants may be unable to complement loss‐of‐function moss mutants because inherently distinct roles have evolved in the two groups. To test this hypothesis, the ability of PpMKN2 to complement loss‐of‐function Arabidopsis mutants was determined. Ideally, both stm and bp mutants would be analysed but loss‐of‐function stm mutants exhibit severe and/or often pleiotropic shoot defects that are extremely difficult to quantify (Barton & Poethig, 1993; Endrizzi et al., 1996). bp mutants can also exhibit a range of different shoot phenotypes; however, the bp‐9 allele has a consistent mutant phenotype whereby cell division defects in the developing inflorescence lead to downward‐pointing siliques (Venglat et al., 2002) (Fig. 4a). Experiments were thus carried out with the bp‐9 allele. Quantification of silique angles demonstrated that the mutant phenotype was fully rescued by a construct comprising the full‐length BP coding sequence driven by 5.474 kb of the endogenous promoter (Figs 4c,i, S3). This observation demonstrated that the 5.474‐kb sequence is sufficient to ensure appropriate temporal and spatial regulation of BP gene expression during shoot development. The same promoter sequence was therefore used to generate constructs for transgenic expression of PpMKN2 (Fig. S3). Analysis of three lines showed that when the pBPpro:MKN2 transgene was expressed (Fig. 4k), the bp‐9 mutant phenotype was fully complemented (Fig. 4d,i). As such, PpMKN2 is not inherently different from BP in that it can activate or repress targets (either directly or indirectly) that are misexpressed in bp‐9 mutants, and in so doing restore normal cell division patterns.
Figure 4.
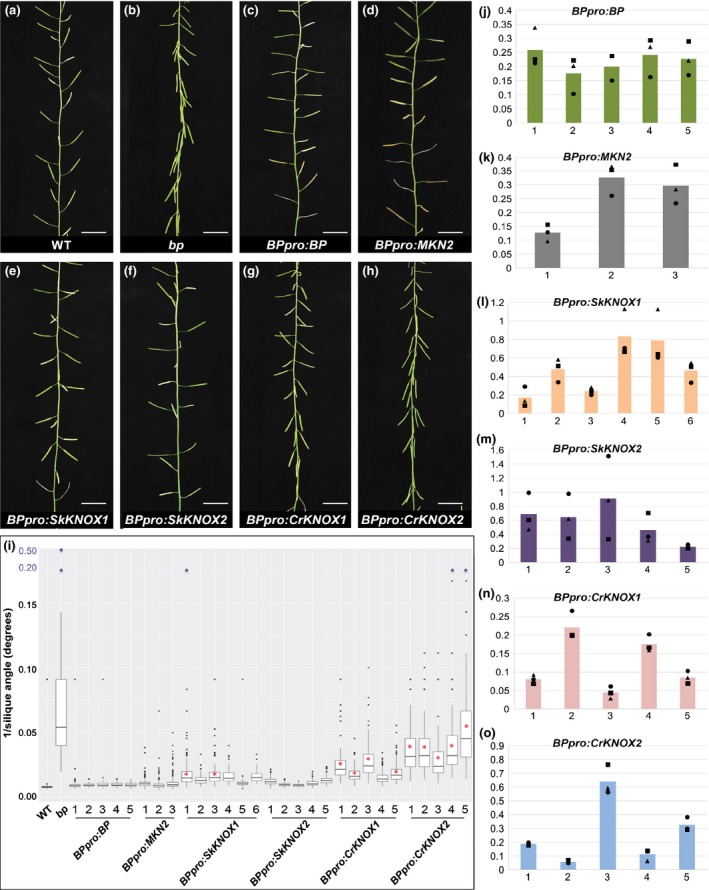
Cross‐species complementation tests with Arabidopsis bp‐9 mutants. (a–h) Representative inflorescence phenotype of (a) wild‐type (WT), (b) bp‐9 mutant, and (c–h) homozygous bp‐9 mutant plants transformed with (c) BPpro:BP, (d) BPpro:MKN2, (e) BPpro:SkKNOX1, (f) BPproSkKNOX2, (g) BPpro:CrKNOX1 and (h) BPpro:CrKNOX2 constructs. Bars, 2 cm. (i) Boxplot showing range of silique angles on the primary inflorescence of lines exemplified in (a–h). Primary inflorescences from 10 plants of at least three independent lines per transgene were analysed. Red asterisks indicate lines that are significantly different (P < 0.05) from WT. All lines are significantly different (P < 0.05) from bp‐9 mutants. Note that blue points on the y‐axis are not to scale relative to those in black. (j–o) Quantitative reverse transcription polymerase chain reaction (qRT‐PCR) analyses of (j) BPpro:BP, (k) BPpro:MKN2, (l) BPpro:SkKNOX1, (m)BPpro:SkKNOX2, (n) BPpro:CrKNOX1 and (o) BPpro:CrKNOX2 transgene expression. Numbers on the x‐axis represent independent transgenic lines as indicated in (i). The y‐axis represents transcript levels (N 0) normalized against the EF1Balpha2 and UBP6 genes. Three technical replicates were performed for three different biological samples of each independent line. Scatter blots represent values of individual biological replicates; bars represent mean of three biological replicates.
Given that PpMKN2 shares only 35% amino acid identity with BP whereas the vascular plant proteins share 43% (SkKNOX2) to 52% (CrKNOX1) identity, we predicted that the lycophyte and fern genes also would complement bp‐9 mutants. To test this prediction, transgenic bp‐9 mutant lines were generated in which expression of full‐length SkKNOX1, SkKNOX2, CrKNOX1 or CrKNOX2 coding sequence was driven from the 5.474‐kb BP promoter (Fig. S3). Analysis of at least five independent lines for each transgene demonstrated that both BPpro:SkKNOX1 and BPpro:SkKNOX2 can fully rescue the mutant phenotype (Fig. 4e,f,i). Quantitative RT‐PCR revealed that the extent of phenotypic rescue roughly correlated with the amount of transcript present in the inflorescence (compare Fig. 4i,l,m), with lines accumulating the most transcript showing a similar level of complementation to lines expressing the BPpro:BP construct. By contrast, BPpro:CrKNOX1 and BPpro:CrKNOX2 constructs showed only partial rescue of the bp‐9 mutant phenotype (Fig. 4g–i), even when relatively high levels of transgene transcripts were detected (compare Fig. 4i,n,o). These results suggest that despite the fern KNOX proteins being more similar in sequence to BP, the lycophyte proteins are better able to regulate BP targets in the context of the Arabidopsis silique development.
Distinct evolutionary trajectories for BELL proteins in lycophytes and euphyllophytes
In order to determine whether the differential ability of lycophyte and fern KNOX proteins to complement the Arabidopsis bp‐9 mutant could reflect conserved vs divergent molecular interactions, existing BELL gene phylogenies (Mukherjee et al., 2009; Furumizu et al., 2015; Sakakibara, 2016) were examined for evidence of coevolution. Unfortunately, the critical lycophyte/fern distinction could not be examined because none of the published phylogenies included fern sequences. BELL gene sequences were therefore aligned and used to infer phylogeny (see the Materials and Methods section; Methods S1; Notes S4, S6). The resultant maximum‐likelihood and Bayesian phylogenies both align well with the species tree and both resolve single clades in each bryophyte group and in the lycophytes, but two distinctive clades in the euphyllophytes (Euphyllophytes I and Euphyllophytes II) (Fig. 5). The topology of the BELL phylogeny indicates that a gene duplication event predated the divergence of the monilophytes and seed plants, and that copies of both duplicates have been retained in each lineage. There is thus a clear lycophyte/fern distinction in the BELL phylogeny.
Figure 5.
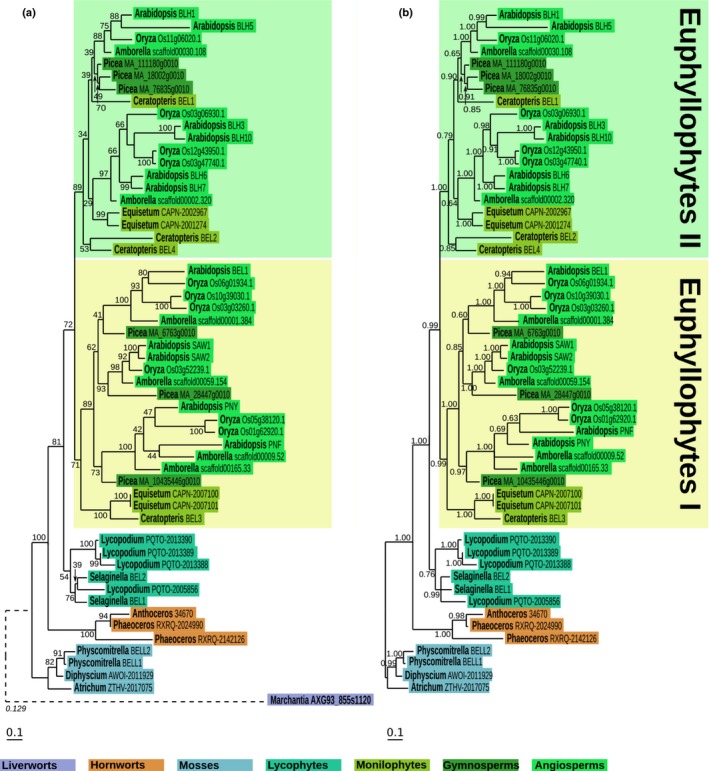
Maximum‐likelihood and Bayesian phylogenetic analysis of BELL homologues. (a, b) Phylogenetic trees inferred with (a) RA x ML or (b) MrBayes, using a protein alignment evolving under the JTT + Γ + I model. Support values (a, bootstrap values; b, posterior probabilities) are indicated next to the corresponding branch. In order to avoid destabilization of the early divergent clades, the identified BELL homologue in Marchantia polymorpha was placed using the evolutionary placement algorithm of RA x ML and the maximum‐likelihood tree as recipient. As a consequence, the M. polymorpha BELL sequence branch is dashed and the associated likelihood weight ratio (Stamatakis, 2014) is indicated in italics. Colour coded boxes correspond to the species phyla. (a) The maximum‐likelihood tree was rooted using the M. polymorpha BELL homologue and (b) the Bayesian tree was rooted using the moss clade.
Discussion
Using newly acquired genome and transcriptome datasets, supported by molecular validation of gene models, we have generated KNOTTED‐LIKE HOMEOBOX (KNOX) and BELL gene phylogenies that have refined the timing of the duplication that led to distinct Class I and Class II KNOX genes, and identified a BELL gene duplication before the divergence of monilophytes and seed plants that distinguishes bryophyte and lycophyte lineages from euphyllophytes. The integration of these phylogenetic data with the results of reciprocal cross‐species complementation experiments reveals an evolutionary trajectory whereby KNOX function becomes more specialized as progressively more gene duplications occur and lineages diverge. Notwithstanding the limitations of heterologous complementation assays, particularly where it is only feasible and/or practical to analyse the activity of single genes from multigene families, this study provides the most comprehensive functional comparison of Class I KNOX genes to date, in that it incorporates genes from all of the major land plant lineages.
Class I and Class II KNOX genes in charophytes
The discovery of Class I and Class II KNOX genes in the charophyte alga Spirogyra pratensis, plus confirmation of single KNOX gene sequences in all chlorophyte algae examined (Fig. 1) refines the timing of the first KNOX duplication event and raises a question about the ancestral function of the two gene classes. It has previously been suggested that the ability to balance activity between Class I and Class II gene function enabled the alternation of generations to evolve in land plants (Sakakibara et al., 2013; Furumizu et al., 2015). Whilst this may be correct, KNOX genes must play a different role in charophyte algae because the diploid phase of the lifecycle is invariantly restricted to the unicellular zygote. Possibly the gene duplication facilitated multicellularity in the gametophyte, a suggestion supported by the observation that transcriptomes of the unicellular charophyte Cylindrocystis contains just a Class II gene whereas both classes are present in the filamentous charophytes S. pratensis and Klebsormidium flaccidum (Figs 1, S4). The presence of the two gene classes in the K. flaccidum genome further infers that the duplication occurred before the divergence of Charales, Zygnemetales and Coleochaetales. As such, the duplication may have occurred concomitant with the chlorophyte/streptophyte divergence, but genome sequence from Clorokybus is needed to confirm or refute this suggestion because existing transcriptome datasets (Timme et al., 2012) do not contain KNOX gene sequences.
Divergent Class I KNOX genes in bryophytes
To date, KNOX gene function in bryophytes has been characterized only in the moss Physcomitrella patens, where PpMKN2 is expressed in the gametophyte, and after fertilization all three Class I genes regulate cell proliferation in the developing sporophyte (Singer & Ashton, 2007; Sakakibara et al., 2008; Frank & Scanlon, 2015a,b; Horst et al., 2016). Class II genes also are expressed in the sporophyte, where they are proposed to suppress the gametophyte developmental program (Sakakibara et al., 2013). Both Class I and Class II genes have been identified in liverworts, although at least in Marchantia polymorpha there is substantial sequence divergence in the Class I gene as evidenced by poor sequence alignment, a very long branch, and unsupported placement in the lycophyte clade (Figs 1, S1, S4). The failure to identify Class I genes in the three hornwort species examined suggests even greater sequence divergence in this group, or even gene loss. These observations are surprising given the phylogenetic position of mosses in relation to the other two bryophyte groups. Possibly the role of Class I KNOX genes in the P. patens sporophyte is a moss (or even P. patens)‐specific trait within the bryophytes. If this is the case, given that Class I KNOX genes also regulate cell proliferation in vascular plant sporophytes, this role either evolved independently in mosses and vascular plants, substantially diverged and/or was lost twice independently in liverworts and hornworts or, contrary to current phylogenetic interpretations (Ligrone et al., 2012b; Cox et al., 2014; Wickett et al., 2014), mosses are the sister group to vascular plants.
Evolution of KNOX function
Reciprocal cross‐complementation experiments have revealed that the Class I KNOX gene PpMKN2 from P. patens is able to complement the Arabidopsis bp‐9 mutant when expressed in the endogenous BP expression domain (Fig. 4), whereas BP cannot substitute for PpMKN2 function in P. patens (Figs 2, 3). As such, it can be concluded that PpMKN2 is able to activate the same downstream gene targets as BP, either on its own or as a heterodimer with Arabidopsis BELL proteins, but that BP is unable to activate PpMKN2 downstream targets. This distinction suggests that sub‐ or neofunctionalization of Class I genes in Arabidopsis has constrained the activity of BP relative to PpMKN2. Examples of both subfunctionalization (stomatal development, MacAlister & Bergmann, 2011; ABA signalling, Marella et al., 2006) and neofunctionalization (LEAFY function, Maizel et al., 2005) during the evolutionary transition from bryophytes to flowering plants have been reported previously. However, in most cases, whereas complementation of Arabidopsis mutants by homologous P. patens genes have been reported (e.g. in root hair development (Menand et al., 2007), stomatal opening (Chater et al., 2011) and photomorphogenesis (Ranjan et al., 2014)), reciprocal experiments with moss mutants were not carried out and thus no trend could be inferred. An exception to this was reported for transcription factors that regulate chloroplast development. In this case, as with Class I KNOX genes, a P. patens GOLDEN2‐LIKE gene complemented an Arabidopsis loss‐of‐function mutant (Yasumura et al., 2005), whereas an Arabidopsis gene could not rescue a P. patens mutant (Bravo‐Garcia et al., 2009).
The inability of Class I KNOX genes from any of the vascular plant lineages to complement the P. patens mkn2;mkn4;mkn5 mutant (Figs 2, 3) is striking because it suggests divergence of gene function after the bryophyte/lycophyte divergence. However, even though the lycophyte genes SkKNOX1 and SkKNOX2 cannot substitute for PpMKN2 in moss, both lycophyte genes complement the bp‐9 mutant to the same extent as PpMKN2 (Fig. 4; Notes S7). This observation indicates an equivalent ability to regulate downstream targets of BP in Arabidopsis (either directly or indirectly), which in the case of promoting appropriate cell division patterns in the silique, requires repression of the Class I genes KNAT2 and KNAT6 (Li et al., 2012). By contrast, the monilophyte fern genes CrKNOX1 and CrKNOX2 only partially complement the bp‐9 mutant phenotype (Fig. 4; Notes S7). Previous reports have suggested that PpMKN2 (Sakakibara et al., 2008), CrKNOX (Sano et al., 2005) and BP (Lincoln et al., 1994) genes function equivalently in Arabidopsis because in each case constitutive expression leads to a lobed leaf phenotype. However, these reports may have overlooked the functional distinction between PpMKN2 and CrKNOX because of the contextual difference between ectopic KNOX activity in the leaf vs loss of endogenous activity in the shoot.
Evolution of KNOX/BELL interactions
Because KNOX proteins function by dimerizing with BELL proteins in both chlorophyte algae (Lee et al., 2008) and flowering plants (reviewed in Hay & Tsiantis, 2010), changes in KNOX protein structure and function must have influenced, and been influenced by, changes in BELL protein structure and function. Despite this assumption, there is little evidence of co‐evolution in the gene phylogenies (Figs 1, 5; Furumizu et al., 2015). KNOX genes underwent a duplication event within the charophytes, and then any subsequent duplications were phylum‐ or species‐specific with structural motifs clearly conserved throughout the green lineage. In contrast to the conserved structure of KNOX proteins, BELL protein structure is highly variant in both chlorophyte and charophyte algae, with clearly recognizable domains only apparent in land plants. Previous phylogenies nested lycophyte BELL sequences within a flowering plant clade (Furumizu et al., 2015) but by increasing the taxon sampling to include ferns, our data have revealed that BELL gene duplications were phylum‐ or species‐specific before the divergence of lycophytes and euphyllophytes. As such, the multiple BELL genes in hornworts, mosses and lycophytes are all derived from a single ancestral sequence at the base of each lineage. By contrast, a gene duplication before the divergence of monilophytes and seed plants resulted in two gene families in euphyllophytes (Euphyllophytes I and II; Fig. 5). Notably the euphyllophyte duplication may have provided the potential for monilophyte and seed plant KNOX proteins to evolve more restricted interactions with BELL proteins, as compared with their lycophyte and bryophyte counterparts. As such, the inability of CrKNOX genes to fully complement bp‐9 mutants could reflect restricted capacity (as compared with BP, SkKNOX1/2 and MKN2) to interact with the Arabidopsis BELL proteins that partner BP. This suggestion may seem counterintuitive given that CrKNOX proteins share the greatest amino acid identity with BP (50%, 48%, 44%, 40% and 44% for CrKNOX1, CrKNOX2, SkKNOX1, SkKNOX2 and MKN2, respectively) in the MEINOX domain that interacts with BELL proteins. However, in silico secondary structure analyses (http://www.sbg.bio.ic.ac.uk/phyre2/; Kelley et al., 2015) predict the folding of two alpha helices in the MEINOX domain of BP, SkKNOX1, SkKNOX2 and MKN2, but predict the presence of just a single helix in CrKNOX2 and only weakly support the presence of a second helix in CrKNOX1 (Fig. S5). The suggestion that differential complementation capacity results from differential ability to form KNOX/BELL dimers needs to be confirmed experimentally, but given that the results of in vitro and heterologous interaction assays are often conflicting depending on the assay used, or have been overturned by in vivo genetic experiments (Furumizu et al., 2015), it is not a trivial hypothesis to validate and thus must remain speculation at this stage.
Author contributions
E.F. conducted Arabidopsis and moss complementation plus hornwort KNOX gene isolation; D.S‐M. conducted Spirogyra and Marchantia transcriptomes/gene isolation plus phylogenetic analysis; L.A.M. conducted BELL gene isolation; E.R. conducted plant culture and maintenance plus RT‐PCR analyses; J.A.L. conducted initial project design; and E.F., L.A.M., D.S‐M. and J.A.L. conducted data analysis and manuscript preparation.
Supporting information
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Fig. S1 Graphical representation of KNOX alignment showing KNOX conserved domains and partitioning used for inferring phylogeny.
Fig. S2 Construct design for homologous recombination of KNOX genes in the Physcomitrella patens MKN2 locus.
Fig. S3 Schematic of BPpro:KNOX constructs for bp complementation experiments.
Fig. S4 KNOX maximum‐likelihood phylogeny including Klebsormidium sequences.
Fig. S5 Predicted secondary structure of the MEINOX domain.
Methods S1 Supplementary details for methods.
Notes S1 List of primers used.
Notes S2 Verified Marchantia polymorpha, Spirogyra pratensis and Anthoceros agrestis KNOX plus Selaginella kraussiana BELL sequences.
Notes S3 Source and accessions of KNOX sequences used for phylogenetic analysis.
Notes S4 Source and accessions of BELL sequences used for phylogenetic analysis.
Notes S5 Edited KNOX alignment.
Notes S6 Edited BELL alignment.
Notes S7 Silique angle measurements and results of Tukey's HSD tests.
Acknowledgements
Thanks to Jill Harrison, Stefanie Mueller, Chris Della Vedova, Yuki Yasumura and Georgios Lagiotis for assistance with initial cloning steps, to Angela Hay for providing advice on the BP promoter sequence and to Michael Niklaus for guidance on R codes. We are grateful to the following for sharing sequence data: Sandy Hetherington, Clement Champion & Liam Dolan (M. polymorpha), Peter Szovenyi (A. agrestis) and the OneKP consortium (www.onekp.com); and to Mitsuyasu Hasebe for providing the P. patens triple mkn mutant and the p35S‐loxP‐BSD (AB537973) plasmid. As always we thank John Baker for photography, Julie Bull for technical assistance and Steve Kelly for bioinformatics advice. This work was funded by ERC AdG (EDIP) and BBSRC (BB/M020517/1) grants to J.A.L., and by a JSPS Fellowship to E.F.
References
- Barton MK. 2010. Twenty years on: the inner workings of the shoot apical meristem, a developmental dynamo. Developmental Biology 341: 95–113. [DOI] [PubMed] [Google Scholar]
- Barton MK, Poethig RS. 1993. Formation of the shoot apical meristem in Arabidopsis thaliana: an analysis of development in the wild type and in the shoot meristemless mutant. Development 119: 823–831. [Google Scholar]
- Belles‐Boix E, Hamant O, Witiak SM, Morin H, Traas J, Pautot V. 2006. KNAT6: an Arabidopsis homeobox gene involved in meristem activity and organ separation. Plant Cell 18: 1900–1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharathan G, Janssen B‐J, Kellogg EA, Sinha N. 1997. Did homeodomain proteins duplicate before the origin of angiosperms, fungi and metazoa. Proceedings of the National Academy of Sciences, USA 94: 13749–13753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravo‐Garcia A, Yasumura Y, Langdale JA. 2009. Specialization of the Golden2‐like regulatory pathway during land plant evolution. New Phytologist 183: 133–141. [DOI] [PubMed] [Google Scholar]
- Burglin T. 1997. Analysis of TALE superclass homeobox genes (MEIS, PBC, KNOX, Iroquois, TGIF) reveals a novel domain conserved between plants and animals. Nucleic Acids Research 25: 4173–4180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chater C, Kamisugi Y, Movahedi M, Fleming A, Cuming AC, Gray JE, Beerling DJ. 2011. Regulatory mechanism controlling stomatal behavior conserved across 400 million years of land plant evolution. Current Biology 21: 1025–1029. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. 1998. Floral dip: a simplified method for Agrobacterium‐mediated transformation of Arabidopsis thaliana . Plant Journal 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Collingridge PW, Kelly S. 2012. MergeAlign: improving multipe sequence alignment performance by dynamic reconstruction of consensus multiple sequence alignments. BMC Bioinformatics 13: 117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox CJ, Li B, Foster PG, Embley TM, Civan P. 2014. Conflicting phylogenies for early land plants are caused by composition biases among synonymous substitutions. Systematic Biology 63: 272–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derelle R, Lopez P, Le Guyader H, Manuel M. 2007. Homeodomain proteins belong to the ancestral molecular toolkit of eukaryotes. Evolution and Development 9: 212–219. [DOI] [PubMed] [Google Scholar]
- Endrizzi K, Moussian B, Haecker A, Levin JZ, Laux T. 1996. The shoot meristemless gene is required for maintenance of undifferentiated cells in Arabidopsis shoot and floral mersitems and acts at a different regulatory level than the meristem genes WUSCHEL and ZWILLE . Plant Journal 10: 967–979. [DOI] [PubMed] [Google Scholar]
- Engel PP. 1968. The induction of biochemical and morphological mutants in the moss Physcomitrella patens . American Journal of Botany 55: 438–446. [Google Scholar]
- Frank MH, Scanlon MJ. 2015a. Transcriptomic evidence for the evolution of shoot meristem function in sporophyte‐dominant land plants through concerted selection of ancestral gametophytic and sporophytic genetic programs. Molecular Biology and Evolution 32: 355–367. [DOI] [PubMed] [Google Scholar]
- Frank MH, Scanlon MJ. 2015b. Cell‐specific transcriptomic analyses of three‐dimensional shoot development in the moss Physcomitrella patens . Plant Journal 83: 743–751. [DOI] [PubMed] [Google Scholar]
- Furumizu C, Alvarez JP, Sakakibara K, Bowman JL. 2015. Antagonistic roles for KNOX1 and KNOX2 genes in patterning the land plant body plan following an ancient gene duplication. PLoS Genetics 11: e1004980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleave AP. 1992. A versatile binary vector system with a T‐DNA organisational structure conducive to efficient integration of cloned DNA into the plant genome. Plant Molecular Biology 20: 1203–1207. [DOI] [PubMed] [Google Scholar]
- Harrison CJ, Corley SB, Moylan EC, Alexander DL, Scotland RW, Langdale JA. 2005. Independent recruitment of a conserved developmental mechanism during leaf evolution. Nature 434: 509–514. [DOI] [PubMed] [Google Scholar]
- Harrison CJ, Langdale JA. 2010. The developmental pattern of shoot apices in Selaginella kraussiana (Kunze) A. Braun. International Journal of Plant Sciences 171: 690–692. [Google Scholar]
- Harrison CJ, Rezvani M, Langdale JA. 2007. Growth from two transient apical initials in the meristem of Selaginella kraussiana . Development 134: 881–889. [DOI] [PubMed] [Google Scholar]
- Hay A, Kaur H, Phillips A, Hedden P, Hake S, Tsiantis M. 2002. The gibberellin pathway mediates KNOTTED1‐type homeobox function in plants with different body plans. Current Biology 12: 1557–1565. [DOI] [PubMed] [Google Scholar]
- Hay A, Tsiantis M. 2010. KNOX genes: versatile regulators of plant development and diversity. Development 137: 3153–3165. [DOI] [PubMed] [Google Scholar]
- Holland PWH. 2013. Evolution of homeobox genes. Wiley Interdisciplinary Reviews: Developmental Biology 2: 31–45. [DOI] [PubMed] [Google Scholar]
- Hori K, Maruyama F, Fujisawa T, Togashi T, Yamamoto N, Seo M, Sato S, Yamada T, Mori H, Tajima N et al 2014. Klebsormidium flaccidum genome reveals primary factors for plant terrestrial adaptation. Nature Communications 5: 3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horst NA, Katz A, Pereman I, Decker EL, Ohad N, Reski R. 2016. A single homeobox gene triggers phase transition, embryogenesis and asexual reproduction. Nature Plants 2: 15 209. [DOI] [PubMed] [Google Scholar]
- Jasinski S, Piazza P, Craft J, Hay A, Woolley L, Rieu I, Phillips A, Hedden P, Tsiantis M. 2005. KNOX action in Arabidopsis is mediated by coordinate regulation of cytokinin and gibberellin activities. Current Biology 15: 1560–1565. [DOI] [PubMed] [Google Scholar]
- Johnson MT, Carpenter EJ, Tian Z, Bruskiewich R, Burris JN, Carrigan CT, Chase MW, Clarke ND, Covshoff S, Depamphilis CW et al 2012. Evaluating methods for isolating total RNA and predicting the success of sequencing phylogenetically diverse plant transcriptomes. PLoS ONE 7: e50226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones CS, Drinnan AN. 2009. The developmental pattern of shoot apices in Selaginella kraussiana (Kunze) A. Braun. International Journal of Plant Sciences 170: 1009–1018. [Google Scholar]
- Kawai J, Tanabe Y, Soma S, Ito M. 2010. Class 1 KNOX gene expression supports the Selaginella rhizophore concept. Journal of Plant Biology 53: 268–274. [Google Scholar]
- Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJE. 2015. The Phyre2 web portal for protein modeling, prediction and analysis. Nature Protocols 10: 845–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerstetter R, Volbrecht E, Lowe B, Veit B, Yamaguchi J, Hake S. 1994. Sequence analysis and expression patterns divide the maize knotted1‐like homebox genes into two classes. Plant Cell 6: 1877–1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langdale JA, Rothermel BA, Nelson T. 1988. Cellular patterns of photosynthetic gene expression in developing maize leaves. Genes & Development 2: 106–115. [DOI] [PubMed] [Google Scholar]
- Lee JH, Lin H, Joo S, Goodenough U. 2008. Early sexual origins of homeoprotein heterodimerization and evolution of the plant KNOX/BELL family. Cell 133: 829–840. [DOI] [PubMed] [Google Scholar]
- Li FW, Villarreal JC, Kelly S, Rothfels CJ, Melkonian M, Frangedakis E, Ruhsam M, Sigel EM, Der JP, Pittermann J et al 2014. Horizontal transfer of an adaptive chimeric photoreceptor from bryophytes to ferns. Proceedings of the National Academy of Sciences, USA 111: 6672–6677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Pi L, Huang H, Xu L. 2012. ATH1 and KNAT2 proteins act together in regulation of plant inflorescence architecture. Journal of Experimental Botany 63: 1423–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ligrone R, Duckett JG, Renzaglia KS. 2012a. The origin of the sporophyte shoot in land plants: a bryological perspective. Annals of Botany 110: 935–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ligrone R, Duckett JG, Renzaglia KS. 2012b. Major transitions in the evolution of early land plants: a bryological perspective. Annals of Botany 109: 851–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lincoln C, Long J, Yamaguchi J, Serikawa K, Hake S. 1994. A knotted1‐like homeobox gene in Arabidopsis is expressed in the vegetative meristem and dramatically alters leaf morphology when overexpressed in transgenic plants. Plant Cell 6: 1859–1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long JA, Moan EI, Medford JI, Barton MK. 1996. A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature 379: 66–69. [DOI] [PubMed] [Google Scholar]
- MacAlister CA, Bergmann DC. 2011. Sequence and function of basic helix‐loop‐helix proteins required for stomatal development in Arabidopsis are deeply conserved in land plants. Evolution & Development 13: 182–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maizel A, Busch MA, Tanahashi T, Perkovic J, Kato M, Hasebe M, Weigel D. 2005. The floral regulator LEAFY evolves by substitutions in the DNA binding domain. Science 308: 260–263. [DOI] [PubMed] [Google Scholar]
- Marella HH, Sakata Y, Quatrano RS. 2006. Characterization and functional analysis of ABSCISIC ACID INSENSITIVE3‐like genes from Physcomitrella patens . Plant Journal 46: 1032–1044. [DOI] [PubMed] [Google Scholar]
- Matasci N, Hung L‐H, Yan Z, Carpenter EJ, Wickett NJ, Mirarab S, Nguyen N, Warnow T, Ayyampalayam S, Barker M et al 2014. Data access for the 1,000 Plants (1KP) project. GigaScience 3: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menand B, Yi K, Jouannic S, Hoffmann L, Ryan E, Linstead P, Schaefer DG, Dolan L. 2007. An ancient mechanism controls the development of cells with a rooting function in land plants. Science 316: 1477–1480. [DOI] [PubMed] [Google Scholar]
- Mukherjee K, Brocchieri L, Bürglin TR. 2009. A comprehensive classification and evolutionary analysis of plant homeobox genes. Molecular Biology and Evolution 26: 2775–2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama T, Hiwatashi Y, Sakakibara I, Kato M, Hasebe M. 2000. Tagged mutagenesis and gene‐trap in the moss, Physcomitrella patens by shuttle mutagenesis. DNA Research 7: 9–17. [DOI] [PubMed] [Google Scholar]
- Ramakers C, Ruijter JM, Deprez RHL, Moorman AF. 2003. Assumption‐free analysis of quantitative real‐time polymerase chain reaction (PCR) data. Neuroscience Letters 339: 62–66. [DOI] [PubMed] [Google Scholar]
- Ranjan A, Dickopf S, Ullrich KK, Rensing SA, Hoecker U. 2014. Functional analysis of COP1 and SPA orthologs from Physcomitrella and rice during photomorphogenesis of transgenic Arabidopsis reveals distinct evolutionary conservation. BMC Plant Biology 14: 178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574. [DOI] [PubMed] [Google Scholar]
- Sakakibara K. 2016. Technological innovations give rise to a new era of plant evolutionary developmental biology In: Rensing SA, ed. Advances in botanical research volume 78 ‘Genomes and evolution of charophytes, bryophytes, lycophytes and ferns’. London, UK: Academic Press, 3–35. [Google Scholar]
- Sakakibara K, Ando S, Yip HK, Tamada Y, Hiwatashi Y, Murata T, Deguchi H, Hasebe M, Bowman JL. 2013. KNOX2 genes regulate the haploid‐to‐diploid morphological transition in land plants. Science 339: 1067–1070. [DOI] [PubMed] [Google Scholar]
- Sakakibara K, Nishiyama T, Deguchi H, Hasebe M. 2008. Class 1 KNOX genes are not involved in shoot development in the moss Physcomitrella patens but do function in sporophyte development. Evolution and Development 10: 555–566. [DOI] [PubMed] [Google Scholar]
- Sakamoto T, Kamiya N, Ueguchi‐Tanaka M, Iwahori S, Matsuoka M. 2001. KNOX homeodomain protein directly suppresses the expression of a gibberellin biosynthetic gene in the tobacco shoot apical meristem. Genes & Development 15: 581–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto T, Sakakibara H, Kojima M, Yamamoto Y, Nagasaki H, Inukai Y, Sato Y, Matsuoka M. 2006. Ectopic expression of KNOTTED1‐like homeobox protein induces expression of cytokinin biosynthesis genes in rice. Plant Physiology 142: 54–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders HL, Darrah PR, Langdale JA. 2011. Sector analysis and predictive modelling reveal iterative shoot‐like development in fern fronds. Development 138: 2925–2934. [DOI] [PubMed] [Google Scholar]
- Sano R, Juarez C, Hass B, Sakakibara K, Ito M, Banks J, Hasebe M. 2005. KNOX homeobox genes potentially have similar function in both diploid unicellular and multicellular meristems, but not in haploid meristems. Evolution and Development 7: 69–78. [DOI] [PubMed] [Google Scholar]
- Schaefer D, Zryd JP, Knight CD, Cove DJ. 1991. Stable transformation of the moss Physcomitrella patens . Molecular and General Genetics 226: 418–424. [DOI] [PubMed] [Google Scholar]
- Serikawa KA, Mandoli DF. 1999. Aaknox1, a kn1‐like homeobox gene in Acetabularia acetabulum, undergoes developmentally regulated subcellular localization. Plant Molecular Biology 41: 785–793. [DOI] [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J et al 2011. Fast, scalable generation of high‐quality protein multiple sequence alignments using Clustal Omega. Molecular Systems Biology 7: 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer SD, Ashton NW. 2007. Revelation of ancestral roles of KNOX genes by a functional analysis of Physcomitrella homologues. Plant Cell Reports 26: 2039–2054. [DOI] [PubMed] [Google Scholar]
- Smith LG, Greene B, Veit B, Hake S. 1992. A dominant mutation in the maize homeobox gene, Knotted‐1, causes its ectopic expression in leaf cells with altered fates. Development 116: 21–30. [DOI] [PubMed] [Google Scholar]
- Smith HM, Hake S. 2003. The interaction of two homeobox genes, BREVIPEDICELLUS and PENNYWISE, regulates internode patterning in the Arabidopsis inflorescence. Plant Cell 15: 1717–1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post‐analysis of large phylogenies. Bioinformatics 30: 1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steeves TA, Sussex IM. 1989. Patterns in plant development. Cambridge, UK: Cambridge University Press. [Google Scholar]
- Sussex IM, Kerk NM. 2001. The evolution of plant architecture. Current Opinion Plant Biology 4: 33–37. [DOI] [PubMed] [Google Scholar]
- Szövényi P, Frangedakis E, Ricca M, Quandt D, Wicke S, Langdale JA. 2015. Establishment of Anthoceros agrestis as a model species for studying the biology of hornworts. BMC Plant Biology 15: 98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timme RE, Bachvaroff TR, Delwiche CF. 2012. Broad phylogenomic sampling and the sister lineage of land plants. PLoS ONE 7: e29696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasco A, Moran RC, Ambrose BA. 2013. The evolution, morphology, and development of fern leaves. Frontiers in Plant Science 4: 345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venglat SP, Dumonceaux T, Rozwadowski K, Parnell L, Babic V, Keller W, Martienssen R, Selvaraj G, Datla R. 2002. The homeobox gene BREVIPEDICELLUS is a key regulator of inflorescence architecture in Arabidopsis. Proceedings of the National Academy of Sciences, USA 99: 4730–4735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HY, Klatte M, Jakoby M, Bäumlein H, Weisshaar B, Bauer P. 2007. Iron deficiency‐mediated stress regulation of four subgroup Ib BHLH genes in Arabidopsis thaliana . Planta 226: 897–908. [DOI] [PubMed] [Google Scholar]
- Wickett NJ, Mirarab S, Nguyen N, Warnow T, Carpenter E, Matasci N, Ayyampalayam S, Barker MS, Burleigh JG, Gitzendanner MA et al 2014. Phylotranscriptomic analysis of the origin and early diversification of land plants. Proceedings of the National Academy of Sciences, USA 111: E4859–E4868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y, Wu G, Tang J, Luo R, Patterson J, Liu S, Huang W, He G, Gu S, Li S et al 2014. SOAPdenovo‐Trans: de novo transcriptome assembly with short RNA‐Seq reads. Bioinformatics 30: 1660–1666. [DOI] [PubMed] [Google Scholar]
- Yasumura Y, Moylan EC, Langdale JA. 2005. A conserved transcription factor mediates nuclear control of organelle biogenesis in anciently diverged land plants. Plant Cell 17: 1894–1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Fig. S1 Graphical representation of KNOX alignment showing KNOX conserved domains and partitioning used for inferring phylogeny.
Fig. S2 Construct design for homologous recombination of KNOX genes in the Physcomitrella patens MKN2 locus.
Fig. S3 Schematic of BPpro:KNOX constructs for bp complementation experiments.
Fig. S4 KNOX maximum‐likelihood phylogeny including Klebsormidium sequences.
Fig. S5 Predicted secondary structure of the MEINOX domain.
Methods S1 Supplementary details for methods.
Notes S1 List of primers used.
Notes S2 Verified Marchantia polymorpha, Spirogyra pratensis and Anthoceros agrestis KNOX plus Selaginella kraussiana BELL sequences.
Notes S3 Source and accessions of KNOX sequences used for phylogenetic analysis.
Notes S4 Source and accessions of BELL sequences used for phylogenetic analysis.
Notes S5 Edited KNOX alignment.
Notes S6 Edited BELL alignment.
Notes S7 Silique angle measurements and results of Tukey's HSD tests.


