Abstract
Fifty years ago, Lynn Margulis, inspiring in early twentieth-century ideas that put forward a symbiotic origin for some eukaryotic organelles, proposed a unified theory for the origin of the eukaryotic cell based on symbiosis as evolutionary mechanism. Margulis was profoundly aware of the importance of symbiosis in the natural microbial world and anticipated the evolutionary significance that integrated cooperative interactions might have as mechanism to increase cellular complexity. Today, we have started fully appreciating the vast extent of microbial diversity and the importance of syntrophic metabolic cooperation in natural ecosystems, especially in sediments and microbial mats. Also, not only the symbiogenetic origin of mitochondria and chloroplasts has been clearly demonstrated, but improvement in phylogenomic methods combined with recent discoveries of archaeal lineages more closely related to eukaryotes further support the symbiogenetic origin of the eukaryotic cell. Margulis left us in legacy the idea of ‘eukaryogenesis by symbiogenesis’. Although this has been largely verified, when, where, and specifically how eukaryotic cells evolved are yet unclear. Here, we shortly review current knowledge about symbiotic interactions in the microbial world and their evolutionary impact, the status of eukaryogenetic models and the current challenges and perspectives ahead to reconstruct the evolutionary path to eukaryotes.
Keywords: eukaryogenesis, symbiosis, syntrophy, archaea, mitochondria, eukaryotic origins
1. Introduction
In 1967, Lynn Margulis (Sagan) published her famous On the origin of mitosing cells (Sagan, 1967), where she set up the basis of her Serial Endosymbiotic Theory for the origin of the eukaryotic cell (Margulis, 1981; Margulis, 1996). In her manuscript, Margulis revived previous ideas proposing the endosymbiotic origin of chloroplasts from 'blue-green algae' (cyanobacteria) (Mereschkowsky, 1905; Mereschkowsky, 1910) and mitochondria from purple bacteria (alphaproteobacteria) (Wallin, 1927); and further put forward the idea that the eukaryotic flagellum derived from symbiotic bacteria (spirochete-like). Although Konstantin Mereschkowsky had also advocated for the endosymbiotic origin of the eukaryotic nucleus (Mereschkowsky, 1910), Margulis discarded this possibility without nonetheless proposing a clear mechanism for the origin of the nucleus in an original heterotrophic amoeboid host (Sagan, 1967). Later, after the recognition of archaea as a third phylogenetic domain of life (Woese and Fox, 1977; Woese et al., 1990) showing significant similarities in terms of informational processes (replication, transcription, translation) with eukaryotes (Rivera et al., 1998; Thiergart et al., 2012), she postulated that the host of the future mitochondrion derived from a thermoacidophilic wall-less archaeon similar to contemporary Thermoplasma spp. (Euryarchaeota) (Margulis, 1981; Margulis, 1996), an idea that she adopted from Dennis Searcy (Searcy, 1992). The nucleus would have evolved autogenously (not by symbiosis) in a chimeric archaeal-bacterial cell (Margulis et al., 2000).
The strength and innovative character of Margulis' ideas lay in the proposal of a unified theory based on symbiosis as evolutionary mechanism for the origin of the eukaryotic cell: eukaryogenesis by symbiogenesis. Strongly criticized at the time, at least part of these ideas became credible with the advent of molecular phylogeny that, among its earliest achievements, allowed demonstrating that conserved genes in chloroplast and mitochondrial genomes clustered with, respectively, cyanobacterial and alphaproteobacterial genes in phylogenetic trees (Schwartz and Dayhoff, 1978). The endosymbiotic origin of these membrane-bound organelles became mainstream science and it is now well established how their bacterial ancestors evolved, undergoing a process of genome reduction, impacting their host and leading to a variety of organelle derivatives (Dyall et al., 2004; Gray, 2012; Muller et al., 2012; Schwartz and Dayhoff, 1978). However, the origin of her eukaryotic 'nucleocytoplasm' was much controversial; neither the Thermoplasma-like nature of the host nor (far less) the symbiotic origin of the flagellum were ever considered seriously. In her view, a metabolic symbiosis based on interspecific sulfur transfer was established between a Thermoplasma-like archaeon, which generated hydrogen sulfide, and a spirochete, which oxidized sulfide to sulfur and, at the same time, provided motility to the symbiotic consortium (Margulis et al., 2000), being at the origin of the eukaryotic cytoskeleton and the mitotic apparatus (Margulis et al., 2006; Sagan, 1967). Margulis argued that, similarly to mitochondria and chloroplasts, which retained reduced genomes, a remnant genome should be found in connection with the (9+2) microtubular basal bodies associated to eukaryotic flagella (Sagan, 1967). But such remnant genomes were never found and phylogenomic analyses in the genomics era did not show any particular similarity between eukaryote and spirochete genomes. Thus, while the endosymbiotic origin of chloroplasts and mitochondria in a eukaryotic host became widely accepted, the symbiotic origin of the eukaryotic cell itself was not and Margulis' ideas stood alone for almost three decades before other symbiogenetic models made it through to the scientific arena.
Lynn Margulis had a classical, exceptional knowledge on the morphology, biology and ecology of microorganisms, which led her to actively participate in attempts to establish global classification systems for microorganisms. She sustained that life should be classified in two superkingdoms or domains (prokaryotes and eukaryotes) and five kingdoms (Margulis, 1992; Margulis and Schwartz, 1998; Whittaker and Margulis, 1978), and co-edited the famous Handbook of Protoctista (Margulis et al., 1990) widely used by eukaryotic microbiologists. Paradoxically, although she recognized the importance of DNA comparisons in establishing a natural (evolutionary) classification system for all organisms (Sagan, 1967), she was reluctant to adopt (Margulis and Guerrero, 1991) the Carl R. Woese's three-domain classification system that resulted from molecular phylogenetic analyses of universally conserved genes (Woese and Fox, 1977; Woese et al., 1990). The molecular revolution initiated by Woese using 16S/18S rRNA genes had three major outcomes that have subsequently impacted many areas in biology, from taxonomy and systematics to microbial ecology and comparative genomics. He showed that it was possible to build universal phylogenetic trees for all cellular life and hence establish a natural biological classification system. In doing so, he incidentally discovered that a group of prokaryotic cells, the archaea, defined a distinct phylogenetic group (a third domain of life) (Woese and Fox, 1977; Woese et al., 1990). And finally, as corollary of his work, it was in principle possible to explore microbial diversity in natural environments by amplifying and sequencing conserved marker genes, thus sidestepping the well-known problem of culture bias (only a tiny fraction of microorganisms are amenable to culture in the laboratory). The amplification, cloning and sequencing of 16S and 18S rRNA genes from many different environments led in the 1990s and early 2000s to the realization that the diversity of both prokaryotic and eukaryotic microorganisms was much more important than ever thought (Moreira and López-García, 2002; Pace, 1997). Today, larger-scale metabarcoding analyses based on amplicon high-throughput sequencing, together with single cell genomics and metagenome-based genome reconstruction not only reinforce the view of an extraordinarily important microbial diversity but at the same time validate the three domains of life (Archaea, Bacteria, Eucarya) that Woese identified forty years ago (Eme and Doolittle, 2015; Hug et al., 2016; Rinke et al., 2013; Yarza et al., 2014).
This possibility to study novel, uncultured organisms in the wild has revealed crucial to advance in the resolution of the long-term query of eukaryotic origins. Notwithstanding Margulis symbiogenetic ideas, the eukaryogenetic model that prevailed in the last part of the twentieth century and until recently was that of an autogenous origin of all typical eukaryotic features but mitochondria and chloroplasts (of demonstrated endosymbiotic origin) in a proto-eukaryotic lineage sister to archaea (de Duve, 2007; Jekely, 2003; Poole and Penny, 2007). The discovery that amitochondriate protists lost (or modified) mitochondria secondarily, implying that the last eukaryotic common ancestor (LECA) possessed mitochondria (Embley, 2006; Embley and Hirt, 1998), left some room for the proposal of new symbiogenetic models for the origin of eukaryotes (for review, see (Embley and Martin, 2006; Keeling, 2014; Lopez-Garcia and Moreira, 2015; McInerney et al., 2014). These models denied the existence of a third, proto-eukaryotic lineage different from classical bacteria and archaea and explained the mixed eukaryotic heritage (archaeal informational genes, bacterial operational genes) by archaea-bacteria merging. Nonetheless, these models remained little accepted, in part by the failure of phylogenomic analyses to identify clear prokaryotic ancestors (other than alphaproteobacteria) of eukaryotes (Gribaldo et al., 2010). However, the recent discovery of Lokiarchaeota (Spang et al., 2015) and other related lineages (collectively, Asgard archaea) (Zaremba-Niedzwiedzka et al., 2017), grouping anaerobic archaea that share more and more similar genes with eukaryotes than the rest of known archaea strongly suggest that the archaeal component of eukaryotes did indeed derive from within archaea, giving fresh credit to symbiogenetic models (Koonin, 2015; Lopez-Garcia and Moreira, 2015; Williams and Embley, 2015).
The seminal On the origin of mitosing cells (Sagan, 1967) did not only stirred evolutionary thinking at the time but today, fifty years later, it is still striking in its modernity and, in many ways, visionary content about the role of symbiosis in eukaryotic evolution. Here, we revisit some of the ideas put forward by Margulis that have made it through time and briefly discuss current knowledge and challenges about eukaryogenesis.
2. Symbiosis in natural ecosystems
One of the most important, yet little appreciated, strengths of Margulis' ideas was her awareness, at a time when molecular biology and reductionist approaches were on the rise, of the intricate, often mutualistic interactions that prevail among microorganisms in natural environments, which she understood as key in fostering evolutionary processes. This allowed her to conceive holistic scenarios about the evolution of life on Earth based in the distribution of energy and carbon metabolism in prokaryotes, the fossil record and her recurrent observation of protist-prokaryote symbioses under the microscope (Margulis et al., 1986; Sagan, 1967). Several decades later, molecular tools for the study of natural microbial communities have nothing but proved her intuition about the importance of mutualistic interactions in ecology and evolution right.
2.1. Symbiosis, cooperation, evolution
The role of cooperation in evolution is being increasingly recognized, particularly in the microbial world. Several recent reviews analyze cooperation in microbes and point to the factors that lead to stable cooperation over time (e.g. (Celiker and Gore, 2013; Mitri and Foster, 2013; Nadell et al., 2016). Some of these principles have been tested experimentally in biofilm populations (Steenackers et al., 2016). Part of this interest has grown exponentially due to studies on human (and other animal) microbiomes and the effect that, more specifically, gut microbial communities have in human metabolism and the development of the immune system (Sonnenburg and Backhed, 2016; Thaiss et al., 2016). Likewise, although the importance of the microorganisms from the rhizosphere has been recognized for a long time (Philippot et al., 2013), interest in microbial endophytes in plants is expanding (Hardoim et al., 2015). This new interest has even led to the qualification, by some authors, of these collective microbial communities (microbiomes) as 'symbionts' and humans, other animals (e.g. corals, insects) or plants as 'holobionts'. Strictly speaking, these terms are stretched inappropriately because the specific interactions amongst all the components of those communities and their multicellular hosts do not necessarily correspond to co-evolutionary mutualistic symbioses but are likely to be largely governed by classical microbial ecology determinants (Moran and Sloan, 2015; Mushegian and Ebert, 2016).
But what is exactly symbiosis? 'Living together' by etymology, symbiosis has two major definitions in biology. One is looser and refers to the co-existence and dependency of at least one organism upon a partner. Symbiosis would then accommodate three types of interactions depending on the effect on the host's fitness: mutualism (fitness increase), commensalism (no fitness change), and parasitism (fitness decrease) (Mushegian and Ebert, 2016). In reality, the line between mutualism and parasitism is thin and these kinds of interactions are often seen as a continuum, sometimes depending on the environmental conditions. Examples of parasite-mutualist transitions are well-documented in plant mycorrhiza (Paszkowski, 2006), Wolbachia-insect interactions (Hosokawa et al., 2010) and even in intrinsic genetic parasites such as plasmids or viruses (Bao and Roossinck, 2013). The other type of definition equals symbiosis and mutualism, so that symbiosis implies a mutually beneficial interaction. This is the type of definition that Margulis used when she referred to symbiosis. In her view, the holobiont properly meant an integrated symbiotic consortium (obligate mutualism) that behaves as a unit of evolution (Guerrero et al., 2013). Eukaryogenesis could only be understood in this context.
We are progressively discovering that mutualistic symbioses (hereafter, symbioses) are plentiful in nature and, in particular environments such as sediments or microbial mats with steep redox gradients, they may be the rule. Some examples follow.
2.2. Eukaryote – prokaryote symbioses
This type of symbioses is often easy to identify since, in many cases, it implies endosymbiosis within a larger host and, in the case of multicellular organisms, the frequent evolution of dedicated structures, such as bacteriocytes in aphids, trophosomes in deep-sea polychaetes or root nodules in plants. Many of these symbioses are the result of long co-evolutionary processes that have shaped the genotypes and phenotypes of the different partners involved.
Prokaryotic symbioses with animals have been widely documented (Moya et al., 2008). They have been particularly well studied in insects, where they can have astounding impacts not only in nutritional but also in reproductive aspects of their biology. Thus, vertically-transmitted alphaproteobacterial endosymbionts of the genus Wolbachia impact sex determination and can induce feminization, parthenogenesis, male killing and sperm-egg incompatibility (Cordaux et al., 2011; Werren et al., 2008). Comparative genomic analyses are starting to reveal the underlying basis for these effects, which is sometimes linked to horizontal gene transfer from the bacterium to the host genome. For instance, a 3-Mb insert of a feminizing Wolbachia genome was recently transferred into the pillbug nuclear genome and its occurrence correlates with the female sex (Leclercq et al., 2016). Also well-studied symbioses are those involving aphids and other phloem-sap feeding insects with different bacteria that complement their imbalanced, sugar-rich diet by supplying amino acids. A famous symbiotic couple is that of aphids with Buchnera aphidicola, an endosymbiotic gammaproteobacterium living in specialized aphid cells (bacteriocytes). These bacterial endosymbionts, being vertically inherited and having low population sizes, are prone to genetic drift. Their genes are fast-evolving and their genomes, AT-biased and progressively reduced (McCutcheon and Moran, 2012; Moran and Bennett, 2014). Genome reduction can lead to the loss of essential genes for amino-acid biosynthesis. When this happens, a second, less reduced endosymbiont that complements the lost biosynthetic pathways can be acquired and maintained in tripartite symbiosis (Perez-Brocal et al., 2006) or it can be simply lost and replaced by a new symbiont (Koga and Moran, 2014). The replacement of a reduced-genome symbiont is not infrequent and can be achieved experimentally (Moran and Yun, 2015).
Less well-known, but equally important are symbioses of bacteria and marine animals. The microbial communities that inhabit sponges, making up to one third of their biomass, are specific to their hosts and many of their bacterial and archaeal members may be true symbionts (Hentschel et al., 2012). Members of the phylum Poribacteria are specifically associated with sponges and seem to be able to fix carbon and contain putative symbiotic factors including adhesins and other proteins potentially mediating host-microbe interactions (Siegl et al., 2011). Similarly, microbial communities in corals are highly diverse and, although several members may be true mutualists (Hernandez-Agreda et al., 2016), the clear established case is that of euphotic-zone corals and dinoflagellate algae of the genus Symbiodinium (Roth, 2014), which also occur in some bivalves and anemones. Climate-change induced ocean acidification and warming appear to correlate with symbiont loss, leading to coral bleaching and death, and seriously affecting coral-reef ecosystems. The counterpart of these phototrophic symbioses, whereby fixed carbon is handed over to the host in exchange for nutrient collection and the availability of a stable environment, is that of chemotrophic symbioses in deep-sea or sediment fauna. Soon after the discovery of deep-sea vents and their cohort of exotic fauna in the late 1970s, symbiotic bacteria filling out completely the modified gut of the giant worm Riftia pachyptila led to propose a chemoautotrophic symbiosis (Cavanaugh et al., 1981). Today the basis for this symbiosis is well known; the gammaproteobacterial symbiont, acquired anew at each generation, fixes carbon gaining energy by the oxidation of H2S, while the polychaete worm transports O2, NO3- and H2S for the needs of its otherwise metabolically versatile symbiont (Robidart et al., 2008). Similar chemotrophic symbioses also abound in the gills of deep-sea bivalves and other deep-sea metazoans and, more generally, ecto- and endosymbionts are frequently associated to animals living in oxygen-deprived marine settings, from the deep-sea to coastal sediments settings (Dubilier et al., 2008). These most often correspond to sulfide- or sulfur-oxidizing Gamma- or Epsilonproteobacteria, methanotrophic Gammaproteobacteria or sulfate-reducing Deltaproteobacteria (Dubilier et al., 2008). In many cases, these symbioses are multiple, involving diverse endosymbionts with distinct metabolic capabilities. For instance, clams of the genus Bathymodiolus harbor dual symbioses with methanotrophic and thiotrophic bacteria (Duperron et al., 2007). The case of the oligochaete Olavius algarvensis is most remarkable; this worm, lacking mouth, gut and nephridia, hosts co-existing sulfide-oxidizing (Gammaproteobacteria) and sulfate-reducing (Deltaproteobacteria) bacteria and benefits from the versatile metabolism of its hosted, sulfur-cycling consortium (Woyke et al., 2006).
Microbial symbioses with plants are also widespread and possibly explain their evolutionary and ecological success. Over 80% plants establish symbioses with specific fungi that provide humidity, nitrogen and phosphorous to the roots in exchange for fixed carbon. The mycorrhizal symbiosis is thought to be at the origin of terrestrial colonization by plants (Field et al., 2015), the algal ancestor of land plants likely being pre-adapted for symbiosis (Delaux et al., 2015). Almost as important are nitrogen-fixing symbioses established with bacteria from various phylogenetic groups, the most successful and best-known of which are the alphaproteobacterial Rhizobiales. Rhizobium-legume symbioses have co-evolved leading to specific signaling processes that involve, among others, immunity suppression during the establishment and maintenance of the symbiosis (Geurts et al., 2016; Gourion et al., 2015).
Prokaryotic symbioses established with protists are more difficult to identify because many protists being grazers, it is difficult to distinguish endosymbionts from prey. Also, although many classical works describe the recurrent presence of endobionts in protists, they often fail to inquire further about their role and many of these potential endosymbioses remain cryptic. Known endosymbioses in microbial eukaryotes often involve photosynthetic cyanobacteria, nitrogen-fixing/recycling bacteria, or methanogenic archaea (in some hydrogenosome-bearing anaerobic protists) (Nowack and Melkonian, 2010). Symbioses with cyanobacteria occur frequently, and one of them, particularly successful, gave rise to the chloroplast more than 1 billion years ago (see below). An intriguing symbiosis occurs in oligotrophic oceans between marine haptophyte algae and the derived, nitrogen-fixing U-CYNA cyanobacteria, which have secondarily lost the ability to make photosynthesis (Zehr et al., 2016). Like in the case of animals, and although not well-known, protist-prokaryote symbioses are particularly abundant in suboxic environments (Bernhard et al., 2000). Hydrogen-transfer appears important, as in methanogenic symbioses of ciliates and other anaerobic protists or the ectosymbiosis established by the breviate Lenisia limosa with Epsilonproteobacteria of the genus Arcobacter (Hamann et al., 2016), also frequent epibionts in deep-sea vent fauna, including alvinellids and shrimps. Similarly, sulfide-oxidizing epsilonproteobacterial epibionts are associated to 'Symbiontida' euglenozoans in anoxic sediments (Edgcomb et al., 2011b). Multiple symbioses, including sometimes ecto- and endosymbionts, such as in the case of oxymonads with spirochetes and Bacteroidales (Noda et al., 2009), have also been described. Some multiple symbioses include three or more partners. For instance, a ciliate from deep, anoxic, sulfide-rich sediments contains at least three different endosymbionts in specialized membrane-bound sub-cellular regions, including two sulfate-reducing Deltaproteobacteria (related to the families Desulfobulbaceae and Desulfobacteraceae) and a methanogenic archaeon; it possibly also includes one endosymbiotic Bacteroidetes and a Type I methanotroph (Edgcomb et al., 2011a). This further supports the idea that multiple symbioses offer to the host the benefit of synergistic metabolisms in this type of anoxic environments.
2.3. Prokaryote-prokaryote syntrophy
Most symbioses imply metabolic interactions. This is particularly true in the case of prokaryote-prokaryote symbioses. Microbial syntrophy implies obligatory mutualistic metabolic cooperation. This type of symbiosis is especially important, sometimes mandatory, in low-energy environments lacking strong electron acceptors and where many endergonic reactions can become exergonic only when one partner acts as an electron sink for the other (Morris et al., 2013). Syntrophic interactions mediated by interspecies hydrogen or formate transfer are possibly the most conspicuous and profuse in the planet, being essential in the anaerobic conversion of organic matter down to methane in anoxic sediments worldwide (McInerney et al., 2009). In addition of playing key roles in the intermediate steps of the carbon cycle, syntrophic interactions can also involve the exchange of organic, sulfur- or nitrogen-containing molecules (Morris et al., 2013).
Symbiotic partners can be so well integrated that it is sometimes possible to isolate and study such consortia (though not the individual partners). Classical examples are 'Methanobacillus omelianski', formed by an ethanol fermenter producing acetate and hydrogen and a methanogen using that hydrogen to reduce CO2 to CH4 (Bryant et al., 1967), or phototrophic consortia of the type 'Chlorochromatium aggregatum', formed by a central motile heterotrophic betaproteobacterium and several peripheral anoxygenic photosynthesizing green sulfur bacteria (Chlorobi) (Overmann and Van Gemerden, 2000). Genomic analysis show that the non-motile chlorobi can fix nitrogen and carbon that it can transfer to the central bacterium which, in turn, can sense and move the consortium towards light and sulfide that is used as electron donor for photosynthesis (Cerqueda-Garcia et al., 2014; Liu et al., 2013). Nonetheless, identifying syntrophic partners in natural ecosystems is difficult due to the high diversity and complexity of microbial communities in this kind of settings, and to the fact that a large part of this diversity corresponds to divergent bacteria and archaeal taxa (including the recently discovered Asgard clades) for which the metabolic potential is only beginning to be explored via metagenomics and single-cell genomics (Baker et al., 2016; Biddle et al., 2011; Biddle et al., 2006; Rinke et al., 2013; Sousa et al., 2016). However, unveiling cryptic microbial symbioses in complex environments like sediments, soils or mats, will require ultimately the visualization of metabolic exchange, which can be achieved by a variety of sophisticated techniques including fluorescent in situ hybridization coupled to secondary ion mass spectrometry or bioorthogonal noncanonical amino acid tagging coupled to fluorescence-activated cell sorting (Hatzenpichler et al., 2016; Orphan, 2009).
Deltaproteobacteria and Gram positive bacteria together with methanogen-related archaea are the better-known and most prominent groups of prokaryotes involved in syntrophic interactions in sediments and similar environments worldwide (McInerney et al., 2008). Deltaproteobacteria are metabolically versatile; most are sulfate-reducing, but some have secondarily lost this ability in favor of anaerobic fermentation or aerobic heterotrophy, while some are predatory, like the bdellovibrios (Madigan et al., 2014). Deltaproteobacterial fermenters are usually engaged in syntrophic partnership with methanogenic archaea, some genera being obligatory syntrophic (Syntrophus, Syntrophobacter), as some Firmicutes are (Syntrophomonas). Interestingly, this type of mutualism can evolve extremely fast, as has been shown by experimental evolution of co-cultures of the sulfate-reducing Desulfovibrio vulgaris and the archaeon Methanococcus maripaludis. When these organisms, with no known history of previous interaction, were grown in co-culture, the deltaproteobacterium stopped reducing sulfate (which consumes hydrogen) and started to ferment, liberating hydrogen that was used by the methanogen for its metabolism. This syntrophy was efficient (evolved co-cultures grew up to 80% faster and were up to 30% more productive in terms of biomass per mole of substrate) and evolved in less than 300 generations (Hillesland and Stahl, 2010).
In addition, sulfate-reducing deltaproteobacteria also establish syntrophic interactions with methanotrophic archaea (methanogen-related ANME lineages that most likely perform partly reversed methanogenesis). Anaerobic oxidation of methane (AOM) was thought to be impossible because energetically unfavorable, until it was discovered that this could be achieved by syntrophic consortia in marine sediments worldwide (Boetius et al., 2000; Orphan et al., 2001; Thauer and Shima, 2008). AOM is also crucial for the global carbon cycle, being responsible for the transformation of a large part of methane in sediments (Wegener et al., 2016). Remarkably, direct cell-cell electron transfer between by nanowire-like structures has been demonstrated in this type of consortia (Wegener et al., 2015). Direct interspecies electron transfer has raised a lot of attention, because some organisms (cable bacteria) can exchange electrons over long distances; it typically occurs in anaerobic methane-producing and methane-consuming communities (Lovley, 2016).
Metagenomic and metatranscriptomic analyses are starting to provide clues into syntrophy for anaerobic metabolic cooperation (Sieber et al., 2012). This kind of studies often reveals the involvement of more than two syntrophic partners in a consortium, as is the case in AOM (Pernthaler et al., 2008). Indeed, it appears that multi-member syntrophic communities are generally favored in nature even in environments not as challenging as anoxic ecosystems, since they reduce the individual metabolic burden in favor of cross-feeding and synergistic growth. This is suggested by experimental evolution of co-existing strains that tend to reduce the biosynthetic cost of amino acid synthesis promoting cooperative interactions, which are stronger for the more costly amino acids (Mee et al., 2014). Although not well understood, multidimensional interactions are also important in natural anaerobic methanogenic communities involved in carbon cycling (Embree et al., 2015).
Most known prokaryote-prokaryote symbioses involve cell-cell contact but not direct endosymbiosis. However, rare cases of endosymbiosis of prokaryotes within prokaryotes are known. Intracellular bacterial endosymbionts exist within endosymbiotic bacteria in mealybugs (von Dohlen et al., 2001), and the symbiont can be replaced in this nested symbiosis (Husnik and McCutcheon, 2016). A similar nested symbiosis involves intracellular alphaproteobacteria residing within tick mitochondria (Sassera et al., 2006).
3. Symbiosis in eukaryotic evolution
If Margulis was well aware of the importance of symbiosis in nature, her major contribution was to propose a symbiogenetic origin of chloroplasts, mitochondria and the eukaryotic cell itself. Except for details regarding the partners and the specific mechanisms, which still remain to be elucidated, she was essentially right.
3.1. Symbiosis and the origin of plastids
Photosynthesis originated very early during the evolution of bacteria, as attested by the phototactic behavior observed in the oldest fossil stromatolites, more than 3,4 Gya (Awramik, 1992) and, especially, by the oxygenation of Earth’s atmosphere that started at least 2,4 Gya (Lyons et al., 2014). Whereas the organisms that built the oldest stromatolites were most likely anoxygenic photosynthesizers, the enormous amount of oxygen that was necessary to oxidize the atmosphere was undoubtedly produced by cyanobacteria, the only bacterial lineage able to carry out oxygenic photosynthesis. Thus, for more than 2 billion years, photosynthetic primary production was exclusively assured by bacteria, as photosynthetic eukaryotes only evolved ~1 Gya (Eme et al., 2014) (see below). Interestingly, eukaryotes did not evolve a brand new oxygenic photosynthetic metabolism but, as Mereschkowsky (Mereschkowsky, 1905; Mereschkowsky, 1910) and, later on, Margulis (Sagan, 1967) posited, they just adopted it from bacteria by the endosymbiosis of a cyanobacterium that became the first photosynthetic plastid. The number of endosymbiotic events and the identity of the partners involved have been the matter of debate for decades (Moreira and Philippe, 2001). Nevertheless, phylogenetic analysis of plastid-encoded genes points to a single origin of all eukaryotic plastids, namely, a single initial cyanobacterial endosymbiosis within a heterotrophic eukaryotic host (an evolutionary event that is known as primary endosymbiosis) (Archibald, 2009; Keeling, 2013; Moreira et al., 2000; Rodriguez-Ezpeleta et al., 2005). Recently, it has been shown that the cyanobacterial endosymbiont most likely belonged to the Gloeomargaritales, a group of unicellular cyanobacteria widely distributed in freshwater systems (Ponce-Toledo et al., 2017). Although virtually all eukaryotic plastids derive from this endosymbiont, a second case of primary endosymbiosis has been identified, involving a cyanobacterium from a different group (the Synechococcus-Prochlorococcus clade) that established a primary symbiosis with testate amoebae of the genus Paulinella (Nakayama and Ishida, 2009). In all these symbioses, a recurrent observation is that genes from the cyanobacterial endosymbiont are massively transferred into the nucleus of the eukaryotic host, a phenomenon called endosymbiotic gene transfer –EGT- (Martin et al., 1998). Since several proteins encoded by those transferred genes remain necessary for plastid function, this implies that a system to address them into the plastid has to evolve.
The initial primary endosymbiosis gave rise to three groups of photosynthetic eukaryotes: red algae, green algae and plants, and glaucophyte algae. They form a monophyletic supergroup called Archaeplastida (Adl et al., 2005) or Plantae (Cavalier-Smith, 1982). However, photosynthetic plastids can be noticed in many other eukaryotic lineages. In contrast with the initial cyanobacterial symbiosis, these plastids originated by the endosymbiosis of red or green algae in other eukaryotic hosts. These eukaryote-within-eukaryote secondary endosymbioses are at the origin of a vast diversity of algae, including the euglenids and chlorarachniophytes (with green plastids) and the cryptophytes, haptophytes, dinoflagellates, stramenopiles (diatoms, brown and golden algae, etc.) and several other groups containing red plastids. In some cases, plastids have lost their photosynthetic activity, but they remain in the cell to carry out other functions, a notorious case being that of the apicoplast, a non-photosynthetic organelle found in the Apicomplexa, a major group of parasites (Keeling, 2013). Several lineages have even more complex histories as they may have acquired secondary plastids by tertiary endosymbioses (e.g., haptophyte and diatom plastids found in some dinoflagellates) or may have replaced a secondary plastid by another one by secondary plastid replacement (e.g., green algal plastids in the dinoflagellate Lepidodinium, which replaced the original red algal one).
Whereas the monophyly of the Archaeplastida is widely accepted, the evolutionary relationships among the lineages containing red algal secondary plastids remain controversial (in the case of euglenids and chlorarachniophytes it is clear that they derive from two independent endosymbioses with two different green algae). As they account for a substantial fraction of the whole eukaryotic diversity, the phylogenetic uncertainty around the secondary red lineages represents a major open question in eukaryotic evolution. It was initially thought that all eukaryotes with red algal plastids were monophyletic (the 'Chromalveolate hypothesis' (Cavalier-Smith, 1999), as supported by the phylogeny of plastid-encoded genes where all red algal-derived secondary plastids form a monophyletic group. However, the phylogeny of nucleus-encoded genes does not support the expected monophyly of the different hosts. To reconcile both observations, it has been proposed that red secondary plastids were acquired by one eukaryotic lineage and this foundational event was followed by an undetermined number of tertiary endosymbioses that spread the red plastids across a variety of lineages (Baurain et al., 2010). As in the case of the primary endosymbiosis, the secondary endosymbioses have been accompanied by massive EGT from the nucleus of the red or green algal secondary plastid to the nuclei of the hosts, which are therefore highly chimeric since they contain genes with very different evolutionary origins. This complicates the study of these organisms as the phylogenies of the different genes may be discordant and difficult to interpret (Moreira and Deschamps, 2014). At any rate, the contemporary diversity and ecological significance of photosynthetic eukaryotes demonstrates the power of symbiosis to generate evolutionary novelty in eukaryotes. Moreover, this is an ongoing phenomenon as many eukaryotes may acquire facultative photosynthetic abilities through the endosymbiosis of a variety of algae. This is very common in animals (e.g., zooxanthellae in corals and other invertebrates) but also in single-celled eukaryotes (e.g., endosymbiotic green algae in some ciliates). Some of these endosymbioses may become obligatory, especially by EGT if essential genes are transferred from the endosymbiont to the host, and enlarge the list of eukaryotic photosynthetic lineages.
3.2. Symbiosis and the origin of mitochondria
In aerobic eukaryotes, mitochondria are the energy factories of the cell, the organelles were oxygen respiration is used to convert biochemical energy from nutrients into ATP. The presence of a small genome within these organelles prompted biologists to speculate that they derive from endosymbiotic organisms, though the diversity of their genome size and physical organization and some other peculiarities (e.g., variations in the genetic code in mitochondria of some eukaryotic lineages) casted doubts on their monophyly and the nature of the endosymbiotic organisms at their origin (Gray and Doolittle, 1982). However, sequence similarities between mitochondrial and bacterial rRNAs (Bonen et al., 1977) and, especially, phylogenetic analysis of c-type cytochrome sequences (Schwartz and Dayhoff, 1978) unequivocally demonstrated the bacterial origin of mitochondria, closely related to the nonsulfur purple bacteria, known today as Proteobacteria. Subsequent analyses showed that all eukaryotic mitochondria originated from a single ancestor. Much more debated was the timing of the endosymbiosis that gave rise to mitochondria during eukaryotic evolution. In fact, a variety of anaerobic eukaryotes lack conventional mitochondria and, in some cases, no trace of a mitochondrial genome was detectable. Moreover, some of these eukaryotes (microsporidia, trichomonads, diplomonads) branched at the base of the first eukaryotic phylogenies reconstructed using 18S rRNA sequences (Sogin, 1991), which led to propose that these organisms were some kind of living fossils that predated the mitochondrial endosymbiosis: the 'Archezoa hypothesis' (Cavalier-Smith, 1989).
This hypothesis did not survive the passing of time when genes of clear mitochondrial origin were discovered in the nuclear genomes of 'amitochondriate' eukaryotes and when alternative phylogenetic markers replaced these lineages far from the base of the eukaryotic tree (Roger, 1999). In fact, as in the case of plastids described above, the mitochondrial endosymbiosis has been followed by massive EGT from the mitochondrial ancestor to the nuclear host genome and several of the proteins encoded by the transferred genes are targeted back into the mitochondria by a specialized translocation mechanism. Thus, even in the case of complete loss of the mitochondrial genome (Dyall et al., 2004; Gray, 2012; Timmis et al., 2004) (Dyall et al., 2004; Gray, 2012; Muller et al., 2012; Schwartz and Dayhoff, 1978), some mitochondrial activities can be maintained thanks to the genes transferred into the nucleus. This is the case for many anaerobic eukaryotes, which do not carry out aerobic respiration any more, but retain mitochondria-derived organelles that may have energy-related activities (such as ATP synthesis in hydrogenosomes (Müller, 1975) or other functions (such as the synthesis of Fe-S clusters (Gill et al., 2007). The presence of conventional mitochondria or of mitochondria-related organelles (MROs) in all major eukaryotic lineages strongly advocates for a mitochondrial endosymbiosis that predated the diversification of all contemporary eukaryotes. Thus, not only photosynthetic eukaryotes possess chimeric genomes because of the EGT of cyanobacterial genes, but all eukaryotes do as they contain genes of mitochondrial origin in their nuclear genomes. A possible outstanding exception has come with the characterization of the anaerobic oxymonad Monocercomonoides sp., which appears to have completely lost all typical mitochondrial genes (Karnkowska et al., 2016). This whole absence of mitochondrial genes reflects a secondary loss, as the phylogenetic position of this species confirms that it derives from mitochondriate ancestors. Nonetheless, this discovery has interesting evolutionary implications as it demonstrates that mitochondria (or any form of MRO) are not a requirement for eukaryotic cell existence despite the fact that, historically, the mitochondrial symbiosis was indissolubly linked to eukaryotic origins.
3.3. Eukaryogenesis by symbiogenesis
As explained above, there is overwhelming evidence supporting that the mitochondrial endosymbiosis occurred before the diversification of all contemporary eukaryotic lineages. Thus, if we define eukaryogenesis as the whole evolutionary process leading from prokaryotic ancestors to the last common ancestor of all eukaryotes, this process is necessarily symbiogenetic at the very least because of the ubiquity of mitochondria in eukaryotes. Mitochondria did not only provide an efficient energy metabolism, oxygen respiration, but genes of mitochondrial origin seem to be involved in a variety of cell activities in eukaryotes, such as DNA repair (Lin et al., 2007) and diverse metabolic functions (Thiergart et al., 2012). Therefore, mitochondrial acquisition not only represents an ancient event in eukaryotic evolution, but it probably entailed important changes at various functional levels of the host cell. Some authors have suggested that the mitochondrial endosymbiosis was the key event that triggered the diversification of contemporary eukaryotes (Philippe et al., 2000) and others have even proposed that it triggered the very origin of the eukaryotic cell itself (see below). At any rate, there is no doubt that contemporary eukaryotes are chimeras as they are formed by the integration of at least two types of cells: an ancient bacterium that transformed into mitochondria plus the host that acquired it. This fact is significant as it allowed to discard a series of classical hypotheses for the origin of eukaryotes that posited that all eukaryotic hallmark features, including mitochondria and the other organelles, evolved by transformation of pre-existing structures of a prokaryotic cell. These hypotheses, collectively called 'autogenous' (Gray and Doolittle, 1982), were instrumental in the early opposition to Margulis’ symbiogenetic ideas but were abandoned when molecular biology and phylogenetics demonstrated the endosymbiotic origin of mitochondria and chloroplasts. Nevertheless, the term autogenous is still used by several authors to refer to the origin of the most idiosyncratic eukaryotic cellular structure: the nucleus (see below).
Whereas the nature of the mitochondrial ancestor was easy to determine by using phylogenetic analysis, which placed mitochondria within Proteobacteria (more specifically, within Alphaproteobacteria (Andersson et al., 1998), the nature of the host and its nucleus have remained much more elusive. Analysis of the macromolecular structure of ribosomes of a variety of organisms revealed interesting similarities between eukaryotes and a group of 'sulfur dependent bacteria', which were called 'eocytes' (Lake et al., 1984). These organisms were later recognized to actually be a major subgroup of Archaea, the Crenarchaeota (Woese et al., 1990). Subsequent phylogenetic analyses of diverse proteins, especially when the first complete genome sequences became available, led to recognize that eukaryotic genomes appeared to be composed of two fractions with different functions and evolutionary origins: eukaryotic genes which function in translation, transcription, and replication (called 'informational genes') are closely related to archaeal homologs, whereas genes involved in energy and intermediary metabolism and in the synthesis of cell components, including amino acids, cofactors, the cell envelope, fatty acids, phospholipids, and nucleotides (referred to as 'operational genes') are more closely related to bacterial homologs (Rivera et al., 1998). The presence of bacterial-like genes in eukaryotic genomes was easy to explain as genes acquired by EGT from the mitochondrial endosymbiont. However, the presence of archaeal-like genes was more intriguing and has been the focus of most debates on eukaryogenesis in recent decades. The hypothesis initially favored was that a third lineage different from the two prokaryotic ones (Archaea and Bacteria) but phylogenetically sister to the Archaea was at the origin of the eukaryotic nucleocytoplasm and, therefore, the host of the mitochondrial endosymbiont. This hypothetical lineage would have evolved 'autogenously' many of the hallmark eukaryotic features (notably a complex cytoskeleton and endomembrane system, including the nucleus). This idea fitted well the rRNA-based Woesian tree of life that supports the division of living beings into three major lineages (or Domains): Archaea, Bacteria and Eucarya (Woese et al., 1990) and by this reason it was called the '3D' hypothesis (Gribaldo et al., 2010). Despite some contradictory results (Lake, 1987), phylogenetic analysis of protein markers also appeared to support this view, as the archaeal-like component of eukaryotes tended to branch deep, before the diversification of contemporary archaeal lineages (Yutin et al., 2008).
This situation has radically changed in recent years thanks to significant improvements in phylogenetic reconstruction methods and in the taxon sampling available. The first ones are mostly related to the implementation of site-heterogeneous mixture models of sequence evolution, which better account for the substitution patterns observed in sequence datasets (Lartillot and Philippe, 2004). The application of those methods to the reconstruction of universal trees has yielded phylogenies where eukaryotes do not form an independent branch but emerge within the Archaea (Williams et al., 2013; Williams et al., 2012). On the other hand, taxon sampling improvement has been made possible by the exploration of a variety of environments using new tools, which have led to the discovery of a vast diversity of archaeal lineages (Pester et al., 2011), some of which are phylogenetically related to the Crenarchaeota (the 'eocytes’) and form a major archaeal supergroup, the TACK or Proteoarchaeota (Guy and Ettema, 2011). Among them, a recently discovered lineage has had a major impact on our view of eukaryogenesis: the Asgard archaeal clades (Thor-, Odin-, Heimdall and Lokiarchaeota) (Spang et al., 2015; Zaremba-Niedzwiedzka et al., 2017). Although not yet cultured in the laboratory, composite genome sequences reconstructed from sediment metagenomes coming from a variety of sources, from deep sea to hot springs, have shown that Asgard lineages appear to contain more genes shared with eukaryotes than any other archaea and, even more importantly, phylogenetic analysis of conserved markers retrieves the monophyly of eukaryotes and Asgard archaea (Spang et al., 2015; Zaremba-Niedzwiedzka et al., 2017). These results provide strong support to the so-called '2D models', which posit that there are only two primary domains (Bacteria and Archaea) and that eukaryotes constitute a derived secondary domain, originated from some sort of mix of lineages of the two prokaryotic domains (Fig. 1). Thus, the archaeal-like component of eukaryotic genomes, namely the informational genes, seems to have originated from within the archaea. These discoveries have led not only to discard the 3D models mentioned above, but also to further discredit other models that proposed that these informational genes, and even the nucleus itself, were acquired from giant viruses, a miscellaneous group of viruses belonging to the nucleocytoplasmic large DNA virus (NCLDV) superfamily (Bell, 2009; Boyer et al., 2010; Forterre and Gaia, 2016; Raoult et al., 2004). Indeed, even before the discovery of Lokiarchaeota and other Asgard archaea, it had already been shown that eukaryotic-like genes found in NCLDV had been acquired by the viruses from their hosts via horizontal gene transfer (HGT) and not the other way round. This can be masked by the high evolutionary rate of viruses that can distort phylogenetic reconstruction, but when accurate phylogenetic methods are applied to retrace the evolutionary history of these genes, their eukaryotic origin becomes clear (Lopez-Garcia and Moreira, 2015; Moreira and Lopez-Garcia, 2005; Williams et al., 2011). Therefore, a NCLDV virus was not at the origin of the informational genes and/or the eukaryotic nucleus, but an archaeon, most likely an ancient member of the Asgard archaea or related lineages. Nonetheless, how this archaeon established a symbiotic relationship with bacteria and the number of its bacterial partners remain open questions that will occupy research in the field of eukaryogenesis in the next years (Lopez-Garcia and Moreira, 2015).
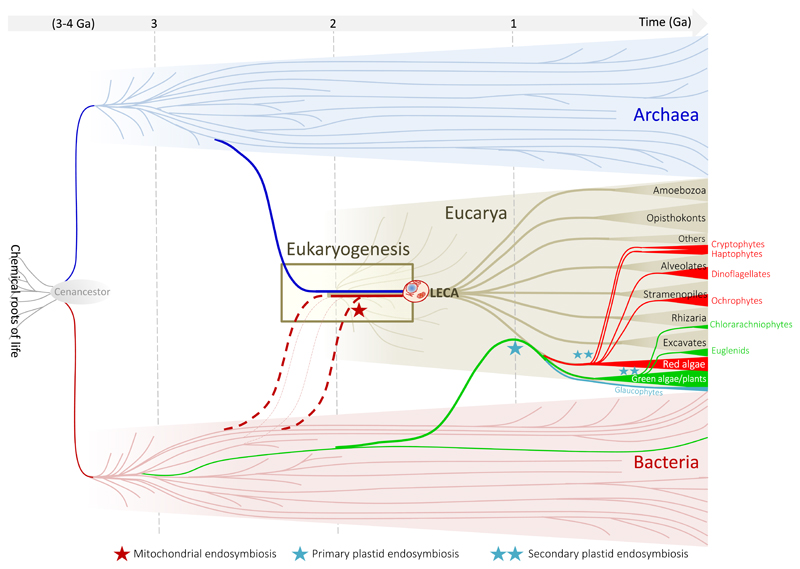
Figure 1.
Schematic representation of the tree of life and the origin of eukaryotes in an approximate historical framework.
4. Eukaryogenesis: open questions
It has become clear now that eukaryogenesis occurred by symbiogenesis of archaea and bacteria (Fig. 1), supporting Margulis' visionary idea, also adopted by later symbiogenetic models, that symbiosis was crucial in eukaryotic evolution, leading to an increase in average cell complexity. Symbiogenesis was an implicit conclusion from recent gene content and phylogenomic analyses suggesting that archaea of the TACK superphylum were more similar to eukaryotes (Martijn and Ettema, 2013; McInerney et al., 2014; Williams and Embley, 2014), which eliminated the possibility that a third, proto-eukaryotic lineage different from archaea ever existed, and became much more explicit when Lokiarchaeota and other Asgard archaea were discovered (Koonin, 2015; Lopez-Garcia and Moreira, 2015; Spang et al., 2015; Zaremba-Niedzwiedzka et al., 2017). However, despite the confirmation of the symbiogenetic origin of eukaryotes, many crucial questions remain open (reviewed in (Lopez-Garcia and Moreira, 2015).
4.1. When did eukaryotes evolve?
The geological record preserves part of life history, including evidence for early (chemical and morphological) traces of the existence of eukaryotes. When considering this evidence, it may be important to distinguish between 'stem' and 'crown' lineages. Briefly, all lineages that descend from LECA are often referred to as crown eukaryotes (Fig. 1). But before a full-fledged LECA evolved, a more or less long period of evolution occurred from the time when, depending on the model, the archaeal ancestor of eukaryotes started to acquire eukaryotic features (Fig 2A) or when the first eukaryogenetic archaea-bacteria consortium started to co-evolve together (Fig. 2B-D). Lineages diverging during that period are considered stem lineages and are now all extinct (but the one that successfully lead to 'crown' eukaryotes). Any characteristic feature of eukaryotes evident in ancient microfossils can, in principle, be a property of either stem or crown organisms. Therefore, unless there are specific features that definitively associate the fossils/biomarkers with particular crown eukaryotic group, it is not possible to distinguish whether they represent stem or crown eukaryotes. Additionally, identifying with confidence microfossils as eukaryotic rather than bacterial is not a trivial task and only fossils combining large size, ornamented walls, complex ultrastructure, and a preservable composition are regarded as probable eukaryotes (Knoll, 2015). As such, while the oldest of microfossils of possible eukaryotic origin were described from 3200 Ma shales, their simple ultrastructure prevents from any definitive conclusion (Javaux et al., 2010). The oldest fossil showing robust structural evidence of eukaryotic affiliation appears in rocks as old as ~1700 Ma (Javaux, 2007; Knoll et al., 2006; Yan and Liu, 1993). However, most of the ancient Proterozoic assemblages (i.e., 1800–1000 Ma) include fossils that are difficult, if not impossible, to associate with crown eukaryotic groups and they could in fact, represent stem eukaryote lineages. A notable exception is Bangiomorpha pubescens from the 1.2-Ga Hunting Formation (Butterfield, 2000) that represents the oldest fossil that some paleontologists confidently assign to a crown eukaryotic lineage, the bangiophyte red algae, setting a lower boundary for eukaryotic diversification.
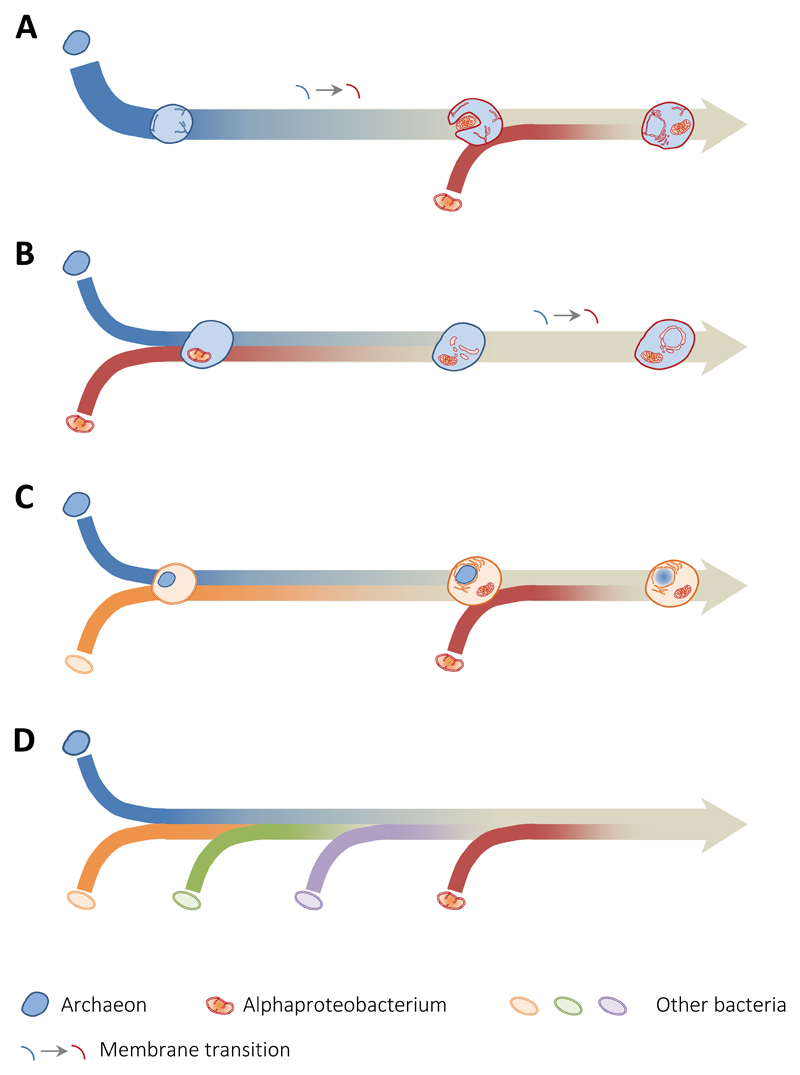
Figure 2.
Schematic representation of current eukaryogenetic models based on symbiosis. A, one archaeon develops endomembranes and the nucleus, acquiring the capacity of phagocytosis and the possibility to engulf the mitochondrial ancestor. B, the mitochondrial ancestor becomes an early endosymbiont in an archaeon, triggering eukaryogenesis. C, endosymbiotic origin of the nucleus (archaeon) within a bacterium; the mitochondrion results from a second endosymbiosis. D, multiple successive symbioses forge the eukaryotic cell.
A complementary approach to determining the timing of eukaryotic evolution is through molecular dating, which allows divergence times to be estimated from genetic distances, based on the simple idea that differences between homologous proteins of different species are proportional to their divergence time (Zuckerkandl and Pauling, 1965). However, variation in substitution rates has been widely documented; to cope with it, 'relaxed molecular clock' (RMC) methods were developed to allow the rate of sequence evolution to vary across different branches (for reviews, see (Ho et al., 2015; Kumar and Hedges, 2016). To estimate divergence times by using RMC approaches, the phylogenetic tree is calibrated with several known dates associated with the available paleobiological data. As our understanding of eukaryote phylogeny has improved, fossil-calibrated molecular-clock-based methods have been applied to date important diversification events, but have yielded vastly different estimates (Berney and Pawlowski, 2006; Douzery et al., 2004; Eme et al., 2014; Hedges and Kumar, 2004; Hedges et al., 2004; Parfrey et al., 2011). These discrepancies can be explained by a myriad of sources of variability and error (Eme et al., 2014; Kumar and Hedges, 2016; Roger and Hug, 2006) such as the relaxed molecular clock models and methods used, and the nature and treatment of fossil calibrations. The most recent analyses provide estimates for the age of LECA in the range of 1000 to 1600 Ma (Eme et al., 2014).
From these results, estimating the age of the mitochondrial endosymbiosis intrinsically depends on the model favored for eukaryogenesis. In the case of 'mitochondria-early' scenarios, which consider the mitochondrial endosymbiosis to be the initial triggering event of eukaryogenesis (Fig. 2B), the timing of the endosymbiosis can be inferred to be older than the oldest evidence for the occurrence of eukaryotes (i.e., 1.7 Ga). It is worth noting that the first organisms belonging to the eukaryotic lineage might have been morphologically indistinguishable from their prokaryotic ancestors. Consequently, any microfossil clearly distinguishable as eukaryotic (i.e., combining all the morphological features listed above) would be considerably more recent than the origin of the eukaryotic lineage itself, and thus, than the mitochondrial endosymbiosis. By contrast, 'mitochondria-late' hypotheses postulate that a significant number of specific eukaryotic-like features predated the acquisition of the mitochondrion (Fig. 2A, C-D). In this case, the timing of the mitochondrial endosymbiosis would be considerably closer to the age of LECA.
The timing of the primary plastid endosymbiosis can be more easily bracketed since it had to occur after LECA and before the last common ancestor of Archaeplastida. Most recent analyses estimate the latter to be 900-1300 Ma (Eme et al., 2014). It is worth noting that the Bangiomorpha fossils seem to be much older than any of the dates estimated from molecular clock data (Berney and Pawlowski, 2006; Eme et al., 2014; Parfrey et al., 2011; Sharpe et al., 2015) and at odds with all other microfossil calibrations for crown eukaryotes. This discrepancy can stem from a number of reasons. For example, either the taxonomic identification of Bangiomorpha fossils, or the estimated age of the rocks in which they were found, could be erroneous (although the latter is thought to be unlikely, see discussion in (Knoll, 2014; Parfrey et al., 2011). Alternatively, it is possible that currently available molecular clock models do not capture properly the evolutionary process occurring in some lineages, leading to incorrect date estimates. The second case of primary stable endosymbiosis identified in Paulinella is considerably younger and has been estimated to have occurred ~60 Mya (Delaye et al., 2016). As to the time when the lineages containing secondary plastids evolved, their confused evolutionary relationships among their heterotrophic hosts currently hampers any serious attempt to date secondary and tertiary endosymbioses. Future phylogenomics progress on the reconstruction of the eukaryotic tree should help constraining the evolution of plastids derived from green and red algae.
4.2. Where did eukaryotes evolve?
One key question concerns the environmental setting where eukaryotes evolved and the metabolic nature of the symbiosis that gave rise to eukaryotes. Metabolic endosymbioses can drive evolution and have explanatory power in evolution (Lopez-Garcia and Moreira, 2015; O'Malley, 2015). Because prokaryotic symbioses essentially involve syntrophy, the eukaryogenetic symbiogenesis was most likely based on metabolic cooperation. Some insight can be obtained from the kind of ecosystems and metabolic networks established in ecosystems where Asgard archaeal lineages occur. Lokiarchaeota, previously known as Deep Sea Archaeal Group (DSAG) or marine benthic group B, have been classically detected in sediments and microbial mats (Biddle et al., 2006; Jorgensen et al., 2013; Jorgensen et al., 2012; Spang et al., 2015). Because their native habitats are anoxic, they must be anaerobic, which would be consistent with a putative dependency on hydrogen deduced from their gene content (Sousa et al., 2016); they have indeed been suggested to be involved in syntrophic interactions leading to dissimilatory CH4 oxidation (Biddle et al., 2006) or iron or manganese reduction (Jorgensen et al., 2012). Likewise, the rest of recently identified Asgard archaea are associated to sediments from various environmental settings (Zaremba-Niedzwiedzka et al., 2017). Obviously, these Asgard archaea constitute a collection of derived lineages that may be quite different from the archaeal ancestor that established the eukaryogenetic symbiosis with bacteria. Also, it might well be that there are other archaeal lineages that are even closer to eukaryotes, since archaeal diversity is far from being completely explored. Actually, a large diversity of deeply divergent archaeal clades occurs precisely in anoxic settings, including the deep subsurface, sediments and different kinds of microbial mats (Baker et al., 2016; Biddle et al., 2006; Harris et al., 2013; Hedlund et al., 2013; Jorgensen et al., 2013; Jorgensen et al., 2012; Kozubal et al., 2013; Lloyd et al., 2013). However, the fact that Asgard and many other deep-branching archaea are systematically found in anoxic settings where syntrophic interactions are the rule supports the idea that eukaryotes emerged from metabolically cooperative consortia in anoxic settings likely close to redox boundaries.
Indeed, symbiotic models that have advanced detailed mechanisms, including metabolic interactions, for the origin of eukaryotes, such as the hydrogen (Martin and Muller, 1998) and the syntrophy (López-García and Moreira, 2006; Moreira and López-García, 1998) hypotheses implied that eukaryotes evolved in anoxic settings where redox gradients occurred and that the mitochondrial ancestor was a facultative anaerobe (López-García and Moreira, 1999). This would offer the possibility, among others, to establish a symbiosis with a facultative aerobic alphaproteobacterium endowed with oxygen respiration but able to thrive in oxygen-depleted environments. A facultative anaerobic alphaproteobacterium, possibly microaerophilic (adapted to low-oxygen concentrations) as mitochondrial ancestor would make sense at a time when oxygen started to accumulate in the atmosphere and most of the oceanic water column was still anoxic (Scott et al., 2008). Both, hydrogen and syntrophy hypotheses proposed hydrogen transfer between symbiotic partners, in the former between a fermentative alphaproteobacterium that established as endosymbiont within a methanogenic archaeon (Martin and Muller, 1998), in the second between an ancestral sulfate-reducing fermentative myxobacterium (Deltaproteobacteria) and an endosymbiotic methanogenic archaeon (future nucleus) to which a versatile alphaproteobacterial methanotroph would have joined in a tripartite symbiosis (López-García and Moreira, 2006; Moreira and López-García, 1998). Thought to be exclusive of the Euryarchaeota branch, methanogenesis has recently been inferred in the Bathyarchaeota, within the TACK superphylum, making methanogenesis likely ancestral in archaea (Evans et al., 2015; Lever, 2016). Nonetheless, as mentioned in section 2.3, the number and type of syntrophic interactions mediated by hydrogen among archaea and bacteria in sediments and other oxygen-depleted environments are far from fully understood. Multiple syntrophic interactions appear to be the rule in natural conditions, so that multiple symbioses at the origin of eukaryotes cannot be excluded on the basis of parsimony criteria because microbial ecology contradicts those arguments. At any rate, better understanding metabolic interactions between archaea and bacteria in this kind of ecosystems will greatly help proposing plausible metabolic interactions at the origin of the eukaryotic cell.
4.3. How did eukaryotes evolve?
Many models proposing symbiotic merging between archaea and bacteria at the origin of eukaryotes avoid providing detailed mechanistic scenarios explaining the origin of the most characteristic features of the eukaryotic cell. However, the most open and contentious questions in eukaryogenesis precisely relate to the mechanisms by which the different eukaryotic features appeared (reviewed in (Lopez-Garcia and Moreira, 2015), such that exposing the details is important to discriminate between existing models, refine them or propose more realistic ones. The different types of models that can currently be considered are schematically shown in Fig. 2. The most popular (Fig. 2.A) would correspond to the transposition of old '3D' models to a '2D' situation. Here, an archaeon would develop an endomembrane system by invagination of the plasma membrane and the nucleus in an 'autogenous' way together with the capacity of phagocytosis, which would allow it to engulf the mitochondrial ancestor (phagocytosing archaeon model, PhAT (Martijn and Ettema, 2013). Many of these models are mechanistically naive but could be nurtured by much more developed character evolution scenarios from previous 3D phagocytosis-based models, including particular variants such as the Neomura hypothesis (Cavalier-Smith, 2014). A second model would correspond to the initial endosymbiosis of the alphaproteobacterial ancestor of mitochondria within an archaeon, the rest of archaeal features would evolve after the symbiosis established (Fig. 2.B) (e.g. hydrogen hypothesis; (Martin and Muller, 1998). A third type of models would correspond to the endosymbiotic origin of the nucleus within one bacterium, the mitochondrion deriving for a second endosymbiosis with an alphaproteobacterium (Fig. 2.C) (e.g. syntrophy hypothesis; (López-García and Moreira, 2006; Moreira and López-García, 1998). Finally, a fourth type of models would imply multiple serial endosymbioses (either within a bacterium or within an archaeon) with the participation of several bacteria that would have left an imprint in the eukaryotic nucleus by successive waves of EGT (Fig. 2.D) (Pittis and Gabaldon, 2016).
Although there is some debate about the specific identity of the mitochondrial ancestor within the Alphaproteobacteria and as to whether it was or not a facultative anaerobe, the most controversial point refers to whether mitochondria arrived early or not (Fig. 2). In the hydrogen hypothesis, mitochondria arrive early because the mitochondrial endosymbiosis is proposed to be the cause of eukaryogenesis (Martin and Muller, 1998). By contrast, in the phagocytosing archaeon model, the mitochondria arrive late because they are, in a way, the consequence of an autogenous eukaryogenetic process that led to phagocytosis and the possibility to engulf exogenous bacteria (Martijn and Ettema, 2013). In the syntrophy hypothesis, the mitochondrial symbiosis is the last, highly successful, endosymbiotic event that determines the eukaryogenetic closure, providing a clear selective metabolic advantage related to the oxygenic respiration (López-García and Moreira, 2006). Recent phylogenomic analyses exploring the relative timing of prokaryotic gene acquisition in eukaryotes indeed suggest that mitochondria arrived late and might additionally support successive, multiple symbioses (Pittis and Gabaldon, 2016).
Another crucial problem relates to the origin of the eukaryotic nucleus, which many eukaryogenetic models leave unexplained. In addition to the selective forces that led to the evolution of this idiosyncratic structure, which remain elusive (Lopez-Garcia and Moreira, 2015), the process by which the nuclear membrane evolved, like the timing of mitochondrial acquisition, also discriminates the hydrogen hypothesis from the rest of symbiogenetic models. The latter propose an autogenous origin of the endoplasmic reticulum and the nuclear membrane (López-García and Moreira, 2006; Martijn and Ettema, 2013), which is consistent with cell biology knowledge and with the presence of many proteins involved in membrane remodeling and vesicle trafficking in archaea including, notably, Asgard archaea, but also other prokaryotes (Devos et al., 2004; Klinger et al., 2016; Spang et al., 2015; Surkont and Pereira-Leal, 2016). By contrast, the hydrogen hypothesis, speculates that the nuclear membrane and the endomembrane system would derive from the formation and progressive fusion of vesicles budding off the alphaproteobacterial ancestor of mitochondria around what would be the future nucleus (Gould et al., 2016). The production of lipid vesicles from that alphaproteobacterial endosymbiont would also explain, according to these authors, another fundamental problem in eukaryogenesis: the bacterial nature of eukaryotic membrane phospholipids.
Indeed, if the host that acquired mitochondria was an archaeon, then the archaeal membranes should have undergone a profound transformation, substituting their membrane phospholipids, very different from bacterial ones, by bacterial phospholipids (Lombard et al., 2012), but also adapting all the integral membrane proteins to a very different physicochemical environment (Lopez-Garcia and Moreira, 2015). In Gould’s et al’s view, the archaeal lipids of the plasma membrane would have been substituted by mitochondria-derived lipids via vesicles protruding from the mitochondria that would fuse with the archaeal membrane and progressively replace archaeal lipids. This still leaves unexplained the selective force that drove that putative membrane transition, as membrane proteins would still be adapted to archaeal lipids. Likewise, bacteria-to-archaea horizontal gene transfer of genes involved in, e.g. fatty acid biosynthesis, has been evoked to explain a putative archaea-to-bacteria phospholipid transition, but some archaea containing those genes (e.g. haloarchaea) have typical archaeal membranes (López-García et al., 2015; Nelson-Sathi et al., 2012), indicating that fatty acids are used for other purpose in the cell. Likewise, the absence of the glycerol-1-phosphate dehydrogenase (G1PDH) gene, which is responsible for the G1P isomer characteristically used to synthesize archaeal phospholipids, seems to be absent from Lokiarchaeota and marine Euryarchaeota groups II/III, leading to propose that these archaea might have chimeric archaea-bacteria or even bacterial-like membrane phospholipids (Villanueva et al., 2016). Failure to identify G1PDH is intriguing. However, this observation cannot be taken for proof in the absence of direct access to the membrane lipids of these organisms. First, the genomes of these organisms have been reconstructed from metagenomes and are not necessarily complete (Deschamps et al., 2014; Iverson et al., 2012; Spang et al., 2015). Second, the gene might have significantly diverged in these organisms and, in any case, the absence of a classical G1PDH does not necessarily imply that G1P is not synthesized in an alternative way. It is indeed worth noting that archaeal lipids were dominant in sediments with abundant Lokiarchaeota (DSAG) (Biddle et al., 2006). Finally, unusual butane- and pentanetriol-based tetraether lipids replacing glycerol-based backbones have been recently detected in a group of methanogens sister to Group II/III archaea and Thermoplasmatales, and these special lipids seem also abundant in sediments (Becker et al., 2016), suggesting a modified, but not bacterial, nature for the lipids of this clade. Clearly, having access to the real biochemistry of the membrane lipids for these novel archaeal groups is required.
5. Conclusion
Fifty years after the publication of On the origin of mitosing cells, and after having endured harsh criticism and important doses of indifference, the legacy of Lynn Margulis seems as solid as ever. Certainly, some of her original ideas, notably the symbiotic origin of flagella, have not withstood the test of time. However, not only the symbiotic origin of mitochondria and chloroplasts but, importantly, that of the eukaryotic cell itself are now beyond any doubt. Eukaryotes are symbiotic mergers forged via cooperative metabolic interactions by progressive physical integration, endosymbiotic gene transfer and the vast evolutionary possibilities that duplicated genes offered to create innovations. A more or less long co-evolutionary path, especially in the case of symbiogenetic models that imply early syntrophy between bacteria and archaea (Fig. 2), offered a 'stem' period during which deep transformations occurred in the prokaryotic ancestors of eukaryotes. By becoming individual units of evolution of increased average complexity, eukaryotes accessed a wealth of new ecological niches and diversified.
Many questions remain still unanswered in terms of the number and nature of the specific prokaryotic partners that took part in the eukaryogenetic process, their metabolic interactions and the historical process that led to LECA. Hopefully, we have left behind the 'phylogenomic impasse'. Combining metagenomic and single-cell genomic analyses of natural microbial communities where Asgard and other deep-branching archaea typically thrive with metabolic modeling and with the use of better phylogenomic tools, we should be able to reconstruct a plausible evolutionary model for the origin of eukaryotes.
Acknowledgements
This work was supported by the European Research Council Advanced Grant ProtistWorld [grant number agreement 322669].
References
- Adl SM, Simpson AG, Farmer MA, Andersen RA, Anderson OR, Barta JR, Bowser SS, Brugerolle G, Fensome RA, Fredericq S, James TY, et al. The new higher level classification of eukaryotes with emphasis on the taxonomy of protists. J Eukaryot Microbiol. 2005;52:399–451. doi: 10.1111/j.1550-7408.2005.00053.x. [DOI] [PubMed] [Google Scholar]
- Andersson SG, Zomorodipour A, Andersson JO, Sicheritz-Ponten T, Alsmark UC, Podowski RM, Naslund AK, Eriksson AS, Winkler HH, Kurland CG. The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature. 1998;396:133–40. doi: 10.1038/24094. [DOI] [PubMed] [Google Scholar]
- Archibald JM. The puzzle of plastid evolution. Curr Biol. 2009;19:R81–88. doi: 10.1016/j.cub.2008.11.067. [DOI] [PubMed] [Google Scholar]
- Awramik SM. The oldest records of photosynthesis. Photosynth Res. 1992;33:75–89. [PubMed] [Google Scholar]
- Baker BJ, Saw JH, Lind AE, Lazar CS, Hinrichs KU, Teske AP, Ettema TJ. Genomic inference of the metabolism of cosmopolitan subsurface Archaea, Hadesarchaea. Nat Microbiol. 2016;1:16002. doi: 10.1038/nmicrobiol.2016.2. [DOI] [PubMed] [Google Scholar]
- Bao X, Roossinck MJ. A life history view of mutualistic viral symbioses: quantity or quality for cooperation? Curr Opin Microbiol. 2013;16:514–518. doi: 10.1016/j.mib.2013.05.007. [DOI] [PubMed] [Google Scholar]
- Baurain D, Brinkmann H, Petersen J, Rodriguez-Ezpeleta N, Stechmann A, Demoulin V, Roger AJ, Burger G, Lang BF, Philippe H. Phylogenomic evidence for separate acquisition of plastids in cryptophytes, haptophytes, and stramenopiles. Mol Biol Evol. 2010;27:1698–1709. doi: 10.1093/molbev/msq059. [DOI] [PubMed] [Google Scholar]
- Becker KW, Elling FJ, Yoshinaga MY, Sollinger A, Urich T, Hinrichs KU. Unusual butane- and pentanetriol-based tetraether lipids in Methanomassiliicoccus luminyensis, a representative of the seventh order of methanogens. Appl Environ Microbiol. 2016;82:4505–4516. doi: 10.1128/AEM.00772-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell PJ. The viral eukaryogenesis hypothesis: a key role for viruses in the emergence of eukaryotes from a prokaryotic world environment. Ann N Y Acad Sci. 2009;1178:91–105. doi: 10.1111/j.1749-6632.2009.04994.x. [DOI] [PubMed] [Google Scholar]
- Berney C, Pawlowski J. A molecular time-scale for eukaryote evolution recalibrated with the continuous microfossil record. Proc Biol Sci. 2006;273:1867–1872. doi: 10.1098/rspb.2006.3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhard JM, Buck KR, Farmer MA, Bowser SS. The Santa Barbara Basin is a symbiosis oasis. Nature. 2000;403:77–80. doi: 10.1038/47476. [DOI] [PubMed] [Google Scholar]
- Biddle JF, White JR, Teske AP, House CH. Metagenomics of the subsurface Brazos-Trinity Basin (IODP site 1320): comparison with other sediment and pyrosequenced metagenomes. ISME J. 2011;5:1038–1047. doi: 10.1038/ismej.2010.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biddle JF, Lipp JS, Lever MA, Lloyd KG, Sorensen KB, Anderson R, Fredricks HF, Elvert M, Kelly TJ, Schrag DP, Sogin ML, et al. Heterotrophic Archaea dominate sedimentary subsurface ecosystems off Peru. Proc Natl Acad Sci U S A. 2006;103:3846–3451. doi: 10.1073/pnas.0600035103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boetius A, Ravenschlag K, Schubert CJ, Rickert D, Widdel F, Gieseke A, Amann R, Jorgensen BB, Witte U, Pfannkuche O. A marine microbial consortium apparently mediating anaerobic oxidation of methane. Nature. 2000;407:623–626. doi: 10.1038/35036572. [DOI] [PubMed] [Google Scholar]
- Bonen L, Cunningham RS, Gray MW, Doolittle WF. Wheat embryo mitochondrial 18S ribosomal RNA: evidence for its prokaryotic nature. Nucleic Acids Res. 1977;4:663–671. doi: 10.1093/nar/4.3.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer M, Madoui MA, Gimenez G, La Scola B, Raoult D. Phylogenetic and phyletic studies of informational genes in genomes highlight existence of a 4 domain of life including giant viruses. PLoS ONE. 2010;5:e15530. doi: 10.1371/journal.pone.0015530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant MP, Wolin EA, Wolin MJ, Wolfe RS. Methanobacillus omelianskii, a symbiotic association of two species of bacteria. Arch Microbiol. 1967;59:20–31. doi: 10.1007/BF00406313. [DOI] [PubMed] [Google Scholar]
- Butterfield NJ. Bangiomorpha pubescens n. gen., n. sp. : implications for the evolution of sex multicellularity, and the Mesoproterozoic/Neoproteorozoic radiation of eukaryotes. Paleobiology. 2000;26:386–404. [Google Scholar]
- Cavalier-Smith T. The origin of plastids. Bull J Linnean Soc. 1982;17:289–306. [Google Scholar]
- Cavalier-Smith T. Molecular phylogeny. Archaebacteria and Archezoa. Nature. 1989;339:l00–1. doi: 10.1038/339100a0. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T. Principles of protein and lipid targeting in secondary symbiogenesis: euglenoid, dinoflagellate, and sporozoan plastid origin and the eukaryote family tree. J Euk Microbiol. 1999;46:347–366. doi: 10.1111/j.1550-7408.1999.tb04614.x. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T. The neomuran revolution and phagotrophic origin of eukaryotes and cilia in the light of intracellular coevolution and a revised tree of life. Cold Spring Harb Perspect Biol. 2014;6:a016006. doi: 10.1101/cshperspect.a016006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanaugh CM, Gardiner SL, Jones ML, Jannasch HW, Waterbury JB. Prokaryotic cells in the hydrothermal vent tube worm Riftia pachyptila Jones: possible chemoautotrophic symbionts. Science. 1981;213:340–341. doi: 10.1126/science.213.4505.340. [DOI] [PubMed] [Google Scholar]
- Celiker H, Gore J. Cellular cooperation: insights from microbes. Trends Cell Biol. 2013;23:9–15. doi: 10.1016/j.tcb.2012.08.010. [DOI] [PubMed] [Google Scholar]
- Cerqueda-Garcia D, Martinez-Castilla LP, Falcon LI, Delaye L. Metabolic analysis of Chlorobium chlorochromatii CaD3 reveals clues of the symbiosis in 'Chlorochromatium aggregatum'. ISME J. 2014;8:991–998. doi: 10.1038/ismej.2013.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordaux R, Bouchon D, Greve P. The impact of endosymbionts on the evolution of host sex-determination mechanisms. Trends Genet. 2011;27:332–341. doi: 10.1016/j.tig.2011.05.002. [DOI] [PubMed] [Google Scholar]
- de Duve C. The origin of eukaryotes: a reappraisal. Nat Rev Genet. 2007;8:395–403. doi: 10.1038/nrg2071. [DOI] [PubMed] [Google Scholar]
- Delaux PM, Radhakrishnan GV, Jayaraman D, Cheema J, Malbreil M, Volkening JD, Sekimoto H, Nishiyama T, Melkonian M, Pokorny L, Rothfels CJ, et al. Algal ancestor of land plants was preadapted for symbiosis. Proc Natl Acad Sci U S A. 2015;112:13390–13395. doi: 10.1073/pnas.1515426112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaye L, Valadez-Cano C, Pérez-Zamorano B. How really ancient is Paulinella chromatophora? PLOS Currents Tree of Life. 2016 doi: 10.1371/currents.tol.e68a099364bb1a1e129a17b4e06b0c6b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deschamps P, Zivanovic Y, Moreira D, Rodriguez-Valera F, Lopez-Garcia P. Pangenome evidence for extensive interdomain horizontal transfer affecting lineage core and shell genes in uncultured planktonic thaumarchaeota and euryarchaeota. Genome Biol Evol. 2014;6:1549–1563. doi: 10.1093/gbe/evu127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devos D, Dokudovskaya S, Alber F, Williams R, Chait BT, Sali A, Rout MP. Components of coated vesicles and nuclear pore complexes share a common molecular architecture. PLoS Biol. 2004;2:e380. doi: 10.1371/journal.pbio.0020380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douzery EJ, Snell EA, Bapteste E, Delsuc F, Philippe H. The timing of eukaryotic evolution: Does a relaxed molecular clock reconcile proteins and fossils? Proc Natl Acad Sci U S A. 2004;19:19. doi: 10.1073/pnas.0403984101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubilier N, Bergin C, Lott C. Symbiotic diversity in marine animals: the art of harnessing chemosynthesis. Nat Rev Microbiol. 2008;6:725–740. doi: 10.1038/nrmicro1992. [DOI] [PubMed] [Google Scholar]
- Duperron S, Sibuet M, MacGregor BJ, Kuypers MM, Fisher CR, Dubilier N. Diversity, relative abundance and metabolic potential of bacterial endosymbionts in three Bathymodiolus mussel species from cold seeps in the Gulf of Mexico. Environ Microbiol. 2007;9:1423–1438. doi: 10.1111/j.1462-2920.2007.01259.x. [DOI] [PubMed] [Google Scholar]
- Dyall SD, Brown MT, Johnson PJ. Ancient invasions: from endosymbionts to organelles. Science. 2004;304:253–257. doi: 10.1126/science.1094884. [DOI] [PubMed] [Google Scholar]
- Edgcomb VP, Leadbetter ER, Bourland W, Beaudoin D, Bernhard JM. Structured multiple endosymbiosis of bacteria and archaea in a ciliate from marine sulfidic sediments: a survival mechanism in low oxygen, sulfidic sediments? Front Microbiol. 2011a;2:55. doi: 10.3389/fmicb.2011.00055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgcomb VP, Breglia SA, Yubuki N, Beaudoin D, Patterson DJ, Leander BS, Bernhard JM. Identity of epibiotic bacteria on symbiontid euglenozoans in O2-depleted marine sediments: evidence for symbiont and host co-evolution. ISME J. 2011b;5:231–243. doi: 10.1038/ismej.2010.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Embley TM. Multiple secondary origins of the anaerobic lifestyle in eukaryotes. Philos Trans R Soc Lond B Biol Sci. 2006;361:1055–1067. doi: 10.1098/rstb.2006.1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Embley TM, Hirt RP. Early branching eukaryotes? Curr Opin Genet Dev. 1998;8:624–9. doi: 10.1016/s0959-437x(98)80029-4. [DOI] [PubMed] [Google Scholar]
- Embley TM, Martin W. Eukaryotic evolution, changes and challenges. Nature. 2006;440:623–630. doi: 10.1038/nature04546. [DOI] [PubMed] [Google Scholar]
- Embree M, Liu JK, Al-Bassam MM, Zengler K. Networks of energetic and metabolic interactions define dynamics in microbial communities. Proc Natl Acad Sci U S A. 2015;112:15450–15455. doi: 10.1073/pnas.1506034112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eme L, Doolittle WF. Microbial diversity: a bonanza of phyla. Curr Biol. 2015;25:R227–30. doi: 10.1016/j.cub.2014.12.044. [DOI] [PubMed] [Google Scholar]
- Eme L, Sharpe SC, Brown MW, Roger AJ. On the age of eukaryotes: evaluating evidence from fossils and molecular clocks. Cold Spring Harb Perspect Biol. 2014;6:a016139. doi: 10.1101/cshperspect.a016139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans PN, Parks DH, Chadwick GL, Robbins SJ, Orphan VJ, Golding SD, Tyson GW. Methane metabolism in the archaeal phylum Bathyarchaeota revealed by genome-centric metagenomics. Science. 2015;350:434–8. doi: 10.1126/science.aac7745. [DOI] [PubMed] [Google Scholar]
- Field KJ, Pressel S, Duckett JG, Rimington WR, Bidartondo MI. Symbiotic options for the conquest of land. Trends Ecol Evol. 2015;30:477–486. doi: 10.1016/j.tree.2015.05.007. [DOI] [PubMed] [Google Scholar]
- Forterre P, Gaia M. Giant viruses and the origin of modern eukaryotes. Current Opinion in Microbiology. 2016;31:44–49. doi: 10.1016/j.mib.2016.02.001. [DOI] [PubMed] [Google Scholar]
- Geurts R, Xiao TT, Reinhold-Hurek B. What does it take to evolve a nitrogen-fixing endosymbiosis? Trends Plant Sci. 2016;21:199–208. doi: 10.1016/j.tplants.2016.01.012. [DOI] [PubMed] [Google Scholar]
- Gill EE, Diaz-Trivino S, Barbera MJ, Silberman JD, Stechmann A, Gaston D, Tamas I, Roger AJ. Novel mitochondrion-related organelles in the anaerobic amoeba Mastigamoeba balamuthi. Mol Microbiol. 2007;66:1306–1320. doi: 10.1111/j.1365-2958.2007.05979.x. [DOI] [PubMed] [Google Scholar]
- Gould SB, Garg SG, Martin WF. Bacterial vesicle Secretion and the evolutionary origin of the eukaryotic endomembrane system. Trends in Microbiology. 2016;24:525–534. doi: 10.1016/j.tim.2016.03.005. [DOI] [PubMed] [Google Scholar]
- Gourion B, Berrabah F, Ratet P, Stacey G. Rhizobium-legume symbioses: the crucial role of plant immunity. Trends Plant Sci. 2015;20:186–194. doi: 10.1016/j.tplants.2014.11.008. [DOI] [PubMed] [Google Scholar]
- Gray MW. Mitochondrial evolution. Cold Spring Harb Perspect Biol. 2012;4:a011403. doi: 10.1101/cshperspect.a011403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray MW, Doolittle WF. Has the endosymbiont hypothesis been proven? Microbiol Rev. 1982;46:1–42. doi: 10.1128/mr.46.1.1-42.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gribaldo S, Poole AM, Daubin V, Forterre P, Brochier-Armanet C. The origin of eukaryotes and their relationship with the Archaea: are we at a phylogenomic impasse? Nat Rev Microbiol. 2010;8:743–752. doi: 10.1038/nrmicro2426. [DOI] [PubMed] [Google Scholar]
- Guerrero R, Margulis L, Berlanga M. Symbiogenesis: the holobiont as a unit of evolution. Int Microbiol. 2013;16:133–143. doi: 10.2436/20.1501.01.188. [DOI] [PubMed] [Google Scholar]
- Guy L, Ettema TJ. The archaeal 'TACK' superphylum and the origin of eukaryotes. Trends Microbiol. 2011;19:580–587. doi: 10.1016/j.tim.2011.09.002. [DOI] [PubMed] [Google Scholar]
- Hamann E, Gruber-Vodicka H, Kleiner M, Tegetmeyer HE, Riedel D, Littmann S, Chen J, Milucka J, Viehweger B, Becker KW, Dong X, et al. Environmental Breviatea harbour mutualistic Arcobacter epibionts. Nature. 2016;534:254–8. doi: 10.1038/nature18297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardoim PR, van Overbeek LS, Berg G, Pirttila AM, Compant S, Campisano A, Doring M, Sessitsch A. The hidden world within plants: ecological and evolutionary considerations for defining functioning of microbial endophytes. Microbiol Mol Biol Rev. 2015;79:293–320. doi: 10.1128/MMBR.00050-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris JK, Caporaso JG, Walker JJ, Spear JR, Gold NJ, Robertson CE, Hugenholtz P, Goodrich J, McDonald D, Knights D, Marshall P, et al. Phylogenetic stratigraphy in the Guerrero Negro hypersaline microbial mat. ISME J. 2013;7:50–60. doi: 10.1038/ismej.2012.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatzenpichler R, Connon SA, Goudeau D, Malmstrom RR, Woyke T, Orphan VJ. Visualizing in situ translational activity for identifying and sorting slow-growing archaeal-bacterial consortia. Proc Natl Acad Sci U S A. 2016;113:E4069–4078. doi: 10.1073/pnas.1603757113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedges SB, Kumar S. Precision of molecular time estimates. Trends Genet. 2004;20:242–247. doi: 10.1016/j.tig.2004.03.004. [DOI] [PubMed] [Google Scholar]
- Hedges SB, Blair JE, Venturi M, Shoe JL. A molecular timescale of eukaryote evolution and the rise of complex multicellular life. BMC Evol Biol. 2004;4:2. doi: 10.1186/1471-2148-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedlund BP, Dodsworth JA, Cole JK, Panosyan HH. An integrated study reveals diverse methanogens, Thaumarchaeota, and yet-uncultivated archaeal lineages in Armenian hot springs. Antonie Van Leeuwenhoek. 2013;104:71–82. doi: 10.1007/s10482-013-9927-z. [DOI] [PubMed] [Google Scholar]
- Hentschel U, Piel J, Degnan SM, Taylor MW. Genomic insights into the marine sponge microbiome. Nat Rev Microbiol. 2012;10:641–654. doi: 10.1038/nrmicro2839. [DOI] [PubMed] [Google Scholar]
- Hernandez-Agreda A, Gates RD, Ainsworth TD. Defining the core microbiome in corals' microbial soup. Trends Microbiol. 2016 doi: 10.1016/j.tim.2016.11.003. [DOI] [PubMed] [Google Scholar]
- Hillesland KL, Stahl DA. Rapid evolution of stability and productivity at the origin of a microbial mutualism. Proc Natl Acad Sci U S A. 2010;107:2124–2129. doi: 10.1073/pnas.0908456107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho SY, Duchene S, Molak M, Shapiro B. Time-dependent estimates of molecular evolutionary rates: evidence and causes. Mol Ecol. 2015;24:6007–6012. doi: 10.1111/mec.13450. [DOI] [PubMed] [Google Scholar]
- Hosokawa T, Koga R, Kikuchi Y, Meng XY, Fukatsu T. Wolbachia as a bacteriocyte-associated nutritional mutualist. Proc Natl Acad Sci U S A. 2010;107:769–774. doi: 10.1073/pnas.0911476107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hug LA, Baker BJ, Anantharaman K, Brown CT, Probst AJ, Castelle CJ, Butterfield CN, Hernsdorf AW, Amano Y, Ise K, Suzuki Y, et al. A new view of the tree of life. Nat Microbiol. 2016;1:16048. doi: 10.1038/nmicrobiol.2016.48. [DOI] [PubMed] [Google Scholar]
- Husnik F, McCutcheon JP. Repeated replacement of an intrabacterial symbiont in the tripartite nested mealybug symbiosis. Proc Natl Acad Sci U S A. 2016;113:E5416–5424. doi: 10.1073/pnas.1603910113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iverson V, Morris RM, Frazar CD, Berthiaume CT, Morales RL, Armbrust EV. Untangling genomes from metagenomes: revealing an uncultured class of marine Euryarchaeota. Science. 2012;335:587–590. doi: 10.1126/science.1212665. [DOI] [PubMed] [Google Scholar]
- Javaux EJ. The early eukaryotic fossil record. Adv Exp Med Biol. 2007;607:1–19. doi: 10.1007/978-0-387-74021-8_1. [DOI] [PubMed] [Google Scholar]
- Javaux EJ, Marshall CP, Bekker A. Organic-walled microfossils in 3.2-billion-year-old shallow-marine siliciclastic deposits. Nature. 2010;463:934–938. doi: 10.1038/nature08793. [DOI] [PubMed] [Google Scholar]
- Jekely G. Small GTPases and the evolution of the eukaryotic cell. Bioessays. 2003;25:1129–1138. doi: 10.1002/bies.10353. [DOI] [PubMed] [Google Scholar]
- Jorgensen SL, Thorseth IH, Pedersen RB, Baumberger T, Schleper C. Quantitative and phylogenetic study of the Deep Sea Archaeal Group in sediments of the Arctic mid-ocean spreading ridge. Front Microbiol. 2013;4:299. doi: 10.3389/fmicb.2013.00299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen SL, Hannisdal B, Lanzen A, Baumberger T, Flesland K, Fonseca R, Ovreas L, Steen IH, Thorseth IH, Pedersen RB, Schleper C. Correlating microbial community profiles with geochemical data in highly stratified sediments from the Arctic Mid-Ocean Ridge. Proc Natl Acad Sci U S A. 2012;109:E2846–2855. doi: 10.1073/pnas.1207574109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karnkowska A, Vacek V, Zubacova Z, Treitli SC, Petrzelkova R, Eme L, Novak L, Zarsky V, Barlow LD, Herman EK, Soukal P, et al. A eukaryote without a mitochondrial organelle. Current Biology. 2016;26:1274–1284. doi: 10.1016/j.cub.2016.03.053. [DOI] [PubMed] [Google Scholar]
- Keeling PJ. The number, speed, and impact of plastid endosymbioses in eukaryotic evolution. Annu Rev Plant Biol. 2013;64:583–607. doi: 10.1146/annurev-arplant-050312-120144. [DOI] [PubMed] [Google Scholar]
- Keeling PJ. The impact of history on our perception of evolutionary events: endosymbiosis and the origin of eukaryotic complexity. Cold Spring Harb Perspect Biol. 2014;6:a016196. doi: 10.1101/cshperspect.a016196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinger CM, Spang A, Dacks JB, Ettema TJ. Tracing the archaeal origins of eukaryotic membrane-trafficking system building blocks. Mol Biol Evol. 2016;33:1528–1541. doi: 10.1093/molbev/msw034. [DOI] [PubMed] [Google Scholar]
- Knoll AH. Paleobiological perspectives on early eukaryotic evolution. Cold Spring Harbor Perspectives in Biology. 2014;6 doi: 10.1101/cshperspect.a016121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoll AH. Paleobiological perspectives on early microbial evolution. Cold Spring Harb Perspect Biol. 2015;7:a018093. doi: 10.1101/cshperspect.a018093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoll AH, Javaux EJ, Hewitt D, Cohen P. Eukaryotic organisms in Proterozoic oceans. Philos Trans R Soc Lond B Biol Sci. 2006;361:1023–1038. doi: 10.1098/rstb.2006.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga R, Moran NA. Swapping symbionts in spittlebugs: evolutionary replacement of a reduced genome symbiont. Isme J. 2014;8:1237–1246. doi: 10.1038/ismej.2013.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV. Archaeal ancestors of eukaryotes: not so elusive any more. BMC Biol. 2015;13:84. doi: 10.1186/s12915-015-0194-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozubal MA, Romine M, Jennings R, Jay ZJ, Tringe SG, Rusch DB, Beam JP, McCue LA, Inskeep WP. Geoarchaeota: a new candidate phylum in the Archaea from high-temperature acidic iron mats in Yellowstone National Park. Isme J. 2013;7:622–634. doi: 10.1038/ismej.2012.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Hedges SB. Advances in time estimation methods for molecular data. Mol Biol Evol. 2016;33:863–869. doi: 10.1093/molbev/msw026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lake JA. Prokaryotes and archaebacteria are not monophyletic: rate invariant analysis of rRNA genes indicates that eukaryotes and eocytes form a monophyletic taxon. Cold Spring Harb Symp Quant Biol. 1987;52:839–46. doi: 10.1101/sqb.1987.052.01.091. [DOI] [PubMed] [Google Scholar]
- Lake JA, Henderson E, Oakes M, Clark MW. Eocytes: a new ribosome structure indicates a kingdom with a close relationship to eukaryotes. Proc Natl Acad Sci U S A. 1984;81:3786–90. doi: 10.1073/pnas.81.12.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lartillot N, Philippe H. A Bayesian mixture model for across-site heterogeneities in the amino-acid replacement process. Mol Biol Evol. 2004;21:1095–1109. doi: 10.1093/molbev/msh112. [DOI] [PubMed] [Google Scholar]
- Leclercq S, Theze J, Chebbi MA, Giraud I, Moumen B, Ernenwein L, Greve P, Gilbert C, Cordaux R. Birth of a W sex chromosome by horizontal transfer of Wolbachia bacterial symbiont genome. Proc Natl Acad Sci U S A. 2016 doi: 10.1073/pnas.1608979113. pii: 201608979. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lever MA. A new era of methanogenesis research. Trends Microbiol. 2016;24:84–86. doi: 10.1016/j.tim.2015.12.005. [DOI] [PubMed] [Google Scholar]
- Lin Z, Nei M, Ma H. The origins and early evolution of DNA mismatch repair genes--multiple horizontal gene transfers and co-evolution. Nucleic Acids Res. 2007;35:7591–603. doi: 10.1093/nar/gkm921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Muller J, Li T, Alvey RM, Vogl K, Frigaard NU, Rockwell NC, Boyd ES, Tomsho LP, Schuster SC, Henke P, et al. Genomic analysis reveals key aspects of prokaryotic symbiosis in the phototrophic consortium "Chlorochromatium aggregatum". Genome Biol. 2013;14:R127. doi: 10.1186/gb-2013-14-11-r127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd KG, Schreiber L, Petersen DG, Kjeldsen KU, Lever MA, Steen AD, Stepanauskas R, Richter M, Kleindienst S, Lenk S, Schramm A, Jorgensen BB. Predominant archaea in marine sediments degrade detrital proteins. Nature. 2013;496:215–218. doi: 10.1038/nature12033. [DOI] [PubMed] [Google Scholar]
- Lombard J, López-García P, Moreira D. The early evolution of lipid membranes and the three domains of life. Nat Rev Microbiol. 2012;10:507–515. doi: 10.1038/nrmicro2815. [DOI] [PubMed] [Google Scholar]
- Lopez-Garcia P, Moreira D. Open questions on the origin of eukaryotes. Trends Ecol Evol. 2015;30:697–708. doi: 10.1016/j.tree.2015.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-García P, Moreira D. Metabolic symbiosis at the origin of eukaryotes. Trends Biochem Sci. 1999;24:88–93. doi: 10.1016/s0968-0004(98)01342-5. [DOI] [PubMed] [Google Scholar]
- López-García P, Moreira D. Selective forces for the origin of the eukaryotic nucleus. Bioessays. 2006;28:525–533. doi: 10.1002/bies.20413. [DOI] [PubMed] [Google Scholar]
- López-García P, Zivanovic Y, Deschamps P, Moreira D. Bacterial gene import and mesophilic adaptation in archaea. Nat Rev Microbiol. 2015;13:447–456. doi: 10.1038/nrmicro3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovley DR. Happy together: microbial communities that hook up to swap electrons. ISME J. 2016 doi: 10.1038/ismej.2016.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons TW, Reinhard CT, Planavsky NJ. The rise of oxygen in Earth's early ocean and atmosphere. Nature. 2014;506:307–315. doi: 10.1038/nature13068. [DOI] [PubMed] [Google Scholar]
- Madigan MT, Martinko JM, Bender KS, Buckley DH, Stahl DA, Brock T, editors. Brock biology of microorganisms. 14th edition. Benjamin Cummings; San Francisco, California, U.S.A: 2014. [Google Scholar]
- Margulis L. Symbiosis in cell evolution. Freeman, W. H.; San Francisco, CA: 1981. [Google Scholar]
- Margulis L. Biodiversity: molecular biological domains, symbiosis and kingdom origins. Biosystems. 1992;27:39–51. doi: 10.1016/0303-2647(92)90045-z. [DOI] [PubMed] [Google Scholar]
- Margulis L. Archaeal-eubacterial mergers in the origin of Eukarya: phylogenetic classification of life. Proc Natl Acad Sci U S A. 1996;93:1071–6. doi: 10.1073/pnas.93.3.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulis L, Guerrero R. Kingdoms in turmoil. New Sci. 1991;1761:46–50. [PubMed] [Google Scholar]
- Margulis L, Schwartz KV. Five kingdoms: An illustrated guide to the phyla of life on Earth. W. H. Freeman and Co; New York: 1998. [Google Scholar]
- Margulis L, Dolan MF, Guerrero R. The chimeric eukaryote: origin of the nucleus from the karyomastigont in amitochondriate protists. Proc Natl Acad Sci U S A. 2000;97:6954–9. doi: 10.1073/pnas.97.13.6954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulis L, Lopez Baluja L, Awramik SM, Sagan D. Community living long before man: fossil and living microbial mats and early life. Sci Total Environ. 1986;56:379–397. doi: 10.1016/0048-9697(86)90342-6. [DOI] [PubMed] [Google Scholar]
- Margulis L, Corliss JO, Melkonian M, Chapman DJ, editors. Handbook of Protoctista. Jones & Bartlett; Boston: 1990. [Google Scholar]
- Margulis L, Chapman M, Guerrero R, Hall J. The last eukaryotic common ancestor (LECA): acquisition of cytoskeletal motility from aerotolerant spirochetes in the Proterozoic Eon. Proc Natl Acad Sci U S A. 2006;103:13080–13085. doi: 10.1073/pnas.0604985103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martijn J, Ettema TJ. From archaeon to eukaryote: the evolutionary dark ages of the eukaryotic cell. Biochem Soc Trans. 2013;41:451–457. doi: 10.1042/BST20120292. [DOI] [PubMed] [Google Scholar]
- Martin W, Muller M. The hydrogen hypothesis for the first eukaryote. Nature. 1998;392:37–41. doi: 10.1038/32096. [DOI] [PubMed] [Google Scholar]
- Martin W, Stoebe B, Goremykin V, Hapsmann S, Hasegawa M, Kowallik KV. Gene transfer to the nucleus and the evolution of chloroplasts. Nature. 1998;393:162–165. doi: 10.1038/30234. [DOI] [PubMed] [Google Scholar]
- McCutcheon JP, Moran NA. Extreme genome reduction in symbiotic bacteria. Nat Rev Microbiol. 2012;10:13–26. doi: 10.1038/nrmicro2670. [DOI] [PubMed] [Google Scholar]
- McInerney JO, O'Connell MJ, Pisani D. The hybrid nature of the Eukaryota and a consilient view of life on Earth. Nat Rev Microbiol. 2014;12:449–455. doi: 10.1038/nrmicro3271. [DOI] [PubMed] [Google Scholar]
- McInerney MJ, Sieber JR, Gunsalus RP. Syntrophy in anaerobic global carbon cycles. Curr Opin Biotechnol. 2009;20:623–632. doi: 10.1016/j.copbio.2009.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInerney MJ, Struchtemeyer CG, Sieber J, Mouttaki H, Stams AJ, Schink B, Rohlin L, Gunsalus RP. Physiology, ecology, phylogeny, and genomics of microorganisms capable of syntrophic metabolism. Ann N Y Acad Sci. 2008;1125:58–72. doi: 10.1196/annals.1419.005. [DOI] [PubMed] [Google Scholar]
- Mee MT, Collins JJ, Church GM, Wang HH. Syntrophic exchange in synthetic microbial communities. Proc Natl Acad Sci U S A. 2014;111:E2149–2156. doi: 10.1073/pnas.1405641111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mereschkowsky C. Über natur und usprung der chromatophoren im pflanzenreiche. Biologisches Centralblatt. 1905;25:593–604. [Google Scholar]
- Mereschkowsky K. Theorie der zwei Plasmaarten als Grundlage der Symbiogenesis, einer neuen Lehre von der Ent-stehung der Organismen. Biol Centralbl. 1910;30:353–367. [Google Scholar]
- Mitri S, Foster KR. The genotypic view of social interactions in microbial communities. Annu Rev Genet. 2013;47:247–273. doi: 10.1146/annurev-genet-111212-133307. [DOI] [PubMed] [Google Scholar]
- Moran NA, Bennett GM. The tiniest tiny genomes. Annu Rev Microbiol. 2014;68:195–215. doi: 10.1146/annurev-micro-091213-112901. [DOI] [PubMed] [Google Scholar]
- Moran NA, Sloan DB. The hologenome concept: helpful or hollow? PLoS Biol. 2015;13:e1002311. doi: 10.1371/journal.pbio.1002311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran NA, Yun Y. Experimental replacement of an obligate insect symbiont. Proc Natl Acad Sci U S A. 2015;112:2093–6. doi: 10.1073/pnas.1420037112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira D, López-García P. Symbiosis between methanogenic archaea and delta-Proteobacteria as the origin of eukaryotes: The syntrophic hypothesis. J Mol Evol. 1998;47:517–530. doi: 10.1007/pl00006408. [DOI] [PubMed] [Google Scholar]
- Moreira D, Philippe H. Sure facts and open questions about the origin and evolution of photosynthetic plastids. Res Microbiol. 2001;152:771–80. doi: 10.1016/s0923-2508(01)01260-8. [DOI] [PubMed] [Google Scholar]
- Moreira D, López-García P. Molecular ecology of microbial eukaryotes unveils a hidden world. Trends Microbiol. 2002;10:31–38. doi: 10.1016/s0966-842x(01)02257-0. [DOI] [PubMed] [Google Scholar]
- Moreira D, Lopez-Garcia P. Comment on “The 1.2-megabase genome sequence of Mimivirus”. Science. 2005;308:1114. doi: 10.1126/science.1111195. [DOI] [PubMed] [Google Scholar]
- Moreira D, Deschamps P. What was the real contribution of endosymbionts to the eukaryotic nucleus? Insights from photosynthetic eukaryotes. Cold Spring Harb Perspect Biol. 2014;6:a016014. doi: 10.1101/cshperspect.a016014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira D, Le Guyader H, Philippe H. The origin of red algae and the evolution of chloroplasts. Nature. 2000;405:69–72. doi: 10.1038/35011054. [DOI] [PubMed] [Google Scholar]
- Morris BE, Henneberger R, Huber H, Moissl-Eichinger C. Microbial syntrophy: interaction for the common good. FEMS Microbiol Rev. 2013;37:384–406. doi: 10.1111/1574-6976.12019. [DOI] [PubMed] [Google Scholar]
- Moya A, Pereto J, Gil R, Latorre A. Learning how to live together: genomic insights into prokaryote-animal symbioses. Nat Rev Genet. 2008;9:218–229. doi: 10.1038/nrg2319. [DOI] [PubMed] [Google Scholar]
- Muller M, Mentel M, van Hellemond JJ, Henze K, Woehle C, Gould SB, Yu RY, van der Giezen M, Tielens AG, Martin WF. Biochemistry and evolution of anaerobic energy metabolism in eukaryotes. Microbiol Mol Biol Rev. 2012;76:444–495. doi: 10.1128/MMBR.05024-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller M. Biochemistry of protozoan microbodies: peroxisomes, alpha-glycerophosphate oxidase bodies, hydrogenosomes. Annu Rev Microbiol. 1975;29:467–483. doi: 10.1146/annurev.mi.29.100175.002343. [DOI] [PubMed] [Google Scholar]
- Mushegian AA, Ebert D. Rethinking "mutualism" in diverse host-symbiont communities. Bioessays. 2016;38:100–108. doi: 10.1002/bies.201500074. [DOI] [PubMed] [Google Scholar]
- Nadell CD, Drescher K, Foster KR. Spatial structure, cooperation and competition in biofilms. Nat Rev Microbiol. 2016;14:589–600. doi: 10.1038/nrmicro.2016.84. [DOI] [PubMed] [Google Scholar]
- Nakayama T, Ishida K. Another acquisition of a primary photosynthetic organelle is underway in Paulinella chromatophora. Curr Biol. 2009;19:R284–285. doi: 10.1016/j.cub.2009.02.043. [DOI] [PubMed] [Google Scholar]
- Nelson-Sathi S, Dagan T, Landan G, Janssen A, Steel M, McInerney JO, Deppenmeier U, Martin WF. Acquisition of 1,000 eubacterial genes physiologically transformed a methanogen at the origin of Haloarchaea. Proc Natl Acad Sci U S A. 2012;109:20537–20542. doi: 10.1073/pnas.1209119109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda S, Hongoh Y, Sato T, Ohkuma M. Complex coevolutionary history of symbiotic Bacteroidales bacteria of various protists in the gut of termites. BMC Evol Biol. 2009;9:158. doi: 10.1186/1471-2148-9-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowack EC, Melkonian M. Endosymbiotic associations within protists. Philos Trans R Soc Lond B Biol Sci. 2010;365:699–712. doi: 10.1098/rstb.2009.0188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Malley MA. Endosymbiosis and its implications for evolutionary theory. Proc Natl Acad Sci U S A. 2015;112:10270–10277. doi: 10.1073/pnas.1421389112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orphan VJ. Methods for unveiling cryptic microbial partnerships in nature. Curr Opin Microbiol. 2009;12:231–237. doi: 10.1016/j.mib.2009.04.003. [DOI] [PubMed] [Google Scholar]
- Orphan VJ, House CH, Hinrichs KU, McKeegan KD, DeLong EF. Methane-consuming archaea revealed by directly coupled isotopic and phylogenetic analysis. Science. 2001;293:484–7. doi: 10.1126/science.1061338. [DOI] [PubMed] [Google Scholar]
- Overmann J, Van Gemerden H. Microbial interactions involving sulfur bacteria: implications for the ecology and evolution of bacterial communities. FEMS Microbiol Ecol. 2000;24:591–599. doi: 10.1111/j.1574-6976.2000.tb00560.x. [DOI] [PubMed] [Google Scholar]
- Pace NR. A molecular view of microbial diversity and the biosphere. Science. 1997;276:734–740. doi: 10.1126/science.276.5313.734. [DOI] [PubMed] [Google Scholar]
- Parfrey LW, Lahr DJ, Knoll AH, Katz LA. Estimating the timing of early eukaryotic diversification with multigene molecular clocks. Proc Natl Acad Sci U S A. 2011;108:13624–13629. doi: 10.1073/pnas.1110633108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paszkowski U. Mutualism and parasitism: the yin and yang of plant symbioses. Curr Opin Plant Biol. 2006;9:364–370. doi: 10.1016/j.pbi.2006.05.008. [DOI] [PubMed] [Google Scholar]
- Perez-Brocal V, Gil R, Ramos S, Lamelas A, Postigo M, Michelena JM, Silva FJ, Moya A, Latorre A. A small microbial genome: the end of a long symbiotic relationship? Science. 2006;314:312–313. doi: 10.1126/science.1130441. [DOI] [PubMed] [Google Scholar]
- Pernthaler A, Dekas AE, Brown CT, Goffredi SK, Embaye T, Orphan VJ. Diverse syntrophic partnerships from deep-sea methane vents revealed by direct cell capture and metagenomics. Proc Natl Acad Sci U S A. 2008;105:7052–7057. doi: 10.1073/pnas.0711303105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pester M, Schleper C, Wagner M. The Thaumarchaeota: an emerging view of their phylogeny and ecophysiology. Curr Opin Microbiol. 2011;14:300–306. doi: 10.1016/j.mib.2011.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippe H, Lopez P, Brinkmann H, Budin K, Germot A, Laurent J, Moreira D, Muller M, Le Guyader H. Early-branching or fast-evolving eukaryotes? An answer based on slowly evolving positions. Proc R Soc Lond B Biol Sci. 2000;267:1213–1221. doi: 10.1098/rspb.2000.1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippot L, Raaijmakers JM, Lemanceau P, van der Putten WH. Going back to the roots: the microbial ecology of the rhizosphere. Nat Rev Microbiol. 2013;11:789–799. doi: 10.1038/nrmicro3109. [DOI] [PubMed] [Google Scholar]
- Pittis AA, Gabaldon T. Late acquisition of mitochondria by a host with chimaeric prokaryotic ancestry. Nature. 2016;531:101–104. doi: 10.1038/nature16941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole AM, Penny D. Evaluating hypotheses for the origin of eukaryotes. Bioessays. 2007;29:74–84. doi: 10.1002/bies.20516. [DOI] [PubMed] [Google Scholar]
- Raoult D, Audic S, Robert C, Abergel C, Renesto P, Ogata H, La Scola B, Suzan M, Claverie JM. The 1.2-megabase genome sequence of Mimivirus. Science. 2004;306:1344–1350. doi: 10.1126/science.1101485. [DOI] [PubMed] [Google Scholar]
- Rinke C, Schwientek P, Sczyrba A, Ivanova NN, Anderson IJ, Cheng JF, Darling A, Malfatti S, Swan BK, Gies EA, Dodsworth JA, et al. Insights into the phylogeny and coding potential of microbial dark matter. Nature. 2013;499:431–437. doi: 10.1038/nature12352. [DOI] [PubMed] [Google Scholar]
- Rivera MC, Jain R, Moore JE, Lake JA. Genomic evidence for two functionally distinct gene classes. Proc Natl Acad Sci USA. 1998;95:6239–44. doi: 10.1073/pnas.95.11.6239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robidart JC, Bench SR, Feldman RA, Novoradovsky A, Podell SB, Gaasterland T, Allen EE, Felbeck H. Metabolic versatility of the Riftia pachyptila endosymbiont revealed through metagenomics. Environ Microbiol. 2008;10:727–737. doi: 10.1111/j.1462-2920.2007.01496.x. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Ezpeleta N, Brinkmann H, Burey SC, Roure B, Burger G, Loffelhardt W, Bohnert HJ, Philippe H, Lang BF. Monophyly of primary photosynthetic eukaryotes: green plants, red algae, and glaucophytes. Curr Biol. 2005;15:1325–1230. doi: 10.1016/j.cub.2005.06.040. [DOI] [PubMed] [Google Scholar]
- Roger AJ. Reconstructing early events in eukaryotic evolution. Am Nat. 1999;154:S146–S163. doi: 10.1086/303290. [DOI] [PubMed] [Google Scholar]
- Roger AJ, Hug LA. The origin and diversification of eukaryotes: problems with molecular phylogenetics and molecular clock estimation. Philos Trans R Soc Lond B Biol Sci. 2006;361:1039–1054. doi: 10.1098/rstb.2006.1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth MS. The engine of the reef: photobiology of the coral-algal symbiosis. Front Microbiol. 2014;5:422. doi: 10.3389/fmicb.2014.00422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagan L. On the origin of mitosing cells. J Theor Biol. 1967;14:255–274. doi: 10.1016/0022-5193(67)90079-3. [DOI] [PubMed] [Google Scholar]
- Sassera D, Beninati T, Bandi C, Bouman EA, Sacchi L, Fabbi M, Lo N. 'Candidatus Midichloria mitochondrii', an endosymbiont of the tick Ixodes ricinus with a unique intramitochondrial lifestyle. Int J Syst Evol Microbiol. 2006;56:2535–2540. doi: 10.1099/ijs.0.64386-0. [DOI] [PubMed] [Google Scholar]
- Schwartz RM, Dayhoff MO. Origins of prokaryotes, eukaryotes, mitochondria, and chloroplasts. Science. 1978;199:395–403. doi: 10.1126/science.202030. [DOI] [PubMed] [Google Scholar]
- Scott C, Lyons TW, Bekker A, Shen Y, Poulton SW, Chu X, Anbar AD. Tracing the stepwise oxygenation of the Proterozoic ocean. Nature. 2008;452:456–459. doi: 10.1038/nature06811. [DOI] [PubMed] [Google Scholar]
- Searcy DG. Origins of mitochondria and chloroplasts from sulfur-based symbioses. In: Hartman H, Matsuno K, editors. The origin and evolution of the cell. World Scientific; Singapore: 1992. pp. 47–78. [Google Scholar]
- Sharpe SC, Eme L, Brown MW, Roger AJ. Timing the origins of multicellular eukaryotes through phylogenomics and relaxed molecular clock analyses. In: Ruiz-Trillo I, Nedelcu AM, editors. Evolutionary Transitions to Multicellular Life. Springer; Dordrecht: 2015. pp. 3–29. [Google Scholar]
- Sieber JR, McInerney MJ, Gunsalus RP. Genomic insights into syntrophy: the paradigm for anaerobic metabolic cooperation. Annu Rev Microbiol. 2012;66:429–452. doi: 10.1146/annurev-micro-090110-102844. [DOI] [PubMed] [Google Scholar]
- Siegl A, Kamke J, Hochmuth T, Piel J, Richter M, Liang C, Dandekar T, Hentschel U. Single-cell genomics reveals the lifestyle of Poribacteria, a candidate phylum symbiotically associated with marine sponges. ISME J. 2011;5:61–70. doi: 10.1038/ismej.2010.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogin ML. Early evolution and the origin of eukaryotes. Curr Opin Genet Dev. 1991;1:457–63. doi: 10.1016/s0959-437x(05)80192-3. [DOI] [PubMed] [Google Scholar]
- Sonnenburg JL, Backhed F. Diet-microbiota interactions as moderators of human metabolism. Nature. 2016;535:56–64. doi: 10.1038/nature18846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa FL, Neukirchen S, Allen JF, Lane N, Martin WF. Lokiarchaeon is hydrogen dependent. Nat Microbiol. 2016;1:16034. doi: 10.1038/nmicrobiol.2016.34. [DOI] [PubMed] [Google Scholar]
- Spang A, Saw JH, Jorgensen SL, Zaremba-Niedzwiedzka K, Martijn J, Lind AE, van Eijk R, Schleper C, Guy L, Ettema TJ. Complex archaea that bridge the gap between prokaryotes and eukaryotes. Nature. 2015 doi: 10.1038/nature14447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steenackers HP, Parijs I, Foster KR, Vanderleyden J. Experimental evolution in biofilm populations. FEMS Microbiol Rev. 2016;40:373–397. doi: 10.1093/femsre/fuw002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surkont J, Pereira-Leal JB. Are there Rab GTPases in archaea? Mol Biol Evol. 2016;33:1833–1842. doi: 10.1093/molbev/msw061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaiss CA, Zmora N, Levy M, Elinav E. The microbiome and innate immunity. Nature. 2016;535:65–74. doi: 10.1038/nature18847. [DOI] [PubMed] [Google Scholar]
- Thauer RK, Shima S. Methane as fuel for anaerobic microorganisms. Ann N Y Acad Sci. 2008;1125:158–170. doi: 10.1196/annals.1419.000. [DOI] [PubMed] [Google Scholar]
- Thiergart T, Landan G, Schenk M, Dagan T, Martin WF. An evolutionary network of genes present in the eukaryote common ancestor polls genomes on eukaryotic and mitochondrial origin. Genome Biol Evol. 2012;4:466–485. doi: 10.1093/gbe/evs018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmis JN, Ayliffe MA, Huang CY, Martin W. Endosymbiotic gene transfer: organelle genomes forge eukaryotic chromosomes. Nat Rev Genet. 2004;5:123–135. doi: 10.1038/nrg1271. [DOI] [PubMed] [Google Scholar]
- Villanueva L, Schouten S, Damste JS. Phylogenomic analysis of lipid biosynthetic genes of Archaea shed light on the 'lipid divide'. Environ Microbiol. 2016 doi: 10.1111/1462-2920.13361. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- von Dohlen CD, Kohler S, Alsop ST, McManus WR. Mealybug beta-proteobacterial endosymbionts contain gamma-proteobacterial symbionts. Nature. 2001;412:433–436. doi: 10.1038/35086563. [DOI] [PubMed] [Google Scholar]
- Wallin IE. Symbionticism and the origin of species. Willliams & Wilkins; Baltimore: 1927. [Google Scholar]
- Wegener G, Krukenberg V, Riedel D, Tegetmeyer HE, Boetius A. Intercellular wiring enables electron transfer between methanotrophic archaea and bacteria. Nature. 2015;526:587–590. doi: 10.1038/nature15733. [DOI] [PubMed] [Google Scholar]
- Wegener G, Krukenberg V, Ruff SE, Kellermann MY, Knittel K. Metabolic capabilities of microorganisms involved in and associated with the anaerobic oxidation of methane. Front Microbiol. 2016;7:46. doi: 10.3389/fmicb.2016.00046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werren JH, Baldo L, Clark ME. Wolbachia: master manipulators of invertebrate biology. Nat Rev Microbiol. 2008;6:741–751. doi: 10.1038/nrmicro1969. [DOI] [PubMed] [Google Scholar]
- Whittaker RH, Margulis L. Protist classification and the kingdoms of organisms. Biosystems. 1978;10:3–18. doi: 10.1016/0303-2647(78)90023-0. [DOI] [PubMed] [Google Scholar]
- Williams TA, Embley TM. Archaeal "dark matter" and the origin of eukaryotes. Genome Biol Evol. 2014;6:474–481. doi: 10.1093/gbe/evu031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams TA, Embley TM. Changing ideas about eukaryotic origins. Philos Trans R Soc Lond B Biol Sci. 2015;370:20140318. doi: 10.1098/rstb.2014.0318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams TA, Embley TM, Heinz E. Informational gene phylogenies do not support a fourth domain of life for nucleocytoplasmic large DNA viruses. PLoS ONE. 2011;6:e21080. doi: 10.1371/journal.pone.0021080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams TA, Foster PG, Cox CJ, Embley TM. An archaeal origin of eukaryotes supports only two primary domains of life. Nature. 2013;504:231–236. doi: 10.1038/nature12779. [DOI] [PubMed] [Google Scholar]
- Williams TA, Foster PG, Nye TM, Cox CJ, Embley TM. A congruent phylogenomic signal places eukaryotes within the Archaea. Proc Biol Sci. 2012;279:4870–4879. doi: 10.1098/rspb.2012.1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese CR, Fox GE. Phylogenetic structure of the prokaryotic domain: the primary kingdoms. Proc Natl Acad Sci USA. 1977;74:5088–90. doi: 10.1073/pnas.74.11.5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese CR, Kandler O, Wheelis ML. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci USA. 1990;87:4576–9. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woyke T, Teeling H, Ivanova NN, Huntemann M, Richter M, Gloeckner FO, Boffelli D, Anderson IJ, Barry KW, Shapiro HJ, Szeto E, et al. Symbiosis insights through metagenomic analysis of a microbial consortium. Nature. 2006;443:950–955. doi: 10.1038/nature05192. [DOI] [PubMed] [Google Scholar]
- Yan Y, Liu ZL. Significance of eukaryotic organisms in the microfossil flora of Changcheng system. Acta Micropalaeontol Sin. 1993;10:167–180. [Google Scholar]
- Yarza P, Yilmaz P, Pruesse E, Glockner FO, Ludwig W, Schleifer KH, Whitman WB, Euzeby J, Amann R, Rossello-Mora R. Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences. Nat Rev Microbiol. 2014;12:635–645. doi: 10.1038/nrmicro3330. [DOI] [PubMed] [Google Scholar]
- Yutin N, Makarova KS, Mekhedov SL, Wolf YI, Koonin EV. The deep archaeal roots of eukaryotes. Mol Biol Evol. 2008;25:1619–1630. doi: 10.1093/molbev/msn108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaremba-Niedzwiedzka K, Caceres EF, Saw JH, Backstrom D, Juzokaite L, Vancaester E, Seitz KW, Anantharaman K, Starnawski P, Kjeldsen KU, Stott MB, et al. Asgard archaea illuminate the origin of eukaryotic cellular complexity. Nature. 2017;541:353–358. doi: 10.1038/nature21031. [DOI] [PubMed] [Google Scholar]
- Zehr JP, Shilova IN, Farnelid HM, Munoz-MarinCarmen MD, Turk-Kubo KA. Unusual marine unicellular symbiosis with the nitrogen-fixing cyanobacterium UCYN-A. Nat Microbiol. 2016;2:16214. doi: 10.1038/nmicrobiol.2016.214. [DOI] [PubMed] [Google Scholar]
- Zuckerkandl E, Pauling L. Evolutionary divergence and convergence in proteins. In: Bryson V, Vogel HJ, editors. Evolving Genes and Proteins. Academic Press; New York: 1965. pp. 97–166. [Google Scholar]