Abstract
CONTEXT
Accurate prediction of who will (or won’t) have high probability of survival benefit from standard treatments is fundamental for individualized cancer treatment strategies.
OBJECTIVE
To develop a predictor of response and survival from chemotherapy for newly diagnosed invasive breast cancer.
DESIGN
Development of different predictive signatures for resistance and response to neoadjuvant chemotherapy (stratified according to estrogen receptor (ER) status) from gene expression microarrays of newly diagnosed breast cancer (310 patients). Then prediction of breast cancer treatment-sensitivity using the combination of signatures for: 1) sensitivity to endocrine therapy, 2) chemo-resistance, and 3) chemo-sensitivity. Independent validation (198 patients) and comparison with other reported genomic predictors of chemotherapy response.
SETTING
Prospective multicenter study to develop and test genomic predictors for neoadjuvant chemotherapy.
PATIENTS
Newly diagnosed HER2-negative breast cancer treated with chemotherapy containing sequential taxane and anthracycline-based regimens then endocrine therapy (if hormone receptor-positive).
MAIN OUTCOME MEASURES
Distant relapse-free survival (DRFS) if predicted treatment-sensitive and absolute risk reduction (ARR, difference in DRFS of the two predicted groups) at median follow-up (3 years), and their 95% confidence intervals (CI).
RESULTS
Patients in the independent validation cohort (99% clinical Stage II–III) who were predicted to be treatment-sensitive (28% of total) had DRFS of 92% (CI 85–100) and survival benefit compared to others (absolute risk reduction (ARR) 18%; CI 6–28). Predictions were accurate if breast cancer was ER-positive (30% predicted sensitive, DRFS 97%, CI 91–100; ARR 11%, CI 0.1–21) or ER-negative (26% predicted sensitive, DRFS 83%, CI 68–100; ARR 26%, CI 4–28), and were significant in multivariate analysis after adjusting for relevant clinical-pathologic characteristics. Other genomic predictors showed paradoxically worse survival if predicted to be responsive to chemotherapy.
CONCLUSION
A genomic predictor combining ER status, predicted chemo-resistance, predicted chemo-sensitivity, and predicted endocrine sensitivity accurately identified patients with survival benefit following taxane-anthracycline chemotherapy.
INTRODUCTION
There is clinical need for predictive tests for patients with newly diagnosed, HER2-negative breast cancer whose clinical-pathologic risk at presentation favors the use of chemotherapy.1 Accurate identification of those with high likelihood of survival following a current standard chemotherapy regimen (then endocrine therapy, if estrogen receptor-positive) would reassure that treatment decision. Conversely, accurate identification of those with significant risk of relapse despite standard chemotherapy could be used to advise participation in an appropriate clinical trial. Also, since neoadjuvant (pre-operative) and adjuvant (postoperative) chemotherapy are equally effective,2 the former provides a clinical model for development of chemo-predictive tests.
Inherent chemo-sensitivity of breast cancers differs according to phenotype, as defined by combined estrogen receptor (ER) and HER2 receptor status.3, 4 However, breast cancer of any phenotype that achieves pathologic complete response (pCR) following a neoadjuvant chemotherapy regimen, has an excellent probability of long-term survival.2, 5, 6 Unfortunately, molecular tests that were specifically developed to predict pCR have not demonstrated any predictive superiority over the combination of standard clinicopathologic parameters (ER status, grade, and age) and have not been compared to a survival endpoint.7–14 Similarly, tests that were designed for molecular classification (including phenotype) or prognosis without chemotherapy, have failed to predict a sufficiently high probability of survival in the patients they classify as chemosensitive.15–19 Additionally, there is currently no clinically useful prognostic or predictive test for patients with ER−/HER2− breast cancer.13, 14, 19
This study addresses our hypothesis that a predictive test for response and survival following sequential taxane-anthracycline chemotherapy for HER2−negative breast cancer would account for each of the following biological characteristics (Figure 1): tumor phenotype (ER+/HER2− or ER−/HER2−), sensitivity to adjuvant endocrine therapy (if ER+), chemo-resistance (extensive residual cancer burden (RCB) or early relapse), and chemo-sensitivity (pCR or minimal RCB).20, 21
Figure 1.
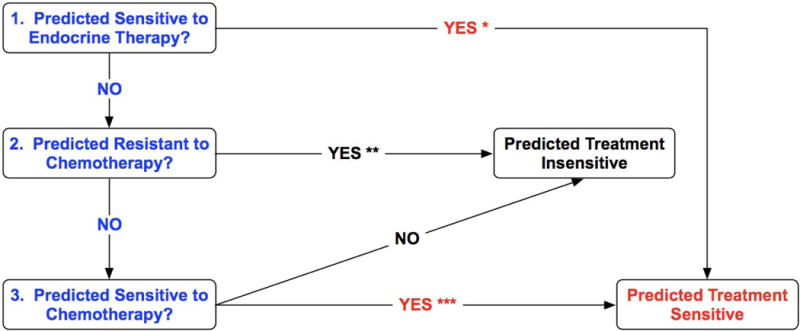
The decision algorithm that was used in the genomic test to predict a patient’s sensitivity to adjuvant chemotherapy or chemo-endocrine therapy from a biopsy of newly diagnosed invasive breast cancer. *, predicted sensitivity to endocrine therapy was defined as high or intermediate genomic sensitivity to endocrine therapy (SET) index; **, predicted resistance to chemotherapy was defined as predicted extensive residual cancer burden (RCB-III) or predicted distant relapse or death within 3 years of diagnosis; ***, predicted sensitivity to chemotherapy was defined as predicted pathologic complete response (pCR) or minimal residual cancer burden (RCB-I).20, 21
METHODS
Patients and Samples
Patients prospectively consented to a research biopsy by fine needle aspiration (FNA) or core biopsy (CBX) prior to any systemic therapy, and to the future assessment of pathologic response and/or survival endpoints.20, 22 Detailed characteristics of the patient and biospecimen cohorts used for test development and validation are provided in Table 1 and Figure 2. In the discovery cohort, there were 227 FNAs (MDACC) and 83 CBXs (I-SPY), and all chemotherapy was administered as neoadjuvant treatment. In the validation cohort, there were 157 FNAs (MDACC, Peru, USO) and 41 CBXs (MDACC, LBJ, Spain), and 165 of 198 patients received all chemotherapy as neoadjuvant treatment. Clinical nodal status was determined before treatment from physical examination, with or without axillary ultrasound, with diagnostic FNA as required. Pathologic HER2 status was defined as negative according to the ASCO/CAP guidelines.23 Details of the type of biopsies collected for genomic analysis and the chemotherapy treatments administered are provided in Table 2. Patients with any nuclear immunostaining for ER in the tumor cells were considered as eligible for adjuvant endocrine therapy.
Table 1.
Pre-treatment Characteristics of the Discovery and Validation Populations
| Discovery Population | Validation Population | ||||||
|---|---|---|---|---|---|---|---|
| MDACC | I-SPY | Total | MDACC | LBJ/IN/GEI | USO | Total | |
| Patients | 227 | 83 | 310 | 86 | 58 | 54 | 198 |
| Age | |||||||
| <= 50 | 112 (49%) | 30 (36%) | 142 (46%) | 48 (56%) | 30 (52%) | 31 (57%) | 109 (55%) |
| >50 | 115 (51%) | 53 (64%) | 168 (54%) | 38 (44%) | 28 (48%) | 23 (43%) | 89 (45%) |
| Mean (SD) | 51 (11) | 47 (8) | 50 (10) | 49 (11) | 51 (11) | 48 (9) | 49 (11) |
| Nodal status | |||||||
| Pos | 165 (73%) | 58 (70%) | 223 (72%) | 52 (60%) | 42 (72%) | 34 (63%) | 128 (65%) |
| Neg | 62 (27%) | 25 (30%) | 87 (28%) | 34 (40%) | 16 (28%) | 20 (37%) | 70 (35%) |
| T stage | |||||||
| 0 | 2 (1%) | – | 2 (1%) | 1 (1%) | – | – | 1 (1%) |
| 1 | 19 (8%) | 1 (1%) | 20 (6%) | 8 (9%) | 1 (1%) | 1 (2%) | 10 (5%) |
| 2 | 131 (58%) | 34 (41%) | 165 (53%) | 52 (61%) | 19 (33%) | 19 (35%) | 90 (45%) |
| 3 | 35 (15%) | 39 (47%) | 74 (24%) | 18 (21%) | 19 (33%) | 34 (63%) | 71 (36%) |
| 4 | 40 (18%) | 9 (11%) | 49 (16%) | 7 (8%) | 19 (33%) | – | 26 (13%) |
| Grade | |||||||
| 1 | 13 (6%) | 6 (7%) | 19 (6%) | 7 (8%) | 5 (8%) | 1 (2%) | 13 (7%) |
| 2 | 92 (40%) | 25 (30%) | 117 (38%) | 28 (33%) | 19 (33%) | 16 (30%) | 63 (32%) |
| 3 | 122 (54%) | 29 (35%) | 151 (49%) | 51 (59%) | 23 (40%) | 34 (63%) | 108 (54%) |
| Unknown | – | 23 (28%) | 23 (7%) | – | 11 (19%) | 3 (5%) | 14 (7%) |
| AJCC Stage | |||||||
| I | 6 (3%) | – | 6 (2%) | 2 (2%) | 0 | 0 | 2 (1%) |
| II | 126 (55%) | 39 (47%) | 165 (53%) | 57 66%) | 18 (31%) | 32 (59%) | 107 (54%) |
| III | 95 (42%) | 44 (53%) | 139 (45%) | 27 (32%) | 40 (69%) | 22 (41%) | 89 (45%) |
| ER Status | |||||||
| Pos | 131 (58%) | 43 (52%) | 174 (56%) | 60 (70%) | 37 (64%) | 27 (50%) | 124 (63%) |
| Neg | 96 (42%) | 35 (42%) | 131 (42%) | 26 (30%) | 21 (36%) | 27 (50%) | 74 (37%) |
| Indeterminate | – | 5 (6%) | 5 (2%) | – | – | – | – |
| PR Status | |||||||
| Pos | 102 (45%) | 40 (48%) | 142 (46%) | 43 (50%) | 31 (53%) | 28 (52%) | 102 (52%) |
| Neg | 125 (55%) | 37 (45%) | 162 (52%) | 43 (50%) | 27 (47%) | 26 (48%) | 96 (48%) |
| Indeterminate | – | 6 (7%) | 6 (2%) | – | – | – | – |
MDACC, M.D. Anderson Cancer Center; I-SPY, I-SPY-1 clinical trial; LBJ, Lyndon B. Johnson Hospital; IN, Instituto Nacional de Enfermedades Neoplásicas (INEN); GE, Grupo Español de Investigación en Cáncer de Mama (GEICAM); USO, US Oncology.
Figure 2.
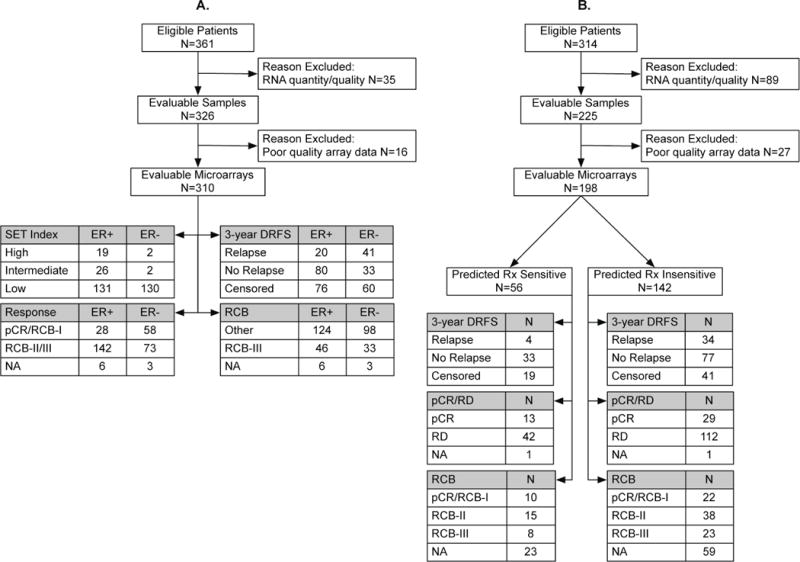
Flow chart of biospecimen accrual and testing in the discovery cohort (A) and validation cohort (B).
Table 2.
Chemotherapy And Pre-treatment Biopsy Details for the Study Cohorts
| Discovery Cohort (N=310) |
Validation Cohort (N=198) |
|
|---|---|---|
| Needle Biopsy for Genomic Testing | ||
| FNA | 227 | 157 |
| CBX | 83 | 41 |
| Chemotherapy Regimen | ||
| Entirely Neoadjuvant | ||
| T × 12 → FAC × 4 → Sx | 227 | 73 |
| AC × 4 → T/Tx × 4 → Sx | 83* | – |
| TxX × 4 → FEC × 4 → Sx | – | 92 |
| Partial Neoadjuvant | ||
| FAC/FEC × 6 → Sx → T × 12 | – | 18 |
| Entirely Adjuvant | ||
| Sx → T × 12 → FAC/FEC × 4 | – | 12 |
| Sx → TxX × 4 → FEC × 4 | – | 2 |
| Sx → Tx × 4 → FEC × 4 | – | 1 |
FNA: fine needle aspiration; CBX: core needle biopsy; T × 12: 12 weekly doses of paclitaxel; FAC/FEC × 4: four cycles of fluorouracil (F), doxorubicin (A) or epirubicin (E), and cyclophosphamide (C); AC × 4: four cycles of doxorubicin and cyclophosphamide; T/Tx × 4: four cycles of paclitaxel or docetaxel (Tx); TxX × 4: four cycles of docetaxel with capecitabine (X); Tx × 4: four cycles of docetaxel; Sx: surgery.
Four cycles of AC followed by T (N=60), Tx (N=18) or taxane not defined (N=5).
All gene expression microarrays were performed in the Department of Pathology at MDACC. Biopsy samples were either collected in 1.5 ml RNAlater™ (Qiagen, Valencia, CA) and stored locally at −70°C and transported to the laboratory on dry ice (MDACC, INEN, LBJ, GEICAM) or couriered overnight in a cooler pack from clinics to the laboratory (USO), or were frozen, cryosectioned and an aliquot of RNA sent to the laboratory on dry ice (I-SPY). Details of our methods for RNA purification and microarray hybridization have been reported previously.9, 21, 24–27 Briefly, a single-round T7 amplification was used to generate biotin-labeled cRNA for hybridization to oligonucleotide microarrays (U133A GeneChip™, Affymetrix, Santa Clara, CA). Details on microarray data processing are provided in the Supplemental Appendix.
Identification of Predictive Signatures For Excellent Pathologic Response and For Extensive Residual Disease
Differentially expressed probe sets in two responder groups: pCR or minimal residual cancer burden (RCB-I) defining excellent response, versus moderate or extensive residual cancer burden (RCB-II/III) defining partial response20 were identified separately in ER+/HER2− and ER−/HER2− training cases using a robust unequal variance t-statistic under a bootstrap scheme. The 209 and 244 probe sets that were significant in at least 30% of the bootstrap replicates in the two cohorts were selected as candidates. Subsequently, a multivariate penalized optimization algorithm, gradient directed regularization, was then used with maximum penalization to select a minimal signature that maximized the area under the ROC curve (AUC) under complete cross-validation.28 The final response predictors used 39 and 55 probe sets for the ER+/HER2− and ER−/HER2− cohorts respectively. Risk scores calculated as the weighted sum of the standardized log2-transformed expression signal of the signature probe sets were dichotomized at zero for both cohorts to predict “responders” (positive scores) or “non-responders” (negative scores).
A similar procedure was followed to develop the predictor for resistance by comparing patients with extensive residual disease (RCB-III) after neoadjuvant chemotherapy treatment versus remaining patients. The final predictor of extensive residual disease used 73 and 54 probe sets for ER+/HER2−and ER−/HER2− subsets respectively (Supplemental Appendix).
Identification of Predictive Signature For Early Relapse Events
To identify this signature of chemo-resistance we included higher risk patients who were clinically lymph node positive at presentation and also predicted to have low sensitivity to adjuvant endocrine therapy21 (SET Low; Figure 1). Probe sets were evaluated in univariate Cox regression analyses under bootstrap, separately in ER+/HER2− and ER−/HER2− training cases, to assess their association with distant relapse or death. A total of 235 and 268 probe sets were deemed significant in at least 20% of the bootstrap replicates in the two cohorts and were selected as candidates for subsequent multivariate Cox regression modeling. Minimal nonredundant signatures were obtained through a univariate shrinkage approach,29 with optimal penalization determined under cross-validation to yield the shortest probe set list that resulted in the biggest incremental improvement in the AUC for predicting 3-year DRFS outcome. The final predictors used 33 and 27 probe sets for the ER+/HER2−and ER−/HER2−subsets respectively. Risk scores calculated as the weighted sum of the standardized log2-transformed expression signal of the signature probe sets were dichotomized using cut points that maximized the predictive accuracy of 3-year distant relapse outcomes in the discovery cohort under cross validation to predict “high-risk” or resistant, versus “low-risk” or non-resistant cases (Supplemental Appendix).
Development of The Predictive Testing Algorithm
We combined the individual predictions into a testing algorithm for predicted sensitivity to adjuvant treatment of HER2-negative breast cancer with taxane-anthracycline chemotherapy: 1) sensitivity to endocrine therapy assessed based on an independently validated 165-gene index of endocrine sensitivity (high or intermediate SET index)21; 2) resistance to chemotherapy predicted either by early distant relapse events or by extensive residual disease after neoadjuvant chemotherapy; and 3) sensitivity (pathologic response) to chemotherapy (Figure 1). Additional methodological details are provided in the Supplemental Appendix.
Comparison With Other Genomic Predictors
We evaluated other published phenotypic predictors reported to be associated with chemotherapy response to neoadjuvant chemotherapy, that have pre-defined thresholds for response classification, are based on Affymetrix microarray data, and we have confirmed to be correctly calculated in our hands: genomic grade index (GGI), intrinsic subtype (PAM50), and a genomic predictor of pCR (DLDA30).9, 19, 30
Statistical Analysis
Distant relapse-free survival (DRFS) was defined as the interval from initial diagnostic biopsy until diagnosis of distant metastasis or death from any cause.31 The primary prediction endpoint was DRFS at 3 years (the median follow-up for validation cohort). Predictive performance was assessed by the positive predictive value (PPV) defined as the probability of distant relapse or death if predicted treatment insensitive, the negative predictive value (NPV) defined as the DRFS if predicted treatment sensitive, and the absolute risk reduction (ARR), defined as the absolute difference in DRFS between the two predicted groups. These were calculated from the Kaplan-Meier estimators of the survival function based on cumulative events following the interval notion for cases and controls.32 Confidence intervals for NPV and PPV were based on Greenwood’s variance estimate and for ARR were estimated by bootstrap using 999 replicates.33 The independent prognostic value of the genomic predictor compared to the full clinical model was assessed in multivariate Cox regression analyses using the likelihood ratio test. Pathologic response to neoadjuvant chemotherapy was defined as pCR for DLDA30 and pCR or RCB-I for GGI, PAM50, and the new predictors.9, 19, 30 Statistical computations were performed in R (v. 2.10.1, R Development Core Team, 2009, Vienna, Austria).
RESULTS
Performance of the Predictive Test in the Independent Validation Cohort
The predictive test (algorithm) was applied to the discovery cohort of 310 samples (Figure 3A) and then evaluated in the independent validation cohort of 198 patients (99% clinical Stage II–III) who received sequential taxane-anthracycline chemotherapy then endocrine therapy (if ER+). The validation cohort had a pathologic response rate of pCR 25% and of pCR or RCB-I 30%, median follow up of 3 years, and an average 3-year baseline DRFS of 79% (95%CI 74 to 85).
Figure 3.
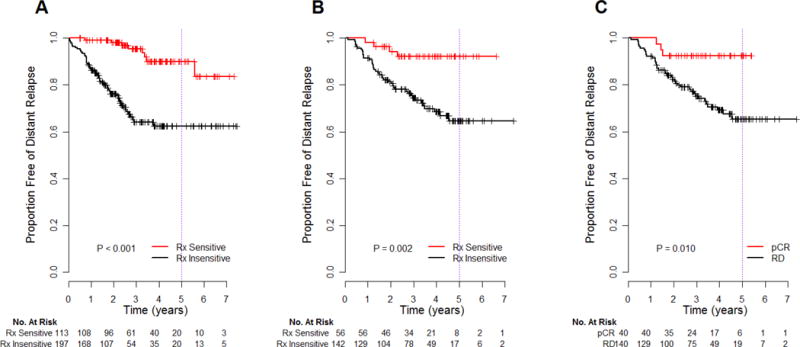
Kaplan-Meier estimates of distant relapse-free survival according to genomic predictions (before treatment) as treatment-sensitive (Rx Sensitive) or treatment-insensitive (Rx Insensitive) in the discovery (A) and independent validation (B) cohorts. For comparison, the prognosis of the groups stratified by actual pathologic response (pathologic complete response vs residual disease) after completion of all chemotherapy is shown for the validation cohort (C). P-values are from the log-rank test.
The 3-year DRFS (NPV) was 92% (95%CI 85 to 100), and there was significant absolute risk reduction (ARR) of 18% (95%CI 6 to 28), in 28% of patients who were predicted to be treatment-sensitive (Table 3). The 3-year point estimate of DRFS for those predicted to be treatment-insensitive was 75% (95%CI 67 to 82). Overall, we observed a significant association between predicted sensitivity to treatment and DRFS (p = 0.002; Figure 3B). In 91 tumors with low SET and evaluated for RCB, excellent response from chemotherapy (pCR or RCB-I) was observed in 56% (95%CI 31 to 78) of those predicted to be treatment-sensitive.
Table 3.
Test Performance in Validation Cohort at 3 Years of Follow Up
| Predicted Cohort | Performance Characteristics | |||||
|---|---|---|---|---|---|---|
|
| ||||||
| Population | N | Baseline 3-yr DRFS | % Predicted Rx Sensitive | % NPV (DRFS) | % PPV | % ARR |
| All Patients | 198 | 79 (74–85) | 28 | 92 (85 – 100) | 25 (18 – 33) | 18 (6 – 28) |
| ER+ | 124 | 89 (83–95) | 30 | 97 (91 – 100) | 14 (6 – 21) | 11 (0.1 – 21) |
| ER− | 74 | 64 (53–76) | 26 | 83 (68 – 100) | 43 (28 – 55) | 26 (4 – 48) |
Estimates of negative predictive value (NPV), i.e. 3-year distant relapse-free survival (DRFS), positive predictive value (PPV) and absolute risk reduction (ARR) of predicted treatment (Rx) sensitive patients compared to predicted treatment-insensitive patients at 3 years (median) follow up (95% confidence interval bounds in parentheses). The results are shown for all patients and for relevant subsets according to positive (ER+) or negative (ER−) pathological estrogen receptor status of the tumor.
Of note, 3-year DRFS in patients predicted to be treatment-sensitive at the time of diagnosis was similar to the 3-year DRFS of 93% (95%CI 85 to 100) in the 21% of patients in the validation cohort who achieved pathologic complete response (pCR) after completion of neoadjuvant chemotherapy. Also, 3-year DRFS for predicted treatment-insensitive was identical to the 3-year DRFS of 75% (95%CI 68 to 83) in those who had residual disease (RD) (Figure 3C). Furthermore, DRFS estimates for the predicted treatment-sensitive and the actual pCR groups were unchanged at 5 years, and were identical at 65% (95%CI 56 to 75) for the predicted treatment-insensitive and for the actual RD groups.
Predicted Treatment Sensitivity According to ER Status
There were 30% and 26% of patients with predicted sensitivity to treatment in the ER+/HER2− and ER−/HER2− subsets, respectively, and both had significantly favorable prognosis (Table 3 and Figure 4A–B). The treatment sensitive patients identified by test in the ER+/HER2− subset had excellent DRFS (NPV) of 97% (95%CI 91 to 100) and a significant ARR of 11% (95%CI 0.1 to 21) at 3 years of follow up (Table 3). In the low SET subset of ER+/HER2−, PPV for pathologic response was 42% (95%CI 15 to 72) in 20% who were predicted treatment-sensitive.
Figure 4.
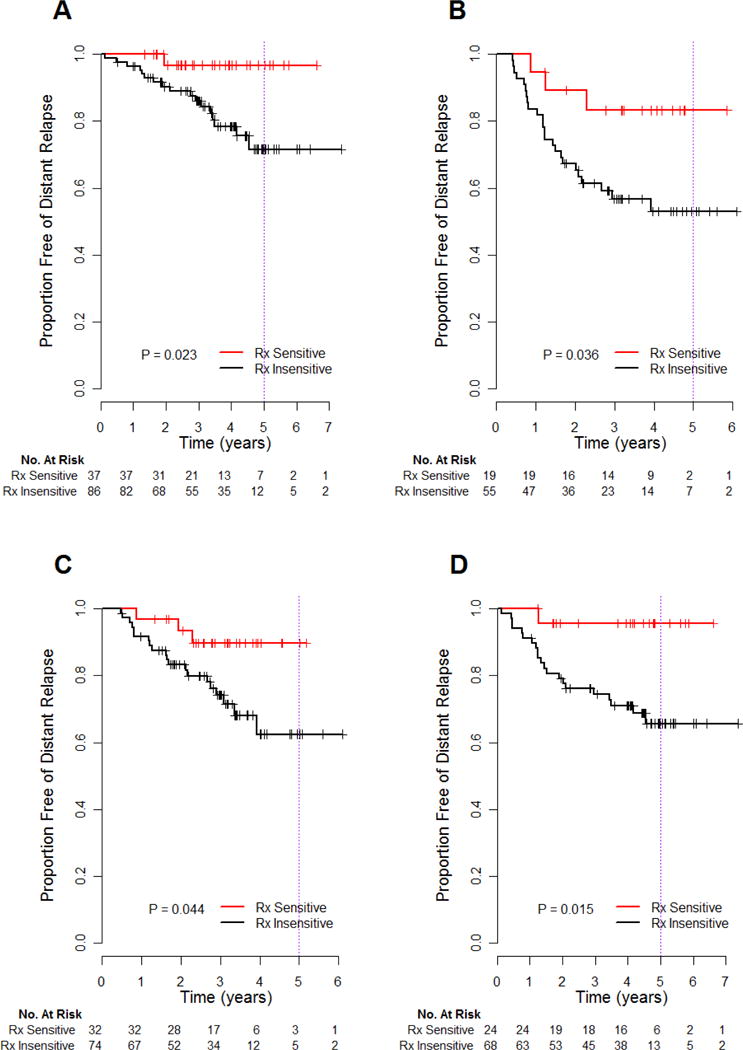
Subset analysis of genomic predictions in the validation cohort: ER+/HER2− (A), ER−/HER2−(B), taxane chemotherapy administered as 12 cycles of weekly paclitaxel (C) or 4 cycles of 3-weekly docetaxel (D). P-values are from the log-rank test.
For ER−/HER2− patients, the PPV for 3-year relapse was 43% (95%CI 28 to 55) if predicted treatment-insensitive (Table 3). Patients predicted to be treatment-sensitive had considerably improved 3-year DRFS (NPV 83% (95%CI 68 to 100)) and significant ARR of 26% (95%CI 4 to 48) overall, and PPV for pathologic response of 83% (95%CI 36–100).
Performance of the Predictive Test in Other Relevant Subsets
The association between predicted treatment sensitivity and DRFS appears to be unrelated to the type of taxane therapy administered (Figure 4C–D). The 3-year DRFS was 90% (95%CI 80 to 100) in the subset who received 12 cycles of weekly paclitaxel, and 96% (95% CI 88 to 100) for 4 cycles of 3-weekly docetaxel with capecitabine. Also, the 3-year DRFS was 93% (95% CI 84 to 100) in 128 clinically node-positive patients, with significantly improved DRFS compared to those predicted to be insensitive (p=0.003). The 3-year DRFS was 91% (95% CI 81 to 100) in 70 clinically node-negative patients, but was not significantly different from predicted insensitivity.
Comparison of the Predictive Test with Clinical-Pathologic Parameters
Genomic predictions were independently and significantly associated with risk of distant relapse or death (sensitive versus insensitive; HR 0.19; 95%CI 0.07 to 0.55; p=0.002), after adjusting for standard clinical-pathologic parameters (Table 4). Addition of the genomic prediction to a multivariate Cox model of the clinical-pathologic factors significantly increased the model’s predictive utility (likelihood ratio of complete model versus clinical model 13.8, p < 0.001). In this model, higher clinical tumor stage (tumor stage T3 or T4 versus T1 or T2; HR 2.13; 95% CI 1.13 to 4.02; p=0.02) and ER-negative status (ER status positive versus negative; HR 0.34; 95% CI 0.18 to 0.65; p=0.001) were associated with statistically significant greater risk of distant relapse or death.
Table 4.
Multivariate Cox Regression Analysis of Association with DRFS
| Validation Cohort (N=183)* | |||
|---|---|---|---|
|
| |||
| Factor | Hazard Ratio | 95% CI | P value |
| Age (>50 vs ≤50) | 0.53 | 0.27 – 1.03 | 0.062 |
| Clinical Nodal Status (pos vs neg) | 1.76 | 0.84 – 3.67 | 0.134 |
| Clinical Tumor Stage (T3 or T4 vs T1 or T2) | 2.13 | 1.13 – 4.02 | 0.020 |
| Histologic Grade (3 vs 1 or 2) | 0.64 | 0.32 – 1.28 | 0.206 |
| ER Status (IHC positive vs negative) | 0.34 | 0.18 – 0.65 | 0.001 |
| Taxane (docetaxel vs paclitaxel) | 0.92 | 0.49 – 1.73 | 0.796 |
| Prediction (Rx Sensitive vs Insensitive) | 0.19 | 0.07 – 0.55 | 0.002 |
Fifteen cases were excluded from the multivariate analysis due to incomplete data. Likelihood ratio test for the addition of Genomic Prediction to the model was 13.8 on one degree of freedom, p = 0.0002.
The Hazard Ratio is a measure of the risk of distant relapse or death; vs, versus; ER, estrogen receptor.
Comparison with Other Predictive Genomic Signatures
Table 5 summarizes the performance of these signatures in the discovery and validation study cohorts for predicting pathologic response and 3-year DRFS following neoadjuvant taxane-anthracycline chemotherapy. All tests were significantly predictive of pathologic response in the discovery cohort (lower 95% confidence limit of the PPV greater than the baseline response rate) with overall pCR rate of 19% and pCR or RCB-I rate of 29%, and the tests had NPV of 84% or greater. Predictor performance in the validation cohort was similar, but not all tests had PPV and NPV that was significantly greater than the baseline response rates (pCR rate of 25% and pCR or RCB-I rate of 30%). The entire predictive test algorithm had PPV of 56% (95%CI 31 to 78) for pathologic response prediction in the validation cohort (Table 5) after excluding patients with predicted endocrine sensitivity (high or intermediate SET).
Table 5.
Performance of Genomic Signatures for Predicting Pathologic Response and 3-year DRFS
| Prediction of Pathologic Response | Prediction of Distant Relapse or Death Within 3 Years | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||||
| Discovery Cohort | Validation Cohort | Discovery Cohort (N=310) | Validation Cohort (N=198) | |||||||||
|
| ||||||||||||
| Predictor | N (%Resp) | %PPV | % NPV | N (%Resp) | %PPV | % NPV | %NPV (DRFS) | %PPV | %ARR | %NPV (DRFS) | %PPV | %ARR |
| Genomic Grade Index (High) |
301 (29) |
36 (30 to 43) |
88 (79 to 93) |
101 (30) |
40 (28 to 54) |
84 (70 to 93) |
72 (65 to 79) |
14 (6 to 22) |
−14 (−25 to −3) |
72 (64 to 81) |
7 (1 to 13) |
−21 (−30 to −10) |
| Genomic Subtype Classifier (Luminal B or Basal-like) |
301 (29) |
40 (32 to 48) |
85 (78 to 90) |
101 (30) |
40 (25 to 56) |
78 (65 to 87) |
66 (58 to 76) |
13 (7 to 19) |
−20 (−31 to −10) |
72 (63 to 81) |
12 (5 to 19) |
−16 (−28 to −5) |
| Genomic Predictor of pCR § |
306 (19)* |
36 (28 to 46) |
93 (88 to 96) |
162 (25)* |
34 (23 to 47) |
82 (72 to 89) |
62 (52 to 73) |
15 (9 to 20) |
−24 (−36 to −12) |
62 (51 to 74) |
10 (4 to 15) |
−28 (−41 to −14) |
| ER-stratified Genomic Predictor of pCR/RCB-I § |
301 (29) |
69 (60 to 77) |
100 (98 to 100) |
101 (30) |
42 (28 to 57) |
81 (68 to 91) |
85 (78 to 93) |
30 (22 to 37) |
15 (4 to 25) |
82 (74 to 90) |
24 (14 to 32) |
5 (−7 to 16) |
| Predictive Test (Rx Sensitive) §¶# |
256 (31) |
78 (66 to 88) |
84 78 to 89) |
91 (33) |
56 (31 to 78) |
73 (61 to 82) |
95 (91 to 100) |
36 (27 to 44) |
31 (22 to 41) |
92 (85 to 100) |
25 (18 to 33) |
18 (6 to 28) |
N, number or patients evaluated; %, percent; Resp, pathologic response rate; PPV, positive predictive value; NPV, negative predictive value; DRFS, distant relapse-free survival estimate at 3 years; ARR, absolute risk reduction for event within 3 years if predicted to be treatment-sensitive (−, any negative risk reduction was in favor of predicted treatment-insensitive). The 95% confidence intervals (parentheses) for PPV and NPV for prediction of pathologic response were based on binomial approximation.
actual pathologic response was defined as pCR, rather than pCR or RCB-I.
Performance of the pCR predictor on the discovery cohort is optimistically biased because the predictor was trained on a subset of these samples. Performance of the pCR/RCB-I predictor and of the overall genomic prediction test on the discovery cohort represents resubstitution performance, since these predictors were trained on the same cohort.
Genomic prediction of pathologic response was evaluated in the SET-Low subset in both cohorts.
Performance of the predictive test is optimistically biased in the discovery cohort because a component of the test was trained on DRFS events to define resistance.
The DRFS of the predicted responder and non-responder groups, as predicted by the same signatures, were evaluated for the discovery and validation cohorts and are shown in Figure 5. Only the ER-stratified predictor of excellent pathologic response (Figure 5D,5H) and the entire prediction algorithm (Figures 1,3A,3B) demonstrated significantly better DRFS for patients predicted to be treatment-sensitive (Table 5). Other tests (GGI, intrinsic subtype, DLDA30) demonstrated worse DRFS for patients predicted to have chemosensitive breast cancer, as indicated by the negative ARR (Figure 5, Table 4).
Figure 5.
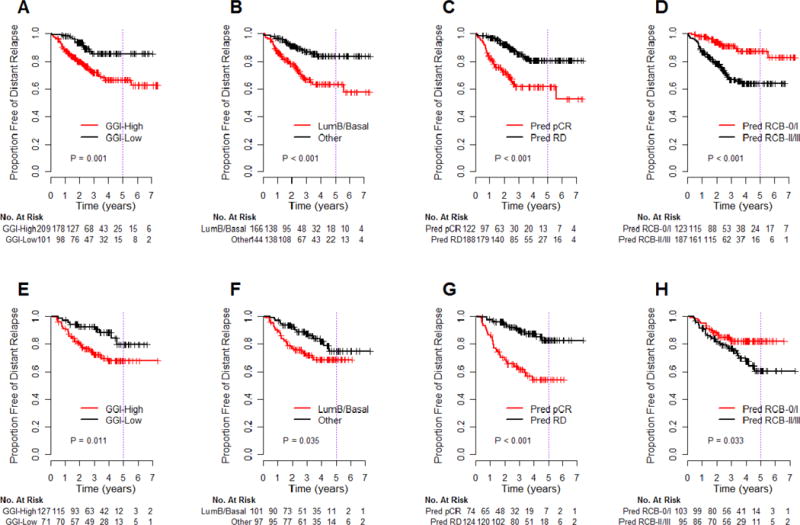
Kaplan-Meier estimates of distant relapse-free survival in the discovery cohort (A–D) and the independent validation cohort (E–H) of patients treated with sequential taxane-anthracycline chemotherapy, then endocrine therapy if hormone receptor-positive, stratified by other signatures reported to be predictive of response to neoadjuvant taxane-anthracycline chemotherapy.9, 19, 30 A prognostic signature for genomic grade index predicts pathologic response if high GGI versus low GGI (A, E)19; the intrinsic subtype classifier predicts pathologic response if basal-like or luminal B versus other subtypes (B, F)30; a genomic predictor of pathologic complete response (pCR) versus residual disease following taxane-anthracycline chemotherapy (C, G)9; and the genomic predictor of excellent pathologic response (pCR or RCB-I) versus other residual disease, according to ER status, that we incorporated in the last step of our prediction algorithm (D, H).9, 19, 30 P-values are from the log-rank test.
COMMENT
Any test that is based on predicted sensitivity and/or resistance to guide the selection of a standard adjuvant treatment regimen should predict a high probability of survival for patients predicted to be treatment-sensitive (NPV, no relapse if predicted to be treatment-sensitive), a clinically meaningful survival difference between predicted treatment-sensitive and insensitive patients (ARR), and improve upon predictions using existing clinical-pathological information. The performance of our predictive test meets these criteria in an independent validation cohort. The 3-year DRFS (NPV) of 92% was higher than in the unselected cohort (79%), and there was significant ARR of 18%. Furthermore, the predictive test added significantly to a multivariate clinical-pathologic model (age, tumor size, nodal status, grade, ER status, and type of taxane administered), wherein patients who were predicted as treatment-sensitive had a 5-fold reduction in the risk of distant relapse (Table 4). It should also be noted that in the validation cohort the a priori test results (predicted treatment-sensitive or insensitive) from a tumor sample obtained before treatment (Figure 3B) were as predictive of DRFS as the pathologic response assessed after the completion of chemotherapy (Figure 3C).
We observed similar performance of this test in patients who received equivalent chemotherapy regimens containing 12 weekly paclitaxel doses or four cycles of docetaxel with capecitabine, in each case sequentially administered before or after four cycles of anthracycline-based chemotherapy.34 Of course, additional studies must address the reproducibility of these results, and other studies should consider whether this predictive test might be generalizable to other regimens that combine taxanes and anthracyclines in sequence or concurrently, or even regimens that do not include a taxane or an anthracycline component. Notably, this chemopredictive test was not prognostic in available data from patients who did not receive chemotherapy (Supplemental Appendix).
A predictive test with this performance could assist medical decision-making. In particular, we could identify patients with Stage II–III, ER+/HER2− breast cancer with excellent 3-year and 5-year DRFS (97%) following a standard adjuvant treatment (Table 3, Figure 4A). This group included the subset of predicted treatment-sensitive patients that were also predicted to have low endocrine-sensitivity (Supplemental Appendix). Similarly, the predicted treatment-sensitive subset with ER−/HER2− cancers had DRFS of 83% and significant ARR of 26% (Table 3, Figure 4B). At issue is whether 83% 3-year DRFS for ER−/HER2− breast cancers is sufficiently good to use this test to choose a currently standard taxane-anthracycline chemotherapy regimen. One clinical strategy might be to perform the test on needle biopsy samples obtained before treatment, and select predicted treatment-sensitive patients for neoadjuvant taxane-anthracycline chemotherapy. Using that approach, additional post-operative adjuvant therapy, preferably on a clinical trial, could be considered if the patient proved to have significant residual disease (RCB-II or RCB-III) at the time of surgery (17% of evaluable cases in our study). Conversely, the PPV for distant relapse (probability of relapse if predicted treatment-insensitive) was 43% at 3 years, sufficiently high to encourage consideration of other therapeutic options, including a clinical trial, for those patients.
The future ability to effectively prioritize and complete prospective clinical trials in breast oncology may be challenged by an increasing number of new treatments to test, questions of synergy in combined treatments, questions of efficacy in biological subsets, and generally low rates of participation in prospective clinical trials of adjuvant treatments.35 It is relevant to consider whether more patients would choose to participate in a clinical trial, from which they might benefit, if they knew in advance that they personally had significant probability of early relapse following a current and intensive adjuvant treatment strategy. Furthermore, any increase in the rate of participation in prospective clinical trials, especially trials that target therapeutic strategies for patients predicted to be insensitive to current standard treatments, could profoundly accelerate clinical investigation of adjuvant treatments.35
It is essential to realize that prediction of excellent pathologic response from neoadjuvant chemotherapy does not necessarily predict survival. This apparent paradox is due to the relationship between the biologic information captured by the predictor and the frequency and prognosis of false positive predictions. Tumors that are less differentiated often have high proliferation, and are generally more likely to: 1) have poor prognosis, 2) respond to cytotoxic chemotherapy, and 3) have poor prognosis if they do not respond to chemotherapy.1, 36 Therefore, tests that are prognostic in the absence of chemotherapy may predict response in higher risk patients, but without an associated high probability of survival. This is likely to be most apparent in Stage II–III breast cancers, as illustrated for genomic grade index and intrinsic subtype (Figure 5A–5B,5E–5F, Table 5),19 and in reports concerning the commercially available 70-gene prognostic signature (that uses a different microarray platform),18 and the recurrence score.15–17 These latter tests could not be directly compared in our study because each uses a different technology lacking direct correlation with our Affymetrix microarray platform. A similar paradox was observed for DLDA30, a predictor trained on pCR in unselected patients,9 and is probably related to different frequency of pCR according to ER status and grade, reflecting differentiation and proliferation (Table 5, Figures 5C,5G).36 We conclude that prediction of pathologic response alone is not sufficient to demonstrate clinical validity of a test. There must also be a survival advantage, with an appropriately high survival estimate, for patients who are predicted to be treatment sensitive.
In this study, overcoming the prediction paradox involved the incorporation of several strategic elements (Figure 1), some of which might also be generalizable to other types of cancer. One element was to identify patients whose excellent survival after chemo-endocrine therapy was due to the endocrine sensitivity of their breast cancer.21 That convinced us to evaluate predictors of chemotherapy response in breast cancers with predicted low SET. Another element was to separately develop predictors within relevant phenotypic subsets using an improved measure of pathologic response (pCR or RCB-I).20 That improvement is sufficient to reverse the prediction paradox, but not to a level of clinical utility (Figures 5D,5H, Table 5). Yet another element was to develop predictors based on clinically relevant definitions of resistance, so that resistant disease could be identified first to avoid misclassification as responsive.20 That further improved predictive accuracy (Figures 3,4, Table 5) to a level of potential clinical utility.
In its current format, this predictive test would be performed on fresh primary tumor sample obtained from a clinical core needle biopsy (2 cores), fine needle aspiration (2 passes) or surgical resection specimen (e.g. tumoral punch biopsy), and placed into a vial containing 1.5 ml of a standard RNA preservative (RNAlater™) at room temperature. Similar methods could become feasibly implemented into diagnostic practice if procurement of an optimal quality tumor sample became a priority for molecular diagnostic tests of proven accuracy and medical utility. Meanwhile, it is also imperative to continue to evaluate the predictive accuracy of this test in additional validation studies.
Supplementary Material
Acknowledgments
Study concept and design: Hatzis, Symmans.
Acquisition of samples or data: All.
Analysis and interpretation of data: Hatzis, Pusztai, Symmans.
Drafting of the manuscript: Hatzis, Symmans.
Critical revision of the manuscript for important intellectual content: All.
Statistical analysis: Hatzis.
Obtained funding: Pusztai, Esserman, Symmans.
Administrative, technical, or material support: Pusztai, Esserman, Hortobagyi, Symmans.
Study supervision: Pusztai, Symmans.
Final approval of the manuscript: All
Dr. Symmans had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis. This research was conducted with approval and waiver of consent from the respective Institutional Research Board (protocols LAB99-402, USO-02-103, 2003-0321, I-SPY-1). Datasets for this study and gene expression weights for signatures are accessible via the GEO repository (http://www.ncbi.nlm.nih.gov/geo/) under accession IDs GSE25066 (combined series), GSE25055 (discovery cohort), and GSE25065 (validation cohort).
This research was supported by grants from Susan G. Komen for The Cure (KG081680) to W.F.S.; the National Cancer Institute (1 R01 CA106290) to L.P.; the Breast Cancer Research Foundation to L.P. and W.F.S.; the Safeway Foundation to L.P. and W.F.S.; faculty funds from Y.G., Y.W., and W.F.S.; and generally by the Nellie B. Connally Breast Cancer Research Fund at MDACC.
Additionally, the ISPY-1 trial was supported, in part, by grants from the National Cancer Institute (CA31946) to the Cancer and Leukemia Group B (Monica M. Bertagnolli, M.D., Chair) and to the CALGB Statistical Center (Daniel J. Sargent, Ph.D., CA33601), and by grants to participating sites: Georgetown University Medical Center, Washington, DC (Minetta C. Liu, M.D., CA77597), Memorial Sloan-Kettering Cancer Center, New York, NY (Clifford A. Hudis, M.D., CA77651), University of California at San Francisco, San Francisco, CA (Charles J. Ryan, M.D., CA60138), University of Chicago, Chicago, IL (Hedy L Kindler, M.D., CA41287), University of Minnesota, Minneapolis, MN (Bruce A. Peterson, M.D., CA16450), University of North Carolina at Chapel Hill, Chapel Hill, NC (Thomas C. Shea, M.D., CA47559), University of Texas Southwestern Medical Center, Dallas, TX (Debasish Tripathy, M.D.). The content of this manuscript is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute.
The sponsors had no influence on the design and conduct of the study; collection, management, analysis, and interpretation of the data; and preparation, review, or approval of the manuscript.
The authors thank the patients who participated in this research and the following colleagues for technical laboratory work in the Department of Pathology: Feng Lin, M.S., Bin Zeng, M.S., Hongxia Sun, Ph.D., and Chunxiao Fu, Ph.D. (MDACC Breast Cancer Pharmacogenomics Laboratory); obtaining biopsy samples: Nour Sneige, M.D. (Cytopathology), or clinical data used in this work: Eva Carrasco (GEICAM), Patricia de los Rios (LBJ Hospital), Marc Lenburg (iSPY), Chingyi Young, M.S. and Anna Kratz (MDACC); and the Departments of Breast Imaging, Pathology, Surgery, and Breast Medical Oncology at M.D. Anderson Cancer Center, Houston, Texas.
Footnotes
The authors have the following potential conflicts of interest to disclose:
Hatzis: Employment (Nuvera Biosciences), Equity (Nuvera Biosciences), Patents
Pusztai: Unpaid scientific advisor (Nuvera Biosciences, uncompensated), Patents
Valero: None
Booser: None
Esserman: None
Lluch: None
Vidaurre: None
Holmes: Honoraria (Novartis, Genentech, Philips) with relevance outside the submitted work
Souchon: None
Martin: None
Cotrina: None
Gomez: Honoraria (Glaxo Smith Kline, Bristol Meyers Squibb) with relevance outside the submitted work
Hubbard: None
Chacón: None
Ferrer-Lozano: None
Dyer: None
Buxton: None
Gong: None
Wu: None
Ibrahim: None
Andreopoulou: None
Ueno: None
Hunt: None
Yang: None
Nazario: None
DeMichele: None
O’Shaughnessy: None
Hortobagyi: Paid consultant (Allergan, Genentech, Novartis, SanofiAventis), research funding (Novartis), with relevance outside the submitted work
Symmans: Unpaid scientific advisor (Nuvera Biosciences), Equity (Nuvera Biosciences), Patents, Honoraria (Agendia BV).
References
- 1.Sotiriou C, Pusztai L. Gene-expression signatures in breast cancer. N Engl J Med. 2009 Feb 19;360(8):790–800. doi: 10.1056/NEJMra0801289. [DOI] [PubMed] [Google Scholar]
- 2.Wolmark N, Wang J, Mamounas E, Bryant J, Fisher B. Preoperative chemotherapy in patients with operable breast cancer: nine-year results from National Surgical Adjuvant Breast and Bowel Project B-18. J Natl Cancer Inst Monogr. 2001;30:96–102. doi: 10.1093/oxfordjournals.jncimonographs.a003469. [DOI] [PubMed] [Google Scholar]
- 3.Hayes DF, Thor AD, Dressler LG, et al. HER2 and response to paclitaxel in node-positive breast cancer. N Engl J Med. 2007 Oct 11;357(15):1496–1506. doi: 10.1056/NEJMoa071167. [DOI] [PubMed] [Google Scholar]
- 4.Rouzier R, Perou CM, Symmans WF, et al. Breast cancer molecular subtypes respond differently to preoperative chemotherapy. Clin Cancer Res. 2005 Aug 15;11(16):5678–5685. doi: 10.1158/1078-0432.CCR-04-2421. [DOI] [PubMed] [Google Scholar]
- 5.Bear HD, Anderson S, Smith RE, et al. Sequential preoperative or postoperative docetaxel added to preoperative doxorubicin plus cyclophosphamide for operable breast cancer:National Surgical Adjuvant Breast and Bowel Project Protocol B-27. J Clin Oncol. 2006 May 1;24(13):2019–2027. doi: 10.1200/JCO.2005.04.1665. [DOI] [PubMed] [Google Scholar]
- 6.Liedtke C, Mazouni C, Hess KR, et al. Response to neoadjuvant therapy and long-term survival in patients with triple-negative breast cancer. J Clin Oncol. 2008 Mar 10;26(8):1275–1281. doi: 10.1200/JCO.2007.14.4147. [DOI] [PubMed] [Google Scholar]
- 7.Chang JC, Wooten EC, Tsimelzon A, et al. Gene expression profiling for the prediction of therapeutic response to docetaxel in patients with breast cancer. Lancet. 2003 Aug 2;362(9381):362–369. doi: 10.1016/S0140-6736(03)14023-8. [DOI] [PubMed] [Google Scholar]
- 8.Ayers M, Symmans WF, Stec J, et al. Gene expression profiles predict complete pathologic response to neoadjuvant paclitaxel and fluorouracil, doxorubicin, and cyclophosphamide chemotherapy in breast cancer. J Clin Oncol. 2004 Jun 15;22(12):2284–2293. doi: 10.1200/JCO.2004.05.166. [DOI] [PubMed] [Google Scholar]
- 9.Hess KR, Anderson K, Symmans WF, et al. Pharmacogenomic predictor of sensitivity to preoperative chemotherapy with Paclitaxel and Fluorouracil, Doxorubicin, and cyclophosphamide in breast cancer. J Clin Oncol. 2006 Sep 10;24(26):4236–4244. doi: 10.1200/JCO.2006.05.6861. [DOI] [PubMed] [Google Scholar]
- 10.Potti A, Dressman HK, Bild A, et al. Genomic signatures to guide the use of chemotherapeutics. Nat Med. 2006 Nov;12(11):1294–1300. doi: 10.1038/nm1491. [DOI] [PubMed] [Google Scholar]
- 11.Bonnefoi H, Potti A, Delorenzi M, et al. Validation of gene signatures that predict the response of breast cancer to neoadjuvant chemotherapy: a substudy of the EORTC 10994/BIG 00-01 clinical trial. Lancet Oncol. 2007 Dec;8(12):1071–1078. doi: 10.1016/S1470-2045(07)70345-5. [DOI] [PubMed] [Google Scholar]
- 12.Salter KH, Acharya CR, Walters KS, et al. An integrated approach to the prediction of chemotherapeutic response in patients with breast cancer. PLoS ONE. 2008;3(4):e1908. doi: 10.1371/journal.pone.0001908. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 13.Lee JK, Coutant C, Kim YC, et al. Prospective comparison of clinical and genomic multivariate predictors of response to neoadjuvant chemotherapy in breast cancer. Clin Cancer Res. 2010 Jan 15;16(2):711–718. doi: 10.1158/1078-0432.CCR-09-2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Popovici V, Chen W, Gallas BG, et al. Effect of training sample size and classification difficulty on the accuracy of genomic predictors. Breast Cancer Res. 2010 Jan 11;12(1):R5. doi: 10.1186/bcr2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gianni L, Zambetti M, Clark K, et al. Gene Expression Profiles in Paraffin-Embedded Core Biopsy Tissue Predict Response to Chemotherapy in Women With Locally Advanced Breast Cancer. J Clin Oncol. 2005 Sep 6;23:7265–7277. doi: 10.1200/JCO.2005.02.0818. [DOI] [PubMed] [Google Scholar]
- 16.Goldstein LJ, Gray R, Badve S, et al. Prognostic utility of the 21-gene assay in hormone receptor-positive operable breast cancer compared with classical clinicopathologic features. J Clin Oncol. 2008 Sep 1;26(25):4063–4071. doi: 10.1200/JCO.2007.14.4501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Albain KS, Barlow WE, Shak S, et al. Prognostic and predictive value of the 21-gene recurrence score assay in postmenopausal women with node-positive, oestrogen-receptor-positive breast cancer on chemotherapy: a retrospective analysis of a randomised trial. Lancet Oncol. 2010 Jan;11(1):55–65. doi: 10.1016/S1470-2045(09)70314-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Straver ME, Glas AM, Hannemann J, et al. The 70-gene signature as a response predictor for neoadjuvant chemotherapy in breast cancer. Breast Cancer Res Treat. 2010 Feb;119(3):551–558. doi: 10.1007/s10549-009-0333-1. [DOI] [PubMed] [Google Scholar]
- 19.Liedtke C, Hatzis C, Symmans WF, et al. Genomic grade index is associated with response to chemotherapy in patients with breast cancer. J Clin Oncol. 2009 Jul 1;27(19):3185–3191. doi: 10.1200/JCO.2008.18.5934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Symmans WF, Peintinger F, Hatzis C, et al. Measurement of residual breast cancer burden to predict survival after neoadjuvant chemotherapy. J Clin Oncol. 2007 Oct 1;25(28):4414–4422. doi: 10.1200/JCO.2007.10.6823. [DOI] [PubMed] [Google Scholar]
- 21.Symmans WF, Hatzis C, Sotiriou C, et al. A genomic index of sensitivity to endocrine therapy of breast cancer. J Clin Oncol. 2010 Sep 20;28:4111–4119. doi: 10.1200/JCO.2010.28.4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Symmans WF, Ayers M, Clark EA, et al. Total RNA yield and microarray gene expression profiles from fine-needle aspiration biopsy and core-needle biopsy samples of breast carcinoma. Cancer. 2003 Jun 15;97(12):2960–2971. doi: 10.1002/cncr.11435. [DOI] [PubMed] [Google Scholar]
- 23.Wolff AC, Hammond ME, Schwartz JN, et al. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for human epidermal growth factor receptor 2 testing in breast cancer. J Clin Oncol. 2007 Jan 1;25(1):118–145. doi: 10.1200/JCO.2006.09.2775. [DOI] [PubMed] [Google Scholar]
- 24.Gong Y, Yan K, Lin F, et al. Determination of oestrogen-receptor status and ERBB2 status of breast carcinoma: a gene-expression profiling study. Lancet Oncol. 2007 Mar;8(3):203–211. doi: 10.1016/S1470-2045(07)70042-6. [DOI] [PubMed] [Google Scholar]
- 25.Loi S, Haibe-Kains B, Desmedt C, et al. Definition of Clinically Distinct Molecular Subtypes in Estrogen Receptor-Positive Breast Carcinomas Through Genomic Grade. J Clin Oncol. 2007 Apr 1;25(10):1239–1246. doi: 10.1200/JCO.2006.07.1522. [DOI] [PubMed] [Google Scholar]
- 26.Desmedt C, Piette F, Loi S, et al. Strong time dependence of the 76-gene prognostic signature for node-negative breast cancer patients in the TRANSBIG multicenter independent validation series. Clin Cancer Res. 2007 Jun 1;13(11):3207–3214. doi: 10.1158/1078-0432.CCR-06-2765. [DOI] [PubMed] [Google Scholar]
- 27.Wang Y, Klijn JG, Zhang Y, et al. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet. 2005 Feb 19;365(9460):671–679. doi: 10.1016/S0140-6736(05)17947-1. [DOI] [PubMed] [Google Scholar]
- 28.Friedman JH, Popescu BE. Gradient directed regularization. Stanford University; 2004. [Google Scholar]
- 29.Tibshirani RJ. Univariate shrinkage in the cox model for high dimensional data. Stat Appl Genet Mol Biol. 2009;8(1) doi: 10.2202/1544-6115.1438. Article21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parker JS, Mullins M, Cheang MC, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol. 2009 Mar 10;27(8):1160–1167. doi: 10.1200/JCO.2008.18.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hudis CA, Barlow WE, Costantino JP, et al. Proposal for standardized definitions for efficacy end points in adjuvant breast cancer trials: the STEEP system. J Clin Oncol. 2007 May 20;25(15):2127–2132. doi: 10.1200/JCO.2006.10.3523. [DOI] [PubMed] [Google Scholar]
- 32.Moskowitz CS, Pepe MS. Quantifying and comparing the accuracy of binary biomarkers when predicting a failure time outcome. Stat Med. 2004 May 30;23(10):1555–1570. doi: 10.1002/sim.1747. [DOI] [PubMed] [Google Scholar]
- 33.Therneau TM, Grambsch PM. Modeling Survival Data: Extending the Cox Model. New York, NY: Springer-Verlag; 2000. [Google Scholar]
- 34.Sparano JA, Wang M, Martino S, et al. Weekly paclitaxel in the adjuvant treatment of breast cancer. N Engl J Med. 2008 Apr 17;358(16):1663–1671. doi: 10.1056/NEJMoa0707056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nass S, Moses H, Mendelsohn J, Committee . A National Cancer Clinical trials system for the 21st Century: reinvigorating the NCI Cooperative Group Program. Institute of Medicine of The National Academies; 2010. [PubMed] [Google Scholar]
- 36.Symmans WF. A Pathologist’s Perspective on Emerging Genomic Tests for Breast Cancer. Semin Oncol. 2007 Apr;34(2 Suppl 3):S4–9. doi: 10.1053/j.seminoncol.2007.03.016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


