Abstract
Background:
Hepatocyte nuclear factor 4 alpha (HNF4α) plays an important role in tumourigenesis. There is growing evidence indicating that HNF4α transcribed by promoter 1 (P1-HNF4α) is expressed at relatively low levels in HCC and its presence predicts a favourable outcome for hepatocellular carcinoma (HCC) patients. However, the role of HNF4α transcribed by promoter 2 (P2-HNF4α) in HCC remains unclear.
Methods:
A total of 615 HCC specimens were obtained to construct tissue microarrays and perform immunohistochemistry. The relationship between P2-HNF4α and clinical features of HCC patients were analysed. Kaplan–Meier analysis was conducted to assess the prognostic value of P2-HNF4α.
Results:
The results showed that the expression of P2-HNF4α in HCC was noticeably increased in HCC tissues compared with the nontumourous tissues. In addition, P1-HNF4α expression was negatively correlated with P2-HNF4α expression (p = 0.023). High P2-HNF4α expression was significantly associated with poor differentiation of HCC (p = 0.002) and vascular invasion (p = 0.017). Kaplan–Meier analysis showed that P2-HNF4α expression was closely correlated with overall survival in the training group (p = 0.01), validation group (p = 0.034), and overall group of patients with HCC (p < 0.001).
Conclusions:
Our data show that the role of HNF4α in cancer development needs to be further refined. P2-HNF4α, different from P1-HNF4α, is markedly upregulated and serves as an oncogene-associated protein in HCC. Our study therefore provides a promising biomarker for prognostic prediction and a potential therapeutic target for HCC.
Keywords: hepatocyte nuclear factor 4α, promoter 1, promoter 2, hepatocellular carcinoma, prognostic biomarker
Introduction
Hepatocyte nuclear factor 4 alpha (HNF4α), a highly conserved member of the nuclear receptor super family of ligand dependent transcription factors, is expressed primarily in liver, kidney, colon, and pancreatic β-cells.1 HNF4α is essential for adult and foetal liver function owing to its regulation of liver-specific gene expression.2 HNF4α-null embryos exhibit severe visceral endoderm defects preventing gastrulation and causing developmental failure.3
There are two differentially utilized promoters regulating HNF4α. During early hepatic development, HNF4α initiates from promoter 2 (P2). The major isoforms of the P2 promoter are HNF4 a7 to a9. During the differentiation of liver, promoter 1 (P1) is favoured for HNF4α transcription and gives rise to the isoforms of HNF4 (a1 to a6). P2 isoforms appear to activate genes involved in early liver development, such as α-fetoprotein (AFP) and transthyretin, and P1 isoforms appear to activate genes involved in hepatic differentiation, such as apoCIII.4,5
The initial evidence suggesting that HNF4α is involved in cancer development came from the observation that HNF4α expression decreases in cancers of multiple organs that normally express HNF4α. Analysis of renal cell carcinoma indicated a downregulation of HNF4α mRNA and protein expression along with suppression of HNF4α DNA-binding activity.6,7 Tanaka and colleagues reported that HNF4α expression is lost in colorectal carcinomas.8 In hepatocellular carcinoma (HCC), Mizuguchi and colleagues found that maintenance of a differentiated phenotype by HNF4α might inhibit hepatocyte proliferation in vitro.9 However, most studies focused on HNF4α isoforms transcribed by P1 (P1-HNF4α). There is no research focused on HNF4α isoforms transcribed by P2 (P2-HNF4α) and its potential effect on HCC.
Here, we investigated the role of P1- and P2-HNF4α isoforms in human HCC. We showed that in HCC, P1- and P2-HNF4α expression was correlated negatively. We also provided evidence indicating that P2-HNF4α expression was closely correlated with overall survival in patients with HCC. Collectively, our data show that the role of HNF4α in cancer development needs to be further refined. P2-HNF4α plays an oncogene-related protein role in HCC and therefore provides a promising biomarker for prognostic prediction and is a potential therapeutic target in HCC clinical management.
Methods
Patients
A total of 615 paraffin-embedded HCC specimens between January 2000 and December 2010 were obtained from the archives of the Department of Pathology of the Sun Yat-sen University Cancer Center, China. The 615 patients with HCC were randomly separated into two groups (training group n = 241 and validation group n = 374) by a random number table. None of the patients received any chemotherapy or radiotherapy prior to the surgery. The follow-up period was defined as the interval from the date of surgery to the date of death or the last follow up. This study was approved by the Institute Research Medical Ethics Committee of Sun Yat-sen University Cancer Center, China (GZR2013-060). All samples were anonymous and the informed consent was waived by the Institute Research Medical Ethics Committee of Sun Yat-sen University Cancer Center.
Tissue microarray construction and immunohistochemistry
The tissue microarray (TMA) slides included 615 HCC and adjacent normal tissue samples. By using a tissue array instrument (Minicore Excilone, Minicore, UK), each tissue core was punched (diameter 0.6 mm) from the marked areas and re-embedded. All specimens were fixed at 4% paraformaldehyde in 0.1 M phosphate buffer for 24 h and embedded in paraffin wax. Then the paraffin-embedded HCC sections were sliced into 4-μm slices and mounted onto glass slides. After dewaxing, the slides were treated with 3% hydrogen peroxide in methanol and blocked using a biotin-blocking kit (DAKO, Germany). After blocking, the slides were incubated with P1-HNF4α (K9218, 1:1000, Abcam, Massachusetts, USA) or P2-HNF4α antibody (H6939, 1:1000, Invitrogen, California, USA) overnight in a moist chamber at 4°C. After being washed in phosphate buffered saline three times, the slides were incubated with biotinylated goat anti-mouse antibodies for 1 h. Then the slides were stained with DAKO liquid 3,′3-diaminobenzidine tetrahydrochloride (DAB). Finally, the slides were counter stained with Mayer’s haematoxylin and observed under a microscope.
The HNF4α protein expression level was determined by semi-quantitative immunohistochemistry (IHC) detection. The positively-stained samples were scored as follows: ‘0’ (<5% positively-stained cells), ‘1’ (6–24% positively-stained cells), ‘2’ (25–49% positively-stained cells), ‘3’ (50–74% positively-stained cells), and ‘4’ (75–100% positively-stained cells). The intensity was scored according to the standard: ‘0’ (negative staining); ‘1’ (weak staining); ‘2’ (moderate staining); and ‘3’ (strong staining). The final score was calculated by multiplying the percentage score by the intensity score. The scores were independently decided by two pathologists (Dr Jing-Ping Yun and Dr Li-Li Liu). The mean IHC score was chosen as the cutoff value for defining high and low expression.
Western blot
Total proteins were extracted and separated by 10% SEMS-PAGE and then transferred onto polyvinylidene difluoride (PVDF) membranes (Millipore, Bedford, MA). Equal amounts of protein (30 μg) were resolved by SDS-PAGE and then electrophoretically transferred onto PVDF membranes. After being blocked in 5% nonfat milk for 1 h at room temperature, the membranes were incubated with appropriately diluted primary antibodies overnight at 4°C. After washing thrice with tris-buffered saline and Tween, the blotted membranes were incubated with the HNF4α antibody (1:1000, Invitrogen, CA, USA). The membranes were incubated with HRP-conjugated anti-mouse antibody at 1:50,000 dilution for 1 h at room temperature. The membranes were visualized by the enhanced Phototope TM-HRP Detection Kit and exposed to a Kodiak medical X-ray processor (Carestream Health, USA). Anti-GAPDH (1:1000, Santa Cruz, CA, USA) was used as a loading control.
Quantitative real-time polymerase chain reaction analysis
Total RNA was extracted from clinical samples and cultured cells using Trizol reagent (BIOO Scientific Co., USA) following the manufacturer’s instructions. The reverse transcription with random primers was done using M-MLV Reverse Transcriptase (Promega Inc., USA) according to the manufacturer’s instructions. SYBR Green-based real-time polymerase chain reaction (PCR) was carried out to measure levels of the corresponding P1- and P2-HNF4α and 18S by the Strata gene Mx3000P. Real-time PCR system and the PCR was performed as previously described.10 Primers were designed as follows: P1-HNF4α, forward: 5′- GGAATTTGAGAATGTGCAGGTGTTG -3′ and reverse: 5′- TGAGGTTGGTGCCTTCTGATG -3′; P2-HNF4α, forward: 5′- GCCATGGTCAGCGTGAAC -3′ and reverse: 5′- CGTTGAGGTTGGTGCCTTCT -3′;11 18S, forward: 5′-TGAGAAACGGCTACCACATCC-3′ and reverse: 5′-ACCAGACTTGCCCTCCAATG-3′.
Statistical analysis
Statistical analysis was performed using SPSS (version 16.0, Chicago, IL, USA). Student’s t test and Pearson’s χ2 test or Fisher’s exact test were chosen for examining the correlations between P2-HNFα expression level and the clinical and pathological variables. Survival curves were carried out by the Kaplan–Meier method (log-rank test). A multivariate Cox proportional hazards regression model was used to evaluate the independence of P2-HNFα in predicting outcomes. Differences were defined as significant for p-values <0.05.
Results
Expression of P1- and P2-HNF4α in HCC cell lines
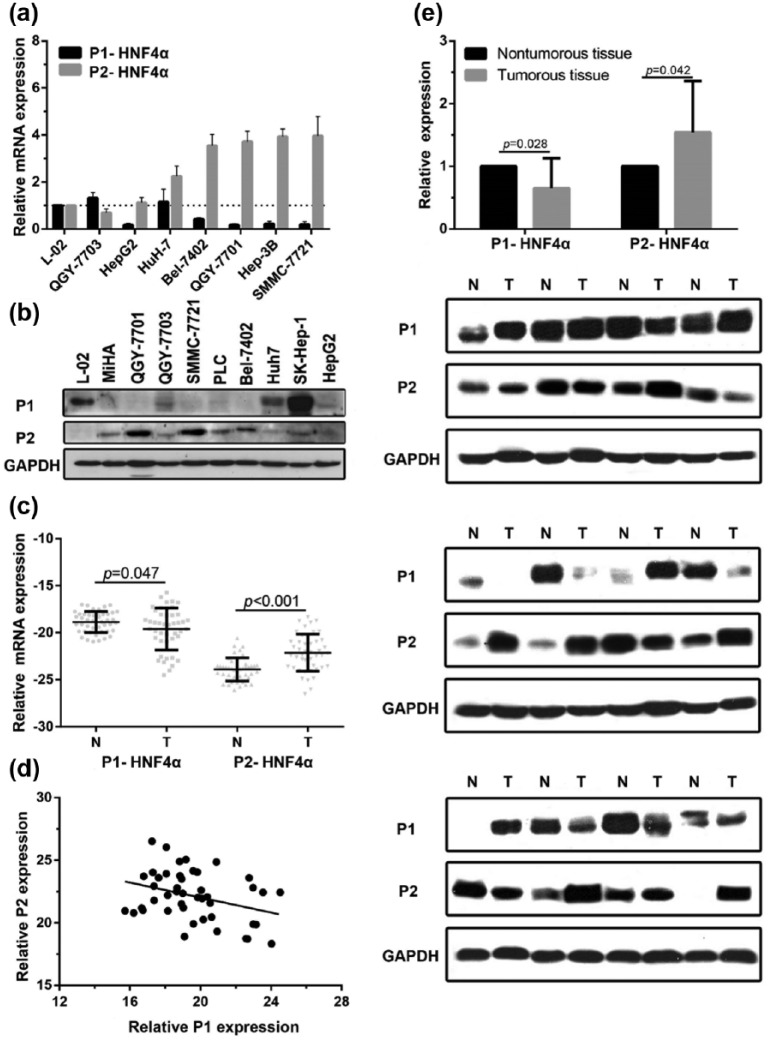
We first determined the expression of P1- and P2-HNF4α in HCC cell lines and fresh liver tissue by qRT-PCR and western blot. The results indicated that P1-HNF4α mRNA levels in most HCC cell lines [Figure 1(a)] and HCC tissues [p = 0.047, n = 45, Figure 1(c)] were downregulated, while P2-HNF4α mRNA levels were upregulated [p < 0.001, n = 45, Figure 1(a) and (c)], compared with those in an immortalized hepatic cell (L-02) and adjacent normal tissues. Consistently, the protein P2-HNF4α levels were significantly increased in HCC cell lines and tumour tissues, and the protein P1-HNF4α was downregulated in HCC cell lines and tumour tissue [Figure 1(b) and Figure 1(e)]. The expression of P1- and P2-HNF4α mRNA was correlated negatively [Figure 1(d)].
Figure 1.
P1-HNFα expression is decreased and P2-HNF4α expression is increased in HCC cell lines by polymerase chain reaction and western blotting (a–b). mRNA and protein levels of P1- and P2-HNF4α were measured in fresh HCC tissues (T) and corresponding adjacent nontumourous tissues (N) (c and e). Correlation of P1- and P2-HNF4α mRNA expression (r = −0.339, p = 0.023; d). Immortalized hepatocytes: L-02, MiHA; HCC cell lines: QGY-7703, HepG2, Huh-7, Bel-7402, QGY-7701, Hep3B, SMMC-7721, PLC, and SK-Hep-1.
HCC, hepatocellular carcinoma; HNF4α, hepatocyte nuclear factor 4 alpha; P1, promoter 1; P2, promoter 2.
Expression of P2-HNF4α in HCC TMA samples
To further confirm the expression of P1- and P2-HNF4α in HCC samples, paraffin-embedded HCC samples were collected to construct TMA to detect P1- (n = 106) and P2-HNF4α expression (n = 615). The P1-HNF4α IHC score in HCC tissue was 2.96 ± 2.50, significantly lower than normal liver samples with 3.87 ± 2.95 (p = 0.013, Supplementary Figure 1). Among the 106 samples, the 39 samples with high P2 expression consisted of 9, 24, and 6 samples with P1 high, low, and negative expression. Whereas in 34 samples with negative P2 expression, 19 of 34 samples had high P1 expression and 13 and 2 samples with low and negative P1 expression (p = 0.026, Supplementary Table 1).
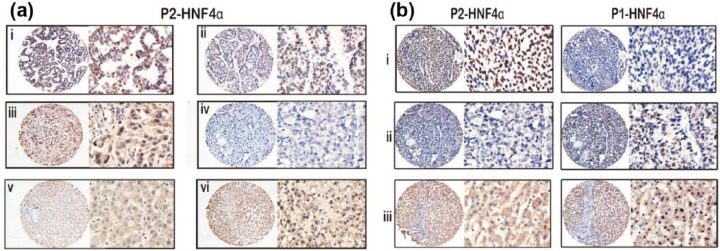
As shown in Figure 2(a), the immunoreactivity of P2-HNF4α was mainly present in the nuclei of cancer cells and barely in adjacent normal tissue. In addition, we observed that P2-HNF4α positive expression samples were always negative for P1-HNF4α expression, and vice versa for liver samples [Figure 2(b)].
Figure 2.
P2-HNF4α expression is increased in HCC samples as shown by immunohistochemistry. Representative images of strong (ai), moderate (aii), weak (aiii), and negative (aiv) immunoreactivities of P2-HNF4α in HCC samples, as well as negative (av) and positive (avi) staining of P2-HNF4α in normal liver tissues are shown (left panel: magnification ×100; right panel: magnification ×400). (b) P2-HNF4α positive expressing HCC tissue is always accompanied by negative P1-HNF4α expression (bi). Negative expression of P2-HNF4α in HCC tissue is accompanied by positive P1-HNF4α expression (bii). Negative expression of P2-HNF4α in nontumourous tissue is accompanied by positive P1-HNF4α expression (biii).
HCC, hepatocellular carcinoma; HNF4α, hepatocyte nuclear factor 4 alpha; P1, promoter 1; P2, promoter 2.
Association of P1- and P2-HNF4α expression and clinical features in HCC
To determine the clinical significance of P2-HNF4α in HCC, the relationship between P2-HNF4 and clinical features were evaluated. According to the mean IHC score in tumour tissue (3.0), high P2-HNF4α expression was identified in 34.9% (159/615) of cases. HCC patients with high P2-HNF4α expression in their tumour tissues had less tumour differentiation (p = 0.048) and more vascular invasion (p = 0.017) (Table 1).
Table 1.
Association of P2-HNF4α expression and clinical features in hepatocellular carcinoma.
| Variable | P2-HNF4α |
p-value | |
|---|---|---|---|
| Low expression | High expression | ||
| Sample size | 456 | 159 | |
| Age, years | 48.76 ± 12.21 | 49.49 ± 11.76 | 0.507 |
| Sex | 0.815 | ||
| Male | 413 | 145 | |
| Female | 43 | 14 | |
| HBsAg | 0.741 | ||
| Positive | 388 | 137 | |
| Negative | 68 | 22 | |
| AFP, ng/ml | 0.081 | ||
| <20 | 133 | 35 | |
| ⩾20 | 323 | 124 | |
| Cirrhosis | 0.942 | ||
| Yes | 374 | 130 | |
| No | 82 | 29 | |
| Tumour size, cm | 0.299 | ||
| <5 | 102 | 42 | |
| ⩾5 | 354 | 117 | |
| Tumour multiplicity | 0.206 | ||
| Single | 270 | 85 | |
| Multiple | 186 | 74 | |
| Differentiation | 0.048 | ||
| Well-moderate | 309 | 94 | |
| Poor-undifferentiated | 147 | 65 | |
| TNM stage | 0.083 | ||
| I–II | 240 | 71 | |
| III–IV | 216 | 88 | |
| Vascular invasion | 0.017 | ||
| Yes | 78 | 41 | |
| No | 378 | 118 | |
| Involucrum | 0.663 | ||
| Complete | 275 | 99 | |
| Incomplete | 181 | 60 | |
AFP: α-fetoprotein; HBsAg: hepatitis B virus surface antigen; HNF4α, hepatocyte nuclear factor 4 alpha; P2, promoter 2.
We also compared samples with low P1-HNF4α expression with samples with high P1-HNF4α expression as shown in Supplemental Table 2. The data indicated that 68.8% of well-moderate samples (11/16) presented with high P1 expression while only 38.9% of poor-undifferentiated samples (35/90) did (p = 0.026). However, the P1 IHC results showed no difference in P1 expression between samples with and without vascular invasion.
Relationship between P1- and P2-HNF4α expression and tumour differentiation
To further explore the relationship between HNF4α expression and tumour differentiation (Figure 3), a total of 129 extra liver biopsy samples were collected and evaluated for P1- and P2-HNF4α IHC. Interestingly, we found that HNF4α expression was significantly associated with tumour differentiation. In those HCC samples with worse differentiation, the proportion with high expression of P2-HNF4α was significantly higher than among those with good differentiation (Table 2), whereas in the HCC samples with good differentiation, P1-HNF4α expression was more common. These results confirm that expression of P1-HNF4α and P2-HNF4α are negatively correlated and closely associated with tumour differentiation.
Figure 3.
Representative images of well-differentiated (a), moderately-differentiated (b) and poorly-differentiated HCC (c) in liver biopsy tissue. In the HCC samples with better differentiation, the proportion of cells with high expression of P1-HNF4α is significantly higher than P2-HNF4α, whereas in HCC samples with poor differentiation, P2-HNF4α expression was more common.
HCC, hepatocellular carcinoma; HNF4α, hepatocyte nuclear factor 4 alpha; P1, promoter 1; P2, promoter 2.
Table 2.
Relationship between P1 and P2-HNF4α expression and tumour differentiation.
| Differentiation | P1-HNF4α expression |
p-value | P2-HNF4α expression |
p-value | ||
|---|---|---|---|---|---|---|
| Low | High | Low | High | |||
| Well | 6 | 14 | 0.001 | 17 | 3 | 0.002 |
| Moderate | 34 | 19 | 33 | 20 | ||
| Poor | 27 | 17 | 29 | 15 | ||
| Undifferentiated | 12 | 0 | 2 | 10 | ||
HNF4α, hepatocyte nuclear factor 4 alpha; P1, promoter 1; P2, promoter 2.
Association of P2-HNF4α expression and clinical outcomes of HCC
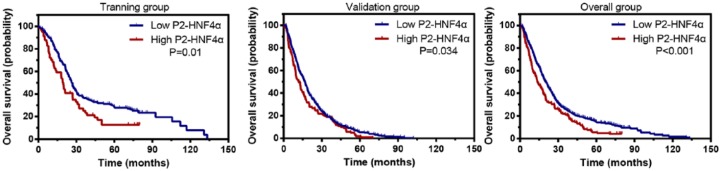
To determine the prognostic effect of P2-HNF4α on HCC patients, Kaplan–Meier survival analysis was conducted. The 615 patients with HCC were randomly separated into two groups (training group n = 241 and validation group n = 374). In the training group, Kaplan–Meier analysis showed that HCC cases with high P2-HNF4α expression had significantly worse outcomes in terms of overall survival (p = 0.01). Consistently, low P2-HNF4α expression was positively correlated with favourable overall survival (p = 0.034) in the validation cohort. The results in the overall group indicated that HCC patients with high P2-HNF4α expression were likely to have a shorter overall survival time (Figure 4).
Figure 4.
High P2-HNF4α expression is correlated with an unfavourable prognosis in both the training and validation cohorts. Kaplan–Meier analysis shows the significant differences in overall survival between postoperative HCC patients with high and low P2-HNF4α expression in training (n = 241), validation (n = 374), and overall cohorts.
HCC, hepatocellular carcinoma; HNF4α, hepatocyte nuclear factor 4 alpha; P2, promoter 2.
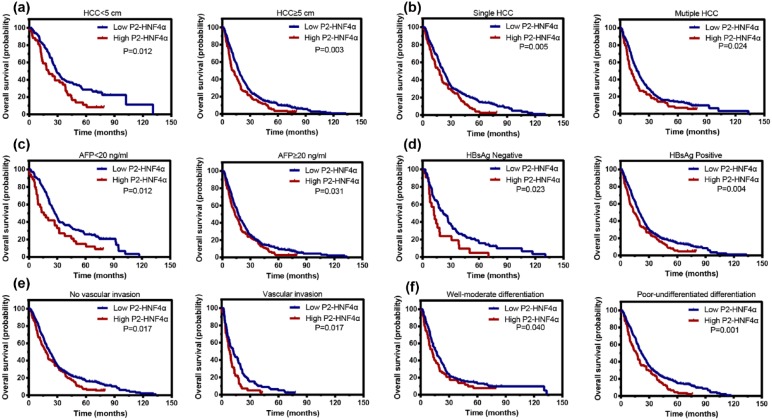
Stratified survival analyses were conducted to further reveal the prognostic significance of P2-HNF4α. The data showed that P2-HNF4α expression was associated with overall survival in small and large HCCs, in single and multiple HCCs, in HCCs with AFP within the upper limit of normal and abnormal, in HCCs negative and positive for hepatitis B surface antigen, in HCCs with or without microvascular invasion, and in HCCs with well-moderate and poor-undifferentiated differentiation (Figure 5).
Figure 5.
High P2-HNF4α expression is associated with unfavourable outcomes in subgroups of HCC patients. Stratified survival analyses show that P2- HNF4α expression is correlated with overall survival in small and large HCCs (a) in single and multinodular HCC (b), in HCCs with normal and abnormal AFP levels (c), in HBsAg positive and negative HCCs (d), in HCCs with or without microvascular invasion (e), and in HCCs with well-moderate differentiation and poor-undifferentiated differentiation (f).
HBsAg, hepatitis B surface antigen; HCC, hepatocellular carcinoma; HNF4α, hepatocyte nuclear factor 4 alpha; P1, promoter 1; P2, promoter 2.
We also performed Kaplan–Meier analysis of groups comparing low versus high P1-HNF4α expression (n = 106) as shown in Supplementary Figure 2. Our results showed HCC cases with low P1 expression had significantly worse outcomes in terms of overall survival (p = 0.043).
Univariate and multivariate analyses of prognostic variables in HCC
To evaluate the independent risk factors for outcomes of HCC, univariate and multivariate analyses were conducted. Tumour size, serum AFP level, tumour differentiation, TNM stage, vascular invasion, and P2-HNF4α expression were shown to be prognostic variables for the outcome of overall survival in HCC patients. While in multivariate analyses, serum AFP levels, tumour size, vascular invasion, and P2-HNF4α were found to be independent prognostic variables for overall survival (Table 3).
Table 3.
Univariate and multivariate analyses of prognostic variables for overall survival.
| Variables | Univariate analysis |
Multivariate analysis |
||||
|---|---|---|---|---|---|---|
| HR | 95% CI | p | HR | 95% CI | p | |
| Age, years | 0.998 | 0.991–1.005 | 0.620 | |||
| Sex | 1.018 | 0.765–1.354 | 0.904 | |||
| HBsAg | 1.052 | 0.833–1.329 | 0.672 | |||
| AFP | 1.679 | 1.383–2.037 | <0.001 | 1.471 | 1.207–1.792 | <0.001 |
| Cirrhosis | 0.952 | 0.764–1.187 | 0.662 | |||
| Tumour size, cm | 1.761 | 1.433–2.163 | <0.001 | 1.641 | 1.332–2.021 | <0.001 |
| Tumour multiplicity | 1.169 | 0.988–1.384 | 0.069 | |||
| Differentiation | 1.199 | 1.005–1.430 | 0.043 | |||
| TNM | 1.504 | 1.273–1.777 | <0.001 | |||
| Vascular invasion | 2.087 | 1.696–2.567 | <0.001 | 1.795 | 1.453–2.218 | <0.001 |
| Involucrum | 1.038 | 0.876–1.230 | 0.665 | |||
| P2-HNF4α | 1.408 | 1.165–1.701 | <0.001 | 1.385 | 1.145–1.675 | 0.001 |
AFP: α-fetoprotein; CI, confidence interval; HBsAg: hepatitis B virus surface antigen; HNF4α, hepatocyte nuclear factor 4 alpha; HR, hazard ratio; P2, promoter 2.
Discussion
It is well established that HNF4α is an essential gene for normal liver development and maintenance of a differentiated phenotype. HNF4α knock-out mice, who lose the expression of many liver genes, fail to develop a functional liver.12,13 Recently, researchers found that HNF4α might play a role in cancer development. Oshima and colleagues found that HNF4α expression decreased in colorectal cancer, and lack of HNF4α expression was correlated with worse outcomes, with a higher probability of liver metastasis.14 Saandi and colleagues demonstrated that low expression of HNF4α in colorectal cancer induced downregulation of CDX2 expression, explaining the role of HNF4α in colorectal carcinoma.15 However, Darsigny and colleagues reported that HNF4α is highly expressed in colorectal samples and promoted the development of tumours in mice by targeting oxidoreductase-related genes.16 The reason for these inconsistent results is that researchers did not distinguish between HNF4α expression driven by P1 or P2 and most studies have focused on P1-HNF4α, ignoring P2-HNF4α. In our study, we showed that HNF4α expression driven by P1 or P2 were different. We confirmed that P1-HNF4α expression was decreased in HCC. In addition, we found that P2-HNF4α was upregulated in liver cancer, and was frequently higher in HCC with worse tumour differentiation and more vascular invasion, indicating that P2-HNF4α might be capable of promoting the development of HCC.
HNF4α is the core component of the HNF pathway.17 By driving the different promoters, nine subtypes of HNF4α splice variants can be produced: subtypes a1–a6 are transcribed from the P1 promoter and subtypes a7–a9 are transcribed from the P2 promoter. P1-HNF4α is mainly expressed in the adult liver and studies have reported that P1-HNF4α is downregulated in about 70% of HCCs;18 P2-HNF4α is expressed mainly in embryonic liver, but the expression of P2-HNF4α in HCC was not previously clear. Here, we confirmed that P1-HNF4α expression was decreased in HCC. Additionally, we found that P2-HNF4α expression was increased. However, the mechanism of regulation of P2-HNF4α is not clear yet. We found the P2-HNF4α was upregulated in HCC tissue and its expression was correlated negatively with P1-HNF4α. The reason why P2-HNF4α, a protein expressed in embryonic liver, is expressed in HCC tissue and why P1-HNF4α expression seems to affect P2-HNF4α expression needs further research.
HNF4α is also known as the master regulator of hepatic differentiation. Postnatal hepatocyte-specific deletion of HNF4α results in a metabolic disorder with accumulation of lipids.19 Furthermore, HNF4α null livers exhibited a decrease in classic hepatocyte gene expression such as apolipoprotein B, microsomal triglyceride transfer protein, and liver fatty acid-binding protein expression.19 Iacob and colleagues reported that after overexpression of HNF4α, albumin secretion and glycogen reserve capacity increased in the liver precursor cells, verifying that HNF4α plays a role in the maintenance of liver cell differentiation and function.20 Hwang-Verslues and colleagues reported HNF4α targets cytochromes and sulfuric acid transferase to maintain drug metabolism.21 Ning found that HNF4α expression gradually decreased in liver cirrhosis and during liver cancer formation.22 We further distinguished the role of HNF4α in hepatocyte differentiation in our study. We found that P1-HNF4α was decreased in less differentiated HCC samples while P2-HNF4α was upregulated. P1 and P2-HNF4α could be used to distinguish different levels of differentiated HCC.
The prognostic implications of P2-HNF4α expression have not been previously reported although low P1-HNF4α expression has been proven to be associated with worse outcomes of cancer. Lazarevich and colleagues found that low expression of HNF4α suggests a poor prognosis in patients with liver cancer.23 In our study, we subdivided HNF4α and P2-HNF4α was identified as an independent factor for overall survival in a large cohort of 615 patients with HCC. Patients with high P2-HNF4α expression usually survived for a shorter time period. These data suggest that P2-HNF4α expression has clinical implications in predicting outcomes of cancer patients.
In summary, our data demonstrate that the role of HNF4α in cancer development needs to be further refined. Our results reveal that P1-HNF4α expression is decreased in HCC samples while P2-HNF4α expression is increased. Thus, expression levels of P1 and P2-HNF4α are correlated negatively. Increases in P2-HNF4α expression were significantly correlated with worse tumour differentiation and vascular invasion, suggesting that P2-HNF4α might play a role in HCC progression. High P2-HNF4α expression was correlated with shorter survival times of HCC patients and served as an independent factor for worse outcomes. Collectively, our data suggest P2-HNF4α is a promising biomarker for the prognosis of patients with HCC.
Supplementary Material
Acknowledgments
Shao-hang Cai and Shi-xun Lu contributed equally to this work.
Footnotes
Funding: The study was supported by grants from the National Natural Science Foundation of China (grant no. 81372572).
Conflict of interest statement: The authors declare that there is no conflict of interest.
Contributor Information
Shao-hang Cai, Sun Yat-sen University Cancer Center, State Key Laboratory of Oncology in South China, Collaborative Innovation Center for Cancer Medicine, Guangzhou, China Department of Pathology, Sun Yat-sen University Cancer Center, Guangzhou, China.
Shi-xun Lu, Sun Yat-sen University Cancer Center, State Key Laboratory of Oncology in South China, Collaborative Innovation Center for Cancer Medicine, Guangzhou, China Department of Pathology, Sun Yat-sen University Cancer Center, Guangzhou, China.
Li-li Liu, Sun Yat-sen University Cancer Center, State Key Laboratory of Oncology in South China, Collaborative Innovation Center for Cancer Medicine, Guangzhou, China Department of Pathology, Sun Yat-sen University Cancer Center, Guangzhou, China.
Chris Zhiyi Zhang, Sun Yat-sen University Cancer Center, State Key Laboratory of Oncology in South China, Collaborative Innovation Center for Cancer Medicine, Guangzhou, China Department of Pathology, Sun Yat-sen University Cancer Center, Guangzhou, China.
Jing-ping Yun, Department of pathology, Sun Yat-sen University Cancer Center, State Key Laboratory of Oncology in South China, Collaborative Innovation Center for Cancer Medicine, No. 651 Dongfeng East Road, Guangzhou, Guangdong Province 510060, China.
References
- 1. Babeu JP, Boudreau F. Hepatocyte nuclear factor 4-alpha involvement in liver and intestinal inflammatory networks. World J Gastroenterol 2014; 20: 22–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hanawa M, Takayama K, Sakurai F, et al. Hepatocyte nuclear factor 4 alpha promotes definitive endoderm differentiation from human induced pluripotent stem cells. Stem Cell Rev 2016; 13: 542–551. [DOI] [PubMed] [Google Scholar]
- 3. Chen WS, Manova K, Weinstein DC, et al. Disruption of the HNF-4 gene, expressed in visceral endoderm, leads to cell death in embryonic ectoderm and impaired gastrulation of mouse embryos. Genes Dev 1994; 8: 2466–2477. [DOI] [PubMed] [Google Scholar]
- 4. Drewes T, Senkel S, Holewa B, et al. Human hepatocyte nuclear factor 4 isoforms are encoded by distinct and differentially expressed genes. Mol Cell Biol 1996; 16: 925–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Torres-Padilla ME, Fougere-Deschatrette C, Weiss MC. Expression of HNF4alpha isoforms in mouse liver development is regulated by sequential promoter usage and constitutive 3’ end splicing. Mech Dev 2001; 109: 183–193. [DOI] [PubMed] [Google Scholar]
- 6. Sel S, Ebert T, Ryffel GU, et al. Human renal cell carcinogenesis is accompanied by a coordinate loss of the tissue specific transcription factors HNF4 alpha and HNF1 alpha. Cancer Lett 1996; 101: 205–210. [DOI] [PubMed] [Google Scholar]
- 7. Lenburg ME, Liou LS, Gerry NP, et al. Previously unidentified changes in renal cell carcinoma gene expression identified by parametric analysis of microarray data. BMC Cancer 2003; 3: 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tanaka T, Jiang S, Hotta H, et al. Dysregulated expression of P1 and P2 promoter-driven hepatocyte nuclear factor-4alpha in the pathogenesis of human cancer. J Pathol 2006; 208: 662–672. [DOI] [PubMed] [Google Scholar]
- 9. Mizuguchi T, Mitaka T, Hirata K, et al. Alteration of expression of liver-enriched transcription factors in the transition between growth and differentiation of primary cultured rat hepatocytes. J Cell Physiol 1998; 174: 273–284. [DOI] [PubMed] [Google Scholar]
- 10. Yi C, Wang Q, Wang L, et al. MiR-663, a microRNA targeting p21(WAF1/CIP1), promotes the proliferation and tumorigenesis of nasopharyngeal carcinoma. Oncogene 2012; 31: 4421–4433. [DOI] [PubMed] [Google Scholar]
- 11. Harries LW, Locke JM, Shields B, et al. The diabetic phenotype in HNF4A mutation carriers is moderated by the expression of HNF4A isoforms from the P1 promoter during fetal development. Diabetes 2008; 57: 1745–1752. [DOI] [PubMed] [Google Scholar]
- 12. Duncan SA, Nagy A, Chan W. Murine gastrulation requires HNF-4 regulated gene expression in the visceral endoderm: tetraploid rescue of Hnf-4(-/-) embryos. Development 1997; 124: 279–287. [DOI] [PubMed] [Google Scholar]
- 13. Li J, Ning G, Duncan SA. Mammalian hepatocyte differentiation requires the transcription factor HNF-4alpha. Genes Dev 2000; 14: 464–474. [PMC free article] [PubMed] [Google Scholar]
- 14. Oshima T, Kawasaki T, Ohashi R, et al. Downregulated P1 promoter-driven hepatocyte nuclear factor-4alpha expression in human colorectal carcinoma is a new prognostic factor against liver metastasis. Pathol Int 2007; 57: 82–90. [DOI] [PubMed] [Google Scholar]
- 15. Saandi T, Baraille F, Derbal-Wolfrom L, et al. Regulation of the tumor suppressor homeogene Cdx2 by HNF4alpha in intestinal cancer. Oncogene 2013; 32: 3782–3788. [DOI] [PubMed] [Google Scholar]
- 16. Darsigny M, Babeu JP, Seidman EG, et al. Hepatocyte nuclear factor-4alpha promotes gut neoplasia in mice and protects against the production of reactive oxygen species. Cancer Res 2010; 70: 9423–9433. [DOI] [PubMed] [Google Scholar]
- 17. Eeckhoute J, Moerman E, Bouckenooghe T, et al. Hepatocyte nuclear factor 4 alpha isoforms originated from the P1 promoter are expressed in human pancreatic beta-cells and exhibit stronger transcriptional potentials than P2 promoter-driven isoforms. Endocrinology 2003; 144: 1686–1694. [DOI] [PubMed] [Google Scholar]
- 18. Lazarevich NL, Shavochkina DA, Fleishman DI, et al. Deregulation of hepatocyte nuclear factor 4 (HNF4) as a marker of epithelial tumors progression. Exp Oncol 2010; 32: 167–171. [PubMed] [Google Scholar]
- 19. Darsigny M, Babeu JP, Seidman EG, et al. Hepatocyte nuclear factor-4alpha promotes gut neoplasia in mice and protects against the production of reactive oxygen species. Cancer Res 2010; 70: 9423–9433. [DOI] [PubMed] [Google Scholar]
- 20. Iacob R, Rudrich U, Rothe M, et al. Induction of a mature hepatocyte phenotype in adult liver derived progenitor cells by ectopic expression of transcription factors. Stem Cell Res 2011; 6: 251–261. [DOI] [PubMed] [Google Scholar]
- 21. Hwang-Verslues WW, Sladek FM. HNF4alpha–role in drug metabolism and potential drug target? Curr Opin Pharmacol 2010; 10: 698–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ning BF, Ding J, Yin C, et al. Hepatocyte nuclear factor 4 alpha suppresses the development of hepatocellular carcinoma. Cancer Res 2010; 70: 7640–7651. [DOI] [PubMed] [Google Scholar]
- 23. Lazarevich NL, Shavochkina DA, Fleishman DI, et al. Deregulation of hepatocyte nuclear factor 4 (HNF4) as a marker of epithelial tumors progression. Exp Oncol 2010; 32: 167–171. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.