Abstract
Background
Rumex patientia L. is consumed as a green vegetable in several parts of the world, and can withstand extremely low temperatures (-35°C). However, little or no available genomic data for this species has been reported to date. Here, we used Illumina Hiseq technology for transcriptome assembly in R. patientia under normal and cold conditions to evaluate how it responds to cold stress.
Results
After an in-depth RNA-Seq analysis, 115,589 unigenes were produced from the assembled transcripts. Based on similarity search analysis with seven databases, we obtained and annotated 60,157 assembled unigenes to at least one database. In total, 1,179 unigenes that were identified as differentially expressed genes (DEGs), including up-regulated (925) and down-regulated ones (254), were successfully assigned GO annotations and classified into three major metabolic pathways. Ribosome, carbon metabolism, oxidative phosphorylation and biosynthesis of amino acids were the most highly enriched pathways according to KEGG analysis. Overall, 66 up-regulated genes were identified as putatively involved in the response to cold stress, including members of MYB, AP2/ERF, CBF, Znf, bZIP, NAC and COR families.
Conclusion
To our knowledge, this investigation was the first to provide a cold-responsive (COR) transcriptome assembly in R. patientia. A large number of potential COR genes were identified, suggesting that this species is suitable for cultivation in northern China. In summary, these data provide valuable information for future research and genomic studies in R. patientia.
Introduction
Temperature substantially influences cytomembrane fluidity and membrane lipid composition, which in turn affect protein folding and gene expression [1]. Temperate plants are tolerant of chilling temperatures (0–15°C) but are usually intolerant of freezing temperatures (< 0°C) [2]. In response to low temperature, the transcriptome and metabolism in plants are strongly altered via regulation of the expression of related genes. After exposure to low temperature, the transcriptome of the model plant Arabidopsis thaliana shows massive changes that culminate in a considerable increase in cold-responsive (COR) genes expression, from 4% to 20% [3,4]. In alfalfa (Medicago sativa), a decrease in membrane fluidity rapidly induces expression of numerous downstream COR genes, completing the process of the response to cold conditions [5]. Gene mutations and chemical agents can restore cell membrane-induced cold acclimatization-specific gene expression at high temperatures [6,7]. In addition, as a second messenger, the Ca2+ signaling pathway is central to the early response to cold stress [8], and many reports have demonstrated that Ca2+ activity is essential for cold-shock gene expression. When calcium ion-chelating agents or materials that block calcium channels are introduced to cells, COR gene expression decreases, and chilling stress is simultaneously mitigated. Additionally, an artificial increase in the cytoplasm Ca2+ concentration can induce cold stress-specific gene expression, even at warm temperatures [9].
Cold stress induces notable changes in C-repeat binding factors (CBFs) and dehydration-responsive element-binding protein 1 (DREB1), which specifically bind to cis-elements in the promoters of COR genes and activate their expression [10,11]. Many genes are regulated by CBFs, which mainly participate in mechanisms that protect cells, including the inositol phospholipid pathway, transcription, reactive oxygen species detoxification, membrane transport, and hormone metabolism. In addition to Arabidopsis, COR homologs have been cloned from other plants, suggesting that CBFs are evolutionarily conserved [10]. Nevertheless, a biochip expression analysis in tomato showed that genes regulated by CBFs in different plants respond in via different pathways [12]. Although CBFs play a crucial role in gene regulation during cold acclimation, according to bioinformatics analysis, they regulate and control only approximately 12% of genes in the COR transcriptome [13]. This finding indicates that most COR genes are governed by CBF-independent regulons.
Rumex patientia L. (2n = 10), a perennial herb belonging to the Polygonaceae family, is consumed as a green vegetable in several parts of the world, particularly in Turkey and India [14]. R. patientia can tolerate abiotic stress and withstand extremely low temperatures (-35°C) by relying on overwintering buds, which become dormant during severe winter months. This tolerance suggests that the species is suitable for cultivation in cold regions and for research on cold tolerance in plants [15,16]. The dried roots of R. patientia have also been employed for many decades as traditional medicines (e.g., as a purgative, antipyretic compound, depurative and tonic) for the treatment of wounds and diverse diseases [17]. The root extract of R. patientia contains anthraquinone, tannin, naphthalene and naphthoquinone derivatives [18]; it was first to North China introduced from Ukraine in 1995 as a foodstuff for animal husbandry [19]. Despite the abovementioned attributes, publicly available genetic databases contain limited reports regarding this species, and no nucleotide sequences from R. patientia have been deposited into the NCBI GenBank database to date.
Therefore, in this investigation, we used Illumina Hiseq technology for transcriptome assembly in R. patientia and compared transcriptomes under normal and cold conditions to evaluate how this species responds to cold stress. Our findings enhance our understanding of the R. patientia gene regulation response to cold stress and reveal novel approaches for improving cold tolerance through genetic engineering.
Materials and methods
Stress treatments and sample preparation
R. patientia seeds were grown in the greenhouse of Northeast Agricultural University (26°C, 16 h light photoperiod). After cultivation for two months, 20 seedlings were transferred to different treatment conditions. For this study, we set -5°C as a cold stress treatment because the temperature of soil is approximately -5°C in winter in Harbin, even though the air temperature is -30°C. Half of the plants were shifted to the cold stress conditions (-5°C for 4 h), and the other half were used as a control. The roots of treated plants were harvested after 4 h of treatment, frozen in liquid nitrogen and prepared for RNA extraction. Three completely independent harvesting experiments were repeated as biological replicates.
Total RNA was extracted from roots using the TRIzol according to the manufacturer’s protocol (TIANGEN, Beijing, China). The RNA purity and integrity were assessed using a Nanophotometer spectrophotometer (IMPLEN, CA, USA). Qualified RNA samples were used for cDNA synthesis with the PrimeScript™ RT Reagent Kit and gDNA Eraser (TaKaRa, Tokyo, Japan). cDNA fragments of 150–200 bp in length were selected for polymerase chain reaction (PCR) amplification, and a cDNA library was used for sequence analysis via Illumina sequencing at Novogene Research Pty. Ltd., Beijing, China.
Transcriptome analysis under cold treatment
Raw data in fastq format were first processed through in-house Perl scripts. In this step, clean data were obtained by removing reads containing adapters, reads containing poly-N and low-quality reads from the raw data. All downstream analyses were based on high-quality clean data. The remaining clean reads were assembled into unigenes using the short-read assembly program SOAPdenovo [20]. All unigenes were then used in a blast search (E-value < 10−5) and annotated against various databases, including the NCBI non-redundant protein (Nr), NCBI nucleotide sequences (Nt), Kyoto Encyclopedia of Genes and Genomes Ortholog (KEGG), SwissProt, Protein family (PFAM), Gene Ontology (GO), and Cluster of Orthologous Groups (COG) databases.
Identification and annotation of differentially expressed genes
Differential expression analysis between two treatments was performed using the DESeq R package (1.10.1). The false discovery rate (FDR) control method was applied in Benjamini and Hochberg method to correct the results for P-values. FDR < 0.01 and FC (fold change) ≥ 2 were set as the threshold to determine the significance of gene expression differences. For differentially expressed genes (DEGs), we considered an adjusted P-value < 0.05 identified by DESeq and a 2-fold or greater change in fragments per kilobase of gene per million mapped reads (FPKM). Regarding the functional annotation of DEGs, the results of GO enrichment analysis were evaluated using topGO R packages based on Wallenius non-central hyper-geometric distribution [21]. Finally, KOBAS software was employed to test the statistical enrichment of DEGs in KEGG pathways [22].
Quantitative reverse transcription PCR (qRT-PCR) validation
Ten representative DEGs identified by RNA sequencing (RNA-Seq) were chosen for experimental validation by qRT-PCR using gene-specific primers (S1 Table). The reaction was performed using SYBR® Premix Ex Taq™ II Kit (Tli RNaseH Plus) (TaKaRa, Tokyo, Japan) in a volume of 20 μl containing 10 μl SYBR Premix Ex Taq (2×), 0.4 μl ROX Reference Dye II (10×), 2 μl cDNA template, and 0.5 μM each primer. Amplification was performed as follows: 95°C for 30 s followed by 45 cycles of 95°C for 5 s and 60°C for 40 s. All reactions were performed in biological triplicate, and the results are expressed relative to the expression levels of β-actin based on the 2-ΔΔCt method [23].
Results
De novo assembly and functional annotation
After filtering adapters, reads containing poly-N and low-quality reads, ~ 2.7 million clean reads were purified from 2.9 million raw reads containing a total of 3.4 G nucleotides with a high percentage of clean reads (81.14%-98.16%) (Table 1). Six sample transcripts (approximately 20.69 Gbp of clean reads) were pooled together to perform de novo transcriptome assembly. Using the short-read assembly program SOAPdenovo, 161,869 transcripts were assembled with an N50 of 1412 bp. A total of 115,589 unigenes were produced from the assembled transcripts (Table 2).
Table 1. Sequencing output statistics of control and cold-treated R. patientia.
| Samples | ||||||
|---|---|---|---|---|---|---|
| Control_1 | Control_2 | Control_3 | Treated_1 | Treated_2 | Treated_3 | |
| Number of raw reads | 30106792 | 26387332 | 28098657 | 35182884 | 26821572 | 30472196 |
| Number of clean reads | 29212660 | 25902104 | 27406323 | 34122498 | 24095683 | 24725848 |
| Percentage of clean reads | 97.03% | 98.16% | 97.54% | 96.99% | 89.84% | 81.14% |
| Size of clean reads | 3.65Gbp | 3.24Gbp | 3.43Gbp | 4.27Gbp | 3.01Gbp | 3.09Gbp |
| Number of total clean reads | 20.69Gbp | |||||
Table 2. Statistics of transcriptome assemble and unigenes.
| Number of transcripts | Number of unigenes | |
|---|---|---|
| Number | 161,869 | 115,589 |
| Total nucleotide | 128,889,868 | 76,040,314 |
| Minimum length | 201 | 201 |
| Median length | 426 | 353 |
| Maximum length | 15363 | 15363 |
| Mean length | 796 | 658 |
| N50 | 1412 | 1101 |
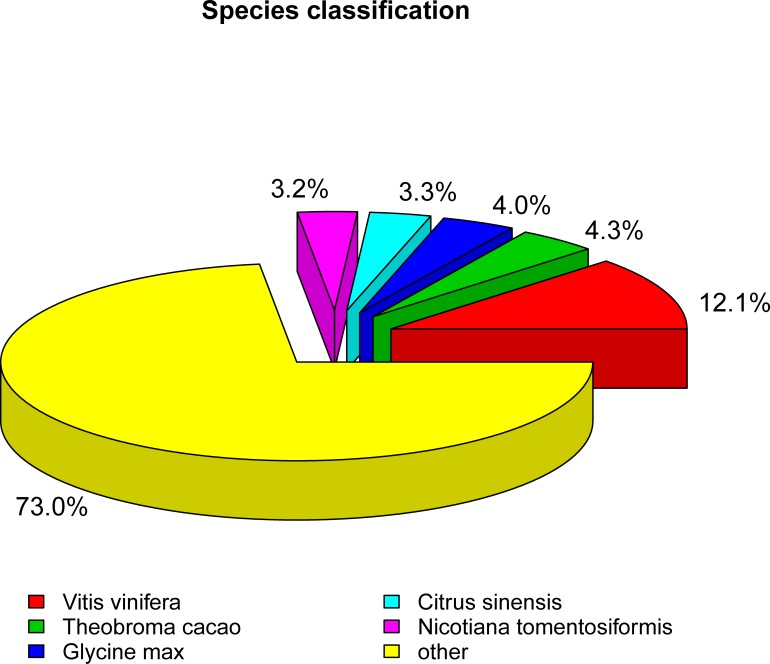
To identify the putative functions of unigenes in R. patientia, all of the assembled unigenes were functionally annotated against seven databases (Table 3 and S2 Table). Of 115,589 unigenes, 44,927 (38.86%), 28,271 (24.45%), 16,653 (14.4%), 35,947 (31.09%), 35,148 (30.4%), 35,784 (30.95%) and 19,129 (16.54%) were aligned to known sequences in the Nt, Nr, KEGG, SwissProt, PFAM, GO, and COG databases, respectively. Overall, 7,444 unigenes showed homology with known genes in all databases. Among these databases, the highest number of unigene annotations was in Nr, whereas the lowest number of annotations was matched in the KO database. As no genomic information on R. patientia has been published to date, we blast searched the unigenes against other species. The results showed the highest similarity to sequences from Vitis vinifera (5415); the next closest matches were Theobroma cacao (1952), Glycine max (1805), Citrus sinensis (1501) and Nicotiana tomentosiformis (1459), with comparable homology among these species (Fig 1 and S1 Fig).
Table 3. The annotation results of unigenes in seven databases.
| Number and percentage of Unigenes | |
|---|---|
| Annotated in Nr | 44927 (38.86%) |
| Annotated in Nt | 28271 (24.45%) |
| Annotated in KEGG | 16653 (14.4%) |
| Annotated in SwissProt | 35947 (31.09%) |
| Annotated in PFAM | 35148 (30.4%) |
| Annotated in GO | 35784 (30.95%) |
| Annotated in COG | 19129 (16.54%) |
| Annotated in all databases | 7444 (6.44%) |
| Annotated in at least one database | 60157 (52.04%) |
Fig 1. Homology search of R. patientia unigenes with other species by BLASTx to the NR database.
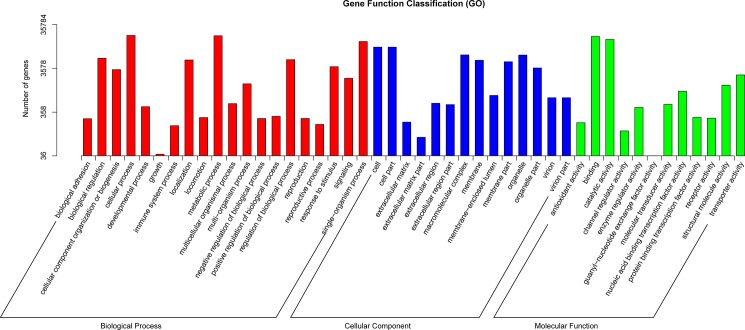
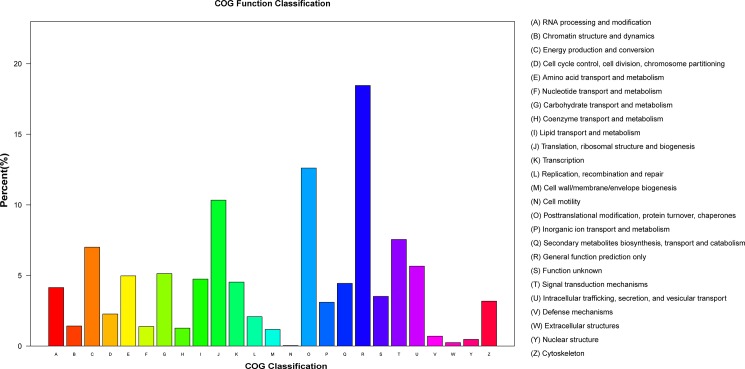
GO analysis fully described the genes and their biological functions in the classification system, which organized the functions of predicted unigenes into three main categories: biological processes, molecular function and cellular component (Fig 2). A total of 35,784 sequences were successfully assigned GO terms, among which 86,291 unigenes were assigned at least one biological process GO term, 54,651 to cellular components, and 42,798 to molecular functions. Furthermore, we performed phylogenetic classification using the COG database. A total of 19,129 unigenes were matched and clustered into 25 functional classes (Fig 3). The groups ‘General function prediction only’ (3,531) and ‘Transcription’ (2,412) were the two largest clusters, accounting for nearly 18.46% and 12.62% of the unigenes, respectively (Table 3 and Fig 3).
Fig 2. Functional annotation of unigenes based on Gene Ontology (GO) classification.
Fig 3. Distribution of genes in the transcriptome with COG functional classification.
A total of 19129 sequences have a COG classification among 25 categories.
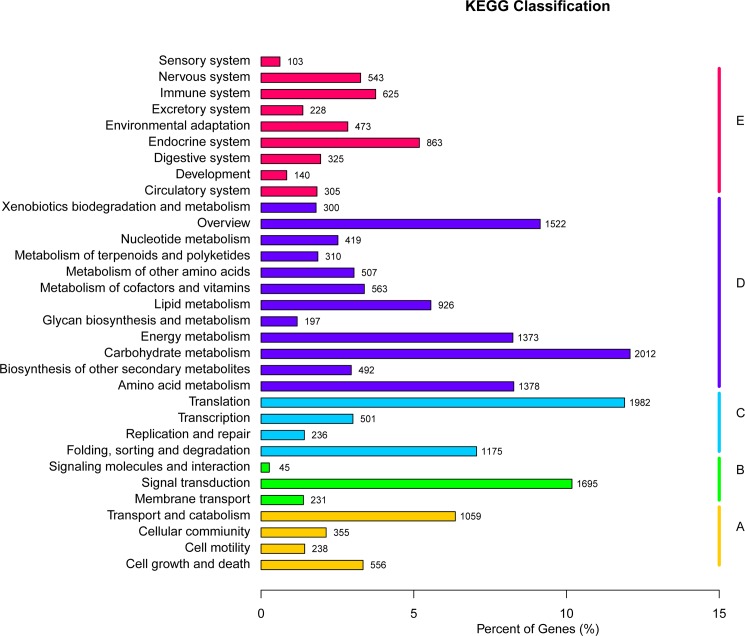
Additionally, all of the assembled unigenes were further classified into KEGG functional subcategories. According to biological processes, a large number of genes involved in metabolic and cellular processes were highly represented, which comprised 20,008 and 20,294, respectively, of the matched unigenes in the subcategory, suggesting that cold temperature affects this cold-tolerant species by altering protein synthesis and metabolism (Fig 4). In the cellular component, the largest subcategory was cells (30.77%), and the second largest was cell parts (30.76%). Regarding molecular function, the largest numbers were found in catalytic activity (46.17%) and binding (53.8%). KEGG analysis was conducted to identify and predict active biochemical pathways in R. patientia, with 16,653 unigenes matching to 280 different KEGG pathways (S3 Table). Among these pathways, the most highly represented were carbon pathways (with 2,012 members), followed by translation (1,982) and signal transduction (1,695).
Fig 4. KEGG pathway classification of R. patientia unigenes.
Changes in gene expression in cold-treated R. patientia
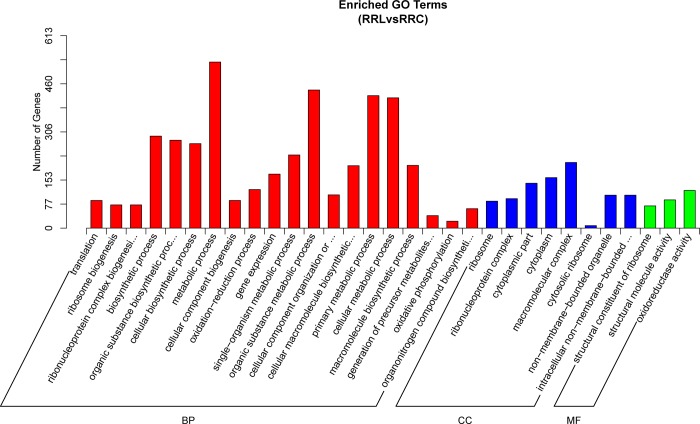
To better understand the genes that respond to cold stress in R. patientia, we identified putative DEGs in control and stress-treated samples using DESeq R package (padj < 0.05). In three control samples, 78,337 putative DEGs were detected. In contrast, 69,638 were found in cold-treated plants, of which 48,246 unigenes in common were detected in six libraries; 21,392 unigenes were identified in cold-treated samples, indicating that 30.72% of genes might be related to chilling stress (S2 Fig). In total, 1179 unigenes that were identified as DEGs, including up-regulated (925) and down-regulated genes (254), were successfully assigned GO annotations and classified into three major metabolic pathways (Fig 5, S2 Table and S3 Fig). Metabolic process (69.06%), macromolecular complex (27.28%) and oxidoreductase activity (15.67%) were the largest subcategories in biological processes, cellular component and molecular function, respectively. To help us better understand the biological function of DEGs under cold stress in R. patientia, KEGG pathway analysis was performed, annotating the DEGs into 10 top pathways (S3 Table and S4 Table). Ribosome, carbon metabolism, oxidative phosphorylation and biosynthesis of amino acids were the most highly enriched pathways according to KEGG analysis.
Fig 5. Functional annotation of differentially expressed genes based on Gene Ontology (GO) classification.
Validation of R. patientia DEGs after cold stress
Table 4 lists the 10 most up- and down-regulated DEGs and their annotations after cold treatment. Among the top 10 up-regulated DEGs, four putative orthologs of the CRT/DRE-binding factor of Arabidopsis were identified, which is one of the CBF transcription factors induced by low, nonfreezing temperatures. Blast hits against known proteins were found for only two down-regulated DEGs; another eight were annotated as unknown functions, and these may constitute novel factors induced by cold stress, which requires further study.
Table 4. The 10 most up- and down-regulated R. patientia genes and annotations after cold stress.
| Gene ID | Log2(fold-change) | Description | Type |
|---|---|---|---|
| c68449_g1 | 8.5563 | CRT/DRE binding factor | up |
| c63591_g1 | 8.2511 | CRT/DRE binding factor | up |
| c76039_g1 | 7.8142 | zinc finger protein CONSTANS-LIKE 1 | up |
| c85484_g1 | 7.2464 | NA | up |
| c76748_g3 | 7.0709 | CRT/DRE binding factor | up |
| c72584_g1 | 6.8482 | Aldo/Keto Reductase | up |
| c74692_g1 | 6.6489 | unsaturated rhamnogalacturonyl hydrolase YesR-like | up |
| c76748_g1 | 6.5762 | CRT/DRE binding factor | up |
| c63095_g1 | 6.5039 | NA | up |
| c76501_g1 | 6.4841 | hypothetical protein COCSUDRAFT_21535 | up |
| c50023_g1 | -7.3423 | LINE-1 retrotransposable element ORF2 protein | down |
| c68197_g6 | -7.0881 | NA | down |
| c57726_g1 | -6.9272 | NA | down |
| c57779_g2 | -6.053 | NA | down |
| c38504_g1 | -5.7153 | NA | down |
| c68060_g2 | -5.1872 | putative non-LTR retroelement reverse transcriptase | down |
| c78553_g1 | -4.8282 | NA | down |
| c58200_g1 | -4.5248 | NA | down |
| c66306_g1 | -4.3839 | NA | down |
| c81134_g2 | -4.3242 | NA | down |
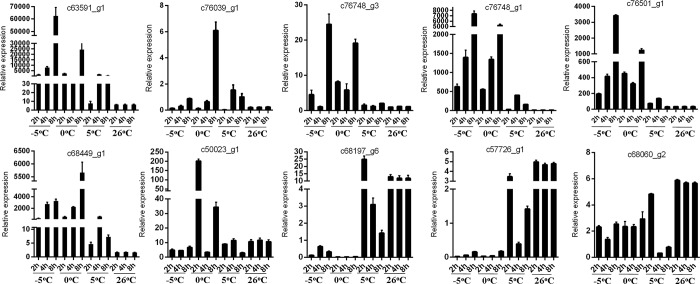
In total, we tested and validated 10 of the top 20 DEGs by qRT-PCR to examine their expression after cold stress for 2, 4, and 8 h. Expression of all 6 up-regulated genes was enhanced by chilling, although their expression changed with the length of chilling treatment and the specific temperature (Fig 6). Consistent with the RNA-Seq results, all four of the most down-regulated genes were repressed by chilling (Fig 6). The consistency between the results of qRT-PCR and the RNA-Seq analyses confirmed the validity of the de novo-assembled transcriptome and our evaluation of the cold stress regulation of the transcriptome.
Fig 6. The expression of R. patientia genes in response to chilling at different temperature for 0 to 8 h as determined by qRT-PCR.
Analysis of R. patientia unigenes involved in the cold response pathway
The CBF transcriptional cascade, a crucial pathway for regulating gene expression under low temperatures, is conserved in diverse plant species. Using our RNA-Seq data, we identified CBF pathway genes to more deeply understand the genes involved in the response to low temperature. Overall, 66 up-regulated genes were identified as putatively involved in the response to cold stress, and their corresponding CBF pathway orthologs are listed in S5 Table. As shown in S5 Table, 25 unigenes were matched with zinc finger family (Znf) members, representing the largest group in R. patientia. The second largest group was COR (15 unigenes), including a large proportion of Ca2+-binding transcription factors involved in the response to cold stimuli. Genes in the AP2/ERF family encode transcriptional regulators with a variety of functions involved in the developmental and physiological processes in plants. In our transcriptome, two subfamilies of AP2/ERF transcriptome factors were identified: DREBs (c76748_g3, c63591_g1, c68449_g1, c53582_g1, and c76748_g4) and ethylene-responsive transcription factors (c74787_g1, c67493_g1, c63394_g1, c32287_g1 and c54408_g1).
Three unigenes (c80957_g2, c81107_g2 and c65606_g1) were found to harbor a domain similar to the MYB-like DNA-binding domain, which can alter transcription when plants are exposed to low temperatures. Three unigenes (c74749_g1, c63109_g2 and c63109_g3) contain the NAC domain, which responds to biotic and abiotic stresses. In addition, 4 unigenes (c71662_g1, c89475_g1, c72305_g1, and c72369_g4) show high similarity to the bZIP transcription factor sequence, which is involved in regulating expression of a subset of COR genes.
Discussion
R. patientia L. is a new type of high-protein forage feed and is also a good ground cover plant for effectively preventing soil erosion and improving the ecological environment [24,25]. The tolerance of R. patientia to extremely low temperatures is attracting research attention, as this species may be suitable for growth in cold regions. In this investigation, we analyzed the transcriptome sequences of R. patientia under cold acclimation and obtained 20.69 Gbp of clean reads from six samples (Table 1). Compared with the control sample, 925 up-regulated genes and 254 down-regulated genes were identified as DEGs, indicating that the altered genes are involved in regulating the response to cold treatment. Similar to previous reports from many other species, the number of DEGs identified under cold stress was increased compared with the control [26–28]. Interestingly, among the 925 up-regulated transcripts, 91 (9.8%) had unknown/unclassified functions (S2 Table). This result suggests that the presence of putative novel genes might be specific to R. patientia, and these genes may be involved in important pathways in cold stress adaptation.
According to functional annotations based on the GO and KEGG databases, the DEGs identified by cold stress are mainly involved in the ribosome, carbon metabolism and oxidative phosphorylation pathways (S4 Table). The largest number of DEGs was related to the ribosome pathway (ko03010), suggesting that transcription and translation are vigorous in R. patientia after cold stress and that there is an additional gradual increase in gene participation during a prolonged stress-response period. Similar findings have been reported in the transcriptomes of many other plants under low temperature [29–31]. Carbon metabolism (ko01200) was the second largest group related to the identified DEGs, suggesting that low, nonfreezing temperatures may increase the concentration of carbon compounds and their primary biosynthetic enzymes. Soluble sugars not only function as a source of metabolic energy but also act as molecular signals regulating different genes associated with stress pathways to cope with abiotic stress conditions [32]. Sucrose was previously shown to be the most abundant soluble sugar in Spinacia oleracea during cold stress via a mechanism that involves increasing sucrose phosphate synthase activity [33]. In the process of cold tolerance in plants, activated starch enzymes hydrolyze starch into sugars, which increases water retention and osmotic potential in plant cells [34].
The oxidative phosphorylation pathway (ko00190) includes enzymes that oxidize nutrients, releasing energy as ATP and producing most of the energy in mitochondria [35]. This pathway is most likely pervasive during a plant’s lifespan. A total of 34 up-regulated DEGs were successfully annotated to this pathway, representing the third largest cluster, and their high levels demonstrated that the energy released by oxidation mainly leads to the reformation of ATP, which may serve as an emergency resistance measure related to a low-temperature environment. This result was consistent with findings for the Anthurium transcriptome, whereby significant enrichment was identified in similar top pathways after cold treatment. However, the opposite results were obtained in chilled corn shoots, in which the efficiency of oxidative phosphorylation was markedly reduced. One possible reason for this low level of efficiency in corn might be that the energy released was consumed to adapt to a cold environment [36]. In addition, accumulation of proline was artificially increased under cold stress, which might act as a signal for reconfiguring gene expression [10]. At low temperatures, ammonium absorption is enhanced by the elevation of glutamine synthetase activity in rice roots, indicating that low temperatures can modulate nitrogen metabolism [37]. One striking finding in our study was the identification of 30 DEGs involved in the biosynthesis of amino acids, indicating that R. patientia can protect itself against cold stress.
During the process of cold adaptation, expression of a large number of COR transcription factor genes is activated by receptor proteins or signal transduction pathways to enhance oxidation resistance and osmotic adjustment ability and to promote reconstruction and material balance in the cell membrane system. In R. patientia, 66 up-regulated genes were annotated as putative CBF transcription factors, including members of the MYB, AP2/ERF, CBF, Znf, bZIP, NAC and COR families, which is consistent with the cold resistance and tolerance of this species (S5 Table). The AP2/ERF family of transcription factors, among the largest groups in R. patientia, contains four major subfamilies: the AP2, ERF, RAV and DREB subfamilies [38,39]. The DREB subfamily consists of a well-known set of transcription factors that have been detected and cloned in numerous species and that are known to activate expression of abiotic stress-responsive genes via specific binding to a DRE/CRT cis-acting element [40,41]. In this investigation, five DREB genes were successfully matched and showed significant increases after cold stress, suggesting that the DREB1/CBF pathway functionally acts as a key transcription factor in the regulation of the cold response in R. patientia.
Zinc-finger transcription factors play a central role in regulating various signal transduction pathways in plants [42,43]. Previous studies have suggested that the overexpression of CBFs that control the transcription of numerous downstream stress-related genes can enhance Zat10 and Zat12 expression in Arabidopsis, and CBF-induced increases in the expression of a set of COR genes were positively correlated with Zat10 and Zat12 in response to cold [10,44]. The large number of up-regulated genes annotated as Znfs in R. patientia indicates that these transcription factors might participate in the regulating the response to cold. Zhang et al. [45] demonstrated that several newly discovered bZIP genes in wheat were strongly induced by multiple abiotic stresses through the abscisic acid (ABA) signaling pathway. However, in the present work, we were unable to verify which signaling pathway might be associated with bZIP in R. patientia based only on the obtained transcriptome sequences. Nonetheless our observations demonstrate that bZIP expression is advantageous for the response to cold conditions.
The COR transcription factor family comprises Ca2+-binding proteins [8]. In this study, the second largest family of cold-stress transcription factors identified in R. patientia was Ca2+-binding proteins, suggesting that these proteins are jointly involved in the response to cold stress. There have been similar discoveries in Medicago sativa cells and Brassica napus leaves, in which cold stress-induced plasma membrane rigidification resulted in actin cytoskeletal rearrangement, induction of Ca2+ channels, and an artificial increase in cytosolic Ca2+ levels. These changes further induce expression of COR genes and cold acclimation in plants [5,46]. Indeed, a transient increase in the cellular Ca2+ concentration in response to dehydration and low temperature appear to stimulate cellular signaling processes [38].
Conclusion
To our knowledge, this investigation is the first to provide a COR transcriptome assembly in R. patientia, a species with little or no available genomic data, which has limited its economic utility for cultivation in cold regions. After an in-depth RNA-Seq analysis, we obtained and annotated 60,157 assembled unigenes to at least one database. A large number of potential COR genes were identified, suggesting that this species is suitable for cultivation in northern China. Our findings of up-regulated DEGs suggested that cold stimuli greatly affect protein translation and cellular metabolism in this species. A large number of the identified unigenes in the CBF pathway and COR genes were induced by low temperatures, which was consistent with the strong cold tolerance of this species. In summary, these data provide valuable information for future research and genomic studies in R. patientia.
Supporting information
(XLSX)
(XLS)
(XLS)
(DOC)
(DOC)
(DOC)
Cluster analysis of expression level (A) and Venn diagram (B) of putative DEGs in R. patientia (fold changes > 2, false discovery rate < 0.01).
(DOC)
(DOC)
Data Availability
All transcriptome data files are available from the NCBI Sequence Read Archive database (accession number SRP116571).
Funding Statement
This research was supported by National Science Foundation for Fostering Talents in Basic Research and International Cooperation Project (Grant no. 2013DFR30270 to BH).
References
- 1.Wang X, Li W, Li M, Welti R (2006) Profiling lipid changes in plant response to low temperatures. Physiologia Plantarum 126: 90–96. [Google Scholar]
- 2.Zhu J, Dong CH, Zhu JK (2007) Interplay between cold-responsive gene regulation, metabolism and RNA processing during plant cold acclimation. Current Opinion in Plant Biology 10: 290–295. doi: 10.1016/j.pbi.2007.04.010 [DOI] [PubMed] [Google Scholar]
- 3.Mirzadeh H, Najafizadeh A (2005) A Global Survey of Gene Regulation during Cold Acclimation in Arabidopsis thaliana. Plos Genetics 1: 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee B, Henderson DA, Zhu JK (2005) The Arabidopsis Cold-Responsive Transcriptome and Its Regulation by ICE1. Plant Cell 17: 3155–3175. doi: 10.1105/tpc.105.035568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Örvar BL, Sangwan V, Omann F, Dhindsa RS (2000) Early steps in cold sensing by plant cells: the role of actin cytoskeleton and membrane fluidity. Plant Journal 23: 785–794. [DOI] [PubMed] [Google Scholar]
- 6.Inaba M, Suzuki I, Szalontai B, Kanesaki Y, Los DA, Hayashi H, et al. (2003) Gene-Engineered Rigidification of Membrane Lipids Enhances the Cold Inducibility of Gene Expression in Synechocystis. Journal of Biological Chemistry 278: 12191–12198. doi: 10.1074/jbc.M212204200 [DOI] [PubMed] [Google Scholar]
- 7.Solanke AU, Sharma AK (2008) Signal transduction during cold stress in plants. Physiology & Molecular Biology of Plants 14: 69–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Knight H, Trewavas AJ, Knight MR (1996) Cold calcium signaling in Arabidopsis involves two cellular pools and a change in calcium signature after acclimation. Plant Cell 8: 489–503. doi: 10.1105/tpc.8.3.489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Monroy AF, Dhindsa RS (1995) Low-temperature signal transduction: induction of cold acclimation-specific genes of alfalfa by calcium at 25 degrees. Plant Cell 7: 321–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chinnusamy V, Zhu J, Zhu JK (2007) Cold stress regulation of gene expression in plants. Trends in Plant Science 12: 444–451. doi: 10.1016/j.tplants.2007.07.002 [DOI] [PubMed] [Google Scholar]
- 11.Zhu JK (2016) Abiotic Stress Signaling and Responses in Plants. Cell 167: 313–324. doi: 10.1016/j.cell.2016.08.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang X, Fowler SG, Cheng H, Lou Y, Rhee SY, Stockinger EJ, et al. (2004) Freezing-sensitive tomato has a functional CBF cold response pathway, but a CBF regulon that differs from that of freezing-tolerant Arabidopsis. Plant Journal 39: 905–919. doi: 10.1111/j.1365-313X.2004.02176.x [DOI] [PubMed] [Google Scholar]
- 13.Fowler S, Thomashow MF (2002) Arabidopsis transcriptome profiling indicates that multiple regulatory pathways are activated during cold acclimation in addition to the CBF cold response pathway. Plant Cell 14: 1675–1690. doi: 10.1105/tpc.003483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qi X, Wang E, Xing M, Zhao W, Chen X (2012) Rhizosphere and non-rhizosphere bacterial community composition of the wild medicinal plant Rumex patientia. World Journal of Microbiology and Biotechnology 28: 2257–2265. doi: 10.1007/s11274-012-1033-2 [DOI] [PubMed] [Google Scholar]
- 15.Singh N, Arya JS, Maurya SB, Srivastava RB (2013) Rumex (Rumex patientia L.)-spinach of high-altitude cold desert. Current Science 104: 574. [Google Scholar]
- 16.Chaurasia, Ahmed, Ballabh (2007) Ethnobotany and plants of Trans-Himalaya. Satish Serial Pub, House.
- 17.Baytop T (1984) Therapy with medicinal plants in Turkey (Past and Present) Nobel Tip Kitabevleri, Istanbul: 243.
- 18.Lin S, Lai W, Ho Cc, Yu F, Chen G, Yang J, et al. (2009) Emodin induces apoptosis of human tongue squamous cancer SCC-4 cells through reactive oxygen species and mitochondria-dependent pathways. Anticancer Research 29: 327–335. [PubMed] [Google Scholar]
- 19.Zhang LS, Wang JF, Cheng YX (2011) Advances of Research on Rumex L. Hubei Agricultural Sciences 50: 865–870. [Google Scholar]
- 20.Li R, Zhu H, Ruan J, Qian W, Fang X, Shi Z, et al. (2010) De novo assembly of human genomes with massively parallel short read sequencing. Genome research 20: 265–272. doi: 10.1101/gr.097261.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Benjamini Y, Hochberg Y (1995) Controlling The False Discovery Rate—A Practical And Powerful Approach To Multiple Testing. Journal of the Royal Statistical Society 57: 289–300. [Google Scholar]
- 22.Mao X (2005) Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 21: 3787–3793. doi: 10.1093/bioinformatics/bti430 [DOI] [PubMed] [Google Scholar]
- 23.Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. methods 25: 402–408. doi: 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 24.Heděnec P, Novotný D, Ust'Ak S, Honzík R, Váňa V, Petříková V, et al. (2015) Effect of long term cropping hybrid sorrel (Rumexpatientia x Rumex tianshanicus) on soil biota. Biomass & Bioenergy 78: 92–98. [Google Scholar]
- 25.Zhang ZS, Li YT, Gao HY, Yang C, Meng QW (2016) Characterization of photosynthetic gas exchange in leaves under simulated adaxial and abaxial surfaces alternant irradiation. Scientific Reports 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aslam M, Sinha VB, Singh RK, Anandhan S, Pande V, Ahmed Z. (2010) Isolation of cold stress-responsive genes from Lepidium latifolium by suppressive subtraction hybridization. Acta Physiologiae Plantarum 32: 205–210. [Google Scholar]
- 27.Jiang Q, Wang F, Tan HW, Li MY, Xu ZS, Tan GF, et al. (2015) De novo transcriptome assembly, gene annotation, marker development, and miRNA potential target genes validation under abiotic stresses in Oenanthe javanica. Molecular Genetics and Genomics 290: 671–683. doi: 10.1007/s00438-014-0953-y [DOI] [PubMed] [Google Scholar]
- 28.Wingler A, Juvany M, Cuthbert C, Munné-Bosch S (2015) Adaptation to altitude affects the senescence response to chilling in the perennial plant Arabis alpina. Journal of Experimental Botany 66: 355–367. doi: 10.1093/jxb/eru426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhan X, Lan Y, Dong W, Jian KZ, Lang Z (2016) De novo assembly and analysis of the transcriptome of Ocimum americanum var. pilosum under cold stress. BMC Genomics 17: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang H, Zou Z, Wang S, Gong M (2013) Global Analysis of Transcriptome Responses and Gene Expression Profiles to Cold Stress of Jatropha curcas L. Plos One 8: e82817–e82817. doi: 10.1371/journal.pone.0082817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ashraf N, Ghai D, Barman P, Basu S, Gangisetty N, Mandal M, et al. (2009) Comparative analyses of genotype dependent expressed sequence tags and stress-responsive transcriptome of chickpea wilt illustrate predicted and unexpected genes and novel regulators of plant immunity. BMC Genomics 10: 1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rosa M, Prado C, Podazza G, Interdonato R, González JA, Hilal MH, et al. (2009) Soluble sugars—Metabolism, sensing and abiotic stress: A complex network in the life of plants. Plant Signaling & Behavior 4: 388–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guy CL, Huber JL, Huber SC (1992) Sucrose phosphate synthase and sucrose accumulation at low temperature. Plant Physiology 100: 502–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chapman HD (1961) Methods of analysis for soils, plants, and waters. University of California, Division of Agricultural Sciences 93: 68. [Google Scholar]
- 35.Saraste M (1999) Oxidative phosphorylation at the fin de siècle. Science 283: págs. 1488–1493. [DOI] [PubMed] [Google Scholar]
- 36.Yang F, Xing J, Chen W, Wang S (1981) Effect of low temperature on oxidative phophorylation and cyanide-insensitive respiration in corn mitochondria. Journal of Integrative Plant Biology 5: 23–27. [Google Scholar]
- 37.Lu B, Yuan Y, Zhang C, Ou J, Zhou W, Lin Q, et al. (2005) Modulation of key enzymes involved in ammonium assimilation and carbon metabolism by low temperature in rice (Oryza sativa L.) roots. Plant Science 169: 295–302. [Google Scholar]
- 38.Shinozaki K, Yamaguchi-Shinozaki K (2000) Molecular response to dehydration and low temperature: Differences and cross-talk between two stress signaling pathways. Curr Opin Plant Biol 3:217–223. Current Opinion in Plant Biology 3: 217–223. [PubMed] [Google Scholar]
- 39.Sakuma Y, Liu Q, Dubouzet JG, Abe H, Shinozaki K, Yamaguchi-Shinozaki K. (2002) DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochemical & Biophysical Research Communications 290: 998–1009. [DOI] [PubMed] [Google Scholar]
- 40.Cruz-Mendívil A, López-Valenzuela JA, Calderón-Vázquez CL, Vega-García MO, Reyes-Moreno C, Valdez-Ortiz A, et al. (2015) Early transcriptional responses to chilling stress in tomato fruit with hot water pre-treatment. Postharvest Biology & Technology 109: 137–144. [Google Scholar]
- 41.Nakashima K, Yamaguchi-Shinozaki K (2006) Regulons involved in osmotic stress-responsive and cold stress-responsive gene expression in plants. Physiologia Plantarum 126: 62–71. [Google Scholar]
- 42.Davletova S, Mittler R (2005) The zinc-finger protein Zat12 plays a central role in reactive oxygen and abiotic stress signaling in Arabidopsis. Plant Physiology 139: 847–856. doi: 10.1104/pp.105.068254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ciftci-Yilmaz S, Mittler R (2008) The zinc finger network of plants. Cellular & Molecular Life Sciences Cmls 65: 1150–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maruyama K, Sakuma Y, Kasuga M, Ito Y, Seki M, Hideki Goda, et al. (2004) Identification of cold-inducible downstream genes of the Arabidopsis DREB1A/CBF3 transcriptional factor using two microarray systems. Plant Journal 38: 982–993. doi: 10.1111/j.1365-313X.2004.02100.x [DOI] [PubMed] [Google Scholar]
- 45.Zhang L, Zhang L, Xia C, Zhao G, Liu J, Jia J, et al. (2015) A novel wheat bZIP transcription factor, TabZIP60, confers multiple abiotic stress tolerances in transgenic Arabidopsis. Physiologia Plantarum 153: 538–554. doi: 10.1111/ppl.12261 [DOI] [PubMed] [Google Scholar]
- 46.Sangwan V, Foulds I, Singh J, Dhindsa RS (2001) Cold-activation of Brassica napus BN115 promoter is mediated by structural changes in membranes and cytoskeleton, and requires Ca2+ influx. Plant Journal for Cell & Molecular Biology 27: 1–12. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
(XLS)
(XLS)
(DOC)
(DOC)
(DOC)
Cluster analysis of expression level (A) and Venn diagram (B) of putative DEGs in R. patientia (fold changes > 2, false discovery rate < 0.01).
(DOC)
(DOC)
Data Availability Statement
All transcriptome data files are available from the NCBI Sequence Read Archive database (accession number SRP116571).