Abstract
The emergence of multidrug-resistant enterococci (MDRE) and particularly vancomycin-resistant enterococci (VRE) is considered a serious health problem worldwide, causing the need for new antimicrobials. The aim of this study was to discover and characterize bacteriocin against clinical isolates of MDRE and VRE. Over 10,000 bacterial isolates from water, environment and clinical samples were screened. E. faecalis strain 478 isolated from human feces produced the highest antibacterial activity against several MDRE and VRE strains. The optimum condition for bacteriocin production was cultivation in MRS broth at 37°C, pH 5–6 for 16 hours. The bacteriocin-like substance produced from E. faecalis strain EF478 was stable at 60°C for at least 1 hour and retained its antimicrobial activity after storage at -20°C for 1 year, at 4°C for 6 months, and at 25°C for 2 months. A nano-HPLC electrospray ionization multi-stage tandem mass spectrometry (nLC-ESI-MS/MS) analysis showed that the amino acid sequences of the bacteriocin-like substance was similar to serine protease of E. faecalis, gi|488296663 (NCBI database), which has never been reported as a bacteriocin. This study reported a novel bacteriocin with high antibacterial activity against VRE and MDRE.
Introduction
Vancomycin-resistant enterococci (VRE) and multidrug-resistant enterococci (MDRE) have emerged as hospital-acquired pathogen over the last three decades [1]. Among various enterococci, Enterococcus faecalis and Enterococcus faecium are known to be the most common causes of nosocomial infections, such as urinary tract infection, surgical wound infection, pneumonia, endocarditis, bacteremia and meningitis [2]. They are intrinsically resistant to many antimicrobial agents used in clinical setting and rapidly acquire resistance genes and mutations [1, 3]. Enterococci can grow in a wide range of pHs and temperatures, high salt concentration and persist on non-living objects for weeks, allowing them to survive in hospitals and extreme environments [4, 5]. Generally, vancomycin is reserved for certain serious infections, which are non-responsive to other antibiotic treatment. Since the first VRE case report in Europe in 1980, the prevalence of VRE has increased dramatically in various countries [6, 7]. VRE can cause serious infections, especially endocarditis and bacteremia, in immunocompromised patients or patients on treatment with antibiotics such as cephalosporins [8]. The search for new effective antimicrobial agents has become a global concern due to the increasing prevalence of MDR-and VRE. In the past three decades, the process of new antibiotic discovery has slowed and only few antibiotics against VRE have been developed [9]. Furthermore, some enterococci have evolved resistance to the most recent antimicrobial agents such as quinupristin-dalfopristin, linezolid, daptomycin and tigecycline during therapy [3]. The number of antibiotics resistant enterococci has been continually rising while the number of effective antibiotics has been declining.
Antimicrobial peptides (AMPs) have drawn attention as alternatives to conventional antibiotics. Bacteriocins are bacterial antimicrobial peptides that have been used in the food industry for over 50 years and are generally recognized as safe (GRAS) [10]. They are also used to treat bacterial infections and control antibiotic-resistant bacteria, either used alone or in combination with conventional antibiotics [11–13]. Bacteriocins kill target bacteria by several mechanisms, including pore formation in the cell membrane, inhibition of cell wall- or protein synthesis, and degradation of cellular DNA [14]. The rise in interest toward bacteriocins is due to their potent inhibitory activities, the fact that they are safe for humans, their stability and various modes of action. Furthermore, resistance toward bacteriocins has been rarely observed [15, 16]. Bacteriocin producing bacteria have been isolated from plants, animals, food, water and soil [17–21].
Although bacteriocins have been extensively studied, it was generally in regard to their application in the food industry as preservative. In addition, there is a scarcity of reports about the efficacy of bacteriocins against vancomycin-resistant and multidrug-resistant enterococci clinical isolates. Therefore, the aims of this study were to discover and characterize bacteriocins active against clinical isolates of antimicrobial resistant E. faecium and E. faecalis.
Materials and methods
Sample collection and bacterial isolation
Bacterial isolates were collected from environmental water, laboratory environment and clinical samples during July 2014 and April 2015 for bacteriocin-production screening. The water samples were collected from Mahasawat Canal, Bangtal Canal, Chao Phraya River Bangkok and Pasak River Saraburi, (S1 Table). The sample sources are communal water supplies, no specific permissions were required for the samples collection. The environmental and clinical samples were collected from the Faculty of Medicine Vajira Hospital, Navamindradhiraj University, Bangkok, Thailand with the approval by the institute’s Ethical Review Committee for Human Research.
The water samples were pre-filtrated using 0.8 μm cellulose nitrate membranes (Nalgene Company, Rochester, USA) to remove large particles and then passed through 0.2 μm Supor membranes to concentrate bacterial cells (Pall Corp, USA). The membranes were then cut and immersed in Luria-Bertani broth (LB) (Merck, KGaA, Darmstadt, Germany) supplemented with 10% fetal bovine serum and incubated with shaking at 37°C for 4 hours (200 rpm, Innova 4300 incubator shaker, New Brunswick Scientific, Edison, NJ). The cultures were then isolated on LB agar.
Environmental samples were collected from laboratory-bench-surfaces (10 cm2) in the hospital using sterile moisten cotton swabs. Each sample was inoculated onto blood and MacConkey agar plates, and incubated at 37°C overnight. Bacterial colonies of all morphology were screened for bacteriocin production. The isolates from stool and rectal swab cultures with Enterococcus-like morphology on blood agar were identified and screened for bacteriocin production (S1 Fig).
Enterococcus faecalis ATCC 51299 and E. faecium ATCC 35677, as well as clinical isolates of MDR-enterococci were used as indicators for antimicrobial activity.
Screening for E. faecalis and E. faecium inhibitors
The inhibitory activity against E. faecalis and E. faecium was preliminary detected using the bacterial inhibition test. Briefly, the indicators were cultured in LB broth overnight at 37°C. Cell density was then adjusted to ~ 107 cells/ml using sterile normal saline solution and swabbed onto the surface of LB agar plates. The tested bacterial isolates were cross streaked onto the lawn of the indicators the plates incubated at 37°C overnight. The zone of inhibition was observed and measured.
Screening for bacteriocin production
The tested isolates that produced large inhibition zone against both indicators were selected to screen for the production of bacteriocin by spot-on-lawn method [22]. Briefly, cell free supernatants (CFSs) of the tested strains were prepared by growing one colony of each tested strains in 10 ml of LB broth at 37°C for 16–18 hours. The cultures were centrifuged at 11,000 x g, 4 °C for 10 minutes (IEC Multi RF, Thermo Electron Corporation, USA) and the supernatant were sterilized by passing through a 0.2 μm pore size filters (Acrodisc® with Supor membrane, Pall Corp., USA). The indicator lawns were prepared by adding each indicator to 3 ml of molten LB medium at the final concentration of 107 cells/ml and overlaid on Muller-Hinton agar plate. After solidification, 10 μl of each CFSs was spotted onto the indicator lawn, incubated at 37°C overnight and the clear inhibition zone were observed.
Phage plaque assay
In order to confirm that the inhibitory activity was not due to bacteriophages, the phage plaque assay was performed with some modification [23]. In brief, overnight cultures of tested isolates were transferred (1:100) to fresh LB broth supplemented with mitomycin C (Ametycin® and TCI, Japan) to a final concentration of 1 μg/ml and incubated at 37°C for 4 hours. The bacterial cells were removed by centrifugation at 11,000 x g for 10 minutes and the supernatants sterilized through a 0.2 μm filter membranes.
The indicators were cultured overnight and diluted with normal saline solution to a final concentration of approximately 108 CFU/ml. One hundred microliters of the indicator strains were either mixed with 10 μl of a mitomycin C-induced CFSs or 10 μl of a mitomycin C (control), and incubated at 37°C for 30 minutes. The treated cells were added into 3 ml of molten LB medium and overlaid on LB plate. The plates were incubated overnight at 37°C and examined for plaque formation.
Bacteriocin production confirmation
Besides bacteriocins, some bacteria can produce organic acids, hydrogen peroxide (H2O2) and other antimicrobial substances that inhibit growth of sensitive bacteria. To eliminate the possibility of the interference from organic acids and H2O2, the CFSs were adjusted to pH 7 and treated with catalase enzyme (1 mg/ml, Sigma Chemical Co., Dorset, UK) for 1 hour at 37°C before antimicrobial activity testing. The CFSs were tested against E. faecalis and E. faecium using the agar well diffusion method [24].
The indicator plates were prepared as described in the spot-on-lawn method. After drying, 6 mm wells were bored and filled with 50 μl of two fold serial dilution of the CFSs. Sterile MRS broth was used as negative control. The activity was expressed in arbitrary unit (AU/ml), the reciprocal of the highest dilution that inhibit the growth of the indicator strains [25]. The assay done in triplicate for at least two independent experiments and the average value was reported.
Inhibitory spectrum of EF478 against MDRE and VRE
To determine the inhibitory spectrum of bacteriocin EF478, the antimicrobial activity of this bacteriocin was evaluated against 68 clinical isolates of MDRE and VRE using the spot-on-lawn method described above.
Identification of bacteriocin-producing strain
The producing strain that showed the strongest inhibitory activity against E. faecalis and E. faecium was identified by conventional method and the automated MicroScan, Walk-Away system (Siemens Healthcare Diagnostics, USA) using 96 well gram-positive combination panels (PC21). Results were interpreted using the Lab Pro program, version 3.01, following the Clinical and laboratory Standards Institute guidelines [26]. The strain was further characterized by MALDI-TOF-MS (autoflex™ speed MALDI-TOF/TOF, Bruker Daltonics, Germany) using flexControl version 3.4. The obtained results were analyzed by comparing the raw spectra with the spectra of the company’s library and expressed as score. The score ranged from 0 to 3 as recommended by the manufacturer. Score values of >1.7 generally indicated relationships at the genus level, and values of >2.0 generally indicated relationships at the species level. The highest score was used for species identification.
Bacteriocin production optimization
The optimal condition for bacteriocin EF478 production was evaluated by varying culture media, medium pH, and incubation temperature and culture length. Three growth media namely Luria-Bertani medium (LB), De Man Rogosa Sharpe medium (MRS) and brain heart infusion broth (BHI) (all were purchased from Merck, Darmstadt, Germany) were used for comparison. After incubation at 37°C overnight, the antimicrobial activity of the CFS derived from each growing medium was tested against E. faecalis ATCC 51299 by agar well diffusion method. The medium with the highest antimicrobial activity was selected for further tests.
Optimal pH for the production of bacteriocin EF478 was evaluated by adjusting culture medium from pH 4 to pH 8 using 1 N HCL or 1 N NaOH. After inoculation and incubation at 37°C overnight, the culture media were readjusted to pH 7 before being measured for its antimicrobial activity against E. faecalis ATCC 51299.
To establish the optimal temperature for bacteriocin production, EF478 was inoculated in the selected medium and pH and incubated with overnight shaking at 25°C, 37°C and 42°C. The CFS derived from each incubation temperature was examined for the antimicrobial activity. To study the kinetics of bacteriocin production, the EF478 was cultivated in the optimal medium, pH and temperature. Aliquots of the sample were analyzed for the antibacterial activity at 0, 8, 16, 24, 32, 40, 48 and 56 hours after incubation.
Mitomycin C, a DNA damaging agent, has been used to induce several bacteriocin production. Mitomycin C (Ametycin®, TCI, Japan) was added to the mid-log phase culture of the EF478 at the final concentration of 0.25 mg/ml, 0.50 mg/ml and 1 mg/ml. After 4 hours incubation, the CFSs were prepared and protein concentration determined by Bradford methods (Bio-Rad Laboratories Inc., Hercules, CA, USA) [27]. Antimicrobial activity against E. faecalis ATCC 51299 was determined by agar well diffusion method using non-induced supernatants as a control. All of the assays were done in triplicate for at least two independent experiments and the average values were reported.
Susceptibility to enzymes, heat and pH treatment
The susceptibility of bacteriocin EF478 to proteolytic cleavage, enzymatic activities, heat treatments and different pH levels was examined. The effect of 1 mg/ml of proteinase K, trypsin, chymotrypsin, amylase and lipase (Sigma Chemical Co., Dorset, UK) on the bacteriocin activity was determined after co-incubation at 37°C for 2 hours. The residual antibacterial activity of the enzyme-treated bacteriocin was then measured against E. faecalis ATCC 51299 by agar well diffusion method. CFS without enzyme treatment was used for comparison.
The heat stability of the bacteriocin EF478 was evaluated by exposing the bacteriocin to various temperatures (37°C, 60°C, 80°C, 100°C) for 30 min or autoclaved at 121°C for 15 min. The remaining antimicrobial activity against E. faecalis ATCC 51299 of the cooled samples was tested by agar well diffusion method.
The effect of pH on the bacteriocin was assessed by adjusting the pH of the crude bacteriocin EF478 from 2 to 12 using either 1 N hydrochloric acid (HCl) or 1 N sodium hydroxide (NaOH). After storage at 25°C for 3 hours, samples were readjusted to pH 7 and the residual antimicrobial activity determined by agar well-diffusion method. All of the assays were done in triplicate for at least two independent experiments and the average values were reported.
Storage condition
Stability of the bacteriocin EF478 after storage at various temperature was determined. The CFS aliquot of 1 ml aliquot tubes at three different temperatures (25°C, 4°C, and -20°C). The antimicrobial activities of stored aliquots and fresh CFS were compared by agar well-diffusion method against E. faecalis ATCC 51299.
Concentration and purification of bacteriocin
Bacteriocin EF478 was size-fractionated and concentrated using Amicon Ultra 30K, 10K (Amicon, Millipore Co., Germany) and Macrosep 3K (Pall Macrosep®, USA), respectively. The concentrated protein from each fraction was resolved in SDS-PAGE with 12% (v/v) separating gel and antibacterial activity was detected by gel overlay method. The active fraction was further applied to reversed-phase chromatography using a LiChrolut RP-18 column (40–63 μm) (Merck KGaA, Darmstadt, Germany). The protein sample was eluted with a linear gradient (20% to 80%) of Milli-Q water-acetonitrile containing 0.1% formic acid. The antimicrobial activity of each purified fraction was examined against E. faecalis ATCC 51299.
Molecular mass and amino acid sequence determination
The protein band with antibacterial activity was cut and subjected to in-gel tryptic digestion. Tryptic peptide sample were analyzed for amino acid sequences using maXis IITM ESI–QTOF (Bruker Daltonics) coupled with an UltiMate 3000 nano-LC system (Dionex, Surrey, UK). The peptide MS fragments were searched against the non-redundant National Center for Biotechnology Information GenBank database (NCBI, www.ncbi.nlm.nih.gov/) and Bacteria (Eubacteria) database using version 2.5.1.2 of the MASCOT search engine (Matrix Science, London, UK, http://www.matrixscience.com). The search parameters were trypsin digest, monoisotopic mass, and allowing a maximum of one missed cleavage. Peptide and fragment mass tolerance were set as 1.2 Da and 0.6 Da, respectively. Variable modifications were set to carbamidomethylation of cysteine and oxidation of methionine. The instrument type was specified as ESI-QUAD-TOF. Proteins matched with scores over 65 were considered to be significant (p< 0.05). Proteins were validated base on protein scores, peptides matches, and percent sequence coverage.
DNA sequence of the bacteriocin gene
PCR amplification and nucleotide sequencing of the gene coding bacteriocin EF478 was carried out. Genomic DNA of E. faecalis 478 was extracted using NucleoSpin® Tissue kit (Macherey-Nagel, Bethlehem, PA) according to the manufacturer’s recommendations. The primers were designed based on the sequence that best matched the peptide mass fingerprinting. The primers SP-EF478- forward: ATGAAAAAACGCCTGTTTGCGA and SP-EF478-reverse: CGCGCTATGGCCCACAAT were synthesized by Sigma-Aldrich (SigmaAldrich Co., St. Louis, MO, USA). The reaction mixture contained 100 ng of DNA template, 1x Phusion HF Buffer, 200 μM of each dNTPs, 500 nM of each primers, 1 U Phusion High-Fidelity DNA Polymerase (Finnzymes Oy, Vantaa, Finland), and deionized water to a final volume of 50 μl. PCR were performed using the following conditions: an initial denaturation at 98°C for 2 minutes, followed by 35 cycles of denaturation at 98°C for 10 seconds, annealing at 64°C for 30 seconds, extension at 72°C for 30 seconds, and a final extension at 72°C for 5 minutes. Nucleotide sequencing of the PCR product was accomplished by Solgent Co. Ltd. (Daejeon, South Korea). The sequence was compared to the non-redundant nucleotide collection of the National Center for Biotechnology Information (NCBI) databases using standard nucleotide BLAST (BLASTN) (http://www.ncbi.nlm.nih.gov/BLAST). The sequence was also searched for homology with bacteriocin published in databases, including BACTIBASE [28] and BAGEL [29].
Protein structure and function prediction
The E. faecalis serine protease WP_002367871 was used to compare with the EF478 bacteriocin. The PDB files and the ribbon structures were created using I-TASSER server [30–33] and CCP4mg molecular graphics software [34], respectively.
Ethics statement
This study was approved by the Ethical Review Committee for Human Research Faculty of Public Health, Mahidol University and Faculty of Medicine Vajira Hospital, Navamindradhiraj University. The sources of water samples were communal water supplies, no specific permissions were required for water collection at these locations. The field studies did not involve endangered or protected species.
Results
Antimicrobial activity of bacterial isolates
From all bacterial isolates, about 1.6% (165/10, 215) inhibited the growth of one or both indicator strains (Table 1) with inhibition zone varying from 2 to 12 mm. The largest inhibition zone (12 mm) was observed from a clinical isolate. Twenty isolates were selected to prepare the CFSs based on their inhibitory activities against both indicator strains.
Table 1. Sources of isolated bacteria and number of isolates (%) that showed inhibition of indicator strains.
| Sources | No. of isolates | Isolates with growth inhibition (%) | |||
|---|---|---|---|---|---|
| Total | Antibacterial activity against | ||||
| E. faecalis *ATCC51299 | E. faecium *ATCC35667 | Both | |||
| Water | 9,900 | 81(0.82) | 40 (0.40) | 38 (0.38) | 3 (0.03) |
| Environment | 180 | 18 (10) | 8 (4.44) | 7 (3.89) | 3 (1.67) |
| Clinical** | 135 | 66 (48.89) | 28 (20.74) | 23 (17.04) | 15 (11.11) |
| Total | 10,215 | 165 (1.62) | 76 (0.74) | 68 (0.67) | 21 (0.21) |
*ATCC, American Type Culture Collection
**Stool and rectal swab samples
The twenty CFSs were examined for their inhibitory activity using the spot-on-lawn assay. Only, 4 out of 20 CFSs exhibited potent inhibitory activity against the indicator strains. For these 4 strains (EF 344, EF 349, EF 355 and EF478), the CFSs minimal inhibitory concentration against E. faecalis ATCC 51299 and E. faecium ATCC 35667 was measured using agar-well diffusion method. EF 355 and EF478 exhibited the highest activity (320 AU/ml) of the 4 isolates, against both indicators (Table 2).
Table 2. Antimicrobial activity of four CFSs against indicators E. faecalis ATCC 51299 and E. faecium ATCC 35667 as measured by agar well diffusion assay.
| Test isolates | CFS activity (AU/ml) against | |
|---|---|---|
| E. faecalis ATCC51299 | E. faecium ATCC35667 | |
| EF344 | 80 | 80 |
| EF355 | 320 | 320 |
| EF349 | 160 | 80 |
| EF478 | 320 | 320 |
AU/ml: arbitrary unit
The antimicrobial activity of both EF 355 and EF478 was then tested against 68 clinical isolates of MDRE and VRE. EF478 was able to inhibit 28 from 68 (41.1%) clinical isolates, whereas EF355 inhibited 26 of 68 isolates (38.2%). Therefore, we selected EF478 for further study and characterization.
Characterization of the bacteriocin EF478
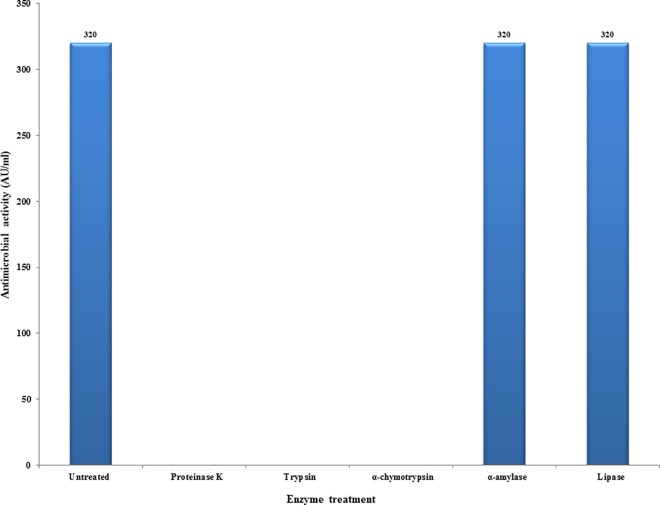
Protease susceptibility test was performed to confirm the nature of EF478 CFS. The antimicrobial activity of the CFS of EF478 was completely eliminated after proteinase K, trypsin and α-chymotrypsin treatment. However, its activity was preserved after incubation with amylase and lipase (Fig 1). The results indicated that the active compounds in EF478 CFS were proteinaceous in nature, and that potential carbohydrate or lipid moieties were not important for the bactericidal activity.
Fig 1. Effect of enzymatic treatments on activity of the EF478 CFS.
The bacteriocin EF478 maintained its antimicrobial activity over a 2 to 8 pH range, with a maximum of 320 AU/ml against E. faecalis ATCC 51299 at pH 5–6. At pH 4 and 7, the activity was reduced to 160 AU/ml whereas there was a significant reduction in the activity at pH 2. The activity was completely lost at pH above 10 (Fig 2).
Fig 2. Effect of pH on antimicrobial activity of the EF478 CFS.
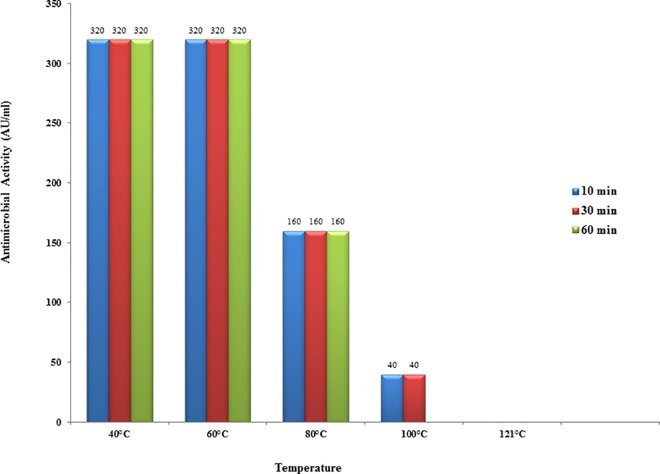
EF478 CFS retained its activity after 1 hour at 60°C, and was reduced to half (50%) and 12.5% after heating at 80°C for 1 hour and 100°C for 30 min, respectively. Its activity was abolished by heating at 100°C for 1 hour or following autoclaving (Fig 3).
Fig 3. Effect of heat on antimicrobial activity of the EF478 CFS.
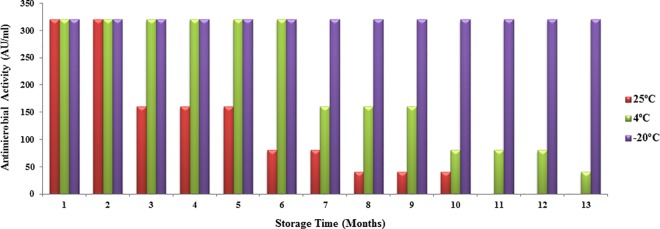
The full antimicrobial activity of EF478 CFS was retained after storage for 1 year at -20°C, 6 months at 4°C, and 2 months at 25°C (Fig 4). The remaining activity was 50% and 25% of its initial value after storage at 4°C for 9 and 12 months, respectively. At room temperature, its activity decreased to 50% after 5 months and 25% after 7 months. The antimicrobial activity was lost after storage at 25°C for 10 months.
Fig 4. Stability after storage of the EF478 CFS.
Identification of the bacteriocin-producing strain
The bacteriocin-producing strain EF478 was isolated from stool samples. It was identified as Enterococcus faecalis by Gram staining and biochemical tests and was confirmed using the automated MicroScan system (Siemens Healthcare Diagnostics) with Lab Pro software database version 3.01 (Beckman Coulter, Sacramento, CA) and MALDI-TOF MS (Microflex, Bruker Daltonics) associated flexControl version 3.4. The highest score obtained after comparison to the predicted protein database was 2.27 and matches Enterococcus faecalis 104575 LDW. In addition, Kirby-Bauer disk diffusion tests showed that this strain was sensitive to antibiotics including gentamycin, teicoplanin, and vancomycin, and resistant to ampicillin, ciprofloxacin and tetracycline.
Optimal condition for bacteriocin production by E. faecalis EF478
To optimize bacteriocin-production and antimicrobial activity, different suitable medium and cultural conditions were tested. EF478 was grown in LB, BHI and MRS broths at 37°C for 24 hours and CFSs from each medium were tested against E. faecalis ATCC 51299. MRS medium showed the highest antimicrobial activity (640 AU/ml). Similar experiments were conducted for optimal medium pH, culture length and temperature. The highest activity (640 AU/ml) was found at pH 5 and 6, after 16–24 hours of culture at 37°C (Fig 5A–5D).
Fig 5. Optimization of bacteriocin-production by E. faecalis EF478.
(A) Culture media (B) Medium pH (C) Incubation temperature (D) Incubation time. LB, Luria-Bertani; BHI, brain heart infusion; MRS, De Man Rogosa Sharpe. AU/ml: arbitrary unit.
Induction of bacteriocin EF478
Mitomycin C is commonly used to enhance the production of bacteriocins. We next measured its ability to increase bacteriocin EF478 activity using the agar well diffusion. As shown in Table 3, the antimicrobial activity of bacteriocin EF 487 doubled after addition of mitomycin C (0.25 mg/ml) in to the culture medium. No further increase was observed for mitomycin C concentration above 0.25 mg/ml, and no induction was observed below this concentration.
Table 3. Total proteins concentration (measured by Bradford assay) in the culture supernatants after mitomycin C treatment [27].
| Mitomycin C (mg/ml) | Protein concentration (mg/ml) | Antimicrobial activity (AU/ml) |
|---|---|---|
| Non-induced | 0.19 | 640 |
| 0.25 mg/ml | 2.96 | 1,280 |
| 0.50 mg/ml | 3.70 | 1,280 |
| 1 mg/ml | 4.02 | 1,280 |
SDS-PAGE and antimicrobial activity testing
EF478 supernatant was then fractionated and concentrated using column with different pore sizes. No antimicrobial activity was detected for the fractions with molecular weight below 10 kDa. However, potent antimicrobial activity was observed in the fraction above 10 kDa, and it corresponded with the presence of a major band of ~ 45 kDa, as determined by SDS-PAGE using protein standard (Fig 6).
Fig 6. SDS-PAGE and antimicrobial activity analysis of EF478 CFS.
(A): Coomassie blue staining from SDS-PAGE of the concentrated CFS of EF478. (B): Inhibition zone on the indicator lawn from the unstained gel of 45 kDa fragment.
Peptide mass fingerprint identification of bacteriocin EF478
The 45 kDa protein band was cut from SDS-PAGE gel and subjected to nLC-ESI-MS/MS analysis to identify its amino acid sequence. The obtained peptide masses and fragmentation spectra were compared with the protein sequence in Genbank NCBI database using BLAST. Bacteriocin EF478 highest similarity was with a serine protease produced by E. faecalis (accession number gi|488296663, NCBI reference sequence: WP002367871.1). The amino acid sequence included 452 residues, and the calculated molecular weight of 47,809 Da corresponded to the mass (~45 KDa) measured by SDS-PAGE analysis (Fig 6). Moreover, 21 peptides were matched, 19 of which were found in the genome database of E. faecalis, covering 43% of the total sequence (S2 Fig). The sequence and some characteristics of each peptide are summarized in Table 4.
Table 4. Peptide fingerprinting report of EF478 using Mascot search engine.
| Observed | Mr(expt) | Mr(calc) | Score | Peptide sequence |
|---|---|---|---|---|
| 459.73 | 917.45 | 917.48 | 37 | K.NQEISSLK.A |
| 474.73 | 947.45 | 947.47 | 23 | K.QSELNVMK.A |
| 482.72 | 963.44 | 963.46 | 41 | K.QSELNVMK.A + Oxidation (M) |
| 600.78 | 1199.56 | 1199.59 | 30 | R.DWEINPGITR.V |
| 681.32 | 1360.63 | 1360.67 | 92 | K.QLEATEAELETK.R |
| 469.89 | 1406.65 | 1406.68 | 87 | K.ASLALEQSSAESSK.A |
| 704.33 | 1406.65 | 1406.68 | 86 | K.ASLALEQSSAESSK.A |
| 719.83 | 1406.65 | 1437.68 | 111 | R.VGFGYSGSTIVGHSA.- |
| 506.58 | 1516.73 | 1516.77 | 54 | K.QLEATEAELETKR.Q |
| 530.27 | 1587.79 | 1587.83 | 53 | K.VKQLEATEAELETK.R |
| 567.60 | 1699.80 | 1699.84 | 75 | K.SEQLQQEITNLNQR.I |
| 850.91 | 1699.80 | 1699.84 | 116 | K.SEQLQQEITNLNQR.I |
| 569.93 | 1706.77 | 1706.81 | 43 | R.AAQVEAGGIPNDHWSR.G |
| 854.39 | 1706.77 | 1706.81 | 23 | R.AAQVEAGGIPNDHWSR.G |
| 633.98 | 1898.93 | 1898.98 | 67 | K.AKSEQLQQEITNLNQR.I |
| 635.98 | 1904.92 | 1904.96 | 62 | K.ASLALEQSSAESSKAGLEK.Q |
| 697.33 | 2088.99 | 2089.04 | 32 | R.VQAVSTIVSANNDLMQQQK.E+ Oxidation (M) |
| 713.71 | 2138.10 | 2138.15 | 99 | R.QSLGLRPVVWDAGLAASATAR.A |
| 758.05 | 2271.15 | 2271.21 | 37 | K.DPIPTPPPATGDVTLDALNVLR.Q |
| 931.42 | 2791.25 | 2790.33 | 54 | R.DVQVNGQSTTMLDAVLDADSVADAISR.V |
| 936.75 | 2807.24 | 2806.32 | 68 | R.DVQVNGQSTTMLDAVLDADSVADAISR.V + Oxidation (M) |
Mr (expt) represented the molecular mass of peptide in experiment; Mr (calc) showed the molecular mass of peptide in calculation.
Identification of the bacteriocin gene
The PCR targeting the EF478 bacteriocin gene generated a single DNA fragment of expected size (1,356 bp) (Fig 7). The nucleotide sequence has been submitted to the GenBank (NCBI/Bankit/GenBank) and assigned the accession number KU641393 (S3 Fig). Its deduced amino acid was 99.53% identity with E. faecalis serine protease accession number WP_002367871 (Fig 8), which confirmed the results of the nLC-ESI-MS/MS.
Fig 7. PCR amplification of E. faecalis EF478 bacteriocin gene.
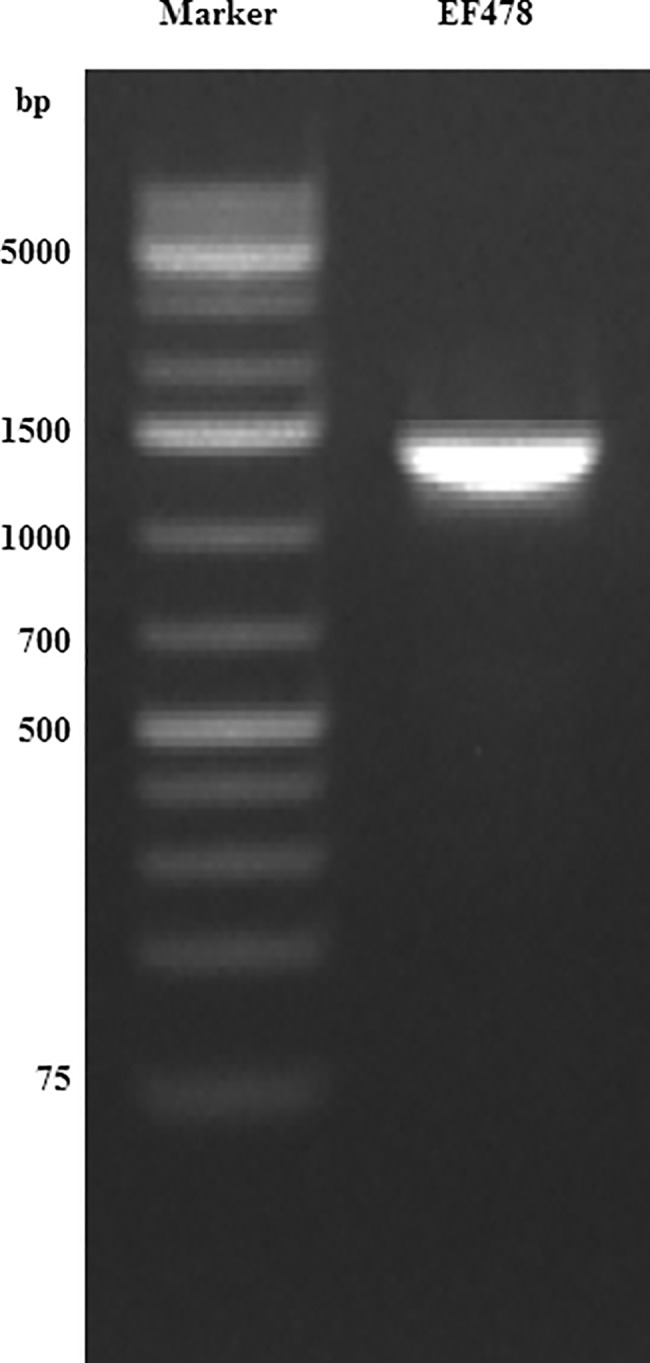
M: 1000-bp ladder plus, Lanes 1: PCR product of bacteriocin gene.
Fig 8. Amino acid sequence alignment of E. faecalis serine proteases WP_002367871 and the predicted EF478, using CLUSTALW.
Identical amino acids are indicated by asterisks. Conserved regions of uncharacterized N-terminal domain of peptidoglycan hydrolase CwlO1 and CAP domains were highlighted with grey and red, respectively. Substituted amino acid residues were highlighted (N to S, and N to G) in yellow.
Predicted protein structure and function
The probable protein functions of the EF478 bacteriocin was predicted by identification of conserved domains using NCBI conserved domain database (CDD) [35]. The CDD analysis revealed that the EF478 bacteriocin contains two conserved domains: a CwlO1 domain (uncharacterized N-terminal domain of peptidoglycan hydrolase CwlO) (E-value 3.82e-37) and a CAP superfamily domain (cysteine-rich secretory proteins, antigen 5, and pathogenesis-related 1 proteins) (E-value 4.02e-10) (Fig 8). The PDB files and ribbon structures of EF478 bacteriocin and the E. faecalis serine protease WP_002367871 were created and compared (Fig 9A and 9B).
Fig 9. Ribbon diagrams of EF478 bacteriocin and E. faecalis serine protease.

WP_002367871: (A) EF478 bacteriocin, (B) E. faecalis serine protease WP_002367871. The zoomed in region is the CAP superfamily domain. There is only one amino acid difference between EF478 bacteriocin and E. faecalis serine protease WP_002367871, but they have a big difference in their secondary structure and perhaps their functions. α-helices (red), β-sheets (blue) and bulge (green).
Discussion
The rapid emergence and spread of vancomycin-resistant enterococci is a threat to public health globally, as vancomycin is often the last drug of choice for severe enterococcal infections. Hence, MDRE and VRE infections are medical challenge with poor prognosis, due to the unavailability of appropriate drugs for treatment [36, 37]. In addition to antimicrobial stewardship program to solve the MDR organisms (MDRO) as well as many preventive measures, public health research focused on identifying novel antimicrobial compounds against MDROs. This study aimed to discover novel bacteriocins effective against clinical isolates of MDR- E. faecium and E. faecalis, which are the two most common causes of MDRE infections.
A total of 10,215 bacteria isolated from water, hospital laboratory environment and clinical samples were screened for their antimicrobial activity using bacterial inhibition, spot on the lawn and agar well diffusion assays. Bacteriocins commonly inhibit the growth of bacteria that are closely related to the producing strain. Therefore, we selected sampling locations where enterococci are commonly present [19, 38–42]. Bacteriocins are usually produced in response to an unfavorable environment; detection rate in the natural habitat would then be lower than in clinical settings. In this study we found 165 isolates with antimicrobial property against two indicator strains, i.e. E. faecalis ATCC 51299 (a high-level aminoglycoside and vancomycin-resistant strain) and E. faecium ATCC 35677 (a vancomycin-susceptible strain). The overall detection rate in our study (1.6%) was slightly low when compared with previous reports [19, 43, 44]. The highest proportion of antimicrobial-producing microorganisms was detected from clinical isolates (48.8%), followed by environmental isolates (10%) and water isolates (0.82%). Previous studies also reported that the highest proportion of bacteriocin-producing bacteria (40%-60%) was found in clinical samples [39, 45, 46]. The detection of bacteriocin-producers has been shown to be influenced by the method used, the sensitivity of the indicator strains to the bacteriocin, the source of bacteriocin-producer [19, 47–49]. Therefore, we used 3 different assays to detect as well as 2 different indicator strains to avoid method-based detection biased.
E. faecalis 478 isolated from stool sample showed the highest antimicrobial activity against indicators and clinical MDRE and VRE samples. Previous studies have reported that bacteriocins produced by Enterococcus spp. have a broad spectrum of activity to a wide range of Gram-positive bacteria [50–52]. The ability to produce bacteriocin might confer a selective advantage in bacteria communities [14, 39]. As the human gastrointestinal tract (GI) flora is one of the most dynamic and complex ecosystems, bacteriocin production could give bacteria a high competitive advantage [53]. Enterococcus species are lactic acid bacteria (LAB) and have been recognized as the most attractive bacteriocin-producers. They are used in food industry for food preservation and in fermentation processes [17, 52]. Culture of these bacteria is very flexible and their bacteriocins are considerably active and stable [41].
Many of bacterial interactions and the subsequent signalings occur efficiently when cells are in very close proximity [54]. Some bacteria can inhibit growth of target cells only through direct cell contact. On the other hand, bacteriocin is secreted extracellularly to kill the susceptible targeted cells, which have specific receptor for the bacteriocin. The antimicrobial activities of the CFSs from the selected isolates were determined using the commonly used methods i.e. the spot on the lawn and agar well diffusion [55].
The spot on the lawn method was a simple, rapid and easy to detect the antimicrobial activity of large number of CFS samples. The possible interfering effects of organic acids, H2O2 and bacteriophages were excluded by adjusting the supernatant to neutral pH, the addition of catalase and by performing plaque assay, respectively.
Furthermore, the agar well diffusion assay used for quantification of the bacteriocin activity also confirmed that the inhibition was not due to the presence of bacteriophages as they cannot diffuse through the agar [56].
The antimicrobial activity of CFS EF478 was inactivated by proteolytic enzymes, demonstrating its proteinaceous nature. Therefore, CFS EF478 was considered a bacteriocin and named bacteriocin EF478. Heat stability at 60°C of bacteriocin EF478 was not as high as the enterocin of E. faecalis BFE 1071 [57] (100°C for 60 min) or anti-listerial bacteriocin of E. faecium (100°C for 30 min) [58]. The pH tolerance was between 2 to 8, with a maximal antimicrobial activity at pH 5 and 6 in MRS medium. This result was comparable to some lactic acid bacteriocins presenting activities at the low pH values, of the gastrointestinal tract [59, 60]. The long-term stability of bacteriocin EF478 makes it an antimicrobial compound of interest.
Three commercial media, namely MRS, BHI and LB, were chosen to optimize EF478 production, and MRS gave the highest bacteriocin EF478. MRS, BHI and LB contain similar major ingredients for general growth requirements of enterococci including peptone, sucrose, yeast extract and sodium chloride [61–64]. The major ingredients found in MRS but absent in BHI and LB include potassium dihydrogen phosphate, magnesium sulfate, tween 80, sodium acetate and manganese sulfate [65]. Potassium dihydrogen phosphate and magnesium sulfate are pH buffering agents that control the pH of medium and provide cations used in its cell metabolism [62]. The surfactant properties of tween 80 help prevents the aggregation of bacteriocin molecules and stimulates protein secretion [66–68]. Tween 80 has also been used as an additive in the culture media of several LAB producing-strains to promote the cellular uptake of nutrients [63, 66, 67, 69]. Sodium acetate has been shown to suppress the growth of many bacteria competing with LAB. Manganese ions, a co-factor of the lactate dehydrogenase, are provided by manganese sulfate [70]. The optimal condition for bacteriocin EF478 production was observed after 16 hours of cultivation in MRS, at pH 5–6 and temperature 37°C. Bacteriocin EF478 expression was also doubled by the addition of mitomycin C (0.25–1 mg/ml).
This bacteriocin was concentrated and separated using three different sizes of ultrafiltration membrane spin-column and purified by reverse phase-chromatography. SDS-PAGE analysis revealed that a major 45 kDa protein was solely responsible for the antimicrobial activity. The band corresponding to bacteriocin EF478 was subjected to nLC-ESI-MS/MS analysis, and the identified peptides corresponded to a serine protease from E. faecalis (accession number WP_00236787). The molecular weight of the predicted protein was 47,809 Da, similar to the molecular weight observed on the SDS-PAGE gel. Moreover, a 1,356 base pair gene encoding the serine protease was amplified from the genomic DNA of E. faecalis 478. This finding strongly confirmed the presence of a serine protease produced by E. faecalis 478.
Serine proteases in prokaryotes and eukaryotes are involved in diverse processes such as digestion, blood clotting, immune responses, inflammation, cell signaling, pathogenesis, cell adhesion, metabolism, protein degradation and post-translation modification [71–78]. Several microorganisms secrete serine protease toxins, such as the subtilase-like serine protease of E. coli [76], HtrA of Streptococcus spp. and E. coli [60, 62, 79, 80], elastolytic of Aspergillus fumigatus [74], BmooSP of Bothrops moojeni [81], and Pro1 of Plasmodiophora brassicae. Therefore, the EF478 protein identified here was likely to be Bacteriocin EF478.
The CDD analysis revealed that Bacteriocin EF 478 contained two putative conserved domains: an uncharacterized N-terminal domain of peptidoglycan hydrolase CwlO (CWlO1) domain and a CAP-superfamily domain. CwlO1 are autolysins with peptidoglycan hydrolases (PHs) activity produced by bacteria. PHs usually contains a lysin motif that can interact with peptidoglycans of various Gram-positive bacteria and therefore are commonly called lysozymes [82]. CwlO plays significant role in the autolysis, cell growth, cell separation, and biofilm formation of bacteria [83, 84]. The CAP protein superfamily contains three major groups of proteins including the cysteine-rich secretory proteins (CRISPs), antigen 5 (Ag5), and pathogenesis-related 1 (PR1) and has been found in fungi, insects, reptiles, mammals and plants [85–87]. The proteins in the CAP superfamily play key roles in important biological such as the host defense, tumor suppression and fertilization [85, 88, 89].
All of the information suggested that bactericidal activity of the bacteriocin EF478 depended on the CwlO domain whereas the CAP domain played a role in protection of the producer.
The deduced amino acid of bacteriocin EF478 was nearly identity (99.53%) with E. faecalis serine protease accession number WP_002367871. There is only one amino acid difference within the CAP domain, but they have a big difference in the predicted secondary structure. As mentioned earlier that CAP domain function as immunity to the bacteriocin producer, the change within this domain might be a critical. We analyzed 25 sequences of E. faecalis serine protease from NCBI database and found that nearly all of them possessed the same amino acid with the WP_002367871. Only one strain that share the same amino acid with bacteriocin EF478 is the strain DD14 (accession number CP021161), which was reported as bacteriocinogenic lactic acid bacteria. To date, there is a scarcity of new potent antimicrobial agents for the treatment of VRE and MDRE. Serine protease from E. faecalis 478 is attractive potential therapeutic agents for the treatment of VRE and MDRE. Bacteriocin EF478 was not present in both bacteriocin databases, Bactibase and Bagel 3. Homology searches revealed that the predicted bacteriocin EF478 was novel to this species. Finally, we have discovered a novel bacteriocin from E. faecalis 478 with a potent antimicrobial activity against clinical strains of VRE. Its high activity in a wide range of pH and temperature, resilience to heat and storage stability make bacteriocin EF478 a promising candidate as an anti-MDRE and VRE antimicrobial.
Supporting information
(DOCX)
The amino acids in bold red indicated the matched peptides obtained by the MASCOT database search with the ESI tandem MS data.
(DOCX)
(DOCX)
(DOCX)
Acknowledgments
We would like to thank the Faculty of Medicine Vajira Hospital, Navamindradhiraj University, Thailand for permissions of sample collections. We are grateful to staff at the Laboratory of Molecular Tropical Medicine and Genetics, Faculty of Tropical Medicine, Mahidol University for helping with protein identification. In particular, we would like to express our deep gratitude to Dr. Fabien Loison for his time to edit our manuscript.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
The study was partially supported for publication by the China Medical Board (CMB), Faculty of Public Health, Mahidol University, Bangkok, Thailand. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Agudelo Higuita N, Huycke M. Enterococcal Disease, Epidemiology, and Implications for Treatment In: Gilmore MS, Clewell DB, Ike Y, Shankar N, editors. Enterococci: From Commensals to Leading Causes of Drug Resistant Infection. Massachusetts Eye and Ear Infirmary; Boston: 2014. [PubMed] [Google Scholar]
- 2.Baldassarri L, Creti R, Montanaro L, Orefici G, Arciola CR. Pathogenesis of implant infections by enterococci. Int J Artif Organs 2005;28(11):1101–9. [DOI] [PubMed] [Google Scholar]
- 3.Kristich CJ, Rice LB, Arias CA. Enterococcal Infection-Treatment and Antibiotic Resistance In: Gilmore MS, Clewell DB, Ike Y, Shankar N, editors. Enterococci: From Commensals to Leading Causes of Drug Resistant Infection. Massachusetts Eye and Ear Infirmary; Boston: 2014. [PubMed] [Google Scholar]
- 4.Bonten MJ, Hayden MK, Nathan C, van Voorhis J, Matushek M, Slaughter S, et al. Epidemiology of colonisation of patients and environment with vancomycin-resistant enterococci. Lancet 1996;348(9042):1615–9. doi: 10.1016/S0140-6736(96)02331-8 [DOI] [PubMed] [Google Scholar]
- 5.Fisher K, Phillips C. The ecology, epidemiology and virulence of Enterococcus. Microbiology 2009;155(Pt 6):1749–57. doi: 10.1099/mic.0.026385-0 [DOI] [PubMed] [Google Scholar]
- 6.Leclercq R, Derlot E, Duval J, Courvalin P. Plasmid-mediated resistance to vancomycin and teicoplanin in Enterococcus faecium. N Engl J Med 1988;319(3):157–61. doi: 10.1056/NEJM198807213190307 [DOI] [PubMed] [Google Scholar]
- 7.Low DE, Keller N, Barth A, Jones RN. Clinical prevalence, antimicrobial susceptibility, and geographic resistance patterns of enterococci: results from the SENTRY Antimicrobial Surveillance Program, 1997–1999. Clin Infect Dis 2001;32 Suppl 2:S133–45. [DOI] [PubMed] [Google Scholar]
- 8.Pallares R, Pujol M, Pena C, Ariza J, Martin R, Gudiol F. Cephalosporins as risk factor for nosocomial Enterococcus faecalis bacteremia. A matched case-control study. Arch Intern Med 1993;153(13):1581–6. [PubMed] [Google Scholar]
- 9.Coates AR, Hu Y. Novel approaches to developing new antibiotics for bacterial infections. Br J Pharmacol 2007;152(8):1147–54. doi: 10.1038/sj.bjp.0707432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cleveland J, Montville TJ, Nes IF, Chikindas ML. Bacteriocins: safe, natural antimicrobials for food preservation. Int J Food Microbiol 2001;71(1):1–20. [DOI] [PubMed] [Google Scholar]
- 11.Joseph B, Dhas B, Hena V, Raj J. Bacteriocin from Bacillus subtilis as a novel drug against diabetic foot ulcer bacterial pathogens. Asian Pac J Trop Biomed 2013;3(12):942–6. doi: 10.1016/S2221-1691(13)60183-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Naghmouchi K, Baah J, Hober D, Jouy E, Rubrecht C, Sane F, et al. Synergistic effect between colistin and bacteriocins in controlling Gram-negative pathogens and their potential to reduce antibiotic toxicity in mammalian epithelial cells. Antimicrob Agents Chemother 2013;57(6):2719–25. doi: 10.1128/AAC.02328-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Riaz S, Kashif Nawaz S, Hasnain S. Bacteriocins produced by L. Fermentum and L. Acidophilus can inhibit cephalosporin resistant E. coli. Braz J Microbiol 2010;41(3):643–8. doi: 10.1590/S1517-83822010000300015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Riley MA. Molecular mechanisms of bacteriocin evolution. Annu Rev Genet 1998;32:255–78. doi: 10.1146/annurev.genet.32.1.255 [DOI] [PubMed] [Google Scholar]
- 15.Giuliani A, Pirri G, Nicoletto SF. Antimicrobial peptides: an overview of a promising class of therapeutics. Cent Eur J Biol 2007;2(1):1–33. [Google Scholar]
- 16.Russell JB, Mantovani HC. The bacteriocins of ruminal bacteria and their potential as an alternative to antibiotics. J Mol Microbiol Biotechnol 2002;4(4):347–55. [PubMed] [Google Scholar]
- 17.Cintas LM, Casaus P, Havarstein LS, Hernandez PE, Nes IF. Biochemical and genetic characterization of enterocin P, a novel sec-dependent bacteriocin from Enterococcus faecium P13 with a broad antimicrobial spectrum. Appl Environ Microbiol 1997;63(11):4321–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Kwaadsteniet M, Ten Doeschate K, Dicks LM. Characterization of the structural gene encoding nisin F, a new lantibiotic produced by a Lactococcus lactis subsp. lactis isolate from freshwater catfish (Clarias gariepinus). Appl Environ Microbiol 2008;74(2):547–9. doi: 10.1128/AEM.01862-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ozdemir GB, Oryasin E, Biyik HH, Ozteber M, Bozdogan B. Phenotypic and genotypic characterization of bacteriocins in enterococcal isolates of different sources. Indian J Microbiol 2011;51(2):182–7. doi: 10.1007/s12088-011-0143-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rajanbabu V, Chen JY. Applications of antimicrobial peptides from fish and perspectives for the future. Peptides 2011;32(2):415–20. doi: 10.1016/j.peptides.2010.11.005 [DOI] [PubMed] [Google Scholar]
- 21.Zendo T, Koga S, Shigeri Y, Nakayama J, Sonomoto K. Lactococcin Q, a novel two-peptide bacteriocin produced by Lactococcus lactis QU 4. Appl Environ Microbiol 2006;72(5):3383–9. doi: 10.1128/AEM.72.5.3383-3389.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ennahar S, Sonomoto K, Ishizaki A. Class IIa bacteriocins from lactic acid bacteria: antibacterial activity and food preservation. J Biosci Bioeng 1999;87(6):705–16. [DOI] [PubMed] [Google Scholar]
- 23.Imaeda T, Rieber M. Mitomycin C-induced phage-like particles in a mutant of Mycobacterium tuberculosis BCG. J Bacteriol 1968;96(2):557–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Belguesmia Y, Choiset Y, Prevost H, Dalgalarrondo M, Chobert JM, Drider D. Partial purification and characterization of the mode of action of enterocin S37: a bacteriocin produced by Enterococcus faecalis S37 isolated from poultry feces. J Environ Public Health 2010;2010:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Daba H, Pandian S, Gosselin JF, Simard RE, Huang J, Lacroix C. Detection and activity of a bacteriocin produced by Leuconostoc mesenteroides. Appl Environ Microbiol 1991;57(12):3450–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wayne P. Clinical and Laboratory Standards Institute; Performance standards for antimicrobial susceptibility testing CLSI M100-S24; 2014. [DOI] [PMC free article] [PubMed]
- 27.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 1976;72:248–54. [DOI] [PubMed] [Google Scholar]
- 28.Hammami R, Zouhir A, Le Lay C, Ben Hamida J, Fliss I. BACTIBASE second release: a database and tool platform for bacteriocin characterization. BMC Microbiol 2010;10:22 doi: 10.1186/1471-2180-10-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Heel AJ, de Jong A, Montalban-Lopez M, Kok J, Kuipers OP. BAGEL3: Automated identification of genes encoding bacteriocins and (non-) bactericidal posttranslationally modified peptides. Nucleic Acids Res 2013;41(Web Server issue):W448–53. doi: 10.1093/nar/gkt391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roy A, Kucukural A, Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nature protocols. 2010;5(4):725–38. doi: 10.1038/nprot.2010.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang J, Yan R, Roy A, Xu D, Poisson J, Zhang Y. The I-TASSER Suite: protein structure and function prediction. Nat Methods. 2015;12(1):7–8. doi: 10.1038/nmeth.3213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang J, Zhang Y. I-TASSER server: new development for protein structure and function predictions. Nucleic Acids Res. 2015;43(W1):W174–81. doi: 10.1093/nar/gkv342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics. 2008;9:40 doi: 10.1186/1471-2105-9-40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McNicholas S, Potterton E, Wilson KS, Noble ME. Presenting your structures: the CCP4mg molecular-graphics software. Acta Crystallogr D Biol Crystallogr. 2011;67(Pt 4):386–94. doi: 10.1107/S0907444911007281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marchler-Bauer A, Derbyshire MK, Gonzales NR, Lu S, Chitsaz F, Geer LY, et al. CDD: NCBI's conserved domain database. Nucleic Acids Res 2015;43(Database issue):D222–6. doi: 10.1093/nar/gku1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Papadimitriou-Olivgeris M, Drougka E, Fligou F, Kolonitsiou F, Liakopoulos A, Dodou V, et al. Risk factors for enterococcal infection and colonization by vancomycin-resistant enterococci in critically ill patients. Infection 2014;42(6):1013–22. doi: 10.1007/s15010-014-0678-1 [DOI] [PubMed] [Google Scholar]
- 37.Salgado CD. The risk of developing a vancomycin-resistant Enterococcus bloodstream infection for colonized patients. Am J Infect Control 2008;36(10):S175 e5–8. [DOI] [PubMed] [Google Scholar]
- 38.Chatterjee S, Chatterjee DK, Jani RH, Blumbach J, Ganguli BN, Klesel N, et al. Mersacidin, a new antibiotic from Bacillus. In vitro and in vivo antibacterial activity. J Antibiot (Tokyo) 1992;45(6):839–45. [DOI] [PubMed] [Google Scholar]
- 39.del Campo R, Tenorio C, Jimenez-Diaz R, Rubio C, Gomez-Lus R, Baquero F, et al. Bacteriocin production in vancomycin-resistant and vancomycin-susceptible Enterococcus isolates of different origins. Antimicrob Agents Chemother 2001;45(3):905–12. doi: 10.1128/AAC.45.3.905-912.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Giraffa G. Functionality of enterococci in dairy products. Int J Food Microbiol 2003;88(2–3):215–22. [DOI] [PubMed] [Google Scholar]
- 41.Nes IF, Diep DB, Ike Y. Enterococcal Bacteriocins and Antimicrobial Proteins that Contribute to Niche Control In: Gilmore MS, Clewell DB, Ike Y, Shankar N, editors. Enterococci: From Commensals to Leading Causes of Drug Resistant Infection. Massachusetts Eye and Ear Infirmary; Boston: 2014. [PubMed] [Google Scholar]
- 42.Valli S, Suvathi SS, Aysha OS, Nirmala P, Vinoth KP, Reena A. Antimicrobial potential of Actinomycetes species isolated from marine environment. Asian Pac J Trop Biomed 2012;2(6):469–73. doi: 10.1016/S2221-1691(12)60078-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nespolo CR, Brandelli A. Production of bacteriocin-like substances by lactic acid bacteria isolated from regional ovine cheese. Braz J Microbiol 2010;41(4):1009–18. doi: 10.1590/S1517-838220100004000020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Simova ED, Beshkova DB, Dimitrov Zh P. Characterization and antimicrobial spectrum of bacteriocins produced by lactic acid bacteria isolated from traditional Bulgarian dairy products. J Appl Microbiol 2009;106(2):692–701. doi: 10.1111/j.1365-2672.2008.04052.x [DOI] [PubMed] [Google Scholar]
- 45.Inoue T, Tomita H, Ike Y. Bac 32, a novel bacteriocin widely disseminated among clinical isolates of Enterococcus faecium. Antimicrob Agents Chemother 2006;50(4):1202–12. doi: 10.1128/AAC.50.4.1202-1212.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tomita H, Fujimoto S, Tanimoto K, Ike Y. Cloning and genetic organization of the bacteriocin 31 determinant encoded on the Enterococcus faecalis pheromone-responsive conjugative plasmid pYI17. J Bacteriol 1996;178(12):3585–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Corsetti A, Settanni L, Van Sinderen D. Characterization of bacteriocin-like inhibitory substances (BLIS) from sourdough lactic acid bacteria and evaluation of their in vitro and in situ activity. J Appl Microbiol 2004;96(3):521–34. [DOI] [PubMed] [Google Scholar]
- 48.Lewus CB, Kaiser A, Montville TJ. Inhibition of food-borne bacterial pathogens by bacteriocins from lactic acid bacteria isolated from meat. Appl Environ Microbiol 1991;57(6):1683–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Navarro L, Zarazaga M, Saenz J, Ruiz-Larrea F, Torres C. Bacteriocin production by lactic acid bacteria isolated from Rioja red wines. J Appl Microbiol 2000;88(1):44–51. [DOI] [PubMed] [Google Scholar]
- 50.Birri DJ, Brede DA, Forberg T, Holo H, Nes IF. Molecular and genetic characterization of a novel bacteriocin locus in Enterococcus avium isolates from infants. Appl Environ Microbiol 2010;76(2):483–92. doi: 10.1128/AEM.01597-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.De Vuyst L, Foulquie Moreno MR, Revets H. Screening for enterocins and detection of hemolysin and vancomycin resistance in enterococci of different origins. Int J Food Microbiol 2003;84(3):299–318. [DOI] [PubMed] [Google Scholar]
- 52.Rodriguez E, Martinez MI, Medina M, Hernandez PE, Rodriguez JM. Detection of enterocin AS-48-producing dairy enterococci by dot-blot and colony hybridization. J Dairy Res 1998;65(1):143–8. [DOI] [PubMed] [Google Scholar]
- 53.Booijink CC, Zoetendal EG, Kleerebezem M, de Vos WM. Microbial communities in the human small intestine: coupling diversity to metagenomics. Future Microbiol 2007;2(3):285–95. doi: 10.2217/17460913.2.3.285 [DOI] [PubMed] [Google Scholar]
- 54.Dunny GM, Leonard BA. Cell-cell communication in gram-positive bacteria. Annual review of microbiology. 1997;51:527–64. doi: 10.1146/annurev.micro.51.1.527 [DOI] [PubMed] [Google Scholar]
- 55.Schlegel R, Slade HD. Alteration of macromolecular synthesis and membrane permeability by a Streptococcus sanguis bacteriocin. Journal of general microbiology. 1974;81(1):275–7. doi: 10.1099/00221287-81-1-275 [DOI] [PubMed] [Google Scholar]
- 56.Tagg JR, McGiven AR. Assay system for bacteriocins. Applied microbiology. 1971;21(5):943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Balla E, Dicks LM, Du Toit M, Van Der Merwe MJ, Holzapfel WH. Characterization and cloning of the genes encoding enterocin 1071A and enterocin 1071B, two antimicrobial peptides produced by Enterococcus faecalis BFE 1071. Applied and environmental microbiology. 2000;66(4):1298–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vijayendra SV, Rajashree K, Halami PM. Characterization of a heat stable anti-listerial bacteriocin produced by vancomycin sensitive Enterococcus faecium isolated from idli batter. Indian J Microbiol 2010;50(2):243–6. doi: 10.1007/s12088-010-0030-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Contreras BG, De Vuyst L, Devreese B, Busanyova K, Raymaeckers J, Bosman F, et al. Isolation, purification, and amino acid sequence of lactobin A, one of the two bacteriocins produced by Lactobacillus amylovorus LMG P-13139. Appl Environ Microbiol 1997;63(1):13–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.du Toit M, Franz CM, Dicks LM, Holzapfel WH. Preliminary characterization of bacteriocins produced by Enterococcus faecium and Enterococcus faecalis isolated from pig faeces. J Appl Microbiol 2000;88(3):482–94. [DOI] [PubMed] [Google Scholar]
- 61.Aunpad R, Na-Bangchang K. Pumilicin 4, a novel bacteriocin with anti-MRSA and anti-VRE activity produced by newly isolated bacteria Bacillus pumilus strain WAPB4. Curr Microbiol 2007;55(4):308–13. doi: 10.1007/s00284-006-0632-2 [DOI] [PubMed] [Google Scholar]
- 62.Li C, Bai J, Cai Z, Ouyang F. Optimization of a cultural medium for bacteriocin production by Lactococcus lactis using response surface methodology. J Biotechnol 2002;93(1):27–34. [DOI] [PubMed] [Google Scholar]
- 63.Nel HA, Bauer R, Vandamme EJ, Dicks LM. Growth optimization of Pediococcus damnosus NCFB 1832 and the influence of pH and nutrients on the production of pediocin PD-1. J Appl Microbiol 2001;91(6):1131–8. [DOI] [PubMed] [Google Scholar]
- 64.Todorov SD, Dicks LM. Optimization of bacteriocin ST311LD production by Enterococcus faecium ST311LD, isolated from spoiled black olives. J Microbiol 2005;43(4):370–4. [PubMed] [Google Scholar]
- 65.Furtado DN, Todorov SD, Landgraf M, Destro MT, Franco BD. Bacteriocinogenic Lactococcus lactis subsp. lactis DF04Mi isolated from goat milk: characterization of the bacteriocin. Braz J Microbiol 2014;45(4):1541–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Aymerich T, Artigas MG, Garriga M, Monfort JM, Hugas M. Effect of sausage ingredients and additives on the production of enterocin A and B by Enterococcus faecium CTC492. Optimization of in vitro production and anti-listerial effect in dry fermented sausages. Journal of applied microbiology. 2000;88(4):686–94. [DOI] [PubMed] [Google Scholar]
- 67.Keren T, Yarmus M, Halevy G, Shapira R. Immunodetection of the bacteriocin lacticin RM: analysis of the influence of temperature and Tween 80 on its expression and activity. Appl Environ Microbiol 2004;70(4):2098–104. doi: 10.1128/AEM.70.4.2098-2104.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Reese ET, Maguire A. Surfactants as stimulants of enzyme production by microorganisms. Appl Microbiol 1969;17(2):242–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang HT, Chen IH, Hsu JT. Production and characterization of a bacteriocin from ruminal bacterium Ruminococcus albus 7. Biosci Biotechnol Biochem 2012;76(1):34–41. doi: 10.1271/bbb.110348 [DOI] [PubMed] [Google Scholar]
- 70.Chauhan K, Trivedi U, Patel KC. Statistical screening of medium components by Plackett-Burman design for lactic acid production by Lactobacillus sp. KCP01 using date juice. Bioresour Technol 2007;98(1):98–103. doi: 10.1016/j.biortech.2005.11.017 [DOI] [PubMed] [Google Scholar]
- 71.Cortes G, de Astorza B, Benedi VJ, Alberti S. Role of the htrA gene in Klebsiella pneumoniae virulence. Infect Immun 2002;70(9):4772–6. doi: 10.1128/IAI.70.9.4772-4776.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hu CC, Dougan SK, Winter SV, Paton AW, Paton JC, Ploegh HL. Subtilase cytotoxin cleaves newly synthesized BiP and blocks antibody secretion in B lymphocytes. J Exp Med 2009;206(11):2429–40. doi: 10.1084/jem.20090782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ibrahim KS, Muniyandi J, Karutha Pandian S. Purification and characterization of manganese-dependent alkaline serine protease from Bacillus pumilus TMS55. J Microbiol Biotechnol 2011;21(1):20–7. [DOI] [PubMed] [Google Scholar]
- 74.Kolattukudy PE, Lee JD, Rogers LM, Zimmerman P, Ceselski S, Fox B, et al. Evidence for possible involvement of an elastolytic serine protease in aspergillosis. Infect Immun 1993;61(6):2357–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lyon WR, Caparon MG. Role for serine protease HtrA (DegP) of Streptococcus pyogenes in the biogenesis of virulence factors SpeB and the hemolysin streptolysin S. Infect Immun 2004;72(3):1618–25. doi: 10.1128/IAI.72.3.1618-1625.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Paton AW, Beddoe T, Thorpe CM, Whisstock JC, Wilce MC, Rossjohn J, et al. AB5 subtilase cytotoxin inactivates the endoplasmic reticulum chaperone BiP. Nature 2006;443(7111):548–52. doi: 10.1038/nature05124 [DOI] [PubMed] [Google Scholar]
- 77.Tripathi LP, Sowdhamini R. Genome-wide survey of prokaryotic serine proteases: analysis of distribution and domain architectures of five serine protease families in prokaryotes. BMC Genomics 2008;9:549 doi: 10.1186/1471-2164-9-549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Watorek W. Azurocidin—inactive serine proteinase homolog acting as a multifunctional inflammatory mediator. Acta Biochim Pol 2003;50(3):743–52. [PubMed] [Google Scholar]
- 79.Dawid S, Sebert ME, Weiser JN. Bacteriocin activity of Streptococcus pneumoniae is controlled by the serine protease HtrA via posttranscriptional regulation. J Bacteriol 2009;191(5):1509–18. doi: 10.1128/JB.01213-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Krojer T, Garrido-Franco M, Huber R, Ehrmann M, Clausen T. Crystal structure of DegP (HtrA) reveals a new protease-chaperone machine. Nature 2002;416(6879):455–9. doi: 10.1038/416455a [DOI] [PubMed] [Google Scholar]
- 81.de Oliveira F, de Sousa BB, Mamede CC, de Morais NC, de Queiroz MR, da Cunha Pereira DF, et al. Biochemical and functional characterization of BmooSP, a new serine protease from Bothrops moojeni snake venom. Toxicon 2016;111:130–8. doi: 10.1016/j.toxicon.2016.01.055 [DOI] [PubMed] [Google Scholar]
- 82.Steen A, Buist G, Leenhouts KJ, El Khattabi M, Grijpstra F, Zomer AL, et al. Cell wall attachment of a widely distributed peptidoglycan binding domain is hindered by cell wall constituents. The Journal of biological chemistry. 2003;278(26):23874–81. doi: 10.1074/jbc.M211055200 [DOI] [PubMed] [Google Scholar]
- 83.Millan-Zambrano G, Chavez S. Nuclear functions of prefoldin. Open Biol. 2014;4(7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Short B, Preisinger C, Schaletzky J, Kopajtich R, Barr FA. The Rab6 GTPase regulates recruitment of the dynactin complex to Golgi membranes. Curr Biol. 2002;12(20):1792–5. [DOI] [PubMed] [Google Scholar]
- 85.Gibbs GM, Roelants K, O'Bryan MK. The CAP superfamily: cysteine-rich secretory proteins, antigen 5, and pathogenesis-related 1 proteins—roles in reproduction, cancer, and immune defense. Endocr Rev 2008;29(7):865–97. doi: 10.1210/er.2008-0032 [DOI] [PubMed] [Google Scholar]
- 86.Milne TJ, Abbenante G, Tyndall JD, Halliday J, Lewis RJ. Isolation and characterization of a cone snail protease with homology to CRISP proteins of the pathogenesis-related protein superfamily. J Biol Chem 2003;278(33):31105–10. doi: 10.1074/jbc.M304843200 [DOI] [PubMed] [Google Scholar]
- 87.van Loon LC, Rep M, Pieterse CM. Significance of inducible defense-related proteins in infected plants. Annu Rev Phytopathol 2006;44:135–62. doi: 10.1146/annurev.phyto.44.070505.143425 [DOI] [PubMed] [Google Scholar]
- 88.Lu G, Villalba M, Coscia MR, Hoffman DR, King TP. Sequence analysis and antigenic cross-reactivity of a venom allergen, antigen 5, from hornets, wasps, and yellow jackets. J Immunol 1993;150(7):2823–30. [PubMed] [Google Scholar]
- 89.Udby L, Cowland JB, Johnsen AH, Sorensen OE, Borregaard N, Kjeldsen L. An ELISA for SGP28/CRISP-3, a cysteine-rich secretory protein in human neutrophils, plasma, and exocrine secretions. J Immunol Methods 2002;263(1–2):43–55. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
The amino acids in bold red indicated the matched peptides obtained by the MASCOT database search with the ESI tandem MS data.
(DOCX)
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.