Torsins are essential AAA+ proteins of the endoplasmic reticulum and nuclear envelope (NE). We now show that Torsin homo-oligomers are essential for Torsin function at the NE and that Torsin’s oligomeric state is negatively modulated by its cofactors, representing a novel regulation mechanism for AAA+ proteins.
Abstract
TorsinA is an essential AAA+ ATPase requiring LAP1 or LULL1 as cofactors. The dynamics of the Torsin/cofactor system remain poorly understood, with previous models invoking Torsin/cofactor assemblies with fixed stoichiometries. Here we demonstrate that TorsinA assembles into homotypic oligomers in the presence of ATP. Torsin variants mutated at the “back” interface disrupt homo-oligomerization but still show robust ATPase activity in the presence of its cofactors. These Torsin mutants are severely compromised in their ability to rescue nuclear envelope defects in Torsin-deficient cells, suggesting that TorsinA homo-oligomers play a key role in vivo. Engagement of the oligomer by LAP1 triggers ATP hydrolysis and rapid complex disassembly. Thus the Torsin complex is a highly dynamic assembly whose oligomeric state is tightly controlled by distinctively localized cellular cofactors. Our discovery that LAP1 serves as a modulator of the oligomeric state of an AAA+ protein establishes a novel means of regulating this important class of oligomeric ATPases.
INTRODUCTION
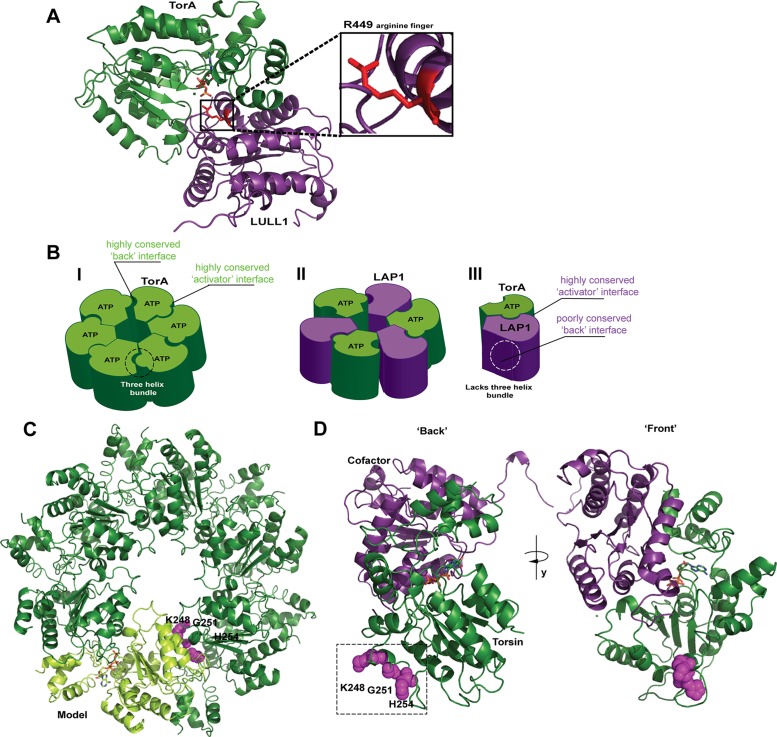
Torsins are highly conserved and essential AAA+ (ATPases associated with various activities) proteins residing in the endoplasmic reticulum (ER) lumen and perinuclear space. Torsins have been implicated in broad functionalities including ER protein quality control (Nery et al., 2011), membrane homeostasis (Rose et al., 2014), nuclear positioning (Saunders et al., 2017), and nuclear pore complex homeostasis (VanGompel et al., 2015; Laudermilch et al., 2016; for recent reviews, see Rose et al., 2015; Laudermilch and Schlieker, 2016; Cascalho et al., 2017). Several studies have shed light on the mechanism by which Torsins’ two distinctively localizing cofactors, the nuclear envelope protein LAP1 (Foisner and Gerace, 1993) and the ER protein LULL1 (Goodchild and Dauer, 2005), activate Torsins’ ATPase activity. Torsin cofactors feature AAA+-like folds that trigger ATP hydrolysis by virtue of an arginine finger-based active site complementation mechanism (Brown et al., 2014; Sosa et al., 2014; Demircioglu et al., 2016; Figure 1A).
FIGURE 1:
Models of Torsin’s functional assembly and identification of a conserved region at the “back” interface. (A) TorsinA forms a tightly apposed, ATP-dependent interface with its cofactors (here, LULL1) LDs whereby an arginine finger (red) from the cofactor can coordinate the γ-phosphate of ATP. Representation based on the LULL1-TorA crystal structure (PDB code 5J1S, nanobody used for crystallization omitted for clarity). (B) Three different models exist for the active assembly of Torsins: (I) a homo-oligomeric (likely hexameric) ring; (II) an alternating trimer of dimers; (III) a Torsin-LAP1LD heterodimer. (C) Model of a homohexameric Torsin ring based on the crystal structure of TorsinA (PDB code 5J1S). A highly conserved region at the “back” interface of each TorA monomer (light green cartoon) contains residues 248, 251, and 254 (magenta spheres) likely contributing to intraprotomer contacts. Bound ATP is shown in stick representation. (D) An ATP-bound Torsin (dark green)-cofactor (purple) complex showing the “back” interface with predicted intraprotomer contact sites at residues K248, G251, and H254 (magenta spheres) and the “front” interface. Representation based on the LULL1-TorA crystal structure (PDB code 5J1S).
One central enigma within the Torsin field has been the composition of the active, functional Torsin complex (Chase et al., 2017). Torsin species consistent with a homohexameric assembly were observed via blue native PAGE (Vander Heyden et al., 2009; Jungwirth et al., 2010) while cross-linking and structural studies led to alternative models invoking mixed ring assemblies in a 3:3 (TorsinA:cofactor) stoichiometry (Brown et al., 2014; Sosa et al., 2014) as well as a Torsin-cofactor heterodimer (Demircioglu et al., 2016; Figure 1B, I–III). Potential limitations inherent to several attempts aimed at a determination of the oligomeric state are the use of stabilizing conditions including ATPase-arresting mutations, stabilizing nanobodies, or cross-linking reagents (Brown et al., 2014; Sosa et al., 2014; Demircioglu et al., 2016), each of which does not recapitulate the dynamic situation encountered in vivo.
In this study, we analyze the formation of the active Torsin assembly using wild-type TorsinA (TorA) to revisit the nucleotide and cofactor dependency of Torsin oligomerization. Using a combination of size-exclusion chromatography and low-resolution electron microscopy (EM), we demonstrate that the TorA/LAP1 assembly is significantly more dynamic than previously appreciated. Contrary to previous belief, LAP1 is not stably integrated into Torsin rings. Instead, LAP1 binding results in rapid disassembly of the Torsin oligomer. Thus the Torsin/cofactor system is in fact a highly dynamic assembly, representing a distinct departure from previous working models.
RESULTS AND DISCUSSION
Considerations on the oligomeric state of the Torsin/cofactor system
The interaction of TorA with its cofactor through the activator binding interface is well-established as a tightly apposed, ATP-dependent interaction by structural and biochemical studies (Zhao et al., 2013; Brown et al., 2014; Sosa et al., 2014; Demircioglu et al., 2016; (Figure 1A). However, the contacts formed at the conserved “back” interface of TorA remain incompletely understood. As discussed previously, distinguishing between existing models for the active complex of Torsin (Figure 1B, I–III) requires closer scrutiny of precisely this face of TorA (Chase et al., 2017). If TorA forms a closed ring similar to related AAA+ ATPases (Kirstein et al., 2009; Figure 1B, I), then homotypic contacts through both the front and back interface would be required. Indeed, both the front and back interface are highly conserved in TorA (Demircioglu et al., 2016), and oligomeric species consistent with a hexamer were observed via blue native PAGE (Vander Heyden et al., 2009; Jungwirth et al., 2010). Alternative models suggested a mixed Torsin/cofactor ring in a 3:3 stoichiometry (Figure 1B, II; Brown et al., 2014; Sosa et al., 2014); however, LAP1 and LULL1 lack the C-terminal α-helical subdomain essential for oligomerization in related AAA+ ATPases (Mogk et al., 2003) and lack sequence conservation on their back interface. These factors called into question whether a 3:3 hetero-oligomer could be populated under equilibrium conditions, motivating a heterodimeric model for the assembly (Figure 1B, III; Demircioglu et al., 2016). Detailed knowledge of the (hetero)oligomeric state of the Torsin/cofactor system thus presents a critical step toward defining working models for Torsin function (Chase et al., 2017).
Mutations at the back interface of Torsin do not affect cofactor binding
As a first step toward discerning between these alternative models (Figure 1, B–D), we identified “back” interface residues on TorA within a conserved area (Supplemental Figure S1) and predicted close proximity to a neighboring subunit within a modeled oligomeric ring (Figure 1, C and D). This yielded residues K248, the highly conserved G251, and H254 (Figure 1, C and D). We reasoned that mutating these residues to aspartates should perturb interactions through the back interface, potentially allowing us to deconvolute Torsin homo-oligomerization and activator binding. Since the luminal domains (LDs) of LAP1 and LULL1 preferentially interact with TorA in the presence of ATP, we also introduced these mutations into a Walker B (E171Q) background that is arrested in the ATP-bound state, allowing us to monitor cofactor binding (Naismith et al., 2009; Zhu et al., 2010; Zhao et al., 2013).
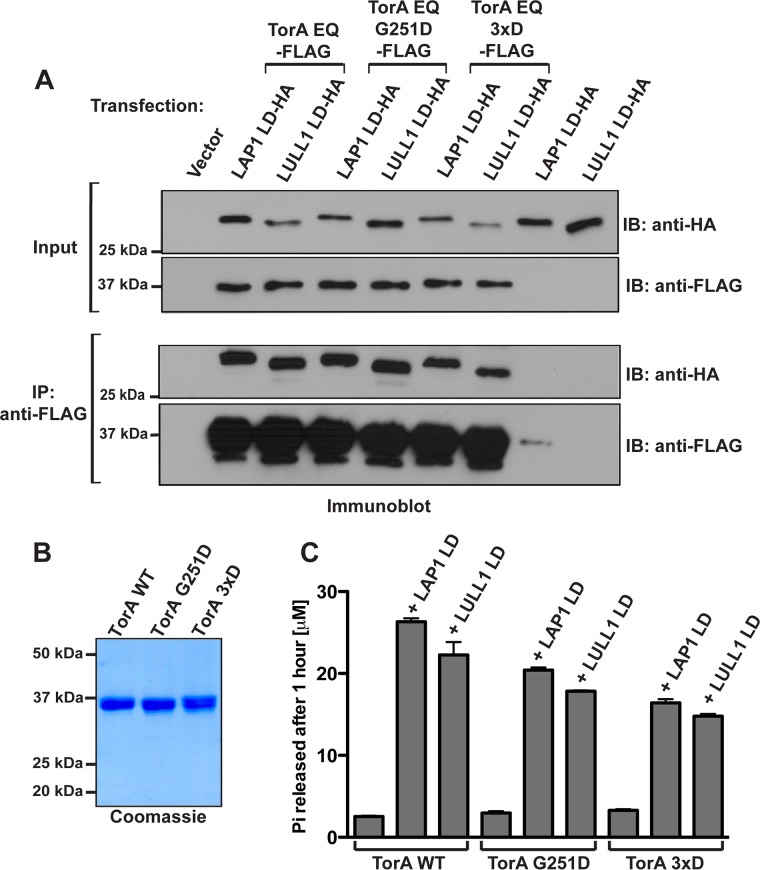
To determine whether cofactor binding is compromised by these mutations–a scenario that we would predict for an assembly in a 3:3 stoichiometry–we cotransfected the LDs of LAP1 and LULL1 (designated LAP1LD and LULL1LD, respectively) with TorA-FLAG WT or TorA E171Q, followed by detergent lysis and anti-FLAG immunoprecipitation in the presence of ATP. We found that TorA’s ability to bind its cofactors is not significantly perturbed by back interface mutations: equivalent levels of LAP1LD and LULL1LD were detected in all immunoprecipitates (Figure 2A).
FIGURE 2:
Effect of mutations of Torsin’s “back” interface residues on LAP1 and LULL1 binding and ATPase activation. (A) Torsin “back” interface mutants G251D and 3xD interact with the LAP1 and LULL1 LDs in vivo. 293T cells were cotransfected with TorsinA E171Q-FLAG or indicated mutant derivatives and LAP1LD-HA or LULL1LD-HA, lysed with Nonidet P-40 (Roche) in the presence of 2 mM ATP, and immunoprecipitated using an M2 FLAG antibody. Immunoprecipitates and input controls were resolved by SDS–PAGE and blotted using the antibodies indicated. (B) Purification of Torsins from Expi293 cells. Wild type and “back” interface mutants were obtained with high purity from Expi293 cells. (C) ATPase activation of “back” interface mutant TorAs relative to wild-type TorsinA. ATP hydrolysis by TorA (3 μM) in the presence of purified LAP1LD and LULL1LD (3 μM) and 2 mM ATP was measured under steady-state conditions by detecting the amount of Pi released after 1 h at 37°C using a malachite green assay. Error bars represent the SD from three independent measurements.
To rule out defects in active site complementation, we next determined whether the ATPase stimulation by cofactors was compromised in these mutants. TorA wild type (WT) and mutant derivatives were purified from Expi293 cells (Figure 2B), and ATPase activity was measured in the absence and presence of LAP1LD and LULL1LD. We found that at equimolar concentrations of cofactor and TorA WT or its mutant derivatives, both interface mutants (TorA G251D; TorA 3xD [K248D, G251D, H254D]) were strongly stimulated by LAP1LD and LULL1LD, albeit with a modest reduction in catalytic efficiency (Figure 2C and Supplemental Figure S2). Remarkably, a substantially higher ATPase activity was observed at lower concentrations of either cofactor for TorA G251D and TorA 3xD relative to TorA WT (Supplemental Figure S2). Given that the interface mutants retain both cofactor binding and cofactor-induced ATPase activity, we conclude that their overall structural integrity is not grossly perturbed.
Structural validation of nucleotide-dependent TorsinA homo-oligomerization
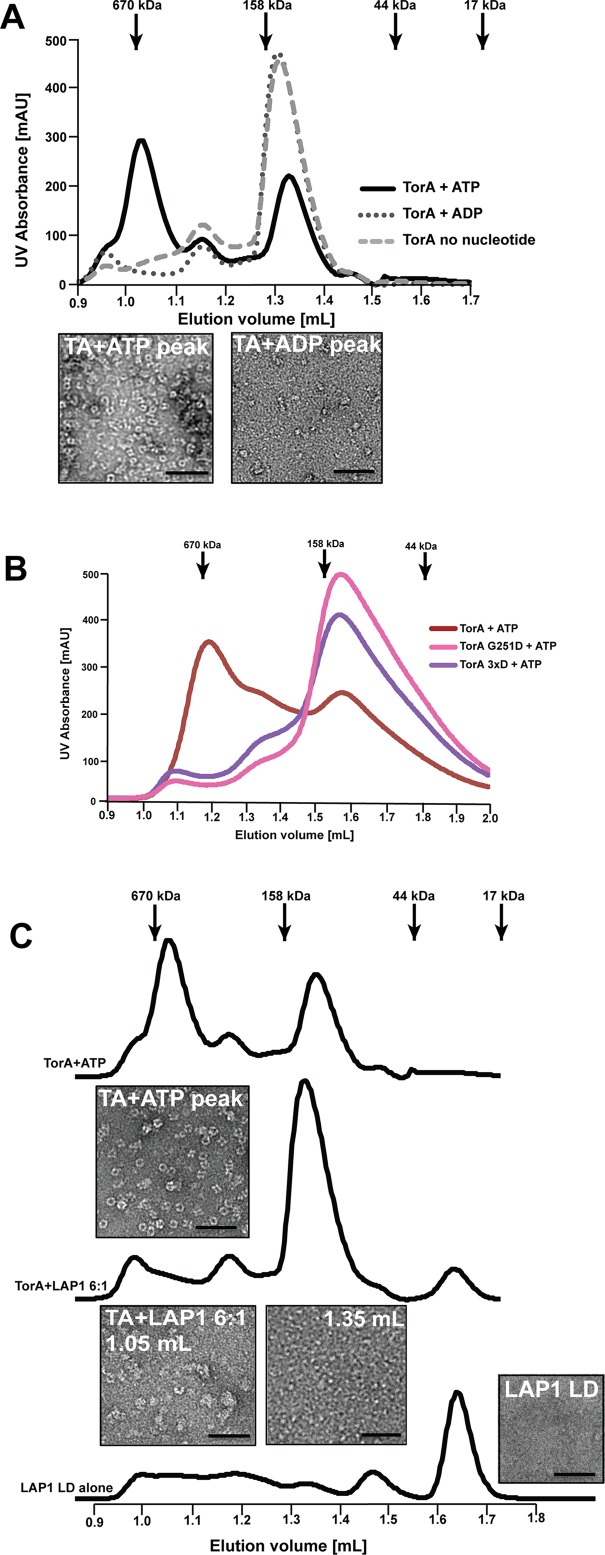
We next asked whether TorA forms oligomeric species in a nucleotide-dependent manner. After incubating purified TorA WT with or without ATP or ADP, we employed size exclusion chromatography (SEC) to resolve oligomeric species. In the presence of ATP, two distinct peaks corresponding to a higher and lower apparent molecular mass emerge, while Torsin without nucleotide or with ADP elutes as one distinct peak (Figure 3A). We next examined the corresponding elution fractions for each of these peaks by negative stain EM. The higher molecular mass peak contains a mixture of ring-shaped and lock washer structures with a diameter of ∼10 nm, while no ordered structures were observed in later fractions (Figure 3, A and B). To our knowledge, homo-oligomeric ring or lock washer assemblies for TorA were not visualized through structural methods previously. Given that our experiments were performed above the critical micelle concentration for dodecyl maltoside (DDM) and assuming that each hydrophobic domain of TorA (37 kDa) is solubilized by a micelle of ∼72 kDa (Strop and Brunger, 2005), we estimate that the observed species in the presence of ATP (∼600 kDa) would be consistent with a hexameric assembly. With a diameter of ∼10 nm, the observed dimensions of the homo-oligomer are in principle consistent with a homohexamer. However, we cannot exclude the formal possibility that more than one TorA subunit is immersed in a shared detergent micelle, which could lead to an underestimation of the apparent mass via SEC. Therefore, additional higher resolution approaches are necessary to establish the precise stoichiometry of the oligomeric state in the future, though we can firmly conclude that TorA forms homo-oligomers in a strictly ATP-dependent manner.
FIGURE 3:
TorsinA forms homo-oligomeric ring structures in the presence of ATP that are disassembled by LAP1LD or mutations of the “back” interface. (A) TorsinA (44 μM) was incubated with 2 mM of ATP or ADP and in the absence of nucleotide and the formation of complexes was assessed by SEC. Fractions were analyzed by negative stain electron microscopy. (B) Purified wild-type, G251D, and 3xD mutant TorsinA (44 μM each) were incubated in the presence of 2 mM ATP and complex formation assessed by SEC. (C) TorsinA (44 μM) was incubated with 2 mM ATP before the addition of 6:1 (mol:mol) Torsin:LAP1LD (5 min. on ice) and complex formation assessed by SEC as before.
We next determined whether mutations designed at Torsin’s back interface affected the formation of Torsin homo-oligomers by SEC. Indeed, none of the purified “back” interface mutants shifts to the homo-oligomer peak. Notably, mutating the strictly conserved G251 alone is sufficient to disrupt ring formation (Figure 3B), suggesting that this residue is indeed critical for interprotomer contacts through the “back” interface. Our observations corroborate previous studies reporting that TorA can form species consistent with a homohexamer in blue native PAGE experiments (Vander Heyden et al., 2009; Jungwirth et al., 2010), which is in excellent agreement with the reported conservation on the TorsinA “back” interface (Demircioglu et al., 2016).
To conclude, we provide unprecedented structural evidence that TorA adopts homo-oligomeric assemblies in a strictly ATP-dependent manner with essential contributions from a conserved glycine residue (Supplemental Figure S1) at the predicted TorA–TorA back interface (Figures 1B and 3, A and B).
LAP1 disrupts Torsin ring structures
We next asked how Torsin’s ATP-bound structure is affected by the addition of the ATPase-activating LAP1. Given 1) that LAP1 binds to TorA in the ATP-bound state (Naismith et al., 2009; Zhu et al., 2010; Zhao et al., 2013; Brown et al., 2014; Sosa et al., 2014; Demircioglu et al., 2016), 2) that LAP1 binding triggers ATP hydrolysis (Zhao et al., 2013), and 3) that ATP is strictly required for TorA homo-oligomerization (Figure 3A), it seems reasonable to propose that LAP1 binding concomitant with ATP hydrolysis could in fact destabilize the Torsin homo-oligomer.
To test this idea, we incubated TorA with ATP to form homo-oligomeric complexes, followed by a brief substoichiometric addition of LAP1LD before SEC. Indeed, we observed a profound reduction of the high molecular mass peak upon addition of LAP1LD, concomitant with a proportional increase of the lower molecular mass species (Figure 3C). Inspection of the elution fractions by EM revealed that the number of Torsin particles is greatly reduced in presence of LAP1, and that the regularity of some remaining particles is perturbed in presence of the cofactor. We also noted the appearance of amorphous structures under these conditions (TorA+LAP1 panel in Figure 3C), which is likely attributable to a reduced protein stability leading to protein aggregation in the absence of the native membrane-associated environment. As previously observed (Sosa et al., 2014), LAP1LD alone does not form oligomeric structures (Figure 3C).
These results are inconsistent with previously proposed hetero-oligomeric assemblies of fixed stoichiometries and highlight the importance of studying the oligomeric state of AAA+ ATPases under conditions that do not restrict the natural dynamics of the system, for example by using ATPase-arrested mutant derivatives. The dynamics of the Torsin/cofactor assembly had not been comprehensively explored in a system based on wild-type TorA and its cofactors, presenting a major obstacle toward developing working models for Torsin function. We find that formation of Torsin-cofactor complexes and ring formation are mutually exclusive: ATP hydrolysis through the cofactor generates ADP-engaged Torsin, which does not form rings (cf. Figure 3A). Thus, previous working models for the active Torsin/cofactor complex (Figure 1B) require a critical reassessment: Torsin assemblies and its interactions with the cofactor LAP1 are much more dynamic and transient than previously appreciated. Indeed, TorA’s oligomeric assembly is modulated not only by the nucleotide state, strictly requiring ATP for oligomerization (Figure 3A), but also by the LAP1 cofactor, as judged by the rapid disassembly of the TorA homo-oligomer upon LAP1 binding (Figure 3C). This finding is to our knowledge the first example of AAA+ protein oligomer disassembly by an activating cofactor, thus representing a new mode for AAA+ modulation. It also offers the first experimental validation of recently proposed arguments against a stable, mixed ring Torsin-cofactor assembly (Rose et al., 2015; Demircioglu et al., 2016; Chase et al., 2017)
Perturbation of homo-oligomerization disrupts TorsinA’s function at the NE
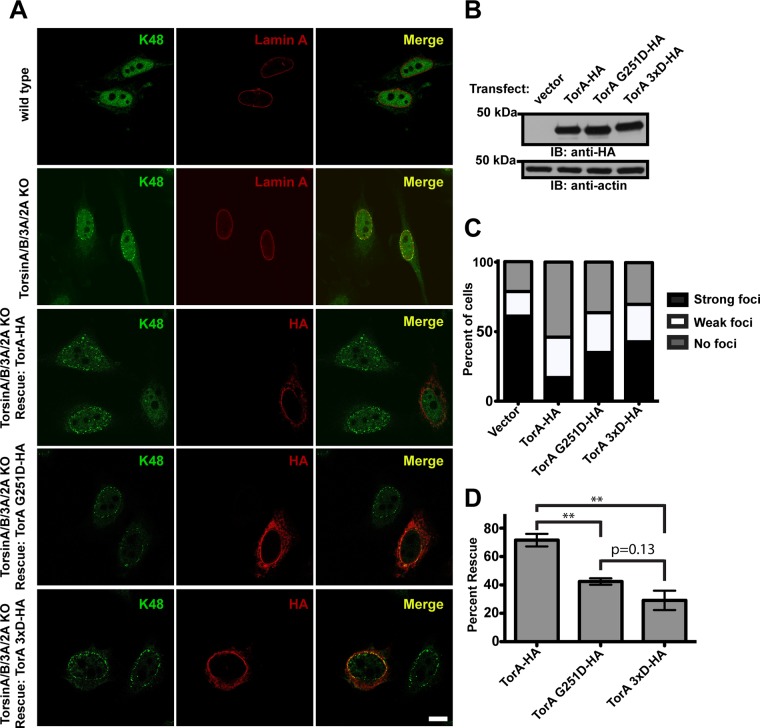
Having demonstrated that Torsins are endowed with the ability to form homo-oligomeric structures in the absence of cofactors, we asked whether ATP-induced homo-oligomerization of Torsin is important in a cellular setting. To this end we utilized a genetic complementation assay with a cell line depleted of all four human Torsins (TorA/B/2A/3A KO cells; Laudermilch et al., 2016). Torsin deletion results in the robust accumulation of perinuclear blebs as assessed by EM, which contain nuclear pore complex (NPC) components and K48-linked ubiquitin (Ub) (Laudermilch et al., 2016). The K48-Ub bleb accumulation phenotype at the nuclear envelope (NE) is readily visible via immunofluorescence and can be rescued by transfecting wild-type TorsinA-HA, as assessed by statistical analysis of cells having either strong, weak, or no K48-Ub foci remaining after rescue (Laudermilch et al., 2016). We therefore used this functional readout to assess the ability of the G251D and 3xD “back” interface Torsin mutants to rescue the K48-Ub foci (perinuclear blebbing) phenotype in TorA/B/2A/3A KO cells (Figure 4A).
FIGURE 4:
Torsin G251D and 3xD “back” interface mutants do not rescue the K48-linked ubiquitin accumulation phenotype in the nuclear periphery. (A) Confocal images of wild-type or Torsin-deficient HeLa cells stained with an anti-K48 ubiquitin antibody. In the top two panels, wild-type or Torsin-deficient cells were costained with lamin A. In the bottom three panels, wild-type or mutant TorA was transfected into Torsin-deficient cells and costained with an anti-HA antibody. Scale bar is 10 μm. (B) Immunoblot of lysates from the bottom four panels of part A using an anti-HA antibody. Actin was used as a loading control. (C) Graph showing the percent of cells exhibiting K48 ubiquitin foci at the nuclear periphery with different rescue constructs. (D) Graph showing the percent rescue of the K48 ubiquitin accumulation with different rescue constructs. Error bars show the SEM. **p < 0.01. In both graphs, the percent reported is an average of three independent experiments of at least 100 cells each.
With all TorA variants expressed at equal levels (Figure 4B), wild-type TorsinA results in a significant reduction in K48-Ub foci compared with the vector control, while the ability of the G251D mutant and the 3xD mutant to rescue is significantly compromised (Figure 4, A and C). Both the G251D mutant and the 3xD mutants exhibit strongly (p = <0.05) reduced efficacy in rescuing bleb accumulation and there is no significant difference (p = 0.13) between the G251D and 3xD mutants’ ability to rescue, consistent with a critical contribution of the strictly conserved G251 that is essential for homo-oligomerization (Figure 3B) but dispensable for cofactor binding or ATPase activation (Figure 2). These results strongly suggest that the formation of Torsin–Torsin homo-oligomers is indeed a prerequisite for its ability to perform critical functions at the NE, clearly suggesting that the TorA homo-oligomer is a functionally significant species in vivo (Figure 4). Though the precise nature of TorsinA’s role(s) in nuclear pore complex and NE homeostasis and its substrate(s) at the NE remain to be determined, our study establishes TorsinA homo-oligomerization to be essential for NE homeostasis and integrity.
Given that TorA homo-oligomerization is strictly ATP-dependent (Figure 3A) and that cofactors are bound by TorA preferentially in the ATP-bound state (Zhao et al., 2013; Brown et al., 2014; Sosa et al., 2014) to trigger rapid ATP hydrolysis upon binding (Zhao et al., 2013), we now validate the proposed dynamic model (Chase et al., 2017) for the Torsin assembly (Figure 5): engagement of the homo-oligomer by the cofactor at the nuclear envelope (NE) triggers ATP hydrolysis concomitant with a dissociation of the cofactor-bound Torsin subunit in the ADP state. Since the cofactor-Torsin interaction is itself ATP-dependent (Naismith et al., 2009; Zhu et al., 2010; Zhao et al., 2013; Brown et al., 2014; Sosa et al., 2014), the cofactor/Torsin heterodimer will also dissociate, allowing the cofactor to be recycled for a second round of hydrolysis, ultimately leading to a complete homo-oligomer disassembly.
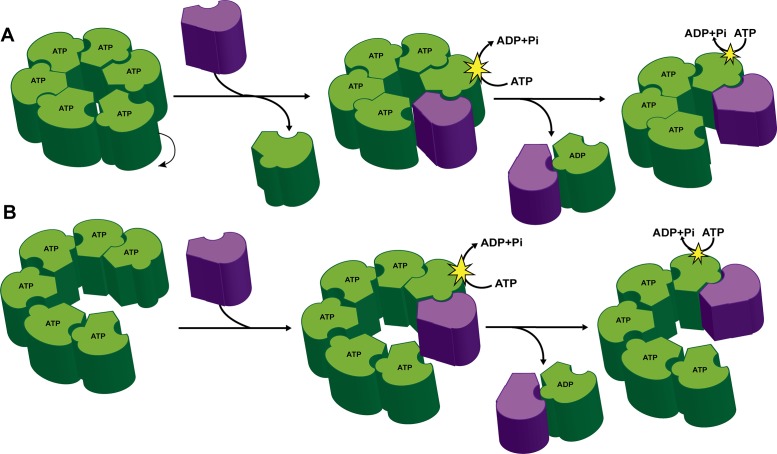
FIGURE 5:
A dynamic model for the Torsin/cofactor assembly. TorA dynamic ring assembly with either a planar (A) or stacked (B) ring conformation. The cofactors (purple) serve to disassemble the Torsin (green) ring structures concurrent with ATP hydrolysis and loss of TorA subunits from the ring.
The initial activation step warrants closer structural investigation in the future: how do the membrane-anchored cofactor LDs access TorA’s activator interface? As proposed previously (Chase et al., 2017), it is possible that flexibility conferred by an unstructured region of TorA C-terminal to the membrane-anchored hydrophobic domain allows Torsin’s AAA+ domain to rotate out of the ring, thus vacating a position that can be occupied by the activator (Figure 5A). Alternatively and similar to related AAA+ ATPases (Blok et al., 2015; Zhao et al., 2015; Yokom et al., 2016; Monroe et al., 2017; Ripstein et al., 2017), TorA could adopt a lock-washer or “spiral” conformation that would expose an activating interface at the raised end of the split washer (Figure 5B). In this model, incremental translation of successive TorA subunits would similarly be made possible by the unstructured region between the hydrophobic domain and the AAA+ domain. We favor the latter possibility, though high-resolution structural studies of ATP-bound Torsin complexes are needed to distinguish between these possibilities. In either case, our models (Figure 5) are in excellent agreement with the observation that Torsin interface mutations result in a higher ATPase activity at low concentrations of cofactor (Supplemental Figure S2). We attribute this to an increase in the number of exposed activator interfaces relative to TorA WT where the majority of activator interfaces would be “shielded” by a neighboring TorsinA protomer in a homo-oligomeric assembly.
In conclusion, our data argue against previously proposed models (Figure 1B) and establish the Torsin system as a highly dynamic assembly whose oligomeric state is regulated by distinctively localizing cofactors (Goodchild and Dauer, 2005; Naismith et al., 2009) in the cell, representing an unprecedented mode of AAA+ protein regulation. As proposed previously (Rose et al., 2015; Chase et al., 2017), it is tempting to speculate that the controlled destabilization of the Torsin ring or lock washer at distinct subcellular locations could afford control over binding or release of Torsin substrates or modulate the properties of closely associated membrane regions. Thus our findings will provide guidance in defining future working models for Torsin-dependent cellular pathways and AAA+ proteins in general.
MATERIALS AND METHODS
Immunoprecipitation and immunoblotting
The binding of TorsinA to the LDS of LAP1 and LULL1 was determined using previously described procedures (Zhao et al., 2013; Brown et al., 2014). Briefly, 80% confluent 10 cm plates of HEK293T cells were transfected with E171Q Torsin construct variants harboring C-terminal FLAG tags in combination with LAP1 or LULL1 LDs (C-terminal HA tags) using Lipofectamine 2000 (Invitrogen) and grown in a humidified incubator at 5% (vol/vol) CO2 in DMEM supplemented with 10% (vol/vol) FBS. After 48 h, cells were harvested and washed twice with ice-cold phosphate-buffered saline (PBS) before lysis for 10 min on ice using Nonidet P-40–containing lysis buffer (50 mM Tris, pH 7.5, 75 mM NaCl, 5 mM MgCl2, and 0.5% Nonidet P-40) supplemented with 2 mM ATP and Complete protease inhibitors (Roche). After sedimentation (20,000 × g, 10 min, 4°C), soluble lysates were precleared using protein A agarose (RepliGen) for 1 h. before immunoprecipitation with M2 FLAG antibody (Sigma Aldrich) added to protein A agarose beads for 3 h. at 4°C. Samples were boiled in SDS–PAGE loading buffer and subjected to SDS–PAGE. Immunoblotting was performed by standard methods: after transferring samples to polyvinylidene fluoride membranes, immunoreactive bands were detected using a Western Lightning Plus Chemiluminescence kit (PerkinElmer) and x-ray film (Kodak).
Protein expression and purification
Expi293 cells (Invitrogen) were transfected with 1 μg/ml culture of C-terminally FLAG-tagged full-length TorsinA construct and expressed according to the instructions provided by the manufacturer. Briefly, cells were grown in suspension (37°C, 8% CO2, 125 rpm), enhancers were added 20 h after transfection, and cells were harvested after 72 h. Cells were lysed in 50 mM Tris, pH 7.5, 150 mM NaCl, 5 mM MgCl2, 1% (wt/vol) DDM (n-dodecyl-β-d-maltopyranoside; Anatrace), 10% glycerol buffer supplemented with complete EDTA-free protease inhibitor cocktail (Roche). Lysate was cleared of cellular debris by centrifugation for 30 min at 20,000 × g (4°C) and cleared lysate was incubated with M2 FLAG beads for at least 3 h or overnight at 4°C. Unbound material was removed by washing with 20 column volumes (CV) of wash buffer I (50 mM Tris-Cl, pH 7.5, 150 mM NaCl, 5 mM MgCl2, 0.05% DDM) followed by 20 CV of Torsin oligomerization buffer (50 mM Tris-Cl, pH 7.5, 30 mM NaCl, 50 mM NaAc, 5 mM MgCl2, 0.05% DDM). Protein was eluted with 0.3 mg/ml FLAG peptide in Torsin oligomerization buffer followed by buffer exchange with PD10 columns (GE) to remove free peptide before concentration with Amicon Ultra-Cel 10,000 MWCO centrifugal units.
The LDs of LAP1 or LULL1 were expressed in Rosetta(DE3)pLysS cells (Novagen) or Origami 2(DE3)pLysS cells (Novagen), respectively, and purified as before (Zhao et al., 2013), except that the step to remove the His-tag with PreScission protease was omitted.
ATPase activity assay
ATPase assays were performed as previously described (Zhao et al., 2013). Briefly, 3 μM of each purified Torsin variant was incubated at 37°C in the presence of 2 mM ATP and 0.1 mg/ml bovine serum albumin (BSA) along with various concentrations of LAP1 or LULL1 LDs purified from bacteria. To determine initial rates, aliquots were removed from each reaction at 5, 10, 15, 30, 60, and 90 min, and the reaction was stopped by adding the aliquot to 20 mM sulfuric acid. The amount of inorganic phosphate (Pi) produced was measured using a colorimetric method. A freshly prepared mixture of malachite green (0.096%), ammonium molybdate (1.48% wt/vol), and Tween-20 (0.173%) in 2.9 M sulfuric acid was added to each sample and incubated for 15 min before determining the absorbance at 620 nm using a 96-well plate reader (BioTek). Concentrations of Pi released during ATP hydrolysis were measured in the linear range of the standard curve used to calibrate Pi concentrations (Na2HPO4 in 20 mM sulfuric acid). The initial rates of the ATPase hydrolysis activity of Torsin for each concentration of LAP1 or LULL1 were determined from the slope of the best-fit line derived by plotting the Pi released from the average of three independent measurements of each reaction versus reaction time. The initial rates were then fitted to Michaelis-Menten kinetics in Prism, yielding the apparent Km, Vmax, and SD for each graph.
For the steady-state measurements in Figure 2C, reactions containing 3 μM Torsin, 3 μM of either LAP1 or LULL1 LD, 2 mM ATP, and 0.1 mg/ml BSA were stopped after 60 min with 20 mM sulfuric acid. The amount of Pi produced was determined with the malachite green assay and was fitted to the standard curve of Pi concentrations to determine the amount of Pi released after 1 h. The average of three independent measurements was then plotted, with error bars representing the SD.
Analytical gel filtration
Analytical gel filtration was performed using a Superdex 200 Increase 3.2/300 (GE) or a Superdex 200 PC 3.2/30 (GE, discontinued) column equilibrated with Torsin oligomerization buffer containing 0.05% DDM. TorA (44 μM) was incubated with 2 mM ATP for 1 h on ice before injection for assessing homo-oligomerization. For experiments with the LAP1 LD, the cofactor was incubated with ATP-bound Torsin for 5 min before sample injection. Fractions of 100 μl were collected and peak fractions (as assessed by measuring A280 using a Nanodrop) used for negative stain analysis undiluted (∼0.1 mg/ml final concentration).
Negative stain electron microscopy and imaging
Continuous carbon grids were glow-discharged for 30 s at 5 mA before applying a 5.5 μl sample, incubation for 1 min, and then staining with three sequential passes through sitting 11 μl 2% (wt/vol) uranyl acetate drops with a 30 s incubation after the final application before blotting and air drying. Grids were imaged using a JEOL JEM-1400 Plus at 120 kV and 60,000× nominal magnification with an EM-14800RUBY charge-coupled device camera.
Immunofluorescence microscopy
Indirect immunofluorescence and confocal microscopy were performed as previously described (Rose et al., 2014; Tsai et al., 2016). Briefly, cells were fixed with 4% paraformaldehyde and permeabilized with 0.1% Triton X-100. Cells were then blocked in 4% BSA in PBS and incubated with primary antibodies for 1 h. After washing, cells were incubated with Alexa 488/568 secondary antibodies, washed with PBS and mounted onto coverslips with Fluoromount-G. The following antibodies were used at 1:500 dilution: anti–K48 Ub (AB_11213655; Millipore); anti–lamin A (AB_306909; Abcam); anti-HA (AB_2314622; Roche).
Statistics and single-cell analysis
For the Torsin complementation experiment (Figure 4), single-cell analysis was performed. Using blinded samples, at least 100 cells from each of three independent experiments were imaged for each sample. Based on the number of K48 foci present in the nuclear periphery of each cell, cells were assessed as strongly exhibiting K48 foci, weakly exhibiting K48 foci, or not exhibiting K48 foci. Statistical analysis was performed in GraphPad Prism using an unpaired t test. To calculate the percent rescue in Figure 4, the following equation was used: Percent rescue = ((Percent of cells strongly exhibiting K48 foci in the vector control – Percent of cells strongly exhibiting K48 foci in the Torsin rescue construct)/Percent of cells strongly exhibiting K48 foci in the vector control)*100.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health (1R01GM114401 to C.S. and CMBTG T32GM007223 to E.L.) and in part by a National Science Foundation GROW award through the Japan Society for the Promotion of Science to A.R.C. for work conducted at the RIKEN Center for Life Science Technologies (CLST). We thank Tomomi Uchikubo-Kamo for help with sample preparation and the group of Mikako Shirouzu (RIKEN CLST) for their support.
Abbreviations used:
- AAA+
ATPases associated with a variety of cellular activities
- EM
electron microscopy
- ER
endoplasmic reticulum
- NE
nuclear envelope
- TorA
TorsinA
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E17-05-0281) on August 16, 2017.
REFERENCES
- Blok NB, Tan D, Wang RY, Penczek PA, Baker D, DiMaio F, Rapoport TA, Walz T. Unique double-ring structure of the peroxisomal Pex1/Pex6 ATPase complex revealed by cryo-electron microscopy. Proc Natl Acad Sci USA. 2015;112:E4017–E4025. doi: 10.1073/pnas.1500257112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown RS, Zhao C, Chase AR, Wang J, Schlieker C. The mechanism of Torsin ATPase activation. Proc Natl Acad Sci USA. 2014;111:E4822–E4831. doi: 10.1073/pnas.1415271111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cascalho A, Jacquemyn J, Goodchild RE. Membrane defects and genetic redundancy: are we at a turning point for DYT1 dystonia. Mov Disord. 2017;32:371–381. doi: 10.1002/mds.26880. [DOI] [PubMed] [Google Scholar]
- Chase A, Laudermilch E, Schlieker C. Torsin ATPases: harnessing dynamic instability for function. Front Mol Biosci. 2017;4:29. doi: 10.3389/fmolb.2017.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demircioglu FE, Sosa BA, Ingram J, Ploegh HL, Schwartz TU. Structures of TorsinA and its disease-mutant complexed with an activator reveal the molecular basis for primary dystonia. eLife. 2016;5:e17983. doi: 10.7554/eLife.17983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foisner R, Gerace L. Integral membrane proteins of the nuclear envelope interact with lamins and chromosomes, and binding is modulated by mitotic phosphorylation. Cell. 1993;73:1267–1279. doi: 10.1016/0092-8674(93)90355-t. [DOI] [PubMed] [Google Scholar]
- Goodchild RE, Dauer WT. The AAA+ protein torsinA interacts with a conserved domain present in LAP1 and a novel ER protein. J Cell Biol. 2005;168:855–862. doi: 10.1083/jcb.200411026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungwirth M, Dear ML, Brown P, Holbrook K, Goodchild R. Relative tissue expression of homologous torsinB correlates with the neuronal specific importance of DYT1 dystonia-associated torsinA. Hum Mol Genet. 2010;19:888–900. doi: 10.1093/hmg/ddp557. [DOI] [PubMed] [Google Scholar]
- Kirstein J, Moliere N, Dougan DA, Turgay K. Adapting the machine: adaptor proteins for Hsp100/Clp and AAA+ proteases. Nat Rev Microbiol. 2009;7:589–599. doi: 10.1038/nrmicro2185. [DOI] [PubMed] [Google Scholar]
- Laudermilch E, Schlieker C. Torsin ATPases: structural insights and functional perspectives. Curr Opin Cell Biol. 2016;40:1–7. doi: 10.1016/j.ceb.2016.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laudermilch E, Tsai PL, Graham M, Turner E, Zhao C, Schlieker C. Dissecting Torsin/cofactor function at the nuclear envelope: a genetic study. Mol Biol Cell. 2016;27:3964–3971. doi: 10.1091/mbc.E16-07-0511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogk A, Schlieker C, Strub C, Rist W, Weibezahn J, Bukau B. Roles of individual domains and conserved motifs of the AAA+ chaperone ClpB in oligomerization, ATP hydrolysis, and chaperone activity. J Biol Chem. 2003;278:17615–17624. doi: 10.1074/jbc.M209686200. [DOI] [PubMed] [Google Scholar]
- Monroe N, Han H, Shen PS, Sundquist WI, Hill CP. Structural basis of protein translocation by the Vps4-Vta1 AAA ATPase. eLife. 2017;6:e24487. doi: 10.7554/eLife.24487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naismith TV, Dalal S, Hanson PI. Interaction of torsinA with its major binding partners is impaired by the dystonia-associated DeltaGAG deletion. J Biol Chem. 2009;284:27866–27874. doi: 10.1074/jbc.M109.020164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nery FC, Armata IA, Farley JE, Cho JA, Yaqub U, Chen P, da Hora CC, Wang Q, Tagaya M, Klein C, et al. TorsinA participates in endoplasmic reticulum-associated degradation. Nat Commun. 2011;2:393. doi: 10.1038/ncomms1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripstein ZA, Huang R, Augustyniak R, Kay LE, Rubinstein JL. Structure of a AAA+ unfoldase in the process of unfolding substrate. eLife. 2017;6:e25754. doi: 10.7554/eLife.25754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose AE, Brown RSH, Schlieker C. Torsins: not your typical AAA+ ATPases. Crit Rev Biochem Mol Biol. 2015;50:632–649. doi: 10.3109/10409238.2015.1091804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose AE, Zhao C, Turner EM, Steyer AM, Schlieker C. Arresting a Torsin ATPase reshapes the endoplasmic reticulum. J Biol Chem. 2014;289:552–564. doi: 10.1074/jbc.M113.515791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders CA, Harris NJ, Willey PT, Woolums BM, Wang Y, McQuown AJ, Schoenhofen A, Worman HJ, Dauer WT, Gundersen GG, et al. TorsinA controls TAN line assembly and the retrograde flow of dorsal perinuclear actin cables during rearward nuclear movement. J Cell Biol. 2017;216:657–674. doi: 10.1083/jcb.201507113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosa BA, Demircioglu FE, Chen JZ, Ingram J, Ploegh HL, Schwartz TU. How lamina-associated polypeptide 1 (LAP1) activates Torsin. eLife. 2014;3:e03239. doi: 10.7554/eLife.03239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strop P, Brunger AT. Refractive index-based determination of detergent concentration and its application to the study of membrane proteins. Protein Sci. 2005;14:2207–2211. doi: 10.1110/ps.051543805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai PL, Zhao C, Turner E, Schlieker CD. The Lamin B receptor is essential for cholesterol synthesis and perturbed by disease-causing mutations. eLife. 2016;5:e16011. doi: 10.7554/eLife.16011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Heyden AB, Naismith TV, Snapp EL, Hodzic D, Hanson PI. LULL1 retargets TorsinA to the nuclear envelope revealing an activity that is impaired by the DYT1 dystonia mutation. Mol Biol Cell. 2009;20:2661–2672. doi: 10.1091/mbc.E09-01-0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanGompel MJ, Nguyen KC, Hall DH, Dauer WT, Rose LS. A novel function for the Caenorhabditis elegans torsin OOC-5 in nucleoporin localization and nuclear import. Mol Biol Cell. 2015;26:1752–1763. doi: 10.1091/mbc.E14-07-1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokom AL, Gates SN, Jackrel ME, Mack KL, Su M, Shorter J, Southworth DR. Spiral architecture of the Hsp104 disaggregase reveals the basis for polypeptide translocation. Nat Struct Mol Biol. 2016;23:830–837. doi: 10.1038/nsmb.3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Brown RS, Chase AR, Eisele MR, Schlieker C. Regulation of Torsin ATPases by LAP1 and LULL1. Proc Natl Acad Sci USA. 2013;110:E1545–E1554. doi: 10.1073/pnas.1300676110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M, Wu S, Zhou Q, Vivona S, Cipriano DJ, Cheng Y, Brunger AT. Mechanistic insights into the recycling machine of the SNARE complex. Nature. 2015;518:61–67. doi: 10.1038/nature14148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L, Millen L, Mendoza JL, Thomas PJ. A unique redox-sensing sensor II motif in TorsinA plays a critical role in nucleotide and partner binding. J Biol Chem. 2010;285:37271–37280. doi: 10.1074/jbc.M110.123471. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.