Osm1 transfers electrons from fumarate to succinate and functions with Mia40 and Erv1 in the redox-regulated import pathway for proteins that form disulfide bonds in the mitochondrial intermembrane space. Expression of Osm1 and cytochrome c is reciprocally regulated, indicating that the cell has strategies to coordinate expression of terminal electron acceptors.
Abstract
Prokaryotes have aerobic and anaerobic electron acceptors for oxidative folding of periplasmic proteins. The mitochondrial intermembrane space has an analogous pathway with the oxidoreductase Mia40 and sulfhydryl oxidase Erv1, termed the mitochondrial intermembrane space assembly (MIA) pathway. The aerobic electron acceptors include oxygen and cytochrome c, but an acceptor that can function under anaerobic conditions has not been identified. Here we show that the fumarate reductase Osm1, which facilitates electron transfer from fumarate to succinate, fills this gap as a new electron acceptor. In addition to microsomes, Osm1 localizes to the mitochondrial intermembrane space and assembles with Erv1 in a complex. In reconstitution studies with reduced Tim13, Mia40, and Erv1, the addition of Osm1 and fumarate completes the disulfide exchange pathway that results in Tim13 oxidation. From in vitro import assays, mitochondria lacking Osm1 display decreased import of MIA substrates, Cmc1 and Tim10. Comparative reconstitution assays support that the Osm1/fumarate couple accepts electrons with similar efficiency to cytochrome c and that the cell has strategies to coordinate expression of the terminal electron acceptors. Thus Osm1/fumarate is a new electron acceptor couple in the mitochondrial intermembrane space that seems to function in both aerobic and anaerobic conditions.
INTRODUCTION
Disulfide bonds are required for the correct folding of many proteins, particularly those that are secreted (Dutton et al., 2008; Mamathambika and Bardwell, 2008). Oxidative folding pathways are present in the periplasmic space of prokaryotes and the endoplasmic reticulum (ER) and mitochondrial intermembrane space (IMS) in eukaryotes (Depuydt et al., 2011). Whereas the components lack conservation at the sequence level, hallmarks of the pathway are similar across these systems.
The prokaryotic oxidative folding pathway has insertion and editing elements. Disulfide bonds are introduced by DsbA in the well-characterized system of Gram-negative bacteria, and DsbA is recycled by DsbB (Mamathambika and Bardwell, 2008). Electrons are then shuttled from DsbB to different aerobic and anaerobic electron acceptors via ubiquinone (Q) and menaquinone (MQ) (Bader et al., 1999; Takahashi et al., 2004). In aerobic conditions, Q passes electrons to cytochrome oxidase and oxygen (O2), whereas in anaerobic conditions, MQ transfers electrons to fumarate or nitrate reductases. DsbC proofreads DsbA activity by removing nonnative disulfide bonds (Rietsch et al., 1996). DsbC is maintained in a reduced state by membrane protein DsbD, and electrons are provided by the thioredoxin system in the cytosol (Katzen and Beckwith, 2000). Finally, secreted glutathione (GSH) and cysteine contribute to the global redox equilibrium in the periplasm (Messens et al., 2007; Ohtsu et al., 2010).
The ER has the oxidoreductase protein disulfide isomerase (PDI), which both inserts and edits disulfide bonds in imported ER proteins (Sevier and Kaiser, 2006). PDI is then maintained in an oxidized state by the sulfhydryl oxidase Ero1, and electrons are subsequently donated to O2, generating hydrogen peroxide (Tu et al., 2000). Yeasts also have a second sulfhydryl oxidase Erv2 that has an auxiliary role in oxidizing ER proteins (Sevier, 2012). In contrast, studies indicate that mammals may use vitamin K epoxide reductase, quiescin sulfhydryl oxidase, and/or peroxiredoxin IV as Ero1 alternatives (Mairet-Coello et al., 2004; Wajih et al., 2007; Zito et al., 2010). The redox environment, which supports the oxidation of cysteines, is −190 mV and is maintained with a ratio of reduced GSH to oxidized glutathione (GSSG) of 3:1 (Sevier et al., 2007). Recently Osm1 has been identified as a potential anaerobic electron acceptor for the ER of Saccharomyces cerevisiae (Williams et al., 2014). The ER form of Osm1 supported growth under anaerobic conditions, suggesting Osm1 may function as a terminal electron acceptor in the ER (Liu et al., 2013; Williams et al., 2014). However, as Osm1 does not seem to be conserved in higher eukaryotes, potential electron acceptors that have been suggested include free flavins or other small molecules (Gross et al., 2006).
The mitochondrial system consists of the oxidoreductase Mia40 and the sulfhydryl oxidase Erv1, termed the mitochondrial intermembrane space assembly (MIA) pathway (Chacinska et al., 2004; Naoe et al., 2004; Allen et al., 2005; Mesecke et al., 2005; Koehler and Tienson, 2009). Both proteins are essential for viability (Becher et al., 1999; Chacinska et al., 2004). Mia40 acts as a receptor in the IMS and inserts disulfide bonds into IMS substrates as they are imported (Grumbt et al., 2007). Substrates include proteins with the CX3C motif (the small Tim proteins) and the CX9C motif (a subset function in cytochrome oxidase assembly) (Gabriel et al., 2007; Longen et al., 2009). Electrons are passed from Mia40 to Erv1. Following this, Erv1 shuttles electrons to either O2 or cytochrome (cyt) c (Bihlmaier et al., 2007; Dabir et al., 2007). O2 subsequently forms hydrogen peroxide, and cyt c donates electrons to the electron transport system or to Ccp1. Whereas the two terminal electron acceptors function in aerobic conditions, a potential anaerobic electron acceptor has not been identified (Dabir et al., 2007; Herrmann and Riemer, 2012).
Considering known anaerobic electron acceptors, two candidates in Saccharomyces cerevisiae are the fumarate reductases Frd1 and Osm1 that use a noncovalently bound FADH2 to reduce fumarate to succinate (Muratsubaki and Enomoto, 1998; Enomoto et al., 2002). Frd1 is a soluble cytosolic enzyme, whereas the Osm1 enzymatic activity has been localized to mitochondria, and Osm1 has a putative mitochondrial-targeting sequence (Muratsubaki and Enomoto, 1998; Enomoto et al., 2002). Expression of Frd1 is induced in anaerobic conditions, but Osm1 is expressed constitutively; Frd1 provides the most fumarate reductase activity under anaerobic conditions. Nevertheless, a Δfrd1Δosm1 strain fails to grow under anaerobic conditions, whereas the single mutants grow (Arikawa et al., 1998; Camarasa et al., 2007). Frd1 and Osm1 likely play an overall role in controlling redox balance, potentially through the reoxidation of NADH or FADH2 for the regeneration of the flavin adenine dinucleotide (FAD)-prosthetic group of essential flavoprotein enzymes (Arikawa et al., 1998; Camarasa et al., 2007).
Given that Osm1 was localized to mitochondria, we considered its potential role as a terminal electron acceptor for Erv1. Here we report that Osm1 showed dual localization to microsomes and the mitochondrial IMS. In addition, FAD-bound Osm1 accepted electrons from Erv1 in vitro reconstitution assays in which reduced Tim13 was oxidized. Both Osm1 and cyt c functioned equally well in accepting electrons from Erv1. Moreover, cyt c expression was induced in cells lacking osm1 and vice versa. Thus Osm1 is a terminal electron acceptor with expression that is reciprocally regulated with cyt c.
RESULTS
osm1 and erv1 are synthetic lethal in anaerobic conditions
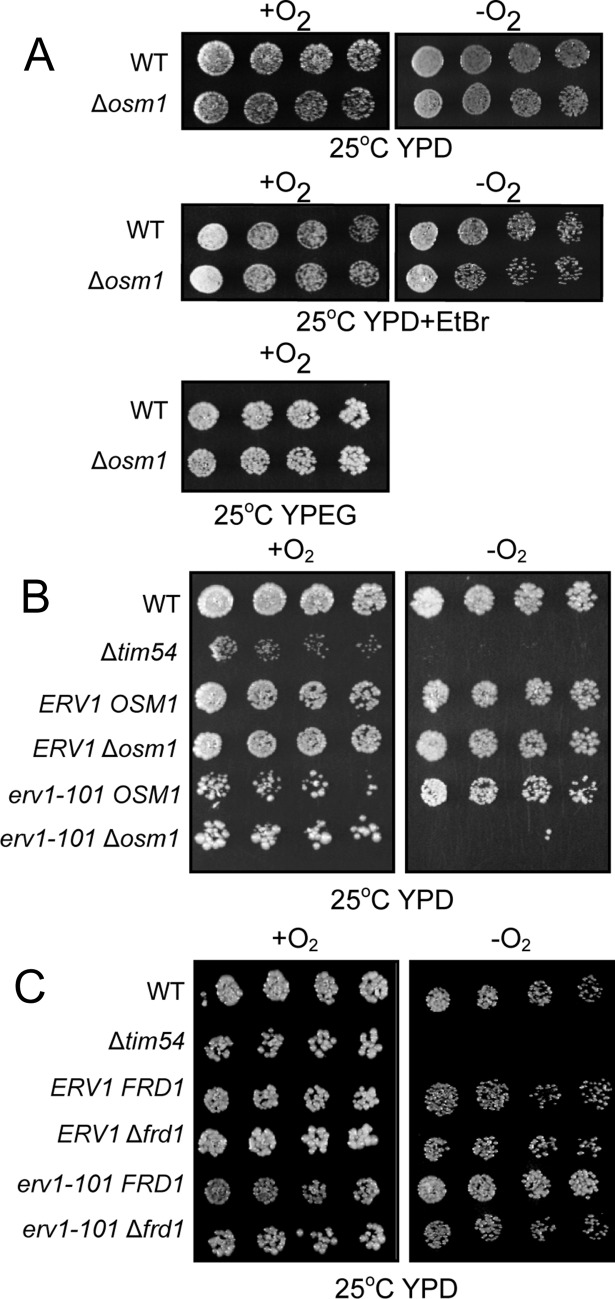
Under aerobic conditions, Erv1 donates electrons to O2 or cyt c. However, the erv1 mutant is viable under anaerobic conditions (Dabir et al., 2007), indicating that an acceptor for anaerobiosis is required. We used a candidate approach and considered the fumarate reductase Osm1 as a potential anaerobic electron receptor, because Osm1 is reported to localize to mitochondria and bypasses a requirement for oxygen in electron transfer from fumarate to succinate (Muratsubaki and Enomoto, 1998). The Δosm1 strain grew slowly under anaerobic conditions, whereas the double Δfrd1Δosm1 mutant failed to grow (Enomoto et al., 2002). Deletion of osm1 in the parental strain did not display any marked growth defects in aerobic or anaerobic conditions (Figure 1A). Treatment with ethidium bromide to remove mitochondrial DNA also did not compromise growth. We predicted that Osm1 would display synthetic lethality with a temperature-sensitive erv1 mutant under anaerobic conditions. We therefore crossed Δosm1 and erv1-101 strains and analyzed growth of the dissected tetrads under anaerobic conditions (Figure 1B). The Δtim54 strain was included as a control because it is petite-negative and does not grow anaerobically (Hwang et al., 2007). In aerobic conditions, all strains including the mutants grew, although strains with the erv1-101 allele showed compromised growth. With the exception of Δtim54 and the double mutant erv1-101Δosm1, all strains grew in anaerobic conditions. We tested similar genetic interactions between erv1-101 and Δfrd1 (Figure 1C); growth was not attenuated in aerobic and anaerobic conditions. Thus the erv1-101Δosm1 strain displayed synthetic lethality, suggesting that Osm1 may function in a pathway with Erv1.
FIGURE 1:
ERV1 and OSM1 are synthetic lethal. (A) Growth analysis of Δosm1 and the parental (WT) strains in aerobic (+O2) and anaerobic (−O2) conditions on glucose (YPD) and ethanol/glycerol (YPEG) carbon sources. Ethidium bromide (EtBr) treatment removed the mitochondrial DNA. (B) A cross between erv1-101 and Δosm1 was sporulated, and the resulting tetrads were serially diluted by a factor of 2 on rich glucose media (YPD) and incubated in aerobic (+O2) and anaerobic (−O2) conditions. The parental strain (WT) from which all strains were derived and tim54-1 mutant (petite-negative) were included as controls. (C) As in B, except erv1-101 was crossed with Δfrd1. Plates were photographed after 3 d at 25°C.
Osm1 localizes to microsomes and the mitochondrial IMS
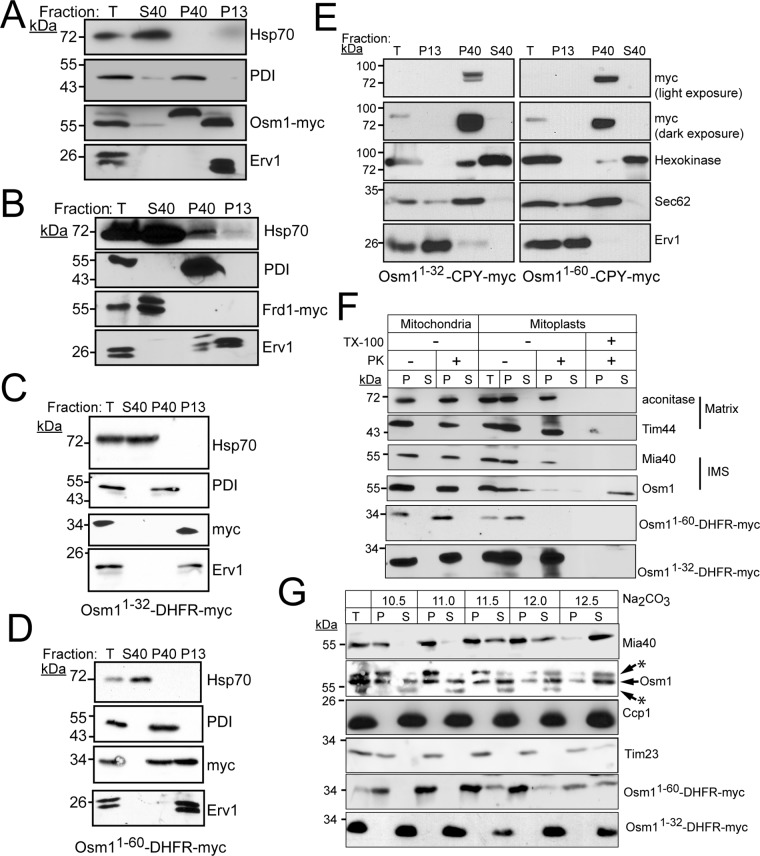
Frd1 is likely cytosolic, but the specific location of Osm1 within mitochondria has not been reported (Muratsubaki and Enomoto, 1998). We localized Frd1 and Osm1 in spheroplasts (Figure 2, A and B). Tagged constructs were generated in which Frd1 and Osm1 contained a C-terminal myc tag. Lysates from spheroplasts were subject to differential centrifugation and analyzed by SDS–PAGE and immunoblotting. Analyzed fractions included the pellet from the 13,000 × g spin (mitochondria), and the supernatant (cytosol) and pellet (microsomes) from the 40,000 × g spin. Immunoblotting with appropriate markers—Erv1 for mitochondria, cytosolic Hsp70 for cytosol, and protein disulfide isomerase (PDI) for ER—verified integrity of the fractionation. Frd1 localized to the cytosol, as expected (Figure 2B). However, Osm1 was detected in both the microsomal and mitochondrial fractions (Figure 2A). The mitochondrial form migrated at a lower molecular mass, likely because the mitochondrial form of Osm1 was generated by translation initiation at position 32 (Williams et al., 2014) or a targeting sequence that started from the first methionine was cleaved by a mitochondrial processing protease (Muratsubaki and Enomoto, 1998). An identical localization pattern for Osm1 was also detected in wild-type (WT) cells using a polyclonal antibody against Osm1 (Supplemental Figure S1A), indicating that the myc tag does not interfere with localization. In contrast, the microsomal form of Osm1 migrated at a higher molecular weight, which is likely due to N-glycosylation that has been reported (Zielinska et al., 2012; Williams et al., 2014; Chen et al., 2014a,b).
FIGURE 2:
Osm1 localizes to the ER and the mitochondrial IMS, whereas Frd1 resides in the cytosol. (A) A yeast transformant expressing Osm1 with a C-terminal myc-tag was grown in YPEG at 30°C, converted to spheroplasts, and fractionated into a total homogenate (T), mitochondrial fraction (P13), microsome fraction (P40), and cytoplasm (S40). An equal amount of each fraction was analyzed by SDS–PAGE and immunoblotted with a monoclonal antibody against the myc tag. As a control, antibodies against cytosolic Hsp70, PDI (microsomes), and Erv1 (mitochondria) were included. (B) As in A, but Frd1 was expressed with a myc tag. (C) Construct Osm11-32-DHFR-myc was expressed in yeast and subcellular fractionation was performed as in A. (D) As in C, except construct Osm11-60-DHFR-myc was expressed. (E) Constructs Osm11-32-CPY-myc, and Osm11-60-CPY-myc were expressed in yeast, and subcellular fractionation was performed as in A. Control antibodies include hexokinase (cytosol), Sec62 (microsomes), and Erv1. Light and dark exposures were included for the myc panels. (F) Submitochondrial localization of Osm1, Osm11-32-DHFR-myc, and Osm11-60-DHFR-myc. Mitochondria were subjected to osmotic shock and the mitoplasts (P) were separated from the soluble IMS fraction (S) by centrifugation. Proteinase K (PK) addition verified localization to the IMS, and detergent (Triton X-100) treatment was included as a control. Aconitase and Tim44, matrix markers; Mia40, IMS marker. (G) Mitochondria were analyzed by alkali extraction (Na2CO3) with 0.1 M carbonate at the indicated pH values. Equal volumes of the pellet (P) and TCA-precipitated supernatant (S) fractions were resolved by SDS–PAGE and analyzed by immunoblotting for Osm1, Osm11-32-DHFR-myc, and Osm11-60-DHFR-myc. Controls include integral membrane proteins Tim23 and Mia40 and soluble Ccp1. Asterisks mark nonspecific bands detected by the Osm1 antibody.
The dual localization of Osm1 to mitochondria and microsomes is atypical (Karniely and Pines, 2005). Osm1 likely functions with Ero1 or the homologue Erv2 that resides in the ER (Gerber et al., 2001; Sevier et al., 2001; Williams et al., 2014). Indeed, a synthetic lethal interaction was reported for ero1-1 and Δosm1 (Costanzo et al., 2010). A recent study showed that Osm1 contains two translational initiation sites (Williams et al., 2014); the ATG at position 1 generates a protein that localizes to the ER, whereas initiation at the second in-frame ATG (methionine at amino acid [aa] 32) generates a shorter protein that localizes to mitochondria. Analysis of the Osm1 N-terminal region using a protein-targeting prediction program supported that the N-terminal region is an ER-targeting sequence (Petersen et al., 2011). However, additional sites indicated that aa 1–32 may have properties of a weak mitochondrial-targeting sequence (Claros and Vincens, 1996; Omura, 1998; Fukasawa et al., 2015), including clustering of the positive residues and clustering of hydrophobic residues (Supplemental Figure S1, B and C). We appended two distinct cargoes, the tightly folded reporter dihydrofolate reductase (DHFR) and the unfolded ER reporter carboxypeptidase Y (CPY) to Osm1 aa 1–32 (Osm11-32-DHFR-myc, Osm11-32-CPY-myc) and aa 1–60 (Osm11-60-DHFR-myc, Osm11-60-CPY-myc). We expressed and localized the resulting fusion proteins in WT yeast (Figure 2, C–E). Amino acids 1–32 targeted DHFR exclusively to the mitochondria, whereas aa 1–60 targeted DHFR to both ER and mitochondria. In contrast, appending N-terminal aa 1–32 or 1–60 to CPY resulted in exclusive targeting to the ER. These results support that the N-terminal 1–32 amino acids of Osm1 mediate ER targeting but also have properties that may facilitate mitochondrial targeting, potentially under suboptimal conditions. Moreover, properties within the mature part of the protein seem to play an additional role in directing Osm1 trafficking. Thus Osm1 is an example of a protein that is dual localized and functions in more than one compartment (Karniely and Pines, 2005).
Osm1 localization within mitochondria was investigated in detail (Figure 2F). Mitochondria from the WT strain were subjected to osmotic shock to disrupt the outer membrane; mitoplasts that contain the matrix and intact inner membrane were separated from the soluble IMS contents by centrifugation, and protease treatment verified localization to the IMS. Osm1 and IMS control Mia40 associated with the mitoplast pellet (Figure 2F) but were sensitive to protease in the mitoplast fraction, indicating a location facing the IMS. In contrast, matrix controls Tim44 and aconitase were protected from protease and recovered in the mitoplast pellet fraction. Mitoplast analysis was also coupled with the in vitro import assay (Supplemental Figure S1D). Imported Osm1 and Mia40 were sensitive to protease, confirming residency in the IMS. However, Su9-DHFR was protected from protease, because of localization to the matrix. Note that some mitochondrial proteins such as Hsp60 and Cpn10 may have tightly folded domains that are protease resistant in the presence of detergent (Rospert et al., 1996). Osm1 seems to fall into this category, because a portion was recovered in the supernatant that was resistant to detergent (Figure 2F); in contrast, imported Osm1 was degraded during protease treatment in the presence of detergent, indicating tight folding was not achieved, but tightly folded DHFR was resistant to protease in the presence of detergent (Supplemental Figure S1B). Note that, in some gel systems, we and others typically find that Erv1 and Osm1 migrate as a doublet, but the underlying reason is not known.
Osm1 association with the membrane was also tested using carbonate extraction over pH range 10.5–12.5 (Figure 2F) (Fujiki et al., 1982). In control reactions, integral membrane protein Tim23 remained in the pellet until pH 12.5, whereas soluble Ccp1 was present in the supernatant at pH 10.5. Mia40, which is anchored by a single transmembrane domain, showed an intermediate pattern with recovery in the pellet until pH 11.5, when Mia40 shifted to the supernatant fraction at pH 12. Osm1 was released to the supernatant at pH 11–11.5, suggesting that Osm1 associates with the membrane, but not as tightly as integral membrane proteins that are released to the supernatant at a pH near 12–12.5. We also investigated mitochondrial localization of the DHFR constructs fused to the Osm1-targeting sequence that were expressed in yeast (Figure 2, F and G). From osmotic shock analysis, Osm11-32-DHFR-myc and Osm11-60-DHFR-myc localized to the matrix and IMS, respectively (Figure 2F). From carbonate extraction analysis, Osm11-32-DHFR-myc was soluble, whereas Osm11-60-DHFR-myc shared the same fractionation distribution as Osm1 (Figure 2G). This analysis supports that Osm1 has a bipartite targeting sequence similar to cytochrome b2, and the hydrophobic region from aa 32 to aa 60 functions as a “stop-transfer” domain to direct import into the IMS (Glick et al., 1992); Osm1, therefore, shares a topology similar to Mia40, in that the N-terminal region anchors the protein to the inner membrane, and most of the protein folds into a soluble domain in the IMS (Chacinska et al., 2004).
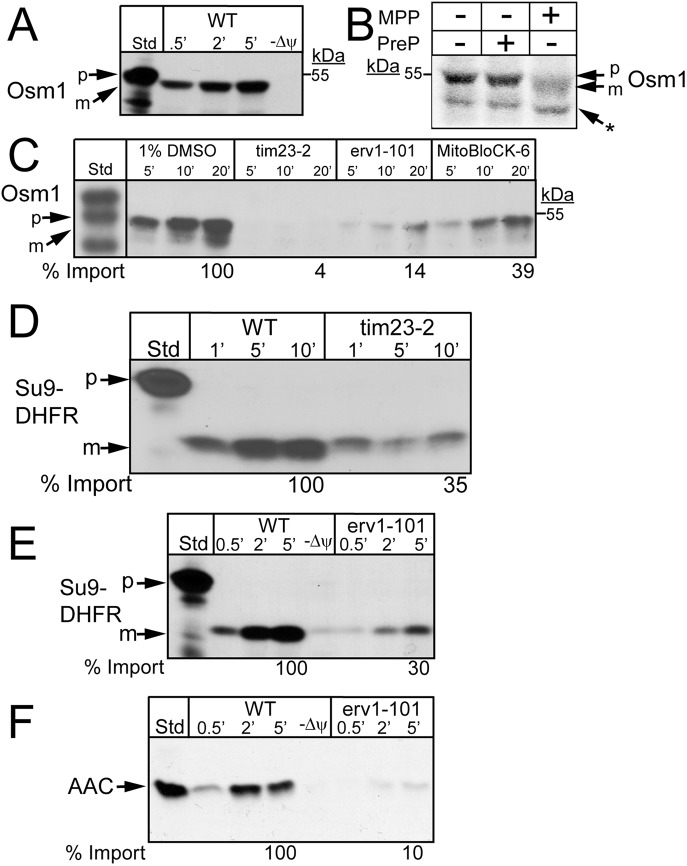
The import pathway of radiolabeled Osm1 into isolated yeast mitochondria was characterized to confirm mitochondrial localization. When these experiments were initially completed, we did not consider that Osm1 had two sites of translation initiation. The construct was designed to begin translation at the first ATG, and this full-length construct was used for the import studies. Osm1 import into mitochondria was dependent on a membrane potential (Δψ), and the N-terminal–targeting sequence seemed to be cleaved (Figure 3A). Recombinant matrix-processing peptidase (MPP) cleaved the Osm1-targeting sequence (Figure 3B) (Rainey et al., 2006). In contrast, the mitochondrial presequence peptidase (PreP), which degrades the processed targeting sequences (Falkevall et al., 2006), did not cleave Osm1. Translocation of Osm1 into mitochondria from the tim23-2 mutant was strongly impaired (Figure 3C), as was the import of Su9-DHFR (Figure 3D). In addition, Osm1 import was compromised in the erv1-101 mutant. However, given that this mutant shows general pleiotropic defects in import (Figure 3, E and F) (Dabir et al., 2013), translocation in the presence of the Erv1 inhibitor MitoBloCK-6 (MB-6) was also tested (Dabir et al., 2013). MB-6 treatment decreased Osm1 import by 60%. Taking the results together, Osm1 requires the TIM23 translocon pathway for import and may require the MIA pathway, potentially for assembly in the IMS, as has been shown for Mia40 (Dabir et al., 2013).
FIGURE 3:
Osm1 is imported via the TIM23 import pathway. (A) Radiolabeled Osm1 was imported into WT mitochondria in the presence and absence of a Δψ. Nonimported precursor was removed by protease treatment, and the imported protein was analyzed by SDS–PAGE and autoradiography. (B) MPP cleavage assay was performed with the addition of 10 µg MPP to radiolabeled Osm1, and samples were resolved on a 10% Tris-Tricine gel. As a negative control, 10 µg of PreP was added to radiolabeled Osm1. p, precursor; m, mature; the asterisk (*) marks a shorter translation product. (C) As in A, Osm1 was imported into tim23-2 and erv1-101 mitochondria. Osm1 was also imported into WT mitochondria in the presence of 25 μM MitoBloCK-6 (Dabir et al., 2013) or a vehicle control (1% dimethyl sulfoxide [DMSO]). Import in the presence and absence (unpublished data) of 1% DMSO is identical. (D) Control showing import of Su9-DHFR into tim23-2 mutant mitochondria. (E,F) Control showing import of (E) Su9-DHFR and (F) AAC into WT and erv1-101 mutant mitochondria.
Osm1 and Erv1 are partner proteins
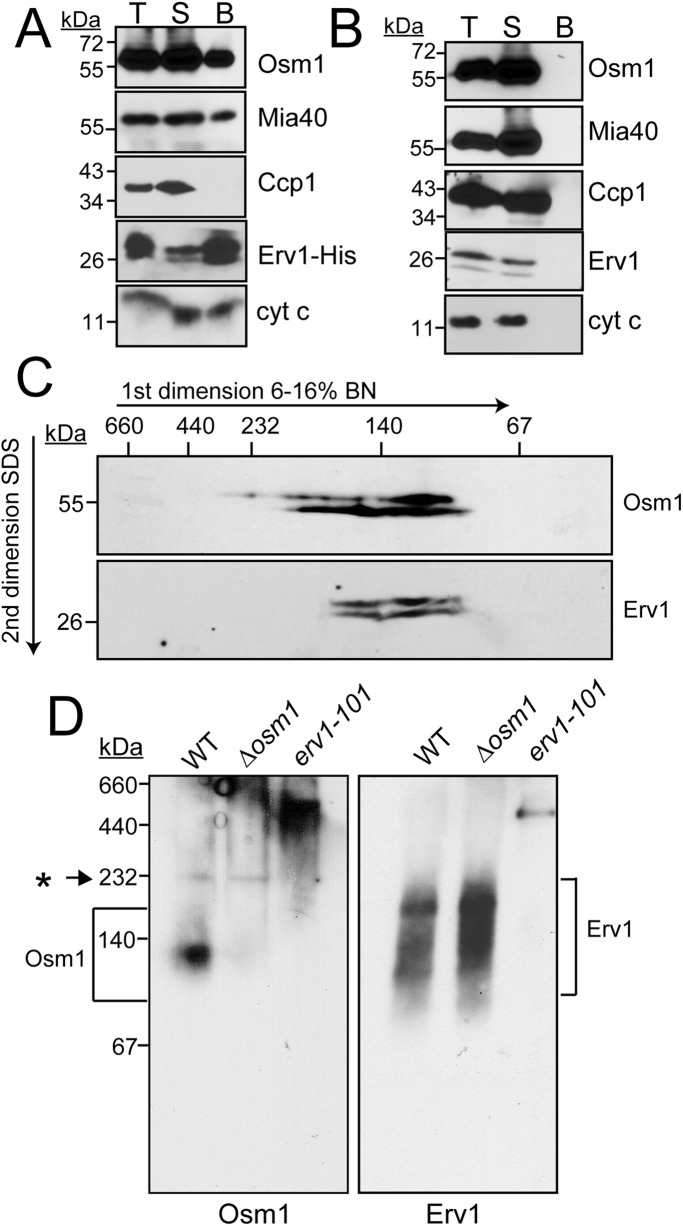
Because Osm1 may accept electrons from Erv1, we investigated whether they might be partner proteins (Figure 4A). We used the Erv1-His strain in which Erv1 contains a C-terminal His tag (Dabir et al., 2007). Mitochondria were purified, solubilized in 1% digitonin, and subjected to pull-down experiments with Ni2+ agarose. Similar to cyt c and Mia40, a fraction (∼10%) of Osm1 copurified with Erv1; as a control, the tested proteins did not bind nonspecifically to the resin in WT mitochondria (Figure 4B). A reciprocal reaction with Osm1-His in pull-down assays failed to show an interaction with Erv1; thus the C-terminal tag on Osm1 may interfere with a stable Erv1 interaction.
FIGURE 4:
Osm1 and Erv1 are partner proteins. (A) Mitochondria from a strain expressing a C-terminal histidine-tagged Erv1 (Erv1-His) were solubilized in 1% digitonin. As a control, 100 μg of extract was withdrawn (T), and 500 μg lysate was incubated with Ni2+-agarose beads. The beads were washed, and bound proteins (B) were eluted with SDS–PAGE sample buffer. For assessment of the effectiveness of binding, 100 μg of the unbound protein fraction (S) was also included. Samples were resolved by SDS–PAGE and analyzed by immunoblotting with specific antibodies against Osm1, Mia40, Ccp1, Erv1, and cyt c. (B) Similar to A, except that WT mitochondria were used. (C) WT mitochondria were solubilized as in A and then separated in the first dimension of a 6–16% BN gel, followed by reducing SDS–PAGE in the second dimension. Osm1 and Erv1 were detected by immunoblotting. (D) WT, Δosm1, and erv1-101 mitochondria were solubilized as in A and then separated in the first dimension of a 6–16% BN gel; Osm1 and Erv1 complexes were detected by immunoblotting. Brackets highlight the region of the complexes and the asterisk (*) highlights a nonspecific band.
In a second experimental set, we used one-dimensional gel analysis (blue-native [BN]-PAGE) (Figure 4D) and two-dimensional gel analysis (BN-PAGE followed by reducing SDS–PAGE) (Figure 4C) to investigate Osm1 and Erv1 complexes in WT mitochondria. Osm1 and Erv1 comigrated in a complex of ∼140 kDa, and Erv1 migrated in complexes of larger size as well (∼160–200 kDa). As a control, we also probed for these complexes in Δosm1 and erv1-101 mitochondria and found that the Erv1 complex is unchanged in the Δosm1 yeast, which is in agreement with steady-state Erv1 levels in this strain (as shown later in Figure 8A). The Osm1 complex, however, was barely detectable in the erv1-101 yeast strain (Figure 4D). Localization of Mia40 into Erv1 complexes using BN gels has been difficult, because Mia40 does not migrate well in BN gels due to its acidic nature (Chacinska et al., 2004). Thus a subset of Osm1 and Erv1 form a complex in the IMS, but Erv1 also seems to assemble in other complexes.
FIGURE 8:
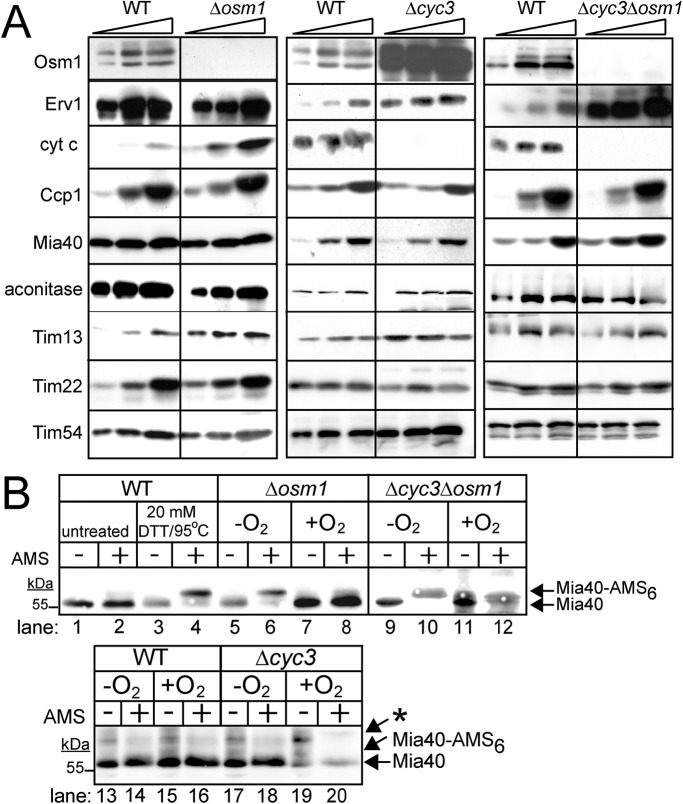
A regulatory pathway for expression of Osm1, cyt c, and Erv1. (A) Analysis of steady-state levels of mitochondrial proteins in the WT, Δosm1, Δcyc3, and Δcyc3Δosm1, strains. Cells were grown aerobically in glucose medium at 25°C, and protein levels were analyzed by immunoblotting. Antibodies against mitochondrial proteins Osm1, Erv1, cyt c, Ccp1, Mia40, aconitase, Tim13, Tim22 and Tim54 were used. (B) Analysis of the Mia40 oxidation state with AMS treatment. Mitochondria were freshly isolated from strains WT, Δosm1, Δcyc3, and Δcyc3Δosm1 that were grown in aerobic or anaerobic conditions in glucose media. Mitochondria were acid precipitated and resuspended in buffer with AMS. Samples were separated by SDS–PAGE followed by immunoblot analysis with antibodies against Mia40. As a control to monitor Mia40 migration on the gel system, cells grown aerobically were either untreated (lanes 1 and 2) or treated with 20 mM DTT and heated to 95°C before acid precipitation and treatment with AMS (lanes 3 and 4). Mia40-AMS6 marks the position of Mia40 that has been fully reduced. Note: an asterisk (*) marks a nonspecific band for Mia40 that is detected by the antibody.
Osm1 facilitates the transfer of electrons from Erv1 to convert fumarate to succinate
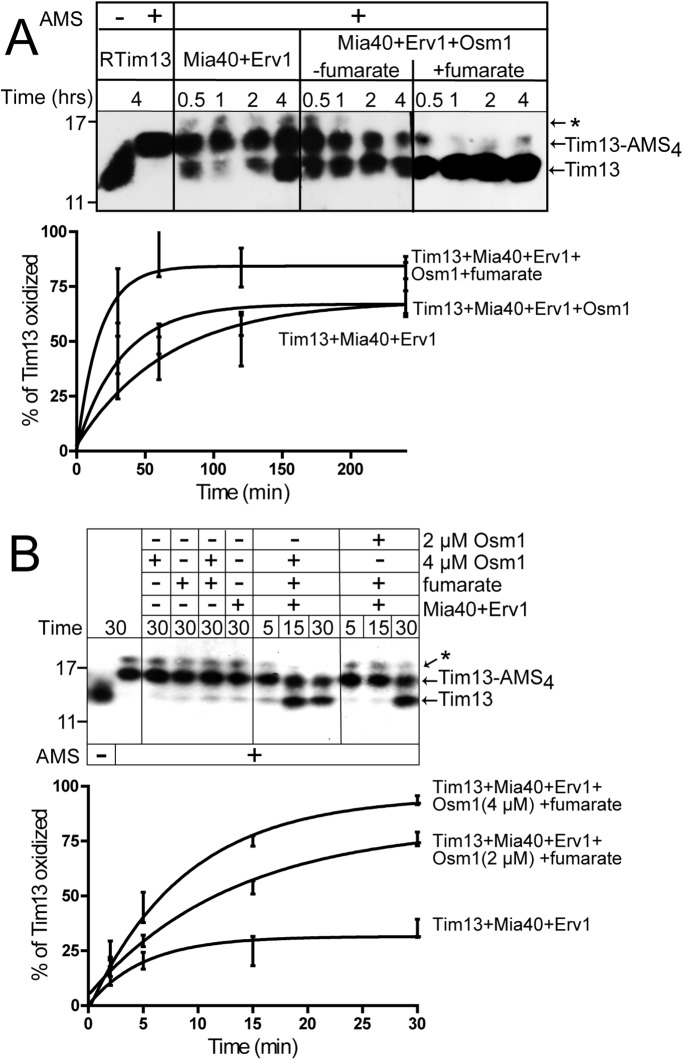
Fumarate may accept electrons from Erv1 via catalysis by Osm1. We used the in vitro reconstitution system with minimal components to test whether the Osm1/fumarate couple facilitated oxidation of Tim13 by Erv1 (Tienson et al., 2009). Tim13 oxidation was monitored over a 4-h time course by the addition of alkylating agent 4-acetamido-4′-maleimidylstilbene-2,2′-disulfonic acid (AMS) followed by nonreducing SDS–PAGE and immunoblot analysis with anti-Tim13 antibodies (Figure 5A). AMS modifies reduced cysteine residues and increases the molecular mass by 0.5 kDa. In the presence of AMS, reduced Tim13 acquired 4 AMS molecules, indicating that the cysteine residues could be modified by AMS and that Tim13 remained reduced during the 4-h assay (Figure 5A). In agreement with published studies (Tienson et al., 2009), excess Tim13 (15 μM) was partially oxidized over a 4-h time period in the presence of 1 μM Mia40 and 1 μM Erv1, with O2 as a terminal electron acceptor. The addition of 1 μM Osm1 resulted in partial oxidation of Tim13, but the inclusion of excess fumarate (200 μM) markedly decreased the time for oxidation of Tim13 to 30 min, because Osm1 catalyzed the transfer of electrons from Erv1 to fumarate, generating succinate. In a series of control reactions, the addition of Osm1, fumarate, and Osm1/fumarate to reduced Tim13 did not directly oxidize Tim13 (Figure 5B). Increasing the concentration of Osm1 in the reconstitution assay from 2 to 4 μM correlated with an increased efficiency in the oxidation of Tim13 (Figure 5B).
FIGURE 5:
The Osm1/fumarate couple increases the rate of Tim13 oxidation in vitro. (A) Reduced Tim13 (15 μM) was incubated with combinations of Mia40 (1 μM), Erv1 (1 μM), Osm1 (1 μM), and fumarate (200 μM) in a reaction buffer (50 mM Tris-HCl, pH 8.0, 150 mM KCl, and 1 mM EDTA) at 25°C in a time-course assay as indicated. Aliquots were removed at the indicated times, and free thiols were blocked by addition of AMS. Oxidized and reduced (Tim13-AMS4) Tim13 were detected by nonreducing SDS–PAGE followed by immunoblotting with antibodies against Tim13. (B) As in A, except different concentrations of Osm1 (2 and 4 μM) were used in the reconstitution, and the time course was decreased to 30 min. The asterisk (*) indicates a nonspecific band that may be a Tim13 aggregate, as Tim13 is prone to forming multimers (Tienson et al., 2009), or a cross-reacting band detected by the antibody that is a carryover from preparation of the recombinant antigen. Bottom panels show quantitation of the Tim13 oxidation reactions.
We also used the O2 electrode to determine whether Osm1/fumarate could effectively compete with O2 during the oxidation of the nonphysiologic substrate dithiothreitol (DTT; Supplemental Figure S2); this technique was used previously to show that the cyt c/Ccp1 couple was more efficient at accepting electrons from Erv1 than O2 (Dabir et al., 2007). Fumarate, Osm1, and Erv1 along with appropriate controls were subsequently added to a buffer that contained excess substrate DTT, and reduction was measured by the consumption of O2. As shown in Supplemental Figure S2, A and D, the rate of O2 consumption was decreased in the presence of Osm1/fumarate, indicating that Osm1/fumarate effectively competes with O2 to accept electrons. Moreover, control reactions that lacked Erv1 but contained Osm1, fumarate, or Osm1/fumarate supported that Osm1 did not directly oxidize DTT and that Erv1 was required (Supplemental Figure S2B). Finally, in a comparison reaction with cyt c/Ccp1 addition, the decreased rate of O2 consumption with cyt c/Ccp1 was similar to that of Osm1/fumarate (Supplemental Figure S2, C and D), confirming that Osm1/fumarate and cyt c are similarly efficient in accepting electrons from Erv1 in vitro. In addition to cyt c, Erv1 thus shuttles electrons to Osm1/fumarate.
We also investigated general Osm1 function in the ER (Supplemental Figure S3). The fractionation results from carbonate extraction of the ER mirrored that of Osm1 in the mitochondria (Figure 2F and Supplemental Figure S3A). Specifically, ER-localized Osm1 fractionated to the pellet until pH 11.5 and then shifted to the supernatant at pH 12.0. The control PDI remained in the supernatant over the entire pH range. With the O2 electrode and the nonphysiologic substrate DTT (as in Supplemental Figure S2), Osm1/fumarate also accepted electrons from ER resident Erv2 and competed with O2 (Supplemental Figure S3B). Osm1 likely has a similar role in mitochondria and ER.
Coordinated regulation of terminal electron acceptors
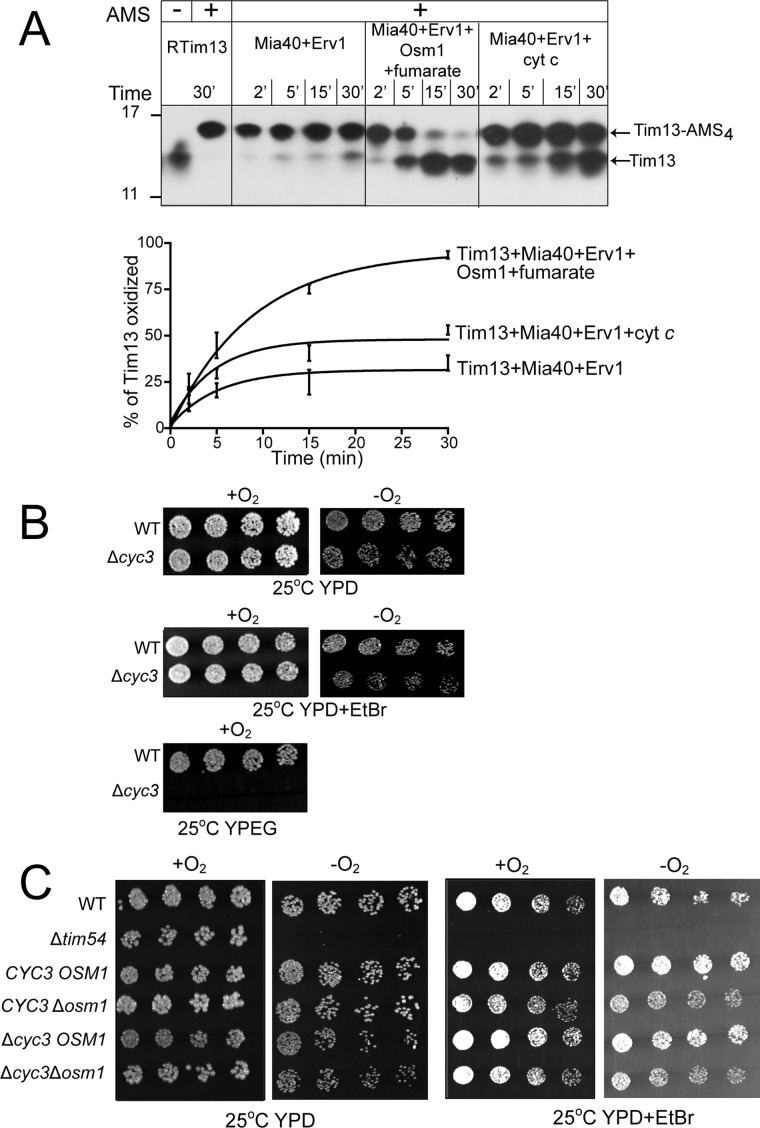
In contrast to Frd1 expression that is induced during anaerobiosis, Osm1 is constitutively expressed, which suggests that Osm1 may operate under aerobic and anaerobic conditions (Camarasa et al., 2007). Because cyt c and Osm1/fumarate are terminal electron acceptors for Erv1, we compared the oxidation of reduced Tim13 in the presence of cyt c versus Osm1/fumarate (Figure 6A). As shown previously, when the terminal electron acceptor was O2, Tim13 remained reduced in the 30-min assay, instead requiring ∼2–4 h for oxidation. However, Osm1/fumarate oxidized Tim13 more quickly than cyt c, suggesting both electron acceptors are poised to accept electrons from Erv1 in vivo (Figure 6A).
FIGURE 6:
Osm1/fumarate and cyt c are poised to accept electrons from Erv1. (A) The reconstitution assay was performed as in Figure 5A, including a reaction series with cyt c (400 μM) as a terminal electron acceptor. The time course was decreased to 30 min. The bottom panel shows quantitation of Tim13 (15 μM) oxidation over the 30-min time course in the reactions with Mia40 (1 μM) and Erv1 (1 μM); Mia40 (1 μM), Erv1 (1 μM), and cyt c (400 μM); and Mia40 (1 μM), Erv1(1 μM), Osm1 (1 μM), and fumarate (200 μM). n = 3. (B) Growth analysis of Δcyc3 in aerobic (+O2) and anaerobic (−O2) conditions on glucose (YPD) and ethanol/glycerol (YPEG) carbon sources. Ethidium bromide (EtBr) treatment removed the mitochondrial DNA. (C) A cross of Δcyc3 with Δosm1 was sporulated, and the resulting tetrads were serially diluted by a factor of 2 onto rich glucose media (YPD) in the presence or absence of EtBr and incubated in aerobic (+O2) and anaerobic (−O2) conditions. The parental strain (WT) and tim54-1 mutant (petite-negative) were included as controls. Plates were photographed after 3 d at 25°C.
We investigated the relationship between cyt c and Osm1 in vivo. Cells that lack Osm1 (Figure 1A) or functional cyt c (achieved in the Δcyc3 strain, which prevents heme addition to apocytochrome c) do not display obvious growth defects (Dabir et al., 2007), except that the Δcyc3 strain may grow slowly in the presence of ethidium bromide under anaerobic conditions (Figure 6B). We crossed the Δcyc3 and Δosm1 strains and performed tetrad analysis (Figure 6C). On glucose media, in the presence or absence of oxygen, a synthetic lethal interaction was not observed. When the cells were treated with ethidium bromide under anaerobic conditions, the Δcyc3Δosm1and Δosm1 strains grew more slowly; however, the strains continued to grow, demonstrating that the genetic interaction was not lethal.
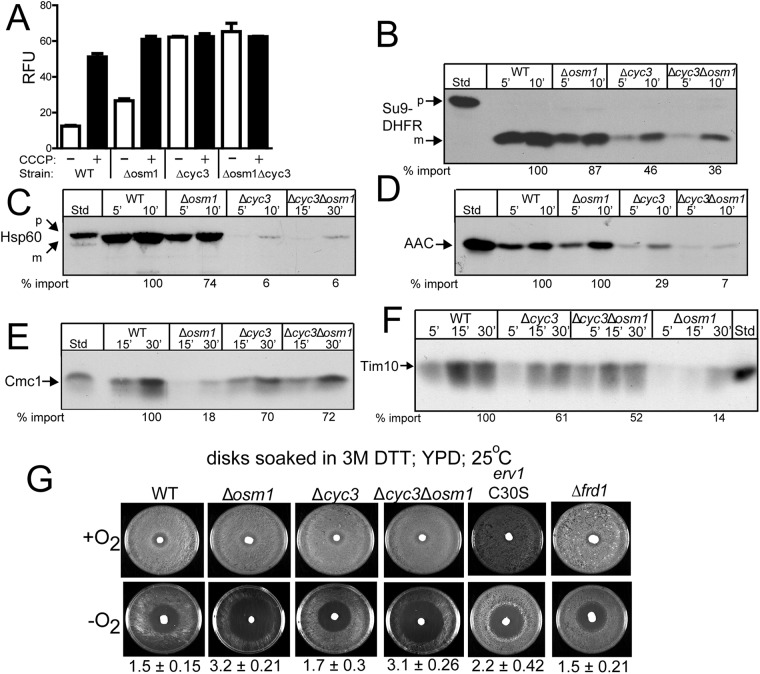
The relationship between cyt c and Osm1 was extended in functional studies with isolated mitochondria. The WT, Δosm1, Δcyc3, and Δcyc3Δosm1 strains were grown to mid–log phase in rich glucose media, and mitochondria were isolated. The Δψ of mitochondria was measured using the fluorescent dye 3,3′-dipropylthiadicarbocyanine iodide (DiSC3(5)), which is taken up by mitochondria and then released when the Δψ is dissipated, typically with the uncoupler carbonyl cyanide m-chlorophenyl hydrazone (CCCP) (Hasson et al., 2010). The relative change in fluorescence between dye uptake and release is a relative measure of the Δψ. The WT mitochondria displayed the largest Δψ, whereas Δosm1 mitochondria had an ∼30% decrease in Δψ (Figure 7A). In contrast, Δcyc3 and Δcyc3Δosm1 mitochondria had a minimal Δψ (Figure 7A); this decreased Δψ was expected, because these strains do not respire (Sanchez et al., 2001).
FIGURE 7:
Osm1 plays a role in the import of MIA substrates. (A) Δψ measurements of purified mitochondria from WT, Δosm1, Δcyc3, and Δcyc3Δosm1 strains were performed with DiSC3(5) using a FlexStation plate reader (Molecular Devices). Coupled mitochondria sequestered and quenched the dye fluorescence; collapse of the Δψ was achieved with CCCP addition. (B) Radiolabeled Su9-DHFR was imported into WT, Δosm1, Δcyc3, and Δcyc3Δosm1 mitochondria for the indicated times. Nonimported precursor was removed by protease treatment. A 10% standard (Std) from the translation reaction was included. Import reactions were quantitated using Image J software; 100% was set as the amount of precursor that imported into WT mitochondria at the endpoint of the time course. p, precursor; m, mature. A representative gel is shown. n = 3. (C) As in B, with Hsp60. (D) As in B, with AAC. Samples were treated with carbonate extraction after protease treatment to confirm that AAC inserted into the inner membrane. (E) As in B, with Cmc1. (F) As in B, withTim10. (G) Equal amounts of cells from WT, Δosm1, Δcyc3, Δcyc3Δosm1, erv1C30S, and Δfrd1 were spread onto YPD plates. Filter disks were placed in the middle of the plates, and 10 μl of 3 M DTT was aliquoted directly onto the filter disks. The plates were grown at 25°C in aerobic and anaerobic conditions for 2 d and photographed. The distance in which growth was inhibited in anaerobic conditions was measured and is listed below the plates, because differences were observed.
The import of radiolabeled precursors into mitochondria from the WT, Δosm1, Δcyc3, and Δcyc3Δosm1 strains was also tested. Matrix precursors Su9-DHFR (Figure 7B) and Hsp60 (Figure 7C) and inner membrane precursor AAC (Figure 7D), which depend on the Δψ, were imported into isolated mitochondria. The import of Su9-DHFR, Hsp60, and AAC was strongly compromised in Δcyc3 and Δcyc3Δosm1 strains, in agreement with the lowered Δψ. In contrast, import was slightly decreased in Δosm1 mitochondria. MIA substrates Cmc1 (Figure 7E) and Tim10 (Figure 7F) were also tested. Here Cmc1 and Tim10 import were slightly decreased in Δcyc3 and Δcyc3Δosm1 mitochondria. In contrast, the import of Cmc1 and Tim10 were decreased by ∼80% in Δosm1 mitochondria. Thus Osm1 seems to play an important role for the import of MIA substrates.
As cells lacking functional Erv1 are sensitive to DTT (Mesecke et al., 2005), we investigated DTT sensitivity of Δcyc3, Δosm1, and Δcyc3Δosm1 strains using a halo assay on plates (Figure 7G) (Frand and Kaiser, 1998). In controls, the erv1C30S mutant showed increased sensitivity to DTT in contrast to the WT strain. Growth inhibition of the Δcyc3 strain was similar to WT, whereas the Δosm1 and Δcyc3Δosm1 strains were more sensitive to DTT than the erv1 mutant. The sensitivity to DTT is specific to the Δosm1 strain, because the Δfrd1 strain displayed a similar phenotype to the WT strain. Because sensitivity to DTT may be representative of a general redox stress to cells, we used the same halo assay to test for sensitivity to oxidants diamide and hydrogen peroxide (Supplemental Figure S4). However, all strains displayed the same sensitivity, indicating that sensitivity was specific to reductant. Given that Osm1 also localizes to the ER, the loss of Osm1 in the ER may contribute to the DTT sensitivity (Frand and Kaiser, 1998).
Previous studies by Herrmann and colleagues indicated that reductants such as GSH stimulate import at concentrations up to 5 mM (Bien et al., 2010). However, our published studies were contradictory, showing that reductants such as GSH and ascorbate inhibit the import of CX9C and CX3C substrates at 2 and 5 mM (Neal et al., 2015). Because the Δosm1 and Δcyc3Δosm1 strains in the halo assays were more sensitive to reductant, we extended our studies to in vitro import assays. The import of Cmc1 and Tim10 into mitochondria from all strains (WT, cyc3, Δosm1, and Δcyc3Δosm1) was impaired in the presence of 1 mM GSH or 1 mM DTT (Supplemental Figure S5). In contrast, the import of Su9-DHFR was not inhibited by the presence of reductant (Supplemental Figure S6). Thus the addition of reductant is a general inhibitor of Cmc1 and Tim10 import that is independent of mutant background.
The steady-state levels of mitochondrial proteins were also determined by immunoblot analysis in the Δcyc3, Δosm1, and Δcyc3Δosm1 strains grown in complete minimal glucose media at 25°C to mid–log phase (Figure 8A). The abundance of aconitase, Mia40, Ccp1, and TIM22 components Tim22, Tim54, Tim13 was similar among the different strains. In contrast, cyt c abundance was increased approximately fivefold in the Δosm1 strain, and similarly, Osm1 was increased ∼10-fold in the Δcyc3 strain. Erv1 steady-state levels were similar in Δosm1 and Δcyc3 strains, but Erv1 abundance increased approximately threefold in the double Δcyc3Δosm1 mutant. The increased abundance of Erv1 in the Δcyc3Δosm1 strain may account for the rescued import of Tim8 and Cmc1 in Δcyc3Δosm1 mitochondria (Figure 7, E and F). Thus cyt c and Osm1 are poised to accept electrons from Erv1, and expression of the terminal electrons acceptors seems to be coordinately regulated.
The redox status of Mia40 in mitochondria from WT, Δcyc3, Δosm1, and Δcyc3Δosm1 strains was investigated using thiol trapping with AMS (Figure 8B) (Sztolsztener et al., 2013; Neal et al., 2015). Mitochondria were purified from cells grown in aerobic or anaerobic conditions. We previously reported that Mia40 can serve as an electron sink and acquire six electrons when presented with reduced substrates (Neal et al., 2015). In control reactions in which Mia40 was treated with reductant at 95°C (Figure 8B, lanes 1–4), 6 AMS molecules bound to Mia40, indicating Mia40 was completely reduced. In mitochondria from Δosm1 cells that were grown under anaerobic conditions, the majority of Mia40 is in a reduced state (Figure 8B, lanes 5, 6, 9, and 10), whereas Mia40 in WT and Δcyc3 mitochondria was oxidized (Figure 8B, lanes 13, 14, 17, and 18). In contrast, in aerobic conditions, Mia40 remains oxidized in all mitochondrial types (Figure 8B, lanes 7, 8, 11, 12, 15, 16, 19, and 20). This thiol trapping analysis suggests that Osm1 plays an important role in maintaining the oxidized state of Mia40 under anaerobic conditions.
DISCUSSION
Prokaryotic cells have a wide range of terminal electron acceptors for both aerobic and anaerobic conditions, enabling the cell to maintain oxidative folding in the periplasm across diverse environmental conditions (Mamathambika and Bardwell, 2008). Whereas electron acceptors have been identified for aerobic conditions in eukaryotes, questions remain about strategies in the anaerobic state. Here we show that yeast can use Osm1/fumarate under aerobic and anaerobic conditions to accept electrons from Erv1.
Osm1 has dual localization to the mitochondria and the ER
Surprisingly, Osm1 has dual localization to the ER and mitochondria. Whereas a large number of proteins show dual localization, most proteins localize to the mitochondria and cytosol or nucleus (Karniely and Pines, 2005). A recent study showed that Osm1 has two alternative start sites at the first ATG codon and an in-frame ATG codon at position 32 (Supplemental Figure S1B) (Williams et al., 2014). As seen with fluorescence microscopy, the mutation of the first ATG codon resulted in Osm1 targeting to mitochondria, and mutation of the second ATG codon at position 32 resulted in targeting to the ER (Williams et al., 2014). A variety of targeting-prediction programs indicated that the first 32 amino acids showed strong targeting for the ER, but the programs also indicated that this region had some properties that supported targeting to mitochondria (Claros and Vincens, 1996; Horton et al., 2007; Petersen et al., 2011; Fukasawa et al., 2015).
To understand the strength of this N-terminal region in differential organellar targeting, we used a different approach in which we appended aa 1–32 or 1–60 either to a tightly folded reporter domain (DHFR) or reporter domain of an ER-translocated protein (CPY) and investigated organellar targeting in yeast using subcellular fractionation. General analysis of the N-terminal–targeting region suggests that the N-terminal 32 amino acids function as a classical ER-targeting sequence (Petersen et al., 2011) and fusions to CPY directed the fusion protein to the ER (Figure 2E). However, the N-terminal 32 amino acids fused to DHFR directed the construct to the mitochondria and aa 1–60 were required for correct sorting within mitochondria (Figure 2, C and F). Indeed, the full-length Osm1 was imported into isolated mitochondria and cleaved by recombinant MPP (Figure 3, A and B), This suggests that the N-terminal stretch of 1–32 amino acids is a suboptimal mitochondrial-targeting sequence, and that differences in the cargoes (DHFR vs. CPY) were responsible for the differential subcellular localization of the fusion proteins. Thus it is likely that additional signals may be present within the mature protein or that different cytosolic factors ultimately dictate the localization of Osm1. Distribution to ER versus mitochondria could also be regulated by other factors such as redox, stress, or nutritional status (Karniely and Pines, 2005).
Osm1 is a potential terminal electron acceptor in the ER
In yeast, Osm1 in the ER likely functions as a terminal electron acceptor, especially in anaerobic conditions, because the only well-characterized acceptor is O2 (Tu and Weissman, 2004; Williams et al., 2014). Electrons may be shuttled from Ero1 or the yeast-specific sulfhydryl oxidase Erv2, which is a homologue of Erv1 (Sevier, 2012). An open question is the mechanism by which fumarate may be transported into the ER, because a specific transporter has not been identified. However, functional studies show a requirement for fumarate in the ER. Liu and colleagues report that fumarate functions as a final electron acceptor in the anaerobic production and secretion of alpha-amylase, a pathway that relies on the ER (Liu et al., 2013). In addition, proteomic studies show that ER proteins such as PDI can be succinated as a posttranslational modification and that fumarate is the substrate (Merkley et al., 2014). Having an anaerobic acceptor in yeast is important, because the organism grows anaerobically. A fumarate reductase has not been identified for mammals. Potentially, an anaerobic acceptor is not required, because cells are not typically anaerobic, except in pathogenic conditions such as ischemia reperfusion (Blaisdell, 2002). However, human embryonic stem cells originate from the blastocyst inner cell mass (Thomson et al., 1998), which is in a hypoxic microenvironment estimated at 1.5–5.3% in the mammalian reproductive tract (Dunwoodie, 2009); thus free O2 may be limiting. As a result, mammals may have alternative oxidases, because ERO-1–deficient cells were surprisingly viable (Zito et al., 2010). In this case, additional oxidases include vitamin K epoxide reductase, quiescin sulfhydryl oxidase, and/or peroxiredoxin IV (Mairet-Coello et al., 2004; Wajih et al., 2007; Zito et al., 2010) that, in addition to O2, may shuttle electrons to potential anaerobic acceptors such as free flavins, which can be reduced in vitro, or other small molecules (Gross et al., 2006).
Osm1 is a terminal electron acceptor in the mitochondrial IMS
Osm1 is a peripheral membrane protein in the mitochondrial IMS and seems to have a complicated assembly pathway. The N-terminal region contains a weak mitochondrial-targeting sequence that is cleaved by MPP, and the TIM23 translocon is required for Osm1 import. However, the MIA pathway may have a secondary role in folding and assembly of Osm1, as shown by decreased import in the presence of the Erv1 inhibitor, MitoBloCK-6 (Figure 3C). This may not be unexpected, because the MIA pathway is required for efficient import of Mia40 (Chacinska et al., 2008; Dabir et al., 2013) and also mediates import of inner membrane proteins Tim22 and AAC (Dabir et al., 2013; Wrobel et al., 2013). Osm1 does not have the typical cysteine motifs of CX3C and CX9C substrates, but does contain 3 cysteine residues that may require the oxidative folding pathway, and Osm1 also binds noncovalently to FAD; thus the MIA pathway may facilitate Osm1 folding or association with Erv1.
Like cyt c, a fraction of Osm1 assembles in a complex with Erv1 (Figure 4, A and B), and Osm1 and Erv1 comigrate in complexes of similar molecular mass (Figure 4C). This is likely a transient interaction, as electrons can be shuttled from Erv1 to FAD-bound Osm1 in vitro (Figure 5). Because Osm1 is expressed in both aerobic and anaerobic conditions and lacks obvious growth defects (Figure 1A), it likely functions in both aerobic and anaerobic conditions. A synthetic lethal interaction between erv1-101 and Δosm1 was observed in anaerobic conditions (Figure 1B), and a similar synthetic lethality was reported with temperature-sensitive erv1 mutants and cyt c (Allen et al., 2005; Dabir et al., 2007). It is difficult to interpret why Erv1 and cyt c showed synthetic lethality under anaerobic conditions, because cyt c transfers electrons to the electron transport chain or Ccp1 and ultimately O2 (Dabir et al., 2007); however, this may be caused by pleiotropic defects associated with the erv1 mutants. Because Osm1 uses fumarate as an electron acceptor to generate succinate, it is poised to function anaerobically.
Yeast has a network to regulate expression of terminal electron acceptors, because cyt c was up-regulated in Δosm1cells and vice versa (Figure 8A). In addition, Δcyc3Δosm1 cells subsequently increased the steady-state level of Erv1. Osm1 may be the preferred terminal electron acceptor in vivo, because the DTT sensitivity of Erv1 is shared in Δosm1 cells (Figure 7G). Moreover, mitochondria lacking Osm1 showed decreased import for the CX9C substrate Cmc1, whereas the import of Cmc1 was decreased by ∼20% (Figure 7, B–E). Finally, the Mia40 pool is mostly reduced in mitochondria that lack Osm1 under anaerobic conditions (Figure 8B), suggesting Osm1/fumarate is critical for efficient oxidation of Mia40 via Erv1. Curiously, yeast must have additional terminal electron acceptors and/or small molecules, because Δcyc3Δosm1 cells were still viable under anaerobic conditions (Figure 6C) and Mia40 was not reduced (Figure 8B).
Because Osm1 is absent in mammals, the presence of other anaerobic electron acceptors is an open question. In trypanosomes, a homologous fumarate reductase has been identified in the glycosome, a peroxisomal-like organelle, which maintains glycosomal NAD+/NADH balance (Besteiro et al., 2002). Again, mammals may not require an acceptor, because cells are never truly anaerobic, but taking clues from the ER, other systems may also function. To this end, the peroxiredoxin family is large, and peroxiredoxin 3 and 5 have been localized to the mitochondrial matrix (Cox et al., 2010). In addition, small molecules such as GSH/GSSG may also mediate disulfide-bond formation in anaerobic conditions (Bien et al., 2010). Yeast has diverse terminal electron acceptors in the IMS.
MATERIALS AND METHODS
Strains and constructs
OSM1 and FRD1 were PCR amplified from yeast genomic DNA isolated from the WT (GA74-6A) strain, and the 3′ primer introduced a myc tag to the C-terminus. The fragment was cloned into vector pRS425 that contains the GPD1 promoter and PGK1 terminator. Osm11-32-DHFR-myc and Osm11-60-DHFR-myc fusions were constructed in the aforementioned pRS425 vector and transformed into the WT yeast strain. Osm11-32-CPY-myc and Osm11-60-CPY-myc fusions were constructed in the low-copy pRS315 vector and transformed into the WT yeast strain. Recombinant Osm1 (starting from aa 23) was cloned in pET28a (Novagen) with an N-terminal His tag.
The parental S. cerevisiae yeast strain used in the study was GA74-6A and GA74-1A (Supplemental Table S1). The ∆osm1::LEU2 strain was generated using the leucine cassette in vector pUG73 with the standard procedure (Gueldener et al., 2002). Standard yeast genetics were used to generate the strains and for synthetic lethal analysis (Guthrie and Fink, 1991).
Recombinant Tim13, Mia40, Erv1, Erv2, and Osm1 were expressed and purified under native conditions as previously described (Dabir et al., 2007, 2013; Tienson et al., 2009). Recombinant Osm1 was transformed into BL21-CodonPlus (DE3)-RIL (Stratagene) and induced with 0.5 mM isopropyl β-d-1-thiogalactopyranoside in 2YT medium (1.6% tryptone, 0.5% yeast extract, 0.5% NaCl, 10 mM Tris, pH 7.4) for 4 h at 30°C. Recombinant Mia40, Erv1, and Osm1 were purified under native conditions with Ni2+-agarose (Qiagen) according to the manufacturer’s instructions. The proteins were dialyzed in buffer (20 mM Tris-HCl, pH 7.0, 150 mM KCl, and 1 mM EDTA) at 4°C overnight. Recombinant Tim13 was purified under denaturing conditions as previously described (Tienson et al., 2009).
Subcellular fractionation and mitochondrial assays
Subcellular fractionation from spheroplasts was performed as previously described (Claypool et al., 2006). The fractions were separated by differential centrifugation, and an equal amount of each fraction was separated by SDS–PAGE followed by immunoblot analysis.
Mitochondria were purified from yeast cells grown in media with glucose or ethanol/glycerol (Glick and Pon, 1995). Proteins were synthesized by the TNT Quick Coupled Transcription/Translation kits (Promega) in the presence of [35S]methionine. The radiolabeled precursor was incubated with isolated mitochondria, and an import time course was performed. Where indicated, the ΔΨ was dissipated with 5 mM FCCP. Nonimported radiolabeled protein was removed by treatment with 100 μg/ml trypsin for 15 min on ice, and trypsin was inhibited with 200 μg/ml soybean trypsin inhibitor for 30 min on ice. Samples were separated by SDS–PAGE and visualized using autoradiography. Mitochondria were purified from the Erv1-His6 strain, and pulldown experiments were performed as previously described (Dabir et al., 2007).
Mitochondrial fractionation was performed as previously described (Claypool et al., 2006). For intact mitochondria, 300 μg of isolated mitochondria was incubated in 0.6 M sorbitol and 20 mM HEPES-KOH (pH 7.4). For generation of mitoplasts, 300 μg of isolated mitochondria was incubated in 0.03 M sorbitol and 20 mM HEPES-KOH (pH 7.4). As indicated, 20 μg/ml Proteinase K and 0.1% Triton X-100 was added. Samples were centrifuged at 14,000 × g for 10 min to separate pellet and supernatant. The supernatants were precipitated with 20% trichloroacetic acid (TCA) and resuspended in SDS sample buffer.
Carbonate extraction was performed as described previously (Claypool et al., 2006). Briefly, 200 μg of mitochondria was incubated in 200 μl of 0.1 M Na2CO3 at the indicated pH for 15 min on ice. Samples were centrifuged at 14,000 × g for 10 min to separate the pellet (P) and supernatant (S). The supernatants were precipitated with 20% (wt/vol) TCA, and the pellet and supernatant fractions were resuspended in equal volumes of SDS sample buffer.
BN-PAGE was performed as described previously (Claypool et al., 2006). Before resolution by second dimension by SDS–PAGE, the BN-PAGE gel was soaked in 1% (wt/vol) SDS, 1% (vol/vol) β-mercaptoethanol for 30 min at 50°C, and individual lanes were isolated with a razor blade and embedded in a 4% stacking gel.
For thiol-trapping assays, cells were grown under anaerobic conditions at 30°C in bioreactors (Photon Systems Instruments) with a working volume of 400 ml with continuous stirring. Traces of oxygen that were present at the beginning of the culture were removed by bubbling nitrogen gas through the medium for 30 min to achieve anaerobiosis. At mid–log phase, mitochondria were immediately isolated and solubilized in buffer containing 1% digitonin, 50 mM Tris (pH 7.0), 150 mM KCl, and 1 mM EDTA supplemented with protease inhibitors: 200 mM phenylmethylsulfonyl fluoride, 10 mM leupeptin, 1 mM chymostatin, and 1 mM pepstatin for 15 min at 4°C followed by centrifugation at 14,000 rpm for 10 min at 4°C. In control reactions, the mitochondrial lysate was reduced with 20 mM DTT and incubated at 25°C or 95°C. The mitochondrial lysate was precipitated with 20% TCA on ice for 15 min, and precipitated proteins were pelleted by centrifugation (14,000 × g for 30 min at 4°C). The pellet was resuspended in 20 μl of 20 mM AMS and incubated for 1 h at 37°C. Samples were separated by nonreducing SDS–PAGE, and Mia40 was detected by immunoblot analysis.
Reconstitution studies and oxygen-consumption assays
In vitro reconstitution studies with recombinant Tim13, Mia40, and Erv1 were performed as previously described (Tienson et al., 2009). Reduced Tim13 (15 μM), Erv1 (1 μM), Osm1 (1–4 μM), and fumarate (200 μM) were mixed in buffer R (50 mM Tris-HCl, pH 8.0, 150 mM KCl, 1 mM EDTA) at 25°C; alternatively, 400 μM cyt c was used in place of Osm1/fumarate. The assay proceeded for up to 4 h, and the free thiols were trapped with addition of AMS. Proteins were resolved using nonreducing 15% SDS–PAGE followed by immunoblot analysis with antibodies against Tim13.
O2 consumption assays were performed as previously described (Dabir et al., 2007). Reduction of O2 by Erv1 was measured with the Hansatech O2 electrode (PP systems). For initiation of O2 consumption, 2 μM of Erv1 was added to 1 ml of Buffer R buffer containing 2 mM DTT (nonphysiologic substrate). Additional components included 2–4 μM Osm1 and 10 μM fumarate, 20 μM cyt c and 20 μM Ccp1, and 2 μM bovine serum albumin (BSA). The rate of O2 consumption was calculated when the reaction was in the linear range.
Assessment of mitochondrial membrane potential
The membrane potential Δψ of isolated yeast mitochondria was assessed by measuring the fluorescence quenching of the potential-sensitive dye DiSC3(5) (Molecular Probes, Eugene, OR) as described previously (Geissler et al., 2000). The measurements were performed using a FlexStation plate reader (Molecular Devices) controlled with the SoftMax Pro software package (Molecular Devices) with excitation at 622 nm and emission at 670 nm at 25°C. Mitochondria (21 μg/ml) in buffer (0.6 M sorbitol, 1% BSA, 10 mM MgCl2, 0.5 mM EDTA, 20 mM KPO4 pH 7.4, and 2 mM NADH) were added to the cuvette, followed by DiSC3(5) (final concentration of 167 nM in ethanol) addition, and the fluorescence was measured. Mitochondria were subsequently uncoupled with CCCP (final concentration of 20 μM in ethanol). The difference in the fluorescence before and after the addition of CCCP represents a relative measurement of Δψ.
Supplementary Material
Acknowledgments
We thank Jesmine Cheung, Jonathan Gonzalez, Tanya Hioe, and Justin Hotter for excellent technical assistance and Steven Claypool for critical discussions. This work was supported by grants to C.M.K. from the California Institute of Regenerative Medicine (RS1-00313 and RB1-01397) and the National Institutes of Health (NIH GM073981 and GM61721). S.E.N. was supported in part by a NIH predoctoral fellowship (5F31GM087108) and D.V.D. was supported in part by postdoctoral fellowships from the United Mitochondrial Disease Foundation and the NIH (1F32GM084568).
Abbreviations used:
- Δψ
membrane potential
- AMS
4-acetamido-4′-maleimidylstilbene-2,2′-disulfonic acid
- BSA
bovine serum albumin
- CCCP
carbonyl cyanide m-chlorophenyl hydrazine
- CPY
carboxypeptidase Y
- cyt c
cytochrome c
- DHFR
dihydrofolate reductase
- DiSC3(5)
3,3′-dipropylthiadicarbocyanine iodide
- DMSO
dimethyl sulfoxide
- DTT
dithiothreitol
- ER
endoplasmic reticulum
- FAD
flavin adenine dinucleotide
- GSH
glutathione
- GSSG
oxidized glutathione
- IMS
intermembrane space
- MIA
mitochondrial intermembrane space assembly
- MPP
matrix-processing peptidase
- MQ
menaquinone
- PDI
protein disulfide isomerase
- PreP
presequence peptidase
- Q
ubiquinone
- TCA
trichloroacetic acid
- WT
wild type.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E16-10-0712) on August 16, 2017.
REFERENCES
- Allen S, Balabanidou V, Sideris DP, Lisowsky T, Tokatlidis K. Erv1 mediates the Mia40-dependent protein import pathway and provides a functional link to the respiratory chain by shuttling electrons to cytochrome c. J Mol Biol. 2005;353:937–944. doi: 10.1016/j.jmb.2005.08.049. [DOI] [PubMed] [Google Scholar]
- Arikawa Y, Enomoto K, Muratsubaki H, Okazaki M. Soluble fumarate reductase isoenzymes from Saccharomyces cerevisiae are required for anaerobic growth. FEMS Microbiol Lett. 1998;165:111–116. doi: 10.1111/j.1574-6968.1998.tb13134.x. [DOI] [PubMed] [Google Scholar]
- Bader M, Muse W, Ballou DP, Gassner C, Bardwell JC. Oxidative protein folding is driven by the electron transport system. Cell. 1999;98:217–227. doi: 10.1016/s0092-8674(00)81016-8. [DOI] [PubMed] [Google Scholar]
- Becher D, Kricke J, Stein G, Lisowsky T. A mutant for the yeast scERV1 gene displays a new defect in mitochondrial morphology and distribution. Yeast. 1999;15:1171–1181. doi: 10.1002/(SICI)1097-0061(19990915)15:12<1171::AID-YEA443>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- Besteiro S, Biran M, Biteau N, Coustou V, Baltz T, Canioni P, Bringaud F. Succinate secreted by Trypanosoma brucei is produced by a novel and unique glycosomal enzyme, NADH-dependent fumarate reductase. J Biol Chem. 2002;277:38001–38012. doi: 10.1074/jbc.M201759200. [DOI] [PubMed] [Google Scholar]
- Bien M, Longen S, Wagener N, Chwalla I, Herrmann JM, Riemer J. Mitochondrial disulfide bond formation is driven by intersubunit electron transfer in Erv1 and proofread by glutathione. Mol Cell. 2010;37:516–528. doi: 10.1016/j.molcel.2010.01.017. [DOI] [PubMed] [Google Scholar]
- Bihlmaier K, Mesecke N, Terziyska N, Bien M, Hell K, Herrmann JM. The disulfide relay system of mitochondria is connected to the respiratory chain. J Cell Biol. 2007;179:389–395. doi: 10.1083/jcb.200707123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaisdell FW. The pathophysiology of skeletal muscle ischemia and the reperfusion syndrome: a review. Cardiovasc Surg. 2002;10:620–630. doi: 10.1016/s0967-2109(02)00070-4. [DOI] [PubMed] [Google Scholar]
- Camarasa C, Faucet V, Dequin S. Role in anaerobiosis of the isoenzymes for Saccharomyces cerevisiae fumarate reductase encoded by OSM1 and FRDS1. Yeast. 2007;24:391–401. doi: 10.1002/yea.1467. [DOI] [PubMed] [Google Scholar]
- Chacinska A, Guiard B, Muller JM, Schulze-Specking A, Gabriel K, Kutik S, Pfanner N. Mitochondrial biogenesis, switching the sorting pathway of the intermembrane space receptor Mia40. J Biol Chem. 2008;283:29723–29729. doi: 10.1074/jbc.M805356200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chacinska A, Pfannschmidt S, Wiedemann N, Kozjak V, Sanjuan Szklarz LK, Schulze-Specking A, Truscott KN, Guiard B, Meisinger C, Pfanner N. Essential role of Mia40 in import and assembly of mitochondrial intermembrane space proteins. EMBO J. 2004;23:3735–3746. doi: 10.1038/sj.emboj.7600389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Smeekens JM, Wu R. Comprehensive analysis of protein N-glycosylation sites by combining chemical deglycosylation with LC-MS. J Proteome Res. 2014a;13:1466–1473. doi: 10.1021/pr401000c. [DOI] [PubMed] [Google Scholar]
- Chen W, Smeekens JM, Wu R. A universal chemical enrichment method for mapping the yeast N-glycoproteome by mass spectrometry (MS) Mol Cell Proteomics. 2014b;13:1563–1572. doi: 10.1074/mcp.M113.036251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claros MG, Vincens P. Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur J Biochem. 1996;241:779–786. doi: 10.1111/j.1432-1033.1996.00779.x. [DOI] [PubMed] [Google Scholar]
- Claypool SM, McCaffery JM, Koehler CM. Mitochondrial mislocalization and altered assembly of a cluster of Barth syndrome mutant tafazzins. J Cell Biol. 2006;174:379–390. doi: 10.1083/jcb.200605043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo M, Baryshnikova A, Bellay J, Kim Y, Spear ED, Sevier CS, Ding H, Koh JL, Toufighi K, Mostafavi S, et al. The genetic landscape of a cell. Science. 2010;327:425–431. doi: 10.1126/science.1180823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox AG, Peskin AV, Paton LN, Winterbourn CC, Hampton MB. Correction to redox potential and peroxide reactivity of human peroxiredoxin 3. Biochemistry. 2010;49:6977. doi: 10.1021/bi900558g. [DOI] [PubMed] [Google Scholar]
- Dabir DV, Hasson SA, Setoguchi K, Johnson ME, Wongkongkathep P, Douglas CJ, Zimmerman J, Damoiseaux R, Teitell MA, Koehler CM. A small molecule inhibitor of redox-regulated protein translocation into mitochondria. Dev Cell. 2013;25:81–92. doi: 10.1016/j.devcel.2013.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabir DV, Leverich EP, Kim SK, Tsai FD, Hirasawa M, Knaff DB, Koehler CM. A role for cytochrome c and cytochrome c peroxidase in electron shuttling from Erv1. EMBO J. 2007;26:4801–4811. doi: 10.1038/sj.emboj.7601909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depuydt M, Messens J, Collet JF. How proteins form disulfide bonds. Antioxid Redox Signal. 2011;15:49–66. doi: 10.1089/ars.2010.3575. [DOI] [PubMed] [Google Scholar]
- Dunwoodie SL. The role of hypoxia in development of the mammalian embryo. Dev Cell. 2009;17:755–773. doi: 10.1016/j.devcel.2009.11.008. [DOI] [PubMed] [Google Scholar]
- Dutton RJ, Boyd D, Berkmen M, Beckwith J. Bacterial species exhibit diversity in their mechanisms and capacity for protein disulfide bond formation. Proc Natl Acad Sci USA. 2008;105:11933–11938. doi: 10.1073/pnas.0804621105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enomoto K, Arikawa Y, Muratsubaki H. Physiological role of soluble fumarate reductase in redox balancing during anaerobiosis in Saccharomyces cerevisiae. FEMS Microbiol Lett. 2002;215:103–108. doi: 10.1111/j.1574-6968.2002.tb11377.x. [DOI] [PubMed] [Google Scholar]
- Falkevall A, Alikhani N, Bhushan S, Pavlov PF, Busch K, Johnson KA, Eneqvist T, Tjernberg L, Ankarcrona M, Glaser E. Degradation of the amyloid beta-protein by the novel mitochondrial peptidasome, PreP. J Biol Chem. 2006;281:29096–29104. doi: 10.1074/jbc.M602532200. [DOI] [PubMed] [Google Scholar]
- Frand AR, Kaiser CA. The ERO1 gene of yeast is required for oxidation of protein dithiols in the endoplasmic reticulum. Mol Cell. 1998;1:161–170. doi: 10.1016/s1097-2765(00)80017-9. [DOI] [PubMed] [Google Scholar]
- Fujiki Y, Hubbard AL, Fowler S, Lazarow PB. Isolation of intracellular membranes by means of sodium carbonate treatment: application to endoplasmic reticulum. J Cell Biol. 1982;93:97–102. doi: 10.1083/jcb.93.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukasawa Y, Tsuji J, Fu SC, Tomii K, Horton P, Imai K. MitoFates: improved prediction of mitochondrial targeting sequences and their cleavage sites. Mol Cell Proteomics. 2015;14:1113–1126. doi: 10.1074/mcp.M114.043083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel K, Milenkovic D, Chacinska A, Muller J, Guiard B, Pfanner N, Meisinger C. Novel mitochondrial intermembrane space proteins as substrates of the MIA import pathway. J Mol Biol. 2007;365:612–620. doi: 10.1016/j.jmb.2006.10.038. [DOI] [PubMed] [Google Scholar]
- Geissler A, Krimmer T, Bomer U, Guiard B, Rassow J, Pfanner N. Membrane potential-driven protein import into mitochondria. The sorting sequence of cytochrome b2 modulates the Δψ-dependence of translocation of the matrix-targeting sequence. Mol Biol Cell. 2000;11:3977–3991. doi: 10.1091/mbc.11.11.3977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber J, Muhlenhoff U, Hofhaus G, Lill R, Lisowsky T. Yeast ERV2p is the first microsomal FAD-linked sulfhydryl oxidase of the Erv1p/Alrp protein family. J Biol Chem. 2001;276:23486–23491. doi: 10.1074/jbc.M100134200. [DOI] [PubMed] [Google Scholar]
- Glick BS, Brandt A, Cunningham K, Muller S, Hallberg RL, Schatz G. Cytochromes c1 and b2 are sorted to the intermembrane space of yeast mitochondria by a stop-transfer mechanism. Cell. 1992;69:809–822. doi: 10.1016/0092-8674(92)90292-k. [DOI] [PubMed] [Google Scholar]
- Glick BS, Pon LA. Isolation of highly purified mitochondria from Saccharomyces cerevisiae. Methods Enzymol. 1995;260:213–223. doi: 10.1016/0076-6879(95)60139-2. [DOI] [PubMed] [Google Scholar]
- Gross E, Sevier CS, Heldman N, Vitu E, Bentzur M, Kaiser CA, Thorpe C, Fass D. Generating disulfides enzymatically: reaction products and electron acceptors of the endoplasmic reticulum thiol oxidase Ero1p. Proc Natl Acad Sci USA. 2006;103:299–304. doi: 10.1073/pnas.0506448103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grumbt B, Stroobant V, Terziyska N, Israel L, Hell K. Functional characterization of Mia40p, the central component of the disulfide relay system of the mitochondrial intermembrane space. J Biol Chem. 2007;282:37461–37470. doi: 10.1074/jbc.M707439200. [DOI] [PubMed] [Google Scholar]
- Gueldener U, Heinisch J, Koehler GJ, Voss D, Hegemann JH. A second set of loxP marker cassettes for Cre-mediated multiple gene knockouts in budding yeast. Nucleic Acids Res. 2002;30:e23. doi: 10.1093/nar/30.6.e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie C, Fink GR. Guide to yeast genetics and molecular biology. Methods Enzymol. 1991;194:1–863. [PubMed] [Google Scholar]
- Hasson SA, Damoiseaux R, Glavin JD, Dabir DV, Walker SS, Koehler CM. Substrate specificity of the TIM22 mitochondrial import pathway revealed with small molecule inhibitor of protein translocation. Proc Natl Acad Sci USA. 2010;107:9578–9583. doi: 10.1073/pnas.0914387107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann JM, Riemer J. Mitochondrial disulfide relay: redox-regulated protein import into the intermembrane space. J Biol Chem. 2012;287:4426–4433. doi: 10.1074/jbc.R111.270678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton P, Park KJ, Obayashi T, Fujita N, Harada H, Adams-Collier CJ, Nakai K. WoLF PSORT: protein localization predictor. Nucleic Acids Res. 2007;35:W585–W587. doi: 10.1093/nar/gkm259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang DK, Claypool SM, Leuenberger D, Tienson HL, Koehler CM. Tim54p connects inner membrane assembly and proteolytic pathways in the mitochondrion. J Cell Biol. 2007;178:1161–1175. doi: 10.1083/jcb.200706195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karniely S, Pines O. Single translation–dual destination: mechanisms of dual protein targeting in eukaryotes. EMBO Rep. 2005;6:420–425. doi: 10.1038/sj.embor.7400394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzen F, Beckwith J. Transmembrane electron transfer by the membrane protein DsbD occurs via a disulfide bond cascade. Cell. 2000;103:769–779. doi: 10.1016/s0092-8674(00)00180-x. [DOI] [PubMed] [Google Scholar]
- Koehler CM, Tienson HL. Redox regulation of protein folding in the mitochondrial intermembrane space. Biochim Biophys Acta. 2009;1793:139–145. doi: 10.1016/j.bbamcr.2008.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Osterlund T, Hou J, Petranovic D, Nielsen J. Anaerobic alpha-amylase production and secretion with fumarate as the final electron acceptor in Saccharomyces cerevisiae. Appl Environ Microbiol. 2013;79:2962–2967. doi: 10.1128/AEM.03207-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longen S, Bien M, Bihlmaier K, Kloeppel C, Kauff F, Hammermeister M, Westermann B, Herrmann JM, Riemer J. Systematic analysis of the twin cx(9)c protein family. J Mol Biol. 2009;393:356–368. doi: 10.1016/j.jmb.2009.08.041. [DOI] [PubMed] [Google Scholar]
- Mairet-Coello G, Tury A, Esnard-Feve A, Fellmann D, Risold PY, Griffond B. FAD-linked sulfhydryl oxidase QSOX: topographic, cellular, and subcellular immunolocalization in adult rat central nervous system. J Comp Neurol. 2004;473:334–363. doi: 10.1002/cne.20126. [DOI] [PubMed] [Google Scholar]
- Mamathambika BS, Bardwell JC. Disulfide-linked protein folding pathways. Annu Rev Cell Dev Biol. 2008;24:211–235. doi: 10.1146/annurev.cellbio.24.110707.175333. [DOI] [PubMed] [Google Scholar]
- Merkley ED, Metz TO, Smith RD, Baynes JW, Frizzell N. The succinated proteome. Mass Spectrom Rev. 2014;33:98–109. doi: 10.1002/mas.21382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesecke N, Terziyska N, Kozany C, Baumann F, Neupert W, Hell K, Herrmann JM. A disulfide relay system in the intermembrane space of mitochondria that mediates protein import. Cell. 2005;121:1059–1069. doi: 10.1016/j.cell.2005.04.011. [DOI] [PubMed] [Google Scholar]
- Messens J, Collet JF, Van Belle K, Brosens E, Loris R, Wyns L. The oxidase DsbA folds a protein with a nonconsecutive disulfide. J Biol Chem. 2007;282:31302–31307. doi: 10.1074/jbc.M705236200. [DOI] [PubMed] [Google Scholar]
- Muratsubaki H, Enomoto K. One of the fumarate reductase isoenzymes from Saccharomyces cerevisiae is encoded by the OSM1 gene. Arch Biochem Biophys. 1998;352:175–181. doi: 10.1006/abbi.1998.0583. [DOI] [PubMed] [Google Scholar]
- Naoe M, Ohwa Y, Ishikawa D, Ohshima C, Nishikawa SI, Yamamoto H, Endo T. Identification of Tim40 that mediates protein sorting to the mitochondrial intermembrane space. J Biol Chem. 2004;279:47815–47821. doi: 10.1074/jbc.M410272200. [DOI] [PubMed] [Google Scholar]
- Neal SE, Dabir DV, Tienson HL, Horn DM, Glaeser K, Ogozalek Loo RR, Barrientos A, Koehler CM. Mia40 protein serves as an electron sink in the Mia40-Erv1 import pathway. J Biol Chem. 2015;290:20804–20814. doi: 10.1074/jbc.M115.669440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsu I, Wiriyathanawudhiwong N, Morigasaki S, Nakatani T, Kadokura H, Takagi H. The l-cysteine/l-cystine shuttle system provides reducing equivalents to the periplasm in Escherichia coli. J Biol Chem. 2010;285:17479–17487. doi: 10.1074/jbc.M109.081356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omura T. Mitochondria-targeting sequence, a multi-role sorting sequence recognized at all steps of protein import into mitochondria. J Biochem. 1998;123:1010–1016. doi: 10.1093/oxfordjournals.jbchem.a022036. [DOI] [PubMed] [Google Scholar]
- Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 2011;8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- Rainey RN, Glavin JD, Chen HW, French SW, Teitell MA, Koehler CM. A new function in translocation for the mitochondrial i-AAA protease Yme1: import of polynucleotide phosphorylase into the intermembrane space. Mol Cell Biol. 2006;26:8488–8497. doi: 10.1128/MCB.01006-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rietsch A, Belin D, Martin N, Beckwith J. An in vivo pathway for disulfide bond isomerization in Escherichia coli. Proc Natl Acad Sci USA. 1996;93:13048–13053. doi: 10.1073/pnas.93.23.13048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rospert S, Looser R, Dubaquie Y, Matouschek A, Glick BS, Schatz G. Hsp60-independent protein folding in the matrix of yeast mitochondria. EMBO J. 1996;15:764–774. [PMC free article] [PubMed] [Google Scholar]
- Sanchez NS, Pearce DA, Cardillo TS, Uribe S, Sherman F. Requirements of Cyc2p and the porin, Por1p, for ionic stability and mitochondrial integrity in Saccharomyces cerevisiae. Arch Biochem Biophys. 2001;392:326–332. doi: 10.1006/abbi.2001.2465. [DOI] [PubMed] [Google Scholar]
- Sevier CS. Erv2 and quiescin sulfhydryl oxidases: erv-domain enzymes associated with the secretory pathway. Antioxid Redox Signal. 2012;16:800–808. doi: 10.1089/ars.2011.4450. [DOI] [PubMed] [Google Scholar]
- Sevier CS, Cuozzo JW, Vala A, Aslund F, Kaiser CA. A flavoprotein oxidase defines a new endoplasmic reticulum pathway for biosynthetic disulphide bond formation. Nat Cell Biol. 2001;3:874–882. doi: 10.1038/ncb1001-874. [DOI] [PubMed] [Google Scholar]
- Sevier CS, Kaiser CA. Conservation and diversity of the cellular disulfide bond formation pathways. Antioxid Redox Signal. 2006;8:797–811. doi: 10.1089/ars.2006.8.797. [DOI] [PubMed] [Google Scholar]
- Sevier CS, Qu HJ, Heldman N, Gross E, Fass D, Kaiser CA. Modulation of cellular disulfide-bond formation and the ER redox environment by feedback-regulation of Ero1. Cell. 2007;129:333–344. doi: 10.1016/j.cell.2007.02.039. [DOI] [PubMed] [Google Scholar]
- Sztolsztener ME, Brewinska A, Guiard B, Chacinska A. Disulfide bond formation: sulfhydryl oxidase ALR controls mitochondrial biogenesis of human MIA40. Traffic. 2013;14:309–320. doi: 10.1111/tra.12030. [DOI] [PubMed] [Google Scholar]
- Takahashi YH, Inaba K, Ito K. Characterization of the menaquinone-dependent disulfide bond formation pathway of Escherichia coli. J Biol Chem. 2004;279:47057–47065. doi: 10.1074/jbc.M407153200. [DOI] [PubMed] [Google Scholar]
- Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- Tienson HL, Dabir DV, Neal SE, Loo R, Hasson SA, Boontheung P, Kim SK, Loo JA, Koehler CM. Reconstitution of the Mia40-Erv1 oxidative folding pathway for the small Tim proteins. Mol Biol Cell. 2009;20:3481–3490. doi: 10.1091/mbc.E08-10-1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu BP, Ho-Schleyer SC, Travers KJ, Weissman JS. Biochemical basis of oxidative protein folding in the endoplasmic reticulum. Science. 2000;290:1571–1574. doi: 10.1126/science.290.5496.1571. [DOI] [PubMed] [Google Scholar]
- Tu BP, Weissman JS. Oxidative protein folding in eukaryotes: mechanisms and consequences. J Cell Biol. 2004;164:341–346. doi: 10.1083/jcb.200311055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wajih N, Hutson SM, Wallin R. Disulfide-dependent protein folding is linked to operation of the vitamin K cycle in the endoplasmic reticulum. A protein disulfide isomerase-VKORC1 redox enzyme complex appears to be responsible for vitamin K1 2,3-epoxide reduction. J Biol Chem. 2007;282:2626–2635. doi: 10.1074/jbc.M608954200. [DOI] [PubMed] [Google Scholar]
- Williams CC, Jan CH, Weissman JS. Targeting and plasticity of mitochondrial proteins revealed by proximity-specific ribosome profiling. Science. 2014;346:748–751. doi: 10.1126/science.1257522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrobel L, Trojanowska A, Sztolsztener ME, Chacinska A. Mitochondrial protein import: Mia40 facilitates Tim22 translocation into the inner membrane of mitochondria. Mol Biol Cell. 2013;24:543–554. doi: 10.1091/mbc.E12-09-0649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielinska DF, Gnad F, Schropp K, Wisniewski JR, Mann M. Mapping N-glycosylation sites across seven evolutionarily distant species reveals a divergent substrate proteome despite a common core machinery. Mol Cell. 2012;46:542–548. doi: 10.1016/j.molcel.2012.04.031. [DOI] [PubMed] [Google Scholar]
- Zito E, Melo EP, Yang Y, Wahlander A, Neubert TA, Ron D. Oxidative protein folding by an endoplasmic reticulum-localized peroxiredoxin. Mol Cell. 2010;40:787–797. doi: 10.1016/j.molcel.2010.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.