Summary
DNA structuring proteins such as CTCF facilitate DNA loop formation and are presumed to be among the major determinants of eukaryotic genome structure. Recent studies, including Rowley et al (this issue), suggest that gene activation and repression play fundamentally important roles in structuring the genome independently of CTCF.
Main text
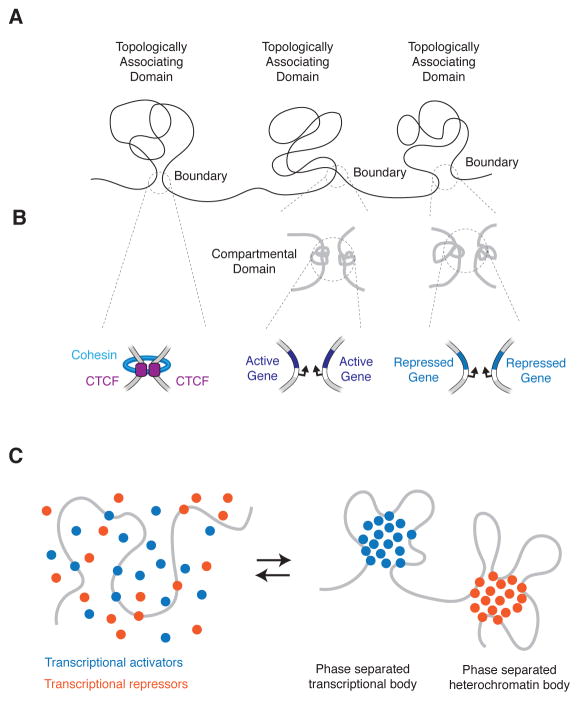
Proper structuring of the genome is essential for chromosome maintenance and gene regulatory activity. Key features of eukaryotic chromosome structure have been deduced in the last few years, but the mechanisms that contribute to specific chromosome structures are not yet well understood. Chromosomes are folded into topologically associating domains (TADs), which in mammals are Mb sized regions of DNA that interact more frequently with each other than with other regions of the genome (Figure 1A) (Dixon et al., 2012; Nora et al., 2012; Sexton et al., 2012). TAD boundaries are thought to be anchored by interactions between DNA binding, chromosome-structuring proteins such as CTCF. In this issue of Molecular Cell, Rowley et al provide evidence that genes in a transcriptionally active or repressed state can form a small folding unit they call a compartmental domain, and these domains can create TAD boundaries by physically interacting with other genes in the same transcriptional state in the absence of CTCF.
Figure 1. Mechanisms structuring the genome.
A) Topologically Associating Domains (TADs).
B) (Top) High resolution Hi-C in Rowley et al reveals small compartmental domains at the boundaries of some TADs in Drosophila. (Bottom) Molecular underpinnings of TAD boundaries: cohesin associated CTCF-CTCF loops, and gene-gene interactions.
C) Model of genome structuring through phase separation of transcriptional activators and repressors.
There is considerable evidence that the highly conserved zinc-finger protein CTCF, which has been implicated in diverse regulatory functions, including gene activation, repression, insulation, parental imprinting, and X chromosome inactivation (Gomez-Diaz and Corces, 2014), has a major role to play at TAD boundaries. CTCF often occupies TAD boundaries, can form homodimers, and together with the Structural Maintenance of Chromosomes (SMC) cohesin complex, can create DNA loops between boundaries (Figure 1B left). Degradation of CTCF can perturb TAD boundaries (Nora et al., 2017). These CTCF-mediated DNA loops play key roles in specific gene control because they can act as “insulators” by constraining enhancer-gene interactions to occur within the CTCF-CTCF loop. Indeed, cancer cells can acquire somatic mutations that alter CTCF binding sites such that proto-oncogenes are no longer insulated from enhancers located outside CTCF-CTCF loops (Hnisz et al., 2016).
Despite this CTCF-centric picture of chromosome structuring, several lines of evidence suggest there are additional mechanisms that contribute to the boundary structures observed in chromosomes. Early studies of DNA interactions suggested that some structural features of chromosomes are linked to transcriptional activity and repression (Dixon et al., 2012; Lieberman-Aiden et al., 2009; Nora et al., 2012; Sexton et al., 2012), and the domains of active and repressed genes tend to be unaffected by CTCF depletion (Nora et al., 2017). SMC complexes are loaded at sites of transcription initiation independently of CTCF (Kagey et al., 2010). And importantly, some eukaryotic species lack an apparent CTCF homologue, yet form TAD-like structures (Rowley et al).
An understanding of the roles of transcription in genome structure has been limited in part by the resolution of the DNA interaction data used to investigate genome structure. Rowley et al, overcame this by generating a high-resolution map of genome structure in Drosophila cells, and found that the boundaries of many TADs are formed by smaller domains, which they termed compartmental domains (Figure 1B, middle and right). A compartmental domain is comprised of one or more genes with either a transcriptionally active or repressed state. Compartmental domains can create a TAD boundary by physically interacting with other genes that share the same transcriptional state in the absence of CTCF(Figure 1B, middle and right). Based on these insights, the authors developed a computational model of genome structure, and found that the transcriptional status of genes, or the chromatin signatures associated with gene activity or repression, are good predictors of overall genome structure both in Drosophila and several other species. The authors propose that gene regulatory activity is a major determinant of genome structure, an attractive model that was further supported by the finding that the computational approach predicted overall genome structure in species such as Arabidopsis that appear to lack the architectural protein CTCF.
An intriguing question arises from the findings of Rowley et al: What is the nature of the forces that drive physical interactions between genes in the same transcriptional state? Recent work at the intersection of cell biology, biochemistry and soft matter physics provides clues to the nature of these forces.
Compartmentalization of biochemical reactions within cells can occur in membraneless bodies that form as a result of liquid-liquid phase separation (LLPS) driven by multivalent macromolecular interactions (Banani et al., 2017). Transcriptional control is likely to involve phase separation events facilitated by transcriptional activators and repressors (Hnisz et al., 2017; Larson et al., 2017). The activation domains of transcriptional activators, and portions of many other proteins involved in transcriptional control, have intrinsically disordered regions characteristic of proteins that participate in LLPS bodies. Thus, the interactions detected between compartmental domains by Rowley at al., and the evidence for enrichment of highly expressed housekeeping genes at a fraction of TAD boundaries in mammals (Dixon et al., 2012), are consistent with a model where LLPS contributes to both transcriptional control and chromosome structuring (Figure 1C). The study of biomolecular condensates is in its infancy, especially in the fields of gene control and chromosome maintenance, so there is still much to learn.
References
- Banani SF, Lee HO, Hyman AA, Rosen MK. Biomolecular condensates: organizers of cellular biochemistry. Nature reviews Molecular cell biology. 2017 doi: 10.1038/nrm.2017.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Diaz E, Corces VG. Architectural proteins: regulators of 3D genome organization in cell fate. Trends in cell biology. 2014;24:703–711. doi: 10.1016/j.tcb.2014.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnisz D, Shrinivas K, Young RA, Chakraborty AK, Sharp PA. A Phase Separation Model for Transcriptional Control. Cell. 2017;169:13–23. doi: 10.1016/j.cell.2017.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnisz D, Weintraub AS, Day DS, Valton AL, Bak RO, Li CH, Goldmann J, Lajoie BR, Fan ZP, Sigova AA, et al. Activation of proto-oncogenes by disruption of chromosome neighborhoods. Science. 2016;351:1454–1458. doi: 10.1126/science.aad9024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467:430–435. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson AG, Elnatan D, Keenen MM, Trnka MJ, Johnston JB, Burlingame AL, Agard DA, Redding S, Narlikar GJ. Liquid droplet formation by HP1alpha suggests a role for phase separation in heterochromatin. Nature. 2017;547:236–240. doi: 10.1038/nature22822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nora EP, Goloborodko A, Valton AL, Gibcus JH, Uebersohn A, Abdennur N, Dekker J, Mirny LA, Bruneau BG. Targeted Degradation of CTCF Decouples Local Insulation of Chromosome Domains from Genomic Compartmentalization. Cell. 2017;169:930–944. e922. doi: 10.1016/j.cell.2017.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nora EP, Lajoie BR, Schulz EG, Giorgetti L, Okamoto I, Servant N, Piolot T, van Berkum NL, Meisig J, Sedat J, et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature. 2012;485:381–385. doi: 10.1038/nature11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sexton T, Yaffe E, Kenigsberg E, Bantignies F, Leblanc B, Hoichman M, Parrinello H, Tanay A, Cavalli G. Three-dimensional folding and functional organization principles of the Drosophila genome. Cell. 2012;148:458–472. doi: 10.1016/j.cell.2012.01.010. [DOI] [PubMed] [Google Scholar]