Abstract
Patients with asthma experience circadian variations in their symptoms. However it remains unclear how specific aspects of this common airway disease relate to clock genes, which are critical to the generation of circadian rhythms in mammals. Here, we used a viral model of acute and chronic airway disease to examine how circadian clock disruption affects asthmatic lung phenotypes. Deletion of the core clock gene bmal1 or environmental disruption of circadian function by jet-lag exacerbated acute viral bronchiolitis caused by Sendai virus (SeV) and influenza A virus (IAV) in mice. Post-natal deletion of bmal1 was sufficient to trigger increased SeV susceptibility and correlated with impaired control of viral replication. Importantly, bmal1−/− mice developed much more extensive asthma-like airway changes post-infection, including mucus production and increased airway resistance. In human airway samples from two asthma cohorts, we observed altered expression patterns of multiple clock genes. Our results suggest a role for bmal1 in the development of asthmatic airway disease via the regulation of lung antiviral responses to common viral triggers of asthma.
INTRODUCTION
Asthma is a complex lung disease whose symptoms are driven by airway remodeling, consisting of inflammation, sub-epithelial fibrosis, smooth muscle proliferation, and goblet cell metaplasia1. Although the pathogenesis of asthma is multifactorial, a key mechanism implicated in its development involves sensitivity to common respiratory viruses. Viruses can be isolated from the airways of 80% of asthmatic children and 60% of asthmatic adults when they present with exacerbations (versus a 30% carriage rate in healthy children and <10% in healthy adults)2. Once viral respiratory infection occurs in asthmatics, their lower respiratory tract symptoms are more severe and of longer duration than those of healthy subjects3. Viral bronchiolitis in infancy is statistically associated with the development of asthma later in childhood, with the number of infections rather than the identity of the virus being the decisive factor4. Interestingly, aberrant interferon secretion has been observed in response to viral infection in asthmatic airway cells5. Based on this, some investigators theorize that antiviral responses in asthma patients may be inherently abnormal and may help to drive disease activity6. However, there is little information about specific antiviral mechanisms that might pertain to asthma development.
An additional important characteristic of asthma is that patients exhibit circadian rhythms in their symptoms7. Circadian rhythms are ubiquitous, daily oscillations in biological function that evolved to anticipate environmental changes brought about by the day-night cycle8. In mammals, circadian rhythms are generated by a group of transcription factors and regulatory proteins collectively called the “circadian clock”8. The core transcription factor is a heterodimer composed of the proteins Bmal1 and Clock, which transactivates genes containing “E-Box” motifs in their promoters8. Among the genes transactivated by Bmal1/Clock are fellow “clock genes” that provide it with negative and positive feedback regulation8. As a unit, the circadian clock regulates the cyclic expression of thousands of genes, proteins, and metabolites organism-wide8. Importantly, lung parenchymal cells express clock genes and harbor a functioning circadian clock9, 10. Data is now emerging that clock genes may contribute to lung inflammation, fibrosis, glucocorticoid responses, and immunity11–13. However, clock gene function has yet to be studied in the context of asthmatic airway disease.
The objective of this study was to examine the role of circadian clock function on respiratory virus susceptibility and on asthma-like features of post-viral chronic airway disease. Our results indicate a role for the circadian clock gene bmal1 in controlling acute and chronic viral airway pathology, thereby linking the circadian clock to features of asthmatic lung remodeling through the regulation of respiratory viral infection.
RESULTS
Deletion of bmal1 worsens acute viral bronchiolitis
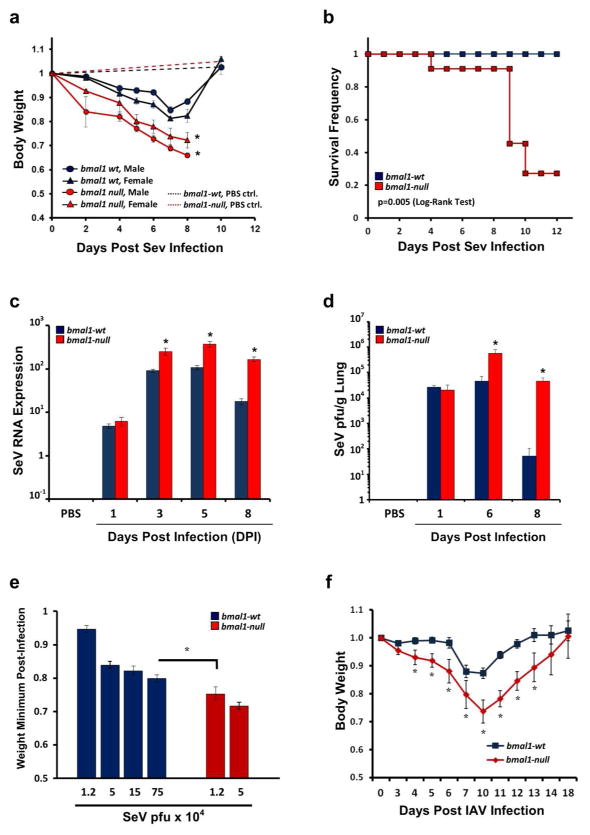
In mice, interactions between viruses and asthmatic airway phenotypes can be studied using Sendai virus (SeV). SeV is a parainfluenza virus native to rodents that causes acute bronchiolitis followed by chronic airway changes reminiscent of asthma, including mucous cell metaplasia, inflammation, and airway hypersensitivity to methacholine14. We challenged bmal1−/− mice, which lack a functional circadian clock, with SeV and compared their responses to wt littermates. When challenged with normally sub-lethal doses of SeV, bmal1−/− mice exhibited more severe infection as gauged by increased weight loss, mortality, viral RNA expression, and viral load (Fig. 1a–d). We observed similar effects at lower SeV doses that did not produce mortality in bmal1−/− mice (Supplementary Figure 1a). The increased SeV RNA expression in bmal1−/− mice localized to the lungs rather than the tracheas of infected mice (Supplementary Figure 1b). Both male and female bmal1−/− mice were similarly vulnerable to SeV (Fig. 1a). By adjusting the dose of SeV in order to produce similar weight loss in bmal1−/− and wt mice, we estimated that bmal1−/− mice are more than 60-fold more sensitive to this virus (Fig. 1e). Bmal1−/− mice were also more vulnerable to influenza A virus (IAV) as indicated by more severe weight loss compared to wt animals, an observation also recently noted by other groups15, 16 (Fig. 1f). To investigate whether our results were specific to bmal1 or were generally reflective of circadian clock gene deletion we challenged per2−/− mice with SeV. Although deletion of per2 is not sufficient to disable circadian clock function, it is a negative regulator of Bmal1 and was previously implicated in the pathogenesis of HCV17. Deletion of per2 did not significantly affect SeV severity as measured by viral RNA levels (Supplementary Fig. 2).
Figure 1.
Deletion of clock gene bmal1 renders mice more vulnerable to viral bronchiolitis. (a) Animal weights (normalized to starting weight) after infection with 1.5x105 pfu SeV. Each point represents the mean weight ± SE. Blue circles: SeV-infected bmal1-wt males (n=19). Blue triangles: SeV-infected bmal1-wt females (n=12). Red circles: SeV-infected bmal1−/− males (n=12). Red triangles: SeV-infected bmal1−/− females (n=12). Blue dashed line: Sham (PBS inoculated) bmal1-wt males (n=5). Red dashed line: Sham (PBS inoculated) bmal1−/− males (n=4). SeV infection data were pooled from 4 independent experiments. (b) Survival after infection with 1.5x105 pfu SeV. Blue symbols: bmal1-wt (n=15). Red symbols: bmal1−/− littermates (n=11). Data is pooled from 2 independent experiments, and each group is composed of near-equal proportions of male and female animals. Note that sham (PBS) inoculation did not produce animal mortality in either genotype (data not shown). Statistical significance by Log-Rank Test is depicted. (c) Viral RNA expression as measured by qPCR at various times after intranasal infection with SeV (1.5x105 pfu). Each bar represents the mean SeV gene expression normalized to tbp ± SE. Blue bars: bmal1-wt (n=4–12 per time point). Red bars: bmal1−/− littermates (n=3–11 per time point). Each group is composed of equal or near-equal proportions of male and female animals, and was pooled from 2 independent rounds of infection. (d) Lung viral load measured by plaque assay at various points after SeV infection (5.0x104 pfu). Each bar represents the mean SeV plaque forming units/g of lung ± SE. Blue bars: bmal1-wt (n=5–6 per time point). Red bars: bmal1−/− littermates (n=4–6 per time point). (e) Comparison of SeV potency in bmal1−/− and wt littermate mice using weight loss as an indicator of severity. Each bar represents the weight minimum (normalized to starting weight) achieved using various doses of SeV (mean ± SE). Blue bars: bmal1-wt (n=5–21 per time point). Red bars: bmal1−/− littermates (n=8–9 per time point). Data were pooled from 4 independent experiments utilizing equal or near equal proportions of male and female mice. Note that sham (PBS) inoculated animals did not exhibit appreciable weight loss (data not shown). (f) Animal weights (normalized to starting weight) after infection with 5 pfu H1N1 IAV. Each point represents the mean weight ± SE. Blue squares: bmal1-wt (n=6). Red diamonds: bmal1−/− littermates (n=8). Equal proportions of male and female animals were used in each group. At the dose of virus tested, we did not encounter mortality in IAV infected bmal1−/− or wt animals during the acute infection. *p<0.05 bmal1−/− vs. wt littermates (Student’s 2-Tailed t-test).
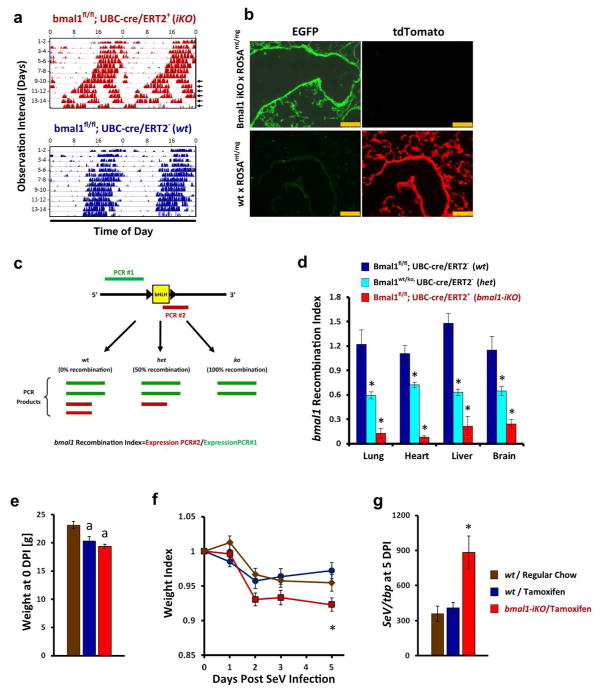
Deletion of bmal1 produces several maladaptive phenotypes in mice, including small stature, accelerated aging, and cardiovascular disease18. We therefore asked whether the viral susceptibility observed in bmal1−/− mice is a product of circadian clock disruption or whether it represents an unrelated effect of deleting this gene. Recently, Yang et al.18 demonstrated one approach to addressing this question by generating tamoxifen-inducible bmal1 knockout (bmal1-iKO) mice, which enabled global deletion of this gene in post-natal life. Yang et al.18 found that post-natal deletion of bmal1 was sufficient to disable circadian clock function without inducing many deleterious phenotypes previously ascribed to this gene, such as short stature, atherosclerosis, and accelerated aging. To determine the impact of post-natal bmal1 deletion in SeV infection, we independently developed a tamoxifen-inducible line of bmal1 conditional knockout mice similar to Yang et al.18 (Fig. 2). In our line of bmal1-iKO mice, cre expression is driven by a tamoxifen inducible ubc promoter rather than chicken b-actin. When fed tamoxifen-containing chow bmal1-iKO mice, but not wt littermates, durably lost circadian patterns of wheel running activity in constant darkness after approximately 10 days of tamoxifen treatment (Fig. 2a). After 3 weeks of tamoxifen chow, we observed Cre recombination activity throughout the lung parenchyma, as detected by crossing bmal1-iKO mice with ROSAmT/mG reporter mice (Fig. 2b). To directly quantify the efficiency of bmal1 deletion in iKO mice, we used qPCR to estimate the relative expression of exon 8, which encodes the bHLH domain of the protein and which is deleted during Cre-induced recombination19 (Fig. 2c). Using this assay we observed that bmal1 recombination efficiency was approximately 75%–90% across multiple organs in our iKO mice (Fig. 2d). As expected from prior literature20, tamoxifen feeding itself led to decreases in body weight in bmal1-iKO mice and wt littermates, although weight loss was similar between genotypes (Fig. 2e). When challenged with SeV, tamoxifen-fed bmal1-iKO mice lost proportionately more weight than wt littermates fed regular or tamoxifen chow (Fig. 2f). Tamoxifen-fed bmal1-iKO mice also had increased SeV RNA levels at 5 DPI compared to wt (Fig. 2g), mirroring our prior results where bmal1 deletion occurred in utero (Fig. 1c). To further validate these results, we independently generated a second line of bmal1-iKO mice where cre expression is driven by the chicken β-actin promoter, similar to Yang et al18. In these mice too, post-natal deletion of bmal1 led to increased SeV virus expression at 5 DPI compared to wt littermates (Supplementary Fig. 3). Taken together, we found that post-natal bmal1 deletion is sufficient to render mice susceptible to acute respiratory viral illness. The virus susceptibility of bmal1 mice therefore is one of the few bmal1 phenotypes that can be recapitulated through post-natal deletion, and suggests our results are not secondary to aberrant embryonic development.
Figure 2.
Post-natal deletion of bmal1 is sufficient to exacerbate acute SeV respiratory infection. (a) Representative actigraphy depicting wheel running behavior of bmal1-iKO (bmal1fl/fl, UBC-cre/ERT2+) and non-iKO (bmal1fl/fl, UBC-cre/ERT2+) littermates, kept in constant darkness and fed tamoxifen-containing chow starting on the first day of recording. Bars represent wheel revolutions per minute and each row represents a 2 day interval of observation. Note that circadian rhythms in wheel running activity begin to break down in bmal1-iKO after 9–10 days of tamoxifen exposure (arrowheads). (b) Representative micrographs depicting cre recombination efficiency in the lungs of bmal1-iKO mice and wt mice. Bmal1-iKO and wt littermate mice were crossed with ROSAmT/mg reporter line, fed tamoxifen for 3 weeks (see Methods), and then frozen lung sections were prepared. Cre recombination efficiency is indicated by the development of EGFP (green) fluorescence and the reciprocal loss of mtTomato (red) fluorescence in cells. Scale bar=250 μm. (c) Schematic of qPCR based assay to quantify bmal1 recombination efficiency in bmal1-iKO mice. The genomic locus of bmal1 for the B6.129S4(Cg)-Arntltm1Weit/J mouse line used in this study is depicted in simplified form (black line) to illustrate the loxP sites flanking exon 8, which encodes the bHLH DNA binding domain. PCR product #1 (green) extends from exon 5–8, terminating upstream of the floxed bHLH coding sequence and therefore its production is insensitive to Cre activity. In contrast, PCR product #2 (red) initiates within the floxed bHLH coding sequence and therefore is not amplified in mRNA templates where Cre recombination has occurred. The efficiency of bmal1 recombination can therefore be expressed as the ratio of bmal1 expression detected by PCR product #2/PCR product #1. (d) Quantification of bmal1 recombination in iKO mice using qPCR. Bars represent mean bmal1 recombination index (see Methods and panel c) ±SE. Dark blue bars, bmal1-wt mice fed tamoxifen chow (no recombination, 100% bmal1gene dosage; n=6). Light-blue bars, global bmal1 heterozygous mice containing a ko/wt, genotype (i.e. 50% gene dosage; n=4). Red bars, bmal1-iKO mice fed tamoxifen chow (n=7). Note that global bmal1-null mice (where recombination occurs in utero) generate a recombination index of 0 in this assay (data not shown). *p<0.05 vs. wt littermates fed tamoxifen chow (Student’s 2-Tailed t-test). (e) Weight of bmal1-iKO and wt mice after 3 weeks of tamoxifen feeding. Bars represent mean weight ±SE. Brown bars: wt mice fed regular chow. Dark blue bars, wt mice fed tamoxifen chow. Red bars: bmal-iKO mice fed tamoxifen chow. ap<0.05 vs. wt littermates fed regular chow (Student’s 2-Tailed t-test). Data is pooled from 2 independent experiments (n=6–10 per group, equal proportions of male and female mice). (f) Animal weights (normalized to starting weight) after infection with 5x104 pfu SeV. Each point represents the mean ± SE. Bar color designations and sample sizes are as described in panel e above. Data is pooled from 2 independent experiments. *p<0.05 bmal1−/− vs. wt littermates fed tamoxifen chow (Student’s 2-Tailed t-test). (g) SeV RNA expression as measured by qPCR 5 days after intranasal infection with SeV (5x104 pfu). Each bar represents the mean SeV gene expression normalized to tbp ± SE. Bar color designations and sample sizes are as described in panel e above. Data is pooled from 2 independent experiments. *p<0.05 bmal1−/− vs. wt littermates fed tamoxifen chow (Student’s 2-Tailed t-test).
Effect of clock gene bmal1 deletion on SeV-induced acute airway inflammation
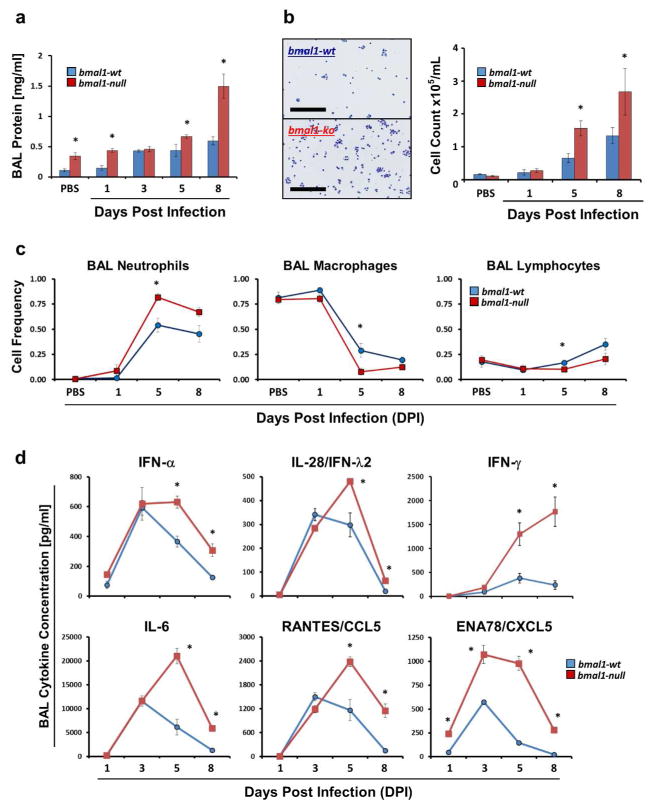
We next compared airway inflammation in SeV-infected bmal1-wt and bmal1−/− mice. Compared to wt, bmal1−/− mice developed more extensive airway inflammation, as evidenced by bronchio-alveolar lavage (BAL) protein concentration and leukocyte counts (Figs. 3a and b). As infection progressed, both genotypes exhibited a similar transition from a macrophage-predominant airway infiltrate to a neutrophil-predominant one, however there was a greater percentage of neutrophils in the BALs of bmal1−/− animals at 5 DPI (Fig. 3c). Peri-bronchial accumulation of macrophages and caspase 3+ cells was similar in bmal1−/− and wt animals, as was the pattern of trafficking for major leukocyte subsets into and out of SeV-infected lungs as indicated by flow cytometry (Supplementary Fig. 4). Overall, bmal1 deletion increased the magnitude of granulocyte-predominant lung inflammation during SeV infection.
Figure 3.
Airway inflammation during acute viral infection is greater in bmal1−/− mice than wt. (a) Bronchio-alveolar lavage (BAL) protein content in mg/ml (mean ± SE). Blue bars: bmal1-wt (n=4–14 per time point). Red bars: bmal1−/− (n=4–6 per time point). Equal or near-equal proportions of male and female animals were included in each group. (b) Representative images of BAL cytospins of bmal1-wt and bmal1−/− mice during acute infection and cell counts at 1, 5, and 8 days after infection with SeV (5x104 pfu). Each bar represents the mean number of cells/mL of BAL fluid ± SE. Blue bars: bmal1-wt (n=4–14 per time point). Red bars: bmal1−/− littermates (n=4–6 per time point). Each group is composed of equal or near-equal proportions of male and female animals. Scale bars within the representative pictures represent 200μm. (c) Frequency of macrophages, neutrophils, and lymphocytes present in BALs of bmal1-wt (blue symbols, n=4–14 per time point) and bmal1−/− (red symbols, n=4–6 per time point) animals at various time points post SeV infection (5x104 pfu). Each bar represents the mean frequency of each cell type ± SE. Groups of mice were composed of equal or near-equal proportions of male and female animals. (d) BAL cytokine concentration in bmal1-wt and bmal1−/− animals at various time points after infection with SeV (5x104 pfu). Each point represents the mean cytokine concentration ± SE. Blue circles: bmal1-wt (n=4–14 per time point). Red squares: bmal1−/− (n=4–6 per time point). Each group is composed of equal or near-equal proportions of male and female animals. *p<0.05 bmal-wt vs bmal−/− (Student’s 2-Tailed t-test).
In models of bacterial infection and endotoxin challenge, bmal1−/− mice were shown to develop exaggerated inflammation because of dysregulation of specific chemotactic factors, such as CCL2 and CXCL511, 21. To examine whether bmal1 affects the pathogenesis of SeV infection through similar means, we profiled BAL cytokine expression (Fig. 3d and Supplementary Table 1). In contrast to other models, bmal1 deletion promoted broad-based increases in pro-inflammatory cytokine expression in response to SeV. Of the 30 cytokines and chemokines we identified as increased in BAL fluid during SeV infection in wt animals, 20 of these exhibited significantly greater levels in bmal1−/− mice for at least 2 time points (Supplementary Table 1). Compared to wt, levels of representative type I (IFN-α), type II (IFN-γ), and type III (IFN-λ2) interferons were all increased in SeV-infected bmal1−/− mice at the protein level, and analogous results were observed at the whole lung RNA level as well (Fig. 3d and Supplementary Figure 5). SeV-infected bmal1−/− mice also produced higher levels of CXCL5 and IL-6 compared to wt11, 21, which has been observed in other disease models18, 24. To understand why bmal1 deletion produces such wide-ranging effects on cytokine expression, we analyzed these results in relation to viral RNA levels, and found that high viral loads in infected bmal1−/− mice could explain their elevated BAL cytokine levels (Supplementary Fig. 6). Taken together, our data indicate that bmal1−/− mice have a primary defect in their ability to control SeV viral replication, leading to more extensive cytokine expression and overall lung inflammation. This represents a different mechanism for bmal1 than what has been described in models of bacterial infection and sterile inflammation.
Effect of clock function and bmal1 deletion on interferon gene expression
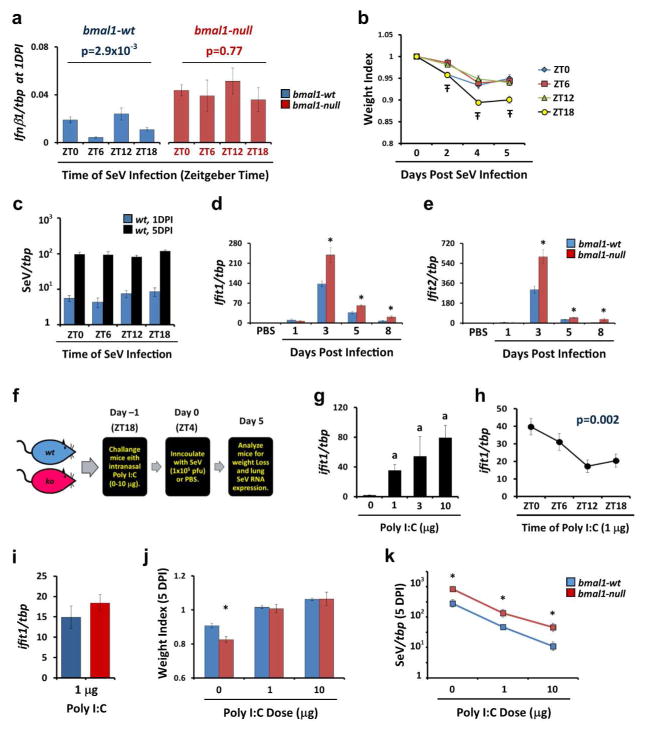
In mammals, interferons are critical to the control and clearance of viruses22. We therefore investigated potential relationships between circadian clock function and interferon-related gene expression, both in wt mice and bmal1−/− mice. We found that lung expression of ifnb1 varies significantly with the time of SeV inoculation in wt mice but not bmal1−/− mice, suggesting that initial type-I interferon responses are subject to circadian regulation (Fig. 4a). Time of SeV infection also affected the degree of weight loss in wt mice, but interestingly viral RNA levels were not impacted by infection time, as observed at 1 DPI and 5 DPI (Figs. 4b, c). Among the downstream effectors of Type-I interferons, the ifit gene family is thought globally important to antiviral defense, and ifit2-null mice were previously shown to be more vulnerable to SeV infection23. Interestingly, expression of ifit1 and ifit2 after SeV infection was increased in bmal1−/− mice compared to wt, which we would have thought would correlate with improved rather than reduced virus protection (Figs. 4d and e). We therefore wondered whether the overall efficacy of interferon antiviral responses in bmal1−/− mice could be diminished. To approach this question, we pretreated mice with intranasal Poly (I:C), a TLR3 agonist previously shown to suppress SeV replication as well as other RNA viruses via the induction of interferon signaling without inducing weight loss in mice24–26 (Fig. 4f). We confirmed that Poly (I:C) treatment led to a dose dependent stimulation of Type-I interferon responses in our model, as evidenced by induction of ifit1 expression (Fig. 4g). Similar to SeV inoculation, time of day impacted the potency of Poly (I:C) treatment in wt mice (Fig. 4h). Poly (I:C) stimulated ifit1 induction to a similar extent in bmal1−/− and wt mice, and was equally able to rescue both genotypes from weight loss after SeV infection (Figs. 4i and j). Despite this, Poly (I:C) pre-treatment did not normalize SeV RNA levels in infected bmal1−/− mice relative to wt littermates (Fig. 4k). Overall, these data indicate an association between the circadian clock, bmal1 function, and interferon antiviral activities. Moreover, control of viral replication via interferon stimulation in the lungs of bmal1−/− mice appears compromised at the whole organ level.
Figure 4.
Interferon-related antiviral responses are altered in bmal1−/− mice. (a) Expression of ifnb1 measured by qPCR 24 hours after inoculation with SeV (5x104 pfu). Inoculation was conducted at various times of day represented in Zeitgeber Time (ZT) where ZT0 represents “lights on” and ZT12 represents “lights off”. Each bar represents the mean ifnb1 expression normalized to tbp ± SE. Blue bars: bmal1-wt (n=9–12 per time point). Red bars: bmal1−/− (n=6–7 per time point). Depicted data was pooled from two independent time series, each showing similar time-dependence in ifnb1 expression. Statistical significance was determined by 1-way ANOVA. Note that lung infb1/tbp ratios in sham (PBS) treated mice were negligible at the ZTs tested, ranging from 2.3–4.4x10−4 in wt mice. (b) Weight loss (mean ± SE, n=10 per group) after inoculating male wt mice with SeV (5x104 pfu) at various ZTs. Data was pooled from 2 independent experiments. Ŧp<0.05, 1-way ANOVA. (c) SeV RNA expression at 1DPI (blue bars) and 5DPI (black bars) after inoculating wt mice with SeV (5x104 pfu) at various ZTs. Data was pooled from 2 independent experiments (n=9–12 per group). (d, e) Gene expression of ifit1 (d), and ifit2 (e) normalized to tbp. Each bar represents the mean expression ± SE at various time points after infection with SeV (5x104 pfu). Blue bars: bmal1-wt (n=4–14 per time point). Red bars: bmal1−/− littermates (n=4–6 per time point). Each group is composed of equal or near-equal proportions of male and female animals pooled from 2 independent rounds of SeV infection. *p<0.05 bmal1-wt vs bmal−/−(Student’s 2-Tailed t-test). (f) Cartoon depicting our protocol for pre-activating interferon responses in mice prior to SeV challenge using the TLR3 agonist Poly (I:C) (also see Methods). Mice received an intranasal challenge of Poly (I:C) and the following morning were inoculated with SeV. The mice were then harvested at 5DPI to assess SeV viral load. (g) Dose response relationship between intranasal Poly (I:C) and stimulation of interferon responses as reflected by induction of ifit1 expression. wt male mice were challenged with the indicated amount of Poly (I:C) and lungs were obtained for qPCR analysis on Day 0 as depicted in panel (f). Bars represent mean ifit1/tbp ratios ± SE (n=3 per group). ap<0.05 vs. PBS treated wt mice (Student’s 2-Tailed t-test). (h) Induction of lung ifit1 expression in wt male mice by 1 μg intranasal Poly (I:C) administered at different ZTs. Data points represent mean ifit1/tbp ratios ± SE (n=9 per data point, pooled from 3 independent experiments). For each data point, lungs were collected 12 hours after Poly (I:C) inoculation. (i) Induction of ifit1 expression by 1 μg Poly (I:C) in bmal1−/− (red bars) and wt (blue bars) littermates. Bars represent mean ifit1/tbp ratios ± SE (n=5–6 per group, pooled from 2 independent experiments). Male and female mice were used in equal or near equal proportion in these experiments. (j) Effect of Poly (I:C) pre-treatment on weight loss after SeV infection (1x105 pfu) in bmal1−/− (red bars) and wt (blue bars) littermates. Bars represent mean weight ± SE at 5 DPI, expressed as a fraction of the starting weight (n=6–30 per group). Data depicted was pooled from 2–4 independent experiments per dose of Poly (I:C) and employed male and female mice in equal or near equal proportion. *p<0.05 bmal1-wt vs bmal−/− (Student’s 2-Tailed t-test). (k) Effect of Poly (I:C) pre-treatment on SeV RNA expression in bmal1−/− (red bars) and wt (blue bars) littermates, 5 days after infection (1x105 pfu). Data points represent mean SeV/tbp ratios ± SE at 5 DPI (n=9–35 per group). Data depicted was pooled from 3–4 independent experiments per dose of Poly (I:C), each showing similar results, and employed male and female mice in equal or near equal proportion. *p<0.05 bmal1-wt vs bmal1−/− (Student’s 2-Tailed t-test).
Deletion of bmal1 exacerbates post-viral lung pathology
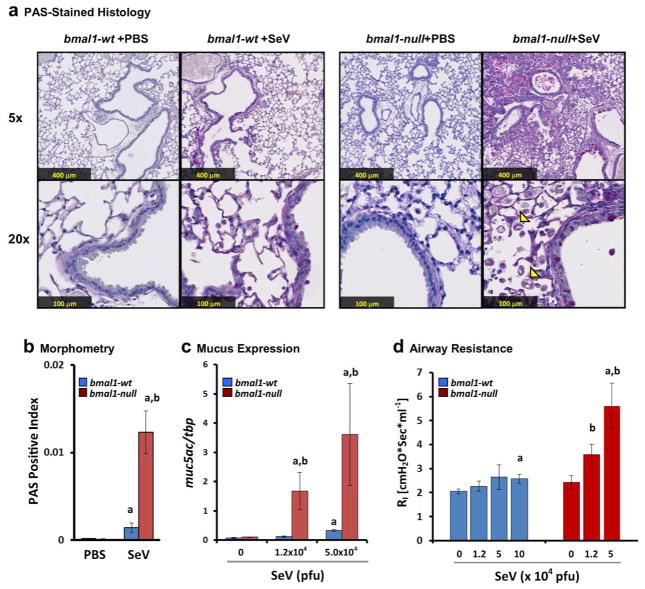
The severity of viral bronchiolitis is of special significance because severe or frequent bouts of bronchiolitis in young children is associated with asthma development4, 27. To examine the effects of bmal1 in asthma-like airway disease we examined the lung histology of bmal1−/− mice and wt mice 49 days after SeV infection, a standard time point for observing chronic airway disease in this model14 (Fig. 5). SeV infection in bmal1−/− mice produced profound airway remodeling and goblet cell metaplasia compared to wt (Figs. 5a and b). This was accompanied by an inoculum-dependent increase in resting airway resistance and SeV-induced mucus production as reflected by muc5ac expression (Figs. 5c and d). The amount of muc5ac expression observed at 49 DPI did not appear sensitive to the time of day SeV infection had been performed or the time of day that lung specimens were obtained, and neither of these factors accounted for the increased muc5ac expression seen in bmal1−/− mice (Supplementary Figs. 7a and b). SeV infection had a subtle effect on the rhythm of bmal1 expression at 49 DPI, producing shifts in phase, mean expression, and amplitude compared to uninfected controls (Supplementary Fig. 7c). These effects were independent of the timing of initial SeV infection (Supplementary Fig. 7d). Previously a pro-fibrotic phenotype was observed in clock mutant mice12. However, trichrome-stained sections from bmal1−/− lungs did not in our hands reveal a significant excess of collagen deposition, either in PBS treated mice or 49 days after SeV infection (Supplementary Fig. 8). We did note a mild but statistically significant increase in collagen staining at 49DPI in wt mice infected with SeV (Supplementary Fig. 8b).
Figure 5.
Deletion of bmal1 exacerbates chronic lung disease after SeV Infection. (a) Representative micrographs of PAS stained lung sections at day 49 after sham (PBS) or SeV (5 x 104 pfu). For each panel the upper picture is at 5x magnification and the lower is at 20x. Note the increased distortion of lung architecture in SeV challenged bmal1−/− mice compared to wt and the infiltration of crystal-laden macrophages (arrowheads). (b) Quantification of PAS+ staining in SeV-infected or sham (PBS treated) bmal1−/− and wt littermate lung sections at 49 DPI (red and blue bars respectively). Bars represent the mean ratio of PAS+ (magenta) pixels to total pixels ± SE. (c) Expression of muc5ac at day 49 as a function of SeV inoculum. Each bar represents the mean expression ± SE normalized to the housekeeping gene tbp. Blue bars: bmal1-wt (n=5–9 per group). Red bars: bmal1−/− littermates (n=3–6 per group). (d) Airway resistance at day 49 as a function of SeV inoculum. Each bar represents the mean airway resistance ± SE. Blue bars: bmal1-wt (n=5–9 per group). Red bars: bmal1−/− littermates (n=3–6 per group). ap<0.05 SeV-infected vs. PBS-control (Student’s 2-Tailed t-Test), bp<0.05 SeV-infected bmal1−/− vs. wt (Student’s 2-Tailed t-test).
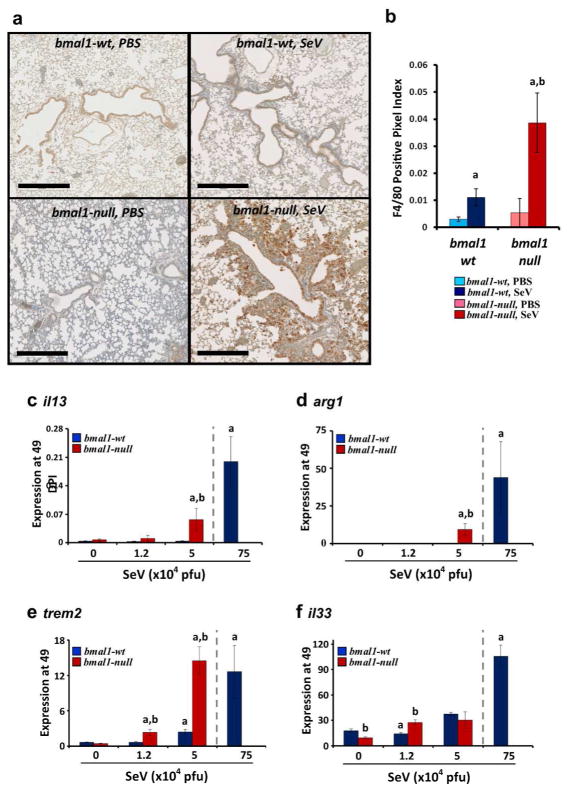
In the SeV post-viral model, chronic airway disease is driven by alternatively activated (M2) macrophages, which accumulate in the lungs in the post-infection period and secrete IL-1328. We found that even low doses of virus that produce minimal lung disease in wt animals resulted in pronounced accumulation of macrophages in bmal1−/− mice around remodeled airways as shown by F4/80 immunohistochemistry (Figs. 6a and b). Lung expression of the M2 markers il13 and arg1 was also magnified in bmal1−/− mice at 49 DPI (Figs. 6c and d). The timing of SeV infection did not significantly affect il13 expression at 49 DPI, similar to muc5ac (Supplementary Fig. 7e). It was previously shown that the accumulation and retention of M2 macrophages in lungs of SeV-infected mice depends on IL-33 secreted from lung epithelial cells29 and soluble Trem-2, a macrophage survival factor30. In post-viral bmal1−/− mice, we found that lung expression of trem2 was significantly enhanced and rivaled levels seen in wt animals only at much higher doses of SeV (Fig. 6e). In contrast, induction of lung il33 expression in bmal1−/− mice was not consistently different compared to wt (Fig. 6f). To summarize, impaired early antiviral responses in bmal1−/− mice translated into more severe chronic lung disease post-infection, characterized by goblet cell metaplasia, increased airway resistance, increased M2 gene expression signatures, and increased expression of the M2 survival factor trem2.
Figure 6.
Expression of M2 signatures is enhanced in bmal1−/− mice post-SeV infection. (a) Representative images of lungs at 49 DPI stained with macrophage marker F4/80. Scale bar=500 μm. (b) Quantification of F4/80+ staining in immunohistochemical sections expressed as a fraction of positive stained pixels (mean expression ± SE, n=3–4 per group). ap<0.05 versus sham (PBS) infected wt controls, bp<0.05 bmal1−/− vs. wt (Student’s 2-Tailed t-test). (c–f). Mice were infected with the indicated dose of SeV, and at 49DPI lungs were harvested and analyzed for gene expression using tbp as a housekeeping gene. As a positive control, lung gene expression in wt mice exposed to a high dose of SeV (75x104 pfu) is depicted to the right in each panel and separated by a dashed line. Each bar represents the mean expression ± SE. (c) il13 expression. (d) arg1 expression. (e) trem2 expression. (f) il33 expression. Red bars indicate bmal1−/− mice (n=3–4 per group), and blue bars denote wt littermates (n=5–9 per group). ap<0.05 versus sham (PBS) infected wt controls, bp<0.05 bmal1−/− vs. wt (Student’s 2-Tailed t-test). Equal or near equal numbers of male and female mice were utilized per group.
Effect of environmental circadian disruption on SeV-induced lung disease
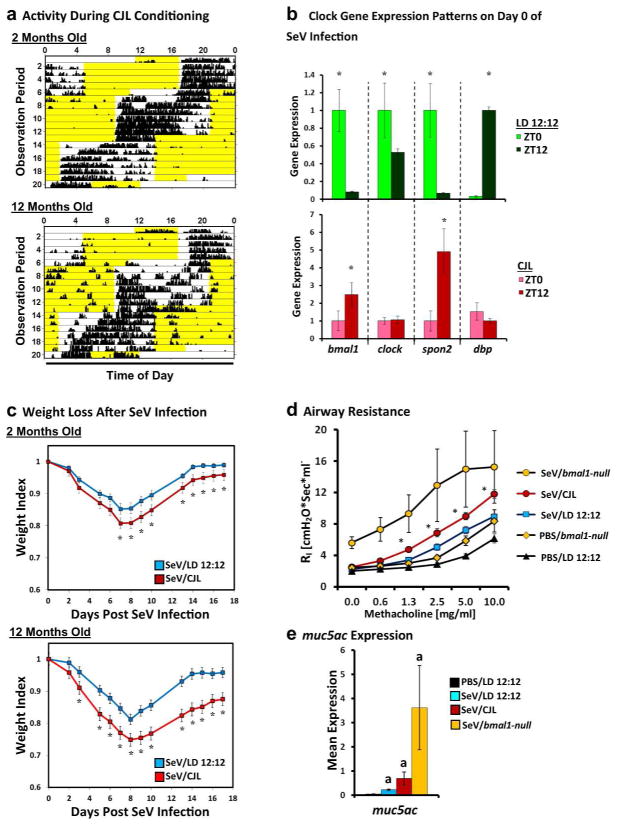
Environmental causes of circadian disruption such as shift-work, artificial lighting, and jet lag are common in society. We therefore examined if the viral phenotypes observed in bmal1-deleted mice could be reproduced with chronic jet lag (CJL), an environmental stress shown to misalign circadian rhythms31. Because the effects of CJL are more pronounced and more durable in older organisms32, we analyzed wt juvenile (6–8 week-old) and middle-aged (9–12 month-old) C57BL6/J mice. (Fig. 7a). As expected, CJL disrupted the temporal expression patterns of bmal1 and other clock genes within the lung (Fig. 7b). Compared to mice kept under standard lighting conditions, weight loss after SeV infection was increased in CJL mice, with especially pronounced effects in middle-aged animals (Fig. 7c). CJL also increased airway resistance and methacholine sensitivity at 49 DPI, and it produced a trend towards higher muc5ac expression (Fig. 7d and e). While the effects of CJL were less potent than that of bmal1 deletion, our results showed that environmental disruption of circadian rhythms in mice can worsen aspects of acute SeV infection and chronic SeV lung pathology. This suggests that real-world disruption of circadian function has ramifications for antiviral responses in the lung.
Figure 7.
Effect of chronic jet lag (CJL) on acute and chronic SeV lung pathology. (a) Representative actigraphs depicting wheel running activity during CJL conditioning, which consisted of weekly 8-hour phase advances in juvenile (2 months old, upper panel) and middle aged (12 months old, lower panel) mice. Black vertical bars represent wheel revolutions per epoch, and each row represents one day of recording. Yellow highlighted regions represent lights-on. (b) Expression of various clock genes at subjective dawn (ZT0) and dusk (ZT12) for mice living under standard 12 hour day-night lighting (LD 12:12, green bars) or CJL (red bars) after the final phase advance in our protocol. Each bar represents the mean expression ± SE (n=3 per group). For each clock gene, expression is normalized to the housekeeping gene tbp and then further standardized to mean expression at ZT0 for ease of viewing. The data depicted was obtained from middle aged mice, but similar changes were observed in juvenile mice subjected to CJL. *p<0.05 ZT0 vs. ZT12 (Student’s 2-Tailed t-test). (c) Weight loss in standard-housed (blue line, n=7–15 per group) and CJL conditioned (red line, n=12–15 per group) mice after infection with SeV (1.5x105 pfu). Data represents the mean weight index ± SE and is representative of two independent experiments. Note that we did not encounter significant mortality in either CJL or control mice during acute SeV infection for these experiments. *p<0.05 CJL vs. LD12:12 (Student’s 2-Tailed t-test). Male mice were used for these experiments. (d) Airway resistance in SeV-infected mice at 49 DPI (1.5x105 pfu) that were conditioned with CJL or standard lighting (LD 12:12). Each line represents the mean airway resistance measured after doubling doses of methacholine ± SE. Orange circles: SeV-infected bmal1-null mice (n=3). Red circles: SeV-infected, CJL conditioned mice (n=11). Blue Squares: SeV-infected mice under normal lighting conditions (n=9). Black triangles: PBS-treated mice under normal lighting conditions (n=5). Orange diamonds: PBS-treated bmal1-null mice (n=6). *p<0.05 SeV/CJL vs. SeV/LD 12:12 (Student’s 2-Tailed t-test). (e) Lung expression of muc5ac at 49 DPI with SeV. Each bar represents the mean muc5ac expression normalized to tbp ± SE. Black bar: PBS-treated mice under normal lighting conditions (n=5). Blue bar: SeV-infected mice under normal lighting conditions (n=9). Red bar: SeV-infected mice subjected to CJL protocol before infection (n=11). Yellow bar: SeV-infected, bmal1-null mice under normal lighting conditions (n=3). Note that in panels (d) and (e), the bmal1−/− mice depicted were inoculated with 5x104 pfu, as this was the highest dose that was below the LD50 in this genotype. ap<0.05 vs PBS/LD 12:12 (Student’s 2-Tailed t-test).
Circadian clock gene expression in asthma patient samples
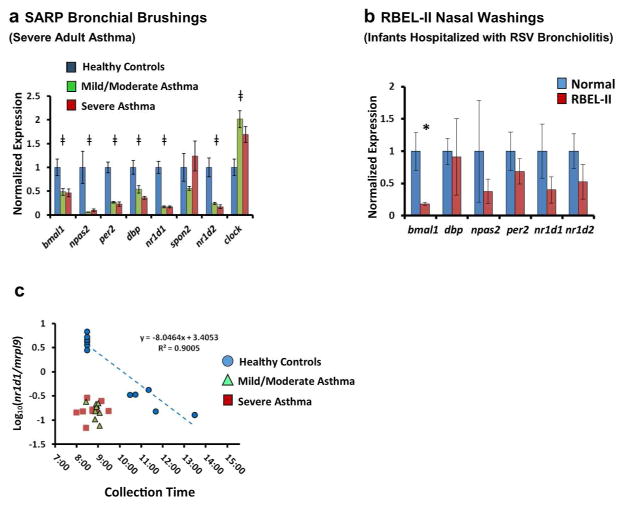
To investigate whether alterations in clock gene expression can is observed in asthma patients, we analyzed respiratory tract expression of these genes in two asthma cohorts. The first cohort comprised bronchial brushings from adult patients with asthma who participated in the Severe Asthma Research Program (SARP, Supplementary Table 2)33. The second group of samples consisted of nasal washes from infants enrolled in the RSV Bronchiolitis in Early Life (RBEL-II) Study34, who had been hospitalized for RSV bronchiolitis and donated samples on follow-up (average length of follow-up 91days ± 96) after hospital discharge. Of note, the majority of RBEL-II subjects we sampled developed persistent wheezing by 2 years of age (Supplementary Table 3). For comparison, we analyzed samples from healthy adult and pediatric subjects, being careful to match the collection times of asthma subjects and healthy volunteers (Supplementary Tables 2 and 3, and see Methods). In the SARP cohort, we noted decreased expression in 6 core clock genes and increased expression in 1 gene (clock) compared to time-matched healthy controls (Fig. 8a). In contrast, the clock controlled gene spon2 did not exhibit consistent differences between asthmatic and healthy subjects. In nasal washes from the RBEL cohort, bmal1 expression was reduced relative to time-matched healthy controls similar to SARP but differences were not observed in the other clock genes tested (Fig. 8b). By their very nature, circadian clock genes are expected to have oscillations in their expression over the course of the day. The SARP and RBEL specimen banks (and indeed most clinical specimen repositories) were not designed to collect serial samples throughout the day that would be ideal for interpreting changes in clock gene expression. Nevertheless, the SARP repository did contain specimens from healthy subjects that were obtained at later times in the morning and early afternoon, allowing us to extrapolate how collection time might influence our results (Fig. 8c). We found that nr1d1 (also known as reverbα) exhibited a linear relationship between log-transformed gene expression and sample collection time in healthy subjects. To observe the same nr1d1 expression in normal subjects that we observed in asthmatics, we estimated that a time delay of 3–4 hours would be required (3.61 ± 0.14 hours relative to mild/moderate asthmatics, and 3.91 ± 0.26 hours relative to severe asthmatics). We obtained similar estimates using bmal1/nr1d1 expression ratios to generate a regression line rather than nr1d1 alone (3.56 ± 0.36 hours relative to mild/moderate asthmatics, and 3.70 ± 0.57 relative to severe asthmatics, Supplementary Fig. 9). Collectively, our results suggest that clock gene expression patterns can be altered in the asthmatic respiratory system.
Figure 8.
Differential expression of clock genes in airway samples from two asthma cohorts. (a) Expression of circadian clock genes in bronchial brush samples obtained from adult patients with severe asthma (n=9, red bars), mild/moderate asthma (n=9, green bars), and healthy volunteers (n=6, blue bars) who participated in SARP. All samples were collected between 0800 and 0930 hours. Bars represent mean expression ratios±s.e. normalized to mrpl9. To simplify the visual depiction of data, expression was then further normalized to the mean for non-asthmatic control subjects. ‡P<0.05 (one-way ANOVA). (b) Clock gene expression in nasal wash samples previously obtained from young children hospitalized for RSV bronchiolitis (n=10, red bars) and control subjects (n=5, blue bars) who participated in RBEL-II. All samples were collected between 1015 1220 hours. Bars represent mean expression ratios±s.e. normalized to mrpl9. To simplify the visual depiction of data, expression was then further normalized to the mean for non-asthmatic control subjects. (c) Log10 normalized nr1d1/mrpl9 expression ratios from SARP participants (Mean±s.e.), graphed as a function of collection time. Blue circles: healthy volunteers (n=11). Green triangles: mild/moderate asthma (n=9). Red squares (severe asthma, n=9). A regression line is depicted for the healthy control samples (blue dashed line). Extrapolating from this line, the time difference that would be needed to reproduce the nr1d1 expression seen in asthmatic subjects using healthy controls is 3.61±0.14 h for mild/moderate asthmatics, and 3.91±0.26 h for severe asthmatics. *P<0.05 (Student’s two-tailed t-test). ANOVA, analysis of variance; RSV, respiratory syncytial virus; SARP, Severe Asthma Research Program.
DISCUSSION
In this study, we identify a role for the clock gene bmal1 in mitigating viral respiratory illness and in the development of chronic lung phenotypes that are pertinent to asthma. We show that acute and chronic viral airway pathology can be exacerbated through environmental as well as genetic manipulation of circadian function. We also show that differential expression of bmal1 can be observed in airway cells of asthmatic adults and children at risk for asthma. To our knowledge, this study is the first to assign a role for clock gene regulation in asthmatic lung phenotypes, at least in mice.
Our data on bmal1 in respiratory viral infection should be viewed in the context of recent studies on this clock gene. Previously, bmal1 deletion has been shown to render mice more susceptible to bacterial infections, to inflammation in response to microbial products like endotoxin, and to sterile agents like bleomycin and cigarette smoke11, 12, 35, 36. Less well understood is how bmal1 or the circadian clock might impact viral infection. Benegiamo et al.17 showed that over-expression of the bmal1 negative regulator per2 in tissue culture cells inhibited replication of hepatitis C virus (HCV). In the case of herpes simplex virus (HSV), Kalamvoki et al. showed that this virus co-opts the Bmal1 interaction partner Clock to drive viral gene expression37. Recently, Sundar et al.16 focused on the ability of H3N1 IAV to disrupt normal clock gene expression patterns in mouse lung and proposed this event synergizes with cigarette smoke to promote chronic obstructive pulmonary disease (COPD). Finally, while this manuscript was in preparation and under review, several groups reported a role for circadian regulation in IAV, HSV, VSV and RSV infection15, 38, 39. Considered along with our results in SeV infection, it would appear that circadian clock function, and bmal1 in particular, is generally important for antiviral defense. Two outstanding questions from the above literature that we begin to address here are the basic mechanism of bmal1 antiviral function and the potential ramifications for asthma, an important complex airway disease previously linked to viral infection. Unlike the case with bacterial infection or endotoxin11, 35, the effects of bmal1 on SeV infection are not likely a function of specific chemotactic factors like CXCL5 or CCL2 being dysregulated. Rather, bmal1−/− mice appear to be more vulnerable to SeV due to defective control of viral replication, as gauged by elevated viral loads, altered ISG expression patterns, and diminished protection by the TLR3 agonist Poly (I:C). Moreover, we show for the first time that post-natal deletion of bmal1 is sufficient to compromise antiviral responses in the lung, making it only the second bmal1 knockout phenotype to date to be inextricably tied to circadian clock function. Further research will be needed to trace the downstream effectors of bmal1 and clock-mediated antiviral activity, and the relationship of those effectors to interferon signaling.
Asthma has long been noted to display circadian variations in symptoms, but previous studies of circadian rhythms in asthma have focused on physiological variables. For example, normal circadian swings in airway resistance tend to be magnified in asthmatics, especially those with prominent nocturnal symptoms40. Our data suggest a novel additional method by which circadian rhythms can impact asthma- through the response to respiratory viral infection. However, this does not preclude a role for circadian regulation in other aspects of asthma pathogenesis. Asthma is a heterogeneous disease, and our Sendai post-viral model of airway remodeling is best at analyzing IL-13 and M2 dependent airway changes that are relevant to some but not all patients. Importantly, our study leaves open the role of clock genes in atopy, which is strongly and independently associated with asthma in adults and children. Circadian rhythms in allergic responses have been documented in humans, and both eosinophils and mast cells harbor circadian clocks. Further research will be needed to evaluate the role of circadian clock genes in atopic asthma and asthma associated with alternative cytokine profiles, such as IL-17.
We present evidence that certain asthma patients may harbor altered expression patterns of bmal1 and other clock genes, and therefore the antiviral and airway remodeling effects of bmal1 deletion in mice could be relevant to asthma. It is important to state that because expression of bmal1 and other clock genes oscillate across the day, our observations of altered gene expression over a narrow time interval might represent a change in the phase, periodicity, or amplitude of clock gene expression in asthmatic airways- or any combination of these factors41. Serial observations of bmal1 expression and other clock genes in respiratory samples from asthmatic patients will be needed to clarify this point. Despite this important caveat, our results are notable because they represent what is possible using currently available human sample repositories and currently available analytical approaches. We expect that future innovations in sample collection and analytics will add to the observations we report here.
In summary, we report a novel role for the circadian clock gene bmal1 in regulating viral bronchiolitis and asthmatic airway phenotypes. Further research into the mechanisms of bmal1 and circadian clock antiviral activity could yield valuable therapeutic targets.
METHODS
Mice
C57BL/6J male mice (4–8 weeks old, 20–25g) were purchased from Jackson Laboratories and acclimatized for two weeks before use. For middle-aged male mice (9–12 months old) we utilized retired C57BL/6J breeders obtained from transgenic core facilities at our institution. To generate global bmal1-null mice where deletion occurs during embryogenesis we crossed Jackson Laboratory strains B6.129S4(Cg)-Arntltm1Weit/J and B6.C-Tg(CMV-cre)1Cgn/J. Heterozygote progeny were mated to each other to obtain wt and bmal1-null littermates as described19. To produce mice with tamoxifen inducible deletion of bmal1 (bmal1-iKO), we crossed Jackson strain B6.129S4(Cg)-Arntltm1Weit/J with either B6.Cg-Tg(UBC-cre/ERT2)1Ejb/J or B6.Cg-Tg(CAG-cre/Esr1)5Amc/J. Heterozygous mice were then bred to yield bmal1fl/flcre+ and bmal1fl/flcre− animals for each strain, which then were crossed to generate mice for experiments. To induce bmal1 recombination mice were fed with tamoxifen-containing chow (Cat# TD.130859, Envigo) for 3 weeks according to the manufacturer’s instructions starting at 4–6 weeks of age, and then returned to a normal chow diet for 1 week prior to starting experiments. To generate fluorescent reporter mice in order to estimate bmal1-iKO Cre activity, we crossed bmal1-iKO mice with the Jackson strain B6.129(Cg)-Gt(ROSA)26Sortm4(ACTB-tdTomato,-EGFP)Luo/J. Resulting heterozygous progeny were fed tamoxifen chow for 3 weeks as above, and frozen lung sections were then obtained for fluorescence microscopy. Our standard lighting conditions were a 12 hour light-dark cycle (lights-on at 6:00 AM and lights-off at 6:00 PM local time). We followed the convention of expressing time of day in “Zeitgeber Time” (ZT), where ZT0 indicated lights-on (subjective dawn) and ZT12 lights-off (subjective dusk). Except when indicated, we utilized male and female mice for experiments in equal proportions. All experiments were approved by the Washington University School of Medicine Animal Studies Committee.
Virus and Mouse Infection
Sendai virus (SeV) was obtained from ATCC (VR-105; Sendai/52). Virus was twice plaque purified, propagated in 10-day-old embryonated chicken eggs, and stored at -70°C. Infectivity of SeV was assayed in VeroE6 cells as described42. Influenza A virus (IAV A/WS/33 H1N1) was obtained from ATCC. Virus was plaque purified using MDCK (ATCC CCL-34) cells and propagated in 9-day-old embryonated chicken eggs. SeV at various doses or 5 pfu of H1N1 influenza A virus was intra-nasally administered to mice as described28. Briefly, animals were anesthetized with ketamine/xylazine HCL cocktail, and virus at various doses was intra-nasally instilled in 30μl of PBS. Mice were then returned to their cages to recover from anesthesia. To measure viral load in SeV-infected lungs, mice were euthanized and lungs were homogenized in PBS, clarified by centrifugation, and filtered through 0.2μm syringe filters. Plaque assay was conducted on clarified homogenates essentially as described42. Unless otherwise stated, SeV infections were performed within a window of ZT4 to ZT8. Our practice was to alternate the order of inoculations between comparison groups for any given experiment (for example bmal1−/− and wt littermates) in order to balance infection times between these groups. For all experiments involving bmal1−/− mice, wt littermates were always assayed in parallel. Moreover, our practice was to co-mingle bmal1−/− mice and wt littermate mice from multiple dams for 1–2 weeks after weaning in order to control for microbiome effects, as suggested by Stappenbeck et al.43
Poly (I:C) Pre-treatment
Poly (I:C) stock solutions (1 mg/ml) were constituted according to manufacturer instructions. To control for manufacturing differences, we independently replicated all experiments using Poly (I:C) sourced from 2 different vendors (InVivoGen Cat# tlr1-puc; Sigma Cat# p1530). At ZT18 mice were anesthetized with ketamine/xylazine, and Poly (I:C) at indicated amounts was diluted into sterile PBS and intransally instilled (30 μl total volume). Mice were then returned to their cages and recovered overnight. The subsequent day mice were re-anesthetized and infected with SeV (1x105 pfu per mouse) as described above.
Chronic Jet Lag (CJL)
To induce CJL, mice were subjected to weekly 8-hour phase advances for 3 weeks analogous to a previously described protocol31. In brief, male C57BL/6J mice were housed in cages that were situated within ventilated, light-tight structures (Gorilla Grow Tent) and were supplied with a 12 hour light/dark cycle via interior compact fluorescent lighting. On the Monday of each week the light cycle was advanced 8 hours, such that by the third phase shift they had processed 24 hours (or back to the standard lighting schedule of our mouse colony). During CJL conditioning mice were group-housed 4–5 to a cage consistent with the standard housing policy of our animal facility. With each CJL run, we also housed 1–2 mice individually in wheel running cages to assist in remote monitoring of animal health, as light hygiene precautions dictated that mice could be accessed only by lab personnel. Actigraphy for these sentinel mice was recorded and analyzed using ClockLab software (Actimetrics). On the day of the final light phase advance, the mice were moved to a satellite location designated for viral infection within our facility, and infected with SeV the following day as described above. We confirmed that CJL had been achieved by sacrificing untreated mice at ZT0 and ZT12 on the day of infection and analyzing clock gene expression patterns relative to mice housed under normal lighting conditions (these ZTs were chosen because they represent the acrophase or bathyphase for most core clock genes). After viral infection mice were maintained under normal lighting conditions in the satellite facility until the end of the experiment.
Gene Expression Analysis
At various time points after infection, lungs were manually excised and immersed in RNAlater solution (Applied Biosystems). Total RNA was isolated from lung homogenates using RNeasy Mini Kits (Qiagen). Reverse transcription was carried out using the Applied Biosystems High Capacity cDNA Reverse Transcription Kit. Real Time PCR was performed on the ABI 7500 Fast Real-Time PCR system. Primers used for RT-qPCR are listed in Supplementary Table 4.
BAL and Cytokine Analysis
SeV infected mice were euthanized and their tracheas were cannulated with a 20G flexible catheter, 1–2 mm distal to the larynx. The lungs were then slowly lavaged in 2 passes with 1 ml sterile phosphate buffered saline (PBS). For downstream analyses, the lungs and trachea were dissected and stored in RNAlater solution. For BAL cell counts, 50 μl of BAL suspension was adsorbed to frosted glass slides using a Cytospin 4 centrifuge (Thermo Scientific) and treated with Kwik-Diff stain (Thermo Scientific). Slides were then digitally scanned at 20x magnification using a Nanozoomer HT imaging system (Hamamatsu). To quantify cell number, scanned slides were analyzed via ImageScope software (Leica) using the “Nuclear v9” algorithm, which counts nuclei in histologically stained samples. For BAL cell differential, sample identifications were blinded and 5 random 40x fields were captured as TIFF files using ImageScope software. Cells in each field were then scored as either lymphocytes, macrophages, or neutrophils by a separate investigator also blinded to the sample ID. A minimum of 50 cells were counted per biological specimen. BAL protein concentration was quantified using Bradford reagent (Sigma) according to standard protocols. BAL cytokine levels were analyzed on a FLEXMAP 3D instrument (Luminex), using the Mouse Cytokine and & Chemokine Panel 1A (Affymetrix), according to the manufacturer’s instructions. We regarded statistically significant increases in log-transformed cytokine levels in wt animals at any time point as evidence of induction by SeV (Student’s 2-Tailed t-Test).
Flow Cytometry
Whole lung flow cytometry was performed essentially as described9. Briefly, lungs from SeV infected mice were dissociated using Liberase-TM and DNase in RPMI (Roche), stained, fixed with 1% paraformaldehyde, and analyzed using a FacsCallibur cytometer (BD). Analysis of data was performed using FlowJo software (Tree Star). Please see Supplementary Table 5 for a list of antibodies used for flow cytometry analysis and Supplementary Fig. 10 for depiction of representative gates. For all samples, 10,000 events were recorded.
Airway Resistance Measurements
At 49 days post infection with SeV, airway resistance was measured using a Buxco Elan RC mouse volume-controlled ventilator as described28. Briefly, mice were anesthetized by i.p. injection of ketamine/xylazine cocktail, and then tracheostomy was performed using a 20G flexible catheter. Mice were then ventilated and given doubling concentrations of methacholine through an in-line nebulizer. Inspiratory resistance measurements were averaged over three minute intervals after each dose. Our practice was to alternate the order of airway resistance measurements between comparison groups for any given experiment (for example bmal1−/− and wt littermates, or CJL vs. non-jet lagged mice) in order to balance out measurement times between the groups. Roughly 30–40 minutes was required to obtain airway resistance measurements per mouse using our apparatus.
Morphometry
Lungs were fixed by inflation with 10% buffered formalin under 20 cmH2O of pressure, paraffin imbedded, and stained with PAS, Mason-trichrome stain, or subjected to immunohistochemistry (IHC) or immune-fluorescence (IF) staining using standard techniques28. PAS, trichrome or IHC-stained slides were scanned at 20x using a Nanozoomer HT imaging system (Hamamatsu) as above. Positively stained pixels were counted across the entire section using ImageScope Software and the “Positive Pixel v9” algorithm as previously described9. For immunofluorescence experiments we utilized antibodies directed against Mac3 (Ebioscience) and cleaved caspase 3 (Cell Signaling) and Alexa Fluor-conjugated secondary antibodies (Invitrogen). Stained slides were visualized using an Olympus BX51 microscope and a Retiga-2000R camera system (QImaging).
Human Samples
Study design and sample collection protocols for the SARP and RBEL-II were previously detailed34, 44. Asthma severity (severe, moderate, and mild) was determined according to ATS/ERS workshop definitions45, and the normal controls had no prior history of lung disease, normal lung function, and negative bronchial hyperreactivity testing. Demographic information for all subjects is detailed in Supplementary Tables 2 and 3. In brief, for SARP participants, bronchial brushing was performed in proximal segmental airways, and epithelial cells obtained were immediately placed in Qiazol reagent (Qiagen). For RBEL-II participants, nasal washes were performed using 5 cc of saline in each nostril; the pellet from this wash was placed in Trizol (Invitrogen, Carlsbad, CA). Samples were collected from SARP and RBEL-II participants when they were not experiencing asthma exacerbations or other illness. For the purposes of matching collection times between cases and controls with the specimens available in the repositories, our primary analysis for SARP utilized samples obtained from 8:00 AM–9:30 AM and 10:15 AM–12:20 PM for RBEL. RNA was extracted using standard protocols recommended by the manufacturer. Gene expression analysis was then performed as described above using the housekeeping gene mrpl946. Use of human samples and de-identified subject information was approved by the Washington University Institutional Review Board.
Statistics
For survival curve analysis (Fig. 1b), we used the log rank test. Throughout this paper, we log-transformed gene expression and cytokine levels prior to performing Student’s 2-Tailed t-tests and 1-way ANOVAs as described47. For all other data we performed Student’s 2-Tailed t-tests or 1-way ANOVAs on non-transformed data. For circadian rhythm analysis of gene expression data we used COSOPT as described9. All statistical analyses were performed in Microsoft Excel.
Methodological Limitations
It is worth noting some limitations to our experimental approach. Although mice are frequently used as a model organism to study airway disease, there is controversy within the literature as to how far results obtained from mouse models can be generalized to human asthma48. Our approach to inducing CJL in mice is based on a well-cited protocol31 however there are in principal numerous potential ways to misalign mice to the solar day. Additionally, we opted to group house our mice during CJL conditioning rather than individually house mice. We did this because of emerging literature that housing mice individually may impose an additional stress on the animals that could confound our interpretation of SeV infection after CJL43, 49. Nevertheless, it is possible that our CJL protocol is milder than previous procedures, and may therefore understate the effects of jet lag on viral bronchiolitis. At the same time, our CJL results are likely to be biased away from false positive findings. Finally, in our analysis of clock gene expression in human samples we included all SARP and RBEL specimens for which we could find reasonably time-matched controls. Our observations will benefit from independent validation in future studies.
Supplementary Material
Acknowledgments
We thank Steven Brody, Derek Byers, Amjad Horani, Erik Herzog, Kangyun Wu and Robyn Haspel for their technical advice, and the participants in the SARP and RBEL-II studies. This study was supported by NIGMS 5K08GM102694 (JH), an ATS Foundation Recognition Award For Early Career Investigators (JH), Children’s Discovery Institute Interdisciplinary Research Initiative PD-II-2016-529 (JH), and NIH grants R01 HL135846 (JH), U10 HL109257, RO1 HL69149 and RO1 HL091762 (SARP); RO1 HL61895 and RO1 HL092486 (RBEL) and 5UL1 TR000448 (MC); S10 RR027552 (Alafi Neuroimaging Core). This work was also supported, in part, by the Center for Human Immunology and Immunotherapy Programs at Washington University, Immuno-monitoring Laboratory.
Footnotes
Conflict of Interest Statement: the authors report no conflicts of interest.
AUTHOR CONTRIBUTIONS
J. H., M. H., M. C., and E. A. conceived and designed the project. A. E., W. X., E. A., S. B., D. S., R. T., G. S., R. S., R. W., X. W., L. B., and J. H. performed the experiments. J. H., A. E., and W. X performed the data analysis. J. H., A. E., and S. B. drafted the manuscript.
References
- 1.Bergeron C, Al-Ramli W, Hamid Q. Remodeling in asthma. Proceedings of the American Thoracic Society. 2009;6(3):301–305. doi: 10.1513/pats.200808-089RM. [DOI] [PubMed] [Google Scholar]
- 2.Meissner HC. Viral Bronchiolitis in Children. The New England journal of medicine. 2016;374(18):1793–1794. doi: 10.1056/NEJMc1601509. [DOI] [PubMed] [Google Scholar]
- 3.Corne JM, Marshall C, Smith S, Schreiber J, Sanderson G, Holgate ST, et al. Frequency, severity, and duration of rhinovirus infections in asthmatic and non-asthmatic individuals: a longitudinal cohort study. Lancet. 2002;359(9309):831–834. doi: 10.1016/S0140-6736(02)07953-9. [DOI] [PubMed] [Google Scholar]
- 4.Bonnelykke K, Vissing NH, Sevelsted A, Johnston SL, Bisgaard H. Association between respiratory infections in early life and later asthma is independent of virus type. The Journal of allergy and clinical immunology. 2015;136(1):81–86. e84. doi: 10.1016/j.jaci.2015.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wark PA, Johnston SL, Bucchieri F, Powell R, Puddicombe S, Laza-Stanca V, et al. Asthmatic bronchial epithelial cells have a deficient innate immune response to infection with rhinovirus. The Journal of experimental medicine. 2005;201(6):937–947. doi: 10.1084/jem.20041901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.James KM, Peebles RS, Jr, Hartert TV. Response to infections in patients with asthma and atopic disease: an epiphenomenon or reflection of host susceptibility? J Allergy Clin Immunol. 2012;130:343–351. doi: 10.1016/j.jaci.2012.05.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clark TJ. Diurnal rhythm of asthma. Chest. 1987;91(6 Suppl):137S–141S. doi: 10.1378/chest.91.6_supplement.137s. [DOI] [PubMed] [Google Scholar]
- 8.Green CB, Takahashi JS, Bass J. The meter of metabolism. Cell. 2008;134(5):728–742. doi: 10.1016/j.cell.2008.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haspel JA, Chettimada S, Shaik RS, Chu JH, Raby BA, Cernadas M, et al. Circadian rhythm reprogramming during lung inflammation. Nature communications. 2014;5:4753. doi: 10.1038/ncomms5753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gibbs JE, Beesley S, Plumb J, Singh D, Farrow S, Ray DW, et al. Circadian timing in the lung; a specific role for bronchiolar epithelial cells. Endocrinology. 2009;150(1):268–276. doi: 10.1210/en.2008-0638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gibbs J, Ince L, Matthews L, Mei J, Bell T, Yang N, et al. An epithelial circadian clock controls pulmonary inflammation and glucocorticoid action. Nature medicine. 2014;20(8):919–926. doi: 10.1038/nm.3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pekovic-Vaughan V, Gibbs J, Yoshitane H, Yang N, Pathiranage D, Guo B, et al. The circadian clock regulates rhythmic activation of the NRF2/glutathione-mediated antioxidant defense pathway to modulate pulmonary fibrosis. Genes & development. 2014;28(6):548–560. doi: 10.1101/gad.237081.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Silver AC, Arjona A, Walker WE, Fikrig E. The circadian clock controls toll-like receptor 9-mediated innate and adaptive immunity. Immunity. 2012;36(2):251–261. doi: 10.1016/j.immuni.2011.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Walter MJ, Morton JD, Kajiwara N, Agapov E, Holtzman MJ. Viral induction of a chronic asthma phenotype and genetic segregation from the acute response. The Journal of clinical investigation. 2002;110(2):165–175. doi: 10.1172/JCI14345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Edgar RS, Stangherlin A, Nagy AD, Nicoll MP, Efstathiou S, O’Neill JS, et al. Cell autonomous regulation of herpes and influenza virus infection by the circadian clock. Proc Natl Acad Sci U S A. 2016;113(36):10085–10090. doi: 10.1073/pnas.1601895113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sundar IK, Ahmad T, Yao H, Hwang JW, Gerloff J, Lawrence BP, et al. Influenza A virus-dependent remodeling of pulmonary clock function in a mouse model of COPD. Scientific reports. 2015;4:9927. doi: 10.1038/srep09927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Benegiamo G, Mazzoccoli G, Cappello F, Rappa F, Scibetta N, Oben J, et al. Mutual antagonism between circadian protein period 2 and hepatitis C virus replication in hepatocytes. PloS one. 2013;8(4):e60527. doi: 10.1371/journal.pone.0060527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang G, Chen L, Grant GR, Paschos G, Song WL, Musiek ES, et al. Timing of expression of the core clock gene Bmal1 influences its effects on aging and survival. Science translational medicine. 2016;8(324):324ra316. doi: 10.1126/scitranslmed.aad3305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Storch KF, Paz C, Signorovitch J, Raviola E, Pawlyk B, Li T, et al. Intrinsic circadian clock of the mammalian retina: importance for retinal processing of visual information. Cell. 2007;130(4):730–741. doi: 10.1016/j.cell.2007.06.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Andersson KB, Winer LH, Mork HK, Molkentin JD, Jaisser F. Tamoxifen administration routes and dosage for inducible Cre-mediated gene disruption in mouse hearts. Transgenic research. 2010;19(4):715–725. doi: 10.1007/s11248-009-9342-4. [DOI] [PubMed] [Google Scholar]
- 21.Gibbs JE, Blaikley J, Beesley S, Matthews L, Simpson KD, Boyce SH, et al. The nuclear receptor REV-ERBalpha mediates circadian regulation of innate immunity through selective regulation of inflammatory cytokines. Proc Natl Acad Sci U S A. 2012;109(2):582–587. doi: 10.1073/pnas.1106750109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schneider WM, Chevillotte MD, Rice CM. Interferon-stimulated genes: a complex web of host defenses. Annual review of immunology. 2014;32:513–545. doi: 10.1146/annurev-immunol-032713-120231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wetzel JL, Fensterl V, Sen GC. Sendai virus pathogenesis in mice is prevented by Ifit2 and exacerbated by interferon. Journal of virology. 2014;88(23):13593–13601. doi: 10.1128/JVI.02201-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao J, Wohlford-Lenane C, Zhao J, Fleming E, Lane TE, McCray PB, Jr, et al. Intranasal treatment with poly(I*C) protects aged mice from lethal respiratory virus infections. Journal of virology. 2012;86(21):11416–11424. doi: 10.1128/JVI.01410-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hill DA, Baron S, Chanock RM. The effect of an interferon inducer on influenza virus. Bulletin of the World Health Organization. 1969;41(3):689–693. [PMC free article] [PubMed] [Google Scholar]
- 26.Gerone PJ, Hill DA, Appell LH, Baron S. Inhibition of respiratory virus infections of mice with aerosols of synthetic double-stranded ribonucleic Acid. Infection and immunity. 1971;3(2):323–327. doi: 10.1128/iai.3.2.323-327.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jackson DJ, Gangnon RE, Evans MD, Roberg KA, Anderson EL, Pappas TE, et al. Wheezing rhinovirus illnesses in early life predict asthma development in high-risk children. American journal of respiratory and critical care medicine. 2008;178(7):667–672. doi: 10.1164/rccm.200802-309OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim EY, Battaile JT, Patel AC, You Y, Agapov E, Grayson MH, et al. Persistent activation of an innate immune response translates respiratory viral infection into chronic lung disease. Nature medicine. 2008;14(6):633–640. doi: 10.1038/nm1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Byers DE, Alexander-Brett J, Patel AC, Agapov E, Dang-Vu G, Jin X, et al. Long-term IL-33-producing epithelial progenitor cells in chronic obstructive lung disease. The Journal of clinical investigation. 2013;123(9):3967–3982. doi: 10.1172/JCI65570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu K, Byers DE, Jin X, Agapov E, Alexander-Brett J, Patel AC, et al. TREM-2 promotes macrophage survival and lung disease after respiratory viral infection. The Journal of experimental medicine. 2015;212(5):681–697. doi: 10.1084/jem.20141732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Castanon-Cervantes O, Wu M, Ehlen JC, Paul K, Gamble KL, Johnson RL, et al. Dysregulation of inflammatory responses by chronic circadian disruption. Journal of immunology. 2010;185(10):5796–5805. doi: 10.4049/jimmunol.1001026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sellix MT, Evans JA, Leise TL, Castanon-Cervantes O, Hill DD, DeLisser P, et al. Aging differentially affects the re-entrainment response of central and peripheral circadian oscillators. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2012;32(46):16193–16202. doi: 10.1523/JNEUROSCI.3559-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moore WC, Fitzpatrick AM, Li X, Hastie AT, Li H, Meyers DA, et al. Clinical heterogeneity in the severe asthma research program. Annals of the American Thoracic Society. 2013;10(Suppl):S118–124. doi: 10.1513/AnnalsATS.201309-307AW. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bacharier LB, Cohen R, Schweiger T, Yin-Declue H, Christie C, Zheng J, et al. Determinants of asthma after severe respiratory syncytial virus bronchiolitis. The Journal of allergy and clinical immunology. 2012;130(1):91–100. e103. doi: 10.1016/j.jaci.2012.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nguyen KD, Fentress SJ, Qiu Y, Yun K, Cox JS, Chawla A. Circadian gene Bmal1 regulates diurnal oscillations of Ly6C(hi) inflammatory monocytes. Science. 2013;341(6153):1483–1488. doi: 10.1126/science.1240636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hwang JW, Sundar IK, Yao H, Sellix MT, Rahman I. Circadian clock function is disrupted by environmental tobacco/cigarette smoke, leading to lung inflammation and injury via a SIRT1-BMAL1 pathway. FASEB J. 2014;28(1):176–194. doi: 10.1096/fj.13-232629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kalamvoki M, Roizman B. Circadian CLOCK histone acetyl transferase localizes at ND10 nuclear bodies and enables herpes simplex virus gene expression. Proc Natl Acad Sci U S A. 2010;107(41):17721–17726. doi: 10.1073/pnas.1012991107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gagnidze K, Hajdarovic KH, Moskalenko M, Karatsoreos IN, McEwen BS, Bulloch K. Nuclear receptor REV-ERBalpha mediates circadian sensitivity to mortality in murine vesicular stomatitis virus-induced encephalitis. Proc Natl Acad Sci U S A. 2016;113(20):5730–5735. doi: 10.1073/pnas.1520489113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Majumdar T, Dhar J, Patel S, Kondratov R, Barik S. Circadian transcription factor BMAL1 regulates innate immunity against select RNA viruses. Innate immunity. 2017;23(2):147–154. doi: 10.1177/1753425916681075. [DOI] [PubMed] [Google Scholar]
- 40.Hetzel MR, Clark TJ. Comparison of normal and asthmatic circadian rhythms in peak expiratory flow rate. Thorax. 1980;35(10):732–738. doi: 10.1136/thx.35.10.732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Refinetti R, Lissen GC, Halberg F. Procedures for numerical analysis of circadian rhythms. Biol Rhythm Res. 2007;38(4):275–325. doi: 10.1080/09291010600903692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sugita K, Maru M, Sato K. A sensitive plaque assay for Sendai virus in an established line of monkey kidney cells. Japanese journal of microbiology. 1974;18(3):262–264. doi: 10.1111/j.1348-0421.1974.tb00955.x. [DOI] [PubMed] [Google Scholar]
- 43.Stappenbeck TS, Virgin HW. Accounting for reciprocal host-microbiome interactions in experimental science. Nature. 2016;534(7606):191–199. doi: 10.1038/nature18285. [DOI] [PubMed] [Google Scholar]
- 44.Balzar S, Fajt ML, Comhair SA, Erzurum SC, Bleecker E, Busse WW, et al. Mast cell phenotype, location, and activation in severe asthma. Data from the Severe Asthma Research Program. American journal of respiratory and critical care medicine. 2011;183(3):299–309. doi: 10.1164/rccm.201002-0295OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chung KF, Wenzel SE, Brozek JL, Bush A, Castro M, Sterk PJ, et al. International ERS/ATS guidelines on definition, evaluation and treatment of severe asthma. The European respiratory journal. 2014;43(2):343–373. doi: 10.1183/09031936.00202013. [DOI] [PubMed] [Google Scholar]
- 46.Eisenberg E, Levanon EY. Human housekeeping genes, revisited. Trends in genetics : TIG. 2013;29(10):569–574. doi: 10.1016/j.tig.2013.05.010. [DOI] [PubMed] [Google Scholar]
- 47.Genser B, Cooper PJ, Yazdanbakhsh M, Barreto ML, Rodrigues LC. A guide to modern statistical analysis of immunological data. BMC immunology. 2007;8:27. doi: 10.1186/1471-2172-8-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Persson CG, Erjefalt JS, Korsgren M, Sundler F. The mouse trap. Trends in pharmacological sciences. 1997;18(12):465–467. doi: 10.1016/s0165-6147(97)01142-5. [DOI] [PubMed] [Google Scholar]
- 49.Messmer MN, Kokolus KM, Eng JW, Abrams SI, Repasky EA. Mild cold-stress depresses immune responses: Implications for cancer models involving laboratory mice. BioEssays : news and reviews in molecular, cellular and developmental biology. 2014;36(9):884–891. doi: 10.1002/bies.201400066. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.