Abstract
Sonic Hedgehog (Shh) signaling regulates the patterning of ventral spinal cord through the effector Gli family of transcription factors. Previous in vitro studies showed that an E3 ubiquitin ligase containing Speckle-type POZ protein (Spop) targets Gli2 and Gli3 for ubiquitination and degradation, but the role of Spop in Shh signaling and mammalian spinal cord patterning remains unknown. Here, we show that loss of Spop does not alter spinal cord patterning, but it suppresses the loss of floor plate and V3 interneuron phenotype of Gli2 mutants, suggesting a negative role of Spop in Gli3 activator activity, Shh signaling and the specification of ventral cell fates in the spinal cord. This correlates with a moderate but significant increase in the level of Gli3 protein in the Spop mutant spinal cords. Furthermore, loss of Spop restores the maximal Shh pathway activation and ventral cell fate specification in the Gli1;Sufu double mutant spinal cord. Finally, we show that loss of Spop-like does not change the spinal cord patterning in either wild type or Spop mutants, suggesting that it does not compensate for the loss of Spop in Shh signaling and spinal cord patterning. Therefore, our results demonstrate a negative role of Spop in the level and activity of Gli3, Shh signaling and ventral spinal cord patterning.
Keywords: neural patterning, ubiquitin ligase, Gli1, Gli2, Sufu, Spopl
1. Introduction
Sonic hedgehog (Shh), a member of the family of Hedgehog (Hh) signaling proteins, plays a central role in the stereotypical arrangement of neural progenitors along the dorsal/ventral (D/V) axis of the ventral spinal cord in vertebrates (Briscoe and Therond, 2013). The Shh protein, produced in the notochord and subsequently in the floor plate, forms a ventral-to-dorsal gradient in the ventral spinal cord, and regulates the expression of a multitude of target genes through a morphogen-like mechanism. These genes, mostly encoding transcription factors, subsequently interact with each other to form an intricate gene regulatory network that helps to sharpen the borders between them and define the locations of various groups of neural progenitors (Cohen et al., 2013).
The direct transcriptional responses to Shh are mediated by the effector Glioma-associated oncogene (Gli) family of transcription factors (Matise and Joyner, 1999). There are three Gli proteins in mammals, Gli1, 2 and 3. In the absence of Hh ligands, Gli2 and Gli3 are phosphorylated by PKA, CK2 and Gsk3, and are ubiquitinated by Cul1/β-TrCP ubiquitin ligase (Pan et al., 2006; Wang et al., 2000). The ubiquitinated Gli3 is partially degraded in the proteasome to produce a strong transcriptional repressor (Gli3R), whereas ubiquitinated Gli2 mostly undergoes complete degradation. Loss of Gli3 repressor activity results in ventralization of the forebrain and severe polydactyly in the limbs, but very subtle defects in the D/V patterning of the spinal cord, suggesting that the requirements for Gli3R activity vary dependent on the developmental contexts (Hui and Joyner, 1993; Persson et al., 2002; Theil et al., 1999; Tole et al., 2000).
Shh inhibits the Cul1/β-TrCP-based proteolytic processing of Gli3 and degradation of Gli2, and turns them into transcriptional activators (Gli2A and Gli3A) (Humke et al., 2010; Pan et al., 2006; Wang et al., 2000). Loss of Gli2 results in the loss of floor plate and great reduction of V3 interneurons, ventral-most spinal cord cell types dependent on high concentrations of Shh, suggesting that Gli2 is the primary activator of the Shh pathway (Ding et al., 1998; Matise et al., 1998). Interestingly, loss of both Gli2 and Gli3 results in more complete loss of all floor plate and V3 interneurons, as well as the mixing of other ventral cell types including the motor neurons, V1 and V2 interneurons, suggesting that Gli3A also contributes to Shh pathway activation in the spinal cord (Bai et al., 2004; Lei et al., 2004). The third member of the Gli family, Gli1, whose transcription is dependent on Hh signaling, contributes to, but is not essential for, ventral spinal cord patterning (Bai et al., 2004; Park et al., 2000).
Suppressor of Fused (Sufu) inhibits Shh signaling and ventral spinal cord development in mammals (Cooper et al., 2005; Svard et al., 2006). In vitro studies suggested that Sufu represses Gli proteins both by sequestering them in the cytoplasm and by inhibiting Gli-mediated transcription in the nucleus (Barnfield et al., 2005; Cheng and Bishop, 2002; Ding et al., 1999; Dunaeva et al., 2003; Kogerman et al., 1999; Lin et al., 2014; Merchant et al., 2004; Murone et al., 2000; Paces-Fessy et al., 2004; Stone et al., 1999). Recent findings suggest that Hh signaling results in the primary cilium-dependent dissociation between Sufu and Gli proteins (Humke et al., 2010; Lin et al., 2014; Tukachinsky et al., 2010). Interestingly, we found that the floor plate and V3 interneurons failed to form in Gli1;Sufu double mutants, in striking contrast to normal spinal cord patterning in Gli1 mutants, suggesting that Sufu is required for the maximal activation of Shh signaling in the absence of Gli1 (Bai et al., 2002; Liu et al., 2012; Park et al., 2000). This positive function of Sufu correlates with the observation that the levels of Gli2 and Gli3 proteins are greatly reduced in the absence of Sufu, suggesting that Sufu may promote Shh signaling by protecting the Gli proteins from degradation (Chen et al., 2009; Jia et al., 2009; Wang et al., 2010).
In addition to Cul1-based proteolytic processing, Gli2 and Gli3 are also subject to degradation mediated by a ubiquitin ligase containing Cul3 and Speckle-type POZ protein (Spop) (Chen et al., 2009; Wang et al., 2010; Wen et al., 2010; Zhang et al., 2009; Zhang et al., 2006). It was reported that Spop specifically targeted activated Gli3 for degradation (Wen et al., 2010), which appears to be consistent with reports suggesting antagonistic roles of Sufu and Spop in Gli2 and Gli3 degradation (Chen et al., 2009; Wang et al., 2010). These in vitro studies raised the possibility that Spop may be responsible for the great reduction in the levels of Gli2 and Gli3 in Sufu mutants. However, the in vivo roles of Spop in Shh signaling and spinal cord development, especially its role in Gli degradation in the absence of Sufu, have not been revealed.
Here we show that loss of Spop does not change the patterning of ventral spinal cord. This lack of spinal cord patterning defects does not result from redundancy with another Speckle-type POZ protein, Spop-like (Spopl), as the Spopl;Spop double mutant spinal cord is also properly patterned along its D/V axis. Interestingly, the level of Gli3 protein increases significantly in the Spop mutant spinal cords, and removing Spop rescues the loss of floor plate and V3 interneurons in Gli2 mutants, suggesting that Spop negatively regulates Gli3A. On the other hand, no change in the level of Gli2 protein was observed in the absence of Spop, and the D/V patterning of Spop;Gli3 double mutant spinal cord resembles that of Gli3 mutants, suggesting that Gli2 is not the major target of Spop in the spinal cord. Furthermore, loss of Spop fails to restore the levels of Gli2 and Gli3 in Sufu mutants, but does exacerbate the ectopic activation of Shh signaling and ventralization of the spinal cord, and restores the formation of the floor plate and V3 interneurons in the Gli1;Sufu double mutant spinal cord. In summary, our data suggest that Spop negatively regulates Gli3A activity and Shh signaling, but does not underlie the drastic decrease in the levels of Gli2 and Gli3 in Sufu mutants.
2. Materials and Methods
2.1 Animal work
Spoptm1a(KOMP)Mbp (SpoplacZKI) and Spopltm1(KOMP)Vlcg were generated by Knockout Mouse Project (http://www.komp.org). The Spop loss-of-function allele used in the current study was generated by breeding SpoplacZKI mutant strain to Tg(ACTFLPe)9205Dym/J (Rodríguez et al., 2000) to remove the lacZ cassette flanked by FRT sites, and subsequently to Tg(EIIa-Cre)C5379Lmgd/J (Lakso et al., 1996) to delete exons 4 and 5. Other mouse strains used in this study were Gli2tm2.1Alj in which a lacZ inserted into exon 2 abolished Gli2 expression (Bai and Joyner, 2001), Gli3Xt-J in which exons 10 to 15 were deleted resulting in a truncation following the first zinc finger motif (Hui and Joyner, 1993; Maynard et al., 2002), Gli1tm2Alj in which a lacZ cassette replaced exons 3 to 11 leading to expression of a non-functional Gli1 mutant that lacked zinc finger domain at best (Bai et al., 2002), and Sufutm1Rto in which a neomycin resistance cassette replaced exon 1 and prevented the expression of Sufu (Svard et al., 2006). All strains were kept on 129S2/SvPasCrl or C3H/HeNCrl (Charles River Lab) background and genotyped as previously described. The animal work in this study was approved by the IACUC at the Penn State University.
2.2 Quantitative real time polymerase chain reaction (qRT-PCR)
RNA was extracted with NucleoSpin RNA kit (Macherey-Nagel), and 1μg of RNA was used to synthesize cDNA with qScript cDNA Synthesis Kit (Quanta Biosciences, #95047-100). qRT-PCR was then performed in a StepOne Plus Real-time PCR system (Applied Biosystems) with PerfeCTa SYBR Green SuperMix (Quanta Biosciences). Primers used: Spopl, 5′-TCAACGTTTTCTTCAGGCCC-3′ and 5′-AAATCCCCAGTCCTTCCCCT-3′; Gapdh, 5′-GTCGGTGTGAACGGATTTGG-3′ and 5′-GACTCCACGACATACTCAGC-3′; Ptch1, 5′-CTCCAAGTGTCGTCCGGTTT-3′ and 5′-ACCCATTGTTCGTGTGACCA-3′; Gli1, 5′-CGTTTGAAGGCTGTCGGAAG-3′ and 5′-GCGTCTTGAGGTTTTCAAGGC-3′.
2.3 Immunoblot analysis
Embryos were homogenized in embryo lysis buffer (10mM Hepes/400mM NaCl/0.1mM EGTA/0.5mM DTT/10% glycerol/1% Triton X-100, pH 7.9) containing protease inhibitors. Protein concentration was measured with Pierce BCA Protein Assay kit (Thermo-Fisher) and equal amount of total proteins were loaded to each well. Nitrocellulose membranes were blocked with PBS/5% chicken serum and subject to primary antibody incubation in PBS/0.1% Tween-20/5% BSA, three washes in PBS/0.1% Tween-20, secondary antibody incubation in PBS/5% chicken serum and three washes in PBS/0.1% Tween-20. Primary antibodies used were Gli2 (R&D systems, AF3635, 1:500), Gli3 (R&D systems, AF3690, 1:200) and β-tubulin. Secondary antibodies were conjugated with fluorophores for detection with Odyssey Imaging Systems (LI-COR).
2.4 Immunohistochemistry
Embryos were lightly fixed with 4% paraformaldehyde (PFA) in PBS and cryosectioned at 10μm thickness. Sections were blocked in blocking buffer (PBS/1% goat serum/0.1% Triton X-100) and subject to primary antibody incubation, three washes, secondary antibody incubation and three washes. Antibody incubation and washes were all performed in blocking buffer. Primary antibodies used were Foxa2 (DSHB, 4C7, 1:40), Nkx2.2 (DSHB, 74.5A5, 1:40), Olig2 (Millipore, AB9610, 1:1000), Nkx6.1 (DSHB, F55A12, 1:500) and Pax6 (DSHB, 1:500). Secondary antibodies were Cy3-conjugated (Jackson Lab, 1:500). Slides were then mounted in DABCO (Sigma-Aldrich). Images were obtained with a Nikon E600 microscope and a QImaging Micropublisher Digital Camera.
3. Results
3.1 Loss of Spop leads to a moderate but significant increase in the level of Gli3 protein
To reveal the roles of Spop in Shh signaling and the D/V patterning of the spinal cord, we characterized two Spop loss-of-function mutant alleles. One mutant allele, SpoplacZKI, contains a lacZ cassette knocked into the third intron of Spop, thus expresses lacZ under the control of Spop cis-regulatory elements (Fig. 1A). The other mutant allele, SpopΔEx, bears a deletion of the 4th and 5th exons of Spop, which was predicted to truncate the Spop protein at residue 26 due to a frame shift. Immunoblot analyses confirmed the absence of Spop protein in embryonic day (E) 10.5 homozygous mutant embryos for both alleles (Cai and Liu, 2016). Both SpoplacZKI and SpopΔEx homozygous mutants exhibited neonatal lethality with skeletal defects (Cai and Liu, 2016). A small number of mutant embryos exhibited exencephaly (n= 11/157 homozygotes and 5/407 heterozygotes, combining both alleles) and spina bifida (n= 4/157 homozygotes, combining both alleles), suggesting that Spop may play a role in the development of the nervous system. For the rest of this study, we used SpopΔEx exclusively to investigate the roles of Spop in spinal cord development, and will abbreviate it as Spop mutant or Spop−.
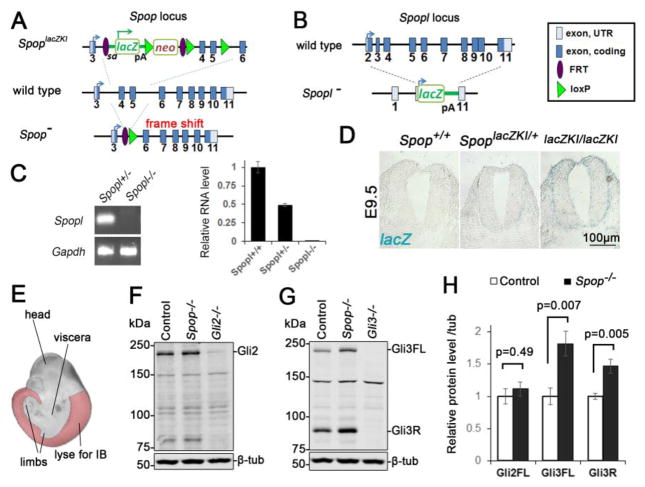
Fig. 1. A moderate increase in Gli3 protein level in the Spop mutant spinal cords.
(A) A schematic illustration of Spop loss-of-function mutant alleles. SpoplacZKI contained a lacZ and neomycin resistance cassette. SpopΔEx was generated from recombination of introduced FRT and loxP sites that deletes the 4th and 5th exons. This deletion also resulted in a frame shift that truncated the protein. (B) A schematic illustration of Spopl null mutant allele, in which a lacZ reporter replaced the entire protein-coding region of Spopl. pA: polyA signal. (C) Quantitative real time PCR showing the absence of Spopl transcript in E9.5 Spopl mutant embryos (mean ± SEM, n=3 wild type, 4 Spopl+/− and 5 Spopl−/− embryos). (D) X-gal staining of E9.5 SpoplacZKI heterozygotes and homozygotes showing Spop expression in the neural tube. (E) A schematic illustration of tissue (highlighted region) used for immunoblot in (F) and (G). The trunks of E10.5 embryos were freed of viscera and limbs to minimize the effect of Gli proteins in these tissues. (F) Immunoblots with antibodies against Gli2 and β-tubulin. (G) Immunoblots with antibodies against Gli3 and β-tubulin. (H) Quantification of (F) and (G) (mean ± SEM from n=6 embryos per group). Student’s t-test showed a significant increase in the levels of Gli3FL and Gli3R, but not Gli2 in Spop mutants.
Previous in vitro studies suggested that Spop targeted overexpressed Gli2 and Gli3 for degradation (Chen et al., 2009; Wang et al., 2010; Wen et al., 2010; Zhang et al., 2009). To investigate whether loss of Spop stabilizes Gli2 and Gli3 in the spinal cord, we examined the levels of these proteins in E10.5 embryos through immunoblotting analyses. We removed the heads, viscera and limbs from E10.5 embryos to specifically determine the protein levels in the spinal cord (Fig. 1E). Consistent with a role of Spop in Gli3 degradation, the levels of both full-length (Gli3FL) and repressor (Gli3R) forms of Gli3 were significantly increased in the Spop mutant spinal cord (Fig. 1G and H). In contrast, we did not observe an increase in the level of Gli2 in Spop mutants (Fig. 1F and H).
3.2 The D/V patterning of the Spop mutant spinal cord was normal
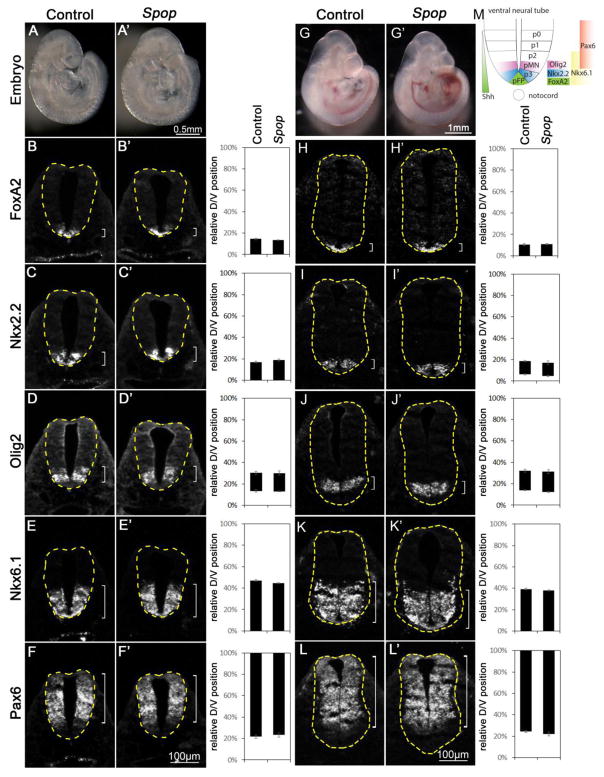
Most Spop mutant embryos resembled wild type morphologically (Fig. 2A, A′, G and G′). To investigate whether the increase in the level of Gli3 disrupted the D/V patterning of the spinal cord, we examined the expression of transcription factors specific for various groups of progenitor cells in the ventral spinal cord. At E9.5, Foxa2 and Nkx2.2 were both expressed in the ventral-most spinal cord (Fig. 2B and C), while at E10.5, Foxa2 and Nkx2.2 labeled the floor plate (FP) and progenitors of V3 interneurons (p3), respectively (Fig. 2H, I and M). Olig2 labeled motor neuron progenitors (pMN) immediately dorsal to p3 (Fig. 2D, J and M). Nearly all ventral progenitor cells (p1, 2, 3 and pMN) expressed Nkx6.1 (Fig. 2E, K and M). Pax6 expression was present in dorsal and lateral regions of the spinal cord (Fig. 2F, L and M). We examined the expression patterns of these genes in both E9.5 and E10.5 Spop mutant spinal cords and found no change compared to their littermate controls (Fig. 2B′–F′ and H′–L′), suggesting that the moderate increase in the level of Gli3 was not sufficient to alter the early patterning of the ventral spinal cord. Consistent with this observation, the levels of Ptch1 and Gli1 expression were comparable between wild type and Spop mutant embryos, suggesting that Shh signaling activity was not affected by loss of Spop (Fig. S1A).
Fig. 2. Normal ventral patterning of the Spop mutant spinal cords.
(A, A′) Lateral views of E9.5 control (A) and Spop (A′) mutant embryos. (B–F′) Immunofluorescent images of transverse sections of E9.5 control (n=4 embryos) and Spop mutant (n=4 embryos) spinal cords at the forelimb level with indicated antibodies. (G, G′) Lateral views of E10.5 control (G) and Spop (G′) mutant embryos. (H–L′) Immunofluorescent images of transverse sections of E10.5 control (n=4 embryos) and Spop mutant (n=4 embryos) spinal cords at the forelimb level. The spinal cords are outlined with dash lines. Brackets indicate the expression domains. The span of each domain along the D/V axis is quantified and shown on the right. Relative D/V position is shown as the distance to the ventral-most point of the spinal cord as a percentage of the entire D/V span of the spinal cord. Student’s t-test suggests no significant difference between Spop mutant and control groups. (M) A schematic illustration of ventral spinal cord patterning at E10.5 depicting the arrangement of various progenitor groups and the expression domains of marker genes in wild type embryos.
3.3 Loss of Spopl does not alter spinal cord patterning in wild type or Spop mutants
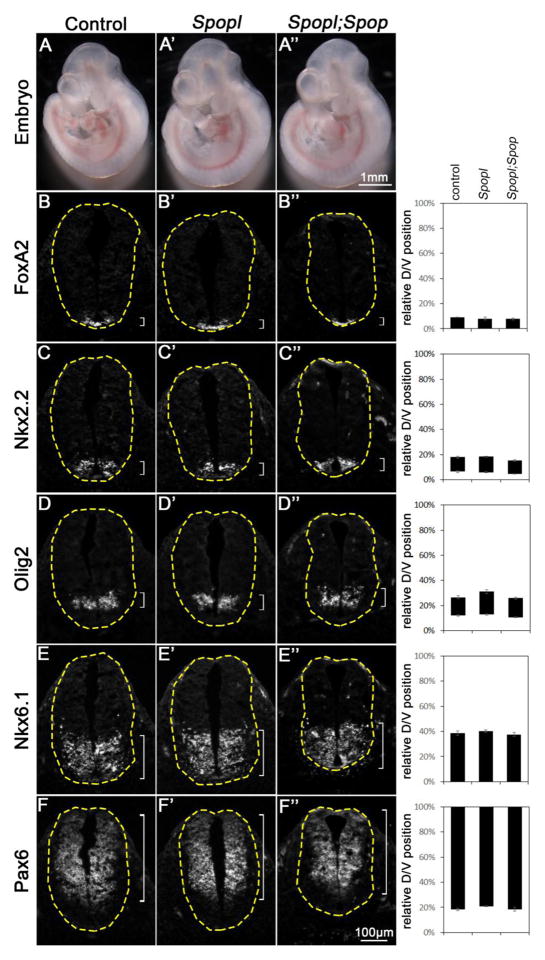
Spopl shares similar substrate specificity with Spop, albeit exhibiting weaker ubiquitin-ligase activity (Choo et al., 2010; Errington et al., 2012). To investigate whether Spopl functionally compensates for the loss of Spop in mouse development, especially the D/V patterning of the spinal cord, we acquired a Spopltm1(KOMP)Vlcg mutant strain in which the entire coding region of Spopl was replaced with a lacZ reporter from KOMP (http://www.komp.org) (Fig. 1B). Since the entire coding region is deleted, Spopltm1(KOMP)Vlcg (hereafter abbreviated as Spopl−) is an absolute null mutant allele. By qRT-PCR, we found that Spopl was expressed in E9.5 mouse embryos, and its expression was reduced by half in Spopl+/− and undetectable in Spopl−/− embryos (Fig. 1C). Spopl homozygous mutants were healthy and fertile (n >55 mutants weaned), and the E10.5 mutant spinal cords were patterned properly along the D/V axis, suggesting that Spopl did not play an essential role in mouse development and spinal cord D/V patterning (Fig. 3). Importantly, Spopl;Spop double mutant mice exhibit postnatal lethality, similar to Spop single mutants. We also found that the spinal cords were properly patterned in Spopl;Spop double mutants (Fig. 3). Furthermore, loss of Spopl did not affect the expression levels of Ptch1 and Gli1 (Fig. S1B). These data suggest that the Speckle-type POZ proteins, Spop and Spopl, are dispensable for the patterning of ventral spinal cord.
Fig. 3. Normal ventral patterning of the Spopl mutant and Spopl;Spop double mutant spinal cords.
(A–A″) Lateral views of E10.5 control (A), Spop mutant (A′) and Spopl;Spop double mutant (A″) embryos. (B–F″) Immunofluorescent images of transverse sections of E10.5 control (n=4 embryos), Spopl mutant (n=4 embryos) and Spopl;Spop double mutant (n=4 embryos) spinal cords at the forelimb level with indicated antibodies are shown. The spinal cords are outlined with dash lines. Brackets indicate the expression domains. The span of each domain along the D/V axis is quantified and shown on the right. Relative D/V position is shown as the distance to the ventral-most point of the spinal cord as a percentage of the entire D/V span of the spinal cord. Student’s t-test suggests no significant difference between Spopl;Spop double mutant, Spopl mutant and control groups. The morphology and spinal cord neural patterning of both Spopl mutant and Spopl;Spop double mutant resemble wild type.
3.4 Loss of Spop restores normal patterning in the Gli2 mutant spinal cord
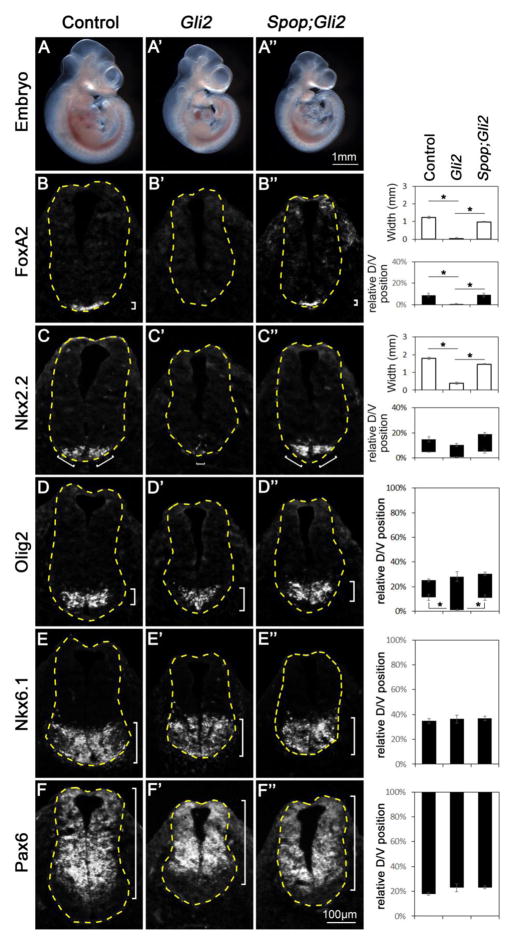
Neither Gli1 nor Gli3 was essential for the mammalian ventral spinal cord development (Bai et al., 2002; Park et al., 2000; Persson et al., 2002); however, the importance of these Gli proteins in supporting maximal Shh signaling and ventral spinal cord development was revealed with simultaneous reduction or ablation of Gli2, suggesting that these three Gli proteins share redundant functions in spinal cord patterning (Bai et al., 2004; Lei et al., 2004; Park et al., 2000). This functional redundancy among the three mouse Gli proteins suggests that a moderate change in the activities of any member of this family may have a more obvious impact in the spinal cord pattering in the absence of another Gli protein. We hence removed Gli2 in Spop mutants to determine whether the increase in the level of Gli3 rescues the spinal cord patterning defects resulting from loss of Gli2. The morphology of Spop;Gli2 double mutant embryos resembled that of Gli2 mutants at E10.5 (Fig. 4A–A″). As reported previously (Ding et al., 1998; Matise et al., 1998), the Foxa2-expressing FP was missing and Nkx2.2-expressing p3 cells were diminished in the Gli2 mutant spinal cord (Fig. 4B′, C′), whereas Olig2 expression was expanded to the ventral midline (Fig. 4D′). Interestingly, Foxa2 was expressed, and Nkx2.2 expression was increased, in the Spop;Gli2 double mutant spinal cord (Fig. 4B″ and C″). Meanwhile, Olig2 expression was excluded from the ventral-most part of the Spop;Gli2 double mutant spinal cord (Fig. 4D″). The Nkx6.1 expression domain, which includes cell types that do not require high levels of Shh pathway activation, and Pax6 expression domain, which includes cell types that are inhibited by high levels of Shh activation, were similar in wild type, Gli2 mutants and Spop;Gli2 double mutants (Fig. 4E–F″). We observed similar restoration of the spinal cord patterning in E9.5 Spop;Gli2 double mutants (Fig. S2). The restored formation of FP and p3 in the Spop;Gli2 double mutant spinal cords suggested an increase in Shh pathway activation. Supporting this hypothesis, the expression levels of Ptch1 and Gli1 were decreased in Gli2 mutant embryos, and restored in Spop;Gli2 double mutants (Fig. S1C), suggesting that loss of Spop enhanced Shh signaling activity, which underlies the rescue of spinal cord patterning defect caused by loss of Gli2. Theoretically, changes in the activities of both Gli1 and Gli3 could contribute to this increase; however, previous in vitro studies suggested that Spop did not target Gli1 for degradation (Chen et al., 2009; Zhang et al., 2009). Therefore, the increase in the activation of the Shh pathway in Spop;Gli2 double mutants likely results from an increase in the Gli3A activity (Fig. 1G), which is better revealed in the absence of Gli2.
Fig. 4. Restored floor plate and V3 interneuron progenitor fates in Spop;Gli2 double mutants.
(A–A″) Lateral views of E10.5 control (A), Gli2 mutant (A′) and Spop;Gli2 double mutant (A″) embryos. (B–F″) Immunofluorescent images of transverse sections of E10.5 control (n=3 embryos), Gli2 mutant (n=3 embryos) and Spop;Gli2 (n=3 embryos) double mutant spinal cords at the forelimb level with indicated antibodies. The spinal cords are outlined with dash lines. Brackets indicate the expression domains. The span of each domain along D/V axis is shown on the right. Relative D/V position is shown as the distance to the ventral-most point of the spinal cord as a percentage of the entire D/V span of the spinal cord. The width of Foxa2 and Nkx2.2 domains is also quantified (hollow columns). (B–B″) Foxa2 was present in the ventral-most region of the control and diminished in the Gli2 mutant, but restored in the Spop;Gli2 double mutant spinal cords. Student’s t-test was employed to compare the width of floor plate. *: p<0.05. (C–C″) Nkx2.2 was present in juxtaposition to floor plate of the control but diminished in the Gli2 mutant, and was restored in the Spop;Gli2 double mutant spinal cords. Student’s t-test was employed to compare the size of V3 interneuron progenitor domain. *: p<0.05. (D–D″) Olig2 expression was excluded from the ventral-most region of control and Spop;Gli2 double mutant but expanded ventrally in Gli2 mutant spinal cords. Student’s t-test was employed to compare the distance from the Olig2 expression domain to the ventral-most point of the spinal cords. *: p<0.05. (E–F″) Nkx6.1 and Pax6 expression domains remained unchanged in the Gli2 mutant and Spop;Gli2 double mutant spinal cords.
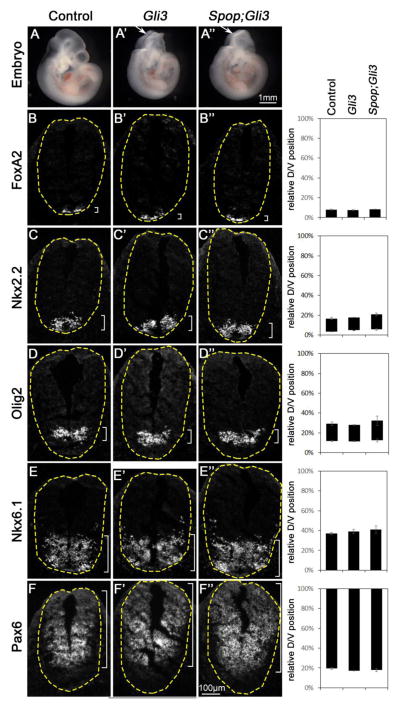
3.5 The ventral spinal cord was properly patterned in Spop;Gli3 double mutants
In contrast to the moderate increase in the level of Gli3, the level of Gli2 was not changed in Spop mutant neural tube (Fig. 1F and H). However, we could not rule out the possibility that Spop might regulate Gli2 activity without affecting its protein level. As the presence of Gli3 repressor in the spinal cord may conceal the effect of any moderate increase in Gli2 activator activity, we sought to reveal such increase by removing Gli3 in Spop mutants. As previously reported, loss of Gli3 led to frequent exencephaly at E10.5 (Fig. 5A and A′), but had no impact on the expression of Foxa2, Nkx2.2, Olig2, Nkx6.1 and Pax6 (Fig. 5B–F′). If loss of Spop increased Gli2 activity, a dorsal expansion of FoxA2, Nkx2.2, Olig2 or Nkx6.1 expression would be expected in Spop;Gli3 double mutants. In contrast, Spop;Gli3 double mutant embryos resembled Gli3 mutants morphologically (Fig. 5A″), and did not exhibit any change in the expression patterns of these genes in the ventral spinal cord (Fig. 5B″–F″). Similarly, Spop;Gli3 double mutants resembled Gli3 mutants both in morphology and in spinal cord patterning at E9.5 (Fig. S3). These data suggest that loss of Spop does not result in a noticeable increase in Gli2 activity, consistent with the unchanged Gli2 level in Spop mutants (Fig. 1F, H).
Fig. 5. Spop;Gli3 double mutants resembled Gli3 mutants in ventral spinal cord patterning.
(A–A″) Lateral views of E10.5 control (A), Gli3 mutant (A′) and Spop;Gli3 double mutant (A″) embryos. Arrows in A′ and A″ indicate exencephaly. (B–F″) Immunofluorescent images of transverse sections of E10.5 spinal cords in Gli3 mutant (n=4 embryos) and Spop;Gli3 double mutant (n=5 embryos) resembled those in the control (n=5 embryos) at the forelimb level with indicated antibodies. The spinal cords are outlined with dash lines. Brackets indicate the expression domains. The span of each domain is quantified and shown on the right. Relative D/V position is shown as the distance to the ventral-most point of the spinal cord as a percentage of the entire D/V span of the spinal cord. Student’s t-test suggests no significant difference between Spop;Gli3 double mutant, Gli3 mutant and control groups.
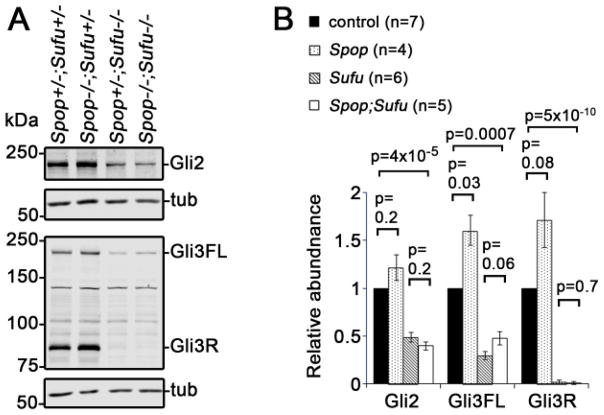
3.6 Loss of Spop exacerbates the spinal cord patterning defects in Sufu mutants
A well-known negative regulator of Shh signaling, Sufu, is expressed in the spinal cord and plays an essential role in spinal cord D/V patterning (Cooper et al., 2005; Stone et al., 1999; Svard et al., 2006). We and others have previously shown that the levels of Gli2 and Gli3 were drastically reduced in Sufu mutants, suggesting that in addition to its inhibitory function, Sufu may also protect Gli2 and Gli3 from degradation (Chen et al., 2009; Jia et al., 2009; Wang et al., 2010). It was suggested that Spop might underlie the degradation of Gli2 and Gli3 in the absence of Sufu, and prevent the full activation of Hh pathway (Chen et al., 2009; Wang et al., 2010). To test this hypothesis, we performed immunoblot analyses on E9.5 Spop;Sufu double mutant embryos. We found a moderate increase in the level of Gli3FL in Spop;Sufu double mutants than in Sufu single mutants, although the difference did not reach statistical significance (Fig. 6). In contrast, the levels of Gli2 and Gli3R were comparable between Spop;Sufu double mutants and Sufu single mutants. Nevertheless, the levels of both Gli2 and Gli3 in Spop;Sufu double mutants were still significantly lower than those in wild type littermates, suggesting that Spop was not solely accountable for the drastic reduction of Gli2 and Gli3 in Sufu mutants (Fig. 6).
Fig. 6. Moderate increase in the level of Gli3, but not Gli2, in Spop;Sufu double mutant embryos.
(A) Immunoblots of E9.5 embryo lysates with indicated antibodies. (B) Quantitative analyses of the levels of Gli2, Gli3FL and Gli3R (normalized to β-tubulin, mean ± SEM) based on the data of immunoblot analyses. Statistical significance was determined with Student’s t-test.
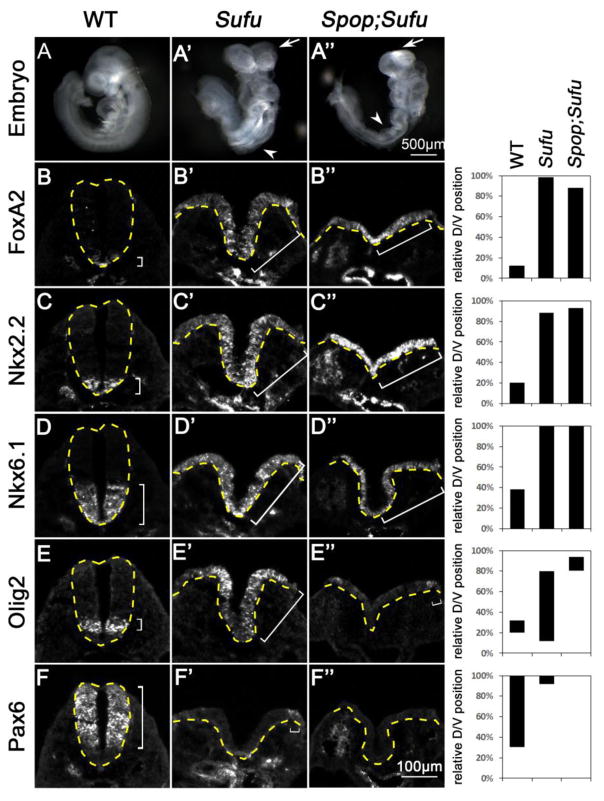
We next sought to investigate whether the moderate increase in the level of Gli3FL had an impact on the morphology and spinal cord patterning of Spop;Sufu double mutants. Consistent with our previous report (Liu et al., 2012), E9.5 Sufu mutant embryos exhibited twisted body axis and severe neural tube closure defects (Fig. 7A′). The Sufu mutant spinal cord was severely ventralized, with strong Foxa2, Nkx2.2 and Nkx6.1 expression throughout the D/V axis (Fig. 7B′–D′). Olig2 expression was strong in the dorsal spinal cord, but appeared weaker and scattered ventrally, consistent with its being activated by low levels of Shh signaling but repressed by high levels of Shh signaling (Fig. 7E′). Consistent with the hyperactivation of Shh signaling, Pax6 expression was restricted to just a few cells in the dorsal spinal cord of Sufu mutants (Fig. 7F′). Spop;Sufu double mutant embryos failed to turn, and exhibited severe neural tube closure defects like Sufu single mutants (Fig. 7A″). Similar to Sufu mutants, the Spop;Sufu double mutant spinal cord exhibited widespread expression of Foxa2, Nkx2.2 and Nkx6.1 (Fig. 7B″–D″). Furthermore, Olig2 expression was only found in a few dorsal cells (Fig. 7E″), and Pax6 expression was completely absent, in the Spop;Sufu double mutant spinal cord (Fig. 7F″). These data indicate that loss of Spop leads to more severe ventralization of the spinal cords in the absence of Sufu.
Fig. 7. Loss of Spop exacerbated the ventralization of the Sufu mutant spinal cord.
(A–C) Lateral views of E9.5 wild type (A), Sufu mutant (A′) and Spop;Sufu double mutant (A″) embryos. Arrows in B and C indicate exencephaly; arrowheads indicate spina bifida. (B–F″) Immunofluorescent images of transverse sections of E9.5 spinal cords at the thoracic level. The spinal cords are outlined with dash lines. Brackets indicate the expression domains. (B–C″) Foxa2 and Nkx2.2 were expressed in the ventral-most region of wild type but expanded to the dorsal region in the Sufu mutant and Spop;Sufu double mutant spinal cords. (D–D″) Nkx6.1 expression domain was expanded to the dorsal region in both Sufu mutant and Spop;Sufu double mutant spinal cords. (E–E″) Olig2 expression was shifted to the dorsal region in Sufu mutant and even more dorsally shifted in Spop;Sufu double mutant spinal cords. (F–F″) Pax6 expression domain was shifted dorsally in Sufu mutant and was absent in Spop;Sufu double mutant spinal cord. n=2 embryos for each genotype were analyzed. The span of each domain is quantified and shown on the right. Relative D/V position is shown as the distance to the ventral-most point of the spinal cord as a percentage of the entire D/V span of the spinal cord.
Although it is formally possible that Spop may regulate an unknown process that indirectly affects the D/V patterning of the spinal cord, the similarity between the morphological and patterning defects between Spop;Sufu double mutants and those of previously reported Ptch1 mutants (Goodrich et al., 1997) and Sufu;Gli3 double mutants (Liu et al., 2012) suggests that the extreme ventralization of the spinal cord was likely the direct result of the hyperactivation of the Shh pathway due to loss of both Spop and Sufu.
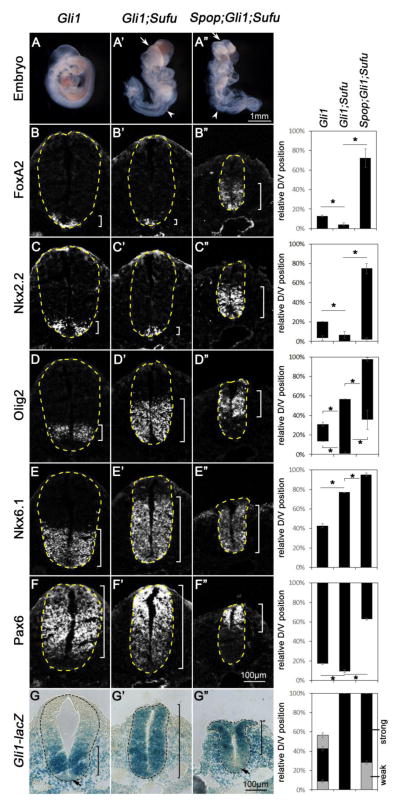
3.7 Loss of Spop restored the ventral-most cell fates in the Gli1; Sufu double mutant spinal cord
We previously found that the FP and p3, the ventral-most cell types in the spinal cord, were lost in Gli1;Sufu double mutants, but not in Gli1 single mutants, indicating a positive role of Sufu in Hh pathway activation (Liu et al., 2012). We hypothesized that Spop-mediated degradation of activated Gli2 and/or Gli3 in the absence of Sufu might underlie the loss of ventral cell types in Gli1;Sufu double mutants. Although the above immunoblot analyses (see Fig. 6) suggested that loss of Spop could not fully restore the levels of Gli2 and Gli3, we were encouraged by the exacerbation of the Sufu mutant spinal cord defects resulting from the additional loss of Spop, and tested the impact of loss of Spop on the spinal cord patterning defects of Gli1;Sufu double mutants. E9.5 Gli1;Sufu double mutant embryos exhibited less severe neural tube closure defects than Sufu single mutants, especially in the anterior part of the embryos (compare Fig. 8A′ to Fig. 7A′). Consistent with our previous report (Liu et al., 2012), fewer Foxa2- and Nkx2.2-expressing cells were present in the E9.5 Gli1;Sufu double mutant spinal cord compared to Gli1 single mutants (Fig. 8B–C′). Meanwhile, the Olig2 and Pax6 expression domains were expanded toward the ventral midline (Fig. 8D, D′, F and F′), suggesting that Sufu was required for the full activation of Hh pathway in the absence of Gli1. The Olig2 and Nkx6.1 domains were also expanded more dorsally in Gli1;Sufu double mutants than in Gli1 single mutants, suggesting that lower levels of ectopic activation of Shh signaling persisted in the double mutants (Fig. 8D–E′). In contrast, Spop;Gli1;Sufu triple mutants exhibited widespread edema in addition to more severe neural tube closure defects than those of Gli1;Sufu double mutants (Fig. 8A″). Interestingly, the expression domains of Foxa2, Nkx2.2 and Nkx6.1 were all dorsally expanded in Spop;Gli1;Sufu triple mutants, indicating higher levels of Shh pathway activation in the absence of Spop (Fig. 8B″, C″ and E″). Moreover, the expression of Olig2 and Pax6, which was inhibited by high levels of Shh signaling, was restricted to the dorsal regions of the spinal cord in Spop;Gli1;Sufu triple mutants (Fig. 8D″ and F″). These results suggest that Spop plays a negative role in Shh pathway activation and ventral spinal cord development, and accounts for the lack of maximal activation of Shh pathway in Gli1;Sufu double mutant embryos.
Fig. 8. Loss of Spop rescued the floor plate and V3 interneuron progenitor fates in Gli1;Sufu double mutant embryos.
(A–A″) Lateral views of E9.5 Gli1 mutant (A), Gli1;Sufu double mutant (A′) and Spop;Gli1;Sufu triple mutant (A″) embryos. Arrows in A′ and A″ indicate exencephaly; white arrowheads indicate spina bifida; red arrowheads indicate edema. (B–G″) Immunofluorescent (B–F″) or Xgal-stained (G–G″) images of transverse sections of the E9.5 Gli1 mutant (n=3 embryos), Gli1;Sufu double mutant (n=3 embryos) and Spop;Gli1;Sufu triple mutant (n=3 embryos) spinal cords at the thoracic level. The spinal cords are outlined with dash lines. Brackets indicate the expression domains. The span of each domain along the D/V axis is shown on the right. Relative D/V position is shown as the distance to the ventral-most point of the spinal cord as a percentage of the entire D/V span of the spinal cord. Student’s t-tests were performed to compare the dorsal and ventral borders of expression domains of various genes. *: p<0.05. (B–C″) The expression of Foxa2 and Nkx2.2 in the ventral-most region of Gli1 mutant was diminished in Gli1;Sufu double mutant, but expanded dorsally in the Spop;Gli1;Sufu triple mutant spinal cords. (D–D″) Olig2 expression was expanded both ventrally and dorsally in Gli1;Sufu double mutant, but was dorsally restricted in the Spop;Gli1;Sufu triple mutant spinal cords. (E–E″) Nkx6.1 expression domain was expanded dorsally in the Gli1;Sufu double mutant and more dorsally in the Spop;Gli1;Sufu triple mutant spinal cords. (F–F″) Pax6 expression domain was expanded ventrally in the Gli1;Sufu double mutant, and restricted to the more dorsal region in the Spop;Gli1;Sufu triple mutant spinal cord. (G) Gli1-lacZ expression formed a ventral-to-dorsal gradient in the Gli1 mutant spinal cord, with the exception of the floor plate, in which Gli1-lacZ has been downregulated. (G′) Strong Gli1-lacZ expression is present throughout the entire spinal cord of Gli1;Sufu double mutant. (G″) Gli1-lacZ is highly expressed in the dorsal, but not ventral region of the Spop;Gli1;Sufu triple mutant spinal cords.
To assess the Shh pathway activity more directly, we examined the expression of the lacZ reporter inserted into the Gli1 locus (Gli1-lacZ). As reported previously, Gli1-lacZ expression formed a ventral-to-dorsal gradient in the Gli1 mutant spinal cord, consistent with the existence of a ventral-to-dorsal Shh signaling gradient (Fig. 8G; Bai et al., 2002). A notable exception was the floor plate, in which prolonged exposure to extremely high Shh activity resulted in a downregulation of Gli1-lacZ expression, as previously reported (Fig. 8G; Ribes et al., 2010). Consistent with the widespread low and intermediate levels of Shh pathway activation but lack of maximal Shh pathway activation, strong Gli1-lacZ expression is present throughout the entire Gli1;Sufu double mutant spinal cord (Fig. 8G′). Interestingly, Gli1-lacZ is downregulated in the ventral region of the Spop;Gli1;Sufu triple mutant spinal cords, suggesting the restoration of maximal activation of Shh signaling (Fig. 8G″). Combined with the above spinal cord D/V patterning analyses, we conclude that loss of Spop allows maximal activation of Shh pathway in the absence of Gli1 and Sufu.
4. Discussion
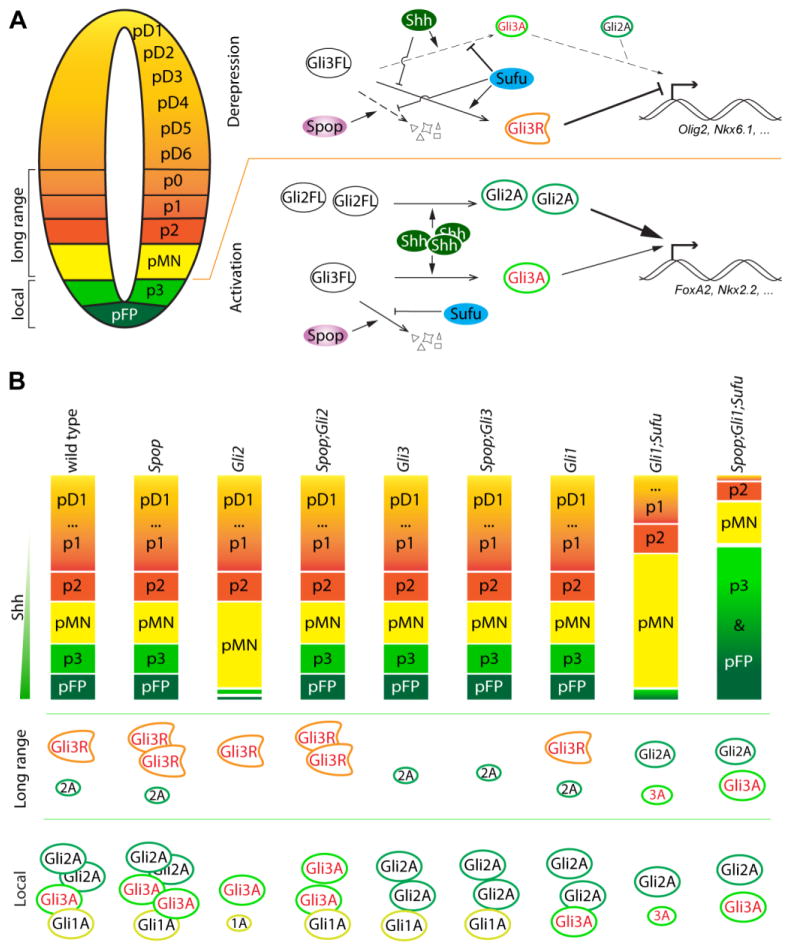
Despite extensive studies in Drosophila and cultured cells (Chen et al., 2009; Kent et al., 2006; Seong et al., 2010; Wang et al., 2010; Wen et al., 2010; Zhang et al., 2009; Zhang et al., 2006), the in vivo function of Spop in Shh signaling and mouse spinal cord ventral patterning has not been revealed. In the present study, we show that Spop negatively regulates the protein level of Gli3, but not Gli2. As a result, ventral spinal cord patterning in Gli2 mutants, in which Gli3 activator acts as the primary effector of the Shh pathway, is subject to Spop regulation. In addition, we reveal a role of Spop in dampening Shh pathway over-activation in the absence of Sufu, especially in preventing the maximal activation of Shh signaling and the ventral-most cell fates in the Gli1;Sufu double mutant spinal cord (Fig. 9).
Fig. 9. A model of the roles of Spop in spinal cord patterning.
(A) In the lateral region of the spinal cord (long range) including p0-pMN, low to intermediate levels of Shh reduce the production of Gli3R from Gli3FL by antagonizing the repressive function of Sufu, allowing the expression of genes such as Olig2 and Nkx6.1. In pFP and p3, high levels of Shh promote the production of Gli2A and Gli3A, which then activate the expression of Foxa2 and Nkx2.2. Gli2A plays a more predominant role than Gli3A in this context. Spop targets Gli3 for degradation, preventing over activation of the Shh pathway. (B) The levels of both Gli3A and Gli3R increase in the absence of Spop, but the effect of the increased Gli3A on the formation of pFP and p3 is only revealed in Spop;Gli2 double mutants, in which the much more potent Gli2A is absent. In Gli1;Sufu double mutants, the reduced levels of Gli2A and Gli3A are insufficient to support pFP and p3 formation. Loss of Spop increases Gli3A and rescues the formation of pFP and p3 in Sufu;Gli1;Spop triple mutants. The moderate increase in Gli3R in the absence of Spop does not show apparent effect in ventral spinal cord patterning in various single and compound mutants.
In Drosophila, the loss of roadkill (hib/rdx), the homolog of Spop, results in an obvious activation of Hh signaling, which is in striking contrast to the normal Shh-dependent D/V patterning of the spinal cord in Spop null mutants (Kent et al., 2006; Seong et al., 2010; Zhang et al., 2006). Two factors may underlie this difference. First, the Drosophila hib/rdx is highly expressed only in cells experiencing high levels of Hh pathway activation, whereas mouse Spop is expressed at low levels throughout the spinal cord. Second, cubitus interruptus (ci) is the only Gli family member in Drosophila and it is targeted by hib/rdx for degradation. In contrast, there are three mammalian Gli family members, and Gli3, better known for its repressor function in Hh signaling, appears to be the only endogenous target for Spop-mediated degradation. Therefore, it is not surprising that loss of Spop only impacts the spinal cord patterning in the absence of Gli2.
Previous studies showed that Spop targets both Gli2 and Gli3 for degradation in vitro (Chen et al., 2009; Wang et al., 2010; Wen et al., 2010; Zhang et al., 2009; Zhang et al., 2006). In contrast, the level of Gli3, but not Gli2, increased in Spop mutant embryos. Since Spop interacts with both the N and C termini of Gli3 but only with the C terminus of Gli2, it is possible that Spop preferentially binds Gli3 but not Gli2 under physiological condition (Wang et al., 2010; Zhang et al., 2009). Alternatively, Gli2 may be associated with other proteins that prevent its interaction with Spop, or protect it from subsequent degradation in vivo.
Previous in vitro studies showed that RNAi knockdown of Spop restored the levels of Gli2 and Gli3 to, or even above the wild type level in Sufu mutant cells (Chen et al., 2009; Wang et al., 2010) or cells treated with Shh (Wen et al., 2010). In contrast, our data show that removal of Spop has no impact on the level of Gli2 and only moderately increases the level of Gli3FL in Sufu mutant embryos (Fig. 6). The difference between our observation and the previous ones could reflect the intrinsic difference between the permanent knockout mutant embryos and cells in culture with transient knockdown of the Spop protein. Recent studies have shown that compensatory mechanisms might be activated only in mutant embryos, but not in transient knockdown assays (e.g. Hall et al., 2013). It will be interesting to investigate whether this is indeed the case in Spop mutants.
Despite the lack of full restoration of the levels of Gli2 and Gli3 (Fig. 6), we show that removing Spop does result in more extreme ventralization of the Sufu mutant spinal cord and rescue the formation of the FP and p3 interneuron progenitors in the Gli1;Sufu double mutant spinal cord, suggesting higher levels of Shh signaling in both cases. It is possible that the moderate increase in Gli3FL is sufficient for this increase in Hh pathway activity in the absence of Spop. Alternatively, Spop may inhibit Gli activator activities through additional, degradation-independent mechanisms. Interestingly, a recent work showed that hib/rdx inhibits the maximal activation of Hh signaling at the border between the anterior and posterior compartments of the wing disc by sequestering ci in the cytoplasm (Seong et al., 2010). It is possible that a similar mechanism may partially underlie Spop function in mammals as overexpression of Spop appears to sequester Gli3 in the cytoplasm in cultured cells (Chen et al., 2009 and data not shown). An antibody that specifically recognizes Gli3FL will allow us to investigate whether this is true with endogenous Gli3.
Due to difficulty in breeding Spop;Sufu double mutants, we analyzed spinal cord patterning in only two Spop;Sufu double mutant embryos. However, these two double mutants both exhibited more extreme ventralization of the spinal cord than Sufu mutants. Importantly, the comparison between Gli1;Sufu double and Spop;Gli1;Sufu triple mutants also show the exacerbation of spinal cord ventralization and Shh signaling activation as the result of loss of Spop. Therefore, we are confident at the conclusion that Spop plays an important negative role in Shh pathway activation in the absence of Sufu.
Spopl shares similar substrate specificity as Spop, but exhibits much weaker ubiquitin-ligase activity in vitro (Errington et al., 2012). It was suggested that Spopl interferes with the more efficient Spop and may have a dominant-negative effect. However, we did not observe any morphological or patterning defects in Spopl mutants, and Spopl;Spop double mutants appear to be identical to Spop mutants. These suggest that functional redundancy between these two speckle-type POZ proteins does not underlie the lack of noticeable patterning defects in the Spop mutant spinal cord.
In recent years, it has become increasingly appreciated that prolonged exposure to Shh ligand changes the sensitivity of the spinal cord cells to the ligand, a phenomenon termed as adaptation (Balaskas et al., 2012; Dessaud et al., 2010; Ribes et al., 2010). A mathematical model has been proposed that involves transcriptional upregulation of Ptch1, transcriptional downregulation of the Gli genes, and differential degradation of the Gli activators as important contributing factors for adaptation (Cohen et al, 2015). As previous in vitro studies suggested that Spop differentially targets the activator form of Gli3 for degradation, the increase in Hh pathway activation in E9.5 Spop;Gli2 double mutants as indicated by the levels of Ptch1 and Gli1 transcription could result, at least partly, from altered adaptation (see Fig. S1C). A closer scrutiny at the temporal profile of Shh pathway activity in this mutant will help to provide a more definitive answer.
In conclusion, we find an in vivo role of Spop in Gli3 degradation and ventral spinal cord patterning better revealed in the absence of Gli2. Loss of Spop in the absence of Sufu exacerbates Hh pathway over-activation and spinal cord ventralization without fully restoring the levels of Gli2 and Gli3 proteins. Our results provide insight into the roles of Spop in the regulation of Gli3 activity and spinal cord development in vivo, and suggest more complexity in the mechanisms of Spop function unappreciated in previous in vitro studies. They also raise new questions such as what, if not Spop, is responsible for the drastic reduction of Gli2 and Gli3 in Sufu mutants, which will continue to be an interesting question awaiting an answer.
Supplementary Material
qRT-PCR was performed to measure the levels of Ptch1 and Gli1 expression in E9.5 embryos with Gapdh as the internal control. (A) Ptch1 and Gli1 were expressed at a similar level in Spop homozygous mutants (n=4 embryos) as in heterozygous (n=4 embryos) and wild type (n=3 embryos) littermates. (B) Ptch1 and Gli1 were expressed at a similar level in Spopl homozygous mutants (n=5 embryos) as in heterozygous (n=4 embryos) and wild type (n=3 embryos) littermates. (C) Ptch1 expression was moderately reduced in Gli2 mutants (n=3 embryos) and rescued in Spop;Gli2 double mutants (n=3 embryos), although the difference did not reach statistical significance. Gli1 was expressed at a significantly lower level in Gli2 mutants, and was significantly increased in Spop;Gli2 double mutants. Student’s t-test was performed to compare the groups. *: p<0.05.
(A–A″) Lateral views of E9.5 control (A), Gli2 mutant (A′) and Spop;Gli2 double mutant (A″) embryos. (B–F″) Immunofluorescent images of transverse sections of E9.5 control (n=2 embryos), Gli2 mutant (n=2 embryos) and Spop;Gli2 (n=2 embryos) double mutant spinal cords at the forelimb level with indicated antibodies. The spinal cords are outlined with dash lines. Brackets indicate the expression domains. (B–B″) Foxa2 was present in the ventral-most region of the control, diminished in the Gli2 mutant, but restored in the Spop;Gli2 double mutant spinal cords. (C–C″) Nkx2.2 was present in juxtaposition to floor plate of the control, diminished in the Gli2 mutant, and was restored in the Spop;Gli2 double mutant spinal cords. (D–D″) Olig2 expression was excluded from the ventral-most region of control and Spop;Gli2 double mutant, but expanded ventrally in Gli2 mutant spinal cords. (E–F″) Nkx6.1 and Pax6 expression domains remained unchanged in the Gli2 mutant and Spop;Gli2 double mutant spinal cords.
(A–A″) Lateral views of E9.5 control (A), Gli3 mutant (A′) and Spop;Gli3 double mutant (A″) embryos. Arrows in A′ and A″ indicate exencephaly. (B–F″) Immunofluorescent images of transverse sections of E9.5 spinal cords at the forelimb level. The expression patterns of genes detected with indicated antibodies were similar between control (n=3 embryos), Gli3 mutant (n=3 embryos) and Spop;Gli3 double mutant (n=2 embryos) embryos. The spinal cords are outlined with dash lines. Brackets indicate the expression domains.
Highlights.
Spop and Spop-like are dispensable for the dorsoventral patterning of the mouse spinal cord.
Loss of Spop leads to a significant increase in the level of Gli3, but not Gli2, in the mouse spinal cord.
Removing Spop rescues the formation of the floor plate and V3 interneuron progenitors, cell types requiring the maximal activation of Shh signaling, in the absence of Gli2, consistent with an increase in Gli3 activator activity.
Removing Spop allows more extreme Hh pathway activation and spinal cord ventralization in the absence of Sufu, and restores the formation of the floor plate and V3 interneuron progenitors in the absence of Sufu and Gli1.
Loss of Spop results in a moderate increase in the level of Gli3 in the absence of Sufu, but fails to restore the level of Gli2 or Gli3 to the wild type level, suggesting additional mechanisms underlying Gli protein level regulation in Sufu mutants.
Acknowledgments
We thank Drs Zhi-Chun Lai and Wendy Hanna-Rose for critically reading the manuscript. We thank Drs Joyner and Toftgard for Gli1, Gli2, Gli3 and Sufu mutant mice. The monoclonal antibodies against Foxa2, Nkx2.2, Nkx6.1 and Pax6 developed by Drs. Jessell, Madsen and Kawakami were obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD and maintained by The University of Iowa, Department of Biological Sciences, Iowa City, IA 52242. A.L. was supported by the National Institutes of Health [grant number HD083625] and a Penn State Start-up Fund.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bai CB, Auerbach W, Lee JS, Stephen D, Joyner AL. Gli2, but not Gli1, is required for initial Shh signaling and ectopic activation of the Shh pathway. Development. 2002;129:4753–61. doi: 10.1242/dev.129.20.4753. [DOI] [PubMed] [Google Scholar]
- Bai CB, Joyner AL. Gli1 can rescue the in vivo function of Gli2. Development. 2001;128:5161–72. doi: 10.1242/dev.128.24.5161. [DOI] [PubMed] [Google Scholar]
- Bai CB, Stephen D, Joyner AL. All mouse ventral spinal cord patterning by hedgehog is Gli dependent and involves an activator function of Gli3. Dev Cell. 2004;6:103–15. doi: 10.1016/s1534-5807(03)00394-0. [DOI] [PubMed] [Google Scholar]
- Balaskas N, Ribeiro A, Panovska J, Dessaud E, Sasai N, Page KM, Briscoe J, Ribes V. Gene regulatory logic for reading the Sonic Hedgehog signaling gradient in the vertebrate neural tube. Cell. 2012;148:273–84. doi: 10.1016/j.cell.2011.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnfield PC, Zhang X, Thanabalasingham V, Yoshida M, Hui CC. Negative regulation of Gli1 and Gli2 activator function by Suppressor of fused through multiple mechanisms. Differentiation. 2005;73:397–405. doi: 10.1111/j.1432-0436.2005.00042.x. [DOI] [PubMed] [Google Scholar]
- Briscoe J, Therond PP. The mechanisms of Hedgehog signalling and its roles in development and disease. Nat Rev Mol Cell Biol. 2013;14:416–29. doi: 10.1038/nrm3598. [DOI] [PubMed] [Google Scholar]
- Cai H, Liu A. Spop promotes skeletal development and homeostasis by positively regulating Ihh signaling. Proc Natl Acad Sci U S A. 2016;113:14751–14756. doi: 10.1073/pnas.1612520114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen MH, Wilson CW, Li YJ, Law KK, Lu CS, Gacayan R, Zhang X, Hui CC, Chuang PT. Cilium-independent regulation of Gli protein function by Sufu in Hedgehog signaling is evolutionarily conserved. Genes Dev. 2009;23:1910–28. doi: 10.1101/gad.1794109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng SY, Bishop JM. Suppressor of Fused represses Gli-mediated transcription by recruiting the SAP18-mSin3 corepressor complex. Proc Natl Acad Sci U S A. 2002;99:5442–7. doi: 10.1073/pnas.082096999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choo KB, Chuang TJ, Lin WY, Chang CM, Tsai YH, Huang CJ. Evolutionary expansion of SPOP and associated TD/POZ gene family: impact of evolutionary route on gene expression pattern. Gene. 2010;460:39–47. doi: 10.1016/j.gene.2010.04.003. [DOI] [PubMed] [Google Scholar]
- Cohen M, Briscoe J, Blassberg R. Morphogen interpretation: the transcriptional logic of neural tube patterning. Curr Opin Genet Dev. 2013;23:423–8. doi: 10.1016/j.gde.2013.04.003. [DOI] [PubMed] [Google Scholar]
- Cooper AF, Yu KP, Brueckner M, Brailey LL, Johnson L, McGrath JM, Bale AE. Cardiac and CNS defects in a mouse with targeted disruption of suppressor of fused. Development. 2005;132:4407–17. doi: 10.1242/dev.02021. [DOI] [PubMed] [Google Scholar]
- Dessaud E, Ribes V, Balaskas N, Yang LL, Pierani A, Kicheva A, Novitch BG, Briscoe J, Sasai N. Dynamic assignment and maintenance of positional identity in the ventral neural tube by the morphogen sonic hedgehog. PLoS Biol. 2010;8:e1000382. doi: 10.1371/journal.pbio.1000382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Q, Fukami S, Meng X, Nishizaki Y, Zhang X, Sasaki H, Dlugosz A, Nakafuku M, Hui C. Mouse suppressor of fused is a negative regulator of sonic hedgehog signaling and alters the subcellular distribution of Gli1. Curr Biol. 1999;9:1119–22. doi: 10.1016/s0960-9822(99)80482-5. [DOI] [PubMed] [Google Scholar]
- Ding Q, Motoyama J, Gasca S, Mo R, Sasaki H, Rossant J, Hui CC. Diminished Sonic hedgehog signaling and lack of floor plate differentiation in Gli2 mutant mice. Development. 1998;125:2533–43. doi: 10.1242/dev.125.14.2533. [DOI] [PubMed] [Google Scholar]
- Dunaeva M, Michelson P, Kogerman P, Toftgard R. Characterization of the physical interaction of Gli proteins with SUFU proteins. J Biol Chem. 2003;278:5116–22. doi: 10.1074/jbc.M209492200. [DOI] [PubMed] [Google Scholar]
- Errington WJ, Khan MQ, Bueler SA, Rubinstein JL, Chakrabartty A, Prive GG. Adaptor protein self-assembly drives the control of a cullin-RING ubiquitin ligase. Structure. 2012;20:1141–53. doi: 10.1016/j.str.2012.04.009. [DOI] [PubMed] [Google Scholar]
- Goodrich LV, Milenkovic L, Higgins KM, Scott MP. Altered neural cell fates and medulloblastoma in mouse patched mutants. Science. 1997;277:1109–13. doi: 10.1126/science.277.5329.1109. [DOI] [PubMed] [Google Scholar]
- Hall EA, Keighren M, Ford MJ, Davey T, Jarman AP, Smith LB, Jackson IJ, Mill P. Acute versus chronic loss of mammalian Azi1/Cep131 results in distinct ciliary phenotypes. PLoS Genet. 2013;9:e1003928. doi: 10.1371/journal.pgen.1003928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui CC, Joyner AL. A mouse model of greig cephalopolysyndactyly syndrome: the extra-toesJ mutation contains an intragenic deletion of the Gli3 gene PG - 241–6. Nat Genet. 1993;3 doi: 10.1038/ng0393-241. [DOI] [PubMed] [Google Scholar]
- Humke EW, Dorn KV, Milenkovic L, Scott MP, Rohatgi R. The output of Hedgehog signaling is controlled by the dynamic association between Suppressor of Fused and the Gli proteins. Genes Dev. 2010;24:670–82. doi: 10.1101/gad.1902910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia J, Kolterud A, Zeng H, Hoover A, Teglund S, Toftgard R, Liu A. Suppressor of Fused inhibits mammalian Hedgehog signaling in the absence of cilia. Dev Biol. 2009;330:452–60. doi: 10.1016/j.ydbio.2009.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent D, Bush EW, Hooper JE. Roadkill attenuates Hedgehog responses through degradation of Cubitus interruptus. Development. 2006;133:2001–10. doi: 10.1242/dev.02370. [DOI] [PubMed] [Google Scholar]
- Kogerman P, Grimm T, Kogerman L, Krause D, Unden AB, Sandstedt B, Toftgard R, Zaphiropoulos PG. Mammalian suppressor-of-fused modulates nuclear-cytoplasmic shuttling of Gli-1. Nat Cell Biol. 1999;1:312–9. doi: 10.1038/13031. [DOI] [PubMed] [Google Scholar]
- Lakso M, Pichel JG, Gorman JR, Sauer B, Okamoto Y, Lee E, Alt FW, Westphal H. Efficient in vivo manipulation of mouse genomic sequences at the zygote stage. Proc Natl Acad Sci U S A. 1996;93:5860–5. doi: 10.1073/pnas.93.12.5860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei Q, Zelman AK, Kuang E, Li S, Matise MP. Transduction of graded Hedgehog signaling by a combination of Gli2 and Gli3 activator functions in the developing spinal cord. Development. 2004;131:3593–604. doi: 10.1242/dev.01230. [DOI] [PubMed] [Google Scholar]
- Lin C, Yao E, Wang K, Nozawa Y, Shimizu H, Johnson JR, Chen JN, Krogan NJ, Chuang PT. Regulation of Sufu activity by p66beta and Mycbp provides new insight into vertebrate Hedgehog signaling. Genes Dev. 2014;28:2547–63. doi: 10.1101/gad.249425.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Heydeck W, Zeng H, Liu A. Dual function of suppressor of fused in Hh pathway activation and mouse spinal cord patterning. Dev Biol. 2012;362:141–53. doi: 10.1016/j.ydbio.2011.11.022. [DOI] [PubMed] [Google Scholar]
- Matise MP, Epstein DJ, Park HL, Platt KA, Joyner AL. Gli2 is required for induction of floor plate and adjacent cells, but not most ventral neurons in the mouse central nervous system PG - 2759–70. Development. 1998:125. doi: 10.1242/dev.125.15.2759. [DOI] [PubMed] [Google Scholar]
- Matise MP, Joyner AL. Gli genes in development and cancer. Oncogene. 1999;18:7852–9. doi: 10.1038/sj.onc.1203243. [DOI] [PubMed] [Google Scholar]
- Maynard TM, Jain MD, Balmer CW, LaMantia AS. High-resolution mapping of the Gli3 mutation extra-toes reveals a 51.5-kb deletion. Mamm Genome. 2002;13:58–61. doi: 10.1007/s00335-001-2115-x. [DOI] [PubMed] [Google Scholar]
- Merchant M, Vajdos FF, Ultsch M, Maun HR, Wendt U, Cannon J, Desmarais W, Lazarus RA, de Vos AM, de Sauvage FJ. Suppressor of fused regulates Gli activity through a dual binding mechanism. Mol Cell Biol. 2004;24:8627–41. doi: 10.1128/MCB.24.19.8627-8641.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murone M, Luoh SM, Stone D, Li W, Gurney A, Armanini M, Grey C, Rosenthal A, de Sauvage FJ. Gli regulation by the opposing activities of fused and suppressor of fused. Nat Cell Biol. 2000;2:310–2. doi: 10.1038/35010610. [DOI] [PubMed] [Google Scholar]
- Paces-Fessy M, Boucher D, Petit E, Paute-Briand S, Blanchet-Tournier MF. The negative regulator of Gli, Suppressor of fused (Sufu), interacts with SAP18, Galectin3 and other nuclear proteins. Biochem J. 2004;378:353–62. doi: 10.1042/BJ20030786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y, Bai CB, Joyner AL, Wang B. Sonic hedgehog signaling regulates Gli2 transcriptional activity by suppressing its processing and degradation. Mol Cell Biol. 2006;26:3365–77. doi: 10.1128/MCB.26.9.3365-3377.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park HL, Bai C, Platt KA, Matise MP, Beeghly A, Hui CC, Nakashima M, Joyner AL. Mouse Gli1 mutants are viable but have defects in SHH signaling in combination with a Gli2 mutation PG - 1593–605. Development. 2000:127. doi: 10.1242/dev.127.8.1593. [DOI] [PubMed] [Google Scholar]
- Persson M, Stamataki D, te Welscher P, Andersson E, Bose J, Ruther U, Ericson J, Briscoe J. Dorsal-ventral patterning of the spinal cord requires Gli3 transcriptional repressor activity. Genes Dev. 2002;16:2865–78. doi: 10.1101/gad.243402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribes V, Balaskas N, Sasai N, Cruz C, Dessaud E, Cayuso J, Tozer S, Yang LL, Novitch B, Marti E, Briscoe J. Distinct Sonic Hedgehog signaling dynamics specify floor plate and ventral neuronal progenitors in the vertebrate neural tube. Genes Dev. 2010;24:1186–200. doi: 10.1101/gad.559910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez CI, Buchholz F, Galloway J, Sequerra R, Kasper J, Ayala R, Stewart AF, Dymecki SM. High-efficiency deleter mice show that FLPe is an alternative to Cre-loxP. Nat Genet. 2000;25:139–40. doi: 10.1038/75973. [DOI] [PubMed] [Google Scholar]
- Seong KH, Akimaru H, Dai P, Nomura T, Okada M, Ishii S. Inhibition of the nuclear import of cubitus interruptus by roadkill in the presence of strong hedgehog signal. PLoS One. 2010;5:e15365. doi: 10.1371/journal.pone.0015365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone DM, Murone M, Luoh S, Ye W, Armanini MP, Gurney A, Phillips H, Brush J, Goddard A, de Sauvage FJ, Rosenthal A. Characterization of the human suppressor of fused, a negative regulator of the zinc-finger transcription factor Gli. J Cell Sci. 1999;112(Pt 23):4437–48. doi: 10.1242/jcs.112.23.4437. [DOI] [PubMed] [Google Scholar]
- Svard J, Heby-Henricson K, Persson-Lek M, Rozell B, Lauth M, Bergstrom A, Ericson J, Toftgard R, Teglund S. Genetic elimination of Suppressor of fused reveals an essential repressor function in the mammalian Hedgehog signaling pathway. Dev Cell. 2006;10:187–97. doi: 10.1016/j.devcel.2005.12.013. [DOI] [PubMed] [Google Scholar]
- Theil T, Alvarez-Bolado G, Walter A, Ruther U. Gli3 is required for Emx gene expression during dorsal telencephalon development. Development. 1999;126:3561–71. doi: 10.1242/dev.126.16.3561. [DOI] [PubMed] [Google Scholar]
- Tole S, Ragsdale CW, Grove EA. Dorsoventral patterning of the telencephalon is disrupted in the mouse mutant extra-toes(J) Dev Biol. 2000;217:254–65. doi: 10.1006/dbio.1999.9509. [DOI] [PubMed] [Google Scholar]
- Tukachinsky H, Lopez LV, Salic A. A mechanism for vertebrate Hedgehog signaling: recruitment to cilia and dissociation of SuFu-Gli protein complexes. J Cell Biol. 2010;191:415–28. doi: 10.1083/jcb.201004108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Fallon JF, Beachy PA. Hedgehog-regulated processing of Gli3 produces an anterior/posterior repressor gradient in the developing vertebrate limb. Cell. 2000;100:423–34. doi: 10.1016/s0092-8674(00)80678-9. [DOI] [PubMed] [Google Scholar]
- Wang C, Pan Y, Wang B. Suppressor of fused and Spop regulate the stability, processing and function of Gli2 and Gli3 full-length activators but not their repressors. Development. 2010;137:2001–9. doi: 10.1242/dev.052126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen X, Lai CK, Evangelista M, Hongo JA, de Sauvage FJ, Scales SJ. Kinetics of hedgehog-dependent full-length Gli3 accumulation in primary cilia and subsequent degradation. Mol Cell Biol. 2010;30:1910–22. doi: 10.1128/MCB.01089-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Shi Q, Chen Y, Yue T, Li S, Wang B, Jiang J. Multiple Ser/Thr-rich degrons mediate the degradation of Ci/Gli by the Cul3-HIB/SPOP E3 ubiquitin ligase. Proc Natl Acad Sci U S A. 2009;106:21191–6. doi: 10.1073/pnas.0912008106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Zhang L, Wang B, Ou CY, Chien CT, Jiang J. A hedgehog-induced BTB protein modulates hedgehog signaling by degrading Ci/Gli transcription factor. Dev Cell. 2006;10:719–29. doi: 10.1016/j.devcel.2006.05.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
qRT-PCR was performed to measure the levels of Ptch1 and Gli1 expression in E9.5 embryos with Gapdh as the internal control. (A) Ptch1 and Gli1 were expressed at a similar level in Spop homozygous mutants (n=4 embryos) as in heterozygous (n=4 embryos) and wild type (n=3 embryos) littermates. (B) Ptch1 and Gli1 were expressed at a similar level in Spopl homozygous mutants (n=5 embryos) as in heterozygous (n=4 embryos) and wild type (n=3 embryos) littermates. (C) Ptch1 expression was moderately reduced in Gli2 mutants (n=3 embryos) and rescued in Spop;Gli2 double mutants (n=3 embryos), although the difference did not reach statistical significance. Gli1 was expressed at a significantly lower level in Gli2 mutants, and was significantly increased in Spop;Gli2 double mutants. Student’s t-test was performed to compare the groups. *: p<0.05.
(A–A″) Lateral views of E9.5 control (A), Gli2 mutant (A′) and Spop;Gli2 double mutant (A″) embryos. (B–F″) Immunofluorescent images of transverse sections of E9.5 control (n=2 embryos), Gli2 mutant (n=2 embryos) and Spop;Gli2 (n=2 embryos) double mutant spinal cords at the forelimb level with indicated antibodies. The spinal cords are outlined with dash lines. Brackets indicate the expression domains. (B–B″) Foxa2 was present in the ventral-most region of the control, diminished in the Gli2 mutant, but restored in the Spop;Gli2 double mutant spinal cords. (C–C″) Nkx2.2 was present in juxtaposition to floor plate of the control, diminished in the Gli2 mutant, and was restored in the Spop;Gli2 double mutant spinal cords. (D–D″) Olig2 expression was excluded from the ventral-most region of control and Spop;Gli2 double mutant, but expanded ventrally in Gli2 mutant spinal cords. (E–F″) Nkx6.1 and Pax6 expression domains remained unchanged in the Gli2 mutant and Spop;Gli2 double mutant spinal cords.
(A–A″) Lateral views of E9.5 control (A), Gli3 mutant (A′) and Spop;Gli3 double mutant (A″) embryos. Arrows in A′ and A″ indicate exencephaly. (B–F″) Immunofluorescent images of transverse sections of E9.5 spinal cords at the forelimb level. The expression patterns of genes detected with indicated antibodies were similar between control (n=3 embryos), Gli3 mutant (n=3 embryos) and Spop;Gli3 double mutant (n=2 embryos) embryos. The spinal cords are outlined with dash lines. Brackets indicate the expression domains.