Abstract
The most dramatic feature of life on Earth is our adaptation to the cycle of day and night. Throughout evolutionary time, almost all living organisms developed a molecular clock linked to the light-dark cycles of the sun. In present time, we know that this molecular clock is crucial to maintain metabolic and physiological homeostasis. Indeed, a dysregulated molecular clockwork is a major contributing factor to many metabolic diseases. In fact, the time of onset of acute myocardial infarction exhibits a circadian periodicity and recent studies found that the light regulated circadian rhythm protein Period 2 (PER2) elicits endogenous cardioprotection from ischemia. Manipulating the molecular clockwork may prove beneficial during myocardial ischemia in humans. MicroRNAs are small non-coding RNA molecules capable of silencing messenger RNA (mRNA) targets. MicroRNA dysregulation has been linked to cancer development, cardiovascular and neurological diseases, lipid metabolism, and impaired immunity. Therefore, microRNAs are gaining interest as putative novel disease biomarkers and therapeutic targets. To identify circadian microRNA-based cardioprotective pathways, a recent study evaluated transcriptional changes of PER2 dependent microRNAs during myocardial ischemia. Out of 352 most abundantly expressed microRNAs, miR-21 was amongst the top PER2 dependent microRNAs and was shown to mediate PER2 elicited cardioprotection. Further analysis suggested circadian entrainment via intense light therapy to be a potential strategy to enhance miR-21 activity in humans. In this review, we will focus on circadian microRNAs in the context of cardioprotection and will highlight new discoveries, which could lead to novel therapeutic concepts to treat myocardial ischemia.
Keywords: PER2, miR-21, circadian, clock, heart, ischemia, intense light, metabolism
Introduction
Our sun, the most prominent mechanism of circadian entrainment, formed 4.5 billion years ago [1]. However, it took another 2 billion years until photosynthetic organisms completely changed the world: 2.4 billion years ago, some of the first organisms to appear on earth, cyanobacteria, acquired the ability to use sunlight via photosynthesis, producing oxygen as a byproduct. Oxygen thereby accumulated in the atmosphere [2] leading to The Great Oxygenation Event and subsequent extinction of almost all anaerobic bacteria [3]. Organisms that could use oxygen for respiration therefore thrived and populated the earth as we see today.
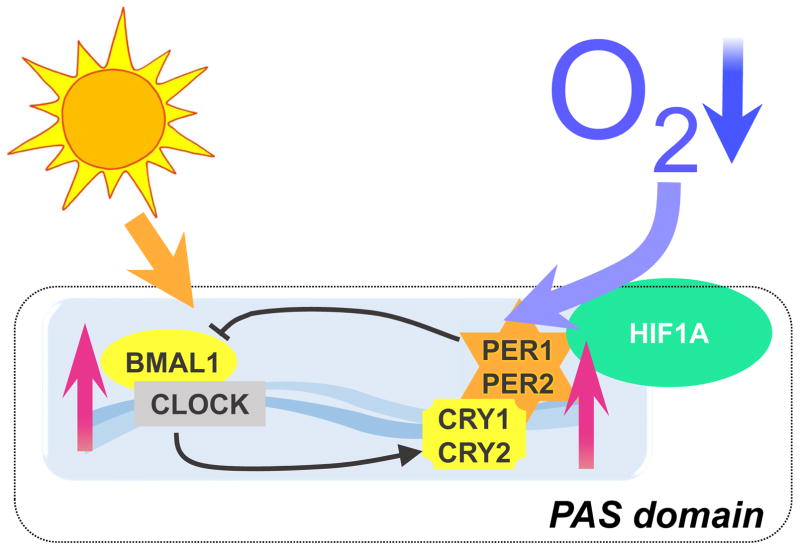
The emergence of sunlight and oxygen were undoubtedly two of the most dramatic environmental changes to influence organismal evolutionary history [2]. Biological systems in all organisms had to adapt to sunlight cycles and oxygen saturation to survive. As a result, almost all organisms on this planet are equipped with light and oxygen sensing proteins [4, 5]. Light sensing proteins are known as circadian rhythm proteins. A single celled organism like cyanobacteria can directly sense light [6], but more complex multi-cellular species developed a light sensing machinery within the interconnected biological system. In mammals, sunlight is sensed by melanopsin receptors in retinal ganglion cells followed by an activation of nerve cells and a signaling cascade through the retino-hypothalamic tract to the SCN (suprachiasmatic nuclei) in the hypothalamus of the brain [7, 8]. In the SCN, core circadian proteins, which are CLOCK, BMAL1, PER1, PER2, CRY1, and CRY2, change their expression according to light-dark cycles [9]. Interestingly, this light sensing pathway is linked to the oxygen sensing system in mammals [10]. In fact, hypoxia inducible factor 1ζ (HIF1A), one of the most commonly known oxygen sensing proteins [11] belongs to the same protein family as CLOCK, BMAL1, PER1/2, and CRY1/2 [12]. All these proteins are part of the PAS domain superfamily of transcriptional regulators [13]. Indeed, it was shown that Hif1ζ mRNA levels cycle in a circadian manner in mouse cardiac tissue [14] and several investigations identified a relationship between HIF1A and the circadian clock [14]. The PAS domain superfamily was first described in Drosophila and abbreviated for PER, ARNT and SIM. The PAS domain enables proteins the ability to sense oxygen, light or metabolism [6]. Based on these fundamental principles, it becomes clear how important a robust and functional circadian system is for health and disease prevention. In fact, numerous diseases are known to have a circadian periodicity. Examples include myocardial ischemia, sudden cardiac death, stroke, thrombosis, and sepsis which vary during a 24-h time suggesting circadian involvement [4,5,15–17]. Recently, studies identified a protective role of circadian rhythm proteins in myocardial ischemia [18–21]. It was found that the light regulated circadian protein PER2 mediates the cardioprotective effects of ischemic preconditioning [15]. Ischemic preconditioning, where the heart is exposed to short, non-lethal episodes of ischemia prior to an extended ischemia time is probably the most powerful cardioprotective strategy at the bench [16]. Very recent studies confirmed the cardioprotective role of PER2 in ischemic preconditioning [17] and additional studies found that hypoxia resets the circadian clock through HIF1A [18] (Figure 1). These observations and the relationship between circadian and oxygen sensing proteins indicates an important role of circadian rhythm proteins in disease states of low oxygen availability, such as myocardial ischemia. Targeting these pathways is inevitable and could lead to the development of novel and powerful cardioprotective strategies.
Figure 1. Mechanisms of circadian rhythms.
The circadian clock is composed of a primary negative feedback loop involving the genes CLOCK, BMAL1, Period homologue 1 (PER1), PER2, Cryptochrome 1 (CRY1) and CRY2. This clockwork is composed of a set of proteins that are synchronized by daylight. Those proteins that belong to the so-called PAS domain superfamily. PAS stands for Period, Arnt and Sim, three drosophila genes in which the PAS domain was discovered. Hypoxia Inducible Factor 1 Alpha (HIF1A) that plays an important role in hypoxic and ischemic disease states also belongs to this family of PAS domain positive proteins. The PAS domain has been described as a binding site to allow interactions between those proteins and to sense oxygen or light. Recent findings indicate that low hypoxia can also regulate the clockwork, like light.
Putative novel disease biomarkers with therapeutic targets include microRNAs, which gained the interest of many researchers in the past several years. To identify circadian microRNA-based cardioprotective pathways, a recent study evaluated transcriptional changes of PER2 dependent microRNAs during myocardial ischemic preconditioning [19]. Out of the 352 most abundantly expressed microRNAs, miR-21 was amongst the top PER2 dependent microRNAs. In depth studies identified miR-21 as a cardioprotective downstream target of PER2 and suggested circadian entrainment via intense light therapy as a potential strategy to enhance miR-21 activity in humans. While this is the first study on cardiac-specific circadian microRNAs to date, other studies showed the impact of microRNAs in controlling general circadian pathways. This current review will summarize recent findings on circadian and cardiac micro RNAs and will evaluate potential therapeutic concepts.
MicroRNAs as cardiovascular therapeutic targets
Only 1% of the human genome codes for genes that function in protein synthesis [20]. The remaining 99% of DNA was initially considered to be non functional. However, it is now well recognized that several noncoding RNAs have important biological functions [21–29]. Among these noncoding RNAs several subcategories exist, including long noncoding and small noncoding RNAs. One type of small noncoding RNAs, microRNAs, have attracted a lot of attention in the past few years [30]. MicroRNAs are short (22 nucleotides), interact with messenger RNAs, and can silence gene expression. The functional domain of microRNAs is the so-called seed region which is only 6–8 nucleotides long [24]. Initially, double-stranded RNA is formed from a precursor transcript. Primary microRNAs (pri-microRNA) are transcribed by RNA polymerase II. The pri-microRNAs are 5′ capped, have a stem loop structure, and are 3′ polyadenylated. The canonical biogenesis of a pri-microRNA transcript is cleaved by the endoribonuclease Drosha in the nucleus and subsequently by endoribonuclease Dicer in the cytoplasm. Noncanonical pathways that are independent of this canonical pathway could produce microRNAs from small nucleolar RNA, transfer RNA (tRNA), or Y RNA, as intermediate products. Later, the microRNA duplex unwinds, whereby only the guide strand, which is usually the functional unit, is loaded in the RNA-induced silencing complex (RISC). The complimentary strand is often degraded. In the RISC, the microRNA binds to its target mRNA, preventing its translation into a protein. The human genome harbors 1881 microRNA loci that encode for 2588 mature microRNAs [31], indicating an important role in gene regulation. Single microRNAs suppress more than one gene, and microRNAs with similar seed regions may suppress a similar, but non-identical set of genes. Gene suppression is usually partial rather than total (supporting that microRNAs function to maintain cellular homeostasis), and a single gene can have binding sites for multiple microRNAs [21]. In general, overexpressing microRNAs via mimetics will suppress target genes, whereas inhibiting an endogenous microRNA will undo the suppression of its target gene. As repression of microRNAs is considered to be safer, current clinical studies mainly use microRNA inhibitors (anti-miRs[21]). Current difficulties for cardiovascular applications include safety issues for systemic applications: generally high doses are required which can compromise efficacy and safety. So far, several microRNAs were identified as cardiovascular therapeutic targets and are abundantly expressed in the heart. For example, mir-133 is highly expressed in cardiomyocytes and overexpression seems to prevent hypertrophic cardiomyopathy through controlling multiple components of the β1AR transduction cascade [32]. Other microRNA manipulation in the heart, like miR-208 inhibition, were shown to be beneficial in animal models of heart failure. It was found that miR-208a inhibition reversed myosin switching during heart failure thereby improving cardiac function and remodeling during heart disease progression [33]. Several other microRNAs were shown to regulate the gene program that controls cardiac fibrosis. As such, miR-15 and miR-30 are negative regulators of fibrosis. Expression of these microRNAs are reduced during myocardial ischemia or upon thoracic aortic constriction surgery [31]. Moreover, miR-15 was found as a regulator of cardiac hypertrophy and fibrosis by inhibition of the TGFβ-pathway [34]. MiR-21, which was recently identified as a potential circadian microRNA [19] has also been studied extensively in the context of heart fibrosis [35]. It was found that in vivo inhibition of miR-21 attenuates the fibrotic response and improves cardiac function in mouse models of heart failure [36]. However, these results were not reproduced in a subsequent study using different anti-miRs [37], indicating current challenges in the therapeutic use of microRNAs.
MicroRNAs in cardioprotection from myocardial ischemia
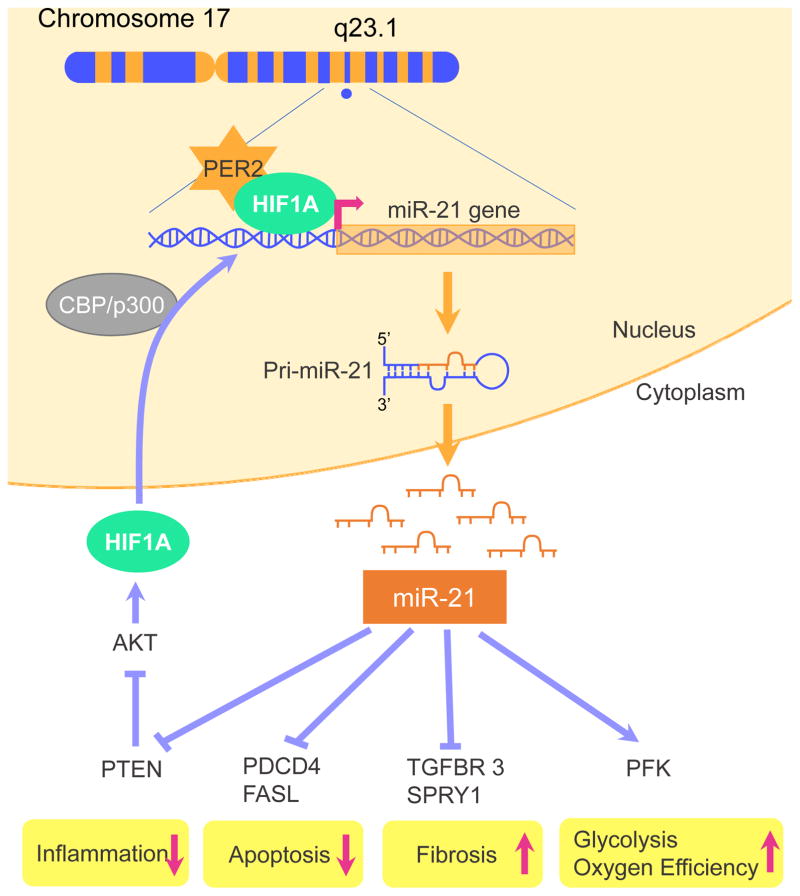
Several microRNAs have been implicated in influencing infarct sizes after ischemia-reperfusion (IR) injury. Examples of microRNAs found to increase cell death in IR-injury are miR-15 [38], miR-34 [39], miR-320 [40], miR-140 [41], miR-1/miR-206 [42], miR-92a [43], miR-122 [44], miR-150 [45], miR-181a [46] and miR-376b-5p [47]. MicroRNAs that can reduce cardiomyocyte cell death or enhance cardiac regeneration are miR-24 [48], miR-29 [49], miR-30 [50], mir-214 [51], miR-7a/b [52], miR-20a [53], miR-132 [54], miR-138 [55], miR-144/451 [56], miR-155 [57], miR-210 [58], miR-499 [59] and miR-874 [60]. In addition, a recent study found a crucial role for miR-21 in preventing cardiomyocyte cell death through targeting the programmed cell death 4 gene (PDCD4 [61], Figure 2). Other miR-21 target genes are Fas ligand (FasL) or tensin homology deleted on chromosome 10 (PTEN) and are also antiapoptotic [62]. Besides regulating apoptosis, miR-21 has been implicated in the attenuation of inflammation or in the angiogenic repair process of ischemic injury via a decrease of NF-kappa B or through the PTEN/AKT/ERK1-VEGF pathway, respectively [62]. Recent studies indicate that miR-21 also reduces hydrogen peroxide-induced apoptosis in cardiac stem cells through PTEN/PI3K/AKT signaling [63]. Moreover, bone marrow-derived mesenchymal stem cells overexpressing miR-21 efficiently repair myocardial damage in rats [64]. Interestingly, HIF1A was identified as an upstream transcriptional regulator of miR-21[18]. Moreover, it was shown that HIF1A and PER2 bind together during hypoxia and that PER2 is essential for the regulation of HIF1A target genes during hypoxia [15].
Figure 2. Potential upstream and downstream targets of miR-21.
MiR-21 is located on Chromosome 17 and has been found to be a hypoxia inducible factor 1 α (HIF1A) target gene. Through AKT1 activation, miR-21 can increase its own transcription via a positive feedback loop. During myocardial ischemia HIF1A and Period 2 (PER2) bind together. PER2 was shown to be essential in the regulation of HIF1A target genes, which explains recent findings on a PER2 dependent regulation of miR-21. The main mechanisms how miR-21 reduces cardiac cell death is through targeting programmed cell death 4 (PDCD4), Fas ligand (FASL), and phosphatase and tensin homology deleted on chromosome 10 (PTEN). Inflammation may also be reduced because of a decrease in NF-κB. Recent studies indicate that miR-21 also protects from ischemia trough upregulation of oxygen efficient glycolysis – probably via stimulation of HIF1 and PER2 dependent phosphofructokinase (PFK). Some data, however, indicate that miR-21, induced by transforming growth factor beta receptor 3 (TGFBR3) stimulation, exacerbates tissue fibrosis via suppression of Sprouty homolog 1(SPRY1).
In vivo studies confirmed many of these findings and identified miR-21 as a top microRNA in anesthetic preconditioning of the heart [65]. Exposure to volatile anesthetics, such as isoflurane, was shown to decrease myocardial infarct size in vivo and increase cell viability after oxidative stress in vitro [66]. Using anesthetic preconditioning in miR-21 deficient mice, however, did not mediate any cardioprotective effects [67]. Interestingly, a recent study on ischemic preconditioning of the heart identified miR-21 as a top circadian protein PER2 dependent microRNA [19]. Similarly, an earlier study found miR-21 as the top microRNA in ischemic preconditioning of the heart [68]. Here, miR-21 knockdown using anti-miRs abolished ischemic preconditioning mediated cardioprotection. As such, ischemic and anesthetic preconditioning of the heart seem to merge on the same microRNA level. Along these lines, a recent review on anesthetic preconditioning indicated that anesthetic and ischemic preconditioning share similar mechanisms [66]. Having miR-21 identified as a major downstream target of anesthetic or ischemic preconditioning of the heart opens up new possibilities to target powerful endogenous cardioprotective mechanisms [69].
Circadian microRNAs
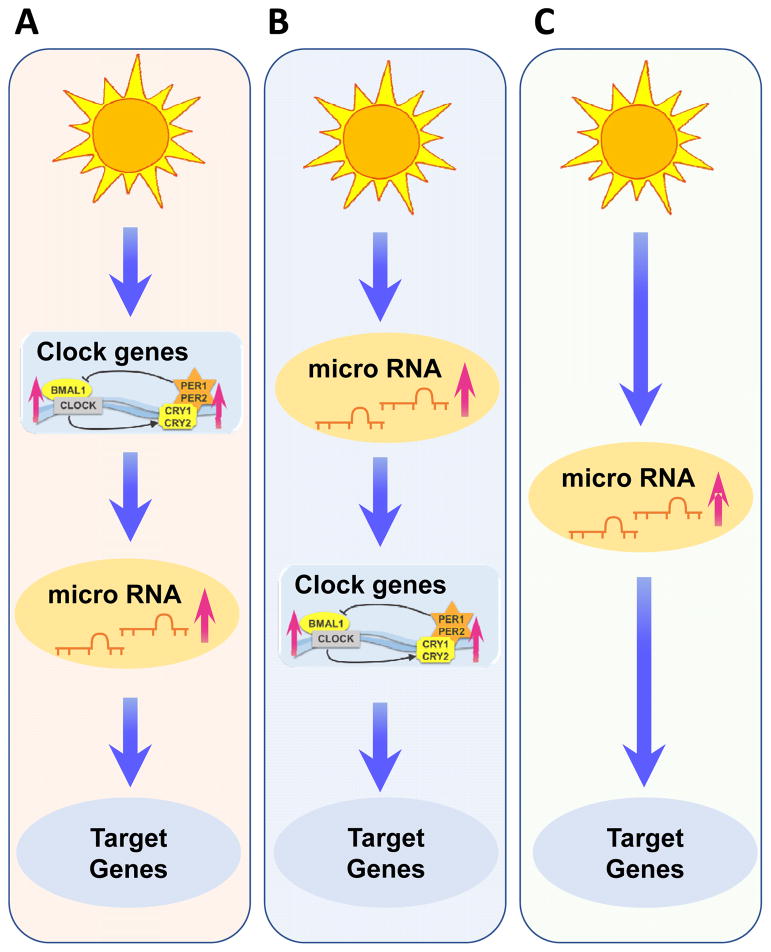
Disease development is strongly influenced by circadian rhythms and therefore researchers have begun to explore the potential role of microRNAs. Rhythmic control of microRNA expression appears to be highly conserved and has significant consequences for circadian timing [70]. As such several rhythmically regulated microRNAs were identified. Surprisingly many biologically relevant time cues, such as the daily light cycle can also drive rhythmic microRNA expression. Studies in Arabidopsis have recently found that miR-167, miR-168, miR-171 and miR-398 oscillate, with higher levels during the daytime than during the night [71]. However, rhythmicity of these microRNAs was not regulated by the circadian clock which became evident when Arabidopsis was transferred to constant light conditions. Under constant light, no oscillation was observed [71]. This suggests that light might control these microRNAs. Within humans, it would not be surprising to observe a similar pattern of microRNA expression driven by exogenous time cues [70]. In fact, findings on miR-21 expression indicated regulation by light in mice and humans [19]. In addition, a diurnal expression pattern and regulation of circadian rhythm pathways was found. However, dysregulation of miR-21 at baseline in circadian rhythm protein deficient mice was not shown. This could mean that oscillating microRNAs could function as circadian rhythm protein independent regulators of circadian rhythm dependent gene regulation (Figure 3).
Figure 3. Possible microRNA pathways for regulating the circadian clock.
(A) External cues control the circadian clock work and thereby regulate clock dependent microRNAs and their target genes. (B) External cues such as sunlight directly control microRNAs and thereby regulate microRNA regulated clock genes and their dependent target genes. (C) External cues regulate microRNAs and their target genes in a circadian manner based on the rhythm of the external cue.
In the mammalian SCN, microRNAs play also an important role in clock timing and entrainment. For example, miR-132 and miR-219 show oscillatory expression in wildtype mice, but not in circadian mutant mice [72]. Both microRNAs have CRE-enhancer sequences allowing for CREB-dependent regulation [73], but only miR-132 was shown to be inducible by light [72]. Moreover, miR-219 regulates the length of the circadian day and miR-132 modulates the phase-shifting capacity of light [72]. As miR-132 is induced by photic entrainment cues via a MAPK/CREB-dependent mechanism, it modulates clock-gene expression, and thereby attenuates the entraining effects of light. In addition, both light-responsive miR-132 and clock-regulated miR-219 influence cellular excitability and thereby probably lead to changes in length and phase of the circadian clock. The expression of miR-122, a known regulator of liver metabolism, is also subject to circadian control. While the mature microRNA does not oscillate, both miR-122 precursors have a robust circadian expression pattern [74]. In fact, genetic deletion of miR-122 revealed changes in the expression of clock-controlled genes. Interestingly, REV-ERBα, a key nuclear receptor regulator of the circadian system, also regulates miR-122 [74]. Another clock controlled microRNA, miR-142–3p, is controlled by the BMAL1/CLOCK heterodimer and, in turn, can target BMAL1 [75, 76]. In a different study evidence found that miR-155, a proinflammatory microRNA induced by TLRs (Toll-like receptors), controls BMAL1 mRNA and protein levels in myeloid cells, leading to alterations in clock function and circadian control of inflammation [77]. Indeed, the free-running period in miR-155−/− mice was shortened when compared to wildtype mice. MicroRNA control of the circadian system is further supported by studies in Dicer deficient mice, in which microRNA processing is globally compromised [78]. These studies demonstrate that RNA interference mediated by microRNAs primarily affect the clock through translational control of PER in the cytoplasm, which delays cytoplasmic PER accumulation and thus generates a time delay in the circadian feedback inhibition. These studies also identified three microRNAs, miR-24, 29a and 30a, which affected the circadian clock through regulation of PER1 and PER2 mRNA stability and translation. Thus, these studies indicate microRNAs as key regulators of the pacemaker genes, PER1 and PER2, and for generating the time delay crucial for the circadian feedback loop. This evidence positions microRNAs as important and dynamic regulators of different aspects of circadian rhythms: Some microRNAs affect and fine-tune the pace of the core clock, while others influence the response to external temporal cues as well as behavioral and peripheral outputs. Therefore, microRNAs have the potential to become novel modulators of circadian rhythms and might be able to positively influence cardiovascular physiology (Figure 3).
MicroRNAs in Circadian Entrainment
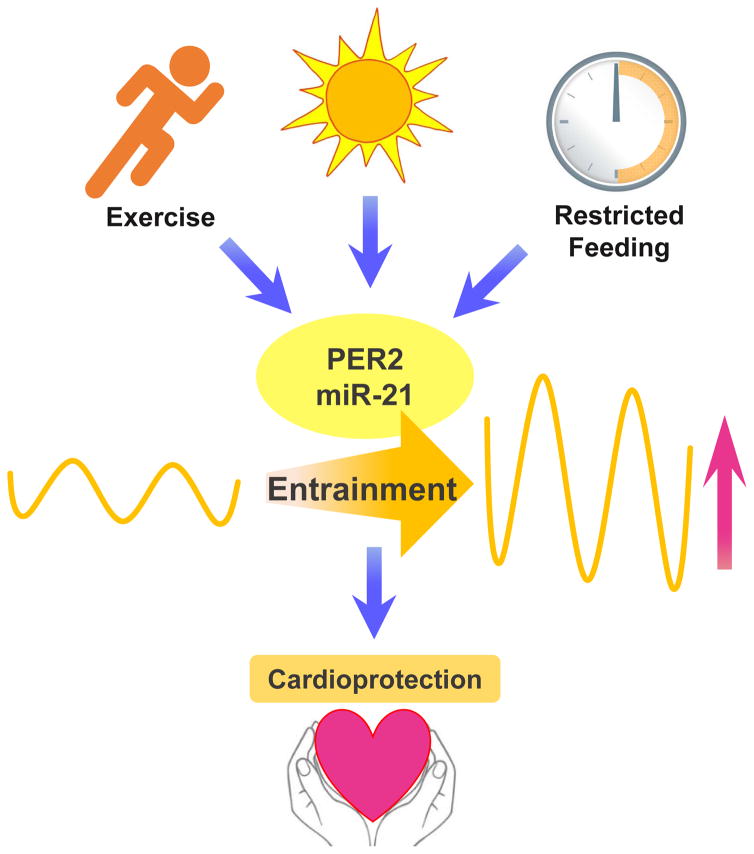
The most concerning illness associated with clock physiology may result from a dampened circadian oscillator, which is observed as part of the aging process [79]. Therefore, researchers believe that a robust circadian timekeeping system is important for human health and well-being [4, 5, 80–83]. Indeed, approaches to increase entrainment and thereby the robustness of the clock timing process have been found to be beneficial in certain disease states. One study found that restricted feeding increased the circadian amplitude and could prevent mice from becoming obese when exposed to a high fat diet [84]. Another study found using short term caloric restricted feeding in mice to be cardioprotective. In addition, cardiac microRNA profiling of short term caloric restricted feeding was associated with the circadian clock [85]. Similarly, an independent study tested whether activity alters or could rescue a disrupted circadian system. Here, they examined the effects of wheel access on vasointestinal polypeptide (VIP)-deficient mice, a model that exhibits circadian deficits [86]. Indeed, voluntary scheduled exercise increased the amplitude of PER2 expression and rescued the disrupted circadian system in VIP deficient mice. Some studies on blue enriched light exposure on rats found similar effects on circadian rhythmicity and amplitude. In addition, marked positive effect on the circadian regulation of neuroendocrine, metabolic, and physiologic parameters associated with the promotion of animal health and wellbeing were observed [87]. Similarly, one study demonstrated that day/night rhythms play a critical role in compensatory remodeling of cardiovascular tissue, and disruption of day/night rhythms exacerbated disease pathophysiology [88]. Here, they used a murine model of pressure overload cardiac hypertrophy in a rhythm-disruptive 20-hour versus 24-hour environment. Echocardiographic studies revealed increased left ventricular end-systolic and -diastolic dimensions and reduced contractility in rhythm-disturbed animals exposed to transverse aortic constriction. Considering these findings, it is compelling that light exposure, restricted feeding or scheduled exercise could also be cardioprotective through strengthening the circadian clock. Since microRNAs can be regulated by external cues such as light, without involvement of the circadian rhythm proteins, it is probable that light exposure also improves a disrupted circadian system through enhancing the amplitude of microRNAs and optimizing their function (Figure 4).
Figure 4. Circadian cardiac entrainment as cardioprotective mechanism.
Apart from light, exercise or restricted feeding have been shown to entrain the circadian system. Increasing the amplitude and the robustness of PER2 or miR-21 expression could represent a possible cardioprotective mechanism.
MiR-21, a cardiac PER2 dependent microRNA
In general, while several circadian microRNAs have been implicated in cardioprotection from ischemia (Table 1), studies on cardiac circadian microRNAs are scarce [85]. Recent profiling of PER2 dependent microRNAs in cardiac ischemia indicated a critical role for miR-21 [19]. In vitro studies revealed that PER2 dependent miR-21 regulates cellular glycolysis during cellular stress [15]. In fact, myocardial ischemia leads to the activation of pathways directed towards enhancing myocardial oxygen efficiency. As such, a metabolic switch from more “energy-efficient” utilization of fatty acids to more “oxygen-efficient” utilization of glucose as the main source for energy generation is pivotal to allow the myocardium to function under ischemic conditions [89].
Table 1.
Micro RNAs implicated in cardioprotective and/or circadian mechanisms
| cardioprotection | circadian | cardioprotection and circadian | function in the heart |
|---|---|---|---|
|
| |||
| miR-1/miR-206 | increase cell death of cardiomyocytes | ||
| miR-15 | increase cell death of cardiomyocytes | ||
| miR-7a/b | reduce cell death of cardiomyocytes | ||
| miR-20a | reduce cell death of cardiomyocytes | ||
| miR-21* | miR-21* | miR-21* | reduce cell death of cardiomyocytes |
| miR-24 | miR-24 | miR-24 | reduce cell death of cardiomyocytes |
| mir-29a | mir-29a | mir-29a | enhanced cardiac regeneration |
| miR-30/miR-30a | miR-30/miR-30a | miR-30/miR-30a | reduce cell death of cardiomyocytes |
| miR-34 | increase cell death of cardiomyocytes | ||
| miR-92a | increase cell death of cardiomyocytes | ||
| miR-122 | miR-122 | miR-122 | increase cell death of cardiomyocytes |
| miR-132 | miR-132 | miR-132 | enhanced cardiac regeneration |
| miR-133 | enhanced cardiac regeneration | ||
| miR-138 | reduce cell death of cardiomyocytes | ||
| miR-140 | increase cell death of cardiomyocytes | ||
| miR-142-3p | unknown | ||
| miR-144/451 | reduce cell death of cardiomyocytes | ||
| miR-150 | increase cell death of cardiomyocytes | ||
| miR-155 | miR-155 | miR-155 | reduce cell death of cardiomyocytes |
| miR-167* | unknown | ||
| miR-168* | unknown | ||
| miR-171* | unknown | ||
| miR-181a | increase cell death of cardiomyocytes | ||
| miR-208 | enhanced cardiac regeneration | ||
| miR-210 | reduce cell death of cardiomyocytes | ||
| miR-214 | reduce cell death of cardiomyocytes | ||
| miR-219 | unknown | ||
| miR-320 | increase cell death of cardiomyocytes | ||
| miR-376b-5p | increase cell death of cardiomyocytes | ||
| miR-398* | unknown | ||
| miR-499 | reduce cell death of cardiomyocytes | ||
| miR-874 | reduce cell death of cardiomyocytes | ||
light regulated microRNAs
Following studies on myocardial ischemia and reperfusion injury found larger infarct sizes and abolished light elicited PER2 cardioprotection [15] in miR-21−/− mice. In addition, intense light exposure in wildtype mice increased miR-21 levels. Here, mice were housed under 14h light and 10h dark conditions. Instead of room light (200LUX), however, intense light (10,000 LUX) was used. The strategy enhanced PER2 or miR-21 in murine hearts. These findings indicate that circadian entrainment through intense light could be the underlying mechanism (Figure 4). Notably, a hallmark of the mammalian circadian pacemaker is its ability to be entrained by light [90]. Humans are primarily entrained by sunlight, regardless of other external cues [91]. The greater the intensity of the light, the more robust the entrainment and the amplitude of the circadian rhythm. This has been shown by studies on melatonin suppression, indicating that humans might need brighter or more intense light for optimal entrainment of circadian rhythms [92]. In fact, intense light exposure was found to increase PER2 and glycolytic enzymes and decrease infarct size and troponin levels during myocardial ischemia in mice [15]. However, intense light therapy in the regulation of cardiac microRNAs has not yet been described. While studies on cardioprotective effects of light exposure in humans are missing, light induced cardioprotective miR-21 could be one mechanism by which intense light exposure reduced myocardial damage in murine studies [15]. Interestingly, the above-mentioned studies on the cardiac circadian microRNA miR-21 used also intense light in human healthy volunteers. Here, intense light exposure for 30 minutes over five days increased miR-21 or PER2 dependent phosphofructokinase activity in plasma samples. To our knowledge, nobody previously analyzed human metabolic changes upon intense light therapy. However, the effects of intense light therapy in humans are recognized and already widely used. As such, intense light therapy is used to treat seasonal affective disorder [93, 94], but also might have effects on preventing delirium [95] or may improve sleep overall [96, 97]. Intense light significantly increases miR-21 in human plasma samples, which is associated with increased phosphofructokinase activity (the key enzyme of glycolysis). These findings indicate that light is a promising strategy to activate PER2 elicited cardioprotection [15] and to increase the robustness of the circadian system in humans [80, 98]. More detailed studies on intense light therapy in humans will help us to further dissect these mechanisms.
Conclusion
Using cardiac circadian microRNAs for cardiac entrainment and concomitant cardioprotection seems compelling considering the reviewed research findings. However, several discrepancies in studies on cardioprotective microRNAs hamper the immediate translation from bench to bedside. One study found significantly increased infarct sizes in miR-21 deficient mice when compared to controls. In contrast, other studies on miR-21 null mice did not find any significant differences in infarct sizes during myocardial ischemia and reperfusion injury [52,53]. While several differences in methodologies might have contributed to the contrary findings, the most prominent differences were the ischemia times. While some studies use 60 minutes of ischemia, others have a 30-minute ischemia protocol. Indeed, marked differences in cardioprotective mechanisms using various ischemia times have been reported [54] and may explain the many differences reported on microRNAs in cardioprotection. In addition, miR-21 is predominantly expressed in cardiac fibroblasts when compared with other cell types of the heart [99] and it was found that miR-21 inhibition attenuates the fibrotic response and improves cardiac function in mouse models of heart failure [36]. However, these results were not reproduced in subsequent studies [37], emphasizing our current challenges in the therapeutic use of microRNAs. In fact, another study on bone marrow-derived mesenchymal stem cells found that overexpressing miR-21 efficiently repaired myocardial damage in rats [64].
Nevertheless, possible therapeutic approaches could target specific microRNAs and thereby limit their ability to suppress components of the core clock timing process or clock output genes. In fact, a robust circadian timekeeping system is important for human health and well-being [4, 5, 80–83]. Circadian entrainment using intense light during the day increases robustness and circadian amplitude of cardiac PER2, which was found to significantly reduce infarct sizes in an in-situ mouse model for myocardial IR-injury [15]. Administration of microRNAs in a time of the day dependent manner could therefore help to restore a weakened circadian system, improve metabolism via the increase of oxygen efficient pathways and thereby promote cardioprotection from ischemia. As numerous microRNAs have also been found to be protective in the acute setting during cardiac ischemia or reperfusion, administration of microRNAs during cardiac revascularization would represent another therapeutic approach.
There are several challenges that need to be addressed for these types of approaches to be implemented. For example, the timing of microRNA administration and organ-specific therapeutic targeting are crucial areas of investigation. In fact, one study uncovered a microRNA tissue-specific regulation of gene expression phase and amplitude [100]. These findings suggest that microRNAs function to adapt clock-driven gene expression to tissue-specific requirements [101]. Further research is required to determine whether similar cardiac microRNAs exist. In general, practical application of these methods will need a deeper understanding of microRNA biology, the clock, and its role in the regulation of a myriad of gene networks that contribute to human physiology and pathology.
Acknowledgments
Source of financial support for the work:
National Heart, Lung, and Blood Institute (NIH-NHLBI) 5R01HL122472 Grant to T.E.; American Heart Association (AHA) Predoctoral Fellowship 16PRE30510006 and
Colorado Clinical & Translational Sciences Institute (CCTSI) TL1 TR001081 to C.M.B.
The original manuscript was published via Current Pharmaceutical Design. The published manuscript is available at EurekaSelect via http://www.eurekaselect.com/openurl/content.php?genre=article&doi=10.2174/1381612823666170707165319
References
- 1.Banerjee P, Qian YZ, Heger A, Haxton WC. Evidence from stable isotopes and 10Be for solar system formation triggered by a low-mass supernova. Nat Commun. 2016;7:13639. doi: 10.1038/ncomms13639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zerkle AL, Poulton SW, Newton RJ, Mettam C, Claire MW, Bekker A, Junium CK. Onset of the aerobic nitrogen cycle during the Great Oxidation Event. Nature. 2017;542:465–467. doi: 10.1038/nature20826. [DOI] [PubMed] [Google Scholar]
- 3.Luo G, Ono S, Beukes NJ, Wang DT, Xie S, Summons RE. Rapid oxygenation of Earth’s atmosphere 2. 33 billion years ago. Sci Adv. 2016;2:e1600134. doi: 10.1126/sciadv.1600134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brainard J, Gobel M, Bartels K, Scott B, Koeppen M, Eckle T. Circadian Rhythms in Anesthesia and Critical Care Medicine: Potential Importance of Circadian Disruptions. Semin Cardiothorac Vasc Anesth. 2014 doi: 10.1177/1089253214553066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brainard J, Gobel M, Scott B, Koeppen M, Eckle T. Health Implications of Disrupted Circadian Rhythms and the Potential for Daylight as Therapy. Anesthesiology. 2015 doi: 10.1097/ALN.0000000000000596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Taylor BL, Zhulin IB. PAS domains: internal sensors of oxygen, redox potential, and light. Microbiol Mol Biol Rev. 1999;63:479–506. doi: 10.1128/mmbr.63.2.479-506.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hankins MW, Peirson SN, Foster RG. Melanopsin: an exciting photopigment. Trends Neurosci. 2008;31:27–36. doi: 10.1016/j.tins.2007.11.002. [DOI] [PubMed] [Google Scholar]
- 8.Panda S, Provencio I, Tu DC, Pires SS, Rollag MD, Castrucci AM, Pletcher MT, Sato TK, Wiltshire T, Andahazy M, Kay SA, Van Gelder RN, Hogenesch JB. Melanopsin is required for non-image-forming photic responses in blind mice. Science. 2003;301:525–7. doi: 10.1126/science.1086179. [DOI] [PubMed] [Google Scholar]
- 9.Takahashi JS. Transcriptional architecture of the mammalian circadian clock. Nat Rev Genet. 2017;18:164–179. doi: 10.1038/nrg.2016.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Edgar RS, Green EW, Zhao Y, van Ooijen G, Olmedo M, Qin X, Xu Y, Pan M, Valekunja UK, Feeney KA, Maywood ES, Hastings MH, Baliga NS, Merrow M, Millar AJ, Johnson CH, Kyriacou CP, O’Neill JS, Reddy AB. Peroxiredoxins are conserved markers of circadian rhythms. Nature. 2012;485:459–64. doi: 10.1038/nature11088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Semenza GL. Hypoxia. Cross talk between oxygen sensing and the cell cycle machinery. Am J Physiol Cell Physiol. 2011;301:C550–2. doi: 10.1152/ajpcell.00176.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu W, Shen SM, Zhao XY, Chen GQ. Targeted genes and interacting proteins of hypoxia inducible factor-1. Int J Biochem Mol Biol. 2012;3:165–78. [PMC free article] [PubMed] [Google Scholar]
- 13.Hogenesch JB, Gu YZ, Jain S, Bradfield CA. The basic-helix-loop-helix-PAS orphan MOP3 forms transcriptionally active complexes with circadian and hypoxia factors. Proc Natl Acad Sci U S A. 1998;95:5474–9. doi: 10.1073/pnas.95.10.5474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Button EL, Bersten DC, Whitelaw ML. HIF has Biff - Crosstalk between HIF1a and the family of bHLH/PAS proteins. Exp Cell Res. 2017 doi: 10.1016/j.yexcr.2017.03.055. [DOI] [PubMed] [Google Scholar]
- 15.Eckle T, Hartmann K, Bonney S, Reithel S, Mittelbronn M, Walker LA, Lowes BD, Han J, Borchers CH, Buttrick PM, Kominsky DJ, Colgan SP, Eltzschig HK. Adora2b-elicited Per2 stabilization promotes a HIF-dependent metabolic switch crucial for myocardial adaptation to ischemia. Nat Med. 2012;18:774–82. doi: 10.1038/nm.2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eckle T. About Dogs, Mice, and Men: From Ischemic Preconditioning to Anesthetic Postconditioning of the Heart. Semin Cardiothorac Vasc Anesth. 2014;18:247–248. doi: 10.1177/1089253214542253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu Y, Tang D, Liu N, Xiong W, Huang H, Li Y, Ma Z, Zhao H, Chen P, Qi X, Zhang EE. Reciprocal Regulation between the Circadian Clock and Hypoxia Signaling at the Genome Level in Mammals. Cell Metab. 2017;25:73–85. doi: 10.1016/j.cmet.2016.09.009. [DOI] [PubMed] [Google Scholar]
- 18.Adamovich Y, Ladeuix B, Golik M, Koeners MP, Asher G. Rhythmic Oxygen Levels Reset Circadian Clocks through HIF1alpha. Cell Metab. 2017;25:93–101. doi: 10.1016/j.cmet.2016.09.014. [DOI] [PubMed] [Google Scholar]
- 19.Bartman CM, Oyama Y, Brodsky K, Khailova L, Walker L, Koeppen M, Eckle T. Intense light-elicited upregulation of miR-21 facilitates glycolysis and cardioprotection through Per2-dependent mechanisms. PLoS One. 2017;12:e0176243. doi: 10.1371/journal.pone.0176243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, Gocayne JD, Amanatides P, Ballew RM, Huson DH, Wortman JR, Zhang Q, Kodira CD, Zheng XH, Chen L, Skupski M, Subramanian G, Thomas PD, Zhang J, Gabor Miklos GL, Nelson C, Broder S, Clark AG, Nadeau J, McKusick VA, Zinder N, Levine AJ, Roberts RJ, Simon M, Slayman C, Hunkapiller M, Bolanos R, Delcher A, Dew I, Fasulo D, Flanigan M, Florea L, Halpern A, Hannenhalli S, Kravitz S, Levy S, Mobarry C, Reinert K, Remington K, Abu-Threideh J, Beasley E, Biddick K, Bonazzi V, Brandon R, Cargill M, Chandramouliswaran I, Charlab R, Chaturvedi K, Deng Z, Di Francesco V, Dunn P, Eilbeck K, Evangelista C, Gabrielian AE, Gan W, Ge W, Gong F, Gu Z, Guan P, Heiman TJ, Higgins ME, Ji RR, Ke Z, Ketchum KA, Lai Z, Lei Y, Li Z, Li J, Liang Y, Lin X, Lu F, Merkulov GV, Milshina N, Moore HM, Naik AK, Narayan VA, Neelam B, Nusskern D, Rusch DB, Salzberg S, Shao W, Shue B, Sun J, Wang Z, Wang A, Wang X, Wang J, Wei M, Wides R, Xiao C, Yan C, Yao A, Ye J, Zhan M, Zhang W, Zhang H, Zhao Q, Zheng L, Zhong F, Zhong W, Zhu S, Zhao S, Gilbert D, Baumhueter S, Spier G, Carter C, Cravchik A, Woodage T, Ali F, An H, Awe A, Baldwin D, Baden H, Barnstead M, Barrow I, Beeson K, Busam D, Carver A, Center A, Cheng ML, Curry L, Danaher S, Davenport L, Desilets R, Dietz S, Dodson K, Doup L, Ferriera S, Garg N, Gluecksmann A, Hart B, Haynes J, Haynes C, Heiner C, Hladun S, Hostin D, Houck J, Howland T, Ibegwam C, Johnson J, Kalush F, Kline L, Koduru S, Love A, Mann F, May D, McCawley S, McIntosh T, McMullen I, Moy M, Moy L, Murphy B, Nelson K, Pfannkoch C, Pratts E, Puri V, Qureshi H, Reardon M, Rodriguez R, Rogers YH, Romblad D, Ruhfel B, Scott R, Sitter C, Smallwood M, Stewart E, Strong R, Suh E, Thomas R, Tint NN, Tse S, Vech C, Wang G, Wetter J, Williams S, Williams M, Windsor S, Winn-Deen E, Wolfe K, Zaveri J, Zaveri K, Abril JF, Guigo R, Campbell MJ, Sjolander KV, Karlak B, Kejariwal A, Mi H, Lazareva B, Hatton T, Narechania A, Diemer K, Muruganujan A, Guo N, Sato S, Bafna V, Istrail S, Lippert R, Schwartz R, Walenz B, Yooseph S, Allen D, Basu A, Baxendale J, Blick L, Caminha M, Carnes-Stine J, Caulk P, Chiang YH, Coyne M, Dahlke C, Mays A, Dombroski M, Donnelly M, Ely D, Esparham S, Fosler C, Gire H, Glanowski S, Glasser K, Glodek A, Gorokhov M, Graham K, Gropman B, Harris M, Heil J, Henderson S, Hoover J, Jennings D, Jordan C, Jordan J, Kasha J, Kagan L, Kraft C, Levitsky A, Lewis M, Liu X, Lopez J, Ma D, Majoros W, McDaniel J, Murphy S, Newman M, Nguyen T, Nguyen N, Nodell M, Pan S, Peck J, Peterson M, Rowe W, Sanders R, Scott J, Simpson M, Smith T, Sprague A, Stockwell T, Turner R, Venter E, Wang M, Wen M, Wu D, Wu M, Xia A, Zandieh A, Zhu X. The sequence of the human genome. Science. 2001;291:1304–51. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 21.Barwari T, Joshi A, Mayr M. MicroRNAs in Cardiovascular Disease. J Am Coll Cardiol. 2016;68:2577–2584. doi: 10.1016/j.jacc.2016.09.945. [DOI] [PubMed] [Google Scholar]
- 22.Biggar KK, Storey KB. The emerging roles of microRNAs in the molecular responses of metabolic rate depression. J Mol Cell Biol. 2010 doi: 10.1093/jmcb/mjq045. [DOI] [PubMed] [Google Scholar]
- 23.Bronze-da-Rocha E. MicroRNAs expression profiles in cardiovascular diseases. Biomed Res Int. 2014;2014:985408. doi: 10.1155/2014/985408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Condorelli G, Latronico MV, Cavarretta E. microRNAs in cardiovascular diseases: current knowledge and the road ahead. J Am Coll Cardiol. 2014;63:2177–87. doi: 10.1016/j.jacc.2014.01.050. [DOI] [PubMed] [Google Scholar]
- 25.De Rosa S, Curcio A, Indolfi C. Emerging role of microRNAs in cardiovascular diseases. Circ J. 2014;78:567–75. doi: 10.1253/circj.cj-14-0086. [DOI] [PubMed] [Google Scholar]
- 26.De Rosa S, Indolfi C. Circulating microRNAs as Biomarkers in Cardiovascular Diseases. EXS. 2015;106:139–49. doi: 10.1007/978-3-0348-0955-9_6. [DOI] [PubMed] [Google Scholar]
- 27.Dimmeler S, Zeiher AM. Circulating microRNAs: novel biomarkers for cardiovascular diseases? Eur Heart J. 2010;31:2705–7. doi: 10.1093/eurheartj/ehq221. [DOI] [PubMed] [Google Scholar]
- 28.Fichtlscherer S, De Rosa S, Fox H, Schwietz T, Fischer A, Liebetrau C, Weber M, Hamm CW, Roxe T, Muller-Ardogan M, Bonauer A, Zeiher AM, Dimmeler S. Circulating microRNAs in patients with coronary artery disease. Circ Res. 2010;107:677–84. doi: 10.1161/CIRCRESAHA.109.215566. [DOI] [PubMed] [Google Scholar]
- 29.Ohtani K, Dimmeler S. Control of cardiovascular differentiation by microRNAs. Basic Res Cardiol. 2011;106:5–11. doi: 10.1007/s00395-010-0139-7. [DOI] [PubMed] [Google Scholar]
- 30.Ferrari D, Bianchi N, Eltzschig HK, Gambari R. MicroRNAs Modulate the Purinergic Signaling Network. Trends Mol Med. 2016;22:905–918. doi: 10.1016/j.molmed.2016.08.006. [DOI] [PubMed] [Google Scholar]
- 31.Gurha P. MicroRNAs in cardiovascular disease. Curr Opin Cardiol. 2016;31:249–54. doi: 10.1097/HCO.0000000000000280. [DOI] [PubMed] [Google Scholar]
- 32.Castaldi A, Zaglia T, Di Mauro V, Carullo P, Viggiani G, Borile G, Di Stefano B, Schiattarella GG, Gualazzi MG, Elia L, Stirparo GG, Colorito ML, Pironti G, Kunderfranco P, Esposito G, Bang ML, Mongillo M, Condorelli G, Catalucci D. MicroRNA-133 modulates the beta1-adrenergic receptor transduction cascade. Circ Res. 2014;115:273–83. doi: 10.1161/CIRCRESAHA.115.303252. [DOI] [PubMed] [Google Scholar]
- 33.Montgomery RL, Hullinger TG, Semus HM, Dickinson BA, Seto AG, Lynch JM, Stack C, Latimer PA, Olson EN, van Rooij E. Therapeutic inhibition of miR-208a improves cardiac function and survival during heart failure. Circulation. 2011;124:1537–47. doi: 10.1161/CIRCULATIONAHA.111.030932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tijsen AJ, van der Made I, van den Hoogenhof MM, Wijnen WJ, van Deel ED, de Groot NE, Alekseev S, Fluiter K, Schroen B, Goumans MJ, van der Velden J, Duncker DJ, Pinto YM, Creemers EE. The microRNA-15 family inhibits the TGFbeta-pathway in the heart. Cardiovasc Res. 2014;104:61–71. doi: 10.1093/cvr/cvu184. [DOI] [PubMed] [Google Scholar]
- 35.van Rooij E, Sutherland LB, Liu N, Williams AH, McAnally J, Gerard RD, Richardson JA, Olson EN. A signature pattern of stress-responsive microRNAs that can evoke cardiac hypertrophy and heart failure. Proc Natl Acad Sci U S A. 2006;103:18255–60. doi: 10.1073/pnas.0608791103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thum T, Gross C, Fiedler J, Fischer T, Kissler S, Bussen M, Galuppo P, Just S, Rottbauer W, Frantz S, Castoldi M, Soutschek J, Koteliansky V, Rosenwald A, Basson MA, Licht JD, Pena JT, Rouhanifard SH, Muckenthaler MU, Tuschl T, Martin GR, Bauersachs J, Engelhardt S. MicroRNA-21 contributes to myocardial disease by stimulating MAP kinase signalling in fibroblasts. Nature. 2008;456:980–4. doi: 10.1038/nature07511. [DOI] [PubMed] [Google Scholar]
- 37.Patrick DM, Montgomery RL, Qi X, Obad S, Kauppinen S, Hill JA, van Rooij E, Olson EN. Stress-dependent cardiac remodeling occurs in the absence of microRNA-21 in mice. J Clin Invest. 2010;120:3912–6. doi: 10.1172/JCI43604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hullinger TG, Montgomery RL, Seto AG, Dickinson BA, Semus HM, Lynch JM, Dalby CM, Robinson K, Stack C, Latimer PA, Hare JM, Olson EN, van Rooij E. Inhibition of miR-15 protects against cardiac ischemic injury. Circ Res. 2012;110:71–81. doi: 10.1161/CIRCRESAHA.111.244442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boon RA, Iekushi K, Lechner S, Seeger T, Fischer A, Heydt S, Kaluza D, Treguer K, Carmona G, Bonauer A, Horrevoets AJ, Didier N, Girmatsion Z, Biliczki P, Ehrlich JR, Katus HA, Muller OJ, Potente M, Zeiher AM, Hermeking H, Dimmeler S. MicroRNA-34a regulates cardiac ageing and function. Nature. 2013;495:107–10. doi: 10.1038/nature11919. [DOI] [PubMed] [Google Scholar]
- 40.Ren XP, Wu J, Wang X, Sartor MA, Qian J, Jones K, Nicolaou P, Pritchard TJ, Fan GC. MicroRNA-320 is involved in the regulation of cardiac ischemia/reperfusion injury by targeting heat-shock protein 20. Circulation. 2009;119:2357–66. doi: 10.1161/CIRCULATIONAHA.108.814145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li J, Li Y, Jiao J, Wang J, Li Y, Qin D, Li P. Mitofusin 1 is negatively regulated by microRNA 140 in cardiomyocyte apoptosis. Mol Cell Biol. 2014;34:1788–99. doi: 10.1128/MCB.00774-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shan ZX, Lin QX, Deng CY, Zhu JN, Mai LP, Liu JL, Fu YH, Liu XY, Li YX, Zhang YY, Lin SG, Yu XY. miR-1/miR-206 regulate Hsp60 expression contributing to glucose-mediated apoptosis in cardiomyocytes. FEBS Lett. 2010;584:3592–600. doi: 10.1016/j.febslet.2010.07.027. [DOI] [PubMed] [Google Scholar]
- 43.Zhang B, Zhou M, Li C, Zhou J, Li H, Zhu D, Wang Z, Chen A, Zhao Q. MicroRNA-92a inhibition attenuates hypoxia/reoxygenation-induced myocardiocyte apoptosis by targeting Smad7. PLoS One. 2014;9:e100298. doi: 10.1371/journal.pone.0100298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang X, Huang F, Yang D, Dong F, Shi X, Wang H, Zhou X, Wang S, Dai S. Expression of microRNA-122 contributes to apoptosis in H9C2 myocytes. J Cell Mol Med. 2012;16:2637–46. doi: 10.1111/j.1582-4934.2012.01577.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li X, Kong M, Jiang D, Qian J, Duan Q, Dong A. MicroRNA-150 aggravates H2O2-induced cardiac myocyte injury by down-regulating c-myb gene. Acta Biochim Biophys Sin (Shanghai) 2013;45:734–41. doi: 10.1093/abbs/gmt067. [DOI] [PubMed] [Google Scholar]
- 46.Wang L, Huang H, Fan Y, Kong B, Hu H, Hu K, Guo J, Mei Y, Liu WL. Effects of downregulation of microRNA-181a on H2O2-induced H9c2 cell apoptosis via the mitochondrial apoptotic pathway. Oxid Med Cell Longev. 2014;2014:960362. doi: 10.1155/2014/960362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pan Z, Guo Y, Qi H, Fan K, Wang S, Zhao H, Fan Y, Xie J, Guo F, Hou Y, Wang N, Huo R, Zhang Y, Liu Y, Du Z. M3 subtype of muscarinic acetylcholine receptor promotes cardioprotection via the suppression of miR-376b-5p. PLoS One. 2012;7:e32571. doi: 10.1371/journal.pone.0032571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Qian L, Van Laake LW, Huang Y, Liu S, Wendland MF, Srivastava D. miR-24 inhibits apoptosis and represses Bim in mouse cardiomyocytes. J Exp Med. 2011;208:549–60. doi: 10.1084/jem.20101547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.van Rooij E, Sutherland LB, Thatcher JE, DiMaio JM, Naseem RH, Marshall WS, Hill JA, Olson EN. Dysregulation of microRNAs after myocardial infarction reveals a role of miR-29 in cardiac fibrosis. Proc Natl Acad Sci U S A. 2008;105:13027–32. doi: 10.1073/pnas.0805038105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Roca-Alonso L, Castellano L, Mills A, Dabrowska AF, Sikkel MB, Pellegrino L, Jacob J, Frampton AE, Krell J, Coombes RC, Harding SE, Lyon AR, Stebbing J. Myocardial MiR-30 downregulation triggered by doxorubicin drives alterations in beta-adrenergic signaling and enhances apoptosis. Cell Death Dis. 2015;6:e1754. doi: 10.1038/cddis.2015.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Aurora AB, Mahmoud AI, Luo X, Johnson BA, van Rooij E, Matsuzaki S, Humphries KM, Hill JA, Bassel-Duby R, Sadek HA, Olson EN. MicroRNA-214 protects the mouse heart from ischemic injury by controlling Ca(2)(+) overload and cell death. J Clin Invest. 2012;122:1222–32. doi: 10.1172/JCI59327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li B, Li R, Zhang C, Bian HJ, Wang F, Xiao J, Liu SW, Yi W, Zhang MX, Wang SX, Zhang Y, Su GH, Ji XP. MicroRNA-7a/b protects against cardiac myocyte injury in ischemia/reperfusion by targeting poly(ADP-ribose) polymerase. PLoS One. 2014;9:e90096. doi: 10.1371/journal.pone.0090096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Frank D, Gantenberg J, Boomgaarden I, Kuhn C, Will R, Jarr KU, Eden M, Kramer K, Luedde M, Mairbaurl H, Katus HA, Frey N. MicroRNA-20a inhibits stress-induced cardiomyocyte apoptosis involving its novel target Egln3/PHD3. J Mol Cell Cardiol. 2012;52:711–7. doi: 10.1016/j.yjmcc.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 54.Katare R, Riu F, Mitchell K, Gubernator M, Campagnolo P, Cui Y, Fortunato O, Avolio E, Cesselli D, Beltrami AP, Angelini G, Emanueli C, Madeddu P. Transplantation of human pericyte progenitor cells improves the repair of infarcted heart through activation of an angiogenic program involving micro-RNA-132. Circ Res. 2011;109:894–906. doi: 10.1161/CIRCRESAHA.111.251546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.He S, Liu P, Jian Z, Li J, Zhu Y, Feng Z, Xiao Y. miR-138 protects cardiomyocytes from hypoxia-induced apoptosis via MLK3/JNK/c-jun pathway. Biochem Biophys Res Commun. 2013;441:763–9. doi: 10.1016/j.bbrc.2013.10.151. [DOI] [PubMed] [Google Scholar]
- 56.Zhang X, Wang X, Zhu H, Zhu C, Wang Y, Pu WT, Jegga AG, Fan GC. Synergistic effects of the GATA-4-mediated miR-144/451 cluster in protection against simulated ischemia/reperfusion-induced cardiomyocyte death. J Mol Cell Cardiol. 2010;49:841–50. doi: 10.1016/j.yjmcc.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu J, van Mil A, Vrijsen K, Zhao J, Gao L, Metz CH, Goumans MJ, Doevendans PA, Sluijter JP. MicroRNA-155 prevents necrotic cell death in human cardiomyocyte progenitor cells via targeting RIP1. J Cell Mol Med. 2011;15:1474–82. doi: 10.1111/j.1582-4934.2010.01104.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hu S, Huang M, Li Z, Jia F, Ghosh Z, Lijkwan MA, Fasanaro P, Sun N, Wang X, Martelli F, Robbins RC, Wu JC. MicroRNA-210 as a novel therapy for treatment of ischemic heart disease. Circulation. 2010;122:S124–31. doi: 10.1161/CIRCULATIONAHA.109.928424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang J, Jia Z, Zhang C, Sun M, Wang W, Chen P, Ma K, Zhang Y, Li X, Zhou C. miR-499 protects cardiomyocytes from H 2O 2-induced apoptosis via its effects on Pdcd4 and Pacs2. RNA Biol. 2014;11:339–50. doi: 10.4161/rna.28300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang K, Liu F, Zhou LY, Ding SL, Long B, Liu CY, Sun T, Fan YY, Sun L, Li PF. miR-874 regulates myocardial necrosis by targeting caspase-8. Cell Death Dis. 2013;4:e709. doi: 10.1038/cddis.2013.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xiao J, Pan Y, Li XH, Yang XY, Feng YL, Tan HH, Jiang L, Feng J, Yu XY. Cardiac progenitor cell-derived exosomes prevent cardiomyocytes apoptosis through exosomal miR-21 by targeting PDCD4. Cell Death Dis. 2016;7:e2277. doi: 10.1038/cddis.2016.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xu X, Kriegel AJ, Jiao X, Liu H, Bai X, Olson J, Liang M, Ding X. miR-21 in ischemia/reperfusion injury: a double-edged sword? Physiol Genomics. 2014;46:789–97. doi: 10.1152/physiolgenomics.00020.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Deng W, Wang Y, Long X, Zhao R, Wang Z, Liu Z, Cao S, Shi B. miR-21 Reduces Hydrogen Peroxide-Induced Apoptosis in c-kit+ Cardiac Stem Cells In Vitro through PTEN/PI3K/Akt Signaling. Oxid Med Cell Longev. 2016;2016:5389181. doi: 10.1155/2016/5389181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zeng YL, Zheng H, Chen QR, Yuan XH, Ren JH, Luo XF, Chen P, Lin ZY, Chen SZ, Wu XQ, Xiao M, Chen YQ, Chen ZZ, Hu JD, Yang T. Bone marrow-derived mesenchymal stem cells overexpressing MiR-21 efficiently repair myocardial damage in rats. Oncotarget. 2017;8:29161–29173. doi: 10.18632/oncotarget.16254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Olson JM, Yan Y, Bai X, Ge ZD, Liang M, Kriegel AJ, Twaroski DM, Bosnjak ZJ. Up-regulation of microRNA-21 mediates isoflurane-induced protection of cardiomyocytes. Anesthesiology. 2015;122:795–805. doi: 10.1097/ALN.0000000000000567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bonney S, Hughes K, Eckle T. Anesthetic cardioprotection: the role of adenosine. Curr Pharm Des. 2014;20:5690–5. doi: 10.2174/1381612820666140204102524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Qiao S, Olson JM, Paterson M, Yan Y, Zaja I, Liu Y, Riess ML, Kersten JR, Liang M, Warltier DC, Bosnjak ZJ, Ge ZD. MicroRNA-21 Mediates Isoflurane-induced Cardioprotection against Ischemia-Reperfusion Injury via Akt/Nitric Oxide Synthase/Mitochondrial Permeability Transition Pore Pathway. Anesthesiology. 2015;123:786–98. doi: 10.1097/ALN.0000000000000807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cheng Y, Zhu P, Yang J, Liu X, Dong S, Wang X, Chun B, Zhuang J, Zhang C. Ischaemic preconditioning-regulated miR-21 protects heart against ischaemia/reperfusion injury via anti-apoptosis through its target PDCD4. Cardiovasc Res. 2010;87:431–9. doi: 10.1093/cvr/cvq082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Frank A, Bonney M, Bonney S, Weitzel L, Koeppen M, Eckle T. Myocardial ischemia reperfusion injury: from basic science to clinical bedside. Semin Cardiothorac Vasc Anesth. 2012;16:123–32. doi: 10.1177/1089253211436350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hansen KF, Sakamoto K, Obrietan K. MicroRNAs: a potential interface between the circadian clock and human health. Genome Med. 2011;3:10. doi: 10.1186/gm224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sire C, Moreno AB, Garcia-Chapa M, Lopez-Moya JJ, San Segundo B. Diurnal oscillation in the accumulation of Arabidopsis microRNAs, miR167, miR168, miR171 and miR398. FEBS Lett. 2009;583:1039–44. doi: 10.1016/j.febslet.2009.02.024. [DOI] [PubMed] [Google Scholar]
- 72.Cheng HY, Papp JW, Varlamova O, Dziema H, Russell B, Curfman JP, Nakazawa T, Shimizu K, Okamura H, Impey S, Obrietan K. microRNA modulation of circadian-clock period and entrainment. Neuron. 2007;54:813–29. doi: 10.1016/j.neuron.2007.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Impey S, McCorkle SR, Cha-Molstad H, Dwyer JM, Yochum GS, Boss JM, McWeeney S, Dunn JJ, Mandel G, Goodman RH. Defining the CREB regulon: a genome-wide analysis of transcription factor regulatory regions. Cell. 2004;119:1041–54. doi: 10.1016/j.cell.2004.10.032. [DOI] [PubMed] [Google Scholar]
- 74.Gatfield D, Le Martelot G, Vejnar CE, Gerlach D, Schaad O, Fleury-Olela F, Ruskeepaa AL, Oresic M, Esau CC, Zdobnov EM, Schibler U. Integration of microRNA miR-122 in hepatic circadian gene expression. Genes Dev. 2009;23:1313–26. doi: 10.1101/gad.1781009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tan X, Zhang P, Zhou L, Yin B, Pan H, Peng X. Clock-controlled mir-142–3p can target its activator, Bmal1. BMC Mol Biol. 2012;13:27. doi: 10.1186/1471-2199-13-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shende VR, Goldrick MM, Ramani S, Earnest DJ. Expression and rhythmic modulation of circulating microRNAs targeting the clock gene Bmal1 in mice. PLoS One. 2011;6:e22586. doi: 10.1371/journal.pone.0022586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Curtis AM, Fagundes CT, Yang G, Palsson-McDermott EM, Wochal P, McGettrick AF, Foley NH, Early JO, Chen L, Zhang H, Xue C, Geiger SS, Hokamp K, Reilly MP, Coogan AN, Vigorito E, FitzGerald GA, O’Neill LA. Circadian control of innate immunity in macrophages by miR-155 targeting Bmal1. Proc Natl Acad Sci U S A. 2015;112:7231–6. doi: 10.1073/pnas.1501327112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chen R, D’Alessandro M, Lee C. miRNAs are required for generating a time delay critical for the circadian oscillator. Curr Biol. 2013;23:1959–68. doi: 10.1016/j.cub.2013.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Schroeder AM, Colwell CS. How to fix a broken clock. Trends Pharmacol Sci. 2013;34:605–19. doi: 10.1016/j.tips.2013.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ritchie H, Stothard ER, Wright KP. Entrainment of the human circadian clock to the light-dark cycle and its impact on patients in the ICU and nursing home settings. Curr Pham Des. 2015 doi: 10.2174/1381612821666150706111155. [DOI] [PubMed] [Google Scholar]
- 81.Arble DM, Ramsey KM, Bass J, Turek FW. Circadian disruption and metabolic disease: findings from animal models. Best Pract Res Clin Endocrinol Metab. 2010;24:785–800. doi: 10.1016/j.beem.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Foster RG, Peirson SN, Wulff K, Winnebeck E, Vetter C, Roenneberg T. Sleep and circadian rhythm disruption in social jetlag and mental illness. Prog Mol Biol Transl Sci. 2013;119:325–46. doi: 10.1016/B978-0-12-396971-2.00011-7. [DOI] [PubMed] [Google Scholar]
- 83.Depner CM, Stothard ER, Wright KP., Jr Metabolic consequences of sleep and circadian disorders. Curr Diab Rep. 2014;14:507. doi: 10.1007/s11892-014-0507-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hatori M, Vollmers C, Zarrinpar A, DiTacchio L, Bushong EA, Gill S, Leblanc M, Chaix A, Joens M, Fitzpatrick JA, Ellisman MH, Panda S. Time-restricted feeding without reducing caloric intake prevents metabolic diseases in mice fed a high-fat diet. Cell Metab. 2012;15:848–60. doi: 10.1016/j.cmet.2012.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Noyan H, El-Mounayri O, Isserlin R, Arab S, Momen A, Cheng HS, Wu J, Afroze T, Li RK, Fish JE, Bader GD, Husain M. Cardioprotective Signature of Short-Term Caloric Restriction. PLoS One. 2015;10:e0130658. doi: 10.1371/journal.pone.0130658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Schroeder AM, Truong D, Loh DH, Jordan MC, Roos KP, Colwell CS. Voluntary scheduled exercise alters diurnal rhythms of behaviour, physiology and gene expression in wild-type and vasoactive intestinal peptide-deficient mice. J Physiol. 2012;590:6213–26. doi: 10.1113/jphysiol.2012.233676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Dauchy RT, Wren-Dail MA, Hoffman AE, Hanifin JP, Warfield B, Brainard GC, Hill SM, Belancio VP, Dauchy EM, Blask DE. Effects of Daytime Exposure to Light from Blue-Enriched Light-Emitting Diodes on the Nighttime Melatonin Amplitude and Circadian Regulation of Rodent Metabolism and Physiology. Comp Med. 2016;66:373–383. [PMC free article] [PubMed] [Google Scholar]
- 88.Martino TA, Tata N, Belsham DD, Chalmers J, Straume M, Lee P, Pribiag H, Khaper N, Liu PP, Dawood F, Backx PH, Ralph MR, Sole MJ. Disturbed diurnal rhythm alters gene expression and exacerbates cardiovascular disease with rescue by resynchronization. Hypertension. 2007;49:1104–13. doi: 10.1161/HYPERTENSIONAHA.106.083568. [DOI] [PubMed] [Google Scholar]
- 89.Aragones J, Fraisl P, Baes M, Carmeliet P. Oxygen Sensors at the Crossroad of Metabolism. Cell Metab. 2009;9:11–22. doi: 10.1016/j.cmet.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 90.Jakubcakova V, Oster H, Tamanini F, Cadenas C, Leitges M, van der Horst GT, Eichele G. Light entrainment of the mammalian circadian clock by a PRKCA-dependent posttranslational mechanism. Neuron. 2007;54:831–43. doi: 10.1016/j.neuron.2007.04.031. [DOI] [PubMed] [Google Scholar]
- 91.Roenneberg T, Kumar CJ, Merrow M. The human circadian clock entrains to sun time. Curr Biol. 2007;17:R44–5. doi: 10.1016/j.cub.2006.12.011. [DOI] [PubMed] [Google Scholar]
- 92.Lewy AJ, Wehr TA, Goodwin FK, Newsome DA, Markey SP. Light suppresses melatonin secretion in humans. Science. 1980;210:1267–9. doi: 10.1126/science.7434030. [DOI] [PubMed] [Google Scholar]
- 93.Terman JS, Terman M, Lo ES, Cooper TB. Circadian time of morning light administration and therapeutic response in winter depression. Arch Gen Psychiatry. 2001;58:69–75. doi: 10.1001/archpsyc.58.1.69. [DOI] [PubMed] [Google Scholar]
- 94.Terman JS, Terman M, Schlager D, Rafferty B, Rosofsky M, Link MJ, Gallin PF, Quitkin FM. Efficacy of brief, intense light exposure for treatment of winter depression. Psychopharmacol Bull. 1990;26:3–11. [PubMed] [Google Scholar]
- 95.Yang J, Choi W, Ko YH, Joe SH, Han C, Kim YK. Bright light therapy as an adjunctive treatment with risperidone in patients with delirium: a randomized, open, parallel group study. Gen Hosp Psychiatry. 2012;34:546–51. doi: 10.1016/j.genhosppsych.2012.05.003. [DOI] [PubMed] [Google Scholar]
- 96.Eckle T. Editorial: Health Impact and Management of a Disrupted Circadian Rhythm and Sleep in Critical Illnesses. Curr Pharm Des. 2015;21:3428–30. doi: 10.2174/1381612821999150709123504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Phipps-Nelson J, Redman JR, Dijk DJ, Rajaratnam SM. Daytime exposure to bright light, as compared to dim light, decreases sleepiness and improves psychomotor vigilance performance. Sleep. 2003;26:695–700. doi: 10.1093/sleep/26.6.695. [DOI] [PubMed] [Google Scholar]
- 98.Wright KP, Jr, McHill AW, Birks BR, Griffin BR, Rusterholz T, Chinoy ED. Entrainment of the human circadian clock to the natural light-dark cycle. Curr Biol. 2013;23:1554–8. doi: 10.1016/j.cub.2013.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Shinde AV, Frangogiannis NG. Fibroblasts in myocardial infarction: a role in inflammation and repair. J Mol Cell Cardiol. 2014;70:74–82. doi: 10.1016/j.yjmcc.2013.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ballesta A, Innominato PF, Dallmann R, Rand DA, Levi FA. Systems Chronotherapeutics. Pharmacol Rev. 2017;69:161–199. doi: 10.1124/pr.116.013441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Du NH, Arpat AB, De Matos M, Gatfield D. MicroRNAs shape circadian hepatic gene expression on a transcriptome-wide scale. Elife. 2014;3:e02510. doi: 10.7554/eLife.02510. [DOI] [PMC free article] [PubMed] [Google Scholar]