Abstract
Numerous variations are known to occur in the chloroplast genomes of parasitic plants. We determined the complete chloroplast genome sequences of two hemiparasitic species, Taxillus chinensis and T. sutchuenensis, using Illumina and PacBio sequencing technologies. These species are the first members of the family Loranthaceae to be sequenced. The complete chloroplast genomes of T. chinensis and T. sutchuenensis comprise circular 121,363 and 122,562 bp-long molecules with quadripartite structures, respectively. Compared with the chloroplast genomes of Nicotiana tabacum and Osyris alba, all ndh genes as well as three ribosomal protein genes, seven tRNA genes, four ycf genes, and the infA gene of these two species have been lost. The results of the maximum likelihood and neighbor-joining phylogenetic trees strongly support the theory that Loranthaceae and Viscaceae are monophyletic clades. This research reveals the effect of a parasitic lifestyle on the chloroplast structure and genome content of T. chinensis and T. sutchuenensis, and enhances our understanding of the discrepancies in terms of assembly results between Illumina and PacBio.
Introduction
The chloroplast is a key plant cell organelle that carries out photosynthesis1. The chloroplast genome is highly conserved and has multiple copies, which means that target genes are expressed at high levels2,3. In recent years, the chloroplast genome has increasingly been used as a source of molecular markers4,5 and barcoding identification6,7, and genomic information from this organelle has been utilized in studies of plant evolution, phylogenetics, and diversity8,9. With the rapid development of sequencing and bioinformation technology, an increasing number of plant chloroplast genomes, including medicinal plants, have been determined, such as Glycine max 10, Soughum bicolor 11, Magnolia officinalis 12, Taxus chinensis var. mairei13 and Astragalus membranaceus 14.
Given that parasitic plants have either lost some or all their photosynthetic capacity, they absorb organic and inorganic nutrients as well as water from their hosts by maintaining a much higher transpiration rate and using specialized parasitic organs called haustoria15. Despite their large known diversity, only a few chloroplast genomes from parasitic plants have been obtained. The first complete parasitic plant chloroplast genome to be sequenced was from Epifagus virginiana 16. All photosynthesis and energy producing genes in this species have been lost, although a few fragments remain as pseudogenes, and the entire chloroplast genome no longer performs photosynthesis17. Subsequently sequenced chloroplast genomes include four species from the holoparasitic genus Cuscuta, including C. reflexa, C. gronovii, C. exaltata and C. obtusiflora 18,19. Previous studies showed that the chloroplast genome of Rafflesia lagascae is completely lost20. The complete chloroplast genomes of several species within the parasitic family Orobanchaceae have been sequenced and analyzed in recent years, including the completely non-photosynthetic plants, Cistanche deserticola 1, Phelipanche ramosa 21, Orobanche austrohispanica 22, and Lathraea squamaria 23. More recently, Petersen et al. sequenced and analyzed the complete chloroplast genome of one species of the genus Osyris and three species of the genus Viscum 24. A number of photosynthetic and photorespiratory genes, some protein-coding genes, ribosomal protein genes, transfer RNA (tRNA) genes from some parasitic plants have either been completely lost or pseudogenized23–25. Horizontal gene transfer also occurs between donor and recipient in some parasitic plants1,26.
Plants within Loranthaceae comprise hemiparasitic species that have retained photosynthesis and have seeds which are widely propagates by birds27. The taxonomy of plants within Loranthaceae is controversial, particularly regarding the branching point between these taxa and Viscaceae. To date, the plants in China classified within Loranthaceae have been studied and the results demonstrated that, apart from the hemiparasitic characteristics, significant differences exist in pollen morphology28, chemical composition29, and DNA molecules30, which nevertheless support the theory that Loranthaceae and Viscaceae are branched independently. However, one medicinal plant within Viscaceae, namely, Viscum coloratum (Kom.) Nakai, is assigned to Loranthaceae in the Chinese Pharmacopoeia31.
Approximately 70 genera comprising more than 900 species are classified within Loranthaceae32. Most of these plant species primarily live in tropical and subtropical regions, with 8 genera and 51 species (18 endemic) found in China33. Of these genera, the hemiparasitic plant genus Taxillus consists of species with degenerated chloroplasts and restricted photosynthetic capacity. Specifically, T. chinensis is used in traditional Chinese herbal medicine and is recorded in the Chinese Pharmacopoeia31. Another species, namely, T. sutchuenensis, is used in folk medicine. These two medicinal plants are commonly used to treat diseases, such as rheumatism, hypertension, and fetal irritability34,35. The recorded hosts of T. chinensis and T. sutchuenensis include species within Moraceae, Rutaceae, Aceraceae, Anacardiaceae, Euphorbiaceae, Rosaceae, Theaceae and rarely Taxodiaceae33.
The third-generation sequencing platform PacBio is based on single-molecule real-time (SMRT) sequencing technology. The main advantage of this sequencing approach is the long read length, generating read lengths of over 10 kb on average, with some reads possibly reaching up to 60 kb36–38. Previous studies have demonstrated that the long read lengths provide many benefits in genome assembly, including generating longer contigs and fewer unresolved gaps39. PacBio has been successfully applied in a number of chloroplast genome sequencing projects involving three species of Fritillaria 38, Aconitum barbatum var. puberulum 40, and Swertia mussotii41. However, PacBio has high rates of random error in single-pass reads37. In this study, the chloroplast genome sequence of T. chinensis was sequenced using second-generation Illumina platform and third-generation PacBio system to verify the accuracy of the genome sequence.
We report the complete chloroplast genome of T. chinensis and T. sutchuenensis, which are the first two sequences completed within Loranthaceae. We also present a comparative analysis of the genetic changes together with chloroplast genomes of five other species, including the previously reported sequence of Viscum minimum, to determine the effect of a parasitic lifestyle on chloroplast structure and the genome content. We also analyzed the phylogenetic relationships of T. chinensis and T. sutchuenensis within Dicotyledoneae based on the complete chloroplast genomes to provide baseline data for systematic classification of Loranthaceae.
Results
Chloroplast Genome Structures of T. chinensis and T. sutchuenensis
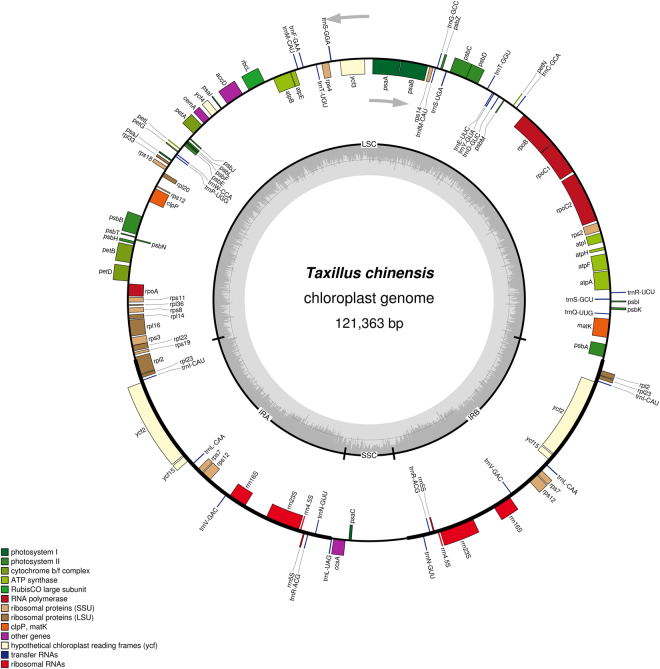
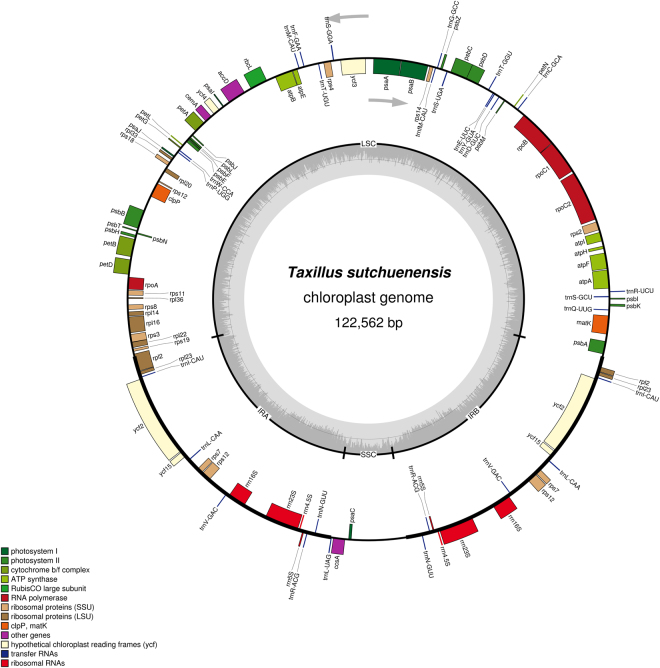
Results show that the chloroplast genome sequence of T. chinensis is a circular molecule that is 121,363 bp in length, which can be divided into a large single-copy (LSC) region of 70,357 bp and a small single-copy (SSC) region of 6,082 bp, and separated by a pair of inverted repeats (IRa and IRb) each 22,462 bp in length (Fig. 1). This sequence, which was assembled using the reads obtained by the Illumina sequencing platform, is 121,363 bp in length. By contrast, the sequence assembled using the reads obtained by the PacBio system is 12 bp shorter than that assembled from the reads obtained by the Illumina platform. After verification using PCR, we found that the complete chloroplast genome of T. chinensis is consistent with the assembly results obtained using the reads from second-generation sequencing. The chloroplast genome of T. sutchuenensis is extremely similar to that of T. chinensis in size and genomic structure; it is 122,562 bp in length and retains a typical structure comprising a LSC (70,630 bp), a SSC (6,102 bp), and two IRs, each having 22,915 bp (Fig. 2). The complete and correct chloroplast genome sequences of T. chinensis and T. sutchuenensis were deposited in GenBank under accession numbers KY996492 and KY996493, respectively.
Figure 1.
Gene map of the complete chloroplast genome of T. chinensis. Genes on the inside of the circle are transcribed clockwise, while those outside are transcribed counter clockwise. The darker gray in the inner circle corresponds to GC content, whereas the lighter gray corresponds to AT content.
Figure 2.
Gene map of the complete chloroplast genome of T. sutchuenensis. Genes on the inside of the circle are transcribed clockwise, while those outside are transcribed counter clockwise. The darker gray in the inner circle corresponds to GC content, whereas the lighter gray corresponds to AT content.
Data reveal that both species have a GC content of 37.3%, which is unevenly distributed across the whole chloroplast genome. In both cases, the GC content of the IR regions exhibits the highest values across the complete chloroplast genome, 43.0% in T. chinensis and 42.8% in T. sutchuenensis, respectively. This high GC content in IR regions is the result of four rRNA genes (rrn16, rrn23, rrn4.5 and rrn5) that occur in this region42. In addition, after the LSC, which has a GC content of 34.7%, lowest values of 26.2% are seen in SSC regions.
A total of 106 genes were identified in each genome, which include 66 protein-coding genes, 28 tRNAs, 8 rRNAs, and 4 pseudogenes. Simultaneously, we compared these two Taxillus species with autotrophic plants, including Nicotiana tabacum and Osyris alba. Genes encoding subunits of the NAD(P)H dehydrogenase complex (ndh genes) were missing from the chloroplast genome of the two species, whereas three genes for ribosomal proteins (rpl32, rps15, and rps16), seven tRNA genes (trnA-UGC, trnG-UCC, trnH-GUG, trnL-GAU, trnK-UUU, trnL-UAA, and trnV-UAC), four ycf genes (ycf1, ycf5, ycf9, and ycf10), and initiation factor gene (infA) were also lost (Table S1). Two ribosomal protein genes (rpl16 and rpl2) and the duplicate gene ycf15 have also been pseudogenized because their gene-coding regions are interrupted by deletion, insertions or internal stop codons, while the pseudogene rpl2 is located in IRb region. We designed the primers to perform PCR to verify the accuracy of the pseudogenes rpl16, ycf15 and rpl2. The primer sequences are listed in Supplementary Table S2.
The basic information and gene contents of the chloroplast genomes of T. chinensis and T. sutchuenensis compared to other five species are presented in Table 1 and Supplementary Table S1.
Table 1.
Comparisons among the chloroplast genome characteristics of T. chinensis, T. sutchuenensis, and other five species.
| Species | Tax i llus ch i nens i s | Tax i llus sutchuenens i s | V i scum m i n i mum | Osyr i s alba | Schoepf i a jasm i nodora | Ep i fagus v i rg i n i ana | N i cot i ana tabacum |
|---|---|---|---|---|---|---|---|
| Family | Loranthaceae | Loranthaceae | Viscaceae | Santalaceae | Olacaceae | Orobanchaceae | Solanaceae |
| Accession No. | KY996492 | KY996493 | KJ512176 | KT070882 | KX775962 | M81884 | Z00044 |
| Genome size(bp) | 121,363 | 122,562 | 131,016 | 147,253 | 118,743 | 70,028 | 155,844 |
| LSC length(bp) | 70,357 | 70,630 | 75,814 | 84,601 | 84,168 | 19,799 | 86,684 |
| SSC length(bp) | 6,082 | 6,102 | 9,014 | 13,972 | 9,763 | 4,759 | 18,482 |
| IR length(bp) | 22,462 | 22,915 | 23,094 | 24,340 | 12,406 | 22,735 | 25,339 |
| GC content(%) | 37.3 | 37.3 | 36.2 | 37.7 | 38.1 | 37.5 | 37.8 |
| Number of genes | 106 | 106 | 104 | 114 | 112 | 53 | 151 |
| Number of protein-coding genes | 66 | 66 | 66 | 67 | 69 | 10 | 112 |
| Number of tRNAs | 28 | 28 | 29 | 30 | 35 | 17 | 30 |
| Number of rRNAs | 8 | 8 | 8 | 8 | 8 | 8 | 8 |
| Number of pseudogenes | 4 | 4 | 1 | 9 | 5 | 18 | 1 |
Introns play an important role in the regulation of gene expression. Introns enhance exogenous gene expression at specific sites within plants at particular times, resulting in desirable agronomic traits43. Introns within these two species are similar to other angiosperms1,44,45. Results reveal the presence of nine genes containing introns in each chloroplast genome, including atpF, rpoC1, ycf3, rps12, rpl2, ψrpl16, clpP, petB, and petD. In addition, the ycf3 gene and rps12 gene each contain two introns and three exons. The ycf3 gene is located within the LSC, as seen in Metasequoia glyptostroboides 45, Aquilaria sinensis 46, while the rps12 gene is specialized for trans-splicing. The 5′ exon is located in the LSC, and the 3′ exon is located in the IR, as is the case in Panax ginseng 44, C. deserticola 1, and L. squamaria 23. Relevant lengths of exons and introns are listed in Table 2.
Table 2.
Genes with introns in the chloroplast genomes of T. chinensis and T. sutchuenensis as well as the lengths of the exons and introns.
| Species | Gene | Location | Exon1(bp) | Intron1(bp) | Exon2(bp) | Intron2(bp) | Exon3(bp) |
|---|---|---|---|---|---|---|---|
| T. chinensis | atpF | LSC | 150 | 779 | 375 | ||
| clpP | LSC | 335 | 621 | 229 | |||
| petB | LSC | 6 | 755 | 642 | |||
| petD | LSC | 9 | 718 | 483 | |||
| rpl2 | LSC; IR | 394 | 645 | 437 | |||
| ψrpl16 | LSC | 10 | 924 | 397 | |||
| rps12 | LSC, IR | 114 | — | 232 | 543 | 26 | |
| rpoC1 | LSC | 456 | 752 | 1617 | |||
| ycf3 | LSC | 127 | 730 | 230 | 771 | 153 | |
| T. sutchuenensis | atpF | LSC | 163 | 753 | 410 | ||
| clpP | LSC | 332 | 634 | 229 | |||
| petB | LSC | 6 | 799 | 642 | |||
| petD | LSC | 6 | 715 | 483 | |||
| rpl2 | LSC; IR | 399 | 712 | 369 | |||
| ψrpl16 | LSC | 9 | 924 | 389 | |||
| rps12 | LSC, IR | 114 | — | 232 | 539 | 26 | |
| rpoC1 | LSC | 450 | 756 | 1602 | |||
| ycf3 | LSC | 127 | 759 | 230 | 785 | 153 |
Comparative genome analyses
Data plotted using mVISTA (Fig. S1) reveal that non-coding regions of the chloroplast genomes of the two Taxillus species are more divergent than their coding counterparts. Moreover, the two IR regions have lower sequence divergence than the LSC and SSC regions. Similar results were obtained in previous research on the complete chloroplast genomes of five Lamiales species42 as well as in a comparative study of five Epimedium chloroplast genomes47. In the present study, rpl16 gene is the most divergent of the coding regions, probably because of pseudogenization. Thus, we conducted a series of linear rearrangement comparisons across the complete chloroplast genome sequences of six species (T. chinensis, T. sutchuenensis, S. jasminodora, V. minimum, O. alba and N. tabacum) aligned in Geneious using the Mauve algorithm (Fig. 3). The comparisons reveal the presence of two structural variants, including an approximately 24-kb-long inversion within the LSC region of the V. minimum chloroplast genome and an approximately 3-kb-long inversion in the SSC region of the O. alba chloroplast genome, which is consistent with a previous report24. The lengths of the IR regions in our two species are also similar to that of other plants, with the exception of S. jasminodora 48 where they are much shorter (at least 10 kb) than the length of the IR regions of the five species considered here, including T. chinensis and T. sutchuenensis.
Figure 3.
Comparison of the complete chloroplast genomes of six species using the MAUVE algorithm. Local collinear blocks are colored in this figure to indicate syntenic regions, while histograms within each block represent the degree of sequence similarity.
Codon Usage
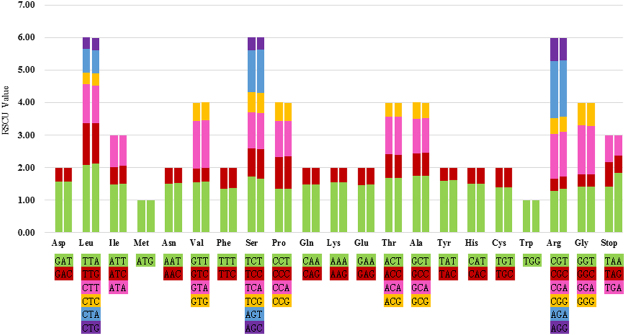
The calculations for the codon usage of protein-coding genes within T. chinensis and T. sutchuenensis chloroplast genomes are summarized in Fig. 4 and Supplementary Table S3. Results reveal the presence of 63 codons encoding 20 amino acids within the chloroplast protein-coding genes of these two species; of these, 1711 encode leucine and 191 encode cysteine, which are respectively the most and least prevalent amino acids in T. chinensis chloroplast genome. Results also reveal that most of the amino acid codons have preferences, with the exception of methionine and tryptophan. Moreover, usage is generally biased toward A or T with high relative synonymous codon usage (RSCU) values, including TTA (2.12) in leucine, TAT (1.62) in tyrosine, and the stop-codon TAA (1.84) in the T. sutchuenensis chloroplast genome (Supplementary Table S3). The data presented in Fig. 4 illustrates that the RSCU value increases with the quantity of codons that code for a specific amino acid. High codon preference, especially a strong AT bias in codon usage, is very common in other land plant chloroplast genomes42,44. The present results are similar to the chloroplast genomes of A. sinensis 46 and species within the genus Ulmus 49 in terms of codon usage.
Figure 4.
Codon content of 20 amino acid and stop codons in all protein-coding genes of the chloroplast genomes of two species. The histogram on the left-hand side of each amino acid shows codon usage within the T. chinensis chloroplast genome, while the right-hand side illustrates the genome of T. sutchuenensis.
Simple Sequence Repeats (SSRs) Analyses
SSRs are ubiquitous throughout genomes and are also known as microsatellites. SSRs comprise tandem repeated DNA sequences that consist of between one and six repeat nucleotide units50. As such, SSRs are widely used as molecular markers in species identification, population genetics, and phylogenetic investigations because they exhibit high levels of polymorphism51–53. In total, 195 and 198 SSRs are identified within the chloroplast genomes of T. chinensis and T. sutchuenensis, respectively (Table 3; Supplementary Tables S4–S5), which mainly comprise mononucleotide repeats encountered 146 and 139 times in each case. In addition, A/T mononucleotide repeats (93.9% and 96.4%, respectively; Table 3) are the most common, while the majority of dinucleotide repeat sequences comprise AT/TA repeats (59.5% and 67.3%, respectively; Table 3). Results show that SSRs within the chloroplast genomes of T. chinensis and T. sutchuenensis are dominated by AT-rich repetitive motifs, which is consistent with the fact that AT content is also very high (62.7%) in these species. This result is also in agreement with previous studies showing higher proportions of polyadenine (polyA) and polythymine (polyT) relative to polycytosine (polyC) and polyguanine (polyG) within the chloroplast SSRs in many plants6.
Table 3.
Types and amounts of SSRs in the T. chinensis and T. sutchuenensis chloroplast genomes.
| SSR type | Repeat unit | Amount | Ratio(%) | ||
|---|---|---|---|---|---|
| T. chinensis | T. sutchuenensis | T. chinensis | T. sutchuenensis | ||
| mono | A/T | 138 | 134 | 93.9 | 96.4 |
| C/G | 9 | 5 | 6.1 | 3.6 | |
| di | AC/GT | 3 | 4 | 7.2 | 7.7 |
| AG/CT | 14 | 13 | 33.3 | 25 | |
| AT/TA | 25 | 35 | 59.5 | 67.3 | |
| tri | AAG/CTT | 2 | 2 | 100 | 50 |
| AAT/ATT | 0 | 2 | 0 | 50 | |
| tetra | AAAC/GTTT | 1 | 0 | 25 | 0 |
| AAAG/CTTT | 1 | 0 | 25 | 0 | |
| AATC/ATTG | 1 | 0 | 25 | 0 | |
| ACAG/CTGT | 1 | 1 | 25 | 50 | |
| AAGT/ACTT | 0 | 1 | 0 | 50 | |
| penta | AATAT/ATATT | 1 | 1 | 100 | 100 |
Phylogenetic Analyses
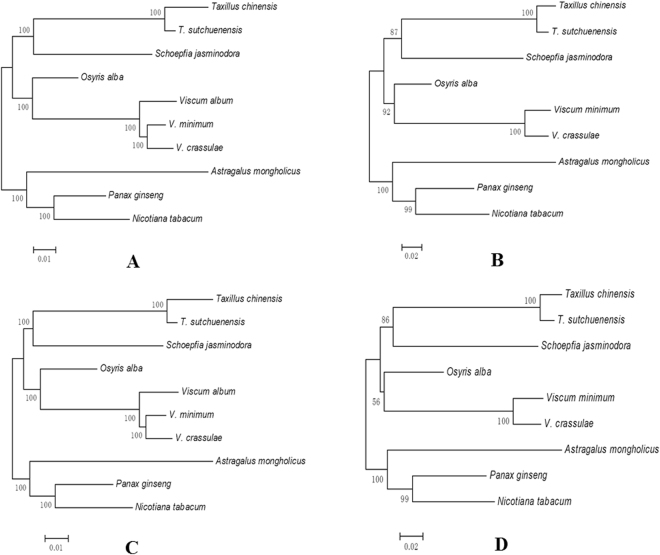
Phylogenetic trees were constructed using two methods based on two datasets from different species (Fig. 5). Results revealed extremely similar tree topologies from each dataset irrespective of the method used, as supported by high bootstrap values. All nodes in our maximum likelihood (ML) and neighbor-joining (NJ) trees based on 54 protein-coding genes have 100% bootstrap support values, whereas four out of six nodes that received bootstrap values of ≥99% were recovered in both sets of trees when matK genes were used for analyses. All nodes in all phylogenetic trees received higher than 50% bootstrap support. All four phylogenetic trees showed that T. chinensis and T. sutchuenensis are sister taxa with respect to S. jasminodora (Olacaceae), whereas the three species within genus Viscum group with Osyris alba (Santalaceae) and all Santalales species are clustered within a lineage distinct from the outgroup.
Figure 5.
Phylogenetic trees constructed using two methods based on two datasets from different species. (A) ML tree based on 54 protein-coding genes; (B) ML tree based on matK genes; (C) NJ tree based on 54 protein-coding genes; (D) NJ tree based on matK genes. Number at nodes are values for bootstrap support.
Discussion
Numerous variations occur in the chloroplast genomes of parasitic plants. To date, however, most investigations on these genomes in parasitic and heterotrophic plants focused on nonphotosynthetic species24. For instance, some complete chloroplast genomes of holoparasitic plants from Orobanchaceae were reported1,21,22. A small number of hemiparasitic plants within Santalales and other groups have been studied24,48. In this study, the complete chloroplast genomes of T. chinensis and T. sutchuenensis from Santalales were assembled, annotated, and analyzed.
Gene loss events have occurred within the chloroplast genomes of most parasitic plants and in a handful of autotrophic species21,54. Previous work has shown that genes of chlB, chlL, chlN, and trnP-GGG have been lost from the chloroplast genomes of most flowering plants55, whereas gene infA, which codes for a translation initiation factor, is either missing or has been transferred in many plants56, including the two species observed in this study. All ndh genes have been lost from the chloroplast genomes of T. chinensis and T. sutchuenensis, similar to the case of Cuscuta gronovii and C. obtusiflora 18,19. Similarly, nine out of eleven ndh genes have been pseudogenized within the chloroplast genome of L. squamaria 23. This degree of ndh gene degradation is not only observed in heterotrophic organisms, but also in many autotrophic plants, including Orchidaceae57,58, Geraniaceae59 and Cactaceae60. Kim et al. reported that losses of ndh genes in angiosperms are usually associated with nutritional status and/or extensive rearrangements of chloroplast structures57. Also, ndh genes loss events and pseudogenization have occurred in reported chloroplast genomes of parasitic plants, regardless of the degree of degradation in photosynthetic capacity1,23,24,48. As a result, studies suggested that ndh genes were first lost in the transformation from autotrophy to heterotrophy18,61.
In this study, seven transfer RNA genes, including trnK-UUU, have been lost from the chloroplast genomes of both species. Although similar tRNA losses have commonly occurred in most plants (Supplementary Table S1), the trnK-UUU gene, which is generally absent from most parasitic plants, is completely preserved (including its intron matK gene) within the chloroplast genome of Cistanche deserticola 1. Li et al. suggested that tRNA genes from the chloroplast genome were lost later than photosynthesis genes1.
A pseudogene, which is a defective copy of the functional gene, is widespread in the chloroplast genome of plants and has lost the normal protein coding function1,18,19,62. Loss of genic normal activity is generally caused by mutations inhibiting gene expression. Pseudogenes not only demonstrate gene mutation accumulation but are also associated with gene expression and regulation63. Four pseudogenes exist in the chloroplast genomes of T. chinensis and T. sutchuenensis; these pseudogenes include rpl16, rpl2 and ycf15 (duplicate gene). The gene of ycf15 has been pseudogenized in many plants, including S. jasminodora 48, C. reflexa 19 and C. exaltata 18. Genes of rpl16 and rpl2 exist in most plants as functional genes, whereas they have been pseudogenized in the current study.
A previous study pointed out that one early response of the chloroplast genomes to the evolution of a parasitic lifestyle was condensation via losses in numerous non-coding and unimportant regions; this event resulted in reduction of chloroplast genome size19. Although gene loss can be regarded as a terminal evolutionary step, accumulation of point mutations leading to pseudogenization nevertheless occurred at previous steps24.
Second-generation sequencing technology provides an efficient, novel, and rapid method for whole-genome sequencing12,64,65. SMRT sequencing, which is combined with circular consensus sequencing (CCS), provides multiple reads of individual templates40. Wu et al.66 have compared three generations of sequencing technologies (Sanger, Illumina and PacBio) on chloroplast genome assembly. Results demonstrated that long reads from PacBio showed potential for highly accurate “finished” genomes. However, the accuracy between second-generation and third-generation sequencing platforms was not compared thoroughly. In the present study, the complete chloroplast genome sequence of T. chinensis was sequenced using Illumina and PacBio platforms. Discrepancies in terms of assembly results between Illumina and PacBio were detected using PCR-based conventional Sanger sequencing, and the quality is very high. Results revealed that in PacBio platform, the error rate is high in homopolymers when the number of repeat units of a mononucleotide is higher than or equal to six. In the chloroplast genome of T. chinensis, polyA/T and polyC/G (repeat higher than or equal to six) included 509 and 72 sites, respectively. Although A/T mononucleotide repeats are the most common types (Table 3), these errors are mainly present in structures of polyC and polyG (Table 4). Among the 14 errors, 12 were G/C deletions, 1 was A/T deletion, and 1 was A/T insertion. All errors differed in terms of only one base.
Table 4.
Discrepancies in assembly results obtained using Illumina and PacBio.
| No. | Sites (bp) | Repeat unit | Number of repeat unit | Location | |
|---|---|---|---|---|---|
| Illumina | PacBio | ||||
| 1 | 4615 | C | 10 | 9 | intergenic region |
| 2 | 17326 | C | 9 | 8 | introns |
| 3 | 17952 | C | 7 | 6 | introns |
| 4 | 27768 | G | 7 | 6 | psbC |
| 5 | 32700 | C | 6 | 5 | psbA |
| 6 | 41040 | C | 10 | 9 | intergenic region |
| 7 | 45020 | G | 8 | 7 | rbcL |
| 8 | 51046 | T | 6 | 7 | intergenic region |
| 9 | 55514 | C | 10 | 9 | intergenic region |
| 10 | 58056 | A | 9 | 8 | intergenic region |
| 11 | 87056 | G | 7 | 6 | intergenic region |
| 12 | 90138 | C | 8 | 7 | intergenic region |
| 13 | 101590 | G | 8 | 7 | intergenic region |
| 14 | 104671 | C | 7 | 6 | intergenic region |
As a result of multiple comparisons (Table 1 and Fig. 3), we observed that complete lengths of the chloroplast genomes of T. chinensis and T. sutchuenensis are similar to those of S. jasminodora and V. minimum, whereas the lengths of SSC regions are much smaller (at least 3 kb). These regions, which contain most ndh genes, also encapsulate the largest variation within the chloroplast genome67 and have undergone dramatic reductions in some parasitic plants, including L. clandestine 68. Previous studies have demonstrated that positions of IR junction and SSC region are correlated with degeneration of ndhF and ycf1 genes57,69. Loss of ycf1 and all ndh genes (including ndhF), as revealed by this study may explain why SSC chloroplast genome regions of the two considered species are shorter than those of others.
Chloroplast genomes have provided significant data for evolutionary, taxonomic, and phylogenetic studies46. Specifically, the chloroplast gene of matK has been widely utilized in plant phylogenetic analyses70,71. In this study, we constructed phylogenetic trees using ML and NJ methods based on matK and 54 protein-coding genes commonly present in the chloroplast genomes of ten species, including two medicinal hemiparasites in the current study. Phylogenetic results are extremely consistent, irrespective of method and dataset. All phylogenetic results strongly support the theory that Loranthaceae and Viscaceae diverged independently from one another. Phylogenetic results discussed in the present study are broadly consistent with those of a previous research, which utilized chloroplast trnL intron sequences to investigate inter-familial relationships within Santalales30.
Conclusions
The complete chloroplast genome sequences of traditional medicinal hemiparasites T. chinensis and T. sutchuenensis were obtained and analyzed. Results of this study revealed effects of parasitic lifestyle on chloroplast structure and genome content in these species and enhanced understanding of phylogenetic positions and relationships of T. chinensis and T. sutchuenensis. This research also showed that sequences assembled using reads obtained by the Illumina platform is more accurate than those from PacBio.
Materials and Methods
Plant Material, DNA Extraction, and Sequencing
Fresh leaves of T. chinensis and T. sutchuenensis were collected from Qinzhou City in Guangxi Province and from Lichuan City in Hubei Province, respectively. All samples were identified by Professor Yulin Lin, who is based at the Institute of Medicinal Plant Development (IMPLAD), Chinese Academy of Medical Sciences & Peking Union Medical College. The voucher specimens were deposited in the herbarium of IMPLAD. Approximately 100 g of samples frozen in −80 °C were used to extract total genomic DNA using DNeasy Plant Mini Kit (Qiagen Co., Germany). DNA quality was assessed based on electrophoresis and optical density results. DNA of two species was used to generate libraries with average insert size of 500 bp and sequenced using Illumina Hiseq X in accordance with standard protocol. Approximately 4.4 Gb of raw data from T. chinensis and 3.7 Gb from T. sutchuenensis using Illumina sequencing platform were generated with 150 bp paired-end read lengths. To compare PacBio with Illumina sequencing technology when employed in chloroplast genome study, we sequenced a PacBio shotgun library of T. chinensis with an insert size of 3 kb on PacBio RS II platform using P6-C4 chemistry (Pacific Biosciences, Menlo Park, CA, USA). A total of 24,590 CCS reads with a length of 67,512,059 bp were obtained from one SMRT cell, and these reads were used for assembly. Assembly results showed that 13.6% of chloroplast sequences were detected in total data, revealing percentage of chloroplast DNA in total DNA during DNA extraction experiment.
Chloroplast Genome Assembly
Low-quality reads resulting from all samples were trimmed using the software Trimmomatic72. The trimmed reads included a mixture of data from nuclear and organelle genomes. We used the chloroplast genome sequence of Viscum minimum, which was downloaded from GenBank to establish a Basic Local Alignment Search Tool (BLASTn) database. Then all trimmed reads were mapped onto this database, and the mapped reads were extracted from raw data based on coverage and similarity. Extracted reads were assembled to contigs using SOAPdenovo273. SSPACE74 was used to construct the scaffold of the chloroplast genome, and GapCloser73 was used to fill gaps. Reads sequenced using PacBio system were used to assemble the chloroplast genome according to the strategy described by Xiang et al.41. Assembly results obtained using Illumina and PacBio (Table 4) differed in terms of 14 sites, which are all homopolymers and mainly located at intergenic regions. To detect these discrepancies, we performed PCR-based conventional Sanger sequencing. The primer sequences are listed in Supplementary Table S6.
Genome Annotation and Structural Analyses
To verify accuracy, including boundaries of single copy and IR regions of assembled sequences, we designed a series of PCR primers (Supplementary Table S7). Annotations of genome sequences of two Taxillus species were performed using the online software Dual Organellar GenoMe Annotator (DOGMA, http://dogma.ccbb.utexas.edu/)75 and CPGAVAS76 with default settings and checked manually. We then used the software tRNAscan-SE77 to annotate tRNA genes. Boundaries of genes, introns/exons and coding regions were verified using BLAST versus reference sequences. Circular chloroplast genome map was constructed using an online program Organellar Genome DRAW (OGDRAW) v1.278, and subsequently modified manually. GC content was analyzed using the software MEGA 6.079. Genome comparisons between T. chinensis and T. sutchuenensis were performed and plotted using the mVISTA program80. The whole-genome alignment for chloroplast genomes of six species, including T. chinensis, T. sutchuenensis, S. jasminodora, V. minimum, O. alba and N. tabacum, was performed using the algorithm MAUVE V2.3.181 in the software Geneious v10.1.2 (Biomatters Ltd., http://www.geneious.com/).
Codon Usage and SSRs Analyses
RSCU value, the ratio between frequency of use and expected frequency of a particular codon, is a simple method for detecting non-uniform synonymous codon usage (SCU) within a coding sequence82. In the present study, utilizing the RSCU ratio, we performed statistical analyses to investigate the distribution of codon usage with the software CodonW (http://codonw.sourceforge.net/), applying a 1.00 value for no preference. In addition, a value less than 1.00 refers to a frequency of use that is less than expected, whereas a value higher than 1.00 indicates codons that are more frequently used than expected. Potential SSRs were exploited using the software MISA (http://pgrc.ipk-gatersleben.de/misa/), with parameters set to encompass the number of repeat units of a mononucleotide SSR higher than or equal to eight; followed by higher than or equal to four repeat units for di- and tri-nucleotide SSRs; and higher than or equal to three repeat units for tetra-, penta- and hexa-nucleotides, respectively. In this study, we mainly searched for complete repetitive SSR loci, treating cycled or reverse complementary SSRs as the same type.
Phylogenetic Analyses
To determine phylogenetic positions of T. chinensis and T. sutchuenensis within Santalales, we analyzed the chloroplast genomes of ten species, encompassing five other taxa within this lineage, V. album (accession number: KT003925), V. crassula (KT070881), V. minimum (KJ512176), O. alba (KT070882), and S. jasminodora (KX775962). We also used the chloroplast genomes of P. ginseng (AY582139), N. tabacum (Z00044), and Astragalus mongholicus (KU666554) as outgroups, and constructed phylogenetic trees using ML and NJ methods in the software MEGA 6.079 with 1000 bootstrap replicates employing 54 protein-coding genes commonly present in the ten species and matK genes. ML analysis was conducted based on the Tamura-Nei model using a heuristic search for initial trees. This most appropriate model was determined by Modeltest 3.783. NJ trees were performed with NJ method84, and evolutionary distances were computed using the Kimura 2-parameter method85.
Electronic supplementary material
SI-Gene losses and partial deletion of small single-copy regions of the chloroplast genomes of two hemiparasitic Taxillus species
Acknowledgements
This work was supported by CAMS Innovation Fund for Medical Sciences (CIFMS) (NO. 2016-I2M-3–016), Major Scientific and Technological Special Project for “Significant New Drugs Creation” (No. 2014ZX09304307001) and Guangxi Natural Science Foundation (NO. 2013GXNSFAA019120).
Author Contributions
J.Z., X.C., and Z.X., performed the experiments; Y.L., J.Z., and Y.C., assembled sequences and analyzed the data; Y.L. and J.Z. wrote the manuscript; Y.H.L., B.D., and J.S. collected plant material; H.Y. conceived the research and revised the manuscript. All authors have read and approved the final manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Ying Li and Jian-guo Zhou contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-017-13401-4.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Li X, et al. Complete chloroplast genome sequence of holoparasite Cistanche deserticola (Orobanchaceae) reveals gene loss and horizontal gene transfer from its host Haloxylon ammodendron (Chenopodiaceae) PloS One. 2013;8:e58747. doi: 10.1371/journal.pone.0058747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Raubeson, L. A. & Jansen, R. K. In: Diversity and Evolution of Plants; Genotypic and Phenotypic Variation in Higher Plants (ed R. Henry) 45–68 (CABI Publishing, 2005).
- 3.Verma D, Daniell H. Chloroplast vector systems for biotechnology applications. Plant Physiol. 2007;145:1129–1143. doi: 10.1104/pp.107.106690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu FH, et al. Complete chloroplast genome of OncidiumGower Ramsey and evaluation of molecular markers for identification and breeding in Oncidiinae. BMC Plant Biol. 2010;10:68. doi: 10.1186/1471-2229-10-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jheng CF, et al. The comparative chloroplast genomic analysis of photosynthetic orchids and developing DNA markers to distinguish Phalaenopsis orchids. Plant Sci. 2012;190:62–73. doi: 10.1016/j.plantsci.2012.04.001. [DOI] [PubMed] [Google Scholar]
- 6.Kuang DY, et al. Complete chloroplast genome sequence of Magnolia kwangsiensis (Magnoliaceae): implication for DNA barcoding and population genetics. Genome. 2011;54:663–673. doi: 10.1139/g11-026. [DOI] [PubMed] [Google Scholar]
- 7.Nock CJ, et al. Chloroplast genome sequences from total DNA for plant identification. Plant Biotechnol. J. 2011;9:328–333. doi: 10.1111/j.1467-7652.2010.00558.x. [DOI] [PubMed] [Google Scholar]
- 8.Tangphatsornruang S, et al. The chloroplast genome sequence of mungbean (Vigna radiata) determined by high-throughput pyrosequencing: structural organization and phylogenetic relationships. DNA Res. 2009;17:11–22. doi: 10.1093/dnares/dsp025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carbonell JC, et al. A phylogenetic analysis of 34 chloroplast genomes elucidates the relationships between wild and domestic species within the genus. Citrus. Mol. Biol. Evol. 2015;32:2015–2035. doi: 10.1093/molbev/msv082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saski C, et al. Complete chloroplast genome sequence of Glycine max and comparative analyses with other legume genomes. Plant Mol. Biol. 2005;59:309. doi: 10.1007/s11103-005-8882-0. [DOI] [PubMed] [Google Scholar]
- 11.Saski C, et al. Complete chloroplast genome sequences of Hordeum vulgare, Sorghum bicolor and Agrostis stolonifera, and comparative analyses with other grass genomes. Theor. Appl. Genet. 2007;115:571–590. doi: 10.1007/s00122-007-0567-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li XW, et al. High-throughput pyrosequencing of the complete chloroplast genome of Magnolia officinalis and its application in species identification. Acta Pharm.Sinica. 2012;47:124–130. [PubMed] [Google Scholar]
- 13.Zhang Y, et al. The complete chloroplast genome sequence of Taxus chinensis var. mairei (Taxaceae): loss of an inverted repeat region and comparative analysis with related species. Gene. 2014;540:201–209. doi: 10.1016/j.gene.2014.02.037. [DOI] [PubMed] [Google Scholar]
- 14.Lei W, et al. Intraspecific and heteroplasmic variations, gene losses and inversions in the chloroplast genome of Astragalus membranaceus. Sci. Rep. 2016;6:21669. doi: 10.1038/srep21669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang XY, Guan KY, Ai-Rong LI. Biological traits and their ecological significances of parasitic plants: A review. Chinese J. of Ecol. 2011;30:1838–1844. [Google Scholar]
- 16.Wolfe KH, Morden CW, Palmer JD. Function and evolution of a minimal plastid genome from a nonphotosynthetic parasitic plant. Proc. Natl. Acad. Sci. USA. 1992;89:10648–10652. doi: 10.1073/pnas.89.22.10648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wolfe KH, Morden CW, Ems SC, Palmer JD. Rapid evolution of the plastid translational apparatus in a nonphotosynthetic plant: loss or accelerated sequence evolution of tRNA and ribosomal protein genes. J. Mol. Evol. 1992;35:304–317. doi: 10.1007/BF00161168. [DOI] [PubMed] [Google Scholar]
- 18.Mcneal JR, Kuehl JV, Boore JL, de Pamphilis CW. Complete plastid genome sequences suggest strong selection for retention of photosynthetic genes in the parasitic plant genus Cuscuta. BMC Plant Biol. 2007;7:57. doi: 10.1186/1471-2229-7-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maier UG, Karin K, Sabine B, Funk HT, Kirsten K. Complete DNA sequences of the plastid genomes of two parasitic flowering plant species, Cuscuta reflexa and Cuscuta gronovii. BMC Plant Biol. 2007;7:1–12. doi: 10.1186/1471-2229-7-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Molina J, et al. Possible Loss of the Chloroplast Genome in the Parasitic Flowering Plant Rafflesia lagascae (Rafflesiaceae) Mol. Biol. Evol. 2014;31:793. doi: 10.1093/molbev/msu051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wicke S, et al. Mechanisms of functional and physical genome reduction in photosynthetic and nonphotosynthetic parasitic plants of the broomrape family. Plant Cell. 2013;25:3711. doi: 10.1105/tpc.113.113373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cusimano N, Wicke S. Massive intracellular gene transfer during plastid genome reduction in nongreen Orobanchaceae. New Phytol. 2015;210:680–693. doi: 10.1111/nph.13784. [DOI] [PubMed] [Google Scholar]
- 23.Samigullin TH, Logacheva MD, Penin AA, Vallejoroman CM. Complete plastid genome of the recent holoparasite Lathraea squamaria reveals earliest stages of plastome reduction in Orobanchaceae. PloS One. 2016;11:e0150718. doi: 10.1371/journal.pone.0150718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Petersen G, Cuenca A, Seberg O. Plastome evolution in hemiparasitic mistletoes. Genome Biol. Evol. 2015;7:2520–2532. doi: 10.1093/gbe/evv165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Etienne D, Sota F, Catherine FS, Mark B, Ian S. Rampant gene loss in the underground orchid Rhizanthella gardneri highlights evolutionary constraints on plastid genomes. Mol. Biol. Evol. 2011;28:2077–2086. doi: 10.1093/molbev/msr028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Park JM, Manen JF, Schneeweiss GM. Horizontal gene transfer of a plastid gene in the non-photosynthetic flowering plants Orobanche and Phelipanche (Orobanchaceae) Mol. Phylogenet. Evol. 2007;43:974–985. doi: 10.1016/j.ympev.2006.10.011. [DOI] [PubMed] [Google Scholar]
- 27.Li Y, et al. Study on medicinal plants of Loranthaceae resources in china. World Science & Technology: Modernization of Traditional Chinese Medicine and Materia Medica. 2009;11:665–669. [Google Scholar]
- 28.Liu LF, Qiu HX. Pollen morphology of Loranthaceae in China. Guihaia. 1993;13:235–245. [Google Scholar]
- 29.Gong Z, et al. A chemotaxonomic study of 27 species of the Loranthaceae plant from China. Guihaia. 2004;24:493–487. [Google Scholar]
- 30.Han R, Hao G, Zhang D. Interfamilial relationships of Santalales as revealed by chloroplast trnL intron sequences. J. Tro. Subtro. Botany. 2004;12:393–398. [Google Scholar]
- 31.Commission, C. P. The Chinese Pharmacopoeia. 51–52 (Beijing: Chemical Industry Press, 2015).
- 32.Vidal-Russell R, Nickrent DL. Evolutionary relationships in the showy mistletoe family (Loranthaceae) Am. J. Bot. 2008;95:1015–1029. doi: 10.3732/ajb.0800085. [DOI] [PubMed] [Google Scholar]
- 33.The Editorial Committee of Flora of China. Flora of China. Vol. 5, 246–269 (Beijing: Science Press, China; Missouri: Missouri Botanical Garden Press, USA, 2003).
- 34.Wang Y, Zhang SY, Ma XF, Tian WX. Potent inhibition of fatty acid synthase by parasitic loranthus [Taxillus chinensis (DC.) Danser] and its constituent avicularin. J. Enzym. Inhib. Med. Ch. 2006;21:87–93. doi: 10.1080/14756360500472829. [DOI] [PubMed] [Google Scholar]
- 35.Liu CY, et al. Antioxidant, anti-inflammatory, and antiproliferative activities of Taxillus sutchuenensis. Am. J. Chinese Med. 2012;40:335–348. doi: 10.1142/S0192415X12500267. [DOI] [PubMed] [Google Scholar]
- 36.Eid J, et al. Real-time DNA sequencing from single polymerase molecules. Science. 2009;323:133–138. doi: 10.1126/science.1162986. [DOI] [PubMed] [Google Scholar]
- 37.Roberts RJ, Carneiro MO, Schatz MC. The advantages of SMRT sequencing. Genome Biology. 2013;14:405. doi: 10.1186/gb-2013-14-6-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li Q, et al. High-accuracy de novo assembly and SNP detection of chloroplast genomes using a SMRT circular consensus sequencing strategy. New Phytol. 2014;204:1041–1049. doi: 10.1111/nph.12966. [DOI] [PubMed] [Google Scholar]
- 39.Ferrarini M, et al. An evaluation of the PacBio RS platform for sequencing and de novo assembly of a chloroplast genome. BMC Genomics. 2013;14:670. doi: 10.1186/1471-2164-14-670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen X, Li Q, Li Y, Qian J, Han J. Chloroplast genome of Aconitum barbatum var. puberulum (Ranunculaceae) derived from CCS reads using the PacBio RS platform. Front. Plant Sci. 2015;6:42. doi: 10.3389/fpls.2015.00042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xiang B, et al. The complete chloroplast genome sequence of the medicinal plant Swertia mussotii using the PacBio RS II platform. Molecules. 2016;21:1029. doi: 10.3390/molecules21081029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Qian J, et al. The complete chloroplast genome sequence of the medicinal plant Salvia miltiorrhiza. PloS One. 2013;8:e57607. doi: 10.1371/journal.pone.0057607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu J, et al. The first intron of rice EPSP synthase enhances expression of foreign gene. Sci. China C. Life Sci. 2003;46:561–569. doi: 10.1360/02yc0120. [DOI] [PubMed] [Google Scholar]
- 44.Kim KJ, Lee HL. Complete chloroplast genome sequences from Korean ginseng (Panax schinseng Nees) and comparative analysis of sequence evolution among 17 vascular plants. DNA Research. 2004;11:247–261. doi: 10.1093/dnares/11.4.247. [DOI] [PubMed] [Google Scholar]
- 45.Curci PL, De PD, Danzi D, Vendramin GG, Sonnante G. Complete chloroplast genome of the multifunctional crop globe artichoke and comparison with other Asteraceae. PloS One. 2015;10:e0120589. doi: 10.1371/journal.pone.0120589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang Y, et al. Complete chloroplast genome sequence of Aquilaria sinensis (Lour.) Gilg and evolution analysis within the Malvales order. Front. Plant Sci. 2016;7:280. doi: 10.3389/fpls.2016.00280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang Y, et al. The complete chloroplast genome sequences of five Epimedium species: lights into phylogenetic and taxonomic analyses. Front. Plant Sci. 2016;7:306. doi: 10.3389/fpls.2016.00306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Su HJ, Hu JM. The complete chloroplast genome of hemiparasitic flowering plant Schoepfia jasminodora. Mitochondr. DNA Part B. 2016;1:767–769. doi: 10.1080/23802359.2016.1238753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zuo LH, et al. The first complete chloroplast genome sequences of Ulmus species by de novo sequencing: genome comparative and taxonomic position analysis. PloS One. 2017;12:e0171264. doi: 10.1371/journal.pone.0171264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Powell W, Morgante M, Mcdevitt R, Vendramin GG, Rafalski JA. Polymorphic simple sequence repeat regions in chloroplast genomes: applications to the population genetics of pines. Proc. Natl Acad. Sci. USA. 1995;92:7759–7763. doi: 10.1073/pnas.92.17.7759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang AH, Zhang JJ, Yao XH, Huang HW. Chloroplast microsatellite markers in Liriodendron tulipifera (Magnoliaceae) and cross-species amplification in L. chinense. Am. J. Bot. 2011;98:e123. doi: 10.3732/ajb.1000532. [DOI] [PubMed] [Google Scholar]
- 52.Jiao Y, et al. Development of simple sequence repeat (SSR) markers from a genome survey of Chinese bayberry (Myrica rubra) BMC Genomics. 2012;13:201. doi: 10.1186/1471-2164-13-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xue J, Wang S, Zhou SL. Polymorphic chloroplast microsatellite loci in Nelumbo (Nelumbonaceae) Am. J. Bot. 2012;99:240–244. doi: 10.3732/ajb.1100547. [DOI] [PubMed] [Google Scholar]
- 54.Bryant N, Lloyd J, Sweeney C, Myouga F, Meinke D. Identification of nuclear genes encoding chloroplast-localized proteins required for embryo development in. Arabidopsis. Plant Physiol. 2011;155:1678–1689. doi: 10.1104/pp.110.168120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gao L, Ying-Juan SU, Wang T. Plastid genome sequencing, comparative genomics, and phylogenomics: current status and prospects. J. Syst. Evol. 2010;48:77–93. doi: 10.1111/j.1759-6831.2010.00071.x. [DOI] [Google Scholar]
- 56.Millen RS, et al. Many parallel losses of infA from chloroplast DNA during angiosperm evolution with multiple independent transfers to the nucleus. Plant Cell. 2001;13:645–658. doi: 10.1105/tpc.13.3.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kim HT, et al. Seven new complete plastome sequences reveal rampant independent loss of the ndh gene family across orchids and associated instability of the inverted repeat/small single-copy region boundaries. PloS One. 2015;10:e0142215. doi: 10.1371/journal.pone.0142215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lin CS, et al. Concomitant loss of NDH complex-related genes within chloroplast and nuclear genomes in some orchids. Plant J. 2017;90:994–1006. doi: 10.1111/tpj.13525. [DOI] [PubMed] [Google Scholar]
- 59.Chris BJ, Guisinger MM, Jansen RK. Recent loss of plastid-encoded ndh genes within Erodium (Geraniaceae) Plant Mol. Biol. 2011;76:263–272. doi: 10.1007/s11103-011-9753-5. [DOI] [PubMed] [Google Scholar]
- 60.Sanderson MJ, et al. Exceptional reduction of the plastid genome of saguaro cactus (Carnegiea gigantea): Loss of the ndh gene suite and inverted repeat. Am. J. Bot. 2015;102:1115–1127. doi: 10.3732/ajb.1500184. [DOI] [PubMed] [Google Scholar]
- 61.Martín M, Sabater B. Plastid ndh genes in plant evolution. Plant Physiol. Bioch. 2010;48:636–645. doi: 10.1016/j.plaphy.2010.04.009. [DOI] [PubMed] [Google Scholar]
- 62.Li WH, Gojobori T, Nei M. Pseudogenes as a paradigm of neutral evolution. Nature. 1981;292:237–239. doi: 10.1038/292237a0. [DOI] [PubMed] [Google Scholar]
- 63.Balakirev ES, Ayala FJ. Pseudogenes: are they “junk” or functional DNA? Annu. Rev. Genet. 2003;37:123–151. doi: 10.1146/annurev.genet.37.040103.103949. [DOI] [PubMed] [Google Scholar]
- 64.Mardis ER, Next-generation DNA. sequencing methods. Annu Rev Genom. Hum G. 2008;9:387. doi: 10.1146/annurev.genom.9.081307.164359. [DOI] [PubMed] [Google Scholar]
- 65.Metzker ML. Sequencing technologies - the next generation. Nat. Rev. Genet. 2010;11:31–46. doi: 10.1038/nrg2626. [DOI] [PubMed] [Google Scholar]
- 66.Wu Z, et al. A precise chloroplast genome of Nelumbo nucifera (Nelumbonaceae) evaluated with Sanger, Illumina MiSeq, and PacBio RS II sequencing platforms: insight into the plastid evolution of basal eudicots. BMC Plant Biol. 2014;14:289. doi: 10.1186/s12870-014-0289-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Walker JF, Jansen RK, Zanis MJ, Emery NC. Sources of inversion variation in the small single copy (SSC) region of chloroplast genomes. Am. J. Bot. 2015;102:1751–1752. doi: 10.3732/ajb.1500299. [DOI] [PubMed] [Google Scholar]
- 68.Delavault PM, Russo NM, Lusson NA, Thalouarn PA. Organization of the reduced plastid genome of Lathraea clandestina, an ahlorophyllous parasitic plant. Physiol. Plantarum. 1996;96:674–682. doi: 10.1111/j.1399-3054.1996.tb00242.x. [DOI] [Google Scholar]
- 69.Logacheva MD, Schelkunov MI, Nuraliev MS, Samigullin TH, Penin AA. The plastid genome of mycoheterotrophic monocot Petrosavia stellaris exhibits both gene losses and multiple rearrangements. Genome Biol. Evol. 2014;6:238–246. doi: 10.1093/gbe/evu001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hilu KW, et al. Angiosperm phylogeny based on matK sequence information. Am. J. Bot. 2003;90:1758–1776. doi: 10.3732/ajb.90.12.1758. [DOI] [PubMed] [Google Scholar]
- 71.Goldman DH, et al. Phylogenetics of Arethuseae (Orchidaceae) based on plastid matK and rbcL sequences. Syst. Bot. 2001;26:670–695. [Google Scholar]
- 72.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Luo R, et al. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. GigaScience. 2012;1:18. doi: 10.1186/2047-217X-1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Boetzer M, Henkel CV, Jansen HJ, Butler D, Pirovano W. Scaffolding pre-assembled contigs using SSPACE. Bioinformatics. 2011;27:578–579. doi: 10.1093/bioinformatics/btq683. [DOI] [PubMed] [Google Scholar]
- 75.Wyman SK, Jansen RK, Boore JL. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 2004;20:3252–3255. doi: 10.1093/bioinformatics/bth352. [DOI] [PubMed] [Google Scholar]
- 76.Liu C, et al. CpGAVAS, an integrated web server for the annotation, visualization, analysis, and GenBank submission of completely sequenced chloroplast genome sequences. BMC Genomics. 2012;13:715. doi: 10.1186/1471-2164-13-715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Schattner P, Brooks AN, Lowe TM. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 2005;33:W686–W689. doi: 10.1093/nar/gki366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lohse M, Drechsel O, Bock R. OrganellarGenomeDRAW (OGDRAW): a tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr. Genet. 2007;52:267–274. doi: 10.1007/s00294-007-0161-y. [DOI] [PubMed] [Google Scholar]
- 79.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Comput. Appl. Biosci. Cabios. 2013;30:2725. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Frazer KA, Pachter L, Poliakov A, Rubin EM, Dubchak I. VISTA: computational tools for comparative genomics. Nucleic Acids Res. 2004;32:W273. doi: 10.1093/nar/gkh458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Darling AE, Mau B, Perna N. T. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PloS One. 2010;5:e11147. doi: 10.1371/journal.pone.0011147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sharp PM, Li WH. The codon Adaptation Index–a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987;15:1281–1295. doi: 10.1093/nar/15.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Posada D, Crandall KA. MODELTEST: testing the model of DNA substitution. Bioinformatics. 1998;14:817. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- 84.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 85.Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SI-Gene losses and partial deletion of small single-copy regions of the chloroplast genomes of two hemiparasitic Taxillus species