Abstract
Oral fluid analysis for herd monitoring is of interest to the commercial pig production in Korea. The aim of this study was to investigate pathogen-positive rates and correlations among eight pathogens associated with porcine respiratory disease complex by analyzing oral fluid samples from 214 pig groups from 56 commercial farms. Samples collected by a rope-chewing method underwent reverse-transcriptase polymerase chain reaction (RT-PCR) or standard polymerase chain reaction (PCR) analysis, depending on the microorganism. Pathogens were divided into virus and bacteria groups. The former consisted of porcine reproductive and respiratory syndrome virus and porcine circovirus type 2 (PCV2), and the latter Pasteurella multocida, Haemophilus parasuis, Actinobacillus pleuropneumoniae, Mycoplasma hyopneumoniae (MHP), Mycoplasma hyorhinis, and Streptococcus suis (SS). All pathogens were detected more than once by PCR. Age-based analysis showed the PCR-positive rate increased with increasing age for PCV2 and MHP, whereas SS showed the opposite. Correlations between pathogens were assessed among 36 different pair combinations; only seven pairs showed statistically significant correlations. In conclusion, the oral fluid method could be a feasible way to detect various swine respiratory disease pathogens and, therefore, could complement current monitoring systems for respiratory diseases in the swine industry.
Keywords: Korea, oral fluid, pathogens, polymerase chain reaction, porcine respiratory disease complex
Introduction
Porcine respiratory disease complex (PRDC) is regarded as one of the more important health issues affecting pig production. Generally, diseases within the PRDC trigger lung damage which, in turn, results in low economic efficiency, poor growth rate, and higher medication and management costs [6]. Coinfection of various pathogens has been frequently detected in PRDC samples [7]. The bacterial pathogens frequently detected in PRDC cases are Actinobacillus pleuropneumoniae (APP), Pasteurella multocida (PM), Streptococcus suis (SS), Haemophilus parasuis (HPS), Mycoplasma hyopneumoniae (MHP), and Mycoplasma hyorhinis (MHR) [4,10,30]. Viral pathogens in PRDC that have effects similar to those mentioned above include porcine reproductive and respiratory syndrome virus (PRRSV) and porcine circovirus type 2 (PCV2) [6].
Despite the development of various monitoring methods, currently used approaches to diagnosis, surveillance, and monitoring of PRDC are labor intensive and time consuming. Previous studies have used individual sampling approaches (e.g., biopsy, swabbing, blood collection) to investigate seroprevalence or presence of specific pathogens [11,19]. However, due to the difficulty of sampling certain targets (e.g., blood, organs, other tissue), and limitations related to time and budget, there is a need to develop new solutions to monitoring issues [3,9].
Some recent studies have proposed that oral fluid (OF) can function as a suitable sample source for the detection, diagnosis, surveillance, and monitoring of various pathogens, and OF has been investigated and used in swine farms in North America [22,27]. The use of OF for such purposes is only recently being tested for application in swine farms in Korea. Although OF studies have been carried out in North America and Europe, there have been no OF study results reported in Korea.
The objectives of this study were to survey the pathogen-positive rate for eight respiratory pathogens in commercially farmed pigs and determine the correlations between pathogen pairs in Korea by using an OF-based sampling method, which has been shown to be a suitable method for monitoring various respiratory pathogens.
Materials and Methods
Experimental design
Animal experiments related to collecting samples from pigs were approved by the Institutional Animal Care and Use Committee of Konkuk University (KU-15090). All animals were kept at commercial domestic pig farms located nationwide in Korea.
In this study, pigs from 56 farms that included 214 pig groups were sampled. The farms were commercial farrow-to-finisher farms, were located in different regions within Korea, and had different environments, herd sizes, and barn sizes. The age groups of sampled animals ranged between 3 and 26 weeks old. Routine vaccination programs for PCV2, classical swine fever virus, foot and mouth disease (FMD) virus, and MHP were conducted on each farm. Porcine infectious diseases such as porcine epidemic diarrhea (PED) and FMD were not detected and mass medication was not included in the pig feed used on the study farms. According to veterinary practitioners familiar with the farms, the pigs from which samples were collected did not show any respiratory signs.
Each farm was assigned four rope sets with one rope set consisting of four ropes. Thus, there were potentially 16 OF samples taken from each farm. A four rope set was assigned to four different age groups within the target age range (3 to 26 weeks old). Each rope was hung in a pen housing a minimum of 20 and a maximum of 25 pigs. The pigs were allowed to chew on the ropes; thus, each rope represented the OF obtained from similarly aged pigs in each pen.
Oral fluid samples
For OF collection, 45 cm long, 1.3 cm diameter cotton ropes were used. The ropes had three strands that were undyed and unbleached [21]. Prior to use, ropes were sealed in oilpaper, autoclaved (121℃, 15 min), and then packed in an airtight bag. On arrival in the farm's target barn, the rope was unsealed and hung in an individual pen, with sterile handling. One OF sample was collected from each pen after a fixed access time. Animal interaction (biting and chewing) with the rope was limited to 20 min in each pen [15,26]. After 20 min, the rope, which was typically saturated with OF, was removed from the pen and directly transferred to the laboratory within 24 h in order to assure limited damage to the pathogens. For transport, the rope samples were sealed in airtight plastic bags and transported in a cool box with ice packs. After arrival in the laboratory, the airtight bag was unsealed in a biosafety cabinet to protect the samples from contamination. OFs were extracted by squeezing the rope while it remained within the airtight bag. The obtained OF was then poured into a 15 mL centrifuge tube [28]. Samples were then centrifuged at 2,000 × g for 10 min at 4℃. Aliquots of each OF sample were placed in several 2 mL microcentrifuge tubes. One microcentrifuge tube was tested immediately, and the others were stored at –70℃ until further analysis.
Laboratory assays
The presence of PRRSV, PCV2, PM, HPS, APP, MHP, MHR, and SS in OF samples were determined by using reverse-transcriptase polymerase chain reaction (RT-PCR) or standard polymerase chain reaction (PCR) assays. For DNA/RNA extraction, samples were processed by using the Viral Gene-spin Viral DNA/RNA Extraction Kit (iNtRON Biotechnology, Korea) according to the manufacturer's protocol. After DNA/RNA extraction, each sample underwent RT-PCR (Maxime RT-PCR PreMix Kit; iNtRON Biotechnology) or PCR (Maxime PCR PreMix Kit; iNtRON Biotechnology) according to the properties of each pathogen. The primer sequences used in this study are listed in Table 1. To detect pathogens, RT-PCR, nested PCR, or PCR was carried out according to previously described protocols. In case of PRRSV, RT-PCR and nested PCR were performed sequentially with nested PCR used as a confirmatory test. The references describing the corresponding protocols for each pathogen are listed in Table 1.
Table 1. Primers sequences used for the survey of porcine respiratory disease complex pathogens in Korea.
PRRSV, porcine reproductive and respiratory syndrome virus; PCV2, porcine circovirus type 2; PM, Pasteurella multocida; HPS, Haemophilus parasuis; APP, Actinobacillus pleuropneumoniae; MHP, Mycoplasma hyopneumoniae; MHR, Mycoplasma hyorhinis; SS, Streptococcus suis; RT-PCR, reverse transcriptase polymerase chain reaction; PCR; polymerase chain reaction. *RT-PCR and nested PCR were performed sequentially and nested PCR was a confirmatory test. †Approximate product size (units: base pairs). ‡RT-PCR or standard PCR conditions were performed according to protocols detailed in the references cited.
The amplicons were separated by performing electrophoresis with 2.0% agarose gels and staining with DNA dye (SafeView Classic; Applied Biological Materials, Canada) and visualized under UV light. The sizes of the PCR products were determined by comparison with a DNA ladder.
Data analysis
Data were analyzed on both per pen (sample-level) and per farm (farm-level) bases. Pathogen-specific PCR results were analyzed as qualitative (positive/negative) data. A farm was considered to be positive when any sample from that farm was positive for a particular pathogen. Differences in the distribution of pathogen combinations among samples were analyzed by using chi-squared tests. Pathogen combinations were considered positive if multiple pathogens were detected in PCR analysis of the same sample. To assess the presence of correlations (positive/negative) among the eight pathogens (PRRSV, PCV2, PM, HPS, APP, MHP, MHR, and SS), pairwise comparisons were performed by applying the Pearson correlation method to the sample-level data. Statistical analyses for pairwise comparison were performed by using MedCalc (ver. 12.7.0.0; MedCalc Software, Belgium). Correlation results with p < 0.05 were considered statistically significant.
Results
Veterinarians did not report any difficulty in installing, collecting, packaging, and submitting the OF-bearing ropes to the laboratory. Most samples arrived at the laboratory in good condition; however, 12 rope samples (5.3% of the total 226 samples) were not saturated with OF and were eliminated from the analyses, leaving a sample size for analysis of 214.
Sample-level and farm-level PCR results
Table 2 presents the overall PCR results for the sample-level and farm-level data for 214 samples and 56 farms, respectively. The first pathogen surveillance group comprised the typical swine respiratory viruses consisting of PRRSV and PCV2. The second pathogen surveillance group comprised the typical swine respiratory bacteria consisting of PM, HPS, APP, MHP, MHR, and SS.
Table 2. Swine oral fluid polymerase chain reaction (PCR) results at the sample and farm levels.
*PRRSV, porcine reproductive and respiratory syndrome virus; PCV2, porcine circovirus type 2; PM, Pasteurella multocida; HPS, Haemophilus parasuis; APP, Actinobacillus pleuropneumoniae; MHP, Mycoplasma hyopneumoniae; MHR, Mycoplasma hyorhinis; SS, Streptococcus suis. *Farms were considered positive if one or more pen-based oral fluid sample had PCR-positive results for a pathogen.
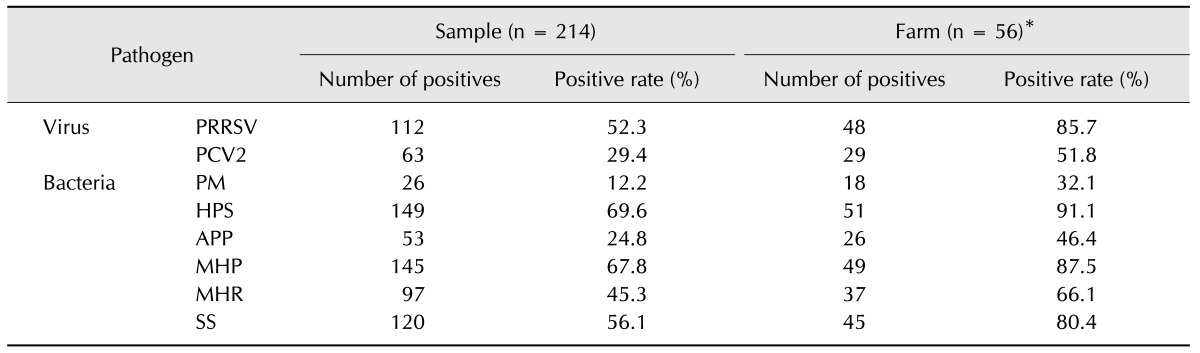
The major pathogen combinations identified from the PCR data in the 214 OF samples are summarized in Fig. 1. Because 96 of 256 possible outcomes (a binary outcome was produced for each of the eight different PCR results, or 28) were observed, all combinations within the pathogen data could not be presented practically; thus, only the most frequent pathogen occurrences in the PCR data are presented in Fig. 1. Overall, multiple pathogen combinations were more common than single pathogen occurrences. Three (1.4%) of the 214 samples were negative for all eight pathogens.
Fig. 1. Proportions of major pathogens and major pathogen combinations in polymerase chain reaction (PCR)-positive porcine oral fluid samples. PRRSV, porcine reproductive and respiratory syndrome virus; PCV2, porcine circovirus type 2; MHP, Mycoplasma hyopneumoniae. *Pathogen combinations were considered positive if the major pathogen and one or more other pathogens were PCR positive. †Pathogen combinations were considered positive if one or more pathogens were PCR positive without PRRSV, PCV2, and MHP.
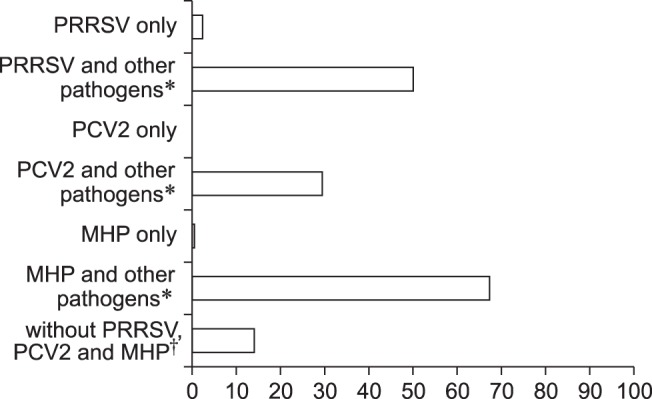
Age-level PCR results
Fig. 2 shows the PCR results obtained at the age level (n = 214). OF samples were divided into four age-based groups: A, aged 3 to 7 weeks old; B, aged 8 to 12 weeks; C, aged 13 to 16 weeks; and D, 17 to 26 weeks. The numbers of OF samples in each group were 51, 77, 39, and 47, respectively. Most pathogens had PCR-positive results among the four groups. The PRRSV-positive rate ranged between 47% and 55% among the age groups. The positive rates for APP and PM were below 30% in all age groups. Negative rates for all eight pathogens were low in the younger pigs (aged 3–12 weeks; groups A and B), whereas negative rates were zero in the older pigs (aged 13–26 weeks; groups C and D). Positive rates for PCV2 and MHP increased with increasing age, but the SS-positive rate decreased with increasing age.
Fig. 2. Pen-level PCR results for 214 oral fluid samples arranged by age group. Oral fluid samples from pigs were analyzed for respiratory pathogen presence by using PCR. Pig age groups: A, aged 3–7 weeks; B, 8–12 weeks old; C, 13–16 weeks old; D, 17–26 weeks old. PRRSV, porcine reproductive and respiratory syndrome virus; PCV2, porcine circovirus type 2; PM, Pasteurella multocida; HPS, Haemophilus parasuis; APP, Actinobacillus pleuropneumoniae; MHP, Mycoplasma hyopneumoniae; MHR, Mycoplasma hyorhinis; SS, Streptococcus suis; All N, negative PCR result for all pathogens.
Correlation between pathogens
Among the pathogen combinations of the eight assessed pathogens, there were significant differences identified by chi-squared testing (p < 0.001) of the sample-level (per pen) data. Pairwise comparisons of the positive and negative presences of various respiratory pathogens stratified by pen identified statistically significant correlations in seven pathogen pairs: PCV2 and MHP (p = 0.0002), PM and MHP (p = 0.02), PM and SS (p = 0.007), HPS and MHP (p = 0.004), HPS and MHR (p = 0.01), HPS and SS (p = 0.005), and MHP and MHR (p = 0.015). No other pathogen-pair correlation results were identified as statistically significant (p < 0.05).
Discussion
Several previous studies have attempted to validate the use of OF sampling for the detection of PRRSV and PCV2 by addressing the issue of sample collection method and establishing the feasibility of performing pathogen identification via OF [15,22,26]. In comparison with serum-based analysis, OF analysis had many advantages, i.e., number of samples required (one per pen vs. one per animal), labor intensity, time required, and budget needed to perform surveillance or monitor disease. Furthermore, during sample collection, other notable advantages of OF collection are its noninvasive stress-free nature and its being highly compliant with animal ethics requirements [27]. Based on these advantages, the current study was performed to survey eight pathogens (both viruses and bacteria) that are associated with PRDC in commercial pig production facilities.
In previous studies, the published positive rates for PRDC pathogens varied depending on sample and sampling types. One such study determined the prevalence of porcine pathogens and reported positive rates for SS (53.7%), PM (27.3%), PRRSV (22.0%), and PCV2 (11.9%) in tonsil samples at slaughter [19]. Previous reports on MHP, HPS, PM, and APP positive rates have reported quite variable results. When analyzed in nasal, tonsillar, and oropharyngeal swab samples, positive rates were: MHP (2.4%), SS (67.1%), APP (30.9%), PM (24.6%), and HPS (23.4%) [5]. However, in another study, the positive rate for MHP was 23.5% in bronchoalveolar lavage fluid and 26.5% in lung tissue [18]. In a study of the association between pathogens in healthy pigs and pigs with pneumonia, regardless of whether pigs had pneumonia, they showed positive rates for PRRSV, PCV2, PM, HPS, MHP, and MHR, despite the absence of clinical symptoms [24].
In the present study, the sample-level results were obtained from the analysis of OF samples from each pen. Pathogens that were most frequently detected by PCR assays were PRRSV (52.3%), HPS (69.6%), MHP (67.8%), and SS (56.1%). The high positive rates for these pathogens suggest that they might be prevalent in herds. As described above, a farm was considered positive for a particular pathogen when any pen sample from that farm tested positive for that pathogen. Considering that pathogens could be transmitted from one pen to another, it is possible that certain pathogens can become widely distributed throughout a farm.
In the results of our pathogen combination analysis, 96 outcome combinations were observed, and the most notable trend was the presence of coinfection with mixed pathogens. There have been previous reports that have referred to coinfection being caused by PRRSV, PCV2, MHP, or APP [1,3,31,32]. In such cases, the hosts infected with those pathogens were then vulnerable to infection by other pathogens via opportunistic infection [23,25]. In this study, a similar tendency was observed. The positive rates for single pathogens were lower than those for combinations of multiple pathogen. The PCR-positive rates for single pathogens ranged from 0% to 2.3%, whereas for the major respiratory pathogens co-infected with one or more pathogens, the PCR-positive rates ranged from 29.4% to 67.3%. The positive rate for samples with one or more pathogens, excluding the presence of PRRSV, PCV2, and MHP, was 14.0%, and this rate could be affected and/or enhanced by the presence of PRRSV, PCV2, or MHP with other respiratory pathogens. It is the statistically significant pathogen correlations that would be of greatest concern in PRDC management, and we noted significant correlations between PCV2 and MHP, PM and MHP, PM and SS, HPS and MHP, HPS and MHR, HPS and SS, and MHP and MHR. The identification of these significant pathogen pairs indicates that if one pathogen is frequently detected within a herd, that herd may also be vulnerable to coinfection with the corresponding paired pathogen.
The positive rates for each pathogen were also analyzed by pig age. Using that approach, it was possible to gain an overview of the age-dependent trends among the pathogens. Several previous studies have focused on longitudinal research and limited condition testing of disease prevalence [26,27]. With these studies in mind, Fig. 2 shows an overview of the pathogen frequency at each age group in our study population. PRRSV maintained a positive rate of 47% to 55% in all age groups, but there were different frequency distributions among the other pathogens. The PRRSV, HPS, MHP, and SS pathogens exhibited a relatively high frequency (about 50%) in pigs aged 3–16 weeks of pigs (groups A–C). Notably, HPS and SS appeared more frequently in the youngest age group (aged 3 to 7 weeks, group A) than in the older age groups. For PCV2 and MHP, PCR-positive rates tended to rise as age increased from group A to group D. These trends indicate differences in the control of pathogenic infections in each age group.
In previous studies, the OF method has been considered a reliable monitoring method for specific pathogens, including PRRSV and the Hepatitis E virus (HEV) [12,14]. While there are many advantages of the OF analysis technique, some weaknesses were reported by the field veterinarians or caretakers involved in this study. Despite the OF samples being theoretically representative of all pen-based pigs, some pigs showed little interest in interacting with the cotton rope. This may be due to selecting an unfavorable installation time or to differences in individual status (e.g., at the same time as feeding or individual health condition). Some mildly depressed pigs were observed to show no interest in interacting with the rope. In such cases, the collected OF sample may not represent the population of the pen or herd. In addition, due to some caretakers being unfamiliar with the OF collection method, some ropes were not saturated with OF upon arrival in the laboratory and were excluded from the study.
In conclusion, this is the first investigation presenting pathogen prevalence data obtained by using an OF method for a large number of pathogens causing PRDC in Korea. The results suggest that OF analysis can be a potentially useful technique in monitoring the major pathogens in PRDC by single sampling of pigs in commercial swine farms in Korea.
Acknowledgments
This study was supported by a grant (No. 112157-3) of the Korea Institute of Planning and Evaluation for Technology in Food, Agriculture, Forestry and Fisheries.
Footnotes
Conflict of Interest: The authors declare no conflict of interests.
References
- 1.Chae C. Porcine circovirus type 2 and its associated diseases in Korea. Virus Res. 2012;164:107–113. doi: 10.1016/j.virusres.2011.10.013. [DOI] [PubMed] [Google Scholar]
- 2.Choi EJ, Lee CH, Song JY, Song HJ, Park CK, Kim B, Shin YK. Genetic diversity of porcine reproductive and respiratory syndrome virus in Korea. J Vet Sci. 2013;14:115–124. doi: 10.4142/jvs.2013.14.2.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dorr PM, Baker RB, Almond GW, Wayne SR, Gebreyes WA. Epidemiologic assessment of porcine circovirus type 2 coinfection with other pathogens in swine. J Am Vet Med Assoc. 2007;230:244–250. doi: 10.2460/javma.230.2.244. [DOI] [PubMed] [Google Scholar]
- 4.Fablet C, Marois C, Dorenlor V, Eono F, Eveno E, Jolly JP, Le Devendec L, Kobisch M, Madec F, Rose N. Bacterial pathogens associated with lung lesions in slaughter pigs from 125 herds. Res Vet Sci. 2012;93:627–630. doi: 10.1016/j.rvsc.2011.11.002. [DOI] [PubMed] [Google Scholar]
- 5.Fablet C, Marois C, Kuntz-Simon G, Rose N, Dorenlor V, Eono F, Eveno E, Jolly JP, Le Devendec L, Tocqueville V, Quéguiner S, Gorin S, Kobisch M, Madec F. Longitudinal study of respiratory infection patterns of breeding sows in five farrow-to-finish herds. Vet Microbiol. 2011;147:329–339. doi: 10.1016/j.vetmic.2010.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fablet C, Marois-Créhan C, Simon G, Grasland B, Jestin A, Kobisch M, Madec F, Rose N. Infectious agents associated with respiratory diseases in 125 farrow-to-finish pig herds: a cross-sectional study. Vet Microbiol. 2012;157:152–163. doi: 10.1016/j.vetmic.2011.12.015. [DOI] [PubMed] [Google Scholar]
- 7.Fraile L, Alegre A, López-Jiménez R, Nofrarías M, Segalés J. Risk factors associated with pleuritis and cranio-ventral pulmonary consolidation in slaughter-aged pigs. Vet J. 2010;184:326–333. doi: 10.1016/j.tvjl.2009.03.029. [DOI] [PubMed] [Google Scholar]
- 8.Gram T, Ahrens P. Improved diagnostic PCR assay for Actinobacillus pleuropneumoniae based on the nucleotide sequence of an outer membrane lipoprotein. J Clin Microbiol. 1998;36:443–448. doi: 10.1128/jcm.36.2.443-448.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hälli O, Ala-Kurikka E, Nokireki T, Skrzypczak T, Raunio-Saarnisto M, Peltoniemi OAT, Heinonen M. Prevalence of and risk factors associated with viral and bacterial pathogens in farmed European wild boar. Vet J. 2012;194:98–101. doi: 10.1016/j.tvjl.2012.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hansen MS, Pors SE, Jensen HE, Bille-Hansen V, Bisgaard M, Flachs EM, Nielsen OL. An investigation of the pathology and pathogens associated with porcine respiratory disease complex in Denmark. J Comp Pathol. 2010;143:120–131. doi: 10.1016/j.jcpa.2010.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hričínová M, Holoda E, Mudroňová D, Ondrašovičová S. Multiplex PCR assay for detection of Actinobacillus pleuropneumoniae, Pasteurella multocida and Haemophilus parasuis in lungs of pigs from a slaughterhouse. Folia Microbiol (Praha) 2010;55:635–640. doi: 10.1007/s12223-010-0103-9. [DOI] [PubMed] [Google Scholar]
- 12.Jones TH, Muehlhauser V. Effect of handling and storage conditions and stabilizing agent on the recovery of viral RNA from oral fluid of pigs. J Virol Methods. 2014;198:26–31. doi: 10.1016/j.jviromet.2013.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kerdsin A, Dejsirilert S, Akeda Y, Sekizaki T, Hamada S, Gottschalk M, Oishi K. Fifteen Streptococcus suis serotypes identified by multiplex PCR. J Med Microbiol. 2012;61:1669–1672. doi: 10.1099/jmm.0.048587-0. [DOI] [PubMed] [Google Scholar]
- 14.Kittawornrat A, Prickett J, Chittick W, Wang C, Engle M, Johnson J, Patnayak D, Schwartz T, Whitney D, Olsen C, Schwartz K, Zimmerman J. Porcine reproductive and respiratory syndrome virus (PRRSV) in serum and oral fluid samples from individual boars: will oral fluid replace serum for PRRSV surveillance? Virus Res. 2010;154:170–176. doi: 10.1016/j.virusres.2010.07.025. [DOI] [PubMed] [Google Scholar]
- 15.Kittawornrat A, Prickett J, Wang C, Olsen C, Irwin C, Panyasing Y, Ballagi A, Rice A, Main R, Johnson J, Rademacher C, Hoogland M, Rowland R, Zimmerman J. Detection of Porcine reproductive and respiratory syndrome virus (PRRSV) antibodies in oral fluid specimens using a commercial PRRSV serum antibody enzyme-linked immunosorbent assay. J Vet Diagn Invest. 2012;24:262–269. doi: 10.1177/1040638711435679. [DOI] [PubMed] [Google Scholar]
- 16.MacInnes JI, Gottschalk M, Lone AG, Metcalf DS, Ojha S, Rosendal T, Watson SB, Friendship RM. Prevalence of Actinobacillus pleuropneumoniae, Actinobacillus suis, Haemophilus parasuis, Pasteurella multocida, and Streptococcus suis in representative Ontario swine herds. Can J Vet Res. 2008;72:242–248. [PMC free article] [PubMed] [Google Scholar]
- 17.Marois C, Cariolet R, Morvan H, Kobisch M. Transmission of pathogenic respiratory bacteria to specific pathogen free pigs at slaughter. Vet Microbiol. 2008;129:325–332. doi: 10.1016/j.vetmic.2007.11.030. [DOI] [PubMed] [Google Scholar]
- 18.Moorkamp L, Nathues H, Spergser J, Tegeler R, Grosse Beilage E. Detection of respiratory pathogens in porcine lung tissue and lavage fluid. Vet J. 2008;175:273–275. doi: 10.1016/j.tvjl.2007.01.008. [DOI] [PubMed] [Google Scholar]
- 19.O'Sullivan T, Friendship R, Blackwell T, Pearl D, McEwen B, Carman S, Slavić D, Dewey C. Microbiological identification and analysis of swine tonsils collected from carcasses at slaughter. Can J Vet Res. 2011;75:106–111. [PMC free article] [PubMed] [Google Scholar]
- 20.Oliveira S, Galina L, Pijoan C. Development of a PCR test to diagnose Haemophilus parasuis infections. J Vet Diagn Invest. 2001;13:495–501. doi: 10.1177/104063870101300607. [DOI] [PubMed] [Google Scholar]
- 21.Olsen C, Karriker L, Wang C, Binjawadagi B, Renukaradhya G, Kittawornrat A, Lizano S, Coetzee J, Main R, Meiszberg A, Panyasing Y, Zimmerman J. Effect of collection material and sample processing on pig oral fluid testing results. Vet J. 2013;198:158–163. doi: 10.1016/j.tvjl.2013.06.014. [DOI] [PubMed] [Google Scholar]
- 22.Olsen C, Wang C, Christopher-Hennings J, Doolittle K, Harmon KM, Abate S, Kittawornrat A, Lizano S, Main R, Nelson E a, Otterson T, Panyasing Y, Rademacher C, Rauh R, Shah R, Zimmerman J. Probability of detecting Porcine reproductive and respiratory syndrome virus infection using pen-based swine oral fluid specimens as a function of within-pen prevalence. J Vet Diagn Invest. 2013;25:328–335. doi: 10.1177/1040638713481471. [DOI] [PubMed] [Google Scholar]
- 23.Opriessnig T, Thacker EL, Yu S, Fenaux M, Meng XJ, Halbur PG. Experimental reproduction of postweaning multisystemic wasting syndrome in pigs by dual infection with Mycoplasma hyopneumoniae and Porcine circovirus type 2. Vet Pathol. 2004;41:624–640. doi: 10.1354/vp.41-6-624. [DOI] [PubMed] [Google Scholar]
- 24.Palzer A, Ritzmann M, Wolf G, Heinritzi K. Associations between pathogens in healthy pigs and pigs with pneumonia. Vet Rec. 2008;162:267–271. doi: 10.1136/vr.162.9.267. [DOI] [PubMed] [Google Scholar]
- 25.Pogranichniy RM, Yoon KJ, Harms PA, Sorden SD, Daniels M. Case-control study on the association of Porcine circovirus type 2 and other swine viral pathogens with postweaning multisystemic wasting syndrome. J Vet Diagn Invest. 2002;14:449–456. doi: 10.1177/104063870201400601. [DOI] [PubMed] [Google Scholar]
- 26.Prickett J, Kim W, Simer R, Yoon KJ, Zimmerman J. Oral-fluid samples for surveillance of commercial growing pigs for porcine reproductive and respiratory syndrome virus and porcine circovirus type 2 infections. J Swine Health Prod. 2008;16:86–91. [Google Scholar]
- 27.Prickett J, Simer R, Christopher-Hennings J, Yoon KJ, Evans RB, Zimmerman JJ. Detection of Porcine reproductive and respiratory syndrome virus infection in porcine oral fluid samples: a longitudinal study under experimental conditions. J Vet Diagn Invest. 2008;20:156–163. doi: 10.1177/104063870802000203. [DOI] [PubMed] [Google Scholar]
- 28.Ramirez A, Wang C, Prickett JR, Pogranichniy R, Yoon KJ, Main R, Johnson JK, Rademacher C, Hoogland M, Hoffmann P, Kurtz A, Kurtz E, Zimmerman J. Efficient surveillance of pig populations using oral fluids. Prev Vet Med. 2012;104:292–300. doi: 10.1016/j.prevetmed.2011.11.008. [DOI] [PubMed] [Google Scholar]
- 29.Shin J, Bautista EM, Kang YB, Molitor TW. Quantitation of porcine reproductive and respiratory syndrome virus RNA in semen by single-tube reverse transcription-nested polymerase chain reaction. J Virol Methods. 1998;72:67–79. doi: 10.1016/s0166-0934(98)00025-1. [DOI] [PubMed] [Google Scholar]
- 30.Stakenborg T, Vicca J, Butaye P, Imberechts H, Peeters J, de Kruif A, Haesebrouck F, Maes D. A multiplex PCR to identify porcine mycoplasmas present in broth cultures. Vet Res Commun. 2006;30:239–247. doi: 10.1007/s11259-006-3226-3. [DOI] [PubMed] [Google Scholar]
- 31.Wieland B, Werling D, Nevel A, Rycroft A, Demmers TG, Wathes CM, Grierson S, Cook AJC, Done SH, Armstrong D. Porcine circovirus type 2 infection before and during an outbreak of postweaning multisystemic wasting syndrome on a pig farm in the UK. Vet Rec. 2012;170:596. doi: 10.1136/vr.100276. [DOI] [PubMed] [Google Scholar]
- 32.Wongnarkpet S, Pfeiffer DU, Morris RS, Fenwick SG. An on-farm study of the epidemiology of Actinobacillus pleuropneumoniae infection in pigs as part of a vaccine efficacy trial. Prev Vet Med. 1999;39:1–11. doi: 10.1016/s0167-5877(98)00146-9. [DOI] [PubMed] [Google Scholar]
- 33.Zhou JY, Chen QX, Ye JX, Shen HG, Chen TF, Shang SB. Serological investigation and genomic characterization of PCV2 isolates from different geographic regions of Zhejiang province in China. Vet Res Commun. 2006;30:205–220. doi: 10.1007/s11259-006-3203-x. [DOI] [PubMed] [Google Scholar]