Abstract
A simple and rapid immunochromatographic test strip incorporating a colloidal gold-labeled recombinant Nsp7 antigen probe was successfully developed for the detection of anti-porcine reproductive and respiratory syndrome virus (PRRSV) antibodies in swine. Recombinant Nsp7 protein of PRRSV labeled with colloidal gold was dispensed on a conjugate pad for use as the detector. Staphylococcal protein A and purified porcine anti-Nsp7 antibodies were blotted on a nitrocellulose membrane to form test and control lines, respectively. A comparison of the strip with standard diagnostic tests, enzyme-linked immunosorbent assays and immunoperoxidase monolayer assay, was also performed. The immunochromatographic test strip was shown to be of high specificity and sensitivity. Furthermore, the strip assay is rapid and easy to perform with no requirement for professional-level skills or equipment. It is suggested that the immunochromatographic test strip can be used to quickly and accurately detect PRRSV antibody and to be suitable for diagnostic purposes in the field.
Keywords: enzyme-linked immunosorbent assay, immunochromatography, porcine reproductive and respiratory syndrome
Introduction
Porcine reproductive and respiratory syndrome (PRRS), first reported in 1987 in the USA, is a global infectious disease that has resulted in widespread economic loss in the swine industry. PRRS is characterized by reproductive failure in pregnant sows (e.g. abortions and stillbirths), respiratory distress in pigs of different ages, and severe immune suppression [13,16]. The PRRS virus (PRRSV) is the recognized causative agent of this syndrome. Two different genotypes of PRRSV have been described: European or type 1, and North American or type 2 [6,21]. In China, most isolated strains are of type 2 [3].
PRRSV is an enveloped, positive, single-stranded RNA virus in the genus Arterivirus, which belongs to the family Arteriviridae within the order Nidovirales [14].The PRRSV genome is 15 kb in length and contains at least 10 open reading frames (ORFs). Of these, ORF1a and ORF1b represent nearly 75% of the viral genome and encode two large polyproteins (pp), pp1a and pp1ab, respectively. These pps can be hydrolyzed into at least 16 small nonstructural proteins (Nsps). Of these 16 Nsps, at least 14 are involved in viral genome replication and transcription [10]. The Nsp1, Nsp2, and Nsp7 proteins elicit a strong immune response in pigs. Fourteen days after pigs are infected with PRRSV, anti-PRRSV Nsp7 antibodies can be detected, and high levels of antibodies in pigs can last for up to 202 days [2].
A diagnosis of PRRSV is important for its prevention and control. The laboratory diagnostic tests commonly used at present include reverse transcriptase polymerase chain reaction (PCR), quantitative real-time PCR [19,22], and four serological detection methods [1,4,5,20]: the indirect fluorescence assay (IFA), the immunoperoxidase monolayer assay (IPMA), the serum neutralization test (SN), and the enzyme-linked immunosorbent assay (ELISA). In recent years, by using the PRRSV N protein as an antigen, the commercial IDEXX ELISA kit (IDEXX Laboratories, USA) has been widely used for the detection of PRRSV antibodies. Recently, an ELISA kit that uses recombinant Nsp7 protein as the antigen has been introduced, and the concurrence rate between that kit and the IDEXX ELISA assay has been reported to be up to 97.6% [2]. Although traditional laboratory tests offer good sensitivity and specificity, these detection methods require professional and technical personnel and specialized equipment. In addition, there are many methodological aspects that must be constantly improved and complemented.
In developing countries, vaccination is an important strategy for preventing and controlling PRRSV. To monitor the titer of anti-PRRSV antibodies after vaccination in order to confirm the effectiveness of the vaccination, quick and easy techniques suitable for field testing are needed. Compared with the traditional detection methods, an immunochromatographic strip method has the advantages of being easy to use, providing an answer rapidly and at a low cost, and not requiring specialized equipment or technical personnel, thus such a method is suitable for field testing for antigens or antibodies. In this study, we developed an immunochromatographic test strip for detecting anti-PRRSV antibodies and conducted a preliminary field study of the strip.
Materials and Methods
Serum samples
Pigs were experimentally inoculated with either of two strains of PRRSV having differing levels of virulence, HN07-1 and BJ-4, in order to provide positive test serum samples. HN07-1 strain was isolated during an atypical PRRSV outbreak in Henan Province, China in 2007 [17]. The BJ-4 strain was a typical North American (VR2332)-like PRRSV isolated in 1996 in China [17]. The field sera for antibody testing were collected from several swine herds (all sampled pigs were more than 2-weeks-old) within Henan Province, China. Positive serum samples for porcine circovirus-2 (PCV-2), pseudorabies virus (PRV), classical swine fever virus (CSFV), porcine parvovirus (PPV), and foot-and-mouth disease virus (FMDV) were collected and stored in the Henan Provincial Key Laboratory of Animal Immunology, Henan Academy of Agricultural Sciences.
Preparation of recombinant Nsp7 protein of PRRSV
For the expression of a recombinant Nsp7 proteins in Escherichia (E.) coli, reverse transcriptase PCR was performed on viral genomic RNA from PRRSV isolate BJ-4, obtained from Dr. Hanchun Yang of China Agricultural University. To amplify the Nsp7 gene the following oligonucleotide primers were used: forward primer, 5′-CGCGGATCCTCTCTGACTGGTGCCCTCGCTATG-3′ and reverse primer, 5′-CCGCTCGAGTTCCCATTGAACTCTTCCAT-3′ (restriction sites in bold font). The amplified gene was then ligated into the pET-28a vector. The recombinant plasmid was transformed to E. coli BL21-competent cells. Single colonies were obtained and tested by PCR and sequencing, and a positive clone was grown at 37℃ in LB broth supplemented with 100 µg/mL ampicillin to an optical density of 0.8 at 600 nm. Expression of the recombinant protein was induced by 100 mM isopropyl-β-D-thiogalactopyranoside (IPTG, TAKARA Bio, China) for 8 h at 37℃. Cells were then harvested by centrifugation (7000 × g for 30 min).
Purification of recombinant Nsp7 protein
The recombinant Nsp7 protein was purified by immobilized-metal affinity chromatography (IMAC) using a polyhistidine tag and further purified by a gel filtration column Superdex200 (GE Healthcare, Sweden). The cell pellet was suspended and lysed by sonication on ice. The lysate was centrifuged at 16,000 × g for 30 min, and the supernatant was collected and transferred to a Ni-NTA His Band Resin column pre-equilibrated with binding buffer (500 mM NaCl, 20 mM Tris, 5 mM imidazole). More than five column-volumes of washing buffer (500 mM NaCl, 20 mM Tris, 20 mM imidazole) was added to remove the nonspecific binding proteins. The target protein was eluted with elution buffer (500 mM NaCl, 20 mM Tris, 400 mM imidazole). The purity and relative concentration of the recombinant Nsp7 was determined by SDS-PAGE. The protein was further fractionated by gel filtration on a column of Superdex200 in a buffer of 50 mM Tris, 150 mM NaCl by using the Bio-Rad BioLogic system (Bio-Rad Laboratories, USA). The protein of interest was collected in different fractions according to its different states of aggregation. The final protein products were examined by SDS-PAGE before storing at −80℃.
Western blot
For western blot analysis, 4 µg purified recombinant Nsp7 protein were subjected to 15% SDS-PAGE gel and transferred to polyvinylidene difluoride membranes. The membrane was washed with phosphate-buffered saline-Tween20 (PBST) and blocked with 5% skimmed milk. After washing three times with PBST, the membranes were reacted with PRRSV-positive sera; a PRRSV-negative serum were used as a negative control. After incubating at 37℃ for 1 h, the resulting blot was treated with secondary antibody horseradish peroxidase-conjugated rabbit-anti-pig IgG (Abbkine; WuHan AmyJet Scientific, China) for 1 h. As the substrate for color development, 3-amino-9-ethylcarbazole (AEC) was used.
The antigenicity of the separated protein fractions compared by ELISA
The antibody binding capability of the monomer, dimer, and larger aggregate of the recombinant Nsp7, which were separated by Superdex200 gel filtration column, were compared by indirect ELISA assay. The separated proteins were diluted to the appropriate concentration in 50 mM sodium carbonate bicarbonate buffer (pH 9.6). After incubation for 14 h at 4℃, antigen-coated plates were washed five times with phosphate-buffered saline (PBS) containing 0.05% Tween 80 then blocked with 5% skimmed milk powder dissolved by PBST for 1 h at 37℃. Then, appropriate dilutions in PBST of PRRSV-positive HN07-1, PRRSV-positive BJ-4, and PRRSV-negative pig sera were incubated in the antigen-coated wells at 37℃ for 30 min. Secondary antibody horseradish peroxidase-conjugated rabbit-anti-pig IgG was added at a final dilution of 1:2,000, and the mixture incubated for a further 30 min at 37℃. Finally, 3′,3′,5′,5′-tetramethylbenzidine was added as a substrate. Color development was stopped with 2 M H2SO4, and the OD value at 450 nm was read on a spectrophotometer.
Conjugation of antigen with colloidal gold
Colloidal gold with an average particle diameter of approximately 20 to 25 nm was obtained by reduction of a HAuCl4 solution with 1% trisodium citrate. Three milliliters of 1% trisodium citrate (w/v) was added to 100 mL of 0.01% HAuCl4·3H2O solution (w/v) with stirring. Then the mixture was heated to 100℃ for 20 min. The colloidal gold solution was then cooled to room temperature and stored at 4℃.
The colloidal gold-labeled antigen was prepared according to a previously reported method [23]. Briefly, 4 mL of purified protein (0.2 mg/mL) was incubated with 80 mL of colloidal gold solution for 30 min at room temperature with the pH adjusted to 9.4. After the addition of 1 mL of 3% casein solution, the mixture was incubated at room temperature for a further 10 min, and the resulting suspension was centrifuged 25,000 × g at 4℃ for 30 min. Finally, the colloidal gold-labeled antigen was suspended in 4 mL of 0.02 M sodium borate buffer (containing 2% bovine serum albumin [BSA], 1% sucrose, 0.1% NaN3) and stored at 4℃.
Preparation of the conjugate pad and sample pad
Ten percent BSA was sprayed onto a fiberglass conjugate pad by using a XYZ-3000 dispensing platform (BioDot, USA). The pad was then dried at 42℃ for 45 min. Conjugate solution was dispensed onto the modified fiberglass at a speed of 25 µL/cm (about 2 µg per pad) by using the dispensing platform. After drying for 1 h at 42℃, the pad was then stored in a desiccator at room temperature.
The sample pad was saturated with a buffer containing 20 mM sodium borate, 2.0% (w/v) sucrose, 2.0% (w/v) BSA, and 0.1% (w/v) NaN3 then dried and stored as described above.
Pig anti-Nsp7 antibody preparation
Two-month-old pigs were injected intramuscularly with 100 µg recombinant Nsp7 protein of PRRSV in 2.5 ml of an oil-in-water emulsion per pig. The injection was repeated two times at two-week intervals. Fifteen days after the third immunization, blood containing a high titer of anti-Nsp7 antibody was collected from the immunized pigs. Porcine anti-Nsp7 IgG was salted out from the hyperimmune sera through successive treatment with 50%, 40%, and 33% saturated ammonium sulfate and dialyzed with 0.01 M PBS. ELISA was carried out to determine the titer of anti-Nsp7 protein IgG. The ELISA plate was coated with 0.2 µg/mL Nsp7 protein at 37℃ for 2 h. After washing with PBST five times and blocking with 5% skimmed milk, the anti-Nsp7 protein IgG was two-fold serially diluted with physiological saline and added to the ELISA plate. The anti-Nsp7 IgG titer level was up to 1:20,000. The purified Nsp7 IgG was aliquoted into tubes (1 mL/tube) and stored at −80℃.
Immobilization of capture reagents
Staphylococcal protein A (SPA; Sigma Chemical, USA) at 1.5 mg/mL and porcine anti-Nsp7 IgG at 1 mg/mL were sprayed by using Quanti 3000 Biojets (BioDot) attached to a XYZ Biostrip Dispenser (BioDot) onto a nitrocellulose filter membrane (HiFlow Cellulose Membrane; Millipore, USA) to form zones of 300 mm × 25 mm at the test line (T line) and quality control line (C line), respectively. The T and C lines were in the middle of the membrane, and 5 mm apart from each other. The flow rate of the liquid was 180 sec/4 cm on the membrane, the use level was 1 µL/cm, and forced air drying was carried out at 40℃ for 1 h. The membrane was then sealed in plastic with desiccant and kept at room temperature.
Assembly principle and use of test strips
Assembly of the test strips used the same method as is previous descriptions [24,25,26]. The strip assembly consists of a sample pad, a conjugate pad, a nitrocellulose membrane (NC membrane), and an absorbent pad. The NC membrane was adhered to the center of the plastic solid support, and the absorbent pad and conjugate pad were adhered to upper and lower sides, respectively of the cellulose nitrate membrane with a 1 mm overlap. The main card was cut to the required width with a paper cutting machine. Each strip was packed in a plastic block and then sealed into a plastic bag with desiccant. When stored at room temperature, the strip's period of validity is one year.
For testing, serum samples were diluted 1/200 in physiological saline. The 100 µL diluted sample was added to the sample pad and allowed to migrate to the absorbent pad. If the serum sample to be tested contained PRRSV specific antibodies, these antibodies will form a “gold-labeled antigen-antibody” complex with the gold-labeled antigen on the conjugate pad. The complex, while migrating to the NC membrane, binds to SPA, thereby showing a red line in the T line area. Some of the gold-labeled antigen and some of the gold-labeled antigen-antibody complex migrates further to react with the anti-Nsp7 antibody, showing another red line in the C line area. Negative samples do not produce a red line in the T area, whereas the C line will always show a red line whether or not the serum contains anti-PRRSV antibodies.
Cross-reactivity and detection limit of the immunochromatographic strip test
Cross-reactivity of the strip was evaluated with 18 PRRSV-positive sera (individual S/P ratios were 0.98, 1.37, 1.57, 1.66, 1.66, 1.70, 1.71, 1.72, 1.75, 1.77, 1.78, 1.80, 2.00, 2.28, 2.46, 2.611, 2.75, and 2.79), 10 standard PRRSV-free/negative sera (S/P < 0.4) from PRRSV-free and non-vaccinated pigs and 32 CSFV-, FMDV-, PCV-2-, PRV-, and PPV-infected serum samples.
The detection limit of the strip was evaluated by using PRRSV-positive sera. The titers of the sera were determined by using a commercial ELISA Kit (IDEXX PRRS X3 ELISA kit; IDEXX Laboratories, USA) as the reference standard. Physiological saline solution was used to dilute sera from 1:100 to 1:3,200. The diluted sera were tested with the test strip and the intensity of the T line was scanned with a BioDot TSR3000 film scanner to provide a numerical value. The whole process was repeated three times by three different operators.
Comparison of the strip with other methods
The strip and the commercial IDEXX PRRS X3 ELISA kit were simultaneously used to test and evaluate 1034 sera obtained from several swine herds in Henan Province, China. A comparison of the results obtained by the two methods was used to evaluate the effectiveness of the test strip.
To further evaluate the immunochromatographic test strip for detection, anti-PRRSV antibody sera were collected at intervals after infection. Piglets from PRRSV-free, specific pathogen-free herds were experimentally inoculated with two PRRSV strains of differing virulence, HN07-1 and BJ-4. Groups of three animals were inoculated with HN07-1 or BJ-4 and a further three uninfected piglets were sham-inoculated with a control cell culture supernatant. Blood samples were collected at 0, 7, 14, 21, 28, and 35 days after infection. Sera from these blood samples were simultaneously tested by the immunochromatographic test strip and two reference standard methods: the IDEXX PRRS X3 ELISA kit and the IPMA.
A similar experiment was undertaken to evaluate sera from PRRSV-vaccinated pigs. Five piglets were intramuscularly vaccinated with highly pathogenic porcine reproductive and respiratory syndrome virus vaccine (HuN4-F112) following the manufacturer's instruction, which were produced by Harbin WEIKE Biological Technology Development Company (batch No. 2014001). Sera were collected at 3, 7, 14, 21, and 28 days post vaccination, and tested with the strip and the IDEXX PRRS X3 ELISA kit.
The IPMA was performed according to standard methods. Marc-145 cells were inoculated into 96-well cell plates and cultured in Dulbecco's modified Eagle medium (DMEM; Shanghai Bioleaf Biotech, China) containing 10% fetal bovine sera (Shanghai Bioleaf Biotech) at 37℃ in 5% CO2 atmosphere for 24 h. When the cells had grown to cover over 90% of the available area, serum samples to be assayed were diluted 1:100 with PBS, and 100 µL of each dilution were transferred to paired wells of HN07-1-infected and HN07-1-uninfected Marc-145 cells. The plates were incubated at 37℃ for 1 h and washed three times with PBST, the secondary antibody, horseradish peroxidase-conjugated rabbit-anti-pig IgG, was added at a final dilution of 1:2,000 in PBST. After washing 3 times, the monolayers were stained with AEC. The plates were observed with a light microscope, and wells containing distinctly stained individual cells were scored as positive.
Results
Expression and purification of the Nsp7 protein in E. coli
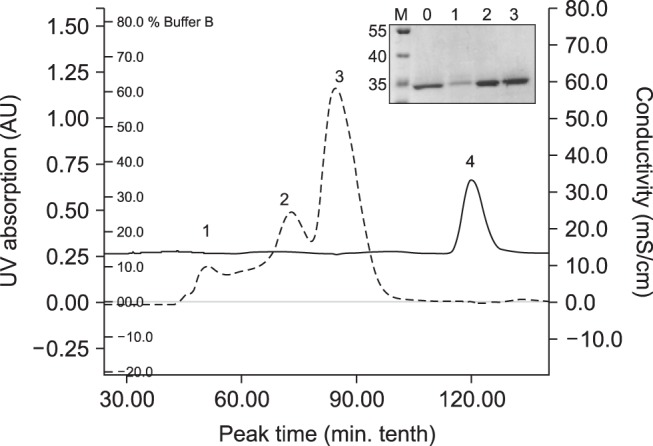
After IPTG induction, the E. coli BL21 (DE3) carrying pET28a-Nsp7 plasmid was shown by using SDS-PAGE to express a recombinant protein with a molecular weight circa 34 kDa. The expressed protein was present in the soluble, as well as in the insoluble, fraction of the bacterial cell pellet. After sonication treatment, the completely soluble target protein was dissolved in phosphate buffer solution and purified by Ni-chelation chromatography. The purified protein was up to 90% pure as determined by SDS-PAGE and western blot analysis revealed that the purified protein reacted with anti-PRRSV sera, confirming its antigenicity (Fig. 1). The target protein self-aggregated and was further fractionated by using molecular sieve chromatography. The different assemblages, referred to as the “larger aggregate”, the “dimer”, and the “monomer” were then identified by SDS-PAGE (Fig. 2).
Fig. 1. Recombinant porcine reproductive and respiratory syndrome virus (PRRSV) Nsp7 preparations in Escherichia coli analyzed by SDS-PAGE and Western blot. M, protein marker; lane 1, supernatant protein of pET 28a-Nsp7 after inducing for 8h; lanes 2, soluble protein purified by Ni-NTA His Band Resin column; and lane 3, soluble protein purified by Western blot.
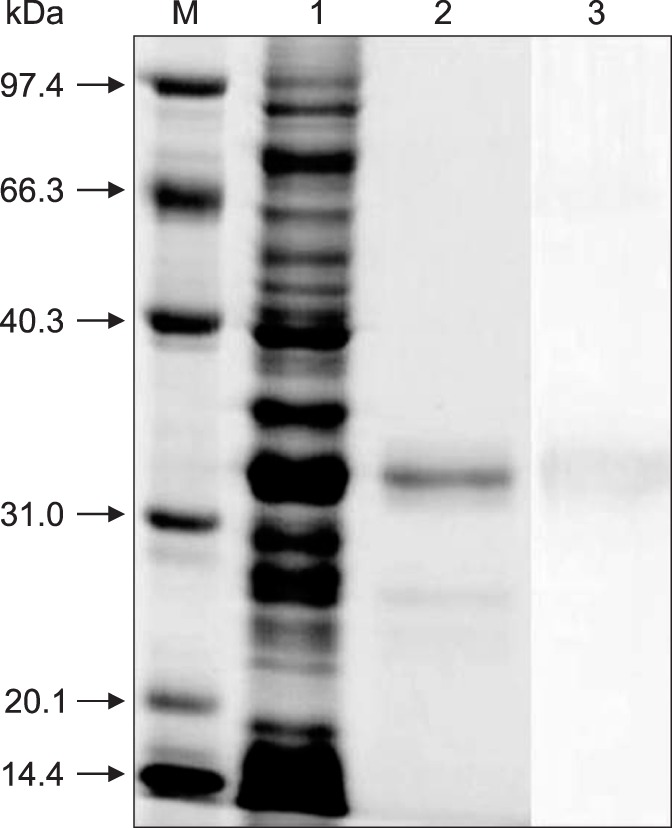
Fig. 2. The different assembling structure proteins identified on SDS-PAGE gels after separation by Superdex 200 gel filtration chromatography (inset). M, the protein molecular mass standard; lane 0, the total protein after purification by Ni-NTA His Band Resin; lanes 1–3, correspond to peaks 1–3 represent the larger aggregate, the dimer, and the monomer. Peak 4 was the elution of imidazole.
Specific immunogenicity of larger aggregate, dimer, and monomer Nsp7
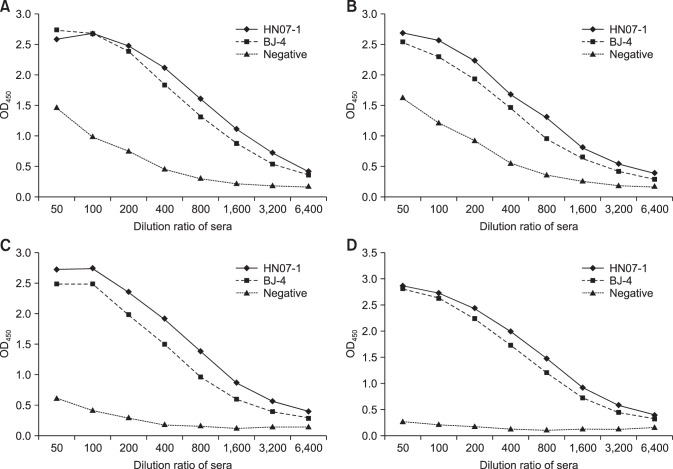
The immunogenicity of the Nsp7 larger aggregate, the dimer, and the monomer assemblages were examined by ELISA. The Nsp7 larger aggregate and dimer proteins showed some reactivity with PRRSV-negative sera. The Nsp7 monomer reacted strongly with pig sera against both classical and atypical PRRSV but reacted weakly with negative sera (Fig. 3). Thus, the monomer was selected for use as the colloidal gold-labeled antigen probe for further study.
Fig. 3. The specific immunogenicity of recombinant Nsp7 tested by enzyme-linked immunosorbent assay. (A) Immunoreaction of the recombinant Nsp7 purified through Ni-chelating affinity chromatography with positive sera to porcine reproductive and respiratory syndrome virus (PRRSV) strain BJ-4 and HN07-1. (B–D) Immunoreaction of larger aggregate, dimer, and monomer separated by a gel filtration chromatography with positive sera to PRRSV strain BJ-4 and HN07-1.
Cross-reactivity of the immunochromatographic strip
Serum samples after 200-fold dilution with physiological saline solution were applied to the immunochromatographic test strip. The combination of antigen and antibody forms an “antigen-antibody complex” which binds to SPA and gives a red line (positive serum) in the test region; no red indicates a negative serum. As a product quality control check, the C line will always show a red line, regardless of whether anti-PRRSV antibodies are present.
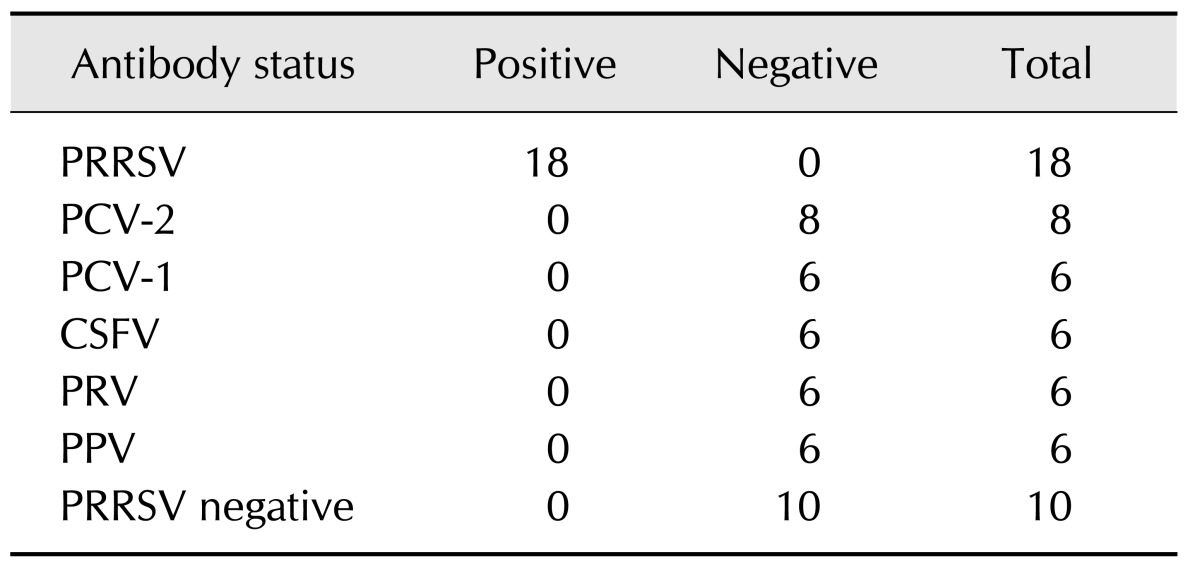
Cross-reactivity of the immunochromatographic test strip was evaluated with 60 reference serum samples, including 18 PRRSV standard positive sera, 10 negative sera samples, and 32 serum samples positive for CSFV, FMDV, PRV, PCV-2, and PPV. As shown in Fig. 4, the positive sera appeared as two visible red bands at the T and C line positions on the strip, while the negative sera and other sera appeared as only one red band at the C line position. As shown in Table 1, the strip gave 18 positive results from the 18 PRRSV-positive samples, and all of the 10 PRRSV-negative samples and the serum samples positive for other non-PRRSV pathogens showed negative results on the immunochromatographic strip. The results indicated that the specificity of the immunochromatographic strip was 100% (42/42).
Fig. 4. Specificity of the immunochromatographic strip test. Positive sera of porcine reproductive and respiratory syndrome virus (PRRSV), porcine circovirus-1 (PCV-1), pseudorabies virus (PRV), classical swine fever virus (CSFV), porcine parvovirus (PPV), porcine circovirus-2 (PCV-2), and PRRSV-negative sera were tested by using the immunochromatographic strip at the same time. Result positivity can be decided by visual judgment.

Table 1. Specificity of the immunochromatographic strip with 200-fold diluted porcine sera.
PRRSV, porcine reproductive and respiratory syndrome virus; PCV-2, porcine circovirus-2; PCV-1, porcine circovirus-1; CSFV, classical swine fever virus; PRV, pseudorabies virus.
The detection limit of the immunochromatographic strip
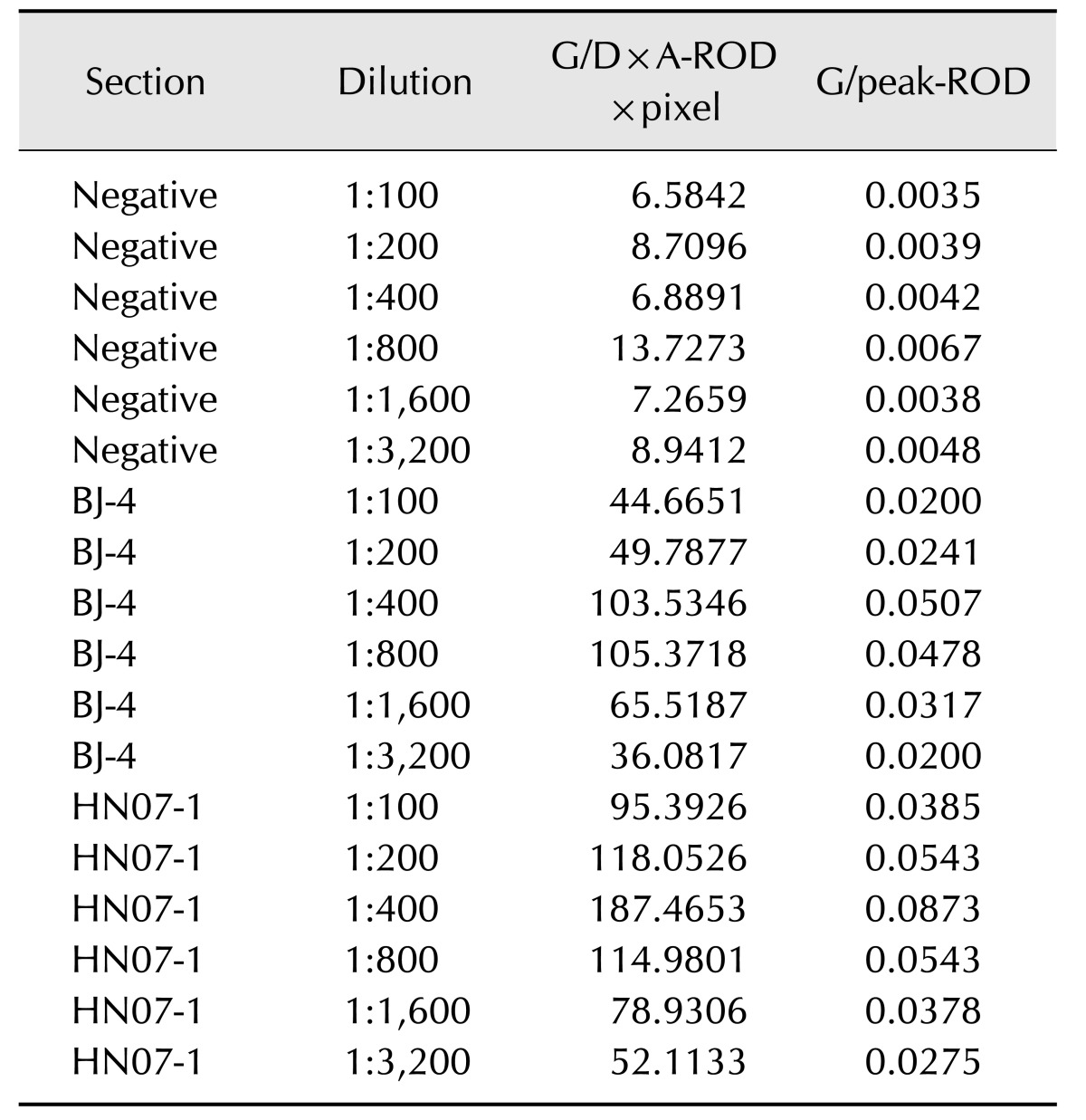
To evaluate the detection limit of the immunochromatographic strip, the PRRSV standard positive sera (S/P ratios were 2.428 and 2.715) were two-fold serially diluted with physiological saline from 1:100 to 1:3,200. A red band could be seen clearly at the T line position when the dilution of the serum sample was 1:3,200 or less, indicating that the strip has a high sensitivity for detecting small amounts of anti-PRRSV antibodies. The strip test lines were scanned with a BioDot TSR3000 membrane strip reader and showed that the G/D×A-ROD×pixel of PRRSV-positive sera for HN07-1 and BJ-4 diluted 1:100, 1:200, 1:400, 1:800, 1:1600, 1:3200 were 95.3926, 118.0526, 187.4653, 114.9801, 78.9306, 52.1133 and 44.6651, 49.7877, 103.5346, 105.3718, 65.5187, 36.0817, respectively. The highest G/D×A-ROD×pixel of the negative/control sera was 13.7273, which was very weak and could not be seen by the naked eye (Table 2). The results indicated that the strip had a high sensitivity for detecting PRRSV antibodies and could detect antibodies in serum samples at low concentrations.
Table 2. G/D×area, and G/peak of the relative optical density (ROD) of test lines of the standard porcine reproductive and respiratory syndrome virus samples*.
*Standard sera samples were tested by using the immunochromatographic test strips and the test lines were scanned with a TSR3000 membrane strip reader. G/D×A indicates graph/density×area, which means density value of the sampled outline, multiplied by its area. G/peak indicates graph peak density, maximum density value of sampled line points. ROD means relative optical density, inverse of gray level value with a logarithmic transformation.
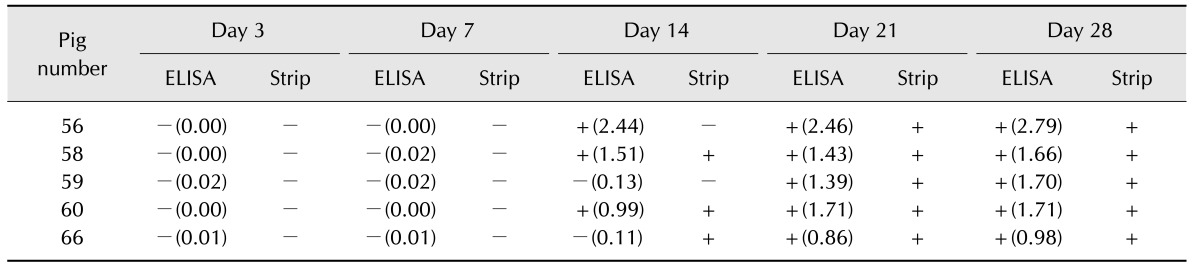
Comparison of the strip with other methods for detection of experimental infection in pigs
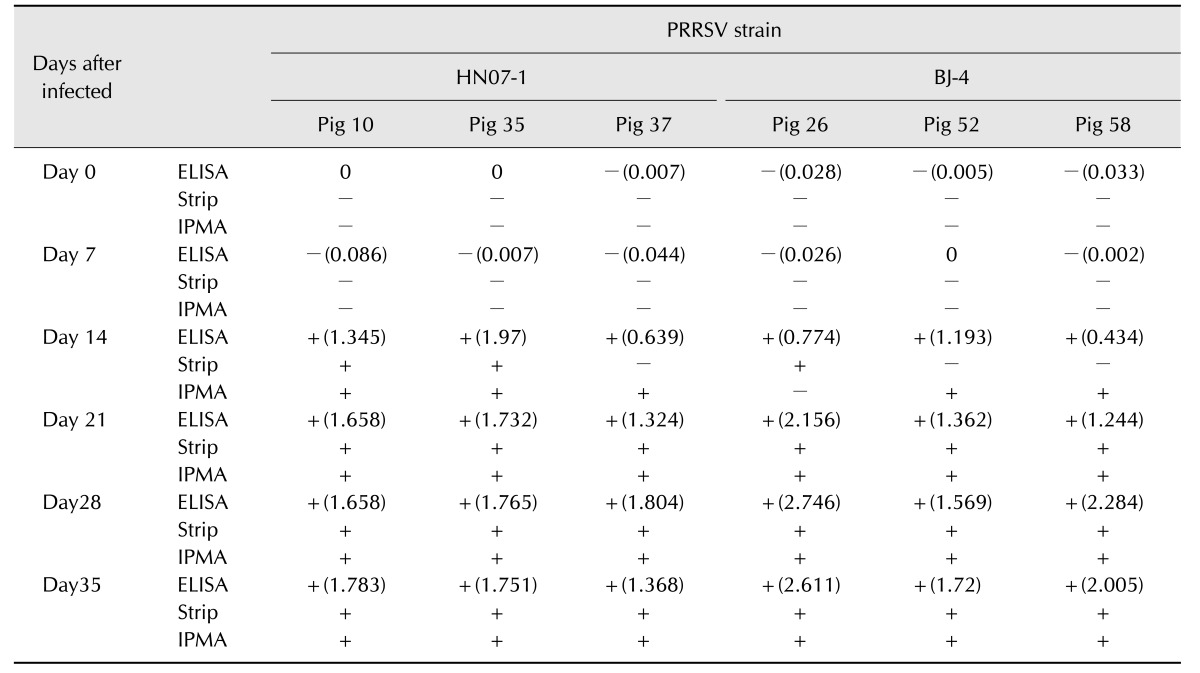
A comparison of the serological reactivity of the strip with the commercial IDEXX PRRS X3 ELISA test and the IPMA was performed by using serum samples that were collected from pigs experimentally infected with isolates HN07-1 and BJ-4 (Table 3). All sera collected prior to 7 days postinoculation were negative in the strip test, ELISA, and IPMA results. At 14 days postinoculation, all samples were detected as seropositive (S/P > 0.4) by the IDEXX ELISA. The strip test detected seroconversion at 14 days post inoculation in pig numbers 10 and 26. At 21 days postinoculation, all samples were identified as positive by all three methods. In summary, 33 of 36 (91.7%) IDEXX results agreed with the results obtained by the strip and 32 of 36 (88.9%) IPMA results were in agreement with the strip.
Table 3. Detection porcine reproductive and respiratory syndrome virus (PRRSV) antibodies in swine sera at different days after infected with PRRSV HN07-1 or BJ-4: test strip vs. IDEXX PRRS 3X ELISA kit and immunoperoxidase monolayer assay (IPMA).
ELISA, enzyme-linked immunosorbent assay.
Comparison of postvaccination PRRSV antibody detection by the strip and IDEXX PRRS X3 ELISA
All sera collected prior to 7 days post vaccination were negative in both the strip and the IDEXX PRRS X3 ELISA results. At 14 days post vaccination, both the strip and ELISA detected seroconversion in the vaccinated pigs, but both tests also recorded animals which were negative. However, at 21 days postvaccination all serum samples were positive in both tests (Table 4).
Table 4. Detection of porcine reproductive and respiratory syndrome virus antibodies in swine sera at different days after vaccination using the strip and the IDEXX PRRS 3X ELISA kit.
ELISA, enzyme-linked immunosorbent assay; −, Negative; + positive. Samples with S/P ratio > 0.4 were considered positive.
Comparison of the strip with the IDEXX PRRS X3 ELISA for the detection pigs infected in the field
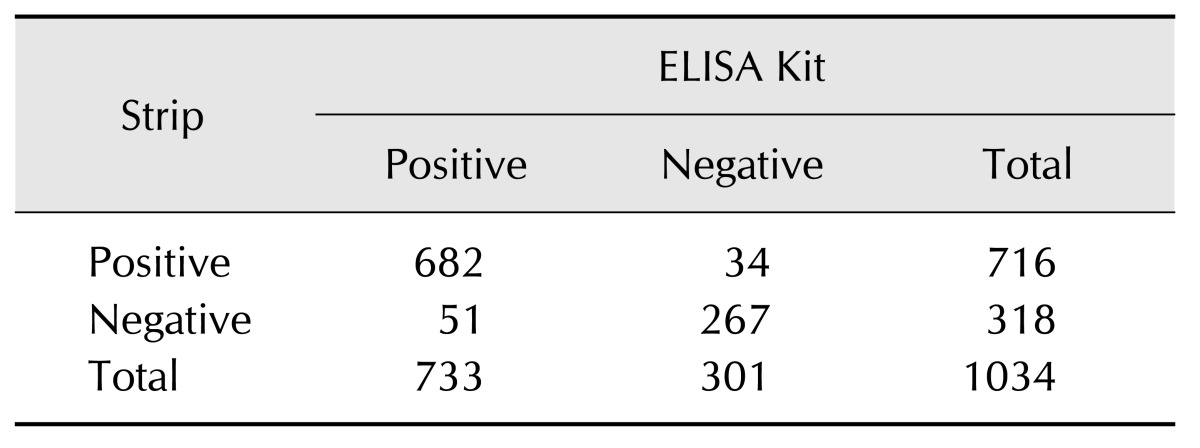
To further demonstrate the usefulness and reliability of the immunochromatographic strip for detecting the serum antibody responses of pigs infected with field strains of the virus a 1034 field serum samples were tested in parallel with the strip and IDEXX PRRS X3 ELISA (Table 5). According to the formula TP/(TP + FN) and TN/(TN + FP): TP, true positive, TN, true negative, FP, false positive, and FN, false negative, which was used to calculate the sensitivity and specificity of the test, the corresponding values were 93.04% (682/733) and 88.70% (267/301), respectively.
Table 5. Comparison of results from the strip and the IDEXX PRRS X3 ELISA from pigs infected with field virus.
ELISA, enzyme-linked immunosorbent assay.
Discussion
Serological testing for antiviral antibodies is a standard diagnostic and surveillance method for determining whether pigs have been exposed to PRRSV. Currently, the IDEXX PRRSV ELISA kit, which is based on the antigen PRRSV N protein, is widely used for determining the viral infection status of swine herds. The recombinant Nsp7 protein of PRRSV has also been used as an antigen in indirect ELISA tests to detect the antibodies against PRRSV [2]. These ELISA methods require professional/technical personnel and are time-consuming. The proposed immunochromatographic test strip described here is simple, easy to operate, cost efficient, and provides rapid results, which is ideal for wide application, including developing countries.
As mentioned, PRRSV has European and North American types. Nucleotide sequences within the same genotype have some variability, whereas the nucleotide sequences between the two different genotypes differ significantly [11,12]. The amino acid sequence identity of the Nsp7 protein in the European-type PRRSV is within 96.7% to 97.4%, whereas that in the North American type is within 84.9% to 100%. However, the amino acid sequence identity of Nsp7 protein between the European and North American types overlapped by only 45% [8,9,15,18]. PRRSV isolates in China are mainly of the North American type, and the BJ-4 strain is a North American type. Thus, we selected the Nsp gene from the BJ-4 strain, a typical North American (VR2332)-like PRRSV, for the preparation of the recombinant antigen for the detection of serum antibodies for the diagnosis of PRRSV infection in China. The developed immunochromatographic strip that uses the recombinant Nsp7 antigen of the BJ-4 strain can detect antibodies against the atypical PRRSV isolate HN07-1, which has been the dominant virus in China after 2006.
Recombinant Nsp7 protein has a number of practical advantages. First, it is expressed in a soluble form, which is convenient for protein purification. The Nsp7 protein is sufficiently immunogenic for antibodies against the Nsp7 protein to be detected 14 days after PRRSV infection, and these antibodies remain at elevated levels for 202 days [2]. Therefore, the recombinant Nsp7 protein can be used as the target antigen for detecting anti-Nsp7 antibodies in serological testing. The immunochromatographic strip for detecting PRRSV antibodies on the basis of N protein was developed in 2008 [7]. The movement to eliminate PRRSV may require a variety of serological diagnostic technologies that can accurately detect persistent infection in pigs. In this study, we used a gold-labeled recombinant Nsp7 protein to bind the anti-PRRSV antibodies. This gold-labeled antigen-antibody complex can be captured by SPA and the result is then seen as a red line indicating the presence of PRRSV antibodies.
The Nsp7 recombinant protein was further purified by gel filtration to yield the monomer form because aggregates of the protein were found to nonspecifically bind IgG. We observed cross reaction of the Nsp7 large aggregate and dimer protein with PRRSV-negative sera, and the Nsp7 monomer reacted strongly and specifically with porcine antibodies against both classical and atypical PRRSV. On this basis, the monomer was selected for a colloidal gold-labeled antigen probe to increase the specificity of the test strip.
The developed test strips have high sensitivity in the detection of antibodies against PRRSV and exhibit no cross reaction with antibodies of other pathogens such as FMDV, PRV, CSFV, PPV, PCV-2, and negative PRRSV sera. We also used the test strips to screen 1034 clinical sera from several swine herds in Henan Province and then compared our results with those from the commercial IDEXX PRRS X3 ELISA kit. The results showed that the sensitivity and specificity of the strip were 93.04% and 88.70%. We used the test strips to detect antibodies in pigs experimentally infected with both classical and atypical PRRSV and compared the results with those of the IDEXX PRRS X3 ELISA kit and the IPMA. The overall coincidence rate was 91.7% and 88.9%, respectively. Post vaccination test results from piglets demonstrated that the strip and the IDEXX PRRS X3 ELISA assay had a high coincidence rate (92%). Considering all of the results, the test strip shows comparable performance to the reference ELISA and should be considered of similar value to the ELISA kit in clinical applications.
In summary, the chromatographic strip developed in this study is a simple, sensitive, and serologically-specific detection method. The study results show that the strip accurately identifies antibodies against PRRSV in experimentally infected swine sera, field-infected porcine sera, and sera from piglets after vaccination. Results using the new method are well within the ranges achieved by the commercial IDEXX ELISA assay and the IPMA test. However, the strip cannot distinguish antibodies induced by vaccination from those resulting from viral infection. Further investigation to develop an antibody differentiation test is required.
Acknowledgments
This work was supported by grants from China Agriculture Research System (CARS-36), the Pig Industry Technology System Innovation Team Project of Henan Province (S2012-06), and the Special Fund for Agro-scientific Research in the Public Interest (201203039). We would like to thank Professor Gregson from IoN University for revising the paper.
Footnotes
Conflict of Interest: The authors declare no conflict of interests.
References
- 1.Albina E, Mesplède A, Chenut G, Le Potier MF, Bourbao G, Le Gal S, Leforban Y. A serological survey on classical swine fever (CSF), Aujeszky's disease (AD) and porcine reproductive and respiratory syndrome (PRRS) virus infections in French wild boars from 1991 to 1998. Vet Microbiol. 2000;77:43–57. doi: 10.1016/s0378-1135(00)00255-8. [DOI] [PubMed] [Google Scholar]
- 2.Brown E, Lawson S, Welbon C, Gnanandarajah J, Li J, Murtaugh MP, Nelson EA, Molina RM, Zimmerman JJ, Rowland RRR, Fang Y. Antibody response to porcine reproductive and respiratory syndrome virus (PRRSV) nonstructural proteins and implications for diagnostic detection and differentiation of PRRSV types I and II. Clin Vaccine Immunol. 2009;16:628–635. doi: 10.1128/CVI.00483-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen J, Liu T, Zhu CG, Jin YF, Zhang YZ. Genetic variation of Chinese PRRSV strains based on ORF5 sequence. Biochem Genet. 2006;44:425–435. doi: 10.1007/s10528-006-9039-9. [DOI] [PubMed] [Google Scholar]
- 4.Cho HJ, Deregt D, Joo HS. An ELISA for porcine reproductive and respiratory syndrome: production of antigen of high quality. Can J Vet Res. 1996;60:89–93. [PMC free article] [PubMed] [Google Scholar]
- 5.Cho HJ, McNab B, Dubuc C, Jordan L, Afshar A, Magar R, Prins S, Eernisse K. Comparative study of serological methods for the detection of antibodies to porcine reproductive and respiratory syndrome virus. Can J Vet Res. 1997;61:161–166. [PMC free article] [PubMed] [Google Scholar]
- 6.Collins JE, Benfield DA, Christianson WT, Harris L, Hennings JC, Shaw DP, Goyal SM, McCullough S, Morrison RB, Joo HS, Gorcyca D, Chladek Dan. Isolation of swine infertility and respiratory syndrome virus (isolate ATCC VR-2332) in North America and experimental reproduction of the disease in gnotobiotic pigs. J Vet Diagn Invest. 1992;4:117–126. doi: 10.1177/104063879200400201. [DOI] [PubMed] [Google Scholar]
- 7.Cui S, Zhou S, Chen C, Qi T, Zhang C, Oh J. A simple and rapid immunochromatographic strip test for detecting antibody to porcine reproductive and respiratory syndrome virus. J Virol Methods. 2008;152:38–42. doi: 10.1016/j.jviromet.2008.05.029. [DOI] [PubMed] [Google Scholar]
- 8.Fang Y, Kim DY, Ropp S, Steen P, Christopher-Hennings J, Nelson EA, Rowland RR. Heterogeneity in Nsp2 of European-like porcine reproductive and respiratory syndrome viruses isolated in the United States. Virus Res. 2004;100:229–235. doi: 10.1016/j.virusres.2003.12.026. [DOI] [PubMed] [Google Scholar]
- 9.Fang Y, Schneider P, Zhang WP, Faaberg KS, Nelson EA, Rowland RRR. Diversity and evolution of a newly emerged North American Type 1 porcine arterivirus: analysis of isolates collected between 1999 and 2004. Arch Virol. 2007;152:1009–1017. doi: 10.1007/s00705-007-0936-y. [DOI] [PubMed] [Google Scholar]
- 10.Fang Y, Snijder EJ. The PRRSV replicase: exploring the multifunctionality of an intriguing set of nonstructural proteins. Virus Res. 2010;154:61–76. doi: 10.1016/j.virusres.2010.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mardassi H, Mounir S, Dea S. Identification of major differences in the nucleocapsid protein genes of a Québec strain and European strains of porcine reproductive and respiratory syndrome virus. J Gen Virol. 1994;75:681–685. doi: 10.1099/0022-1317-75-3-681. [DOI] [PubMed] [Google Scholar]
- 12.Mardassi H, Wilson L, Mounir S, Dea S. Detection of porcine reproductive and respiratory syndrome virus and efficient differentiation between Canadian and European strains by reverse transcription and PCR amplification. J Clin Microbiol. 1994;32:2197–2203. doi: 10.1128/jcm.32.9.2197-2203.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mateu E, Diaz I. The challenge of PRRS immunology. Vet J. 2008;177:345–351. doi: 10.1016/j.tvjl.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meulenberg JJ, Hulst MM, de Meijer EJ, Moonen PL, den Besten A, de Kluyver EP, Wensvoort G, Moormann RJ. Lelystad virus, the causative agent of porcine epidemic abortion and respiratory syndrome (PEARS), is related to LDV and EAV. Virology. 1993;192:62–72. doi: 10.1006/viro.1993.1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nelsen CJ, Murtaugh MP, Faaberg KS. Porcine reproductive and respiratory syndrome virus comparison: divergent evolution on two continents. J Virol. 1999;73:270–280. doi: 10.1128/jvi.73.1.270-280.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Neumann EJ, Kliebenstein JB, Johnson CD, Mabry JW, Bush EJ, Seitzinger AH, Green AL, Zimmerman JJ. Assessment of the economic impact of porcine reproductive and respiratory syndrome on swine production in the United States. J Am Vet Med Assoc. 2005;227:385–392. doi: 10.2460/javma.2005.227.385. [DOI] [PubMed] [Google Scholar]
- 17.Qiao S, Feng L, Bao D, Guo J, Wan B, Xiao Z, Yang S, Zhang G. Porcine reproductive and respiratory syndrome virus and bacterial endotoxin act in synergy to amplify the inflammatory response of infected macrophages. Vet Microbiol. 2011;149:213–220. doi: 10.1016/j.vetmic.2010.11.006. [DOI] [PubMed] [Google Scholar]
- 18.Ropp SL, Wees CEM, Fang Y, Nelson EA, Rossow KD, Bien M, Arndt B, Preszler S, Steen P, Christopher-Hennings J, Collins JE, Benfield DA, Faaberg KS. Characterization of emerging European-like porcine reproductive and respiratory syndrome virus isolates in the United States. J Virol. 2004;78:3684–3703. doi: 10.1128/JVI.78.7.3684-3703.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Spagnuolo-Weaver M, Walker IW, McNeilly F, Calvert V, Graham D, Burns K, Adair BM, Allan GM. The reverse transcription polymerase chain reaction for the diagnosis of porcine reproductive and respiratory syndrome: comparison with virus isolation and serology. Vet Microbiol. 1998;62:207–215. doi: 10.1016/s0378-1135(98)00212-0. [DOI] [PubMed] [Google Scholar]
- 20.Takashima H, Takai K, Goto S. A modified immunoperoxidase assay for detection of antibody porcine reproductive and respiratory syndrome (PRRS) virus in swine sera. J Vet Med Sci. 1999;61:195–196. doi: 10.1292/jvms.61.195. [DOI] [PubMed] [Google Scholar]
- 21.Wensvoort G, de Kluyver EP, Pol JM, Wagenaar F, Moormann RJ, Hulst MM, Bloemraad R, den Besten A, Zetstra T, Terpstra C. Lelystad virus, the cause of porcine epidemic abortion and respiratory syndrome: a review of mystery swine disease research at Lelystad. Vet Microbiol. 1992;33:185–193. doi: 10.1016/0378-1135(92)90046-v. [DOI] [PubMed] [Google Scholar]
- 22.Xiao S, Chen Y, Wang L, Gao J, Mo D, He Z, Liu X. Simultaneous detection and differentiation of highly virulent and classical Chinese-type isolation of PRRSV by real-time RT-PCR. J Immunol Res. 2014;2014:809656. doi: 10.1155/2014/809656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang S, Yang J, Zhang G, Wang X, Qiao S, Zhao D, Zhi Y, Li X, Xing G, Luo J, Fan J, Bao D. Development of an immunochromatographic strip for the detection of antibodies against foot-and-mouth disease virus serotype O. J Virol Methods. 2010;165:139–144. doi: 10.1016/j.jviromet.2010.01.001. [DOI] [PubMed] [Google Scholar]
- 24.Zhang G, Wang X, Zhi A, Bao Y, Yang Y, Qu M, Luo J, Li Q, Guo J, Wang Z, Yang J, Xing G, Chai S, Shi T, Liu Q. Development of a lateral flow immunoassay strip for screening of sulfamonomethoxine residues. Food Addit Contam Part A Chem Anal Control Expo Risk Assess. 2008;25:413–423. doi: 10.1080/02652030701561452. [DOI] [PubMed] [Google Scholar]
- 25.Zhang GP, Guo JQ, Wang XN, Yang JX, Yang YY, Li QM, Li XW, Deng RG, Xiao ZJ, Yang JF, Xing GX, Zhao D. Development and evaluation of an immunochromatographic strip for trichinellosis detection. Vet Parasitol. 2006;137:286–293. doi: 10.1016/j.vetpar.2006.01.026. [DOI] [PubMed] [Google Scholar]
- 26.Zhang GP, Wang XN, Yang JF, Yang YY, Xing GX, Li QM, Zhao D, Chai SJ, Guo JQ. Development of an immunochromatographic lateral flow test strip for detection of beta-adrenergic agonist Clenbuterol residues. J Immunol Methods. 2006;312:27–33. doi: 10.1016/j.jim.2006.02.017. [DOI] [PubMed] [Google Scholar]