Abstract
Aim:
Identification of pathogenic clinical bacterial isolates is mainly dependent on phenotypic and genotypic characteristics of the microorganisms. These conventional methods are costive, time-consuming, and need special skills and training. An alternative, mass spectral (proteomics) analysis method for identification of clinical bacterial isolates has been recognized as a rapid, reliable, and economical method for identification. This study was aimed to evaluate and compare the performance, sensitivity and reliability of traditional bacteriology, phenotypic methods and matrix-assisted laser desorption-ionization-time-of-flight mass spectrometry (MALDI-TOF MS) in the identification of clinical Escherichia coli and Salmonella isolates recovered from chickens.
Materials and Methods:
A total of 110 samples (cloacal, liver, spleen, and/or gall bladder) were collected from apparently healthy and diseased chickens showing clinical signs as white chalky diarrhea, pasty vent, and decrease egg production as well as freshly dead chickens which showing postmortem lesions as enlarged liver with congestion and enlarged gall bladder from different poultry farms.
Results:
Depending on colonial characteristics and morphological characteristics, E. coli and Salmonella isolates were recovered and detected in only 42 and 35 samples, respectively. Biochemical identification using API 20E identification system revealed that the suspected E. coli isolates were 33 out of 42 of colonial and morphological identified E. coli isolates where Salmonella isolates were represented by 26 out of 35 of colonial and morphological identified Salmonella isolates. Serological identification of isolates revealed that the most predominant E. coli serotypes were O1 and O78 while the most predominant Salmonella serotype of Salmonella was Salmonella Pullorum. All E. coli and Salmonella isolates were examined using MALDI-TOF MS. In agreement with traditional identification, MADI-TOF MS identified all clinical bacterial samples with valid scores as E. coli and Salmonella isolates except two E. coli isolates recovered from apparently healthy and diseased birds, respectively, with recovery rate of 93.9% and 2 Salmonella isolates recovered from apparently healthy and dead birds, respectively, with recovery rate of 92.3%.
Conclusion:
Our study demonstrated that Bruker MALDI-TOF MS Biotyper is a reliable rapid and economic tool for the identification of Gram-negative bacteria especially E. coli and Salmonella which could be used as an alternative diagnostic tool for routine identification and differentiation of clinical isolates in the bacteriological laboratory. MALDI-TOF MS need more validation and verification and more study on the performance of direct colony and extraction methods to detect the most sensitive one and also need using more samples to detect sensitivity, reliability, and performance of this type of bacterial identification.
Keywords: ABI, Bruker Daltonics, colibacillosis, Escherichia coli, matrix-assisted laser desorption ionization time-of-flight mass spectrometry, Salmonella, Salmonella pullorum
Introduction
Escherichia coli infection in poultry is one of the principal causes of mortality and morbidity in chickens and turkeys resulting in great economic losses to poultry industry due to, retardation of growth, decreased feed conversion rate, decreased egg production, decreased fertility, reduced hatchability, downgraded carcasses and condemnation of whole affected carcasses or organs after slaughter and finally the high cost of wide range of antibacterial agents used to control E. coli infection in many poultry farms [1]. Colibacillosis in chickens refers to local and systematic (extraintestinal) infections caused mainly by avian pathogenic E. coli [2], which are commonly belong to certain O groups, particularly O1, O2, O8, O15, O18, O35, O78, O88, O109, and O115 [3]. E. coli infection in poultry is responsible for a variety of disease conditions such as colisepticemia, air sac disease, serositis (peritonitis, pericarditis, and perihepatitis), omphalitis, panophthalmitis, synovitis, salpingitis, coligranuloma, swollen head syndrome, cellulitis, yolk sac infection, and enteritis [4].
One of most common economically important bacterial disease in poultry industry is Salmonellosis particularly fowl typhoid and pullorum disease [5]. Avian Salmonella infection is caused by different Salmonella species [6]. More than 2500 Salmonella serotypes have been mentioned under the species but only about 10% of these serotypes have been isolated from poultry [7]. Among this, Salmonella Pullorum (SP) species (S. enterica subsp. enterica serovar pullorum) which causing pullorum disease and Salmonella Enterica serovar Gallinarum is main causative agent of fowl typhoid.
The bacteriological method for detecting clinical bacterial isolates as Salmonella and E. coli involves culturing the organism in different specific and selective media and identifying isolates using traditional and conventional bacteriological methods is time-consuming. Therefore a rapid, sensitive, specific, reliable, and cost effective method for identification of pathogens in clinical samples is required. As an alternative to various other identification methods, mass spectral (proteomics) analysis for identification of clinical bacterial isolates has been recognized. Matrix-assisted laser desorption-ionization-time-of-flight mass spectrometry (MALDI-TOF MS) can be used as a sensitive, reliable and rapid procedures for identification of various clinical bacterial isolates [8], such as Gram-positive bacteria [9], mycobacteria [10], Brucella [11], Enterobacteriaceae [8], yeast [12], mold [13], and non-fermenting bacteria [14].
The aim of this study is to evaluate and compare the performance, reliability, and sensitivity of classical bacteriological and phenotypic methods in comparison to MALDI-TOF MS in identification of E. coli and Salmonella recovered from chickens.
Materials and Methods
Ethical approval
All samples were collected as per standard sample collection procedure without giving any stress or harm to the animals. Such type of study do not require any specific ethical approval.
Sampling
A total of 110 samples collected from different poultry farms including apparently healthy (31 cloacal swabs), and diseased (49 cloacal swabs) chickens which showing clinical signs as white chalky diarrhea, pasty vent, and decrease egg production and also from freshly dead chickens (30 liver, spleen, and gallbladder samples) which showing postmortem lesions as enlarged liver with congestion and enlarged gallbladder. The samples were transferred immediately to sterile buffered peptone water, then wrapped with ice, kept in box and transferred directly to the lab [15].
Isolation of E. coli and Salmonella isolates
Isolation of E. coli and Salmonella was carried out on three successive stages which are pre-enrichment in non-selective liquid broth [15], enrichment in selective liquid media [16] and plating onto solid selective agar media as MacConkey agar, SS agar and eosin methylene blue (EMB) agar media [17].
Identification of E. coli and Salmonella isolates
Colonial and microscopical examination E. coli and Salmonella isolates
The suspected colonies were examined for their colonial morphology [15] on nutrient agar, EMB agar, MacConkey agar, xylose lysine decarboxylase agar (XLD), and Salmonella-Shigella agar (S-S). Microscopical examination was performed according to Merchant and Packer [18]. Isolates were preserved for further examination by growing and spreading of the microorganism by stabbing in semisolid agar [19]. Isolates were tested for motility [20].
Biochemical identification of E. coli and Salmonella isolates
Biochemical identification of isolates was done using pure cultures of each of the suspected isolates using API 20E plate system (Biomerieux –France cat# 20-100).
Serological identification of E. coli and Salmonella isolates
Serological identification of the isolates was conducted according to Kauffmann [21]. Smooth colonies of E. coli isolates that were preliminary identified biochemically as E. coli were subjected to serological identification according to Sojka [22], Edward and Ewing [23] against the polyvalent 1, 2, 3, and 4 antisera using the agglutination test. These polyvalent antisera are:
•Polyvalent (1): O1, O26, O86, O111, O119, O127, O128
•Polyvalent (2): O2, O11, O87, O127, O142
•Polyvalent (3): O6, O27, O78, O148, O159, O168
•Polyvalent (4): O44, O55, O125, O126, O146, O166.
The positive agglutinating isolates with the polyvalent antisera was retested with corresponding specific monovalent antisera. These monovalent antisera are:
O1, O26, O86, O111, O119, O127, O128. O2, O11, O87, O127, O142, O6, O27, O78, O148, O159, O168, O44, O55, O125, O126, O146, O166.
Smooth culture of biochemically identified Salmonella isolates was further tested using polyvalent and monovalent Salmonella antisera O and H factor using slide agglutination [21,23].
MALDI-TOF MS (extraction method) [24,25]
One to 2 pure colonies of E. coli or Salmonella were suspended in 300 ul of molecular grade water (Sigma-Aldrich, St. Louis, MO) and vortexed. Then, 900 ul of absolute ethanol was added, vortexed, and centrifuged at 20,800 ×g for 3 min. The supernatant was decanted, and the pellet was dried at room temperature then, 50 ul of 70% formic acid and 50 ul of acetonitrile were added and mixed by pipetting, followed by centrifugation at 20,800 ×g for 2 min. 2 ul of supernatant was applied into the 24 spot plate and left to dry at room temperature followed by the addition of 2 ul of MALDI matrix (a saturated solution of -cyano-4-hydroxycinnamic acid in 50% acetonitrile and 2.5% trifluoroacetic acid). For each plate, a bacterial test standard (Bruker Daltonics) was included to calibrate the instrument and validate the run. Spectra were analyzed using MALDI Biotyper automation control and the Bruker Biotyper 2.0 software and library (version 2.0, 3,740 entries; Bruker Daltonics). Identification score criteria were performed as recommended by Bruker Daltonics which evaluated as follow:
•A score of 2.000 indicated species level identification
•A score of 1.700-1.999 indicated identification to the genus level
•A score of 1.700 was interpreted as no identification.
With respect of direct isolation of causative agents as a gold standard test, API 20A and MALDI-TOF MS sensitivity, relative sensitivity and specificity in identification of causative agents were calculated using (https://www.medcalc.org/calc/diagnostic_test.php) as shown in Table-1.
Table-1.
Calculation of sensitivity and specificity with respect of gold standard test (https://www.medcalc.org/calc/diagnostic_test.php).
| Results | Gold standard test (cft) | Total | |
|---|---|---|---|
|
| |||
| Positive | Negative | ||
| Test under evaluation | |||
| Positive | A | B | A+b |
| Negative | C | D | C+d |
| Total | A+c | B+d | n (264) |
Relative sensitivity=A/A+C, specificity=D/D+B, true positive (positive predictive value)=A/A+B, false positive (B)=B/A+B, true negative (negative predictive value)=D/D+C, false negative (C)=C/D+C
Results and Discussion
Isolation and identification of E. coli and Salmonella isolates
In birds, E. coli infections cause many clinical manifestations; the most common is being airsacculitis, pericarditis, septicemia, and death [26]. Colibacillosis due to virulent E. coli in chickens is characterized by a respiratory disease which is frequently followed by a generalized infection [27]. Salmonellae are widespread in human and animals worldwide. In industrialized countries, non-typhoid Salmonellae is an important cause of bacterial gastroenteritis. Zoonotic Salmonella Enterica serovars are among the most important agents of food-borne infections throughout the world. Poultry is one of the major sources of Salmonella-contaminated food products that cause human Salmonellosis [28].
In this study, a total of 110 samples were collected from apparently healthy (31 cloacal), diseased (49 cloacal), and freshly dead (30 liver and hearts) chickens from different poultry farms and examined microbiologically.
Colonial characteristics and morphological characteristics of the E. coli and Salmonella isolates
Depending on colonial characteristics and morphological characteristics, E. coli was detected in only 42 clinical specimens. These isolates were 11 out of 31 isolates recovered from apparently healthy chickens, 17 out of 49 isolates recovered from diseased chickens, and 14 out of 30 isolates recovered from freshly dead chickens. Suspected E. coli isolates when cultured on different media were showed rounded, non-pigmented colonies on nutrient agar medium, while on MacConkey agar medium showed rounded, non-mucoid pink colonies (lactose fermenter). At the same time, the same isolates on SS agar appeared as rounded, non-mucoid pink colonies and on EMB agar showed a distinctive yellow-green metallic sheen. These isolates were Gram-negative, motile, non-sporulated, and medium-sized bacilli (Table-2). Whereas, 35 suspected isolates were behaved as Salmonella spp. and were aerobic and facultatively anaerobic, have a wide temperature range and like all enterobacteria grow readily on all ordinary media. On MacConkey agar, Salmonella colonies were 2-4 mm in diameter and pale since lactose was not fermented after 18-24 h incubation at 37°C while on SS agar, Salmonella appeared transparent with black centers. In the same time on XLD agar, Salmonella appeared pink with black pigment indicating H2S production. These isolates were Gram-negative non-spore-forming medium size straight rods and usually motile (Table-2). All above-mentioned results agree with Antunes et al. [29] and Ozbey and Ertas [15].
Table-2.
E. coli and Salmonella isolates recovered from different samples.
| Source | Number of samples | Number of suspected E. coli isolates | Number of suspected Salmonella isolates |
|---|---|---|---|
| Apparently healthy | 31 | 11 | 9 |
| Diseased | 49 | 17 | 17 |
| Freshly dead | 30 | 14 | 9 |
| Total | 110 | 42 | 35 |
E. coli=Escherichia coli
Biochemical identification of E. coli and Salmonella isolates
Depending on the results of API 20E identification system, the suspected E. coli isolates were 8 out of 11 apparently healthy samples, 14 out of 17 diseased samples, and 11 out of 14 freshly dead samples representing recovery rates of 73%, 82%, and 79%, respectively (Table-3), where 6 suspected Salmonella isolates were recovered from 9 of apparently healthy samples, 13 isolates out of 17 diseased samples, and 7 isolates out of 9 freshly dead samples representing recovery rates of 67%, 76%, and 78%, respectively (Table-4).
Table-3.
Biochemical characteristics of the suspected E. coli isolates using API20E system.
| Type of samples | Number of samples | API 20E results | Number of recovered isolates | Recovery rate (%) | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||||||||||||||||||
| ONPG | ADH | LDC | ODC | CIT | H2S | URE | TDA | IND | VP | GEL | GLU | MAN | INO | SOR | RHA | SAC | MEL | AMY | ARA | OX | |||||
| Apparently healthy | 11 | + | - | + | + | - | - | - | - | + | - | - | + | + | - | + | + | + | + | - | + | - | 4 | 8 | 73 |
| + | - | + | - | - | - | - | - | + | - | + | + | - | + | + | - | + | - | + | - | 2 | |||||
| + | + | + | + | - | - | - | - | + | - | - | + | + | - | + | + | + | + | - | + | - | 2 | ||||
| Diseased | 17 | + | - | + | + | - | - | - | - | + | - | - | + | + | - | + | + | + | + | - | + | - | 5 | 14 | 82 |
| + | - | + | - | - | - | - | - | + | - | - | + | + | - | + | + | - | + | - | + | - | 3 | ||||
| + | + | + | + | - | - | - | - | + | - | - | + | + | - | + | + | + | + | - | + | - | 4 | ||||
| + | - | - | - | - | - | - | - | + | - | - | + | + | - | + | + | + | + | - | + | - | 2 | ||||
| Freshly dead | 14 | + | - | + | + | - | - | - | - | + | - | - | + | + | - | + | + | + | + | - | + | - | 3 | 11 | 79 |
| + | - | + | - | - | - | - | - | + | - | - | + | + | - | + | + | - | + | - | + | - | 2 | ||||
| + | + | + | + | - | - | - | - | + | - | - | + | + | - | + | + | + | + | - | + | - | 3 | ||||
| + | - | - | - | - | - | - | - | + | - | - | + | + | - | + | + | + | + | - | + | - | 3 | ||||
| Total | 42 | 33 | |||||||||||||||||||||||
E. coli=Escherichia coli, ONPG=Ortho nitro phenyl-βD-galactopyranosidase, ADH=Arginine dihydrolase, LDC=Lysine decarboxylase, ODC=Ornithine decarboxylase, CIT=Citrate utilization, H2S=Hydrogen sulfide, URE=Urease, TDA=Tryptophan deaminase, IND=Indole, VP=Voges Proskauer, GEL=Gelatinase, GLU=Glucose (fermentation/oxidation), MAN=Mannitol (fermentation/oxidation), INO=Inositol (fermentation/oxidation), SOR=Sorbitol (fermentation/oxidation), RHA=Rhamnose (fermentation/oxidation), SAC=Saccharose (fermentation/oxidation), MEL=Melibiose (fermentation/oxidation), AMY=Amygdalin (fermentation/oxidation), ARA=Arabinose (fermentation/oxidation), OX=Oxidase
Table-4.
Biochemical characteristics of the suspected Salmonella isolates using API20E system.
| Type of samples | Number of samples | API results | Recovered number of isolates | Recovery rate (%) | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||||||||||||||||||
| ONPG | ADH | LDC | ODC | CIT | H2S | URE | TDA | IND | VP | GEL | GLU | MAN | INO | SOR | RHA | SAC | MEL | AMY | ARA | OX | |||||
| Apparently healthy | 9 | - | - | + | + | + | + | - | - | - | - | - | + | + | + | + | + | - | + | - | + | - | 2 | 6 | 67 |
| - | + | + | + | + | + | - | - | - | - | - | + | + | - | + | + | - | + | - | + | - | 4 | ||||
| Diseased | 17 | - | - | + | + | + | + | - | - | - | - | - | + | + | + | + | + | - | + | - | + | - | 4 | 13 | 76 |
| - | + | + | + | + | + | - | - | - | - | - | + | + | - | + | + | - | + | - | + | - | 3 | ||||
| - | + | + | + | + | + | - | - | - | - | - | + | + | + | + | + | - | + | - | + | - | 6 | ||||
| Freshly dead | 9 | - | - | + | + | + | + | - | - | - | - | - | + | + | + | + | + | - | + | - | + | - | 2 | 7 | 78 |
| - | + | + | + | + | + | - | - | - | - | - | + | + | - | + | + | - | + | - | + | - | 2 | ||||
| - | + | + | + | + | + | - | - | - | - | - | + | + | + | + | + | - | + | - | + | - | 3 | ||||
| Total | 35 | 26 | |||||||||||||||||||||||
ONPG=Ortho nitro phenyl-βD-galactopyranosidase, ADH=Arginine dihydrolase, LDC=Lysine decarboxylase, ODC=Ornithine decarboxylase, CIT=Citrate utilization, H2S=Hydrogen sulfide, URE=Urease, TDA=Tryptophane deaminase, IND=Indole, VP=Vagous Proskauer, GEL=Gelatinase, GLU=Glucose (fermentation/oxidation), MAN=Mannitol (fermentation/oxidation), INO=Inositol (fermentation/oxidation), SOR=Sorbitol (fermentation/oxidation), RHA=Rhamnose (fermentation/oxidation), SAC=Saccharose (fermentation/oxidation), MEL=Melibiose (fermentation/oxidation), AMY=Amygdalin (fermentation/oxidation), ARA=Arabinose (fermentation/oxidation), OX=Oxidase
Serological identification of E. coli and Salmonella isolates
Tables-5 and 6 summarized serotyping of E. coli and Salmonella isolates using polyvalent and monovalent antisera. Most of E. coli strains were belonging to serotype O1 and O78 were the most predominant serotype of Salmonella strains was SP.
Table-5.
Serogrouping of the suspected E. coli isolates.
| Source | Apparently healthy | Diseased | Freshly dead | Total | Recovery rates (%) |
|---|---|---|---|---|---|
| Number of isolates | 8 | 14 | 11 | 33 | |
| Polyvalent antisera | |||||
| 1 | 4 | 2 | 3 | 9 | |
| 2 | 0 | 2 | 1 | 3 | |
| 3 | 4 | 7 | 5 | 16 | |
| 4 | 0 | 2 | 3 | 5 | |
| Monovalent antisera | |||||
| O1 | 4 | 3 | 2 | 9 | 27.3 |
| O2 | 0 | 2 | 1 | 3 | 9.1 |
| O6 | 2 | 3 | 2 | 7 | 21.2 |
| O78 | 2 | 4 | 3 | 9 | 27.3 |
| O126 | 0 | 2 | 3 | 5 | 15.1 |
E. coli=Escherichia coli
Table-6.
Serotyping of the suspected Salmonella isolates.
| Source | Apparently healthy | Diseased | Freshly dead | Total | Recovery rates (%) |
|---|---|---|---|---|---|
| Number of isolates | 6 | 13 | 7 | 26 | |
| SP | 2 | 4 | 2 | 8 | 30.8 |
| SM | 1 | 1 | 0 | 2 | 7.7 |
| SE | 3 | 2 | 2 | 7 | 26.9 |
| SG | 0 | 2 | 1 | 3 | 11.5 |
| ST | 0 | 4 | 2 | 6 | 23.1 |
SP=Salmonella Pullorum, SM=Salmonella Montevideo, SE=Salmonella Enteritidis, SG=Salmonella Gallinarum, ST=Salmonella Typhimurium
It was surprising that the identified E. coli samples of the same source showed variations in their biochemical reactions, this may be due to difference in serotypes of these identified samples. Kwon et al. [30] identified E. coli isolates by screening biochemical traits using API 20E identification system. Regarding serodifferentiation, chicken may harbor many different serotypes in their gastrointestinal tract, in this study, only a restricted number of serotypes O1, O2, O6, O78, and O126 have been recovered. These results were confirmed by Salama et al. [31] who recovered 5 different E. coli serotypes identified as O1, O2, O6, O78, and O126. Pathogenic E. coli isolates for poultry commonly belong to certain serogroups, particularly the serogroups O78, O1, and O2, and sometimes O15 [32,33]. The relation between biochemical and serological identification of E. coli confirmed that the variation of reactions in between the same source of samples was related to the difference in serotypes and also revealed the similarity between serotypes O1 and O2 in their biochemical reactions [34]. Similar serotypes (O1, O2, and O78) were obtained by Chart et al. [33], McPeake et al. [35]. In addition, Peighambari et al. [36], Lafont et al. [37], Dho-Moulin et al. [38], and Gross [39] recorded that the most common serogroups of E. coli from avian diseases were O78, O2, and O1 which were associated with septicemic E. coli infection in poultry. Furthermore, Cloud et al. [40] and Orajaka and Mohan [41] recorded a high incidence of serovars O1, O2, and O78 in case of colibacillosis. Furthermore, Hossain et al. [42] recorded that out of 110 bird samples, 66 samples were found to be positive for E. coli meanwhile Robab and Azadeh [43], isolated 50 E. coli strains from bile and liver of poultry. All the isolated and identified bacteria possess the morphological, biochemical and serological characteristics of E. coli and the O1 and O78 serotypes are the most predominated. On the other hand, Raji et al. [44] isolated E. coli from hatcheries and the most common serovares were O8, O9 and O78 among poultry cases. Kilic et al. [45] isolated E. coli from 110 samples collected from colibacillosis suspicious hens at different poultry farms in a recovery rate of 48%. Serogroup O1 is known pathogen in poultry and usually isolated from birds with colibacillosis [46]. Rosenberger et al. [47] reported that O2 serovars of avian origin are among virulent avian E. coli in colibacillosis. The isolation of O6 serotype which usually cause septicemic diarrhea in newborn and enteritis in domestic animals is evidence that the water sources of the farms were probably contaminated with sewage and/or the farms laborers did not observe sanitary measures [48].
For Salmonella isolation and identification, Moustafa [49] reported that the predilection seats for isolation of Salmonella were the genital organs, spleen, gallbladder, and liver while intestinal contents or feces were not reliable for Salmonella isolation. Furthermore, Bygrave and Gallagher [50] isolated Salmonella Enteritidis (SE) from pooled samples of liver, lungs, testes, cecum, and intestine. Zahraei et al., [17] isolated 30 Salmonella species from intestine and liver of chicken in poultry farms using SS agar and xylose-lysine deoxycholate agar after enriching on selenite-f broth. Further, serological identification of the suspected colonies was applied using the polyvalent and monovalent antisera. The results revealed that five serotypes of Salmonella were isolated represented by SP, Salmonella Typhimurium (ST), SE, Salmonella Gallinarum, and Salmonella Montevideo (SM). These results were confirmed by Chaiba et al. [51] who used poultry samples and identified four different Salmonella serotypes which are ST, Salmonella Newport, SM, and Salmonella Heidelberg using polyvalent O and H antisera.
MADI-TOF MS identification of E. coli and Salmonella isolates
Using MADI-TOF MS, all microscopical, morphological, biochemical and serological identified E. coli, and Salmonella isolates were tested. MADI-TOF MS identified all clinical bacterial samples as E. coli and Salmonella except two E. coli isolates recovered from apparently healthy and diseased birds, respectively, with recovery rate of 93.9% and 2 Salmonella isolates recovered from apparently healthy and dead birds, respectively, with recovery rate of 92.3%. 3 out of these 4 isolates were had un-valid score (red color) where the 4th sample which isolated from apparently healthy bird and bacteriologically identified as E. coli were identified with a valid score as Pseudomonas fragi using MALDI-TOF MS (Table-7). For more accuracy of the results, the samples being processed and spotted in duplicates and consequences the reproducibility of MALDI-TOF MS apparatus was evaluated and found to be consistent for all bacterial clinical samples [52,53]. Preparatory extraction is superior to direct colony method for the bacterial identification by MALDI-TOF MS using the Bruker system also using the extraction method increased identification to the species level [28,54].
Table-7.
Identification of E. coli and Salmonella field isolates using MALDI-TOF.
| Analyte ID | Organism (best matched) | Matched pattern | Score value | NCBI identifier |
|---|---|---|---|---|
| EA1 | E. coli | E. coli DH5alpha BRL+E. cloacae MB_8779_05 THL | 2.362 | 562 |
| EA2 | E. coli | E. coli DH5alpha BRL+E. kobei DSM 13645T DSM | 2.493 | 562 |
| EA3 | E. coli | E. coli W3350 MMG+E. fergusonii DSM 13698T HAM | 2.1 | 562 |
| EA4 | E. coli | E. coli W3350 MMG+K. cowanii DSM 18146T DSM | 2.448 | 562 |
| EA5 | P. fragi | P. fragi DSM 3456T HAM+P. jessenii CIP 105274T HAM | 2.325 | 296 |
| EA6 | E. coli | E. coli ATCC 25922 THL+C. koseri DSM 4570 DSM | 2.57 | 562 |
| EA7 | E. coli | E. coli W3350 MMG+C. farmeri CCUG 29877 CCUG | 2.36 | 562 |
| EA8 | E. coli | E. coli ATCC 25922 THL+C. koseri DSM 4570 DSM | 2.66 | 562 |
| EDS1 | E. coli | E. coli DH5alpha BRL+E. hormaechei ssp hormaechei DSM 12409T DSM | 2.573 | 562 |
| EDS2 | E. coli | E. coli W3350 MMG+E. fergusonii DSM 13698T HAM | 2.494 | 562 |
| EDS3 | E. coli | E. coli DH5alpha BRL+E. cloacae MB_8779_05 THL | 2.095 | 562 |
| EDS4 | Not reliable identification | E. coli ATCC 25922 CHB | 1.585 | 562 |
| EDS5 | E. coli | E. coli W3350 MMG+K. cowanii DSM 18146T DSM | 2.345 | 562 |
| EDS6 | E. coli | E. coli ATCC 25922 THL+C. koseri DSM 4570 DSM | 2.675 | 562 |
| EDS7 | E. coli | E. coli DH5alpha BRL+E. cloacae MB_8779_05 THL | 2.278 | 562 |
| EDS8 | E. coli | E. coli DH5alpha BRL+E. kobei DSM 13645T DSM | 2.354 | 562 |
| EDS9 | E. coli | E. coli W3350 MMG+K. cowanii DSM 18146T DSM | 2.476 | 562 |
| EDS10 | E. coli | E. coli ATCC 25922 THL+C. koseri DSM 4570 DSM | 2.133 | 562 |
| EDS11 | E. coli | E. coli DH5alpha BRL+E. cloacae MB_8779_05 THL | 2.464 | 562 |
| EDS12 | E. coli | E. coli DH5alpha BRL+E. cloacae MB_8779_05 THL | 2.565 | 562 |
| EDS13 | E. coli | E. coli DH5alpha BRL+E. kobei DSM 13645T DSM | 2.467 | 562 |
| EDS14 | E. coli | E. coli ATCC 25922 THL+C. koseri DSM 4570 DSM | 2.423 | 562 |
| EDE1 | E. coli | E. coli DH5alpha BRL+E. kobei DSM 13645T DSM | 2.575 | 562 |
| EDE2 | E. coli | E. coli W3350 MMG+K. cowanii DSM 18146T DSM | 2.257 | 562 |
| EDE3 | E. coli | E. coli ATCC 25922 THL+C. koseri DSM 4570 DSM | 2.165 | 562 |
| EDE4 | E. coli | E. coli ATCC 25922 THL+C. koseri DSM 4570 DSM | 2.298 | 562 |
| EDE5 | E. coli | E. coli DH5alpha BRL+E. kobei DSM 13645T DSM | 2.376 | 562 |
| EDE6 | E. coli | E. coli ATCC 25922 THL+C. koseri DSM 4570 DSM | 2.256 | 562 |
| EDE7 | E. coli | E. coli W3350 MMG+E. fergusonii DSM 13698T HAM | 2.237 | 562 |
| EDE8 | E. coli | E. coli W3350 MMG+E. fergusonii DSM 13698T HAM | 2.237 | 562 |
| EDE9 | E. coli | E. coli DH5alpha BRL+E. kobei DSM 13645T DSM | 2.312 | 562 |
| EDE10 | E. coli | E. coli ATCC 25922 THL+C. koseri DSM 4570 DSM | 2.296 | 562 |
| EDE11 | E. coli | E. coli ATCC 25922 THL+C. koseri DSM 4570 DSM | 2.276 | 562 |
| SA1 | Not reliable identification | Salmonella sp. (choleraesuis) 08 LAL | 1.328 | 591 |
| SA2 | Salmonella | Salmonella sp. (enterica st Dublin) Sa05_188 VAB | 2.134 | 98,360 |
| SA3 | Salmonella | Salmonella sp. (enterica st Enterica) DSM 17058T HAM+E. coli MB11464_1 CHB | 2.328 | 59,201 |
| SA4 | Salmonella | Salmonella sp. (enterica st Hadar) Sa05_506 VAB+E. coli W3350 MMG | 2.425 | 149,385 |
| SA5 | Salmonella | Salmonella sp. (enterica st Enterica) DSM 17058T HAM+E. hormaechei ssp hormaechei DSM 12409T DSM | 2.294 | 59,201 |
| SA6 | Salmonella | Salmonella sp. (choleraesuis) 08 LAL+E. coli ATCC 25922 CHB | 2.118 | 591 |
| SDS1 | Salmonella | Salmonella sp. (enterica st Gallinarum) FLR+C. sakazakii DSM 4485T DSM | 2.051 | 594 |
| SDS2 | Salmonella | Salmonella sp. (enterica st Enterica) DSM 17058T HAM+K. pneumoniae ssp pneumoniae 9295_1 CHB | 2.366 | 59,201 |
| SDS3 | Salmonella | Salmonella sp. (enterica st Gallinarum) FLR+K. pneumoniae ssp pneumoniae 9295_1 CHB | 2.361 | 594 |
| SDS4 | Salmonella | Salmonella sp. (enterica st Anatum) 11 LAL+C. koseri 9553_1 CHB | 2.386 | 58,712 |
| SDS5 | Salmonella | Salmonella sp. (enterica st Hadar) Sa05_506 VAB+E. coli ATCC 25922 THL | 2.346 | 149,385 |
| SDS6 | Salmonella | Salmonella sp. (enterica st Enterica) DSM 17058T HAM+K. cowanii DSM 18146T DSM | 2.413 | 59,201 |
| SDS7 | Salmonella | Salmonella sp. (enterica st Enterica) DSM 17058T HAM+K. cowanii DSM 18146T DSM | 2.333 | 59,201 |
| SDS8 | Salmonella | Salmonella sp. (enterica st Hadar) Sa05_506 VAB+E. coli ATCC 25922 THL | 2.268 | 149,385 |
| SDS9 | Salmonella | Salmonella sp. (enterica st Anatum) 11 LAL+C. koseri 9553_1 CHB | 2.236 | 58,712 |
| SDS10 | Salmonella | Salmonella sp. (enterica st Hadar) Sa05_506 VAB+E. coli W3350 MMG | 2.578 | 149,385 |
| SDS11 | Salmonella | Salmonella sp. (enterica st Gallinarum) FLR+C. sakazakii DSM 4485T DSM | 2.378 | 594 |
| SDS12 | Salmonella | Salmonella sp. (enterica st Enterica) DSM 17058T HAM+E. hormaechei ssp hormaechei DSM 12409T DSM | 2.319 | 59201 |
| SDS13 | Salmonella | Salmonella sp. (enterica st Enterica) DSM 17058T HAM+K. pneumoniae ssp pneumoniae 9295_1 CHB | 2.372 | 59201 |
| SDE1 | Salmonella | Salmonella sp. (enterica st Enterica) DSM 17058T HAM+E. hormaechei ssp hormaechei DSM 12409T DSM | 2.333 | 59201 |
| SDE2 | Salmonella | Salmonella sp. (enterica st Dublin) Sa05_188 VAB | 2.224 | 98,360 |
| SDE3 | Not reliable identification | Salmonella sp. (choleraesuis) 08 LAL | 1.211 | 591 |
| SDE4 | Salmonella | Salmonella sp, (enterica st Anatum) 11 LAL+C. koseri 9553_1 CHB | 2.328 | 58,712 |
| SDE5 | Salmonella | Salmonella sp. (enterica st Enterica) DSM 17058T HAM+E. coli MB11464_1 CHB | 2.239 | 59,201 |
| SDE6 | Salmonella | Salmonella sp. (enterica st Dublin) Sa05_188 VAB | 2.334 | 98,360 |
| SDE7 | Salmonella | Salmonella sp. (enterica st Hadar) Sa05_506 VAB+E. coli W3350 MMG | 2.106 | 149,385 |
EA=E. coli isolate recovered from apparently healthy birds, EDS=E. coli isolate recovered from diseased birds, EDE=E. coli isolate recovered from dead birds, SA=Salmonella isolate recovered from apparently healthy birds, SDS=Salmonella isolate recovered from diseased birds, SDE=Salmonella isolate recovered from dead birds, E. cloacae=Enterobacter cloacae, E. kobei=Enterobacter kobei, E. fergusonii=Escherichia fergusonii, K. cowanii=Kosakonia cowanii, P. fragi=Pseudomonas fragi, P. jessenii=Pseudomonas jessenii, C. koseri=Citrobacter koseri, C. farmeri=Citrobacter farmeri, E. hormaechei=Enterobacter hormaechei, E. cloacae=Enterobacter cloacae, C. sakazakii=Cronobacter sakazakii, K. pneumoniae=Klebsiella pneumoniae, E. coli=Escherichia coli
Valid identification scores as explained by Bruker Daltonik MALDI Biotyper is 2.0 or more were enough for a reliable identification to the species level (green color) which mean highly probable species identification (2.300-3) or secure genus identification, probable species identification (2-2.299) where score 1.700-1999 and 0.000-1.699 means probable genus identification (yellow color) and not reliable identification (red color), respectively [55,56]. By examination of E. coli and Salmonella isolates and strains revealed from apparently healthy, diseased and dead chickens by MALDI-TOF MS, 10-20 prominent ion peaks were identified in the mass spectra. Range of these prominent ion peaks were from the 3000 and 10,500 m/z, with the highest-intensity peaks being in the range of 4375-9625 m/z with E. coli isolates while in the case of Salmonella isolates, range of these spectra peaks were from the 3000 and 11,000 m/z, with the highest-intensity spectra peaks being in the range of 4350-9500 m/z. On this basis, the score values achieved by MALDI-TOF MS correctly identified all E. coli and Salmonella isolates at the species level (score ≥2.0). Inspection of mass spectra reveals strain-specific peaks at 4375, 5375, 6650, 7190, and 9625 m/z for all E. coli isolates which agree with Christner et al. [57] and also reveals strain-specific peaks at 4350, 5300, 5600, 6090, 6200, 6300, 7200, 7750, 8500, and 9500 m/z for all Salmonella isolates which agree to large extent with Dieckmann and Malorny [58] and Leuschner et al. [59], respectively (Figures-1 and 2).
Figure-1.
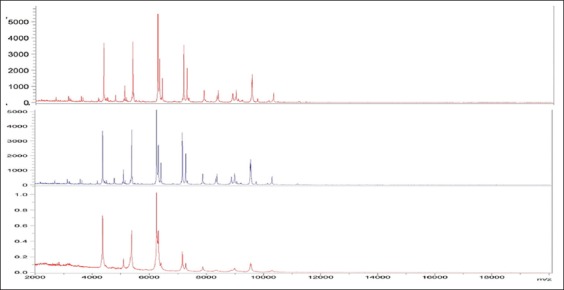
Overview of the matrix-assisted laser desorption-ionization-time-of-flight mass spectra of 3 Escherichia coli field isolates.
Figure-2.
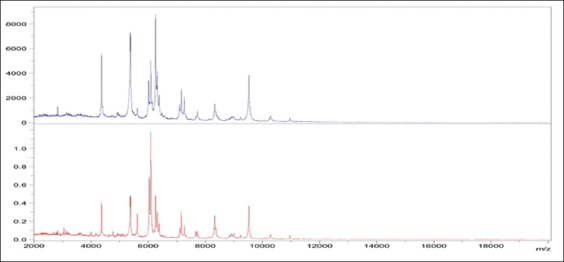
Overview of the matrix-assisted laser desorption-ionization-time-of-flight mass spectra of 3 Salmonella Gallinarum field isolates.
In our study, MALDI-TOF MS gave a valid score for genus and species identification of 93.94% when used in identification of previously identified E. coli culture using ABI system and conventional methods this agrees with Ge et al. [60], Jesumirhewe et al. [61], and Naiara et al. [62] which achieved species identification of E. coli isolates using MALDI-TOF MS of 94.7%, 80%, and 83%, respectively, when compared with traditional methods of identification. All this studies not identified E. coli to sub species level. On the other hand, Huixia et al. [63] was developed a rapid method to identify E. coli at subspecies level (identifying flagellar (H) antigen) using a MALDI-TOFMS platform with high sensitivity and specificity which could identify 100% of reference strains containing H types (53 strains) and could detect 75 out of 85 clinical isolates representing matched results obtained from traditional serotyping.
Furthermore, pure colonies previously identified as Salmonella isolates using ABI system and traditional methods gave valid score of 91.66% using MALDI-TOF MS assay. This results agrees with Ulrich et al. [64] which reported that no positive sample was missed by this novel approach which allowed detection of pure Salmonella culture after just 1 day of incubation and also agrees with Rebecca et al. [65] which found that MALDI-TOF MS could identified 98% of Salmonella clinical samples that previously identified by traditional methods. Public Health England [66], Clark et al. [67] and Kuhns et al. [68] reported that MALDI-TOF MS has been used to help in both detection and species-level identification of Salmonella and also has been utilized in discriminating Salmonella Enterica serovar Typhi from other Salmonella serovars (subspecies level).
Results revealed that there is no satisfactory differences were observed in and sensitivity (positive cases/total number of suspected cases × 100) of 20A and MALDI-TOF MS when compared with direct isolation of causative agents as sensitivity in case of E. coli were 78.57% and 73.8%, respectively, wherein case of Salmonella 74.29% and 68.57%, respectively, where sensitivity of MALDI-TOF MS in compression of API 20A was 93.93% and 92.3% in case of E. coli and Salmonella isolates, respectively. With respect of direct isolation of causative agents as a gold standard test, relative sensitivity, and specificity were 100% and 88.31% with API 20A and 100% and 86.08% with MALDI-TOF, respectively, in case of E. coli isolates where in case of Salmonella isolates, relative sensitivity, and specificity of API 20A were 100% and 89.29% and of MALDI-TOF MS were 100% and 87.21%, respectively. With respect of API 20A, relative sensitivity, and specificity of MALDI-TOF MS were 100% and 81.82%, respectively, in the case of E. coli and Salmonella isolates.
MALDI-TOF MS showed significant promise in E. coli and Salmonella identification on genus and species levels and can be also used as a tool for sub species and serovar typing, but it will require additional studies and modifications to existing protocols and commercial and the extended database. The identification using MALDI-TOF MS method could analyze pure positive culture rapidly (may be within minutes especially when direct cultural identification methods used rather than ethanol: Formic acid extraction method) and also reliable manner. However, identification by traditional methods needs more facilities, media, chemicals, experiences, and time and this in contrast with the non-requirement of high technical expertise, the simple extraction procedure and low running cost identification using MALDI-TOF MS which provide more advantages over other methods for identification. However, the applications have to be carried out with cautions because the accuracy decreases using of too much of chemicals and materials and the samples have to be spotted with the matrix solution with care to avoid the presence of the liquid smear between spots, which increase possibility of cross-contamination [69,70]. The sample size used for this study is low as it is a preliminary study to use this technique in diagnostic laboratories in Egypt, but anyhow, more samples are needed in future studies to detect sensitivity, reliability, and performance of this type of bacterial identification.
Conclusion
This study demonstrated that Bruker MALDI-TOF MS Biotyper is a reliable fast and economic tool for the identification of Gram-negative bacteria, especially E. coli and Salmonella which could be used as alternative regular diagnostic tool for routine identification and differentiation of clinical isolates in the bacteriological laboratory to provide more precise identification on clinical specimens. MALDI-TOF MS need more validation and verification and more study on the performance of direct colony and extraction methods to detect the most sensitive one and also need using more samples to detect sensitivity, reliability, and performance of this type of bacterial identification.
Authors’ Contributions
All authors designed and planned this research work. Isolation of causative agents from field and preparation of samples for MALDI-TOF analysis were done by WSS, MLS and AAS. Biochemical and serological identification were done by FMGA, FEMG and AAK. All authors contributed equally in preparation and revision of the manuscript and collection of scientific papers related to the subject of this research. All authors read and approved the final manuscript.
Acknowledgments
Central Laboratory for Evaluation of Veterinary Biologics, Egypt funded all materials used in this study. Appreciation is expressed to Microbiology Department, Medicine Faculty, Alexandria University, and all the technical staff of Microbiology Departments of CLEVB and AHRI, for their contribution in the practical part of this study.
Competing Interests
The authors declare that they have no competing interests.
References
- 1.Jordan F.T, Williams N.J, Wattret A, Jones T. Observations on salpingitis, peritonitis and salpingoperitonitis in a layer breeder flock. Vet. Rec. 2005;157:573–577. doi: 10.1136/vr.157.19.573. [DOI] [PubMed] [Google Scholar]
- 2.Sun H, Liu P, Nolan L.K, Lamont S.J. Thymus transcriptome reveals novel pathways in response to avian pathogenic Escherichia coli infection. Poult. Sci. 2016;95(12):2803–2814. doi: 10.3382/ps/pew202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.da Silveira W.D, Ferreira A, Brocchi M, de Hollanda L.M, de Castro A.P, Yamada A.T, Lancellotti M. Biological characteristics and pathogenicity of avian Escherichia coli strains. Vet. Microbiol. 2002;85:4753. doi: 10.1016/s0378-1135(01)00482-5. [DOI] [PubMed] [Google Scholar]
- 4.Glisson J.R, Hofacre C.L, Christensen J.P. Fowl cholera. In: Saif Y.M, Barnes H.J, Glisson J.R, Fadly A.M, McDougald L.R, Swayne D.E, editors. Diseases of Poultry. 12th ed. Ames, IA: Blackwell Publishing; 2008. pp. 739–758. [Google Scholar]
- 5.Teferi M, Nejash A. Epidemiology and economic importance of pullorum disease in poultry: A review. Glob. Vet. 2016;17(3):228–237. [Google Scholar]
- 6.Endris M, Taddesse F, Geloye M, Degefa T, Jibat T. Sero and media culture prevalence of salmonellosis in local and exotic chicken, Debre Zeit, Ethiopia. Afr. J. Microbiol. Res. 2013;7(12):1041–1044. [Google Scholar]
- 7.Bidhendi M, Khaki P, Cheraghchi N. Study on phenotypic characteristics of Salmonella gallinarum and Sallmonella pullorum isolates based on biochemical and antimicrobial susceptibility tests in Iran. Arch. Razi Inst. 2015;70:171–177. [Google Scholar]
- 8.Singhal N, Kumar M.P.K, Virdi J.S. MALDI-TOF mass spectrometry: An emerging technology for microbial identification and diagnosis. Front. Microbiol. 2015;6:791. doi: 10.3389/fmicb.2015.00791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schulthess B, Brodner K, Bloemberg G.V, Zbinden R, Bottger E.C, Hombach M. Identification of gram-positive cocci using MALDI-TOF MS: Comparison of different preparation methods and implementation of a practical algorithm for routine diagnostics. J. Clin. Microbiol. 2013;51:1834–1840. doi: 10.1128/JCM.02654-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Panda A, Kurapati S, Samantaray J.C, Myneedu V.P, Verma A, Srinivasan A. Rapid identification of clinical mycobacterial isolates by protein profiling using matrix assisted laser desorption ionization-time of flight mass spectrometry. Indian J. Med. Microbiol. 2013;31:117–122. doi: 10.4103/0255-0857.115217. [DOI] [PubMed] [Google Scholar]
- 11.Jennifer M, Sébastien R, Valérie M, Victoria G, Sandrine A, Martin W, David O, Jean-Philippe L, Anne K. A simple and safe protocol for preparing Brucella samples for matrix-assisted laser desorption ionization-time of flight mass spectrometry analysis. J. Clin. Microbiol. 2016;54(2):449–452. doi: 10.1128/JCM.02730-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blattel V, Petri A, Rabenstein A, Kuever J, Konig H. Differentiation of species of the genus Saccharomyces using biomolecular fingerprinting methods. Appl. Microbiol. Biotechnol. 2013;97:4597–4606. doi: 10.1007/s00253-013-4823-z. [DOI] [PubMed] [Google Scholar]
- 13.Lau A.F, Drake S.K, Calhoun L.B, Henderson C.M, Zelazny A.M. Development of a clinically comprehensive database and a simple procedure for identification of molds from solid media by matrix assisted laser desorption ionization-time of flight mass spectrometry. J. Clin. Microbiol. 2013;51:828–834. doi: 10.1128/JCM.02852-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Degand N, Carbonnelle E, Dauphin B, Beretti J.L, Le Bourgeois M, Sermet-Gaudelus I. Matrix-assisted laser desorption ionization-time of flight mass spectrometry for identification of nonfermenting gram-negative bacilli isolated from cystic fibrosis patients. J. Clin. Microbiol. 2008;46:3361–3367. doi: 10.1128/JCM.00569-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ozbey G, Ertas H.B. Salmonella spp. Isolation from chicken samples and identification by polymerase chain reaction. Bulg. J. Vet. Med. 2005;9(1):67–73. [Google Scholar]
- 16.Hossain M.S, Chowdhury E.H, Islam M.M, Haider M.G, Hossain M.M. Avian Salmonella infection: Isolation and identification of organisms and histopathological study. Bangladesh J. Vet. Med. 2006;4(1):7–12. [Google Scholar]
- 17.Zahraei S.T, Mahzounieh M, Saeedzadeh A. The isolation of antibiotic-resistant Salmonella from intestine and liver of poultry in Shiraz Province of Iran. Int. J. Poult. Sci. 2005;4(5):320–322. [Google Scholar]
- 18.Merchant I.A, Packer R.A. Veterinary Bacteriology and Virology. 7th ed. Ames, Iowa, USA: The Iowa University Press; 1967. pp. 286–306. [Google Scholar]
- 19.Abdel H.M.A. Isolation, Identification and Characterization of Salmonella from Laying Farms. M. V. Sc. Thesis (Microbiology) Faculty of Veterinary Medicine, Cairo University. 2007 [Google Scholar]
- 20.Mohamed Z.A. Identification and Classification of Salmonella Strains by the Use of Protein Profile Analysis, Antimicrobial Susceptibility and DNA Fingerprinting. Ph.D. Thesis (Microbiology) Veterinary Medicine, Cairo University; 1999. [Google Scholar]
- 21.Kauffmann F. Serological Diagnosis of Salmonella Species. Denmark: Kauffman White Sceme Minkagaard Copenhagen; 1972. [Google Scholar]
- 22.Sojka W.J. 1st ed. Royal Buck, England: Commonwealth Agriculture, Bureau, Farnham; 1965. E. coli in Domestic Animals and Poultry. [Google Scholar]
- 23.Edward P.R, Ewing W.H. Edwards and Ewing's Identification of Enterobacteriacae. 3rd ed. Minneapolis: Burgess; 1972. [Google Scholar]
- 24.Sara J.B, Steven K.D, Andrasko J.L, Christina M.H, Kamal K, Stella A, Lilia M, Patricia C, Karen M.F, Susan M.H, Joan-Miquel B, Adrian M.Z. Multi-center MALDI-TOF MS study for the identification of clinically-relevant Nocardia spp. J. Clin. Microbiol. 2016;54:1251–1258. doi: 10.1128/JCM.02942-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Adnan A.A, Scott A.C, Sherry M.I, Jayawant M, Robin P. Comparison of direct colony method versus extraction method for identification of gram-positive cocci by use of Bruker Biotyper matrix-assisted laser desorption ionization-time of flight mass spectrometry. J. Clin. Microbiol. 2011;49(8):2868–2873. doi: 10.1128/JCM.00506-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tripti D, Reena V, Vijaylatha R. Prevalence, bacteriology, pathogenesis and isolation of E. coli in sick layer chickens in Ajmer region of Rajasthan, India. Int J. Curr. Microbiol. Appl. Sci. 2016;5(3):129–136. [Google Scholar]
- 27.Melha M. Human and Avian extraintestinal pathogenic Escherichia coli: Infections, zoonotic risks, and antibiotic resistance trends. Foodborne Pathog. Dis. 2013;10(11):916–932. doi: 10.1089/fpd.2013.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Noori T.E, Alwan M.J. Isolation and Identification of zoonotic bacteria from poultry meat. Int. J. Adv. Res. Biol. Sci. 2016;3(8):57–66. [Google Scholar]
- 29.Antunes P, Reu C, Sousa J.C, Peixe L, Pestana N. Incidence of Salmonella from poultry products and their susceptibility to antimicrobial agents. Int. J. Food Microbiol. 2003;82:97–103. doi: 10.1016/s0168-1605(02)00251-9. [DOI] [PubMed] [Google Scholar]
- 30.Kwon S.G, Cha S.Y, Choi E.J, Kim B, Song H.J, Jang H.K. Epidemiological prevalence of avian pathogenic E. coli differentiated by multiplex PCR from commercial chickens and hatchery in Korea. J. Bacteriol. Virol. 2008;38(4):179–188. [Google Scholar]
- 31.Salama S.S, Afaf A.K, Elham A.E, Taha M.M. Molecular strategies for the differentiation and identification of local E. coli isolated from chicken: I. Characterization of protein profile. BS Vet. Med. J. 2007;17(1):25–28. [Google Scholar]
- 32.Gross W.B. Diseases due to Escherichia coli in poultry. In: Gyles C.L, editor. Escherichia coli in Domestic Animals and Humans. Wallingford, United Kingdom: CAB International Library; 1994. pp. 237–260. [Google Scholar]
- 33.Chart H, Smith H.R, La Ragione R.M, Woodward M.J. An investigation into the pathogenic properties of Escherichia coli strains BLR, BL21, DH5α, and EQ1. J Appl. Microbiol. 2000;89:1048–1058. doi: 10.1046/j.1365-2672.2000.01211.x. [DOI] [PubMed] [Google Scholar]
- 34.Ibrahim I.S. Prevalence of E. coli in Slaughtered Broilers and Their Products. Ph.D. Thesis (Meat Hygiene), Faculty of Veterinary Medicine, Cairo University. 1997 [Google Scholar]
- 35.McPeake S.J.W, Smyth J.A, Ball H.J. Characterization of avian pathogenic E. coli(APEC) associated with colicepticemia compared to fecal isolates from healthy birds. Vet. Microbiol. 2005;110:245–253. doi: 10.1016/j.vetmic.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 36.Peighambari S.M, Vaillancourt J.P, Wilson R.A, Gyles C.L. Characteristics of E. coli isolates from avian cellulites. Avian Dis. 1995;39:116–124. [PubMed] [Google Scholar]
- 37.Lafont J.P, Dho H, D'Hauteville H.M, Bree A, Sansonetti P.J. Presence and expression of aerobactin genes in virulent avian strains of E. coli. Infect. Immun. 1987;55:192–197. doi: 10.1128/iai.55.1.193-197.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dho-Moulin M, Vandenboseh J.F, Girardeau J.P, Bree A, Barat T, Lafont J.P. Surface antigens from E. coli O2, and O78 strains. Infect. Immun. 1990;58:740–745. doi: 10.1128/iai.58.3.740-745.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gross W.B. Colibacillosis. Dis. Poult. 1991;9:138–144. [Google Scholar]
- 40.Cloud S.S, Rosenberger J.K, Fries P.A, Wilson R.A, Odor E.M. In vitro and in vivo characterization of avian E. coli serotypes, metabolic activity and antibiotic sensitivity. Avian Dis. 1985;29:1084–1093. [PubMed] [Google Scholar]
- 41.Orajaka L.J.E, Mohan K. E. coli serotypes isolated from dead-in-shell embryos from Nigeria. Bull. Anim. Health Prod. Afr. 1986;34:139–141. [Google Scholar]
- 42.Hossain M.T, Siddique M.P, Hossain F.M.A, Zinnah M.A, Hossain M.M, Alam M.K, Rahman M.T, Choudhury K.A. Isolation, identification, toxin profile and anti-biogram of E. coli isolated from broilers and layers in Mymensingh district of Bangladesh. Bangladesh J. Vet. Med. 2008;6(1):1–5. [Google Scholar]
- 43.Robab R.T, Azadeh N. Isolation, identification and antimicrobial resistance patterns of E. coli isolated from chicken flock. Iran. J. Pharmacol. Ther. 2003;2:39–42. [Google Scholar]
- 44.Raji M, Adekeye J, Kwaga J, Bale J, Henton M. Serovars and biochemical characterization of Escherichia coli isolated from colibacillosis cases and dead-in-shell embryos inpoultry in Zaria-Nigeria. Vet. Arh. 2007;77(6):495–505. [Google Scholar]
- 45.Kilic A, Muz A, Ertaşh B, Özbey G. Random amplified polymorphic DNA (RAPD) analysis of Escherichia coli isolated from chickens. Fırat Üniv. Sağlık Bilimleri Vet. Derg. 2009;23(1):1–4. [Google Scholar]
- 46.Allan B.J, van den Hurk J.V, Potter A.A. Characterization of E. coli isolated from cases of Avian colibacillosis. Can. J. Vet. Res. 1993;57(3):146–151. [PMC free article] [PubMed] [Google Scholar]
- 47.Rosenberger J.K, Fries P.A, Cloud S.S, Wilson R.A. In vitro and in vivo characterization of avian E. coli. Factors associated with pathogenicity. Avian Dis. 1985;29:1094–1107. [PubMed] [Google Scholar]
- 48.Blanco J.E, Blanco M, Mora A, Blanco J. Prevalence of bacterial resistence to quinolones and other antimicrobials among avian E. coli strains isolated from septicemic and healthy chickens in Spain. J. Clin. Microbiol. 1997b;35:2184–2185. doi: 10.1128/jcm.35.8.2184-2185.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moustafa F.M. Microbiological and Serological Studies on Avian Salmonellosis. Ph.D. Thesis (Microbiology) Veterinary Medicine, Cairo University; 1982. [Google Scholar]
- 50.Bygrave A.C, Gallagher J. Transmission of S. enteritidis in poultry. Vet. Rec. 1989;124(21):571–575. doi: 10.1136/vr.124.21.571-c. [DOI] [PubMed] [Google Scholar]
- 51.Chaiba A, Rhazi F.F, Chahlaoui A, Soulaymani B.R, Zerhouni M. Prevalence and anti-microbial susceptibility of Salmonella isolates from chicken carcass and giblets in Meknes, Morocco. Afr. J. Microbiol. Res. 2009;3(5):215–219. [Google Scholar]
- 52.Ashutosh P, Sravya K, Jyotish C.S, Alagiri S, Shehla K. MALDI-TOF mass spectrometry proteomic based identification of clinical bacterial isolates. Indian J. Med. Res. 2014;140:770–777. [PMC free article] [PubMed] [Google Scholar]
- 53.Mari L.D, Carey-Ann D.B. Diafiltration MALDI-TOF mass spectrometry method for culture-independent detection and identification of pathogens directly from urine specimens. Am. J. Clin. Pathol. 2014;141:204–212. doi: 10.1309/AJCPQYW3B6JLKILC. [DOI] [PubMed] [Google Scholar]
- 54.Seng P, Drancourt M, Gouriet F, La Scola B, Fournier P.E, Rolain J.M, Raoult D. Ongoing revolution in bacteriology: Routine identification of bacteria by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin. Infect. Dis. 2009;49:543–551. doi: 10.1086/600885. [DOI] [PubMed] [Google Scholar]
- 55.Belén R, María J.R, Mercedes M, Paula L.R, Marta R.C, Emilio B. Evaluation of matrix-assisted laser desorption ionization-time of flight mass spectrometry for identification of nontuberculous mycobacteria from clinical isolates. J. Clin. Microbiol. 2015;53(8):2737–2740. doi: 10.1128/JCM.01380-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Abdessalam C, Stephane E, Jose F, Didier S, Jacques S.S. Evaluation of matrix-assisted laser desorption ionization-time of flight mass spectrometry for rapid identification of beta-hemolytic streptococci. J. Clin. Microbiol. 2011;49(8):3004–3005. doi: 10.1128/JCM.00240-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Christner M, Trusch M, Rohde H, Kwiatkowski M, Schluter H, Wolters M, Aepfelbacher M, Hentschke M. Rapid MALDI-TOF mass spectrometry strain typing during a large outbreak of Shiga-toxigenic Escherichia coli. PLoS One. 2014;9(7):e101924. doi: 10.1371/journal.pone.0101924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dieckmann R, Malorny B. Rapid screening of epidemiologically important Salmonella enterica subsp Enterica serovars by whole-cell matrix-assisted laser desorption ionization-time of flight mass spectrometry. Appl. Environ. Microbiol. 2011;77(12):4136–4146. doi: 10.1128/AEM.02418-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Leuschner R.G.K, Beresford-Jones N, Robinson C. Difference and consensus of whole cell Salmonella enterica subsp Enterica serovars matrix-assisted laser desorption/ionization time-of-flight mass spectrometry spectra. Lett. Appl. Microbiol. 2004;38:24–31. doi: 10.1046/j.1472-765x.2003.01436.x. [DOI] [PubMed] [Google Scholar]
- 60.Ge M, Kuo A, Liu K, Wen Y, Chia J, Chang P, Lee M, Wu T, Chang S, Lu J. Routine identification of microorganisms by matrix-assisted laser desorption ionization time-of-flight mass spectrometry: Success rate, economic analysis, and clinical outcome. J. Microbiol. Immunol. Infect. 2016;XX:1–7. doi: 10.1016/j.jmii.2016.06.002. [DOI] [PubMed] [Google Scholar]
- 61.Jesumirhewe C, Ogunlowo P.O, Olley M, Springer B, Allerberger F, Ruppitsch W. Accuracy of conventional identification methods used for Enterobacteriaceae isolates in three Nigerian hospitals. Peer J. 2016;4:e2511. doi: 10.7717/peerj.2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Naiara M.B.R, Greiciane F.B, Gabrielli S.S, Larissa A.B.B, Beatriz M.M, Irene D.C, Miliane M.S.D, Shana D.D.C. The matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) identification versus biochemical tests: A study with enterobacteria from a dairy cattle environment. Braz. J. Microbiol. 2017;48:132–138. doi: 10.1016/j.bjm.2016.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Huixia C, Michael C, Drexler H, Patrick C, Stuart M, Alyssia R, Matthew W, Lorea A.M.P, Sam R, David J.M.H, Sadjia B, John W, Linda C, Garrett W, Bianli X, Mike D, Celine N, David K.J, Gehua W, Keding C. Rapid, sensitive, and specific Escherichia coli H antigen typing by matrix-assisted laser desorption ionization-time of flight-based peptide mass fingerprinting. J. Clin. Microbiol. 2015;53(8):2480–2485. doi: 10.1128/JCM.00593-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ulrich W, Katrin S, Christiane B, Leith F, Markus K. Rapid detection of Salmonella from clinical specimen by MALDI-TOF MS. Pathology. 2011;43(Supp1):S74. [Google Scholar]
- 65.Rebecca L.B, Karen G.J, Andrea R.O, Melinda A.M, Eric W.B. Recent and emerging innovations in Salmonella detection: A food and environmental perspective. Microb. Biotechnol. 2016;9(3):279–292. doi: 10.1111/1751-7915.12359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Public Health England. Identification of Salmonella Species. UK Standards for Microbiology Investigations. ID 24 Issue 3. 2015. [Accessed on 26-11-2016]. Available from: https://www.gov.uk/uk-standards-for-microbiology-investigations-smi-quality-and-consistency-in-clinical-laboratories .
- 67.Clark A.E, Kaleta E.J, Arora A, Wolk D.M. Matrix-assisted laser desorption ionization-time of flight mass spectrometry: A fundamental shift in the routine practice of clinical microbiology. Clin. Microbiol. Rev. 2013;26:547–603. doi: 10.1128/CMR.00072-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kuhns M, Zautner A.E, Rabsch W, Zimmermann O, Weig M, Bader O. Rapid discrimination of Salmonella enterica serovar Typhi from other serovars by MALDI-TOF mass spectrometry. PLoS One. 2012:e40004. doi: 10.1371/journal.pone.0040004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Markus K, Elisabeth N. How MALDI-TOF mass spectrometry can aid diagnosis of hard-to-identify pathogenic bacteria. Exp. Rev. Mol. Diagn. 2016;16(5):509–511. doi: 10.1586/14737159.2016.1157019. [DOI] [PubMed] [Google Scholar]
- 70.Andrew E.C, Erin J.K, Amit A, Donna M.W. Matrix-assisted laser desorption ionization-time of flight mass spectrometry: A fundamental shift in the routine practice of clinical microbiology. Clin. Microbiol. Rev. 2013;26(3):547–603. doi: 10.1128/CMR.00072-12. [DOI] [PMC free article] [PubMed] [Google Scholar]


