Abstract
Aim:
Wild birds are considered silent vectors of some zoonotic water and food borne pathogens of public health significance. Owing to the importance of Shiga toxin producing Escherichia coli (STEC) as the most pathogenic among the emerging diarrheagenic E. coli groups that can infect man; the present study was designed to detect the occurrence of STEC among wild birds in Egypt.
Materials and Methods:
A total of 177 intestinal content swab samples originating from five wild bird species were investigated for the presence of E. coli and STEC by standard culture methods. Suspect STEC isolates were further characterized by serotyping, random amplified polymorphic DNA polymerase chain reaction (RAPD PCR), antimicrobial resistance pattern and PCR detection of stx1, stx2, and eae genes.
Results:
A total of 30 suspect STEC isolates from 30 positive birds’ samples were detected and identified on STEC CHROMagar (semi-captive pigeons, 15; house crows, 8; cattle egrets, 3; moorhens, 2; and house teals, 2). 25 isolates were grouped into 13 serogroups (O:20, O:25, O:26, O:27, O:63, O:78, O:111, O:114, O:125, O:128, O:142, O:153, and O:158), while five were rough strains. The distribution of STEC virulence genes among wild birds was as follows: 16 birds carried stx1 gene only (nine pigeons [28.1%], six crows [7.1%], and one cattle egret [5.6%]). Stx1 and stx2 genes together were detected in four birds (one cattle egret [5.6%], two moorhens [6.1%], and one house teal, [10%]). Only one pigeon (3.1%) possessed the three alleles. Disk diffusion test results showed that cefixime was the most effective against STEC serotypes with (93.3%) sensitivity, followed by gentamycin (56.7%), and amoxicillin (50%). On the other hand, all the recovered STEC isolates were resistant to cefotaxime, doxycycline, cephalothin, and sulfisoxazole. RAPD fingerprinting using primers OPA-2 and OPA-9 showed that STEC isolates were heterogeneous; they yielded 30 and 27 different clusters, respectively.
Conclusions:
Wild birds carry STEC and may add to the contamination of the surrounding environment.
Keywords: antibiotic, eae, random-amplified polymorphic DNA polymerase chain reaction, Shiga toxin producing Escherichia coli, stx1, stx2, wild birds
Introduction
Diarrheagenic Escherichia coli (DEC) strains are leading causes of diarrheal illnesses throughout the world [1,2]. There are five pathotypes (groups) of DEC. Shiga toxin-producing E. coli (STEC) is the only zoonotic among these groups. STEC are ubiquitous food and water borne pathogens inhabiting different animals, wildlife, humans as well as the environment [3-5]. STEC may also be referred to as verocytotoxin-producing E. coli (VTEC). They produce toxins that are variously known as verotoxins, verocytotoxins, or Shiga toxins. They are named for their similarity to the Shiga toxin produced by the bacterium Shigella dysenteriae. Enterohemorrhagic E. coli is a subset of VTEC that carries an additional virulence factor called intimin, which is encoded by the eae gene. Intimin assists in colonization and attachment to intestinal epithelial cells and effacement of microvilli. STEC strains are characterized by the production of one or both of two toxins, Shiga toxin 1 or Shiga toxin 2, which is encoded by the genes stx1 and stx2. Combinations of these virulence genes (or their variants) are associated with life-threatening damage to major organ systems [5-7]. There are 300-400 known STEC serotypes; of which approximately 200 are able to cause disease in humans [8]. When infecting humans, they often cause bloody diarrhea, hemorrhagic colitis (HC), hemolytic uremic syndrome (HUS), and thrombotic thrombocytopenia purpura [4,5,8]. The most renowned example is E. coli O157:H7 which has been incriminated in human outbreaks since the 1980’s [9-12]. On the other hand, the recovery rate of non-O157:H7 STEC is the same or even exceeds that of O157:H7 [6]. The epidemiology of some STEC serotypes is well understood, while for others needs more investigation. The transmission routes of these pathogens may involve the direct fecal-oral route either from infected persons, animals, birds or by the consumption of contaminated food and water. The findings of Santaniello et al. [9], Cernicchiaro et al. [11], Nielsen et al. [13], Foster et al. [14], Persad and LeJeune [15], and Kobayashi et al. [16,17] lent support to the claim that wild birds are vectors and reservoirs for the maintenance and spread of STEC infections. The interactions between humans and wild birds are obvious; they reside in human habitats, migrate between waste collection areas, cattle, pig and poultry farms and deposit their droppings in soil and water and hence allowing the transmission of these zoonoses to man and animals [9,11,13-20]. The long survival time of STEC in soil (for up to 7 months) may give the opportunity for such transmission [10,21-23]. Furthermore, the emergence and dissemination of multi-drug resistant (MDR) bacteria in the environments constitute a global risk to human and animal health [24,25]. Aquatic wild birds, in particular, are often considered indicators for this environmental pollution [18,24,26]. Wild birds can serve as reservoirs of antibiotic resistant bacteria including E. coli and may contribute to the global spread of MDR E. coli in natural ecosystems [18,24,27,28].
After all, typing methods for discriminating different bacterial strains of the same species have become urgent epidemiological tools in disease prevention and control [29]. Traditional typing systems that are based on phenotypes, such as serotype, biotype, or antibiogram have been used for many years. However, other methods that examine the relatedness of isolates at a molecular level have reformed researchers’ capability to differentiate among bacterial types and subtypes [29]. One of the useful means that is used for this purpose is Random-Amplified Polymorphic DNA PCR (RAPD PCR) analysis. Unlike traditional polymerase chain reaction (PCR) analysis, RAPD analysis does not require any specific knowledge of the DNA sequence of the target organism. RAPD is an inexpensive and relatively powerful typing tool for many bacterial species. It has a high discriminatory capacity for typing E. coli isolates in the case of suspected cross infection or epidemic spread [30,31].
Due to the lack of information about the role of wild birds in the dissemination of STEC in the study area, this study was planned to elucidate the role of wild birds as reservoirs of antibiotic-resistant, stx- and eae-producing strains of STEC. The prevalence of these STEC strains in wild birds was examined using standard culture methods. STEC isolates were characterized by serotyping, antibiotic susceptibility, multiplex PCR, and RAPD PCR tests.
Materials and Methods
Ethical approval
The study protocol was approved by the Council of the Department of Animal Hygiene and Zoonoses.
Sampling
The work was conducted in Egypt, namely, at Ismailia (Latitude: 30°36′15″ N and Longitude: 32°16′20″ E) and Damietta Cities (Latitude: 31°24′59″ N and Longitude: 31°48′47″E). 177 intestinal content swab samples originating from five wild bird species were collected over the years (2013 and 2016). They comprised (84) house crows (Corvus splendens), (33) moorhens (Gallinula chloropus), (32) semi-captive pigeons (Columba livia), (18) cattle egrets (Bubulcus ibis), and (10) house teals (Anas crecca). The moorhens were purchased from different retailers at live wild bird markets in Damietta City. Pigeons (semi-captive) and house teals were purchased from Ismailia’s live bird markets. Cattle egrets were hunted from different parks at Ismailia City using traps. A professional hunter was hired to shoot crows that were present near human residence areas in Ismailia City. The selected spp. were chosen because they either approach human habitats (pigeons, cattle egrets, and crows) or because they are commonly raised and/or consumed in the study area (pigeons, house teals, and moorhens). The birds’ handling, transportation and euthanization were performed in compliance with the American Veterinary Medical Association guidelines on the euthanasia of animals [32].
Isolation and identification of E. coli and STEC
Dissection of the euthanized birds was undertaken under aseptic conditions. The intestine was opened, sterile cotton swabs were saturated with about 1 g of the intestinal contents and immediately put into sterile tubes containing 9 ml of 1% tryptone broth (Oxoid, Basingstoke, UK) and incubated at 37°C for 24 h. Two loopfuls of the incubated broth were aseptically streaked onto eosin methylene blue (EMB; Oxoid) and STEC CHROMagar™ (Paris, France) that were prepared according to manufacturers’ instructions and incubated at 40°C and 37°C, respectively, for 24-48 h. Two of suspect colonies on EMB that were green to deep red purple colored with a green metallic tinge sheen and those on STEC CHROMagar (entirely mauve, or mauve with white edge) were selected and biochemically identified according to USFDA [33].
Serological identification of STEC isolates
E. coli isolates selected from STEC CHROMagar were serogrouped on the basis of their “O” antigen from the Reference Lab for Veterinary Quality Control on Poultry Production, Animal Health Research Institute, Dokki, Egypt. The identified isolate was preserved in tryptone broth 1% with adding glycerol to a final concentration of 15%. The tubes were kept at −20°C for further analysis.
Antibiogram susceptibility pattern of STEC
Disk diffusion method was used to identify suspect STEC isolates. Seven antibiotics (Oxoid) that are commonly used in veterinary and human medicine were chosen. Amoxicillin (AML 10 µg), gentamycin (CN 10 µg), cefixime (CFM 5 µg), cefotaxime (CTX 30 µg), doxycycline (Do 30 µg), cephalothin (KF 30 µg), and sulfisoxazole (300 µg). An inoculated 24 h broth with visible turbidity that is equal to or greater than that of the 0.5 McFarland Standard was used. An inoculum was spread evenly over the entire surface of dry Iso-Sensitest agar plate (ISA; Oxoid, Basingstoke, UK) by swabbing in three directions. The inoculum was semi-confluent with no gaps between the swab streaks. Discs were applied to the surface of the agar within 15 min of inoculation. The plate was inverted and incubated at 37°C for 24-48 h. The diameters of zones of inhibition to the nearest millimeters were measured with a graduated ruler and were translated to sensitive, intermediate, and resistant categories according to the guidelines of Andrews [34].
Measurement of multiple antibiotic resistance (MAR) index
The MAR index for STEC isolates was calculated according to the formula of Krumperman [35]: MAR = a/b, where a is a number of antibiotics to which the isolate was resistant, b is a number of antibiotics to which the isolate was subjected. Intermediate and resistant categories were both considered resistant when calculating MAR as suggested by Luczkiewicz et al. [36].
DNA extraction
The suspect STEC isolates were cultured on nutrient agar. A single colony was used for PCR. The DNA was isolated from a colony sweep by suspending it in 500 µl of sterile Milli-Q water. The suspension was boiled for 10 min at 95°C and centrifuged at 10,000 × g for 10 min. The supernatant was then used as the DNA template [37].
Multiplex PCR assay
The multiplex PCR assays were standardized for the detection of stx1, stx2 [38], and eae genes [39]. The DNA templates were subjected to multiplex PCR with the following primers: stx1 F: 5’CTG GAT TTA ATG TCG CAT AGT G3’, stx1 R: 5’AGA ACG CCC ACT GAG ATC ATC3’, stx2 F: 5’ GGC ACT GTC TGA AAC TGC TCC 3’ and stx2 R: 5’ TCG CCA GTT ATC TGA CAT TCT G 3’, eae F: 5’ GAC CCG GCA CAA GCA TAA GC 3’, and eae R: 5’ CCA CCT GCA GCA ACA AGA GG 3’ yielding 150, 255 and 384 (bp) products, respectively. The primers were ordered from Bio basic Inc., Canada. The specificity of each primer was confirmed by monoplex PCR. The total reaction volume was 25 µl containing 5 µl of the extracted DNA from STEC isolates, 12.5 µl of 2× PCR master mix (GeneDirex, USA and Taiwan), 0.5 µl of each primer (20 pmol), and 4.5 µl of sterile Milli-Q water. An Applied Biosystems GeneAmp® PCR System was used for the PCR thermal cycling conditions with an initial denaturation step at 95°C for 3 min, 35 cycles (denaturation at 95°C for 1 min, annealing at 57°C for 1 min, extension at 72°C for 1 min) and a final extension step at 72°C for 7 min. The amplified products were then run along a 0.1-0.5 µg/ml ethidium bromide-stained agarose gel 1.5% with a 100 bp DNA ladder (GeneDirex, USA and Taiwan) in 1× TBE buffer for 30 min at 100 V and then recorded using the SynGene Gel Documentation System.
RAPD PCR assay
Preliminary trials were done using 10 random decamer oligonucleotide primers (OPA1-OPA 10, Eurofins Genomics, Brussels - Belgium) and different PCR conditions. The most yielding primers for discrimination of STEC were OPA-2 (5’TGCCGAGCTG3’), OPA-9 (5’GGGTAACGCC3’) followed by OPA-10 (5’GTGATCGCAG3’) (data were not shown). The most optimum PCR condition was that described by Hopkins and Hilton [40] with modifications. The PCR was carried out in a 25-µl volume containing 12.5 µl PCR master mix (GeneDirex, USA, and Taiwan), 1 µl of primer (OPA-2, OPA-9, or OPA-10) (30 pmol), 3 µl of DNA and 8.5 µl of sterile Milli-Q water. The reactions were run using Techne thermal cycler (Techne, Cambridge, UK). The reactions consisted of one cycle of 4.5 min at 94°C followed by five low stringency cycles of 30 s at 94°C, 1 min at 22°C, 2 min at 72°C and 35 high stringency cycles of 30 s at 94°C, 30 s at 28°C, and 3 min at 72°C. A final extension at 72°C for 5 min and the reactions were hold at 4°C until analysis. The amplified products (8 µl) were separated by electrophoresis in 1.5% agarose stained with ethidium bromide (0.1-0.5 µg/ml) with two DNA ladders (100-1500 bp and 100-3000 bp) and recorded using the SynGene Gel Documentation System.
Analysis of RAPD data
RAPD data were analyzed using computer software (SynGene GeneTools - File version: 4.03.05.0). A scoring Excel sheet was made. Each isolate was scored for the presence (1) or absence (0) of each band on agarose gel. Different banding patterns were recorded. A difference of >2 bands were considered different strains, while isolates with ≤2 bands difference were regarded as the same strain [30].
Statistical analysis
The percentages of colonization were compared using Chi-square test, using SPSS version (20). The p value was set at p≤0.05. The molecular relatedness and genotypic clustering of isolates were analyzed by converting the data to binary code, creating triangular similarity matrix and dendrograms using primer (5) software.
Results
The bacteriological analysis indicated that the percentages of E. coli and STEC colonization were highest in pigeons (90.6 and 46.9), followed by cattle egrets (44.4 and 16.7), crows (41.7 and 9.5), moorhens (39.4 and 6.1), and house teals (20 and 20), respectively (Table-1). Chi-square values for E. coli and STEC colonization were χ2=28.723 and χ2=26.496, at p<0.0001, respectively. Both were considered significant.
Table-1.
Distribution of STEC serotypes among wild bird species.
| Species | Ex. No. | n (%) | STEC serotypes n (%) | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Positive E. coli | Positive STEC | O:20 | O:25 | O:26 | O:27 | O:63 | O:78 | O:111 | O:114 | O:125 | O:128 | O:142 | O:153 | O:158 | Rough | ||
| Cattle egrets | 18 | 8 (44.4) | 3 (16.7) | - | - | - | 1 (33.3) | - | - | 1 (33.3) | - | - | - | - | - | - | 1 (33.3) |
| Pigeons | 32 | 29 (90.6) | 15 (46.9) | 1 (6.7) | - | - | 4 (26.7) | 1 (6.7) | 1 (6.7) | - | 1 (6.7) | - | 1 (6.7) | - | - | 4 (26.7) | 2 (13.3) |
| Crows | 84 | 35 (41.7) | 8 (9.5) | - | 1 (12.5) | - | - | - | - | - | 2 (25) | 2 (25) | - | 1 (12.5) | 1 (12.5) | - | 1 (12.5) |
| Moorhens | 33 | 13 (39.4) | 2 (6.1) | - | - | - | 1 (50) | - | - | - | - | - | - | - | - | - | 1 (50) |
| House teals | 10 | 2 (20) | 2 (20) | - | - | 1 (50) | 1 (50) | - | - | - | - | - | - | - | - | - | - |
| Total no. | 177 | 87 | 30 | 1 | 1 | 1 | 7 | 1 | 1 | 1 | 3 | 2 | 1 | 1 | 1 | 4 | 5 |
For E. coli, χ2=28.723, p<0.0001. For STEC, χ2=26.496, p<0.0001. E. coli=Escherichia coli, STEC=Shiga toxin producing Escherichia coli
The data that were shown in Table-1 revealed that 30 suspect STEC isolates were recovered from (30) positive birds’ samples. By serotyping, (25) isolates were identified as follows: From pigeons, serotypes (O20, [1]; O27, [4]; O63, [1]; O78, [1]; O114 [1]; O128, [1]; and O158, [4]) were detected. The isolates from crows were identified as O25, (1); O114, (2); O125 (2); O142, (1); and O153, (1). Two serotypes were recovered from cattle egrets (O27 and O111), one from moorhens (O27), and two from house teals (O26 and O27). In addition to, five rough strains. However, none of the STEC isolates belonged to the O157 serogroup.
PCR screening of the virulence genes of STEC revealed that stx1 gene only was detected in 16 birds as follows: (28.1%, 9/32) from pigeons, (7.1%, 6/84) from crows, in addition to (5.6%, 1/18) from cattle egrets. There were four birds that possessed both stx1 and stx2 genes in the following manner: (5.6%, 1/18) from cattle egrets, (6.1%, 2/33) from moorhens, and (10%, 1/10) from house teals. Only one pigeon (1/32, 3.1%) possessed the three alleles (Data were retrieved from Table-2).
Table-2.
Epidemiologic data, phenotypic and genotypic traits of STEC serotypes.
| Serial number | Code | Source | Serotype | Antibiotic type | Phenotypic pattern | a | MAR index | Source | Serotype | Virulence factors | RAPD patterns | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | G | C | CT | D | K | S | stx1 | stx2 | eae | Primer 2 A | Primer 9 B | ||||||||||
| 17 P | 9E | Pigeon | O27 | Type 2 | R | R | S | R | R | R | R | 6 | 0.857 | Pigeon | O27 | - | - | - | A9 | B9 | |
| 13 P | 8E | Pigeon | O27 | Type 2 | Pigeon | O27 | + | - | - | A8 | B8 | ||||||||||
| 3 P | 24E | Pigeon | O114 | Type 3 | R | S | S | R | R | R | R | 5 | 0.714 | Pigeon | O114 | - | - | - | A24 | - | |
| 16 p | 10E | Pigeon | O78 | Type 3 | Pigeon | O78 | + | - | - | A10 | B10 | ||||||||||
| 21 P | 7E | Pigeon | O27 | Type 3 | Pigeon | O27 | + | - | A7 | B7 | |||||||||||
| 11 P | 4E | Pigeon | Rough | Type 4 | I | R | S | R | R | R | R | 6 | 0.857 | Pigeon | Rough | + | + | + | A4 | B4 | |
| 7 P | 3E | Pigeon | O128 | Type 4 | Pigeon | O128 | + | - | - | A3 | B3 | ||||||||||
| 2 P | 5E | Pigeon | O20 | Type 6 | S | R | S | R | R | R | R | 5 | 0.714 | Pigeon | O20 | - | - | - | A5 | B5 | |
| 18 P | 6E | Pigeon | O27 | Type 6 | Pigeon | O27 | - | - | - | A6 | B6 | ||||||||||
| 31 P | 16E | Pigeon | O63 | Type 8 | S | S | S | R | R | R | R | 4 | 0.571 | Pigeon | O63 | + | - | - | A16 | B16 | |
| 5 P | 12E | Pigeon | O158 | Type 8 | Pigeon | O158 | + | - | - | A12 | B12 | ||||||||||
| 10 p | 11E | Pigeon | O158 | Type 8 | Pigeon | O158 | + | - | - | A11 | B11 | ||||||||||
| 29 P | 14E | Pigeon | O158 | Type 8 | Pigeon | O158 | - | - | - | A14 | B14 | ||||||||||
| 30 P | 15E | Pigeon | O158 | Type 8 | Pigeon | O158 | + | - | - | A15 | B15 | ||||||||||
| 28 P | 13E | Pigeon | Rough | Type 8 | Pigeon | Rough | + | - | - | A13 | B13 | ||||||||||
| 5 Eg | 2E | Cattle egret | O27 | Type 2 | Cattle egret | O27 | + | + | - | A2 | B2 | ||||||||||
| 7 Eg | 1E | Cattle egret | O111 | Type 6 | Cattle egret | O111 | - | - | - | A1 | B1 | ||||||||||
| 11 Eg | 29E | Cattle egret | Rough | Type 7 | S | S | R | R | R | R | R | 5 | 0.714 | Cattle egret | Rough | + | - | - | A29 | B26 | |
| 76 C | 25E | Crow | O125 | Type 2 | Crow | O125 | + | - | - | A25 | B22 | ||||||||||
| 42 C | 17E | Crow | O114 | Type 3 | Crow | O114 | + | - | - | A17 | B17 | ||||||||||
| 80 C | 27E | Crow | O 153 | Type 3 | Crow | O 153 | - | - | - | A27 | B24 | ||||||||||
| 84 C | 28E | Crow | O25 | Type 5 | I | S | S | R | R | R | R | 5 | 0.714 | Crow | O25 | + | - | - | A28 | B25 | |
| 81 C | 30E | Crow | O125 | Type 6 | 81 C | Crow | O125 | - | - | - | A30 | B27 | |||||||||
| 71 C | 23E | Crow | O114 | Type 8 | 71 C | Crow | O114 | + | - | - | A23 | B21 | |||||||||
| 32 C | 22E | Crow | O142 | Type 8 | 32 C | Crow | O142 | + | - | - | A22 | - | |||||||||
| 48 C | 18E | Crow | Rough | Type 8 | Crow | Rough | + | - | - | A18 | B18 | ||||||||||
| 8 M | 19E | Moorhen | O27 | Type 2 | 8 M | Moorhen | O27 | + | + | - | A19 | B19 | |||||||||
| 9 M | 26E | Moorhen | Rough | Type 6 | Moorhen | Rough | + | + | - | A26 | B23 | ||||||||||
| 5 T | 20E | House teal | O27 | Type 1 | R | R | R | R | R | R | R | 7 | 1 | House teal | O27 | - | - | - | A20 | B20 | |
| 6 T | 21E | House teal | O26 | Type 3 | House teal | O26 | + | + | - | A21 | - | ||||||||||
A=Amoxicillin. G=Gentamycin, C=Cefixime, CT=Cefotaxime, D=Doxycycline, K=Cephalothin, S=Sulfisoxazole. Total number of antibiotics to which the isolate is resistant (a). Total number of antibiotics to which the isolate is subjected (b)=7. MAR index=a/b
Positive growth on STEC CHROMagar that was confirmed by stx gene-detecting PCR indicated that the specificity of STEC CHROMagar medium for detecting STEC was (70%), as nine non-stx-producing isolates out of (30) STEC isolates grew as mauve colonies.
The antibiotic susceptibility patterns showed that cefixime was the most effective against STEC serotypes with 93.3% sensitivity and 6.7% resistance, while the sensitivity and resistance percentages of gentamycin were 56.7 and 43.3 and for amoxicillin were 50 and 40, respectively. All the recovered serotypes were resistant to cefotaxime, doxycycline, cephalothin, and sulfisoxazole (Table-3). The recovered STEC serotypes were organized into eight phenotypic groups according to their antibiogram susceptibility patterns. 30% of STEC isolates (n=9) belonged to phenotype (8); they were resistant to four antibiotics and had MAR index of 0.571. Types 3, 5, 6, and 7 comprised (43.3%, n=13) of STEC isolates; they were resistant to five antibiotics and had MAR index of 0.714. Types 2 and 4 (23.3% of STEC, n=7) were resistant to six antibiotics, having MAR index of 0.857. Type 1 was resistant to all antibiotics tested (Table-2).
Table-3.
Antibiogram susceptibility pattern of different STEC serotypes.
| Antibiotic tested | STEC serotype | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| O:20 | O:25 | O:26 | O:27 | O:63 | O:78 | O:111 | O:114 | O:125 | O:128 | O:142 | O:153 | O:158 | Rough | Total | |
| S (%) I (%) R (%) |
S (%) I (%) R (%) |
S (%) I (%) R (%) |
S (%) I (%) R (%) |
S (%) I (%) R (%) |
S (%) I (%) R (%) |
S (%) I (%) R (%) |
S (%) I (%) R (%) |
S (%) I (%) R (%) |
S (%) I (%) R (%) |
S (%) I (%) R (%) |
S (%) I (%) R (%) |
S (%) I (%) R (%) |
S (%) I (%) R (%) |
S (%) I (%) R (%) |
|
| Amoxicillin | 1 (6.7) | - | - | 1 (6.7) | 1 (6.7) | - | 1 (6.7) | 1 (6.7) | 1 (6.7) | - | 1 (6.7) | - | 4 (26.7) | 4 (26.7) | 15 (50) |
| - | 1 (33.3) | - | - | - | - | - | - | - | 1 (33.3) | - | - | - | 1 (33.3) | 3 (10) | |
| - | - | 1 (8.3) | 6 (50) | - | 1 (8.3) | - | 2 (16.7) | 1 (8.3) | - | - | 1 (8.3) | - | - | 12 (40) | |
| Gentamycin | - | 1 (5.9) | 1 (5.9) | 1 (5.9) | 1 (5.9) | 1 (5.9) | - | 3 (17.6) | - | - | 1 (5.9) | 1 (5.9) | 4 (23.5) | 3 (17.6) | 17 (56.7) |
| - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| 1 (7.7) | - | - | 6 (46.2) | - | - | 1 (7.7) | - | 2 (15.4) | 1 (7.7) | - | - | - | 2 (15.4) | 13 (43.3) | |
| Cefixime | 1 (3.6) | 1 (3.6) | 1 (3.6) | 6 (21.4) | 1 (3.6) | 1 (3.6) | 1 (3.6) | 3 (10.7) | 2 (7.1) | 1 (3.6) | 1 (3.6) | 1 (3.6) | 4 (14.3) | 4 (14.3) | 28 (93.3) |
| - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| - | - | - | 1 (50) | - | - | - | - | - | - | - | - | - | 1 (50) | 2 (6.7) | |
| Cefotaxime | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| 1 (3.3) | 1 (3.3) | 1 (3.3) | 7 (23.3) | 1 (3.3) | 1 (3.3) | 1 (3.3) | 3 (10) | 2 (6.7) | 1 (3.3) | 1 (3.3) | 1 (3.3) | 4 (13.3) | 5 (16.7) | 30 (100) | |
| Doxycycline | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| 1 (3.3) | 1 (3.3) | 1 (3.3) | 7 (23.3) | 1 (3.3) | 1 (3.3) | 1 (3.3) | 3 (10) | 2 (6.7) | 1 (3.3) | 1 (3.3) | 1 (3.3) | 4 (13.3) | 5 (16.7) | 30 (100) | |
| Cephalothin | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| 1 (3.3) | 1 (3.3) | 1 (3.3) | 7 (23.3) | 1 (3.3) | 1 (3.3) | 1 (3.3) | 3 (10) | 2 (6.7) | 1 (3.3) | 1 (3.3) | 1 (3.3) | 4 (13.3) | 5 (16.7) | 30 (100) | |
| Sulfisoxazole | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| 1 (3.3) | 1 (3.3) | 1 (3.3) | 7 (23.3) | 1 (3.3) | 1 (3.3) | 1 (3.3) | 3 (10) | 2 (6.7) | 1 (3.3) | 1 (3.3) | 1 (3.3) | 4 (13.3) | 5 (16.7) | 30 (100) | |
| Total examined | 1 | 1 | 1 | 7 | 1 | 1 | 1 | 3 | 2 | 1 | 1 | 1 | 4 | 5 | 30 |
S=Sensitive, I=Intermediate, R=Resistant
The computer aided RAPD PCR analysis using (OPA-2) primer yielded 122 different bands. The molecular masses of the fragments ranged between 133 and 1777 bp. Primer (OPA-2) could differentiate all the STEC strains from one another yielding 30 different RAPD profiles. At 80% similarity, the 30 STEC isolates lied into 30 different clusters (Figure-1). Using (OPA-9), a total of 27 RAPD profiles were observed. 94 bands ranged between 156 and 1997 bp were distinguished among the 27 STEC isolates. At 80% similarity, the STEC isolates yielded 27 different clusters (Data were not shown). Primer (OPA-9) was unable to amplify serotypes O 26 from teals, O 142 from crows and O 114 from pigeons. Primer (OPA-10) could not amplify most of the isolates (Data were not shown).
Figure-1.
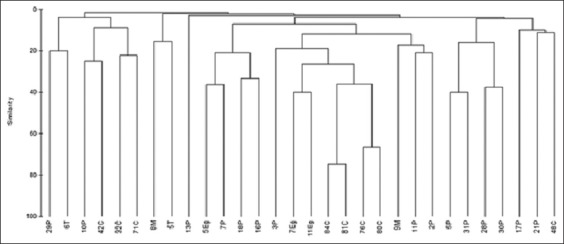
Dendrogram based on random amplified polymorphic DNA profile of Shiga toxin producing Escherichia coli using primer OPA-2.
Discussion
Zoonoses with a wildlife reservoir represent a global public health problem [41]. The migratory nature of birds, their ability to cover vast distances within a relatively short period of time, their residence near livestock areas, farms, waste disposal sites, and human habitats made them important vectors of some zoonoses [7,23,42]. Human infections with STEC are increasingly recognized as causes of HC and HUS [10]. The most renowned example is E. coli O157:H7, however the prevalence of non-O157 STEC can be the same or even exceeds that of O157: H7 [6]. Among patients with non-O157 STEC infections, the “Big Six” strains; O26, O45, O103, O111, O121, and O145 are the most common cause of hospitalizations. These types can cause the most severe sequelae as kidney failure (HUS) and death [43]. The incidence of DEC can vary from one region to another due to varying principles and methods for detection [2]. For example, the annual detection rate of non-O157 STEC infection in the USA exceeds that caused by O157 STEC [5]. In developing countries, the detection and identification of non-O157 STEC infections are complex and underestimated. First, most efforts are directed to detect O157. Yet, non-O157 STEC is not recognized on the media that are used to isolate this organism. Second, clinical laboratories must detect Shiga toxins in stool samples and then, the positive samples must be sent to the public health laboratories for serotyping [44]. The financial cost is another keystone factor that may hinder such diagnosis.
Earlier studies confirmed the importance of wild birds in the maintenance of E. coli. In Germany, E. coli was isolated from birds of prey, waterfowls and passerines [24]. In Denmark, the frequency of VTEC-positive wild birds was 1.6%, as judged by the PCR screening, showing that wild birds may become infected from farm animals or vice versa [13]. Foster et al. [14] reported that one (0.43%) out of 231 composite feces collected from a bird table in southwest Scotland was positive for E. coli O157. They attributed the low isolation rate to either the storage process, the sensitivity of the test method, or other factors. Kobayashi et al. [17] recorded that the prevalences of stx- and eae-positive strains of E. coli among wild birds in Japan were 5% (23/447) and 25% (113/447), respectively. In Egypt, E. coli was isolated from white ibis at the rate of 43.6% [45]. In Germany, E. coli was the second most common isolated pathogen, being present in 18% (46/251) of diseased wild birds [19]. Koochakzadeh et al. [20] found that 0.45% of wild and pet birds in Iran carry EPEC strains. Gioia-Di Chiacchio et al. [7] found that E. coli isolated from psittacine birds in Brazil was eae+ and stx2+.
Stx-PCR confirmed results revealed that the specificity of STEC CHROMagar medium for detecting STEC was (70%). However, STEC isolates might contain other stx variants, and this needs further investigation.
The main virulence factor of STEC group is the production of stx1 or stx2 proteins. It is worth mentioning that Byrne et al. [46] reported that HUS is significantly associated with STEC strains possessing eae and/or stx2. In this study, most of the isolates possessed stx1 gene. The significance of stx1 was underscored by Hedican et al. [6] who indicated that non-O157 isolates that had only stx1 can cause severe bloody diarrhea or HUS. Furthermore, Käppeli et al. [4] depicted the stx gene distribution among 97 non-O157 STEC strains isolated from HUS cases in Switzerland. They showed that 45 (46.4%) strains had only stx2, 36 (37.1%) had only stx1, and 16 (16.5%) had both genes.
In contrast, other researchers could not detect stx1 or stx2 in wild birds. Kobayashi et al. [16] found no stx+ samples among fecal samples from gulls, pigeons, and chickens that were examined in Finland. Koochakzadeh et al. [20] found that 1.8% of wild and pet birds carry stx2f+ STEC. Schmidt et al. [47] in their study in Italy reported the isolation of STEC strains from the feces of feral pigeons which contained a new stx2 variant gene designated stx2f. Wani et al. [48] showed that none of the isolated E. coli strains (O9, O18, O25, O60, O77, O147, O157, O168, O169, rough [R], untypable [UT]) from free flying pigeons in India were stx1+ or stx2+.
The current research develops the claim that pigeons can be a source of STEC for humans. Among the examined wild bird species, they had the highest STEC colonization rate (46.9%) of which 10/32, 31.3% carried virulence genes. They were also colonized with a diverse number of STEC serotypes. It is noteworthy to mention that pigeons are commonly found in human residence areas and parks, moreover, they are commonly raised and/or consumed in Egypt; an issue that may allow such transmission. Along similar lines, Pedersen and Clark [23] reported that pigeons and sparrows are important in recirculation of STEC. In Italy, stx genes were detected in 10.8% of the stool enrichment cultures collected from feral pigeons [42]. In another study in Italy [9], four E. coli O157:H7 strains from pigeons were isolated; all strains carried eae and stx2 genes, whereas only one strain carried the stx1 gene. In Germany, 67% of the examined pigeon feces harbored stx genes [49]. Dutta et al. [12] found that out of 150 pigeons subjected to microbiological investigation in India, 91 (60.67%) samples were found positive for E. coli. The most frequently occurring serotypes were O157, followed by O68, O121, O9, O75, O131, O2, O13, and O22.
Concerning crows, cattle egrets and water fowls, they can contaminate water and pasture with their fecal droppings; moreover, they have been implicated as a source of DEC infection [15,44,45]. The noteworthy point is that people living in Egyptian coastal cities are accustomed to eating different spp. of wild water fowls. They purchase them either eviscerated or even process them at home. Thus, the increased prevalence of STEC colonization in moorhens (6.1%) and house teals (20%) may represent a hazard to human contacts. The following section will highlight the importance of wild birds as indicators, long-distance vectors, reservoirs, and potential spreaders of MDR E. coli and this was previously confirmed by various researchers [18,24-28,50]. In this study, STEC had MAR index that ranged from 0.571 to 1. MAR indexing has been used as an indicator to identify high-risk contamination that may pose a hazard to humans. MAR index values which are higher than 0.2 were considered to have originated from high-risk sources where antibiotics are often used [35]. Evidence for antibiotic-resistant E. coli was borne out by many researchers. Guenther et al. [24] showed that nine of the 187 E. coli isolates (4.8%) exhibited multiresistant phenotypes including resistances against beta-lactams, aminoglycosides, fluoroquinolones, tetracyclines, and sulfonamides. Similarly, Käppeli et al. [4] investigated the antimicrobial pattern of non-O157 STEC strains isolated from HUS cases in Switzerland; all the recovered strains were susceptible to five antimicrobial drugs (ceftazidime, ciprofloxacin, cefotaxime, cefepime, and cefoxitin). In Spain, 41% out of 581 non-O157 STEC strains showed resistance to at least one of the 26 antimicrobial agents tested and sulfisoxazole (36%) had the most common antimicrobial resistance [51]. They found also an association between a higher level of multiple resistances to antibiotics and the presence of the virulence genes eae and stx1 among non-O157 strains. Hasan et al.’s [18] results provided confirmatory evidence that wild ducks and domestic poultry harbored the same ESBL-producing E. coli isolates. They attributed this commonality to be caused by a common use of natural water resources. They concluded also that E. coli that produces CTX-M-15 is endemic to birds in Bangladesh. Similarly, Dutta et al. [12] concluded that antimicrobial-resistant pathogenic E. coli is present in pigeons. In Spain, cefotaxime-resistant E. coli isolates were identified in 16 of the 100 tested wild birds’ species samples [28]. In summary, the present study reveals that a high percentage of wild birds carry antibiotic-resistant STEC and this may pose a threat to human and animal health.
The importance of typing methods for zoonotic disease prevention and control is increasingly esteemed. One of the useful means that is used for discrimination of bacterial strains within the same spp. is RAPD Analysis. The usefulness of RAPD PCR for tracing the clonal relations of E. coli serovars has been underscored by several researchers [30,40,52,53]. The results of our study demonstrated that RAPD PCR analysis of the E. coli strains in conjunction with serotyping may fulfill these criteria. The use of computerized analysis aided in the differentiation of 30 and 27 different DNA fingerprinting profiles for primers OPA-2 and OPA-9, respectively. Most of these profile bands were impossible to be correctly sized using naked eye analysis. Correspondingly, in India, primers OPAC 04, OPAC 07, OPAC 09, OPAC 11, and OPAC 12 yielded entirely different banding pattern for each E. coli serotype and were able to differentiate all the serotypes from one another [52]. The view that at least two independent primers should be used to maximize the discriminatory capacity of RAPD PCR is in line with Idil and Bilkay [54]. There are some previous reports on RAPD genotyping of E. coli strains using one or two primers and agarose electrophoresis, but none of these studies have used computer-aided analysis of bands [55,56]. Our results showed that non-O157 STEC are heterogeneous; they were grouped into 30 different clusters at 80% similarity. The finding that certain isolates could not be amplified using OPA-9 and OPA-10 primers might be attributed to the absence of sequence in the bacterial DNA which is complementary to the sequence of the primer and the fact that some primers may amplify only a small sequence of the genome, while other primers may amplify different sites of the genome and so differentiate between strains [53,56].
Overall, the investigated birds which were sampled from live bird markets, parks and areas near human residence areas carry STEC that harbor virulence genes and are resistant to multiple antibiotics. We also found that RAPD PCR when complemented with serotyping become useful means for discrimination of STEC strains.
Conclusions
Wild birds must be monitored for MDR zoonotic pathogens including STEC. Therefore, the development of local centers that are globally connected for the early detection, prevention, and control of such infections must be prioritized.
Authors’ Contributions
This research was designed and funded by HMF and RA. HMF was responsible for collection of samples, performance of the experiments, writing and revising of the manuscript. RA collected samples and shared in isolation and identification of E. coli. DMA performed PCR for detection of stx1, stx2, and eae genes. The manuscript has been seen and approved by all authors.
Acknowledgments
This research was funded by HMF and RA. The two authors equally shared the purchase of reagents, media, and the PCR running fees.
Competing Interests
The authors declare that they have no competing interests.
References
- 1.WHO. E. coli Fact Sheet. 2016. [Viewed on 02-02-2017]. Available from: http://www.who.int/mediacentre/factsheets/fs125/en .
- 2.Espenhain L.E. Master of Public Health Science, Faculty of the Health and Medical Sciences. Denmark: University of Copenhagen; 2013. Epidemiology and surveillance of Three Diarrheagenic Escherichia coli in Denmark between 2000-2012-Can Surveillance Be Improved? [Google Scholar]
- 3.Garcia A, Fox J.G, Besser T.E. Zoonotic enterohemorrhagic Escherichia coli: A one health perspective. ILARJ. 2010;51(3):221–232. doi: 10.1093/ilar.51.3.221. [DOI] [PubMed] [Google Scholar]
- 4.Käppeli U, Hächler H, Giezendanner N, Beutin L, Stephan R. Human infections with non-O157 Shiga toxin-producing Escherichia coli Switzerland, 2000-2009. Emerg. Infect. Dis. 2011;17(2):180–185. doi: 10.3201/eid1702.100909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.CDC-Centers for Disease Control and Prevention. Multistate outbreaks of Shiga toxin-producing Escherichia coli O26 infections linked to Chipotle Mexican grill restaurants. 2015. [Viewed on 14-05-2016]. Available from: http://www.cdc.gov/ecoli/2015/O26-H11-15/signs-symptoms.html .
- 6.Hedican E.B, Medus C, Besser J.M, Juni B.A, Koziol B, Taylor C, Smith K.E. Characteristics of O157 versus non-O157 Shiga toxin-producing Escherichia coli infections in Minnesota, 2000-2006. Clin. Infect. Dis. 2009;49(3):358–364. doi: 10.1086/600302. [DOI] [PubMed] [Google Scholar]
- 7.Gioia-Di Chiacchio R.M, Cunha M.P.V, Sturn R.M, Moreno L.Z, Moreno A.M, Pereira C.B.P, Martins F.H, Franzolin R.M, Piazza R.M.F, Knöbl T. Shiga toxin-producing Escherichia coli(STEC): Zoonotic risks associated with psittacine pet birds in home environments. Vet. Microbiol. 2016;184:27–30. doi: 10.1016/j.vetmic.2016.01.004. [DOI] [PubMed] [Google Scholar]
- 8.USDA-United States Department of Agriculture. Risk profile for pathogenic non-O157 Shiga toxin-producing Escherichia coli (non-O157 STEC) Food Safety and Inspection Service. 2012. [Viewed on 12-02-2016]. Available from: http://www.aphis.usda.gov .
- 9.Santaniello A, Gargiulo A, Borrelli L, Dipineto L, Cuomo A, Sensale M, Fontanella M, Calabria M, Musella V, Menna L.F, Fioretti A. Survey of Shiga toxin-producing Escherichia coli O157: H7 in urban pigeons (Columba livia) in the city of Napoli, Italy. Ital. J. Anim. Sci. 2007;6:313–316. [Google Scholar]
- 10.CFSPH-The Center for Food Security and Public Health. Enterohemorrhagic Escherichia coli and other E. coli causing hemolytic uremic syndrome. 2016. [Viewed on 20-02-2017]. Available from: http://www.cfsph.iastate.edu/Factsheets/pdfs/e_coli.pdf .
- 11.Cernicchiaro N, Pearl D.L, McEwen S.A, Harpster L, Homan H.J, Linz G.M, LeJeune J.T. Association of wild bird density and farm management factors with the prevalence of E. coli O157 in dairy herds in Ohio (2007-2009) Zoonoses Public Health. 2012;59(5):320–329. doi: 10.1111/j.1863-2378.2012.01457.x. [DOI] [PubMed] [Google Scholar]
- 12.Dutta P, Borah M.K, Sarmah R, Gangil R. Isolation, histopathology and antibiogram of Escherichia coli from pigeons (Columba livia) Vet. World. 2013;6(2):91–94. [Google Scholar]
- 13.Nielsen E.M, Skov M.N, Madsen J.J, Lodal J, Jespersen J.B, Baggesen D.L. Verocytotoxin-producing Escherichia coli in wild birds and rodents in close proximity to farms. Appl. Environ. Microbiol. 2004;70(11):6944–6947. doi: 10.1128/AEM.70.11.6944-6947.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Foster G, Evans J, Knight H.I, Smith A.W, Gunn G.J, Allison L.J, Synge B.A, Pennycott T.W. Analysis of feces samples collected from a wild-bird garden feeding station in Scotland for the presence of verocytotoxin producing Escherichia coli O157. Appl. Environ. Microbiol. 2006;72(3):2265–2267. doi: 10.1128/AEM.72.3.2265-2267.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Persad A.K, LeJeune J.T. Animal reservoirs of Shiga toxin-producing E. coli. Microbiol. Spectr. 2014;2(4) doi: 10.1128/microbiolspec.EHEC-0027-2014. EHEC-0027-2014. [DOI] [PubMed] [Google Scholar]
- 16.Kobayashi H, Pohjanvirta T, Pelkonen S. Prevalence and characteristics of intimin-and Shiga toxin-producing Escherichia coli from gulls, pigeons and broilers in Finland. J. Vet. Med. Sci. 2002;64(11):1071–1073. doi: 10.1292/jvms.64.1071. [DOI] [PubMed] [Google Scholar]
- 17.Kobayashi H, Kanazaki M, Hata E, Kubo M. Prevalence and characteristics of eae- and stx-positive strains of Escherichia coli from wild birds in the immediate environment of Tokyo Bay. Appl. Environ. Microbiol. 2009;75(1):292–295. doi: 10.1128/AEM.01534-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hasan B, Sandegren L, Melhus Å, Drobni M, Hernandez J, Waldenström J, Alam M, Olsen B. Antimicrobial drug-resistant Escherichia coli in wild birds and free-range poultry, Bangladesh. Emerg. Infect. Dis. 2012;18(12):2055–2058. doi: 10.3201/eid1812.120513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stenkat J, Krautwald-Junghanns M.E, Schmidt V. Causes of morbidity and mortality in free-living birds in an urban environment in Germany. Ecohealth. 2013;10(4):352–365. doi: 10.1007/s10393-013-0868-9. [DOI] [PubMed] [Google Scholar]
- 20.Koochakzadeh A, Askari B.M, Zahraei S.T, Aghasharif S, Soltani M, Ehsan M.R. Prevalence of Shiga toxin-producing and enteropathogenic Escherichia coli in wild and pet birds in Iran. Braz. J. Poult. Sci. 2015;17(4):445–450. [Google Scholar]
- 21.Fukushima H, Hoshina K, Gomyoda M. Long-term survival of Shiga toxin-producing Escherichia coli O26, O111, and O157 in bovine feces. Appl. Environ. Microbiol. 1999;65:5177–5181. doi: 10.1128/aem.65.11.5177-5181.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Neher S, Hazarika A.K, Barkalita L.M, Borah P, Bora D.P, Sharma R.K. Isolation and characterization of Shiga toxigenic Escherichia coli of animal and bird origin by multiplex polymerase chain reaction. Vet. World. 2016;9(2):123–127. doi: 10.14202/vetworld.2016.123-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pedersen K, Clark L. A review of Shiga toxin Escherichia coli and Salmonella enterica in cattle and free-ranging birds: Potential association and epidemiological links. Hum. Wildl. Confl. 2007;1(1):68–77. [Google Scholar]
- 24.Guenther S, Ewers C, Wieler L.H. Extended-spectrum beta-lactamases producing E. coli in wildlife, yet another form of environmental pollution? Front Microbiol. 2010;2:246–259. doi: 10.3389/fmicb.2011.00246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shobrak M.Y, Abo-Amer E. Role of wild birds as carriers of multi-drug resistant Escherichia coli and Escherichia vulneris. Braz. J. Microbiol. 2014;45(4):1199–1209. doi: 10.1590/s1517-83822014000400010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Veldman K, van Tulden P, Kant A, Testerink J, Mevius D. Characteristics of cefotaxime-resistant Escherichia coli from wild birds in the Netherlands. Appl. Environ. Microbiol. 2013;79(24):7556–7561. doi: 10.1128/AEM.01880-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cole D, Drum D.J, Stalknecht D.E, White D.G, Lee M.D, Ayers S, Sobsey M, Maurer J.J. Free-living Canada geese and antimicrobial resistance. Emerg. Infect. Dis. 2005;11:935–938. doi: 10.3201/eid1106.040717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alcalá L, Alonso C.A, Simón C, González-Esteban C, Orós J, Rezusta A, Ortega C, Torres C. Wild Birds, frequent carriers of extended-spectrum β-lactamase (ESBL) producing Escherichia coli of CTX-M and SHV-12 types. Microb. Ecol. 2016;72(4):861–869. doi: 10.1007/s00248-015-0718-0. [DOI] [PubMed] [Google Scholar]
- 29.Sabat A.J, Budimir A, Nashev D, Sá-Leão R, van Dijl J.M, Laurent F, Grundmann H, Friedrich A.W. Overview of molecular typing methods for outbreak detection and epidemiological surveillance. Eur. Surveill. 2013;18(4):20380–20395. doi: 10.2807/ese.18.04.20380-en. [DOI] [PubMed] [Google Scholar]
- 30.Vogel L, van Oorschot E, Maas H.M.E, Minderhoud B, Dijkshoorn L. Epidemiologic typing of Escherichia coli using RAPD analysis, ribotyping and serotyping. Clin. Microbiol. Infect. 2000;6(2):82–87. doi: 10.1046/j.1469-0691.2000.00013.x. [DOI] [PubMed] [Google Scholar]
- 31.Mahanti A, Samanta I, Bandyopadhyay S, Joardar S.N. Molecular characterization and antibiotic susceptibility pattern of caprine Shiga toxin producing-Escherichia coli(STEC) isolates from India. Iran. J. Vet. Res. 2015;16(1):31–35. [PMC free article] [PubMed] [Google Scholar]
- 32.AVMA Guidelines on Euthanasia. Edition American Veterinary Medical Association 1931. IL: N Meacham Road Schaumburg; 2007. [Google Scholar]
- 33.USFDA. United States Food and Drug Administration for Detection, Enumeration and Identification to Species Level of Individual Organisms, Bacteriological Analytical Manual. 8th ed. Washington, DC: U.S. Food and Drug Administration; 2001. [Google Scholar]
- 34.Andrews J.M. BSAC standardized disc susceptibility testing method (version 4) J. Antimicrob. Chemother. 2005;56:60–76. doi: 10.1093/jac/dki124. [DOI] [PubMed] [Google Scholar]
- 35.Krumperman P.H. Multiple antibiotic resistance indexing of Escherichia coli to identify high-risk sources of fecal contamination of foods. Appl. Environ. Microbiol. 1983;46(1):165–170. doi: 10.1128/aem.46.1.165-170.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Luczkiewicz A, Jankowska K, Kurlenda J, Olanczuk-Neyman K. Identification and antimicrobial resistance of Enterococcus spp. isolated from surface water. Water Sci. Technol. 2010;62(2):466–473. doi: 10.2166/wst.2010.909. [DOI] [PubMed] [Google Scholar]
- 37.Madico G, Akopyants N.S, Berg D.E. Arbitrarily primed PCR DNA fingerprinting of Escherichia coli O157: H7 strains by using templates from boiled cultures. J. Clin. Microbiol. 1995;33(6):1534–1536. doi: 10.1128/jcm.33.6.1534-1536.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee G.Y, Jang H.I, Hwang I.G, Rhee M.S. Prevalence and classification of pathogenic Escherichia coli isolated from fresh beef, poultry, and pork in Korea. Int. J. Food Microbiol. 2009;134(3):196–200. doi: 10.1016/j.ijfoodmicro.2009.06.013. [DOI] [PubMed] [Google Scholar]
- 39.López-Saucedo C, Cerna J.F, Villegas-Sepulveda N, Thompson R, Velazquez F.R, Torres J, Tarr P.I, Estrada-García T. Single multiplex polymerase chain reaction to detect diverse loci associated with diarrheagenic Escherichia coli. Emerg. Infect. Dis. 2003;9(1):127–131. doi: 10.3201/eid0901.01-0507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hopkins K.L, Hilton A.C. Optimization of random amplification of polymorphic DNA analysis for molecular subtyping of Escherichia coli O157. Lett. Appl. Microbiol. 2001;32(3):126–130. doi: 10.1046/j.1472-765x.2001.00871.x. [DOI] [PubMed] [Google Scholar]
- 41.Hassell J.M, Begon M, Ward M.J, Fèvre E.M. Urbanization and disease emergence: Dynamics at the wildlife-livestock-human interface. Trends Ecol. Evol. 2017;32(1):55–67. doi: 10.1016/j.tree.2016.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morabito S, Dell'Omo G, Agrimi U, Schmidt H, Karch H, Cheasty T, Caprioli A. Detection and characterization of Shiga toxin-producing Escherichia coli in feral Pigeons. Vet. Microbiol. 2001;82(3):275–283. doi: 10.1016/s0378-1135(01)00393-5. [DOI] [PubMed] [Google Scholar]
- 43.Gould L.H, Mody R.K, Ong K.L, Clogher P, Cronquist A.B, Garman K.N, Lathrop S, Medus C, Spina N.L, Webb T.H, White P.L, Wymore K, Gierke R.E, Mahon B.E, Griffin P.M. Increased recognition of non-O157 Shiga toxin-producing Escherichia coli infections in the United States during 2000-2010: Epidemiologic features and comparison with E. coli O157 infections. Foodborne Pathog. Dis. 2013;10(5):453–460. doi: 10.1089/fpd.2012.1401. [DOI] [PubMed] [Google Scholar]
- 44.Friesema I, van der Zwaluw K, Schuurman T, Kooistra-Smid M, Franz E, van Duynhoven Y, van Pelt W. Emergence of Escherichia coli encoding Shiga toxin 2f in human Shiga toxin producing E. coli(STEC) infections in the Netherlands, January 2008 to December 2011. Eur., Surveill. 2014;19(17):26–32. [PubMed] [Google Scholar]
- 45.Awad-Alla M.E, Abdien H.M.F, Dessouki A.A. Prevalence of bacteria and parasites in white ibis in Egypt. Vet. Ital. 2010;46(3):277–286. [PubMed] [Google Scholar]
- 46.Byrne L, Vanstone G.L, Perry N.T, Launders N, Adak G.K, Godbole G, Grant K.A, Smith R, Jenkins C. Epidemiology and microbiology of Shiga toxin-producing Escherichia coli other than serogroup O157 in England, 2009-2013. J. Med. Microbiol. 2014;63:1181–1188. doi: 10.1099/jmm.0.075895-0. [DOI] [PubMed] [Google Scholar]
- 47.Schmidt H, Scheef J, Morabito S, Caprioli A, Wieler L.H, Karch H. A new Shiga toxin 2 variant (Stx2f) from Escherichia coli isolated from pigeons. Appl. Environ. Microbiol. 2000;66(3):1205–1208. doi: 10.1128/aem.66.3.1205-1208.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wani S.A, Samanta I, Bhat M.A, Nishikawa Y. Investigation of Shiga toxin-producing Escherichia coli in avian species in India. Lett. Appl. Microbiol. 2004;39(5):389–394. doi: 10.1111/j.1472-765X.2004.01586.x. [DOI] [PubMed] [Google Scholar]
- 49.Grobmann K.M. Isolation and Characterization of Shiga Toxin-Producing Escherichia coli (STEC) in Pigeons. Doctoral Thesis. Universitätsbibliothek, Garystr. 39, 14195 Berlin Institutional Repository of the Freie Universität Berlin. 2010 [Google Scholar]
- 50.Bonnedahl J, Järhult J.D. Antibiotic resistance in wild birds. Ups. J. Med. Sci. 2014;119(2):113–116. doi: 10.3109/03009734.2014.905663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mora A, Blanco J.E, Blanco M, Alonso M.P, Dhabi G, Echeita A, González E.A, Bernárdez M.I, Blanco J. Antimicrobial resistance of Shiga toxin (verotoxin)-producing Escherichia coli O157: H7 and non-O157 strains isolated from humans, cattle, sheep and food in Spain. Res. Microbiol. 2005;156(7):793–806. doi: 10.1016/j.resmic.2005.03.006. [DOI] [PubMed] [Google Scholar]
- 52.Gomes A.R, Muniyappa L, Krishnappa G, Suryanarayana V.V.S, Isloor S, Prakash B, Hugar P.G. Genotypic characterization of avian Escherichia coli by random amplification of polymorphic DNA. Int. J. Poult. Sci. 2005;4(6):378–381. [Google Scholar]
- 53.Kumar V, Gupta R.K, Kewalramani H, Sikhwal Y, Sharma M, Nayar S. Genetic differentiation of Escherichia coli strains isolated from the urinary tract infected patients. Int. J. Microbiol. Allied Sci. 2015;2(1):17–22. [Google Scholar]
- 54.Idil N, Bilkay I.S. Application of RAPD-PCR for determining the clonality of methicillin resistant Staphylococcus aureus isolated from different hospitals. Braz. Arch. Biol. Technol. 2014;57(4):548–553. [Google Scholar]
- 55.Dhanashree B, Mallya S.P. Molecular typing of enteropathogenic Escherichia coli from diarrheagenic stool samples. J. Clin. Diagn. Res. 2012;6(3):400–404. [Google Scholar]
- 56.Wang G, Whittam T.S, Berg C.M, Berg D.E. RAPD (arbitrary primer) PCR is more sensitive than multilocus enzyme electrophoresis for distinguishing related bacterial strains. Nucleic Acids Res. 1993;21(25):5930–5933. doi: 10.1093/nar/21.25.5930. [DOI] [PMC free article] [PubMed] [Google Scholar]


