Abstract
Approximately 70 human RNA-binding proteins (RBPs) contain a prion-like domain (PrLD). PrLDs are low-complexity domains that possess a similar amino acid composition to prion domains in yeast, which enable several proteins, including Sup35 and Rnq1, to form infectious conformers, termed prions. In humans, PrLDs contribute to RBP function and enable RBPs to undergo liquid–liquid phase transitions that underlie the biogenesis of various membraneless organelles. However, this activity appears to render RBPs prone to misfolding and aggregation connected to neurodegenerative disease. Indeed, numerous RBPs with PrLDs, including TDP-43 (transactivation response element DNA-binding protein 43), FUS (fused in sarcoma), TAF15 (TATA-binding protein-associated factor 15), EWSR1 (Ewing sarcoma breakpoint region 1), and heterogeneous nuclear ribo-nucleoproteins A1 and A2 (hnRNPA1 and hnRNPA2), have now been connected via pathology and genetics to the etiology of several neurodegenerative diseases, including amyotrophic lateral sclerosis, frontotemporal dementia, and multisystem proteinopathy. Here, we review the physiological and pathological roles of the most prominent RBPs with PrLDs. We also highlight the potential of protein disaggregases, including Hsp104, as a therapeutic strategy to combat the aberrant phase transitions of RBPs with PrLDs that likely underpin neurodegeneration.
Protein misfolding unites diverse neurodegenerative diseases
The problem of neurodegeneration remains a pressing public health concern and a biologic black box [1,2]. Age-related neurodegenerative diseases such as Alzheimer’s disease (AD), Parkinson’s disease (PD), amyotrophic lateral sclerosis (ALS), frontotemporal dementia [FTD, the clinical disorder resulting from frontotemporal lobar degeneration (FTLD) [3]], and Huntington’s disease (HD) lead to cell death within the central nervous system (CNS) and progressive CNS dysfunction [4–9]. ALS pathology also extends to the peripheral nervous system [5,10]. Our lack of understanding of the mechanisms and risk factors governing the development and progression of neurodegenerative diseases has largely precluded the development of disease-reversing therapeutics [4,11,12]. Symptomatic treatments are available for PD and AD, but the efficacy of these can be modest or limited by problematic side effects, and they do not address the root cause of disease [12,13].
Despite dramatic differences in characteristic age of onset, symptomatology, and regional involvement of CNS tissue, neurodegenerative disorders are united on a cellular and biochemical level by the accumulation of misfolded proteins in the brain [5–7,14–16]. Cytoplasmic inclusions of α-synuclein in the neurons of the substantia nigra pars compacta and other brain regions are a hallmark feature of PD [8,16,17]. In AD, intracellular tangles of misfolded tau protein in conjunction with extracellular plaques of aggregated amyloid-β are defining features found in the neocortex and hippocampus [9,16,18,19]. In HD, a genetic trinucleotide repeat expansion leads to an elongated polyglutamine tract in the protein huntingtin, causing it to form both nuclear and cytoplasmic amyloid inclusions [18,19]. In addition, repeat-associated non-ATG (RAN) translation occurs in several diseases caused by repeat expansions, including spinocerebellar ataxia type 8 (SCA8), myotonic dystrophy type 1, fragile X-associated tremor ataxia syndrome, ALS, and HD [20–23]. RAN translation in HD, which occurs in multiple reading frames from both sense and antisense transcripts, leads to the accumulation of aggregated polyalanine, polyserine, polyleucine, and polycysteine in the brains of HD patients [21].
ALS and FTD are related disorders
ALS, also known as Lou Gehrig’s disease in homage to the prominent baseball player who was diagnosed in 1939 and died 2 years later, is a devastating neurodegenerative disorder that affects the upper and lower motor neurons of the brain and spinal cord [5]. The widespread and relentlessly progressive destruction of motor neurons causes muscle weakness and atrophy with hyperreflexia and spasticity, ultimately leading to paralysis and death within 2–5 years of disease onset in most cases [5,24]. FTD is a leading cause of early-onset dementia, second only to AD [25]. It results in the selective degeneration of the frontal and temporal lobes of the brain, which typically manifests as primarily behavioral dysfunction, including changes in personality and executive function or loss of volition, or language deficits [5,25]. It has become increasingly clear that there is a significant overlap between ALS and FTD clinically, genetically, and neuropathologically [3,5,10,25,26].
It is now estimated that up to 50% of ALS patients also suffer from cognitive impairment or behavioral changes associated with FTLD, and while in many cases these symptoms do not reach a clinical severity that meets criteria for dementia, ~15–20% of those with ALS also carry a diagnosis of FTD [3,25,27,28]. Similarly, a study of FTD patients found that ~50% had motor neuron involvement evident via examination or electromyography [3,27]. The idea that purely motor ALS and purely cognitive FTD exist at the two ends of a spectrum of disease is not surprising when it is considered that the two clinical entities are known to share genetic causes in their familial forms and have commonalities in their cellular signatures [15,29]. Like other neurodegenerative disorders, ALS and FTD are characterized by pathologic protein aggregation in the cytoplasm of affected neurons [15,30]. Among the proteins that have been genetically linked to these diseases and identified in cytoplasmic inclusions in patient neurons are several RNA-binding proteins (RBPs) that have low-complexity domains (LCDs), termed prion-like domains (PrLDs), because of their similarity in amino acid composition to yeast prion domains [31].
Prions are self-replicating protein conformers
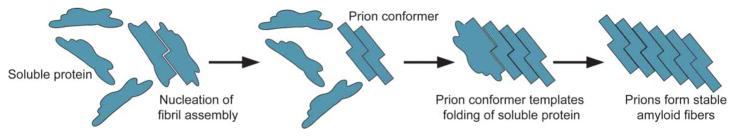
Prions are the cause of devastating human neurodegenerative diseases including Creutzfeldt–Jakob disease, Gerstmann–Sträussler–Scheinker syndrome, and fatal familial insomnia, but confer heritable traits that can be beneficial in yeast [18,31–36]. Prions are infectious protein conformers capable of self-replication, which occurs as the prion templates the folding of soluble proteins comprised of the same amino acid sequence (Figure 1) [37,38]. In the prion conformation, these proteins typically form stable amyloid fibers that are often sodium dodecyl sulfate (SDS) insoluble and resistant to proteases and heat denaturation [18,37]. Amyloid is a polymeric ‘cross-β’ structure in which the strands of the β-sheets run perpendicular to the axis of the fiber [18,35]. The ability of yeast prions to form amyloid is dependent on a prion domain rich in glycine and uncharged polar amino acids, including glutamine, asparagine, tyrosine, and serine [31,39–41]. Deletion of this prion domain precludes access to the prion state [42], and addition of this region to otherwise innocuous proteins is sufficient to confer prion behavior [43–45]. Importantly, randomization of the primary amino acid sequence of the prion domain does not affect prion formation [46,47]. Identification of several prion domains that confer bona fide prion behavior has led to the development of bioinformatics algorithms that scan amino acid composition to screen the human genome for proteins with PrLDs [31,39,40,48].
Figure 1. Prions self-replicate conformation by templating the folding of soluble protein to the prion conformation.
Prions are protein conformers that self-replicate by templating the folding of natively folded proteins of the same amino acid sequence to the prion conformation [18,35]. Prions typically form stable amyloid fibers with a hallmark ‘cross-β’ structure in which β-strands run perpendicular to the axis of the fiber [18,35]. These amyloid assemblies are typically resistant to denaturation by heat, proteases, and detergents [18,37].
Human RBPs with PrLDs cause neurodegenerative diseases
Interestingly, a disproportionate number of the ~240 human proteins with PrLDs are RNA- or DNA-binding proteins, many of which contain a canonical RNA recognition motif (RRM) [41,49]. Gene ontology (GO) annotations indicate that ~30% of human proteins with PrLDs function in RNA binding and ~33% function in DNA binding [41]. While RRM-containing genes represent only ~1% of the human protein-coding genome, they comprise >10% of all genes containing PrLDs [31]. One by one, RNA-/DNA-binding proteins with PrLDs are being implicated in neurodegenerative disease [41,49]. This association began with the identification of a trinucleotide repeat expansion in the gene encoding ataxin 1 (ATXN1) that leads to a polyglutamine protein product and causes SCA1 [50,51]. The expansion is now recognized to occur within the PrLD and promotes aggregation of ATXN1 [41,52]. A similar expansion in ataxin 2 (ATXN2) causes SCA2 [51,53]. The SCAs are a group of autosomal dominantly inherited disorders characterized by ataxia, tremors, and dysarthria with profound cerebellar atrophy [51]. It would be almost a decade before the misfolding of another RBP with a PrLD was linked to the pathogenesis of ALS and FTD.
Transactivation response element DNA-binding protein 43
The first of the RRM- and PrLD-containing proteins to be implicated in neurodegeneration was TDP-43 (trans-activation response element DNA-binding protein 43, see domain architecture in Figure 2) [29,54]. TDP-43 was identified in 2006 as the predominant protein component of the ubiquitinated inclusions observed in ALS patients and a subset of cases of FTD in which there was no observable tau or α-synuclein aggregation [29,55]. TDP-43 is a primarily nuclear protein that shuttles between the nucleus and the cytoplasm, and plays a role in mRNA transport, transcriptional repression, splicing regulation, miRNA biogenesis, stress granule formation, and the stabilization of long intron-containing RNA and long noncoding RNA [56,57]. TDP-43 favors binding to long UG repeats or UG-enriched RNA sequences [58–61]. We now know that TDP-43 is mislocalized to cytoplasmic aggregates in degenerating neurons and glia in roughly 97% of sporadic ALS cases and ~45% of sporadic FTD cases [56,62]. Its mislocalization has been identified as the primary histologic abnormality in cases of inclusion body myositis and a familial form of parkinsonism known as Perry syndrome [29,63]. TDP-43 inclusions are also present in many cases of AD, PD, and HD [29]. Mutations in the gene encoding TDP-43 (TARDBP) have been identified in cases of both familial and sporadic ALS, with mutations segregating with disease in the former, further implicating TDP-43 in the pathogenesis of neurodegeneration [29,64–68]. TARDBP mutations are also found in rare instances of FTD [56,69,70].
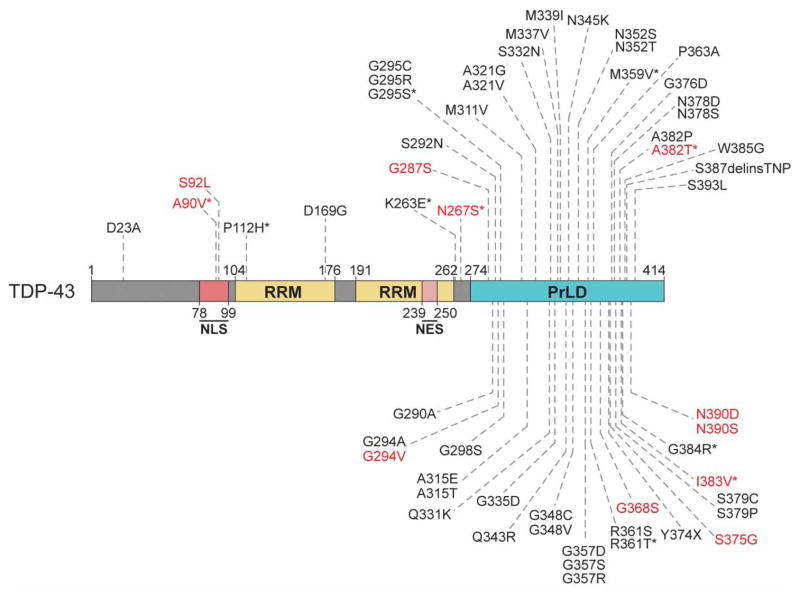
Figure 2. Mutations that cause ALS and FTD cluster in the PrLD of TDP-43.
TDP-43 is an RBP with two canonical RRMs and a C-terminal PrLD [48,77,78,196]. Mutations that have been identified in patients with ALS and FTD are shown, and cluster in the PrLD [71,196]. Mutations identified in patients reported to have features of FTD, with or without a clinical ALS phenotype, are denoted by asterisks [69,219–227]. Mutations in red have also been observed in healthy control individuals [135,226,228–230]. Disease-associated mutations were identified from Buratti [231], Cady et al. [228], Floris et al. 2015 [69], Lagier-Tourenne et al. [71], Peters et al. [196], the ALS data browser (http://alsdb.org) [134], and the ALS Online Genetics Database (http://alsod.iop.kcl.ac.uk/) [232].
The vast majority of these observed mutations are found in the C-terminal PrLD of TDP-43 (Figure 2) [71], which is critical for elements of normal protein function [41]. The PrLD facilitates miRNA biogenesis by mediating interactions with the nuclear Drosha complex, which cleaves pri-miRNAs into pre-miRNAs, and the cytoplasmic Dicer complex, which then cleaves these pre-miRNAs into mature miRNAs [72]. The TDP-43 PrLD mediates protein–protein interactions with other splicing factors, including heterogeneous nuclear ribonucleo-protein A1 (hnRNPA1), hnRNPA2B1, and fused in sarcoma (FUS), and is essential for the regulation of splicing of certain mRNA transcripts [41,73,74]. The PrLD is essential for recruitment of TDP-43 to stress granules [75]. The TDP-43 PrLD is also crucial for aberrant protein aggregation in vitro and in model systems, and select disease-linked mutations accelerate protein aggregation in vitro and in vivo [31,76–79]. Deletion of the PrLD eliminates protein toxicity in model organisms, as does disruption of the RNA-binding ability of TDP-43, suggesting roles for both misfolding and RNA engagement in disease pathogenesis [76,77,80,81].
Fused in sarcoma
Shortly after the connection was made between TDP-43 and disease, another protein with a canonical RRM and a low-complexity PrLD, FUS (see Figure 3 for domain architecture), was linked to both ALS and FTD. Similar to TDP-43 in many ways, FUS, also sometimes known as translocated in liposarcoma (TLS), is a primarily nuclear protein that functions in transcriptional regulation, pre-mRNA splicing, and other elements of mRNA processing and metabolism [56,82]. Notably, though, the most common FUS-binding motif is GUGGU, and the repertoires of RNAs bound by TDP-43 and FUS have little overlap [56]. FUS-binding sites are enriched for 5′-untranslated regions (UTRs), and it has been suggested that FUS also preferentially binds 3′-UTRs and intronic sequences [83,84]. FUS participates in the shuttling of RNA between the nucleus and the cytoplasm, miRNA processing, and the stabilization of long intronic sequences and long noncoding RNAs [56]. FUS interacts with RNA polymerase II and Transcription Factor II D, in addition to other transcription factors, and is thought to have both transcriptional activation and repression activity [56,83]. FUS is recruited to sites of DNA damage and plays an essential role in cellular recovery, including the recruitment of other DNA repair factors [56,83].
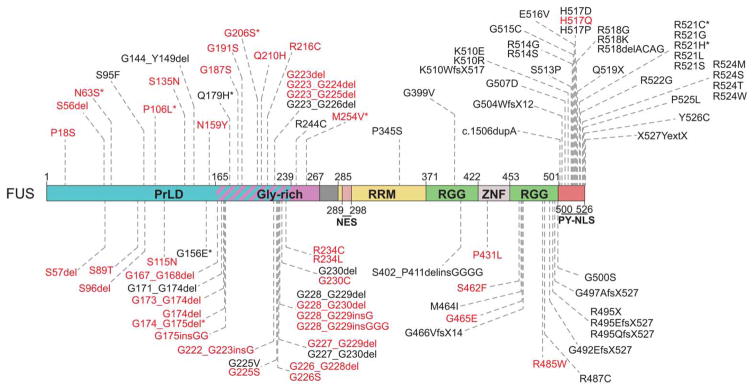
Figure 3. ALS- and FTD-causing mutations in FUS cluster in LC domains and the PrLD.
FUS has an N-terminal PrLD (residues 1–239) that is rich in glutamine, serine, tyrosine, and glycine. Bioinformatic analysis predicts that the PrLD also includes a portion of an adjacent glycine-rich region [93]. FUS has a single RRM, two RGG-rich regions, and a zinc-finger domain [93,196]. Mutations in FUS that have been associated with ALS and FTD cluster in the PrLD, RGG-rich region, and PY-NLS [196,233]. Mutations identified in patients reported to have symptoms of FTD, with or without a clinical ALS phenotype, are denoted by asterisks [234–239]. Mutations in red have also been observed in healthy control individuals [85,86,135,229,236,238,240–244]. Disease-associated mutations were identified from Belzil et al. [240], Corrado et al. [85], Huey et al. [236], Kwiatkowski et al. [86], Lagier-Tourenne et al. [71], Lattante et al. [233], Mackenzie et al. [30], Peters et al. [196], Rademakers et al. [87], Yan et al. [239], the ALS Online Genetics Database (http://alsod.iop.kcl.ac.uk/) [232], and the ALS Data Browser (http://alsdb.org) [135].
Mutations in FUS have been linked to sporadic and familial cases of ALS, and these patients demonstrate the accumulation of FUS-positive inclusions in the cytoplasm of degenerating neurons and glia, and decreased nuclear FUS [5,15,30,85–88]. FUS mutations have caused the earliest reported onset of juvenile-onset ALS reported in children as young as 11 years old [89]. Neuronal and glial FUS aggregates have also been observed in ~9% of FTD cases, and rare mutations in FUS have been identified in FTD patients [5,30,56,90–94]. Of note, nuclear FUS inclusions have been identified in patient neurons in cases of polyglutamine diseases including HD, SCA1, and SCA2 without FUS mutations [56,71].
Putative pathogenic mutations in FUS cluster in the C-terminal proline-tyrosine nuclear localization signal (PY-NLS), the RGG-rich region, and the PrLD (Figure 3) [56,82,93,95]. Studies in model systems have indicated that RNA binding is essential for the toxic effect of FUS, as is the case for TDP-43, but in addition to the RRM and PrLD, a portion of the RGG-rich region is crucial for the aggregation and toxicity of FUS [82,93,96]. ALS-linked FUS mutations confer both gain- and loss-of-function phenotypes [97]. FUS interacts with the U1 snRNP (small nuclear ribonucleoprotein) of the spliceosome and the survival motor neuron (SMN) protein, a component of the complex that enables snRNP biogenesis [71,97]. SMN deficiency causes a childhood motor neuron disease known as spinal muscular atrophy, which is characterized by a reduction in nuclear SMN-containing bodies known as Gems [97]. Similarly, ALS-linked mutations in FUS increase the association of FUS with SMN, leading to a reduction in the abundance of Gems and altered snRNA levels [97]. These pathologic mutations simultaneously decrease FUS binding to the U1 snRNP, resulting in splicing disruptions that phenocopy a partial loss of FUS activity [97].
TATA-binding protein-associated factor 15 and Ewing sarcoma breakpoint region 1
Studies of FUS and TDP-43 pathogenicity highlight not only the fact that ALS and FTD are closely related entities, but also the potential importance of other RBPs with PrLDs in the pathogenesis of neurodegeneration [31,49]. When all human proteins with RRMs were screened for cytoplasmic aggregation and toxicity in yeast, as is seen upon over expression of TDP-43 or FUS in yeast, then filtered based on bioinformatically predicted PrLDs, two proteins, TAF15 (TATA-binding protein-associated factor 15) and EWSR1 (Ewing sarcoma breakpoint region 1), emerged with structural and functional similarities to TDP-43 and FUS [31,48]. TAF15 and EWSR1, along with FUS, belong to a family of proteins known as FET proteins (see Figure 4 for domain architecture) [31,98,99]. As their names imply, FET proteins were originally described as components of pathogenic fusion oncogenes in certain human cancers [99]. Further investigation identified mutations in TAF15 and EWSR1 in patients with sporadic ALS (Figure 4) and revealed that either protein may be found depleted from the nucleus and mislocalized to cytoplasmic neuronal inclusions in ALS and FTD [48,98–100]. Additional evidence for pathogenicity came from in vitro studies demonstrating that both proteins are intrinsically aggregation prone, and ALS-linked TAF15 and EWSR1 mutations accelerate aggregation [48,98]. In addition, both proteins are toxic when overexpressed in the Drosophila nervous system and disease-associated TAF15 mutations cause a more severe phenotype [48,98]. Finally, in cultured mammalian neurons, disease-linked TAF15 and EWSR1 mutations induced formation of cytoplasmic TAF15 and EWSR1 inclusions [48,98].
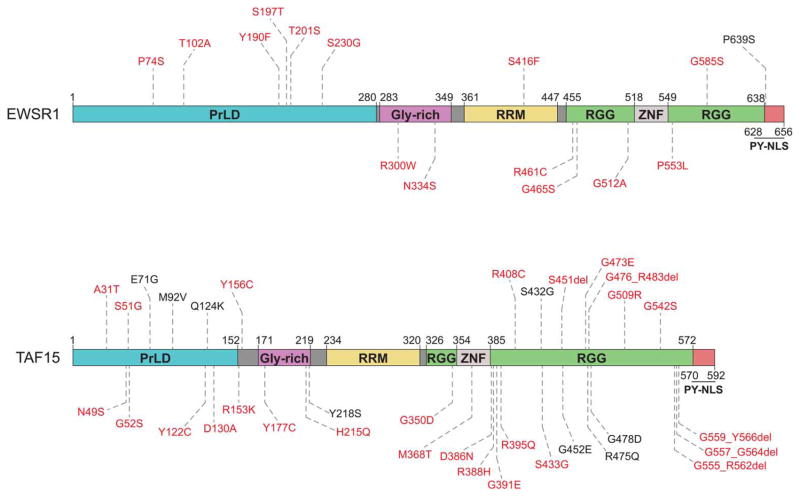
Figure 4. FET proteins EWSR1 and TAF15 have domain architectures similar to the domain architecture of FUS.
FUS, TAF15, and EWSR1 are members of the FET protein family and are similar in domain structure and function [83,98,99]. Like FUS (Figure 3), EWSR1 and TAF15 each have an N-terminal PrLD, a glycine-rich region, and a single RRM, with C-terminal RGG-rich regions, a zinc-finger domain, and a PY-NLS [31,48,98,136,196,245–247]. Mutations shown were identified in ALS patients and compiled from Cady et al. [228], Couthouis et al. [48], Couthouis et al. [98], Couthouis et al. [136], Ticozzi et al. [100], and the ALS Data Browser (http://alsdb.org) [135]. Those in red have also been observed in healthy control individuals [98,135,136,228,229,241].
Mutations in hnRNPA1 and hnRNPA2B1 cause multisystem proteinopathy
More recent information linking PrLDs in the context of RBPs to neurodegeneration has emerged from the study of a rare degenerative syndrome known as multisystem proteinopathy (MSP) [101]. This autosomal, dominantly inherited disorder was formerly known as inclusion body myopathy with Paget’s disease of bone, FTD, and ALS (IBMPFD/ALS) [101,102]. MSP is a heterogeneous, adult-onset disorder that is characterized by a variable presentation, even within the families that it affects [101–103]. Patients may suffer from degeneration of the muscle, bone, brain, motor neurons, or several of these tissues concurrently [101,104]. The most common feature of disease is inclusion body myopathy (IBM), which occurs in ~80–90% of MSP patients and leads to progressive weakness and atrophy, primarily of proximal muscle groups [103,105]. Roughly half of MSP patients will develop Paget’s disease of bone (PDB), a disorder of increased osteoclast activity and bone turnover that is clinically marked by bone pain, pathologic fractures, and skeletal deformities, most often of the skull, vertebrae, and pelvis [103,106]. Cognitive changes and language deficits that define FTD can be observed in a subset of MSP patients, as can be the signs of upper and lower motor neuron dysfunction and electromyo-graphic findings that are hallmarks of ALS [103,104].
There are currently three known genetic causes of MSP [104]. The first identified was valosin-containing protein (VCP), a AAA+ protein (ATPase associated with diverse cellular activities) that participates in many cellular processes including the cell cycle, DNA damage repair, apoptosis, the proteotoxic stress response, post-mitotic Golgi reassembly, endoplasmic reticulum-associated degradation, and ubiquitin-dependent protein degradation [101,102,104,107]. VCP mutations have subsequently been identified in patients with isolated ALS, IBM, and PDB [101,105,106,108]. VCP plays a critical role in the clearance of stress granules via autophagy, and disease-associated VCP variants cause the constitutive formation of stress granules in cell culture, suggesting that aberrant stress granule persistence may contribute to neurodegenerative disease pathogenesis [109].
Exome sequencing and linkage analysis of two MSP-affected families without VCP mutations uncovered pathogenic mutations in the genes encoding heterogeneous nuclear ribonucleoproteins (hnRNPs) A1 and A2B1 (hnRNPA1 and hnRNPA2B1), two RBPs with PrLDs [101,104]. MSP can be caused by a D262V substitution in hnRNPA1 or a D290V substitution in hnRNPA2 [101]. hnRNPA1 and hnRNPA2 (the shorter of two hnRNPA2B1 isoforms by 12 amino acids, which constitutes roughly 90% of hnRNPA2B1 expression in most human tissues) share a domain structure consisting of two N-terminal RRMs and a PY-NLS-containing C-terminal PrLD (Figure 5) [39,101].
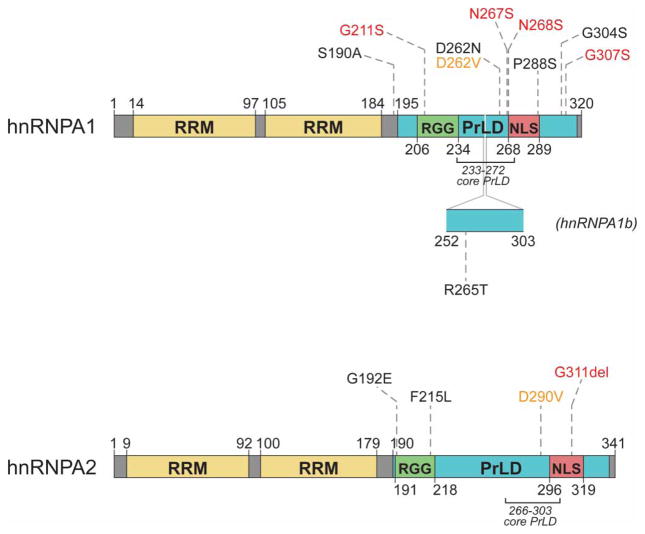
Figure 5. MSP-causing mutations affect a conserved aspartate residue in the hnRNPA1 and hnRNPA2 PrLDs.
hnRNPA1 and hnRNPA2 contain two N-terminal RRMs and a C-terminal PrLD [101]. The PrLDs contain an RGG motif and a PY-NLS that mediates nuclear import [101,110,248]. A 52 amino acid stretch that occurs in the longer isoform of hnRNPA1 (hnRNPA1b) is depicted [249]. Missense mutations in hnRNPA1 and hnRNPA2 that cause MSP are noted in orange [101]. All other mutations were identified in patients with sporadic or familial ALS and compiled from Couthouis et al. [136], Kim et al. [101], Liu et al. [137], and the ALS Data Browser (http://alsdb.org) [135]. Those in red have been observed in healthy control individuals [229].
hnRNPA1 is an abundantly and ubiquitously expressed, primarily nuclear RBP that functions widely in nucleic acid processing [110]. hnRNPA1 binds to promoter sequences or transcription factors to either activate or repress transcription and contributes to the regulation of alternative splicing and splice-site selection, often promoting exon skipping [110–113]. It can shuttle between the nucleus and cytoplasm, facilitating nuclear mRNA export [110]. In addition to showing affinity for specific motifs including UAGGGA, UAGA, UAGG, and UGGGGU [110,114,115], hnRNPA1 binds AU-rich elements (containing AUUUA motifs) that are known to modulate the stability and degradation of mature mRNA transcripts [110,116]. hnRNPA1 also binds to internal ribosomal entry sites to regulate translation [117,118], is critical for telomere biogenesis and length maintenance [110,119], and participates in miRNA processing [120,121]. Like hnRNPA1, hnRNPA2B1 is one of the most abundantly expressed proteins in the cell and is predominantly nuclear with the ability to shuttle between the nucleus and cytoplasm [114]. It has functional similarities to hnRNPA1, including roles in the regulation of alternative pre-mRNA splicing and translation [122–124], mRNA stability [124], and telomere maintenance [125,126]. Distinct from hnRNPA1, hnRNPA2B1 also plays a crucial role in mRNA trafficking in neurons and oligodendrocytes [127,128]. Like hnRNPA1, hnRNPA2B1 has a significant binding preference for UAG motifs [115].
Recent studies of hnRNPA2B1 function in mouse spinal cord, patient fibroblasts, and motor neurons derived from human induced pluripotent stem cells (iPSCs) identified an enriched UAGG-binding motif in CNS tissue [124]. hnRNPA2B1-binding sites were particularly enriched in 3′-UTRs in vivo and in cultured cells, and hnRNPA2B1 was found to contribute to polyadenylation site selection [124]. The importance of hnRNPA2B1 to pre-mRNA splicing was illustrated by altered proportions of the long and short isoforms of the murine protein Dao upon depletion of hnRNPA2B1 in the mouse CNS [124]. The human homolog, DAO, which encodes D-amino acid oxidase, is highly expressed in the CNS and has been implicated in familial ALS [124,129,130]. Loss of hnRNPA2B1 expression in the mouse model causes increased proportional expression of a short Dao isoform that is degraded by the proteasome and has ~85% less enzymatic activity than the longer isoform [124]. Importantly, the splicing changes that result from the MSP-causing substitution, D290V, in hnRNPA2B1 in patient fibroblasts are distinct from those that occur due to loss of hnRNPA2B1 function [124]. In contrast, the splicing changes caused by the D290V substitution in hnRNPA2B1 have a ~66% overlap with splicing alterations observed in fibroblasts from patients with an MSP-causing mutation in VCP [101]. This finding suggests a possible etiology for the shared disease phenotype caused by mutations in VCP and hnRNPA2B1.
MSP-linked hnRNPA1 and hnRNPA2B1 mutations enhance protein aggregation
hnRNPA1 and hnRNPA2 have a common domain architecture consisting of two N-terminal RRMs and a C-terminal PrLD containing a PY-NLS that mediates nuclear import (Figure 5) [101,110]. Interestingly, both MSP-linked mutations involve a valine substitution at a conserved gatekeeper aspartate residue in the PrLD that is computationally predicted, by two separate algorithms, to increase prionogenicity (Figure 5) [39,40,101]. Additionally, an algorithm that scores the ability of hexapeptides to form amyloid fibrils primarily based on structural information rather than amino acid sequence predicts that each of these mutations lies within a ‘steric-zipper’ motif (Figure 6) [101,131]. Steric zippers are defined as two self-complementary β-sheets with the ability to act as the backbone of an amyloid fibril [131]. The aspartate-to-valine substitution in this region is predicted to strengthen a steric zipper, making the protein more prone to fibrillization (Figure 6) [101]. Indeed, both hnRNPA1 and hnRNPA2 form fibrils in vitro that are self-seeding (i.e. can nucleate the aggregation of soluble protein), thereby reducing the lag phase of assembly, and the disease-associated mutations greatly accelerate fibrillization [101,132]. In vitro, the mutant proteins are capable of seeding their own assembly and the assembly of the corresponding wild-type protein [101], providing a potential explanation for the genetic dominance of MSP mutations. A heterozygous individual would produce both wild-type and mutant protein. However, if the presence of the aspartate-to-valine substitution accelerates the misfolding of the mutant protein, and the misfolding of the mutant protein can nucleate the misfolding of the wild-type protein, the presence of the wild-type allele would not be protective against the development of a disease phenotype.
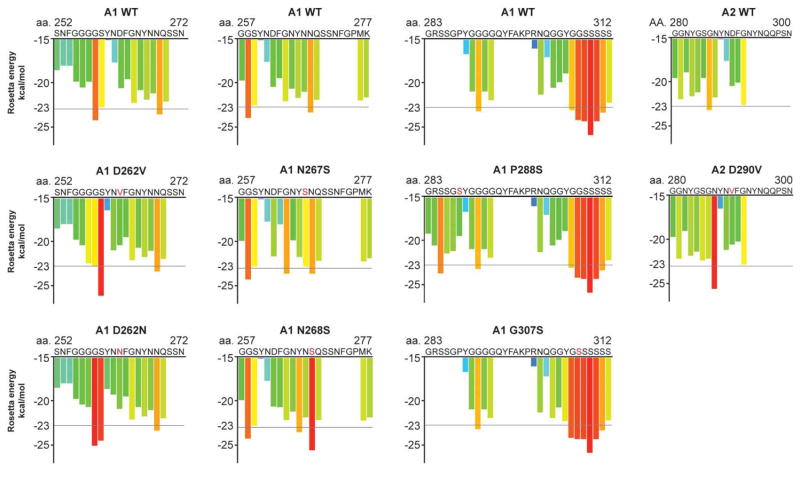
Figure 6. MSP- and ALS-associated mutations are predicted to increase the fibrillization propensity of hnRNPA1 and hnRNPA2.
ZipperDB, a structure-based algorithm, calculates the propensity of hexapeptide fragments to form steric zippers [131]. Steric zippers, which are self-complementary β-sheets that form the backbone of an amyloid fibril, are predicted to form when the Rosetta energy of a hexapeptide is below the empirically determined ‘high fibrillization propensity’ threshold of −23 kcal/mol [131]. Several of the described mutations in hnRNPA1 and hnRNPA2 introduce a predicted steric zipper motif or strengthen an existing zipper [101,131]. For example, the D262V substitution in hnRNPA1 creates a potent SYNVFG zipper, whereas the D262N substitution also strengthens the GSYNDF zipper [101,131].
Muscle biopsies from MSP patients with mutations in VCP, hnRNPA1, or hnRNPA2B1 share cytopathologic features including the cytoplasmic aggregation of TDP-43, which has also been observed in sporadic IBM in addition to ALS and FTD [101,103,105,133]. A biopsy from an affected individual in the family harboring the hnRNPA2D290V variant also demonstrated mislocalization of hnRNPA2 from the nucleus to cytoplasmic inclusions, and in muscle fibers obtained from a patient expressing hnRNPA1D262V, both hnRNPA1 and hnRNPA2 were cleared from myonuclei and localized to sarcoplasmic inclusions [101]. Motor neurons differentiated from iPSCs from MSP patients with hnRNPA2D290V or VCPR155H variants demonstrate nuclear hnRNPA2B1 aggregation [124]. Concurrent mislocalization and partial colocalization of TDP-43 and hnRNPA1 or TDP-43 and hnRNPA2 could be observed in muscle fibers of MSP-affected patients [101]. Cytoplasmic hnRNPA1- and hnRNPA2-positive aggregates have also been identified in sporadic cases of IBM [101,134]. The intersection of protein pathologies in MSP and IBM underscores the fact that there is much to be learned about common degenerative diseases from more rare, familial disorders.
Sequencing efforts to uncover pathogenic mutations in familial and sporadic ALS patients have identified additional mutations in hnRNPA1 and hnRNPA2 linked to ALS [101,135,136]. A substitution (D262N) occurring in a familial case of ALS affects the same aspartate residue implicated in the pathogenesis of MSP [101]. The D262N substitution in hnRNPA1 introduces a strong steric zipper and strengthens an existing steric zipper (Figure 6) [101,131]. Similar to the D262V substitution, D262N significantly reduced the lag phase of fibrillization and accelerated hnRNPA1 aggregation in vitro [101]. Several other mutations in hnRNPA1 that have been identified in patients with ALS also introduce or strengthen steric zipper motifs (Figures 5 and 6) [131]. One of these, a substitution in the PY-NLS of hnRNPA1 (P288S) was recently identified as the cause of a familial case of flail-arm ALS (Figure 5) [137]. The location of this mutation suggests that hnRNPA2P288S may have impaired nuclear import, leading to increased cytoplasmic mislocalization in addition to increased fibrillization propensity.
Many questions remain, however, about the extent and prevalence of hnRNPA1 and hnRNPA2 pathology in patients with MSP and sporadic forms of ALS and FTD. Mislocalized hnRNPA1 and hnRNPA2 inclusions have been observed in muscle fibers of patients with MSP, but can the clearance of these proteins from the nucleus to cytoplasmic foci be observed also in motor neurons of the brain and spinal cord and in the frontal and temporal cortical lobes of these patients? It remains unclear how this disease manifests in such a heterogeneous way among patients with the same mutation, and it would be informative to investigate, via postmortem biopsy, whether patients who developed muscle and bone pathology, for example, but no clinical dementia demonstrated evidence of asymptomatic protein pathology in the frontal cortex. Also of relevance would be a study of ALS patients with TDP-43 or FUS mutations and pathology to look for co-occurrence of hnRNPA1 or hnRNPA2 pathology. Wild-type TDP-43 aggregates along with hnRNPA1 and hnRNPA2 in MSP [101], suggesting the possibility that wild-type hnRNPA1 and hnRNPA2 may be present in the inclusions driven by mutations in other RBPs in ALS and FTD patients. A single study of frontal cortex from 10 patients with FTD and TDP-43 pathology showed no mislocalization of hnRNPA1 or hnRNPA2 [138]. Importantly, one of these patients harbored a familial VCP mutation [138]. Thus, VCP mutations are not always accompanied by hnRNPA1 and hnRNPA2 pathology as they can be in MSP.
Finally, the contribution of hnRNPA1 and hnRNPA2 mutations to the overall landscape of neurodegeneration is currently unknown in that we do not yet know how frequently these mutations occur or how penetrant they are. The discovery of hnRNPA1 and hnRNPA2 mutations in MSP was rapidly followed by the identification of additional hnRNPA1 and hnRNPA2 mutations in patients with sporadic and familial ALS [101,135], and we expect the number of patients suffering from neurodegenerative phenotypes with identified mutations in hnRNPA1 or hnRNPA2 to grow as our knowledge of disease increases. We also anticipate that additional RBPs with PrLDs will emerge in degenerative diseases [31,41]. Indeed, mutations in the PrLD of hnRNPDL, leading to D378N or D378H substitutions, have now been linked to limb-girdle muscular dystrophy type 1G [139].
Disease-associated RBPs are involved in the formation of RNP granules
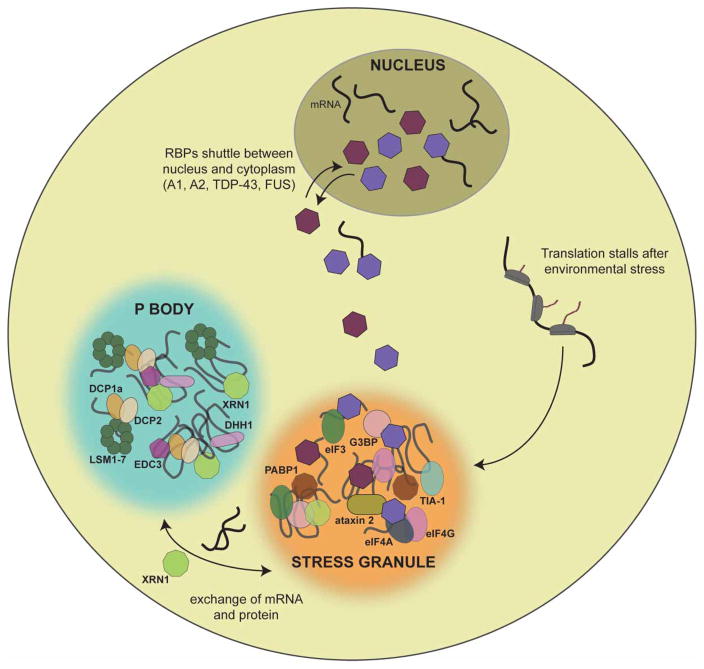
An important shared feature of ATXN2, TDP-43, FUS, hnRNPA1, hnRNPA2, EWSR1, and TAF15 is their recruitment to stress granules upon cellular exposure to environmental stresses like heat shock, infection, ischemia, or oxidative stress [49,101,140]. Stress granules are RNP granules that assemble in the cytoplasm in stress conditions and incorporate nontranslating polyadenylated mRNA transcripts, translation initiation factors, small ribosome subunits, and RBPs (Figure 7) [49,141]. They are sites of translation suppression, consisting of stalled translation–initiation complexes and translational-silencing proteins in addition to other regulators of RNA metabolism, and serve to redirect cellular energy and resources towards the production of cytoprotective proteins that will be essential for survival and recovery after stress [49,140,142,143]. Processing bodies (P bodies) are a related class of RNP granules that are constitutively assembled in addition to being induced by cellular stress (Figure 7) [140]. P bodies are cytosolic sites of mRNA decay that interact with stress granules, allowing for possible exchange of mRNAs and proteins between assemblies [49,140,142,144,145]. Crucial to the reversible assembly of RNP granules is the intermolecular association of PrLDs or other LCDs via multiple weak, transient interactions as target RNAs are engaged, primarily via RNA-binding domains [49,101,143,146]. In some cases, as with hnRNPA1, PrLDs can also bind RNA, frequently via RGG motifs [147,148]. In other cases, as with FUS, the PrLD does not bind to RNA directly [149]. The PrLD of the mammalian stress granule protein T-cell intracellular antigen 1 (TIA1) [150] is required for incorporation into chemically induced stress granules [151]. In yeast, a reduction in the recruitment of prion-like proteins Lsm4 and Pop2 to P-bodies is observed in the absence of their PrLDs [152]. Therefore, despite their propensity for misfolding events, PrLDs have likely been preserved throughout evolution in part because they enable essential protein–protein interactions that provide the fluid architecture of membraneless cellular compartments [41]. In addition to stress granules and P bodies, germ granules are cytoplasmic RNP bodies found in the cytoplasm [153]. Membraneless organelles that contribute to nuclear organization include nucleoli, paraspeckles, gems, Cajal bodies, and promyelocytic leukemia (PML) bodies [41,125].
Figure 7. Cytoplasmic RNP granules include stress granules and P bodies.
Stress granules are cytoplasmic assemblies that form in response to environmental stress and are sites of stalled translation initiation [49,140,144]. They contain polyadenylated mRNA transcripts, RBPs, translation initiation factors, and small ribosomal subunits [141]. P bodies are constitutively present but also form in response to stressful conditions [140]. They serve as sites of mRNA degradation and are characterized by the elements of the mRNA decapping and decay machinery [49,140]. Shown are many protein components of mammalian stress granules and P bodies [49,140,144].
Remarkably, many RBPs with PrLDs, which have not yet been connected to disease, are emerging as critical scaffolds for the formation of these membraneless organelles. For example, the PrLD of RBM14 (as well as FUS) is critical for paraspeckle formation [154]. Likewise, the PrLD of hnRNPD plays an important role in Sam68 nuclear body formation [155], whereas the PrLD of Xvelo is critical for Balbiani body formation [156,157]. Finally, PrLDs in DAZ1-4 and DAZL are predicted to have important roles in the formation of amyloid-like structures that regulate key meiotic events [158,159]. We anticipate that PrLDs in RNA/DNA-binding proteins will continue to surface as key scaffolds for various membraneless organelles. PrLDs in proteins that do not bind nucleic acids will also likely serve as scaffolds in other contexts. For example, the PrLD of Pin2 can function as a trans-Golgi network retention motif by driving the assembly of higher order complexes [160].
A role for the alteration of RNP granule dynamics in neurodegenerative pathology is suggested by studies showing that disease-associated mutant proteins are recruited differently to RNP granules than their wild-type counterparts [95,101,141,150,161–162]. Moreover, changes in the expression of RNP granule components modify the effects of toxic neurodegenerative disease RBPs in model systems [163]. hnRNPA1 and hnRNPA2 are nuclear when expressed in HeLa cells, but are incorporated into cytoplasmic stress granules upon arsenite stress, and recruitment of hnRNPA1D262V and hnRNPA2D290V occurs more rapidly than relocalization of the wild-type proteins [101]. The D290V substitution also enhances hnRNPA2 recruitment to stress granules in motor neurons derived from MSP-patient iPSCs [124]. A VCP mutation that also causes MSP has the same effect on hnRNPA2 [124]. The fact that these mutations promote the targeting of RBPs to stress granules, while VCP mutations can also decrease stress granule clearance [108], suggests a model in which MSP can be caused by any perturbation that shifts the equilibrium of dynamic stress granule formation and dissolution towards granule formation or persistence. In cultured cells, familial ALS mutations cause increased formation of TDP-43 inclusions that are also positive for stress granule markers after exposure to environmental stress [49,150]. FUS variants, too, show enhanced association with stress granule markers in cytoplasmic inclusions [49,95,141,161,162]. In a yeast model of TDP-43 proteinopathy, overexpression of several RNP granule components, including Tis11, Hrp1, Vts1, Kem1, and Pbp1, either enhanced or suppressed the toxicity of TDP-43 expression [163].
Pbp1 is a stress granule protein that interacts with Pab1, also a component of stress granules, and regulates mRNA polyadenylation [80,164]. Interestingly, Pbp1 is the yeast homolog of human ATXN2, which bears a polyglutamine expansion in SCA2 [80]. Deletion of Pbp1 diminishes stress granule formation and suppresses TDP-43 toxicity in yeast, whereas overexpression of Pbp1 enhances TDP-43 toxicity in yeast [80]. The Drosophila homolog, Atx2, also has a dose-dependent effect on TDP-43 toxicity in the fly nervous system, with a reduction in Atx2 expression reducing the toxic TDP-43 phenotype [80]. Further analysis revealed that TDP-43 and ATXN2 physically interact in yeast and humans in an RNA-dependent manner [80]. Furthermore, ATXN2 forms abnormal cytoplasmic foci in ALS and FTD patient neurons, and TDP-43 inclusions can be found in cerebellar Purkinje cells and brainstem nuclei in SCA2 [80]. Genetically, mutations in ATXN2 are the most common known risk factor for ALS [80]. Polyglutamine expansions of >34 repeats cause SCA2, but intermediate length expansions from 27 to 33 glutamines in length were found to increase the likelihood of developing ALS by a factor of ~2.8 [80,165–167].
In Drosophila, increased expression of the stress granule protein polyA-binding protein (PABP) causes more severe TDP-43-induced retinal degeneration [163]. The cytoplasmic human homolog PABPC1 was observed in cytoplasmic inclusions in the motor neurons of ALS patients, despite having a predominantly diffuse pattern of localization in healthy controls [163]. RNP granule markers have also been found to modify FUS toxicity in model systems [93]. Overexpression of stress granule proteins Pab1, Tif2, Tif3, and Tis11 in a yeast model suppressed the toxic effect of FUS overexpression [93,163,164]. The human homolog of Tif2, EIF4A1, is similarly able to suppress FUS toxicity in cultured mammalian cells [93]. FUS toxicity in yeast is also mitigated by over-expression of the P body protein Edc3 or Sbp1, which localizes to both stress granules and P bodies [93,164,168]. Both Edc3 and Sbp1 promote mRNA decapping prior to 5′-to-3′ degradation [164,169]. Deletion of the stress granule protein Pub1 or the P-body protein Lsm7 decreases FUS toxicity in yeast [93,164]. In P bodies, Lsm7 is part of a heteroheptameric complex consisting of Lsm proteins 1–7 [170,171]. The Lsm1–7 proteins activate mRNA decapping and protect mRNA from trimming, a process by which transcripts are shortened by 10–20 nucleotides at the 3′-end [170–172]. Lsm7 also participates in pre-mRNA splicing as a component of a nuclear complex consisting of Lsm proteins 2–8 [170,173,174]. This heptamer stabilizes newly synthesized U6 snRNA by binding to its 3′-end [170,173,174]. The Lsm2–8 complex also contributes to mRNA degradation in the nucleus by targeting nuclear RNAs for decapping [170,175].
Stress granule formation in yeast is diminished by deletion of Pbp1, the yeast homolog of ATXN2, or Pub1, the yeast homolog of the human protein TIA1 [176]. TIA1 is required for mammalian stress granule formation, and reduced ATXN2 expression results in reduced stress granule assembly [177,178]. TIA1 and another stress granule marker, EIF3, have been identified in the proteinaceous inclusions in the brain and spinal cord tissue of patients with ALS and FTD [95,150]. TIA1 is a protein containing RRMs and a PrLD that is essential for stress granule formation in cultured mammalian cells [49,151]. A mutation in TIA1 causes Welender distal myopathy, and mutant TIA1 expression leads to increased stress granule abundance in cultured cells, suggesting that altered stress granule dynamics may underpin this slowly progressive, adult-onset disorder [49,179,180].
Phase transitions underpin RNP granule formation and misregulation
It is now thought that RNP granule components coalesce into membraneless compartments through phase transitions that drive the reversible formation of liquid droplets or more solid hydrogel states [41,49,181–183]. Several RNP granules have been shown to have liquid-like properties, including P granules in Caenorhabditis elegans, P bodies in Saccharomyces cerevisiae, PML nuclear bodies, and mammalian stress granules and P bodies [184–186]. These compartments are spherical, can fuse with one another and relax into a new sphere, and undergo rapid internal rearrangement as demonstrated by half-bleaching experiments [184,185,187]. Liquid droplets form via liquid–liquid phase separation (LLPS), or the ‘demixing’ of the granule components and the cytoplasm, a process modeled by the separation of standing oil and vinegar [187]. Recent work has shown that the liquid droplet environment promotes certain biochemical reactions, including the stabilization of RNA hairpins and the unwinding of double-stranded nucleic acids [188]. The liquid interior is therefore a specialized microcosm for certain nucleic acid remodeling reactions [182]. Liquid droplets create a controlled environment by permitting or restricting entry of proteins based on amino acid sequence [188].
The transition from soluble protein to liquid droplet is characteristically driven by intrinsically disordered proteins and can be mediated by a multitude of intermolecular interactions [146,189]. Disordered LCDs, including PrLDs, associate with each other via weak, nonspecific interactions in a manner that can be concentration-dependent [146,149,190]. RNA binding via RRMs or PrLDs can facilitate additional multivalent interactions, explaining the observation that the protein concentration required for the formation of hnRNPA1 droplets is decreased in the presence of RNA [146,190]. Interactions between disordered regions of the P-granule protein, Ddx4, are mediated by electrostatic interactions resulting from patterned blocks of residues of alternating net charge [146,191]. Structural analysis of the LCD of FUS in the liquid phase-separated state demonstrates that it retains a disordered character within droplets, suggesting that interactions among PrLDs within liquid droplets are likely to be transient with frequent reorientations [41,149].
Hydrogels have solid-like properties, a cross-linked structure, a high water content, and water-soluble components [187]. Stress granules in yeast are gel-like, highlighting the biological relevance of this form of protein assembly [146,185]. LCDs can also facilitate the transition to the gel phase [181,192,193]. In vitro, the PrLDs of FUS, hnRNPA1, and hnRNPA2 all form hydrogels that are composed of amyloid-like fibrils [41,190,192]. These hydrogel structures are capable of trapping homotypic and heterotypic LCDs [192]. FUS LCD hydrogels, for example, bind and retain, with varying avidities, soluble FUS LCDs and the LCDs of hnRNPA1, hnRNPA2, TDP-43, and TIA1 [192]. The role of hydrogel structures in normal RNP granule assembly in mammalian cells has been controversial [185,194]. One recent model of mammalian stress granules suggests that, rather than being pure liquid droplets, stress granules are composed of a liquid-like exterior containing an internal gel-like core [41,194].
Recent evidence suggests that inappropriate phase transitions nucleated by RNP granules may represent a crucial element of the pathogenesis of neurodegenerative disease [190,195]. In vitro experiments exploring LLPS of FUS and hnRNPA1 indicate that, over time, liquid droplets are prone to ‘mature’ and undergo a liquid-to-solid transition involving protein fibrillization [190,195]. This process is accelerated by pathologic PrLD mutations [190,195]. Mutations in the PrLD of FUS also reduce the reversibility of FUS hydrogel formation [193]. This suggests a model in which disease-causing FUS mutations, which tend to cluster in the PrLD, RGG-rich regions, and NLS [196], enhance fiber formation within droplets via one of two mechanisms. First, PrLD mutations likely serve to directly increase the propensity of FUS liquids to transition into irreversible aggregates [193,195]. Second, NLS mutations, or frameshift mutations that disrupt the NLS (Figure 3), may function to increase cytoplasmic FUS concentration by decreased nuclear import, driving liquid droplet formation, persistence, and maturation to fibrous structures [41,195]. Importantly, though, mutations in the FUS NLS can also directly alter the dynamics of phase transitions [193]. When purified FUS with and without mutations in the PY-NLS was induced by a temperature shift to form liquid droplets in vitro, mutant FUS droplets persisted longer than those composed of wild-type FUS [193]. Thus, mutations in regions outside the LCD may contribute to pathologic persistence of RNP granules leading to aberrant fibril formation.
The most common cause of ALS and FTD is a hexanucleotide repeat expansion in a noncoding region of C9ORF72 [197,198]. This expansion leads to the RAN translation of several dipeptide repeat proteins, including poly-(Pro-Arg) (PR) and poly-(Gly-Arg) (GR), which form nuclear and cytoplasmic inclusions in the brain and spinal cord of ALS/FTD patients harboring this expansion [198]. LCDs, such as those containing hnRNPA1, hnRNPA2, and other RNP granule components, are a preferred binding target of PR and GR, which can disrupt granule dynamics [198,199]. GR50 or PR50 expression in cultured cells caused spontaneous assembly of persistent stress granules [198]. GR20 or PR20 reduced the concentration required for hnRNPA1 LLPS and led to the formation of droplets with reduced fluidity [198].
Therapeutic protein disaggregases to counter aberrant phase transitions
A therapeutic agent with the ability to counteract pathologic phase transitions could have tremendous utility across neurodegenerative diseases caused by misfolding events related to RNP granule dysfunction. One approach would identify a small molecule or RNA that could preserve the liquid–granule state by preventing the transition to solid aggregates. An agent that could actively reverse the liquid-to-solid phase transition would be especially appealing for patients with active disease. Hsp104 is a hexameric protein disaggregase in the AAA + ATPase family [200–202]. It is found in yeast and has homologs across eubacteria and eukaryotic species, but no metazoan ortholog exists [200,203]. Hsp104 preserves proteostasis and promotes survival in S. cerevisiae by renaturing aggregated proteins and returning them to their native conformations after exposure to environmental stress [200,203,204]. It also has the ability to rapidly remodel amyloid fibers and prefibrillar oligomers and, in doing so, regulates prionogenesis and the propagation and elimination of yeast prion conformers [201,204–208]. In S. cerevisiae, Hsp104 also functions in the dissolution of stress granules and the maintenance of the liquid-like properties of P bodies [185]. Hsp104 contributes to the proper targeting of P body components, which mislocalize to stress granules in its absence [185]. As a potential therapeutic, Hsp104 has shown promise in several models of neurodegenerative disease [8,204,209]. In a rat model of PD, expression of Hsp104 decreased dopaminergic neuron loss and accumulation of α-synuclein aggregates in the substantia nigra of animals expressing a PD-linked α-synuclein variant [8]. Hsp104 increased lifespan and reduced the number of cortical polyglutamine inclusions in a mouse model of HD [209]. Potentiated Hsp104 variants with enhanced ATPase activity reduce protein aggregation and suppress toxicity of TDP-43, FUS, and α-synuclein in S. cerevisiae [203,204,210–212]. Enhanced Hsp104 variants also protect against dopaminergic neuron loss in a C. elegans model of PD [204]. These studies suggest that Hsp104 has broad activity against neurodegenerative disease substrates, and its substrate repertoire can be expanded or sharpened using engineering strategies.
Finally, it will also be important to determine whether endogenous human protein disaggregases, including Hsp110, Hsp70, Hsp40, and small heat-shock proteins [213–215]; HtrA1 [216]; and NMNAT2 plus Hsp90 [217], also display activity against disease-linked RBPs with PrLDs. These protein disaggregase systems could also be engineered to possess enhanced disaggregase activity against disease-linked RBPs with PrLDs [218]. Moreover, small-molecule drugs that enhance the activity of these systems could be useful therapeutics aimed at restoring homeostasis of RBPs with PrLDs [218]. We anticipate that harnessing the power of protein disaggregases could lead to important advances in treating several devastating diseases caused by aberrant phase transitions of RBPs with PrLDs [218].
Acknowledgments
We thank Korrie Mack, Zachary March, and Lin Guo for comments on the manuscript. We acknowledge the NHLBI GO Exome Sequencing Project and its ongoing studies which produced and provided exome variant calls for comparison: the Lung GO Sequencing Project [HL-102923], the WHI Sequencing Project [HL-102924], the Broad GO Sequencing Project [HL-102925], the Seattle GO Sequencing Project [HL-102926], and the Heart GO Sequencing Project [HL-103010]. A.F.H. was supported by a Center for Neurodegenerative Disease Research T32 Training Grant [National Institutes of Health/National Institute on Aging AG00255] and an F31 fellowship [National Institute of Neurological Disorders and Stroke F31NS087676]. J.S. is supported by the National Institutes of Health [R01GM099836 and R21NS090205], the Life Extension Foundation, a Sanofi Innovation award, the ALS Association, the Muscular Dystrophy Association, Target ALS, and the Robert Packard Center for ALS Research at Johns Hopkins.
Abbreviations
- AD
Alzheimer’s disease
- ALS
amyotrophic lateral sclerosis
- ATXN1
ataxin 1
- ATXN2
ataxin 2
- CNS
central nervous system
- EWSR1
Ewing sarcoma breakpoint region 1
- FTD
frontotemporal dementia
- FTLD
frontotemporal lobar degeneration
- FUS
fused in sarcoma
- GO
gene ontology
- GR
poly-(Gly-Arg)
- HD
Huntington’s disease
- hnRNPs
heterogeneous nuclear ribonucleoproteins
- IBM
inclusion body myopathy
- iPSCs
induced pluripotent stem cells
- LCDs
low-complexity domains
- LLPS
liquid–liquid phase separation
- MSP
multisystem proteinopathy
- P bodies
processing bodies
- PABP
polyA-binding protein
- PD
Parkinson’s disease
- PDB
Paget’s disease of bone
- PML
promyelocytic leukemia
- PR
poly-(Pro-Arg)
- PrLD
prion-like domain
- PY-NLS
proline-tyrosine nuclear localization signal
- RAN
repeat-associated non-ATG
- RBPs
RNA-binding proteins
- RRM
RNA recognition motif
- SCA8
spinocerebellar ataxia type 8
- SDS
sodium dodecyl sulfate
- SMN
survival motor neuron
- snRNP
small nuclear ribonucleoprotein
- TAF15
TATA-binding protein-associated factor 15
- TDP-43
transactivation response element DNA-binding protein 43
- TIA1
T-cell intracellular antigen 1
- TLS
translated in liposarcoma
- UTRs
untranslated regions
- VCP
valosin-containing protein
Footnotes
Competing Interests
The Authors declare that there are no competing interests associated with the manuscript.
References
- 1.Khurana V, Lindquist S. Modelling neurodegeneration in Saccharomyces cerevisiae: why cook with baker’s yeast? Nat Rev Neurosci. 2010;11:436–449. doi: 10.1038/nrn2809. [DOI] [PubMed] [Google Scholar]
- 2.Lepesant JA. The promises of neurodegenerative disease modeling. C R Biol. 2015;338:584–592. doi: 10.1016/j.crvi.2015.06.018. [DOI] [PubMed] [Google Scholar]
- 3.Swinnen B, Robberecht W. The phenotypic variability of amyotrophic lateral sclerosis. Nat Rev Neurol. 2014;10:661–670. doi: 10.1038/nrneurol.2014.184. [DOI] [PubMed] [Google Scholar]
- 4.Li YQ, Tan MS, Yu JT, Tan L. Frontotemporal lobar degeneration: mechanisms and therapeutic strategies. Mol Neurobiol. 2016;53:6091–6105. doi: 10.1007/s12035-015-9507-5. [DOI] [PubMed] [Google Scholar]
- 5.Ferrari R, Kapogiannis D, Huey ED, Momeni P. FTD and ALS: a tale of two diseases. Curr Alzheimer Res. 2011;8:273–294. doi: 10.2174/156720511795563700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bunner KD, Rebec GV. Corticostriatal dysfunction in Huntington’s disease: the basics. Front Hum Neurosci. 2016;10:317. doi: 10.3389/fnhum.2016.00317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gitler AD. Beer and bread to brains and beyond: can yeast cells teach us about neurodegenerative disease? Neurosignals. 2008;16:52–62. doi: 10.1159/000109759. [DOI] [PubMed] [Google Scholar]
- 8.Lo Bianco C, Shorter J, Régulier E, Lashuel H, Iwatsubo T, Lindquist S, et al. Hsp104 antagonizes α-synuclein aggregation and reduces dopaminergic degeneration in a rat model of Parkinson disease. J Clin Invest. 2008;118:3087–3097. doi: 10.1172/JCI35781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Skovronsky DM, Lee VMY, Trojanowski JQ. Neurodegenerative diseases: new concepts of pathogenesis and their therapeutic implications. Annu Rev Pathol Mech Dis. 2006;1:151–170. doi: 10.1146/annurev.pathol.1.110304.100113. [DOI] [PubMed] [Google Scholar]
- 10.Liscic RM, Grinberg LT, Zidar J, Gitcho MA, Cairns NJ. ALS and FTLD: two faces of TDP-43 proteinopathy. Eur J Neurol. 2008;15:772–780. doi: 10.1111/j.1468-1331.2008.02195.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mathis S, Couratier P, Julian A, Vallat JM, Corcia P, Le Masson G. Management and therapeutic perspectives in amyotrophic lateral sclerosis. Expert Rev Neurother. 2017;17:263–276. doi: 10.1080/14737175.2016.1227705. [DOI] [PubMed] [Google Scholar]
- 12.Cummings J, Aisen PS, DuBois B, Frölich L, Jack CR, Jr, Jones RW, et al. Drug development in Alzheimer’s disease: the path to 2025. Alzheimers Res Ther. 2016;8:39. doi: 10.1186/s13195-016-0207-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Farzanehfar P. Towards a better treatment option for Parkinson’s disease: a review of adult neurogenesis. Neurochem Res. 2016;41:3161–3170. doi: 10.1007/s11064-016-2053-3. [DOI] [PubMed] [Google Scholar]
- 14.Bose S, Cho J. Targeting chaperones, heat shock factor-1, and unfolded protein response: promising therapeutic approaches for neurodegenerative disorders. Ageing Res Rev. 2017;35:155–175. doi: 10.1016/j.arr.2016.09.004. [DOI] [PubMed] [Google Scholar]
- 15.Da Cruz S, Cleveland DW. Understanding the role of TDP-43 and FUS/TLS in ALS and beyond. Curr Opin Neurobiol. 2011;21:904–919. doi: 10.1016/j.conb.2011.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Forman MS, Trojanowski JQ, Lee VMY. Neurodegenerative diseases: a decade of discoveries paves the way for therapeutic breakthroughs. Nat Med. 2004;10:1055–1063. doi: 10.1038/nm1113. [DOI] [PubMed] [Google Scholar]
- 17.Braak H, Del Tredici K, Rüb U, de Vos RAI, Jansen Steur ENH, Braak E. Staging of brain pathology related to sporadic Parkinson’s disease. Neurobiol Aging. 2003;24:197–211. doi: 10.1016/S0197-4580(02)00065-9. [DOI] [PubMed] [Google Scholar]
- 18.Cushman M, Johnson BS, King OD, Gitler AD, Shorter J. Prion-like disorders: blurring the divide between transmissibility and infectivity. J Cell Sci. 2010;123:1191–1201. doi: 10.1242/jcs.051672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Giasson BI, Lee VMY, Trojanowski JQ. Interactions of amyloidogenic proteins. Neuromol Med. 2003;4:49–58. doi: 10.1385/NMM:4:1-2:49. [DOI] [PubMed] [Google Scholar]
- 20.Ash PEA, Bieniek KF, Gendron TF, Caulfield T, Lin WL, Dejesus-Hernandez M, et al. Unconventional translation of C9ORF72 GGGGCC expansion generates insoluble polypeptides specific to c9FTD/ALS. Neuron. 2013;77:639–646. doi: 10.1016/j.neuron.2013.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bañez-Coronel M, Ayhan F, Tarabochia AD, Zu T, Perez BA, Tusi SK, et al. RAN translation in Huntington disease. Neuron. 2015;88:667–677. doi: 10.1016/j.neuron.2015.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Todd PK, Oh SY, Krans A, He F, Sellier C, Frazer M, et al. CGG repeat-associated translation mediates neurodegeneration in fragile X tremor ataxia syndrome. Neuron. 2013;78:440–455. doi: 10.1016/j.neuron.2013.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zu T, Gibbens B, Doty NS, Gomes-Pereira M, Huguet A, Stone MD, et al. Non-ATG-initiated translation directed by microsatellite expansions. Proc Natl Acad Sci USA. 2011;108:260–265. doi: 10.1073/pnas.1013343108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Robberecht W, Philips T. The changing scene of amyotrophic lateral sclerosis. Nat Rev Neurosci. 2013;14:248–264. doi: 10.1038/nrn3430. [DOI] [PubMed] [Google Scholar]
- 25.Belzil VV, Katzman RB, Petrucelli L. ALS and FTD: an epigenetic perspective. Acta Neuropathol. 2016;132:487–502. doi: 10.1007/s00401-016-1587-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rippon GA, Scarmeas N, Gordon PH, Murphy PL, Albert SM, Mitsumoto H, et al. An observational study of cognitive impairment in amyotrophic lateral sclerosis. Arch Neurol. 2006;63:345–352. doi: 10.1001/archneur.63.3.345. [DOI] [PubMed] [Google Scholar]
- 27.Lillo P, Hodges JR. Frontotemporal dementia and motor neurone disease: overlapping clinic-pathological disorders. J Clin Neurosci. 2009;16:1131–1135. doi: 10.1016/j.jocn.2009.03.005. [DOI] [PubMed] [Google Scholar]
- 28.Strong MJ, Yang W. The frontotemporal syndromes of ALS. Clinicopathological correlates. J Mol Neurosci. 2011;45:648–655. doi: 10.1007/s12031-011-9609-0. [DOI] [PubMed] [Google Scholar]
- 29.Chen-Plotkin AS, Lee VMY, Trojanowski JQ. TAR DNA-binding protein 43 in neurodegenerative disease. Nat Rev Neurol. 2010;6:211–220. doi: 10.1038/nrneurol.2010.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mackenzie IRA, Rademakers R, Neumann M. TDP-43 and FUS in amyotrophic lateral sclerosis and frontotemporal dementia. Lancet Neurol. 2010;9:995–1007. doi: 10.1016/S1474-4422(10)70195-2. [DOI] [PubMed] [Google Scholar]
- 31.King OD, Gitler AD, Shorter J. The tip of the iceberg: RNA-binding proteins with prion-like domains in neurodegenerative disease. Brain Res. 2012;1462:61–80. doi: 10.1016/j.brainres.2012.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Halfmann R, Alberti S, Lindquist S. Prions, protein homeostasis, and phenotypic diversity. Trends Cell Biol. 2010;20:125–133. doi: 10.1016/j.tcb.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Halfmann R, Lindquist S. Epigenetics in the extreme: prions and the inheritance of environmentally acquired traits. Science. 2010;330:629–632. doi: 10.1126/science.1191081. [DOI] [PubMed] [Google Scholar]
- 34.Prusiner SB. Prions. Proc Natl Acad Sci USA. 1998;95:13363–13383. doi: 10.1073/pnas.95.23.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shorter J. Emergence and natural selection of drug-resistant prions. Mol Biosyst. 2010;6:1115–1130. doi: 10.1039/c004550k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.True HL, Lindquist SL. A yeast prion provides a mechanism for genetic variation and phenotypic diversity. Nature. 2000;407:477–483. doi: 10.1038/35035005. [DOI] [PubMed] [Google Scholar]
- 37.Shorter J, Lindquist S. Prions as adaptive conduits of memory and inheritance. Nat Rev Genet. 2005;6:435–450. doi: 10.1038/nrg1616. [DOI] [PubMed] [Google Scholar]
- 38.Wiltzius JJW, Landau M, Nelson R, Sawaya MR, Apostol MI, Goldschmidt L, et al. Molecular mechanisms for protein-encoded inheritance. Nat Struct Mol Biol. 2009;16:973–978. doi: 10.1038/nsmb.1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alberti S, Halfmann R, King O, Kapila A, Lindquist S. A systematic survey identifies prions and illuminates sequence features of prionogenic proteins. Cell. 2009;137:146–158. doi: 10.1016/j.cell.2009.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Toombs JA, McCarty BR, Ross ED. Compositional determinants of prion formation in yeast. Mol Cell Biol. 2010;30:319–332. doi: 10.1128/MCB.01140-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.March ZM, King OD, Shorter J. Prion-like domains as epigenetic regulators, scaffolds for subcellular organization, and drivers of neurodegenerative disease. Brain Res. 2016;1647:9–18. doi: 10.1016/j.brainres.2016.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Masison DC, Maddelein ML, Wickner RB. The prion model for [URE3] of yeast: spontaneous generation and requirements for propagation. Proc Natl Acad Sci USA. 1997;94:12503–12508. doi: 10.1073/pnas.94.23.12503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li L, Lindquist S. Creating a protein-based element of inheritance. Science. 2000;287:661–664. doi: 10.1126/science.287.5453.661. [DOI] [PubMed] [Google Scholar]
- 44.Osherovich LZ, Weissman JS. Multiple Gln/Asn-rich prion domains confer susceptibility to induction of the yeast [PSI+] prion. Cell. 2001;106:183–194. doi: 10.1016/S0092-8674(01)00440-8. [DOI] [PubMed] [Google Scholar]
- 45.Tyedmers J, Treusch S, Dong J, McCaffery JM, Bevis B, Lindquist S. Prion induction involves and ancient system for the sequestration of aggregated proteins and heritable changes in prion fragmentation. Proc Natl Acad Sci USA. 2010;107:8633–8638. doi: 10.1073/pnas.1003895107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ross ED, Baxa U, Wickner RB. Scrambled prion domains form prions and amyloid. Mol Cell Biol. 2004;24:7206–7213. doi: 10.1128/MCB.24.16.7206-7213.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ross ED, Edskes HK, Terry MJ, Wickner RB. Primary sequence independence for prion formation. Proc Natl Acad Sci USA. 2005;102:12825–12830. doi: 10.1073/pnas.0506136102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Couthouis J, Hart MP, Shorter J, DeJesus-Hernandez M, Erion R, Oristano R, et al. A yeast functional screen predicts new candidate ALS disease genes. Proc Natl Acad Sci USA. 2011;108:20881–20890. doi: 10.1073/pnas.1109434108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li YR, King OD, Shorter J, Gitler AD. Stress granules as crucibles of ALS pathogenesis. J Cell Biol. 2013;201:361–372. doi: 10.1083/jcb.201302044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Banfi S, Servadio A, Chung M-y, Kwiatkowski TJ, Jr, McCall AE, Duvick LA, et al. Identification and characterization of the gene causing type 1 spinocerebellar ataxia. Nat Genet. 1994;7:513–520. doi: 10.1038/ng0894-513. [DOI] [PubMed] [Google Scholar]
- 51.Orr HT, Zoghbi HY. Trinucleotide repeat disorders. Annu Rev Neurosci. 2007;30:575–621. doi: 10.1146/annurev.neuro.29.051605.113042. [DOI] [PubMed] [Google Scholar]
- 52.Cummings CJ, Mancini MA, Antalffy B, DeFranco DB, Orr HT, Zoghbi HY. Chaperone suppression of aggregation and altered subcellular proteasome localization imply protein misfolding in SCA1. Nat Genet. 1998;19:148–154. doi: 10.1038/502. [DOI] [PubMed] [Google Scholar]
- 53.Lorenzetti D, Bohlega S, Zoghbi HY. The expansion of the CAG repeat in ataxin-2 is a frequent cause of autosomal dominant spinocerebellar ataxia. Neurology. 1997;49:1009–1013. doi: 10.1212/WNL.49.4.1009. [DOI] [PubMed] [Google Scholar]
- 54.Guo L, Shorter J. Biology and pathobiology of TDP-43 and emergent therapeutic strategies. Cold Spring Harb Perspect Med. 2016:a024554. doi: 10.1101/cshperspect.a024554. [DOI] [PMC free article] [PubMed]
- 55.Neumann M, Sampathu DM, Kwong LK, Truax AC, Micsenyi MC, Chou TT, et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314:130–133. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- 56.Ling SC, Polymenidou M, Cleveland DW. Converging mechanisms in ALS and FTD: disrupted RNA and protein homeostasis. Neuron. 2013;79:416–438. doi: 10.1016/j.neuron.2013.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Polymenidou M, Lagier-Tourenne C, Hutt KR, Bennett CF, Cleveland DW, Yeo GW. Misregulated RNA processing in amyotrophic lateral sclerosis. Brain Res. 2012;1462:3–15. doi: 10.1016/j.brainres.2012.02.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bhardwaj A, Myers MP, Buratti E, Baralle FE. Characterizing TDP-43 interaction with its RNA targets. Nucleic Acids Res. 2013;41:5062–5074. doi: 10.1093/nar/gkt189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lukavsky PJ, Daujotyte D, Tollervey JR, Ule J, Stuani C, Buratti E, et al. Molecular basis of UG-rich RNA recognition by the human splicing factor TDP-43. Nat Struct Mol Biol. 2013;20:1443–1449. doi: 10.1038/nsmb.2698. [DOI] [PubMed] [Google Scholar]
- 60.Polymenidou M, Lagier-Tourenne C, Hutt KR, Huelga SC, Moran J, Liang TY, et al. Long pre-mRNA depletion and RNA missplicing contribute to neuronal vulnerability from loss of TDP-43. Nat Neurosci. 2011;14:459–468. doi: 10.1038/nn.2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tollervey JR, Curk T, Rogelj B, Briese M, Cereda M, Kayikci M, et al. Characterizing the RNA targets and position-dependent splicing regulation by TDP-43. Nat Neurosci. 2011;14:452–458. doi: 10.1038/nn.2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ling JP, Pletnikova O, Troncoso JC, Wong PC. TDP-43 repression of nonconserved cryptic exons is compromised in ALS-FTD. Science. 2015;349:650–655. doi: 10.1126/science.aab0983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wider C, Dickson DW, Stoessl AJ, Tsuboi Y, Chapon F, Gutmann L, et al. Pallidonigral TDP-43 pathology in Perry syndrome. Parkinsonism Relat Disord. 2009;15:281–286. doi: 10.1016/j.parkreldis.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kabashi E, Valdmanis PN, Dion P, Spiegelman D, McConkey BJ, Vande Velde C, et al. TARDBP mutations in individuals with sporadic and familial amyotrophic lateral sclerosis. Nat Genet. 2008;40:572–574. doi: 10.1038/ng.132. [DOI] [PubMed] [Google Scholar]
- 65.Pesiridis GS, Lee VMY, Trojanowski JQ. Mutations in TDP-43 link glycine-rich domain functions to amyotrophic lateral sclerosis. Hum Mol Genet. 2009;18:R156–R162. doi: 10.1093/hmg/ddp303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rutherford NJ, Zhang YJ, Baker M, Gass JM, Finch NA, Xu YF, et al. Novel mutations in TARDBP (TDP-43) in patients with familial amyotrophic lateral sclerosis. PLoS Genet. 2008;4:e1000193. doi: 10.1371/journal.pgen.1000193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sreedharan J, Blair IP, Tripathi VB, Hu X, Vance C, Rogelj B, et al. TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science. 2008;319:1668–1672. doi: 10.1126/science.1154584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Van Deerlin VM, Leverenz JB, Bekris LM, Bird TD, Yuan W, Elman LB, et al. TARDBP mutations in amyotrophic lateral sclerosis with TDP-43 neuropathology: a genetic and histopathological analysis. Lancet Neurol. 2008;7:409–416. doi: 10.1016/S1474-4422(08)70071-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Borroni B, Bonvicini C, Alberici A, Buratti E, Agosti C, Archetti S, et al. Mutation within TARDBP leads to frontotemporal dementia without motor neuron disease. Hum Mutat. 2009;30:E974–E983. doi: 10.1002/humu.21100. [DOI] [PubMed] [Google Scholar]
- 70.Floris G, Borghero G, Cannas A, Di Stefano F, Murru MR, Corongiu D, et al. Clinical phenotypes and radiological findings in frontotemporal dementia related to TARDBP mutations. J Neurol. 2015;262:375–384. doi: 10.1007/s00415-014-7575-5. [DOI] [PubMed] [Google Scholar]
- 71.Lagier-Tourenne C, Polymenidou M, Cleveland DW. TDP-43 and FUS/TLS: emerging roles in RNA processing and neurodegeneration. Hum Mol Genet. 2010;19:R46–R64. doi: 10.1093/hmg/ddq137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kawahara Y, Mieda-Sato A. TDP-43 promotes microRNA biogenesis as a component of the Drosha and Dicer complexes. Proc Natl Acad Sci USA. 2012;109:3347–3352. doi: 10.1073/pnas.1112427109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Buratti E, Brindisi A, Giombi M, Tisminetzky S, Ayala YM, Baralle FE. TDP-43 binds heterogeneous nuclear ribonucleoprotein A/B through its C-terminal tail: an important region for the inhibition of cystic fibrosis transmembrane conductance regulator exon 9 splicing. J Biol Chem. 2005;280:37572–37584. doi: 10.1074/jbc.M505557200. [DOI] [PubMed] [Google Scholar]
- 74.D’Ambrogio A, Buratti E, Stuani C, Guarnaccia C, Romano M, Ayala YM, et al. Functional mapping of the interaction between TDP-43 and hnRNP A2 in vivo. Nucleic Acids Res. 2009;37:4116–4126. doi: 10.1093/nar/gkp342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bentmann E, Neumann M, Tahirovic S, Rodde R, Dormann D, Haass C. Requirements for stress granule recruitment of fused in sarcoma (FUS) and TAR DNA-binding protein of 43 kDa (TDP-43) J Biol Chem. 2012;287:23079–23094. doi: 10.1074/jbc.M111.328757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ash PEA, Zhang YJ, Roberts CM, Saldi T, Hutter H, Buratti E, et al. Neurotoxic effects of TDP-43 overexpression in C. elegans. Hum Mol Genet. 2010;19:3206–3218. doi: 10.1093/hmg/ddq230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Johnson BS, McCaffery JM, Lindquist S, Gitler AD. A yeast TDP-43 proteinopathy model: exploring the molecular determinants of TDP-43 aggregation and cellular toxicity. Proc Natl Acad Sci USA. 2008;105:6439–6444. doi: 10.1073/pnas.0802082105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Johnson BS, Snead D, Lee JJ, McCaffery JM, Shorter J, Gitler AD. TDP-43 is intrinsically aggregation-prone, and amyotrophic lateral sclerosis-linked mutations accelerate aggregation and increase toxicity. J Biol Chem. 2009;284:20329–20339. doi: 10.1074/jbc.M109.010264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Saini A, Chauhan VS. Delineation of the core aggregation sequences of TDP-43 C-terminal fragment. ChemBioChem. 2011;12:2495–2501. doi: 10.1002/cbic.201100427. [DOI] [PubMed] [Google Scholar]
- 80.Elden AC, Kim HJ, Hart MP, Chen-Plotkin AS, Johnson BS, Fang X, et al. Ataxin-2 intermediate-length polyglutamine expansions are associated with increased risk for ALS. Nature. 2010;466:1069–1075. doi: 10.1038/nature09320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Voigt A, Herholz D, Fiesel FC, Kaur K, Müller D, Karsten P, et al. TDP-43-mediated neuron loss in vivo requires RNA-binding activity. PLoS ONE. 2010;5:e12247. doi: 10.1371/journal.pone.0012247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ju S, Tardiff DF, Han H, Divya K, Zhong Q, Maquat LE, et al. A yeast model of FUS/TLS-dependent cytotoxicity. PLoS Biol. 2011;9:e1001052. doi: 10.1371/journal.pbio.1001052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Schwartz JC, Cech TR, Parker RR. Biochemical properties and biological functions of FET proteins. Annu Rev Biochem. 2015;84:355–379. doi: 10.1146/annurev-biochem-060614-034325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Qiu H, Lee S, Shang Y, Wang WY, Au KF, Kamiya S, et al. ALS-associated mutation FUS-R521C causes DNA damage and RNA splicing defects. J Clin Invest. 2014;124:981–999. doi: 10.1172/JCI72723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Corrado L, Del Bo R, Castellotti B, Ratti A, Cereda C, Penco S, et al. Mutations of FUS gene in sporadic amyotrophic lateral sclerosis. J Med Genet. 2010;47:190–194. doi: 10.1136/jmg.2009.071027. [DOI] [PubMed] [Google Scholar]
- 86.Kwiatkowski TJ, Bosco DA, LeClerc AL, Tamrazian E, Vanderburg CR, Russ C, et al. Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science. 2009;323:1205–1208. doi: 10.1126/science.1166066. [DOI] [PubMed] [Google Scholar]
- 87.Rademakers R, Stewart H, Dejesus-Hernandez M, Krieger C, Graff-Radford N, Fabros M, et al. Fus gene mutations in familial and sporadic amyotrophic lateral sclerosis. Muscle Nerve. 2010;42:170–176. doi: 10.1002/mus.21665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Vance C, Rogelj B, Hortobagyi T, De Vos KJ, Nishimura AL, Sreedharan J, et al. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science. 2009;323:1208–1211. doi: 10.1126/science.1165942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Fecto F, Siddique T. Making connections: pathology and genetics link amyotrophic lateral sclerosis with frontotemporal lobe dementia. J Mol Neurosci. 2011;45:663–675. doi: 10.1007/s12031-011-9637-9. [DOI] [PubMed] [Google Scholar]
- 90.Neumann M, Rademakers R, Roeber S, Baker M, Kretzschmar HA, Mackenzie IRA. A new subtype of frontotemporal lobar degeneration with FUS pathology. Brain. 2009;132:2922–2931. doi: 10.1093/brain/awp214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Neumann M, Roeber S, Kretzschmar HA, Rademakers R, Baker M, Mackenzie IRA. Abundant FUS-immunoreactive pathology in neuronal intermediate filament inclusion disease. Acta Neuropathol. 2009;118:605–616. doi: 10.1007/s00401-009-0581-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Seelaar H, Klijnsma KY, de Koning I, van der Lugt A, Chiu WZ, Azmani A, et al. Frequency of ubiquitin and FUS-positive, TDP-43-negative frontotemporal lobar degeneration. J Neurol. 2010;257:747–753. doi: 10.1007/s00415-009-5404-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sun Z, Diaz Z, Fang X, Hart MP, Chesi A, Shorter J, et al. Molecular determinants and genetic modifiers of aggregation and toxicity for the ALS disease protein FUS/TLS. PLoS Biol. 2011;9:e1000614. doi: 10.1371/journal.pbio.1000614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Urwin H, Josephs KA, Rohrer JD, Mackenzie IR, Neumann M, Authier A, et al. FUS pathology defines the majority of tau- and TDP-43-negative frontotemporal lobar degeneration. Acta Neuropathol. 2010;120:33–41. doi: 10.1007/s00401-010-0698-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Dormann D, Rodde R, Edbauer D, Bentmann E, Fischer I, Hruscha A, et al. ALS-associated fused in sarcoma (FUS) mutations disrupt Transportin-mediated nuclear import. EMBO J. 2010;29:2841–2857. doi: 10.1038/emboj.2010.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Daigle JG, Lanson NA, Jr, Smith RB, Casci I, Maltare A, Monaghan J, et al. RNA-binding ability of FUS regulates neurodegeneration, cytoplasmic mislocalization and incorporation into stress granules associated with FUS carrying ALS-linked mutations. Hum Mol Genet. 2013;22:1193–1205. doi: 10.1093/hmg/dds526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sun S, Ling SC, Qiu J, Albuquerque CP, Zhou Y, Tokunaga S, et al. ALS-causative mutations in FUS/TLS confer gain and loss of function by altered association with SMN and U1-snRNP. Nat Commun. 2015;6:6171. doi: 10.1038/ncomms7171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Couthouis J, Hart MP, Erion R, King OD, Diaz Z, Nakaya T, et al. Evaluating the role of the FUS/TLS-related gene EWSR1 in amyotrophic lateral sclerosis. Hum Mol Genet. 2012;21:2899–2911. doi: 10.1093/hmg/dds116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Neumann M, Bentmann E, Dormann D, Jawaid A, DeJesus-Hernandez M, Ansorge O, et al. FET proteins TAF15 and EWS are selective markers that distinguish FTLD with FUS pathology from amyotrophic lateral sclerosis with FUS mutations. Brain. 2011;134:2595–2609. doi: 10.1093/brain/awr201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ticozzi N, Vance C, Leclerc AL, Keagle P, Glass JD, McKenna-Yasek D, et al. Mutational analysis reveals the FUS homolog TAF15 as a candidate gene for familial amyotrophic lateral sclerosis. Am J Med Genet B Neuropsychiatr Genet. 2011;156:285–290. doi: 10.1002/ajmg.b.31158. [DOI] [PubMed] [Google Scholar]
- 101.Kim HJ, Kim NC, Wang YD, Scarborough EA, Moore J, Diaz Z, et al. Mutations in prion-like domains in hnRNPA2B1 and hnRNPA1 cause multisystem proteinopathy and ALS. Nature. 2013;495:467–473. doi: 10.1038/nature11922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Watts GDJ, Wymer J, Kovach MJ, Mehta SG, Mumm S, Darvish D, et al. Inclusion body myopathy associated with Paget disease of bone and frontotemporal dementia is caused by mutant valosin-containing protein. Nat Genet. 2004;36:377–381. doi: 10.1038/ng1332. [DOI] [PubMed] [Google Scholar]
- 103.Nalbandian A, Donkervoort S, Dec E, Badadani M, Katheria V, Rana P, et al. The multiple faces of valosin-containing protein-associated diseases: inclusion body myopathy with Paget’s disease of bone, frontotemporal dementia, and amyotrophic lateral sclerosis. J Mol Neurosci. 2011;45:522–531. doi: 10.1007/s12031-011-9627-y. [DOI] [PubMed] [Google Scholar]
- 104.Benatar M, Wuu J, Fernandez C, Weihl CC, Katzen H, Steele J, et al. Motor neuron involvement in multisystem proteinopathy: implications for ALS. Neurology. 2013;80:1874–1880. doi: 10.1212/WNL.0b013e3182929fc3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Shi Z, Hayashi YK, Mitsuhashi S, Goto K, Kaneda D, Choi YC, et al. Characterization of the Asian myopathy patients with VCP mutations. Eur J Neurol. 2012;19:501–509. doi: 10.1111/j.1468-1331.2011.03575.x. [DOI] [PubMed] [Google Scholar]
- 106.Chung PYJ, Beyens G, de Freitas F, Boonen S, Geusens P, Vanhoenacker F, et al. Indications for a genetic association of a VCP polymorphism with the pathogenesis of sporadic Paget’s disease of bone, but not for TNFSF11 (RANKL) and IL-6 polymorphisms. Mol Genet Metab. 2011;103:287–292. doi: 10.1016/j.ymgme.2011.03.021. [DOI] [PubMed] [Google Scholar]
- 107.Kim NC, Tresse E, Kolaitis RM, Molliex A, Thomas RE, Alami NH, et al. VCP is essential for mitochondrial quality control by PINK1/Parkin and this function is impaired by VCP mutations. Neuron. 2013;78:65–80. doi: 10.1016/j.neuron.2013.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Johnson JO, Mandrioli J, Benatar M, Abramzon Y, Van Deerlin VM, Trojanowski JQ, et al. Exome sequencing reveals VCP mutations as a cause of familial ALS. Neuron. 2010;68:857–864. doi: 10.1016/j.neuron.2010.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Buchan JR, Kolaitis RM, Taylor JP, Parker R. Eukaryotic stress granules are cleared by autophagy and Cdc48/VCP function. Cell. 2013;153:1461–1474. doi: 10.1016/j.cell.2013.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Jean-Philippe J, Paz S, Caputi M. hnRNP A1: the Swiss army knife of gene expression. Int J Mol Sci. 2013;14:18999–19024. doi: 10.3390/ijms140918999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Campillos M, Lamas JR, García MA, Bullido MJ, Valdivieso F, Vázquez J. Specific interaction of heterogeneous nuclear ribonucleoprotein A1 with the -219T allelic form modulates APOE promoter activity. Nucleic Acids Res. 2003;31:3063–3070. doi: 10.1093/nar/gkg435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Hay DC, Kemp GD, Dargemont C, Hay RT. Interaction between hnRNPA1 and IκBα is required for maximal activation of NF-κB-dependent transcription. Mol Cell Biol. 2001;21:3482–3490. doi: 10.1128/MCB.21.10.3482-3490.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Xia H. Regulation of γ-fibrinogen chain expression by heterogeneous nuclear ribonucleoprotein A1. J Biol Chem. 2005;280:13171–13178. doi: 10.1074/jbc.M414120200. [DOI] [PubMed] [Google Scholar]
- 114.Han SP, Tang YH, Smith R. Functional diversity of the hnRNPs: past, present and perspectives. Biochem J. 2010;430:379–392. doi: 10.1042/BJ20100396. [DOI] [PubMed] [Google Scholar]
- 115.Huelga SC, Vu AQ, Arnold JD, Liang TY, Liu PP, Yan BY, et al. Integrative genome-wide analysis reveals cooperative regulation of alternative splicing by hnRNP proteins. Cell Rep. 2012;1:167–178. doi: 10.1016/j.celrep.2012.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Barreau C, Paillard L, Osborne HB. AU-rich elements and associated factors: are there unifying principles? Nucleic Acids Res. 2005;33:7138–7150. doi: 10.1093/nar/gki1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Bonnal S, Pileur F, Orsini C, Parker F, Pujol F, Prats AC, et al. Heterogeneous nuclear ribonucleoprotein A1 is a novel internal ribosome entry site trans-acting factor that modulates alternative initiation of translation of the fibroblast growth factor 2 mRNA. J Biol Chem. 2005;280:4144–4153. doi: 10.1074/jbc.M411492200. [DOI] [PubMed] [Google Scholar]
- 118.Cammas A, Pileur F, Bonnal S, Lewis SM, Leveque N, Holcik M, et al. Cytoplasmic relocalization of heterogeneous nuclear ribonucleoprotein A1 controls translation initiation of specific mRNAs. Mol Biol Cell. 2007;18:5048–5059. doi: 10.1091/mbc.E07-06-0603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.LaBranche H, Dupuis S, Ben-David Y, Bani MR, Wellinger RJ, Chabot B. Telomere elongation by hnRNP A1 and a derivative that interacts with telomeric repeats and telomerase. Nat Genet. 1998;19:199–202. doi: 10.1038/575. [DOI] [PubMed] [Google Scholar]
- 120.Guil S, Cáceres JF. The multifunctional RNA-binding protein hnRNP A1 is required for processing of miR-18a. Nat Struct Mol Biol. 2007;14:591–596. doi: 10.1038/nsmb1250. [DOI] [PubMed] [Google Scholar]
- 121.Michlewski G, Cáceres JF. Antagonistic role of hnRNP A1 and KSRP in the regulation of let-7a biogenesis. Nat Struct Mol Biol. 2010;17:1011–1018. doi: 10.1038/nsmb.1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Goina E, Skoko N, Pagani F. Binding of DAZAP1 and hnRNPA1/A2 to an exonic splicing silencer in a natural BRCA1 exon 18 mutant. Mol Cell Biol. 2008;28:3850–3860. doi: 10.1128/MCB.02253-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Martinez-Contreras R, Cloutier P, Shkreta L, Fisette JF, Revil T, Chabot B. hnRNP proteins and splicing control. Adv Exp Med Biol. 2007;623:123–147. doi: 10.1007/978-0-387-77374-2_8. [DOI] [PubMed] [Google Scholar]
- 124.Martinez FJ, Pratt GA, Van Nostrand EL, Batra R, Huelga SC, Kapeli K, et al. Protein-RNA networks regulated by normal and ALS-associated mutant HNRNPA2B1 in the nervous system. Neuron. 2016;92:780–795. doi: 10.1016/j.neuron.2016.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Moran-Jones K, Wayman L, Kennedy DD, Reddel RR, Sara S, Snee MJ, et al. hnRNP A2, a potential ssDNA/RNA molecular adapter at the telomere. Nucleic Acids Res. 2005;33:486–496. doi: 10.1093/nar/gki203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Tanaka E, Fukuda H, Nakashima K, Tsuchiya N, Seimiya H, Nakagama H. HnRNP A3 binds to and protects mammalian telomeric repeats in vitro. Biochem Biophys Res Commun. 2007;358:608–614. doi: 10.1016/j.bbrc.2007.04.177. [DOI] [PubMed] [Google Scholar]
- 127.Hoek KS, Kidd GJ, Carson JH, Smith R. hnRNP A2 selectively binds the cytoplasmic transport sequence of myelin basic protein mRNA. Biochemistry. 1998;37:7021–7029. doi: 10.1021/bi9800247. [DOI] [PubMed] [Google Scholar]
- 128.Shan J, Munro TP, Barbarese E, Carson JH, Smith R. A molecular mechanism for mRNA trafficking in neuronal dendrites. J Neurosci. 2003;23:8859–8866. doi: 10.1523/JNEUROSCI.23-26-08859.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Mitchell J, Paul P, Chen HJ, Morris A, Payling M, Falchi M, et al. Familial amyotrophic lateral sclerosis is associated with a mutation in D-amino acid oxidase. Proc Natl Acad Sci USA. 2010;107:7556–7561. doi: 10.1073/pnas.0914128107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Sasabe J, Miyoshi Y, Suzuki M, Mita M, Konno R, Matsuoka M, et al. D-amino acid oxidase controls motoneuron degeneration through D-serine. Proc Natl Acad Sci USA. 2012;109:627–632. doi: 10.1073/pnas.1114639109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Goldschmidt L, Teng PK, Riek R, Eisenberg D. Identifying the amylome, proteins capable of forming amyloid-like fibrils. Proc Natl Acad Sci USA. 2010;107:3487–3492. doi: 10.1073/pnas.0915166107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Paul KR, Molliex A, Cascarina S, Boncella AE, Taylor JP, Ross ED. The effects of mutations on the aggregation propensity of the human prion-like protein hnRNPA2B1. Mol Cell Biol. 2017;37:MCB.00652-16. doi: 10.1128/MCB.00652-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Salajegheh M, Pinkus JL, Taylor JP, Amato AA, Nazareno R, Baloh RH, et al. Sarcoplasmic redistribution of nuclear TDP-43 in inclusion body myositis. Muscle Nerve. 2009;40:19–31. doi: 10.1002/mus.21386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Pinkus JL, Amato AA, Taylor JP, Greenberg SA. Abnormal distribution of heterogeneous nuclear ribonucleoproteins in sporadic inclusion body myositis. Neuromuscul Disord. 2014;24:611–616. doi: 10.1016/j.nmd.2014.04.012. [DOI] [PubMed] [Google Scholar]
- 135.Cirulli ET, Lasseigne BN, Petrovski S, Sapp PC, Dion PA, Leblond CS, et al. Exome sequencing in amyotrophic lateral sclerosis identifies risk genes and pathways. Science. 2015;347:1436–1441. doi: 10.1126/science.aaa3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Couthouis J, Raphael AR, Daneshjou R, Gitler AD, Gibson G. Targeted exon capture and sequencing in sporadic amyotrophic lateral sclerosis. PLoS Genet. 2014;10:e1004704. doi: 10.1371/journal.pgen.1004704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Liu Q, Shu S, Wang RR, Liu F, Cui B, Guo XN, et al. Whole-exome sequencing identifies a missense mutation in hnRNPA1 in a family with flail arm ALS. Neurology. 2016;87:1763–1769. doi: 10.1212/WNL.0000000000003256. [DOI] [PubMed] [Google Scholar]
- 138.Neumann M, Igaz LM, Kwong LK, Nakashima-Yasuda H, Kolb SJ, Dreyfuss G, et al. Absence of heterogeneous nuclear ribonucleoproteins and survival motor neuron protein in TDP-43 positive inclusions in frontotemporal lobar degeneration. Acta Neuropathol. 2007;113:543–548. doi: 10.1007/s00401-007-0221-x. [DOI] [PubMed] [Google Scholar]
- 139.Vieira NM, Naslavsky MS, Licinio L, Kok F, Schlesinger D, Vainzof M, et al. A defect in the RNA-processing protein HNRPDL causes limb-girdle muscular dystrophy 1G (LGMD1G) Hum Mol Genet. 2014;23:4103–4110. doi: 10.1093/hmg/ddu127. [DOI] [PubMed] [Google Scholar]
- 140.Anderson P, Kedersha N. Stress granules: the Tao of RNA triage. Trends Biochem Sci. 2008;33:141–150. doi: 10.1016/j.tibs.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 141.Bosco DA, Lemay N, Ko HK, Zhou H, Burke C, Kwiatkowski TJ, Jr, et al. Mutant FUS proteins that cause amyotrophic lateral sclerosis incorporate into stress granules. Hum Mol Genet. 2010;19:4160–4175. doi: 10.1093/hmg/ddq335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Buchan JR, Parker R. Eukaryotic stress granules: the ins and outs of translation. Mol Cell. 2009;36:932–941. doi: 10.1016/j.molcel.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Wolozin B. Regulated protein aggregation: stress granules and neurodegeneration. Mol Neurodegener. 2012;7:56. doi: 10.1186/1750-1326-7-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Kedersha N, Stoecklin G, Ayodele M, Yacono P, Lykke-Andersen J, Fritzler MJ, et al. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J Cell Biol. 2005;169:871–884. doi: 10.1083/jcb.200502088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Jain S, Parker R. The discovery and analysis of P bodies. In: Chan LEK, Fritzler JM, editors. Ten Years of Progress in GW/P Body Research. Springer; New York, NY: 2013. pp. 23–43. [Google Scholar]
- 146.Guo L, Shorter J. It’s raining liquids: RNA tunes viscoelasticity and dynamics of membraneless organelles. Mol Cell. 2015;60:189–192. doi: 10.1016/j.molcel.2015.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Mayeda A, Munroe SH, Cáceres JF, Krainer AR. Function of conserved domains of hnRNP A1 and other hnRNP A/B proteins. EMBO J. 1994;13:5483–5495. doi: 10.1002/j.1460-2075.1994.tb06883.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Siomi H, Dreyfuss G. A nuclear localization domain in the hnRNP A1 protein. J Cell Biol. 1995;129:551–560. doi: 10.1083/jcb.129.3.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Burke KA, Janke AM, Rhine CL, Fawzi NL. Residue-by-residue view of in vitro FUS granules that bind the C-terminal domain of RNA polymerase II. Mol Cell. 2015;60:231–241. doi: 10.1016/j.molcel.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Liu-Yesucevitz L, Bilgutay A, Zhang YJ, Vanderweyde T, Citro A, Mehta T, et al. Tar DNA binding protein-43 (TDP-43) associates with stress granules: analysis of cultured cells and pathological brain tissue. PLoS ONE. 2010;5:e13250. doi: 10.1371/journal.pone.0013250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Gilks N, Kedersha N, Ayodele M, Shen L, Stoecklin G, Dember LM, et al. Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol Biol Cell. 2004;15:5383–5398. doi: 10.1091/mbc.E04-08-0715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Reijns MAM, Alexander RD, Spiller MP, Beggs JD. A role for Q/N-rich aggregation-prone regions in P-body localization. J Cell Sci. 2008;121:2463–2472. doi: 10.1242/jcs.024976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Seydoux G, Braun RE. Pathway to totipotency: lessons from germ cells. Cell. 2006;127:891–904. doi: 10.1016/j.cell.2006.11.016. [DOI] [PubMed] [Google Scholar]
- 154.Hennig S, Kong G, Mannen T, Sadowska A, Kobelke S, Blythe A, et al. Prion-like domains in RNA binding proteins are essential for building subnuclear paraspeckles. J Cell Biol. 2015;210:529–539. doi: 10.1083/jcb.201504117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Mannen T, Yamashita S, Tomita K, Goshima N, Hirose T. The Sam68 nuclear body is composed of two RNase-sensitive substructures joined by the adaptor HNRNPL. J Cell Biol. 2016;214:45–59. doi: 10.1083/jcb.201601024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Boke E, Mitchison TJ. The balbiani body and the concept of physiological amyloids. Cell Cycle. 2017;16:153–154. doi: 10.1080/15384101.2016.1241605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Boke E, Ruer M, Wühr M, Coughlin M, Lemaitre R, Gygi SP, et al. Amyloid-like self-assembly of a cellular compartment. Cell. 2016;166:637–650. doi: 10.1016/j.cell.2016.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Ford AF, Shorter J. Fleeting amyloid-like forms of Rim4 ensure meiotic fidelity. Cell. 2015;163:275–276. doi: 10.1016/j.cell.2015.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Berchowitz LE, Kabachinski G, Walker MR, Carlile TM, Gilbert WV, Schwartz TU, et al. Regulated formation of an amyloid-like translational repressor governs gametogenesis. Cell. 2015;163:406–418. doi: 10.1016/j.cell.2015.08.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Ritz AM, Trautwein M, Grassinger F, Spang A. The prion-like domain in the exomer-dependent cargo Pin2 serves as a trans-Golgi retention motif. Cell Rep. 2014;7:249–260. doi: 10.1016/j.celrep.2014.02.026. [DOI] [PubMed] [Google Scholar]
- 161.Gal J, Zhang J, Kwinter DM, Zhai J, Jia H, Jia J, et al. Nuclear localization sequence of FUS and induction of stress granules by ALS mutants. Neurobiol Aging. 2011;32:2323.e27–2323.e40. doi: 10.1016/j.neurobiolaging.2010.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Lenzi J, De Santis R, de Turris V, Morlando M, Laneve P, Calvo A, et al. ALS mutant FUS proteins are recruited into stress granules in induced pluripotent stem cell-derived motoneurons. Dis Model Mech. 2015;8:755–766. doi: 10.1242/dmm.020099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Kim HJ, Raphael AR, LaDow ES, McGurk L, Weber RA, Trojanowski JQ, et al. Therapeutic modulation of eIF2α phosphorylation rescues TDP-43 toxicity in amyotrophic lateral sclerosis disease models. Nat Genet. 2014;46:152–160. doi: 10.1038/ng.2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Swisher KD, Parker R. Localization to, and effects of Pbp1, Pbp4, Lsm12, Dhh1 and Pab1 on stress granules in Saccharomyces cerevisiae. PLoS ONE. 2010;5:e10006. doi: 10.1371/journal.pone.0010006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Daoud H, Belzil V, Martins S, Sabbagh M, Provencher P, Lacomblez L, et al. Association of long ATXN2 CAG repeat sizes with increased risk of amyotrophic lateral sclerosis. Arch Neurol. 2011;68:739–742. doi: 10.1001/archneurol.2011.111. [DOI] [PubMed] [Google Scholar]
- 166.Lu HP, Gan SR, Chen S, Li HF, Liu ZJ, Ni W, et al. Intermediate-length polyglutamine in ATXN2 is a possible risk factor among Eastern Chinese patients with amyotrophic lateral sclerosis. Neurobiol Aging. 2015;36:1603.e11–1603.e14. doi: 10.1016/j.neurobiolaging.2014.10.015. [DOI] [PubMed] [Google Scholar]
- 167.Ross OA, Rutherford NJ, Baker M, Soto-Ortolaza AI, Carrasquillo MM, DeJesus-Hernandez M, et al. Ataxin-2 repeat-length variation and neurodegeneration. Hum Mol Genet. 2011;20:3207–3212. doi: 10.1093/hmg/ddr227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Mitchell SF, Jain S, She M, Parker R. Global analysis of yeast mRNPs. Nat Struct Mol Biol. 2013;20:127–133. doi: 10.1038/nsmb.2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Segal SP, Dunckley T, Parker R. Sbp1p affects translational repression and decapping in Saccharomyces cerevisiae. Mol Cell Biol. 2006;26:5120–5130. doi: 10.1128/MCB.01913-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Beggs JD. Lsm proteins and RNA processing. Biochem Soc Trans. 2005;33:433–438. doi: 10.1042/BST0330433. [DOI] [PubMed] [Google Scholar]
- 171.Chowdhury A, Mukhopadhyay J, Tharun S. The decapping activator Lsm1p-7p-Pat1p complex has the intrinsic ability to distinguish between oligoadenylated and polyadenylated RNAs. RNA. 2007;13:998–1016. doi: 10.1261/rna.502507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.He W, Parker R. The yeast cytoplasmic LsmI/Pat1p complex protects mRNA 3′ termini from partial degradation. Genetics. 2001;158:1445–1455. doi: 10.1093/genetics/158.4.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Luhtala N, Parker R. LSM1 over-expression in Saccharomyces cerevisiae depletes U6 snRNA levels. Nucleic Acids Res. 2009;37:5529–5536. doi: 10.1093/nar/gkp572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Pannone BK, Kim SD, Noe DA, Wolin SL. Multiple functional interactions between components of the Lsm2-Lsm8 complex, U6 snRNA, and the yeast La protein. Genetics. 2001;158:187–196. doi: 10.1093/genetics/158.1.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Kufel J, Bousquet-Antonelli C, Beggs JD, Tollervey D. Nuclear pre-mRNA decapping and 5′ degradation in yeast require the Lsm2-8p complex. Mol Cell Biol. 2004;24:9646–9657. doi: 10.1128/MCB.24.21.9646-9657.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Buchan JR, Muhlrad D, Parker R. P bodies promote stress granule assembly in Saccharomyces cerevisiae. J Cell Biol. 2008;183:441–455. doi: 10.1083/jcb.200807043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Kedersha N, Anderson P. Chapter 4 Regulation of translation by stress granules and processing bodies. In: Hershey JWB, editor. Progress in Molecular Biology and Translational Science. Academic Press; 2009. pp. 155–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Nonhoff U, Ralser M, Welzel F, Piccini I, Balzereit D, Yaspo ML, et al. Ataxin-2 interacts with the DEAD/H-box RNA helicase DDX6 and interferes with P-bodies and stress granules. Mol Biol Cell. 2007;18:1385–1396. doi: 10.1091/mbc.E06-12-1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Hackman P, Sarparanta J, Lehtinen S, Vihola A, Evilä A, Jonson PH, et al. Welander distal myopathy is caused by a mutation in the RNA-binding protein TIA1. Ann Neurol. 2013;73:500–509. doi: 10.1002/ana.23831. [DOI] [PubMed] [Google Scholar]
- 180.Klar J, Sobol M, Melberg A, Mäbert K, Ameur A, Johansson ACV, et al. Welander distal myopathy caused by an ancient founder mutation in TIA1 associated with perturbed splicing. Hum Mutat. 2013;34:572–577. doi: 10.1002/humu.22282. [DOI] [PubMed] [Google Scholar]
- 181.Elbaum-Garfinkle S, Brangwynne CP. Liquids, fibers, and gels: the many phases of neurodegeneration. Dev Cell. 2015;35:531–532. doi: 10.1016/j.devcel.2015.11.014. [DOI] [PubMed] [Google Scholar]
- 182.Shorter J. Membraneless organelles: phasing in and out. Nat Chem. 2016;8:528–530. doi: 10.1038/nchem.2534. [DOI] [PubMed] [Google Scholar]
- 183.Shin Y, Berry J, Pannucci N, Haataja MP, Toettcher JE, Brangwynne CP. Spatiotemporal control of intracellular phase transitions using light-activated optoDroplets. Cell. 2017;168:159–171. e14. doi: 10.1016/j.cell.2016.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Brangwynne CP, Eckmann CR, Courson DS, Rybarska A, Hoege C, Gharakhani J, et al. Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science. 2009;324:1729–1732. doi: 10.1126/science.1172046. [DOI] [PubMed] [Google Scholar]
- 185.Kroschwald S, Maharana S, Mateju D, Malinovska L, Nüske E, Poser I, et al. Promiscuous interactions and protein disaggregases determine the material state of stress-inducible RNP granules. eLife. 2015;4:e06807. doi: 10.7554/eLife.06807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Banani SF, Rice AM, Peeples WB, Lin Y, Jain S, Parker R, et al. Compositional control of phase-separated cellular bodies. Cell. 2016;166:651–663. doi: 10.1016/j.cell.2016.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Hyman AA, Weber CA, Jülicher F. Liquid-liquid phase separation in biology. Annu Rev Cell Dev Biol. 2014;30:39–58. doi: 10.1146/annurev-cellbio-100913-013325. [DOI] [PubMed] [Google Scholar]
- 188.Nott TJ, Craggs TD, Baldwin AJ. Membraneless organelles can melt nucleic acid duplexes and act as biomolecular filters. Nat Chem. 2016;8:569–575. doi: 10.1038/nchem.2519. [DOI] [PubMed] [Google Scholar]
- 189.Brangwynne CP, Tompa P, Pappu RV. Polymer physics of intracellular phase transitions. Nat Phys. 2015;11:899–904. doi: 10.1038/nphys3532. [DOI] [Google Scholar]
- 190.Molliex A, Temirov J, Lee J, Coughlin M, Kanagaraj AP, Kim HJ, et al. Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell. 2015;163:123–133. doi: 10.1016/j.cell.2015.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Nott TJ, Petsalaki E, Farber P, Jervis D, Fussner E, Plochowietz A, et al. Phase transition of a disordered nuage protein generates environmentally responsive membraneless organelles. Mol Cell. 2015;57:936–947. doi: 10.1016/j.molcel.2015.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Kato M, Han TW, Xie S, Shi K, Du X, Wu LC, et al. Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell. 2012;149:753–767. doi: 10.1016/j.cell.2012.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Murakami T, Qamar S, Lin JQ, Schierle GSK, Rees E, Miyashita A, et al. ALS/FTD mutation-induced phase transition of FUS liquid droplets and reversible hydrogels into irreversible hydrogels impairs RNP granule function. Neuron. 2015;88:678–690. doi: 10.1016/j.neuron.2015.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Jain S, Wheeler JR, Walters RW, Agrawal A, Barsic A, Parker R. ATPase-modulated stress granules contain a diverse proteome and substructure. Cell. 2016;164:487–498. doi: 10.1016/j.cell.2015.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Patel A, Lee HO, Jawerth L, Maharana S, Jahnel M, Hein MY, et al. A liquid-to-solid phase transition of the ALS protein FUS accelerated by disease mutation. Cell. 2015;162:1066–1077. doi: 10.1016/j.cell.2015.07.047. [DOI] [PubMed] [Google Scholar]
- 196.Peters OM, Ghasemi M, Brown RH., Jr Emerging mechanisms of molecular pathology in ALS. J Clin Invest. 2015;125:2548. doi: 10.1172/JCI82693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.DeJesus-Hernandez M, Mackenzie IR, Boeve BF, Boxer AL, Baker M, Rutherford NJ, et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72:245–256. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Lee KH, Zhang P, Kim HJ, Mitrea DM, Sarkar M, Freibaum BD, et al. C9orf72 dipeptide repeats impair the assembly, dynamics, and function of membrane-less organelles. Cell. 2016;167:774–788. e17. doi: 10.1016/j.cell.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Lin Y, Mori E, Kato M, Xiang S, Wu L, Kwon I, et al. Toxic PR polydipeptides encoded by the C9orf72 repeat expansion target LC domain polymers. Cell. 2016;167:789–802. e12. doi: 10.1016/j.cell.2016.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Vashist S, Cushman M, Shorter J. Applying Hsp104 to protein-misfolding disorders. Biochem Cell Biol. 2010;88:1–13. doi: 10.1139/O09-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Sweeny EA, Shorter J. Prion proteostasis: Hsp104 meets its supporting cast. Prion. 2008;2:135–140. doi: 10.4161/pri.2.4.7952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Sweeny EA, Shorter J. Mechanistic and structural insights into the prion-disaggregase activity of Hsp104. J Mol Biol. 2016;428:1870–1885. doi: 10.1016/j.jmb.2015.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Jackrel ME, Shorter J. Potentiated Hsp104 variants suppress toxicity of diverse neurodegenerative disease-linked proteins. Dis Model Mech. 2014;7:1175–1184. doi: 10.1242/dmm.016113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Jackrel ME, DeSantis ME, Martinez BA, Castellano LM, Stewart RM, Caldwell KA, et al. Potentiated Hsp104 variants antagonize diverse proteotoxic misfolding events. Cell. 2014;156:170–182. doi: 10.1016/j.cell.2013.11.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Halfmann R, Jarosz DF, Jones SK, Chang A, Lancaster AK, Lindquist S. Prions are a common mechanism for phenotypic inheritance in wild yeasts. Nature. 2012;482:363–368. doi: 10.1038/nature10875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Shorter J, Lindquist S. Hsp104 catalyzes formation and elimination of self-replicating Sup35 prion conformers. Science. 2004;304:1793–1797. doi: 10.1126/science.1098007. [DOI] [PubMed] [Google Scholar]
- 207.DeSantis ME, Leung EH, Sweeny EA, Jackrel ME, Cushman-Nick M, Neuhaus-Follini A, et al. Operational plasticity enables hsp104 to disaggregate diverse amyloid and nonamyloid clients. Cell. 2012;151:778–793. doi: 10.1016/j.cell.2012.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.DeSantis ME, Shorter J. Hsp104 drives ‘protein-only’ positive selection of Sup35 prion strains encoding strong [PSI+] Chem Biol. 2012;19:1400–1410. doi: 10.1016/j.chembiol.2012.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Vacher C, Garcia-Oroz L, Rubinsztein DC. Overexpression of yeast hsp104 reduces polyglutamine aggregation and prolongs survival of a transgenic mouse model of Huntington’s disease. Hum Mol Genet. 2005;14:3425–3433. doi: 10.1093/hmg/ddi372. [DOI] [PubMed] [Google Scholar]
- 210.Jackrel ME, Shorter J. Reversing deleterious protein aggregation with re-engineered protein disaggregases. Cell Cycle. 2014;13:1379–1383. doi: 10.4161/cc.28709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Jackrel ME, Shorter J. Engineering enhanced protein disaggregases for neurodegenerative disease. Prion. 2015;9:90–109. doi: 10.1080/19336896.2015.1020277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Jackrel ME, Yee K, Tariq A, Chen AI, Shorter J. Disparate mutations confer therapeutic gain of Hsp104 function. ACS Chem Biol. 2015;10:2672–2679. doi: 10.1021/acschembio.5b00765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Shorter J. The mammalian disaggregase machinery: Hsp110 synergizes with Hsp70 and Hsp40 to catalyze protein disaggregation and reactivation in a cell-free system. PLoS ONE. 2011;6:e26319. doi: 10.1371/journal.pone.0026319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Torrente MP, Shorter J. The metazoan protein disaggregase and amyloid depolymerase system: Hsp110, Hsp70, Hsp40, and small heat shock proteins. Prion. 2013;7:457–463. doi: 10.4161/pri.27531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Duennwald ML, Echeverria A, Shorter J. Small heat shock proteins potentiate amyloid dissolution by protein disaggregases from yeast and humans. PLoS Biol. 2012;10:e1001346. doi: 10.1371/journal.pbio.1001346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Poepsel S, Sprengel A, Sacca B, Kaschani F, Kaiser M, Gatsogiannis C, et al. Determinants of amyloid fibril degradation by the PDZ protease HTRA1. Nat Chem Biol. 2015;11:862–869. doi: 10.1038/nchembio.1931. [DOI] [PubMed] [Google Scholar]
- 217.Ali YO, Allen HM, Yu L, Li-Kroeger D, Bakhshizadehmahmoudi D, Hatcher A, et al. NMNAT2:HSP90 complex mediates proteostasis in proteinopathies. PLoS Biol. 2016;14:e1002472. doi: 10.1371/journal.pbio.1002472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Shorter J. Engineering therapeutic protein disaggregases. Mol Biol Cell. 2016;27:1556–1560. doi: 10.1091/mbc.E15-10-0693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Benajiba L, Le Ber I, Camuzat A, Lacoste M, Thomas-Anterion C, Couratier P, et al. TARDBP mutations in motoneuron disease with frontotemporal lobar degeneration. Ann Neurol. 2009;65:470–473. doi: 10.1002/ana.21612. [DOI] [PubMed] [Google Scholar]
- 220.Borroni B, Archetti S, Del Bo R, Papetti A, Buratti E, Bonvicini C, et al. TARDBP mutations in frontotemporal lobar degeneration: frequency, clinical features, and disease course. Rejuvenation Res. 2010;13:509–517. doi: 10.1089/rej.2010.1017. [DOI] [PubMed] [Google Scholar]
- 221.Chiò A, Calvo A, Moglia C, Restagno G, Ossola I, Brunetti M, et al. Amyotrophic lateral sclerosis-frontotemporal lobar dementia in 3 families with p. Ala382Thr TARDBP mutations. Arch Neurol. 2010;67:1002–1009. doi: 10.1001/archneurol.2010.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Driver-Dunckley E, Connor D, Hentz J, Sabbagh M, Silverberg N, Hernandez J, et al. No evidence for cognitive dysfunction or depression in patients with mild restless legs syndrome. Mov Disord. 2009;24:1843–1847. doi: 10.1002/mds.22701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Millecamps S, Salachas F, Cazeneuve C, Gordon P, Bricka B, Camuzat A, et al. SOD1, ANG, VAPB, TARDBP, and FUS mutations in familial amyotrophic lateral sclerosis: genotype-phenotype correlations. J Med Genet. 2010;47:554–560. doi: 10.1136/jmg.2010.077180. [DOI] [PubMed] [Google Scholar]
- 224.Moreno F, Rabinovici GD, Karydas A, Miller Z, Hsu SC, Legati A, et al. A novel mutation P112H in the TARDBP gene associated with frontotemporal lobar degeneration without motor neuron disease and abundant neuritic amyloid plaques. Acta Neuropathol Commun. 2015;3:19. doi: 10.1186/s40478-015-0190-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Soong BW, Lin KP, Guo YC, Lin CC, Tsai PC, Liao YC, et al. Extensive molecular genetic survey of Taiwanese patients with amyotrophic lateral sclerosis. Neurobiol Aging. 2014;35:2423.e1–2423.e6. doi: 10.1016/j.neurobiolaging.2014.05.008. [DOI] [PubMed] [Google Scholar]
- 226.Winton MJ, Van Deerlin VM, Kwong LK, Yuan W, Wood EM, Yu CE, et al. A90v TDP-43 variant results in the aberrant localization of TDP-43 in vitro. FEBS Lett. 2008;582:2252–2256. doi: 10.1016/j.febslet.2008.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Chiang HH, Andersen PM, Tysnes OB, Gredal O, Christensen PB, Graff C. Novel TARDBP mutations in Nordic ALS patients. J Hum Genet. 2012;57:316–319. doi: 10.1038/jhg.2012.24. [DOI] [PubMed] [Google Scholar]
- 228.Cady J, Allred P, Bali T, Pestronk A, Goate A, Miller TM, et al. Amyotrophic lateral sclerosis onset is influenced by the burden of rare variants in known amyotrophic lateral sclerosis genes. Ann Neurol. 2015;77:100–113. doi: 10.1002/ana.24306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Quadri M, Cossu G, Saddi V, Simons EJ, Murgia D, Melis M, et al. Broadening the phenotype of TARDBP mutations: the TARDBP Ala382Thr mutation and Parkinson’s disease in Sardinia. Neurogenetics. 2011;12:203–209. doi: 10.1007/s10048-011-0288-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Buratti E. Chapter One — functional significance of TDP-43 mutations in disease. In: Theodore Friedmann JCD, Stephen FG, editors. Advances in Genetics. Academic Press; 2015. pp. 1–53. [DOI] [PubMed] [Google Scholar]
- 232.Abel O, Shatunov A, Jones AR, Andersen PM, Powell JF, Al-Chalabi A. Development of a smartphone app for a genetics website: the amyotrophic lateral sclerosis online genetics database (ALSoD) JMIR Mhealth Uhealth. 2013;1:e18. doi: 10.2196/mhealth.2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Lattante S, Rouleau GA, Kabashi E. TARDBP and FUS mutations associated with amyotrophic lateral sclerosis: summary and update. Hum Mutat. 2013;34:812–826. doi: 10.1002/humu.22319. [DOI] [PubMed] [Google Scholar]
- 234.Blair IP, Williams KL, Warraich ST, Durnall JC, Thoeng AD, Manavis J, et al. FUS mutations in amyotrophic lateral sclerosis: clinical, pathological, neurophysiological and genetic analysis. J Neurol Neurosurg Psychiatry. 2010;81:639–645. doi: 10.1136/jnnp.2009.194399. [DOI] [PubMed] [Google Scholar]
- 235.Broustal O, Camuzat A, Guillot-Noel L, Guy N, Millecamps S, Deffond D, et al. FUS mutations in frontotemporal lobar degeneration with amyotrophic lateral sclerosis. J Alzheimers Dis. 2010;22:765–769. [PubMed] [Google Scholar]
- 236.Huey ED, Ferrari R, Moreno JH, Jensen C, Morris CM, Potocnik F, et al. FUS and TDP43 genetic variability in FTD and CBS. Neurobiol Aging. 2012;33:1016.e9–1016.e17. doi: 10.1016/j.neurobiolaging.2011.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Ticozzi N, Silani V, LeClerc AL, Keagle P, Gellera C, Ratti A, et al. Analysis of FUS gene mutation in familial amyotrophic lateral sclerosis within an Italian cohort. Neurology. 2009;73:1180–1185. doi: 10.1212/WNL.0b013e3181bbff05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Van Langenhove T, van der Zee J, Sleegers K, Engelborghs S, Vandenberghe R, Gijselinck I, et al. Genetic contribution of FUS to frontotemporal lobar degeneration. Neurology. 2010;74:366–371. doi: 10.1212/WNL.0b013e3181ccc732. [DOI] [PubMed] [Google Scholar]
- 239.Yan J, Deng HX, Siddique N, Fecto F, Chen W, Yang Y, et al. Frameshift and novel mutations in FUS in familial amyotrophic lateral sclerosis and ALS/dementia. Neurology. 2010;75:807–814. doi: 10.1212/WNL.0b013e3181f07e0c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Belzil VV, Valdmanis PN, Dion PA, Daoud H, Kabashi E, Noreau A, et al. Mutations in FUS cause FALS and SALS in French and French Canadian populations. Neurology. 2009;73:1176–1179. doi: 10.1212/WNL.0b013e3181bbfeef. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Exome Variant Server. NHLBI GO Exome Sequencing Project. Seattle, WA: http://evs.gs.washington.edu/EVS/ [Google Scholar]
- 242.Groen EJN, van Es MA, van Vught PWJ, Spliet WGM, van Engelen-Lee J, de Visser M, et al. FUS mutations in familial amyotrophic lateral sclerosis in the Netherlands. Arch Neurol. 2010;67:224–230. doi: 10.1001/archneurol.2009.329. [DOI] [PubMed] [Google Scholar]
- 243.Hewitt C, Kirby J, Highley JR, Hartley JA, Hibberd R, Hollinger HC, et al. Novel FUS/TLS mutations and pathology in familial and sporadic amyotrophic lateral sclerosis. Arch Neurol. 2010;67:455–461. doi: 10.1001/archneurol.2010.52. [DOI] [PubMed] [Google Scholar]
- 244.Belzil VV, Daoud H, St-Onge J, Desjarlais A, Bouchard JP, Dupre N, et al. Identification of novel FUS mutations in sporadic cases of amyotrophic lateral sclerosis. Amyotroph Lateral Scler. 2011;12:113–117. doi: 10.3109/17482968.2010.536840. [DOI] [PubMed] [Google Scholar]
- 245.Lee BJ, Cansizoglu AE, Süel KE, Louis TH, Zhang Z, Chook YM. Rules for nuclear localization sequence recognition by karyopherinβ2. Cell. 2006;126:543–558. doi: 10.1016/j.cell.2006.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Zakaryan RP, Gehring H. Identification and characterization of the nuclear localization/retention signal in the EWS proto-oncoprotein. J Mol Biol. 2006;363:27–38. doi: 10.1016/j.jmb.2006.08.018. [DOI] [PubMed] [Google Scholar]
- 247.Marko M, Vlassis A, Guialis A, Leichter M. Domains involved in TAF15 subcellular localisation: dependence on cell type and ongoing transcription. Gene. 2012;506:331–338. doi: 10.1016/j.gene.2012.06.088. [DOI] [PubMed] [Google Scholar]
- 248.Thandapani P, O’Connor TR, Bailey TL, Richard S. Defining the RGG/RG motif. Mol Cell. 2013;50:613–623. doi: 10.1016/j.molcel.2013.05.021. [DOI] [PubMed] [Google Scholar]
- 249.Jean-Philippe J, Paz S, Lu ML, Caputi M. A truncated hnRNP A1 isoform, lacking the RGG-box RNA binding domain, can efficiently regulate HIV-1 splicing and replication. Biochim Biophys Acta, Gene Regul Mech. 2014;1839:251–258. doi: 10.1016/j.bbagrm.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]