Summary
Satellite cells are skeletal-muscle-specific stem cells that are activated by injury to proliferate, differentiate, and fuse to enable repair. SOX7, a member of the SRY-related HMG-box family of transcription factors is expressed in quiescent satellite cells. To elucidate SOX7 function in skeletal muscle, we knocked down Sox7 expression in embryonic stem cells and primary myoblasts and generated a conditional knockout mouse in which Sox7 is excised in PAX3+ cells. Loss of Sox7 in embryonic stem cells reduced Pax3 and Pax7 expression. In vivo, conditional knockdown of Sox7 reduced the satellite cell population from birth, reduced myofiber caliber, and impaired regeneration after acute injury. Although Sox7-deficient primary myoblasts differentiated normally, impaired myoblast fusion and increased sensitivity to apoptosis in culture and in vivo were observed. Taken together, these results indicate that SOX7 is dispensable for myogenesis but is necessary to promote satellite cell development and survival.
Keywords: SOX7, myogenesis, muscle satellite cells, PAX3, muscle regeneration, PAX7, conditional knockout, embryonic stem cell
Highlights
-
•
Loss of Sox7 in embryonic stem cells impairs myogenic precursor production
-
•
SOX7 regulates Pax7 expression
-
•
SOX7 protects satellite cells from apoptosis after injury
-
•
Loss of Sox7 in satellite cells impairs myoblast fusion and muscle regeneration
In this article, Wiper-Bergeron and colleagues demonstrate that the transcription factor SOX7 is required for skeletal muscle satellite cell development and maintenance. SOX7 regulates Pax7 expression, and loss of SOX7 leads to fewer satellite cells that are more sensitive to apoptosis. Furthermore, loss of Sox7 in satellite cells results in smaller myotubes and impaired regeneration after injury in post-natal muscle.
Introduction
With the exception of a few muscles (mostly in the head), skeletal muscle is derived from the somites, specifically the dermomyotome (George-Weinstein et al., 1996, Gros et al., 2004). Within the dermomyotome, muscle progenitor cells expressing the transcription factors Paired box 3 (PAX3), PAX7, Mesenchyme Homeobox 1 (MEOX1), and GLI2 migrate ventrally to form the myotome, which is the site where the first fully differentiated embryonic myocytes are found (Hollway and Currie, 2005). A pool of muscle precursor cells expressing PAX3, PAX7, and MEOX1 (premyogenic mesoderm) become committed to myogenesis by expressing the myogenic regulatory factors (MRFs) MYF5, MYOD, and myogenin driving terminal differentiation into myocytes that express structural and contractile proteins such as myosin heavy chain (MyHC) (Relaix et al., 2005).
The double-positive PAX3+PAX7+ population of cells present in the central dermomyotome also gives rise to adult muscle stem cells, known as satellite cells, which have an important role in maintaining skeletal muscle throughout the life of an organism (Buckingham and Relaix, 2007, Relaix et al., 2006). At approximately E16, which marks the end of fetal myogenic development, PAX3+PAX7+ progenitor cells that do not express myogenic regulatory factors begin to accumulate under the basal lamina that forms around muscle fibers, where they enter a state of quiescence as satellite cells, and downregulate PAX3 expression (Relaix et al., 2005). These satellite cells remain dormant until the post-natal growth period, characterized by the fusion of differentiated satellite cells to a fixed number of muscle fibers established during development. In adult mice, satellite cells can exit their dormant state after muscle injury and become activated to differentiate and fuse to existing muscle fibers or to one another to form new fibers (Mauro, 1961, Yin et al., 2013).
Although PAX7 activity is not essential for embryonic myogenesis in the presence of PAX3, Pax7−/− mice lack these precursors (Mansouri et al., 1996, Relaix et al., 2006, Seale et al., 2000). While PAX3 can be detected in quiescent satellite cells, though less predominantly in the hindlimb, in the absence of Pax7, these cells fail to survive the post-natal growth period (Relaix et al., 2006). Further, ablation of cells expressing PAX7 blocks regenerative myogenesis following injury, underscoring an essential role for satellite cells in acute injury-induced muscle regeneration (Lepper et al., 2011, Murphy et al., 2011, Sambasivan et al., 2011), and PAX7 has been shown to be required for satellite cell survival (von Maltzahn et al., 2013). Alternatively, in mice lacking PAX3 expression, the hypaxial somite is lost due to increased apoptosis, and the formation of limb muscles is impaired (Franz et al., 1993, Relaix et al., 2004).
The SOX family of proteins are characterized by a 79-amino acid High-Mobility Group (HMG) domain that shares more than 60% sequence similarity to the HMG box of SRY (sex determining region Y protein) (Capel, 2000). SOX genes are involved in the regulation of embryonic development, chondrogenesis, neural progenitor cell fate, and cardiovascular development (Sarkar and Hochedlinger, 2013). The 30 known SOX genes are classified into nine subgroups (A–I) based on the degree of homology within the HMG domain and the presence of conserved motifs outside the HMG box (Badis et al., 2009, Bowles et al., 2000, Kondoh and Kamachi, 2010, Soullier et al., 1999, Wegner, 1999). Of these, SoxG member SOX15 has been shown to be dispensable for embryonic myogenesis, although in its absence, post-natal muscle regeneration is impaired and myogenic differentiation is delayed (Lee et al., 2004). SoxE family members SOX8 and SOX9 are expressed in satellite cells and act as inhibitors of myogenesis (Schmidt et al., 2003).
SOX7 is a member of subgroup F along with Sox17 and Sox18. Sox7 mRNA is expressed during embryonic development, most abundantly in the brain, heart, lung, kidney, prostate, colon and spleen (Takash et al., 2001). Loss of Sox7 is embryonic lethal between days E10.5 and 14.5, with delayed development and failure of organogenesis observed (Wat et al., 2012). While myogenin expression can be detected in the early myotome at E8.5, no specific muscle phenotype is described in Sox7−/− mice (Sassoon et al., 1989, Wat et al., 2012). SOX7 is a transcription factor and can regulate the expression of Fgf3, Lama1, and Cdh5, among others, as well as activate transcription from a synthetic reporter containing SOX consensus motifs (Costa et al., 2012, Murakami et al., 2004, Niimi et al., 2004).
SOX7 has been implicated in the regulation of vascular development and cardiogenesis (Cermenati et al., 2008, Costa et al., 2012, Shimoda et al., 2007, Zhang et al., 2005) and during normal development is expressed beginning at the 4-somite stage (8 d.p.c.) in the somites and head regions. In the post-natal animal, SOX7 expression is found in quiescent adult muscle satellite cells, and its expression is decreased with activation and differentiation (Ikemoto et al., 2007, Pallafacchina et al., 2010), suggesting a role for SOX7 in this stem cell population.
In P19 embryonal carcinoma cells, which can be differentiated into skeletal myotubes, SOX7 expression is both necessary and sufficient to trigger the formation of skeletal myocytes in conditions that do not typically support myogenesis (Savage et al., 2009). Ectopic SOX7 expression stimulates PAX3 and PAX7 expression, leading to enhanced myogenic regulatory factor expression (MYF5, MYOD, and myogenin) and better terminal myogenic differentiation (Savage et al., 2009), suggesting that SOX7 is important for embryonic myogenesis.
Given that SOX7 is expressed in satellite cells and can regulate PAX3 and PAX7 expression in embryonal carcinoma cells, we examined SOX7 function during myogenesis in embryonic stem cell cultures and through the creation of a conditional null mouse in which Sox7 was excised in PAX3-expressing myogenic precursors and their derivatives in vivo and in vitro. Here, we show that while SOX7 expression is dispensable for myogenic differentiation downstream of PAX3 induction, it is necessary for the development and maintenance of the satellite cell pool.
Results
Knockdown of Sox7 in Embryonic Stem Cells Impairs Pax7 Expression
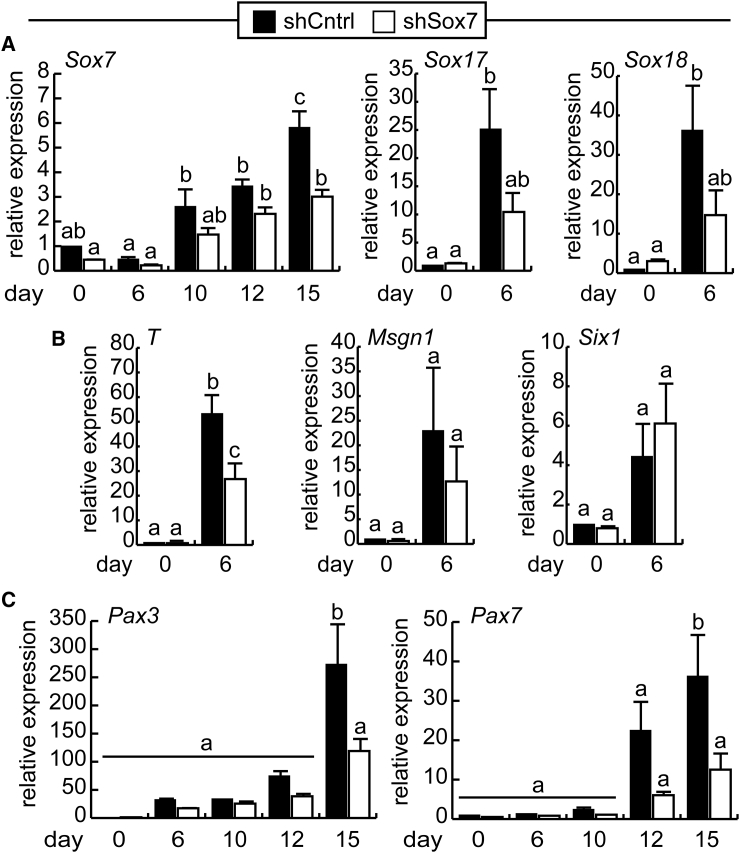
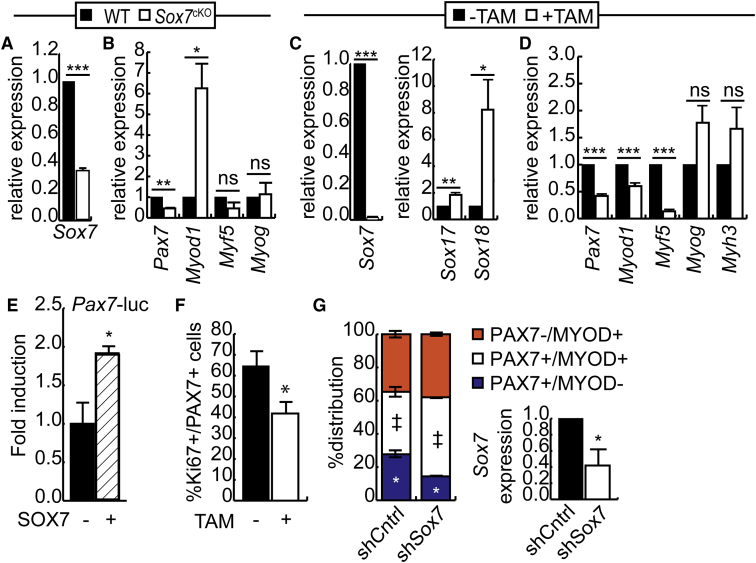
To assess the role of SOX7 in the embryonic development of muscle, we first used a loss-of-function approach in mouse embryonic stem cells (ESCs). We transduced mouse ESCs with a lentiviral vector that expresses a short hairpin RNA (shRNA) construct directed against Sox7 (shSox7) or a scrambled control (shCntrl) and cultured them to develop into myogenic precursors (Figure 1A). Knockdown of Sox7 expression was verified by RT-qPCR at day 0 to 15 of culture. Sox7 expression was most highly expressed at day 15 in shCntrl cells, and expression of the shSox7 construct resulted in statistically significant reduction in Sox7 expression at this time point (Figure 1A). To determine if reduced Sox7 expression influenced the expression of subgroup members, the expression of Sox17 and Sox18 was verified. At day 0, when both factors were weakly expressed, knockdown of Sox7 did not affect Sox17 and Sox18 expression (Figure 1A). On day 6, both Sox17 and Sox18 expression increased relative to day 0, and while trending toward decreased expression in the shSox7 cells, this failed to achieve statistical significance.
Figure 1.
Knockdown of Sox7 Expression in Mouse ESCs Reduces Pax7 Expression and Promotes Myogenic Differentiation
(A) Sox7 and subgroup member mRNA expression in a mouse embryonic stem cell line stably expressing a shRNA targeting Sox7 (shSox7) or a scrambled control sequence (shCntrl) measured by RT-qPCR, shown relative to expression in controls at day 0. n = 5.
(B) Expression of mesoderm markers and Six1 on day 0 and day 6. n = 5.
(C) Expression of Pax3 and Pax7 from day 0 to day 15 of differentiation, shown relative to expression in shCntrl cells on day 12. n = 3.
For all experiments, bars represent the mean and error bars are the SEM for biological replicates. Bars indicated with distinct letters are statistically different from one another at a minimum cut-off of p < 0.05.
The formation of mesoderm (BraT, Msgn1), premyogenic mesoderm (Six1), and myogenic cells (Pax3, Pax7) was assessed next in shSox7 cultures and scrambled controls (Figure 1B). While BraT expression was reduced at day 6 in shSox7 cultures compared with shCntrl cultures, Msgn1 expression was unaffected by changes in Sox7 expression. Further, the formation of premyogenic mesoderm was equally unaffected at day 6 as expression of Six1 was comparable with controls, suggesting that Sox7 is dispensable for the formation of these tissues. However, expression of both Pax3 and Pax7 was significantly reduced in shSox7 cultures on day 15 of culture compared with controls, suggesting that the formation of myogenic precursors is impaired in cells with reduced Sox7 expression (Figure 1C). Taken together, these results suggest that SOX7 is required for normal expression of Pax3 and Pax7 during embryonic myogenic differentiation and promotes their differentiation.
Sox7cKO Mice Have Fewer PAX7+ Satellite Cells at Birth
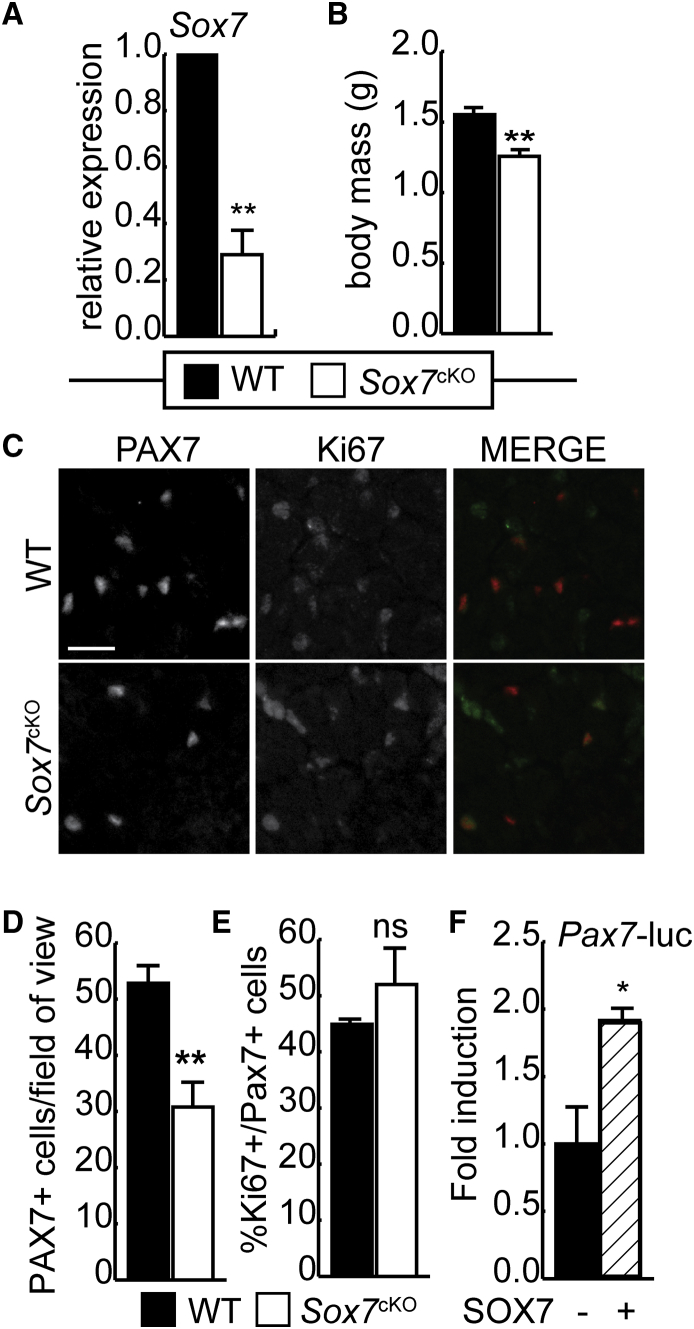
Given that knockdown of Sox7 results in reduced Pax7 expression during differentiation of ESCs, we sought to determine if the satellite cell population developed normally during embryogenesis in the absence of Sox7. In order to generate a mouse strain in which Sox7 is excised in skeletal muscle precursors, Sox7fl/fl mice (Wat et al., 2012) were crossed with mice expressing the Pax3-Cre driver (Pax3tm1(cre)Joe/J; Jackson Laboratories) to produce mice in which Sox7 is excised in all PAX3+ cells and their derivatives. Conditional Sox7 knockout animals (Sox7cKO, Sox7fl/flPax3Cre/+) were produced at Mendelian ratios, with the majority of animals displaying the white belly splotch characteristic of Pax3 haploinsufficiency but otherwise free from overt phenotypes related to limb development or mobility (Auerbach, 1954). We did notice a small increase in perinatal death for the Sox7cKO genotype, occurring at a frequency of 1:46 for wild-type (WT) pups and 1:10 Sox7cKO pups. Increased perinatal death is consistent with what is observed in mice lacking Pax7+ cells (Hutcheson et al., 2009). To verify excision of the Sox7 gene in Cre-expressing mice, primary myoblasts were isolated from Sox7cKO and non-Cre-expressing control mice (WT, Sox7fl/flPax3+/+), and isolated mRNA was subjected to RT-qPCR analysis (Figure 2A). Sox7 expression was reduced by approximately 71% in the Sox7cKO cells compared with control cultures. Sox7cKO mice were significantly smaller at birth, weighing approximately 80% of WT control littermates (Figure 2B) without any gross abnormalities. To assess the size of the satellite cell population, the number of PAX7+ cells was scored in hindlimb muscle (whole-leg cross-sections). Consistent with what was observed during ESC differentiation, there were significantly fewer PAX7+ cells in Sox7cKO mice than in littermate controls (Figures 2C and 2D), suggesting that the defect in satellite cell numbers is established prior to birth. Despite the lower number of satellite cells at birth, the percentage of PAX7+ cells that were proliferating (Ki67+) was equivalent in both genotypes (Figure 2E).
Figure 2.
Loss of Sox7 in PAX3+ Cells Reduces Satellite Cell Numbers at Birth
(A) RT-qPCR analysis of Sox7 expression in primary myoblasts isolated from Sox7fl/fl;Pax3Cre/+ (Sox7cKO) and Sox7fl/fl;Pax3+/+ (WT) mice aged 6 weeks in growth medium. ∗∗p < 0.01. n = 3 biological replicates.
(B) Body mass of Sox7cKO and WT mice at birth. ∗∗p < 0.01. n = 3 mice per genotype.
(C) Immunohistochemistry of PAX7 expression and Ki67 staining in hindlimb muscle from Sox7cKO and WT mice at birth. Scale bar, 50 μm.
(D) Number of PAX7+ cells per field of view. ∗∗p < 0.01. n = 3 mice per genotype.
(E) Percentage of Ki67+/PAX7+ cells relative to total PAX7+ cells. ns, not significant.
(F) Reporter assay measuring Pax7 promoter activity in primary myoblasts in the presence or absence of ectopic SOX7. Data are represented as fold induction over the activity obtained in the absence of SOX7. Error bars are the SEM, n = 3. ∗p < 0.05.
Loss of Sox7 Leads to a Decrease in the Average Muscle Fiber Size
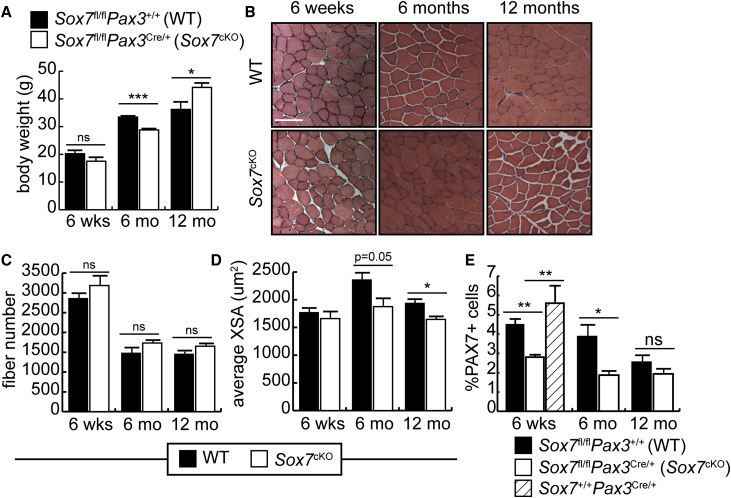
Despite the reduced satellite cell compartment in Sox7cKO mice, post-natal growth of skeletal muscle was normal. At 6 weeks of age, both Sox7cKO and control mice had the same body weight (Figure 3A), and muscle histology was comparable in the Sox7cKO mice and littermate controls (Figures 3B–3D).
Figure 3.
Conditional Knockout of Sox7 in PAX3+ Cells Results in Smaller Muscle Fibers and Fewer Satellite Cells
(A) Mean body mass of Sox7fl/fl;Pax3Cre/+ (Sox7cKO) and Sox7fl/fl;Pax3+/+ (WT) mice aged 6 weeks, 6 months (6 mo), and 12 months (12 mo). ∗p < 0.05, ∗∗∗p < 0.001; ns, not significant. n = 3 mice per group.
(B) Representative H&E-stained cross-sections of the TA muscle from Sox7fl/fl;Pax3Cre/+ (Sox7cKO) and Sox7fl/fl;Pax3+/+ (WT) mice. Scale bar, 50 μm.
(C) Mean fiber number in the TA muscle.
(D) Average cross-sectional area (XSA) of fibers in the TA muscle of Sox7cKO and WT mice. ∗p < 0.05. n = 3 mice per group.
(E) Percentage of PAX7+ cells relative to total nuclei in TA muscle from WT, Sox7cKO, and Sox7+/+;Pax3Cre/+ mice as indicated. ∗p < 0.05, ∗∗p < 0.01; ns, not significant. n = 3 mice per group. Error bar is the SEM.
When Sox7cKO and WT mice were allowed to age, a defect in myofiber cross-sectional area became apparent. At 6 months of age, Sox7cKO animals were significantly lighter than littermate controls, while at 12 months of age, the Sox7 cKO mice were obese, characterized by an increase in subcutaneous white adipose tissue (Figures 3A and S1). Assessment of muscle histology revealed no significant differences in fiber number at either time point, although a significant decrease in the mean fiber cross-sectional area was observed for Sox7cKO mice at both 6 and 12 months compared with controls (Figures 3B–3D).
Using immunohistochemistry, we evaluated the number of PAX7+ cells present in tibialis anterior (TA) muscle sections from Sox7cKO mice and compared it with control mice (Figure 3E). While PAX7+ cells made up approximately 4.5% of nuclei in control muscle at 6 weeks, this was reduced significantly by approximately 50% in Sox7cKO muscle sections. With aging, the percentage of Pax7+ cells slowly decreased in control mice, with a significantly smaller population observed in Sox7cKO animals at 12 months of age that was not significantly different from Sox7cKO mice (Figure 3E). These results suggest that loss of SOX7 leads to a decrease in the number of satellite cells and smaller muscle fibers with age. The decrease in PAX7+ cells was not due to haploinsufficiency at the Pax3 locus due to insertion of the Cre recombinase, as Sox7+/+Pax3Cre/+ mice had a satellite cell population equivalent to Sox7fl/fl Pax3+/+ controls (Figure 3E).
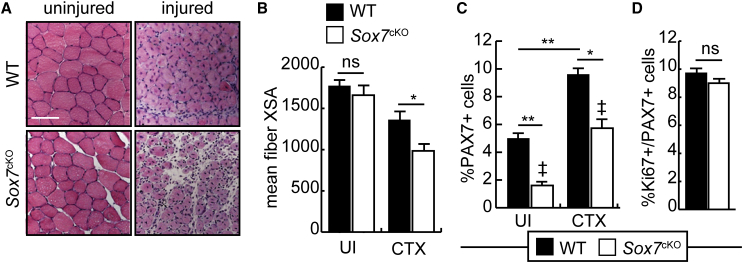
Impaired Regeneration in Sox7cKO Mice
Given the decrease in satellite cell numbers in Sox7cKO mice, we evaluated muscle regenerative capacity after acute injury in 6-week-old mice. Seven days after acute injury with cardiotoxin, littermate control animals (WT) demonstrated robust repair with regenerating fibers (characterized by centrally located nuclei) regaining a fiber cross-sectional area 77% that of uninjured muscle (Figures 4A and 4B). While the mean fiber cross-sectional area was equivalent in uninjured muscle from WT and Sox7cKO mice, regenerating Sox7cKO muscles only recovered to 59% of uninjured controls and were approximately 28% smaller than regenerating WT myofibers, suggesting an impaired repair response (Figures 4A and 4B).
Figure 4.
Loss of Sox7 Impairs Muscle Regeneration after Acute Injury
(A) Representative H&E-stained cross-sections of the TA from injured or uninjured Sox7fl/fl;Pax3Cre/+ (Sox7cKO) and Sox7fl/fl;Pax3+/+ (WT) mice aged 6 weeks. Scale bar, 50 μm.
(B) Average myofiber cross-sectional area 7 days after cardiotoxin injury (CTX) or in uninjured (UI) muscle. ∗p < 0.05; ns, not significant. n = 3 mice per group.
(C) Percentage of PAX7+ cells relative to total nuclei 7 days after injury with CTX or in uninjured muscle. ∗p < 0.05, ∗∗p < 0.01, samples marked with ‡ are significantly different from one another, p < 0.01. n = 3 mice per group.
(D) Percentage of Ki67+/PAX7+ cells (relative to total PAX7+ cells) 1 week after injury with CTX. ns, not significant. Error bar is the SEM.
While 6-week-old Sox7cKO mice had significantly fewer PAX7+ cells in uninjured muscle compared with littermate controls, this population expanded after injury (Figure 4C). Despite expansion of the PAX7+ populations in both WT and Sox7cKO mice after injury (1.9-fold and 3.6-fold, respectively), the percentage of PAX7+ cells remained significantly lower in Sox7cKO muscle compared with WT controls (Figure 4C). Consistent with these observations, the percentage of PAX7+ cells that were also cycling (Ki67+) was similar in both WT and Sox7cKO muscle, suggesting that Sox7cKO PAX7+ cells are able to expand normally after injury (Figure 4D), although their numbers after expansion remain significantly lower than WT.
Satellite Cell Dysfunction with Loss of SOX7
Given the impaired regenerative response in Sox7cKO mice, we sought to determine if satellite cell dysfunction contributed to the phenotype. To evaluate if the PAX7+ population lacking SOX7 function normally, primary myoblasts were isolated from the hindlimb of 5- to 6-week-old Sox7cKO and littermate controls, and Sox7 expression was verified by RT-qPCR in proliferating cells (Figure 5A). Sox7 expression was reduced by approximately 70% in freshly isolated satellite cells compared with control cultures, comparable with satellite cells isolated from older mice (Figure 2A). Freshly isolated Sox7cKO cells expressed significantly less Pax7 and 6-fold more Myod1 than WT controls, without changes in Myf5 or Myog expression (Figure 5B).
Figure 5.
Loss of Sox7 in Primary Myoblasts Results in Loss of Pax7 Expression
(A) RT-qPCR analysis of Sox7 expression in freshly isolated primary myoblasts isolated from Sox7fl/fl;Pax3Cre/+ (Sox7cKO) and Sox7fl/fl;Pax3+/+ (WT) mice aged 5 weeks. ∗∗∗p < 0.001.
(B) RT-qPCR analysis of Pax7, Myod1, Myf5, and Myog expression in freshly isolated primary myoblasts from Sox7cKO mice relative to expression in control littermates. Error bars are the SEM, ∗∗p < 0.01, ∗p < 0.05; ns, not significant. n = 3 biological replicates.
(C) RT-qPCR analysis of Sox7 expression and Sox17 and Sox18 expression in primary myoblasts isolated from the Sox7fl/fl mouse and retrovirally transduced to express the CreER recombinase following treatment with 4-OH tamoxifen (+TAM) to excise Sox7 or with vehicle (−TAM). ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
(D) RT-qPCR analysis of Pax3, Pax7, Myod1, Myf5, Myog, and Myhc3 expression in Sox7 knockdown cells (+TAM) relative to expression in vehicle-treated controls. Error bars are the SEM. ∗∗∗p < 0.001; ns, not significant. n = 5 biological replicates.
(E) Luciferase reporter assay in primary myoblasts measuring activity from the Pax7 promoter in the presence or absence of SOX7. Data are shown as fold over reporter alone and internally controlled for transfection efficiency using a constitutively active β-galactosidase reporter. ∗p < 0.05. n = 4. Error bars are the SEM.
(F) Percentage of proliferating (Ki67+) PAX7+ cells following Sox7 excision compared with vehicle-treated controls. ∗p < 0.05. n = 3 biological replicates.
(G) Distribution of PAX7+/MYOD−, PAX7+/MYOD+, and PAX7-/MYOD+ cells in myofiber-associated satellite cells 72 hr post isolation as determined by immunocytochemistry. Myofibers from the Sox7fl/fl mouse were isolated and transduced with lentivirus to express a shRNA against Sox7 or a scrambled control (shCntrl). Error bars are the SEM. Populations indicated with ‡ are significantly different from one another (p < 0.05). Populations indicated by ∗ are significantly different from one another (p < 0.01). Results are from an average of 15 fibers per animal with three animals per group. Myofibers were cultured on Matrigel to induce satellite cell growth and migration for confirmation of Sox7 knockdown (inset). ∗p < 0.05.
Since excision in the Sox7cKO model was not complete and resulted in a heterogeneous population of primary myoblasts, we confirmed the molecular phenotype of primary myoblasts lacking Sox7 in a second model system. Primary myoblasts were isolated from the hindlimb of adult Sox7fl/fl and retrovirally transduced to express the CreER recombinase. Sox7 excision was induced with tamoxifen treatment, while control cultures were generated with vehicle treatment. Sox7 expression was verified by RT-qPCR in proliferating cells (Figure 5C) and was reduced by approximately 98% compared with control cultures. The expression of the remaining subgroup F members was also verified, and in contrast with what was observed in ESCs, both Sox17 and Sox18 were upregulated in primary myoblasts lacking Sox7 (Figure 5C). Depletion of Sox7 resulted in significantly less Pax7, Myod1, and Myf5 expression than vehicle-treated controls, while Myog and Myhc3 expression trended toward increased expression, without reaching statistical significance (Figure 5D).
Given that Pax7 expression was reduced in both ESCs and primary myoblasts in which Sox7 was knocked down, we performed a reporter assay measuring the activity of a Pax7 promoter-luciferase construct in primary myoblasts in the presence or absence of ectopic SOX7. Addition of ectopic SOX7 expression in these cells resulted in a 2-fold increase in promoter activity, suggesting that SOX7 can regulate Pax7 expression (Figure 5E).
As the higher levels of myogenin could indicate precocious differentiation under growth conditions, we performed Ki67 staining to determine the proportion of proliferating cells. Consistent with cell-cycle exit for differentiation, the cultures lacking Sox7 had significantly fewer proliferating myoblasts than vehicle-treated controls (Figure 5F).
To better understand the phenotype, we isolated myofibers from the Sox7fl/fl extensor digitorum longus (EDL) muscle and immediately after isolation used lentivirus to transduce the fibers to express an shRNA directed against Sox7 (shSox7) or with a scrambled control (shCntrl). Knockdown of Sox7 expression was verified in primary myoblasts that were allowed to grow off the transduced myofibers using RT-qPCR, revealing a ∼60% reduction in Sox7 expression in the shSox7 cells compared with controls (Figure 5G, inset). We performed immunocytochemistry for PAX7 and MYOD expression on transduced myofibers 72 hr after isolation. Expression of PAX7 and MYOD can be used to distinguish three populations of muscle precursors: PAX7+/MYOD− cells are quiescent or self-renewing, PAX7+/MYOD+ cells are proliferating and PAX7-/MYOD+ cells are differentiating. Knockdown of Sox7 expression resulted in a reduction of the PAX7+/MYOD− population, and a concomitant increase in the PAX7+/MYOD+ cells, suggesting that primary myoblasts with reduced Sox7 progress toward differentiation more readily than controls under growth conditions (Figure 5G). However, we failed to observe an increase in PAX7−MYOD+ cells, suggesting that either the cells stall during differentiation or progress rapidly toward a MYOG+ state (Figure 5G).
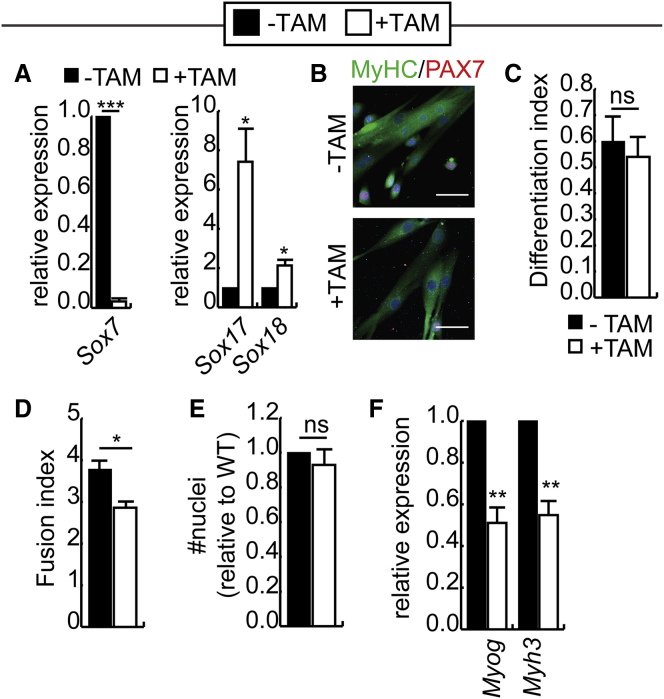
To evaluate the importance of SOX7 in the differentiation of primary myoblasts, primary myoblasts were differentiated for 2 days in low serum conditions. Sox7, Sox17, and Sox18 expression was verified by RT-qPCR after differentiation, and similar to under growth conditions, both Sox17 and Sox18 were upregulated in cells lacking Sox7 (Figure 6A). The extent of differentiation was quantified by immunocytochemistry for MyHC. While the differentiation index (% of nuclei found inside MyHC+ cells relative to total nuclei) was unchanged with loss of SOX7, the myosin heavy-chain-expressing myotubes were smaller in tamoxifen-treated cultures compared with control cultures (Figures 6B and 6D). Indeed, average myotube size, measured as the fusion index (no. of nuclei in MyHC+ myotubes/no. of myotubes) was significantly reduced in the absence of Sox7 (Figure 6D). Since efficient cell fusion relies on high culture density, we ensured that the total number of nuclei in both tamoxifen- and vehicle-treated differentiated cultures were equivalent (Figure 6E). However, it should be noted that cultures lacking Sox7 had to be plated at approximately 2-fold higher densities to ensure equal plating by the end of the experiment, indicating a cell growth defect or increased cell death. Finally, the expression of the late myogenesis markers Myog and Myhc3 was analyzed in differentiated cultures, revealing that the expression of both was significantly decreased in cultures lacking Sox7, consistent with incomplete myogenic differentiation and poor fusion (Figure 6F).
Figure 6.
Sox7 Is Not Required for Post-natal Myogenic Differentiation
(A) RT-qPCR analysis of Sox7, Sox17, and Sox18 expression in Sox7fl/fl primary myoblasts retrovirally transduced to express CreER, treated with TAM or vehicle (−TAM), and induced to differentiate for 2 days under low serum conditions.
(B) Representative images of primary myoblasts isolated and differentiated for 2 days in low serum conditions and stained for myosin heavy chain expression (MyHC; green), PAX7 (red), and counterstained with DAPI (blue) to reveal fused myotubes. Scale bar, 50 μm.
(C) Differentiation index (no. of nuclei in MyHC+ cells/total nuclei) for cultures differentiated as in (A). n = 3.
(D) Fusion index (no. of nuclei in myotubes/total number of myotubes), where a myotube is defined as having 2 or more nuclei. n = 3.
(E) Number of nuclei per field of view in images used for counting in (C and D), shown relative to –TAM controls. n = 3.
(F) RT-qPCR analysis of Myog and Myh3 expression in differentiated primary myoblasts from cells cultured as in (A).
For all experiments, error bars are the SEM. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001; ns, not significant. For RT-qPCR experiments, n = 6 biological replicates. For all other experiments, n = 3 biological replicates.
SOX7 Promotes Myoblast Survival
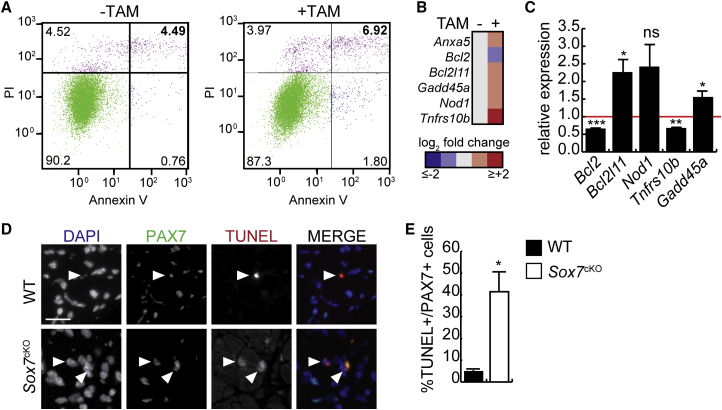
Given the decreased cell density in primary myoblasts lacking Sox7, we examined cell death as a possible mechanism for the reduced cell numbers. In growth medium, in the absence of any apoptotic stimulus, we observed more dead cells in proliferating cultures and confirmed this observation using flow cytometry. Propidium iodide (PI) and Annexin V staining revealed an increased percentage of Annexin V+/PI+ cells in tamoxifen-treated cultures in the absence of apoptotic stimuli (Figure 7A). We collected mRNA from proliferating tamoxifen- and vehicle-treated myoblast cultures to perform a PCR array for regulators of apoptosis. Concentrating on the genes that were regulated at least 1.5-fold in cells lacking Sox7, we identified six genes (Figure 7B) of which three were validated by subsequent RT-qPCR analysis (Bcl2, Bcl2l11, and Gadd45a) (Figure 7C).
Figure 7.
Sox7 Is Required for Myoblast Survival
(A) Flow cytometric evaluation of cell death using propidium iodide (PI) and Annexin V staining in proliferating Sox7fl/fl primary myoblasts retrovirally transduced to express CreER and treated with TAM or vehicle. The percentages of events recorded for Annexin V−/PI− (live cells), Annexin V+/PI− (early apoptotic cells), Annexin V−/PI+ (necrotic cells), and Annexin V+/PI + cells (dead cells) are indicated in the respective quadrants.
(B) Heatmap of genes that regulate apoptosis that are differentially regulated in proliferating primary myoblasts in the absence of Sox7 as measured by PCR array.
(C) Validation of hits from (B) by RT-qPCR. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001; ns, not significant. n = 3 biological replicates.
(D) Representative images of TUNEL-stained and PAX7-immunostained muscle sections from TA muscle isolated from Sox7fl/fl;Pax3Cre/+ (Sox7cKO) and Sox7fl/fl;Pax3+/+ (WT) mice aged 6 weeks 7 days after injury with cardiotoxin. White arrowheads indicate TUNEL+/PAX7+ cells. Scale bar, 20 μm.
(E) Quantification of TUNEL+/PAX7+ cells from (D) shown as a percentage of total PAX7+ cells. ∗p < 0.05. n = 3 animals per group.
To assess whether the increased sensitivity to apoptosis in myoblasts lacking Sox7 was relevant in vivo, we performed TUNEL staining on muscle sections from Sox7cKO and littermate control (WT) mice 7 days after acute injury with cardiotoxin (Figure 7D). Consistent with our observation in culture, Sox7cKO muscle had significantly increased TUNEL+/Pax7+ cells compared with controls (Figures 7D and 7E), suggesting that SOX7 promotes the survival of PAX7+ cells in vivo. Taken together, these data indicate that SOX7 is required for satellite cell survival, and that in its absence, the PAX7+ population is reduced, resulting in lowered regenerative capacity.
Discussion
We found that mice lacking Sox7 in the skeletal muscle-specific lineage have normal muscle architecture with smaller muscle fiber cross-sectional areas. When challenged with acute injury, the Sox7cKO muscle repaired inefficiently, and isolated primary myoblasts, although differentiation competent, were more sensitive to cell death and fused inefficiently. Indeed, the primary defect in the Sox7fl/flPax3Cre/+ conditional null model is a significant reduction in the satellite cell compartment, which is observed at birth and persists until 12 months of age, suggesting a role for SOX7 in the development of muscle stem cells and their maintenance in the post-natal animal. Interestingly, in addition to a smaller satellite cell compartment, Sox7cKO mice develop obesity with age. This phenotype, which is unlikely to be related to the satellite cell defect, suggests a role for SOX7 in other Pax3-derived cell populations. Embryonic PAX3+ cells contribute to the development of the CNS and the neural crest, which is the origin for numerous tissues including the mesenchyme (Monsoro-Burq, 2015). As such, loss of SOX7 may affect central behavior driving weight gain, may promote adipogenesis in white fat depots, or alternatively, negatively affect the formation of brown fat, which promotes energy expenditure.
The induction of PAX7 is required for the normal development of satellite cells and their survival. Our results suggest that SOX7 is required for the induction of PAX7 during embryonic development, and that failure to do so results in a smaller muscle stem cell pool that is less able to support muscle regeneration postnatally. Motif analysis of the Pax7 regulatory region revealed the presence of two putative SOX7 DNA binding motifs in the 2,500 base pairs upstream of the transcriptional start site, and paired with our reporter assay findings, suggest that SOX7 is a transcriptional regulator of Pax7 expression.
In addition to the smaller satellite cell population, primary myoblasts lacking Sox7 were more sensitive to apoptosis, even in the absence of apoptotic stimuli, suggesting that SOX7 may be anti-apoptotic. Indeed, PAX7 has been shown to be important for satellite cell survival, and it remains possible that SOX7 acts to promote survival through potentiation of PAX7 expression. However, we also demonstrate that loss of Sox7 decreases the expression of the pro-survival gene Bcl2 and leads to an increase in the expression of two pro-apoptotic genes (Bcl2l11 and Gadd45a). This mechanism appears to be unique to skeletal muscle tissues as SOX7 has been shown to be pro-apoptotic in colorectal cancers and non-small-cell lung cancers, supporting the notion that regulation of Pax7 contributes to the pro-survival actions (Hayano et al., 2013, Zhang et al., 2009). Despite an increased susceptibility to cell death in vivo after injury, the satellite cell population in Sox7cKO animals is very stable with aging. Given the incomplete excision of Sox7, it is likely that PAX7+ cells that retain Sox7 expression would have a survival advantage over those lacking Sox7, and would therefore persist long term. Further, while Sox7-deficient primary myoblasts are very sensitive to cell death in the absence of apoptotic stimuli, it is unclear whether cell death occurs more frequently in Sox7cKO muscle in the absence of injury, and thus may allow the smaller population to remain relatively stable without muscle insult.
In normal tissues, SOX7 expression promotes erythroid precursor cell maintenance and self-renewal, while differentiation is promoted in its absence (Gandillet et al., 2009). A role for SOX7 in the maintenance of the undifferentiated state is consistent with our own findings demonstrating that, in the absence of Sox7, isolated primary myoblasts have a larger PAX7+/MYOD+ population than WT controls. Paradoxically, myogenin expression trended toward an increase in growth medium but was reduced compared with controls under differentiation conditions. Thus, SOX7 is unlikely to be required for myogenic differentiation per se but does promote efficient cell fusion.
While the PAX7+ population is smaller in Sox7cKO cells, these cells are able to enter the cell cycle and expand their numbers after injury and participate in repair, suggesting that myogenesis itself is not adversely affected by the loss of SOX7. Rather, we postulate that the smaller PAX7+ population size is insufficient to elicit repair as rapidly as the WT controls. Given that the defect in the PAX7+ cell population is established at birth and Pax7 expression is abnormally low in ESCs with reduced Sox7 expression, the smaller satellite population in Sox7cKO is primarily due to a failure to produce sufficient precursors during development but may be exacerbated by post-injury apoptosis of satellite cells.
Experimental Procedures
Mouse Embryonic Stem Cell Culture
D3 mouse ESCs (ATCC, no. CRL-1934) were grown in 100 mm culture dishes (Corning) at 37°C and 5% CO2 in mESC complete medium in the presence of 1,000 U/mL leukemia inhibitory factor (LIF) (Millipore) to prevent differentiation. Cultures were lentivirally transduced to express an shRNA against Sox7 (shSox7; Santa Cruz Biotechnology, sc-38417-V) or a scrambled control shRNA (shCntrl). Following transduction, cells were cultured in mESC complete medium overnight, supplemented with 2 μg/mL puromycin (Invitrogen) and LIF (Millipore) on day 2 post transduction. Medium was changed every day for the first 3 days followed by every alternate day thereafter.
To induce the differentiation of myogenic precursors, shSox7 and shCntrl cell lines were diluted to a concentration of 4 × 104 cells/mL in DMEM and 20 μL drops were placed on the lid of a 150 mm Petri dish (Corning) containing PBS to prevent droplet evaporation. After 2 days in suspension, culture aggregates were moved onto 100 mm Petri dishes (Corning) coated with 1% agarose to prevent adhesion. Cells were allowed to grow for 5 additional days and the medium was replaced with fresh DMEM every 2 days. On day 7, aggregates were resuspended in DMEM and moved into 12-well tissue culture plates for culture in medium containing serum to support the proliferation of myogenic precursors but not their differentiation.
Conditional Sox7 Knockout Mice
B6:129-Pax3 tm1(cre) Joe/J (The Jackson Laboratory) and Sox7fl/fl mice (Wat et al., 2012) were bred to obtain Sox7fl/fl;Pax3Cre/+ (conditional knockouts, Sox7cKO) and Sox7fl/fl;Pax3+/+ non-Cre-expressing control (WT) mice. All animals were maintained at 22°C with 30% relative humidity on a 12 hr light/dark cycle and provided food and water ad libitum.
For cardiotoxin (CTX) injury, mice were anesthetized with isofluorane, and 30 μL of 10 μM CTX in PBS or PBS alone was injected intramuscularly into the TA muscle. Mice were killed 7 days post injury, and the dissected muscle was flash frozen in isobutanol cooled in liquid nitrogen for analysis. All animal experimentation was approved by the University of Ottawa Animal Care Committee and conformed to the guidelines set out by the Canadian Council on Animal Care.
Analysis of Muscle Histology and Immunohistochemistry
Eight-micrometer-thick sections of TA muscle were stained with H&E following fixation in 10% formalin for 15 min. A minimum of 350 fibers were counted for analysis of the average fiber cross-sectional area and quantified using ImageJ.
Indirect immunofluorescence was performed on frozen sections of TA muscle fixed in 4% paraformaldehyde (PFA). Detection was performed according to standard procedures using the following primary antibodies: PAX7 (DSHB), and Ki67 (Abcam, catalog no. ab15580). Detection of immunoconjugates was achieved using biotinylated anti-mouse Fab fragments (Jackson ImmunoResearch Laboratories, catalog no. 715-066-150) and Alexa Fluor 594-streptavidin (Jackson ImmunoResearch, catalog no. 016-580-084) or Dylight 488 donkey anti-rabbit (Jackson ImmunoResearch). For detection of apoptotic cells, muscle sections were air-dried for 30 min at 60°C and fixed for 10 min in 4% fresh PFA. After washes with PBS, sections were permeabilized for 2 min in 0.1% Triton X-100 + 0.1% sodium citrate in PBS at 4°C and then washed with PBS.
In situ TUNEL assays were performed according to the manufacturer's instructions (In Situ Cell Death Detection Kit, TMR Red; Roche), immunostained for Pax7 expression and counterstained with DAPI (0.5 μg/mL) essentially as described (Marchildon et al., 2016). Muscle sections were visualized with a Zeiss AxioObserverZ1 microscope (Carl Zeiss Microscopy) and quantified manually using ImageJ.
Isolation and Myogenic Differentiation of Primary Myoblasts
Primary myoblasts were isolated as described previously (Motohashi et al., 2014). Briefly, hindlimb muscles of adult Sox7fl/fl mice were dissected and digested with collagenase/dispase (Roche), and enriched for satellite cells by magnetic-activated cell sorting. Myoblasts were retrovirally transduced with CreER and selected based on puromycin resistance and maintained in growth medium (DMEM containing 20% fetal bovine serum [FBS], 10% horse serum [HS]) supplemented with 10 ng/mL basic fibroblast growth factor and 2 ng/mL human growth factor (Peprotech). To induce CreER activity in culture, primary myoblasts were treated with 4-OH tamoxifen (2 μM dissolved in 100% ethanol; Sigma-Aldrich) for 48 hr. To induce differentiation, myoblasts were cultured in differentiation medium (DMEM containing 2% FBS and 10% HS).
Isolation and Culture of Single EDL Myofibers
Myofibers were isolated from EDL muscle as described previously (Pasut et al., 2013). Briefly, EDLs were removed from Sox7fl/fl mice and digested with collagenase type I (2 mg/mL in DMEM; Sigma-Aldrich). Muscles were transferred to horse-serum-coated plates and myofibers were separated by trituration using heat-polished glass Pasteur pipettes. Fibers were incubated in control shRNA or Sox7 shRNA lentiviral particles (Santa Cruz Biotechnology) and cultured for 72 hr in DMEM supplemented with 15% FBS and 2% chick embryo extract at 37°C, 5% CO2.
Immunocytochemistry
Myofibers were fixed in 4% PFA in PBS, 1% glycine, and blocked in PBS containing 0.2% Triton X-100 (BioShop), 2% BSA, 5% horse serum (Cedarlane), 1% azide. Myoblasts were fixed in 2% PFA in PBS and blocked in PBS containing 0.3% Triton X-100 and 10% goat serum. The primary antibodies used for detection were mouse anti-PAX7 (DSHB, 1/100), rabbit anti-MYOD (c-20) (Santa Cruz, catalog no. sc-304), MYH (Santa Cruz, catalog no. sc-20641), or anti-Ki67 (Abcam). Cells were washed with PBS and incubated in mouse anti-biotin and streptavidin-Cy3 or secondary antibodies conjugated to a fluorescent dye (Cy3, Alexa 488 or Alexa Flour 647; all from Jackson ImmunoResearch). Nuclei were counterstained with DAPI (0.5 μg/mL). Pictures were acquired using a Leica DM 3000B microscope and Infinity-3 camera (Lumenera).
Flow Cytometry
Primary myoblasts were collected using trypsin, counted, and washed in ice-cold PBS before resuspension in Annexin V buffer (10 mM HEPES, 140 mM NaCl, 2.5 mM CaCl2, pH 7.4). Cells were labeled with Annexin V and PI (Life Technologies) and analyzed by flow cytometry on a BD FACSCelesta instrument (BD Biosciences), and dot plots were made and analyzed using Kaluza v1.2 software (Brea).
Quantitative Real-Time PCR
Total RNA was isolated using the E.N.Z.A. Micro-elute Total RNA Kit I as per the manufacturer's protocol (Omega Bio-tek) and reverse transcribed using a Quantitect Reverse Transcription Kit (QIAGEN). For qPCR analysis, a cDNA template was amplified with the KAPA SYBR FAST Universal qPCR Master Mix (Kapa Biosystems) as per the manufacturer's protocol using the primers listed in the Supplemental Information. Reactions performed in duplicate were performed using a Mastercycler Realplex2 qPCR machine (Eppendorf, Mississauga, ON). Data were analyzed using the delta-delta CT method to quantify gene expression (Schefe et al., 2006) and expressed as means ± SEM of three independent experiments.
Analysis of Reporter Gene Expression
For Pax7 reporter assays, primary myoblasts isolated from C57BL/6 mice were transfected with a −3.8 kb Pax7 promoter-luciferase reporter construct (Lang et al., 2009) and a constitutively active RSV-β-galactosidase reporter in the presence or absence of mammalian expression plasmids for SOX7 using FuGene HD (Promega). After transfection, cells were cultured under growth conditions for 24 hr and then collected for the luciferase assay. Luciferase assays were performed using luciferase assay reagent II (Promega) according to the standard protocol and corrected for transfection efficiency with β-gal enzyme activity.
Statistical Analysis
Statistical differences between means were calculated using the two-tailed Student's t test or ANOVA with post hoc test. p values less than 0.05 were considered significant.
Author Contributions
R.R., conception and design of experiments, collection and/or assembly of data, data analysis or interpretation, manuscript writing. J.K.M., conception and design of experiments, final approval of manuscript. D.A.S., provision of materials required for research, final approval of manuscript. N.L.T., F.M., and E.L., conception and design of experiments, data analysis and interpretation, final approval of manuscript. A.B., conception and design, manuscript writing, final approval of manuscript. N.W.B. and I.S.S., conception and design of experiments, data interpretation, financial support, manuscript writing.
Acknowledgments
This work was supported by grants from the Canadian Institutes of Health Research (MOP-84458 to I.S.S. and 115029 to N.W.B.) and the Heart and Stroke Foundation (000231 to I.S.S.). We thank Dr. Vera Tang, Operations Manager of the Flow Cytometry & Virometry Core Facility at the University of Ottawa, for her assistance with the flow cytometry experiment. We also thank Dabo Yang, John Girgis, Megan Tu, Andrew Wight, and Firoz and Rehana Rajgara for technical assistance.
Published: September 21, 2017
Footnotes
Supplemental Information includes Supplemental Results, two figures, and a list of primers and can be found with this article online at http://dx.doi.org/10.1016/j.stemcr.2017.08.014.
Supplemental Information
References
- Auerbach R. Analysis of the developmental effects of a lethal mutation in the house mouse. J. Exp. Zool. 1954;127:305–329. [Google Scholar]
- Badis G., Berger M.F., Philippakis A.A., Talukder S., Gehrke A.R., Jaeger S.A., Chan E.T., Metzler G., Vedenko A., Chen X. Diversity and complexity in DNA recognition by transcription factors. Science. 2009;324:1720–1723. doi: 10.1126/science.1162327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowles J., Schepers G., Koopman P. Phylogeny of the SOX family of developmental transcription factors based on sequence and structural indicators. Dev. Biol. 2000;227:239–255. doi: 10.1006/dbio.2000.9883. [DOI] [PubMed] [Google Scholar]
- Buckingham M., Relaix F. The role of Pax genes in the development of tissues and organs: Pax3 and Pax7 regulate muscle progenitor cell functions. Annu. Rev. Cell Dev. Biol. 2007;23:645–673. doi: 10.1146/annurev.cellbio.23.090506.123438. [DOI] [PubMed] [Google Scholar]
- Capel B. The battle of the sexes. Mech. Dev. 2000;92:89–103. doi: 10.1016/s0925-4773(99)00327-5. [DOI] [PubMed] [Google Scholar]
- Cermenati S., Moleri S., Cimbro S., Corti P., Giacco Del L., Amodeo R., Dejana E., Koopman P., Cotelli F., Beltrame M. Sox 18 and Sox7 play redundant roles in vascular development. Blood. 2008;111:2657–2666. doi: 10.1182/blood-2007-07-100412. [DOI] [PubMed] [Google Scholar]
- Costa G., Mazan A., Gandillet A., Pearson S., Lacaud G., Kouskoff V. SOX7 regulates the expression of VE-cadherin in the haemogenic endothelium at the onset of haematopoietic development. Development. 2012;139:1587–1598. doi: 10.1242/dev.071282. [DOI] [PubMed] [Google Scholar]
- Franz T., Kothary R., Surani M.A., Halata Z., Grim M. The Splotch mutation interferes with muscle development in the limbs. Anat. Embryol. (Berl) 1993;187:153–160. doi: 10.1007/BF00171747. [DOI] [PubMed] [Google Scholar]
- Gandillet A., Serrano A.G., Pearson S., Lie-A-Ling M., Lacaud G., Kouskoff V. Sox7-sustained expression alters the balance between proliferation and differentiation of hematopoietic progenitors at the onset of blood specification. Blood. 2009;114:4813–4822. doi: 10.1182/blood-2009-06-226290. [DOI] [PubMed] [Google Scholar]
- George-Weinstein M., Gerhart J., Reed R., Flynn J., Callihan B., Mattiacci M., Miehle C., Foti G., Lash J.W., Weintraub H. Skeletal myogenesis: the preferred pathway of chick embryo epiblast cells in vitro. Dev. Biol. 1996;173:279–291. doi: 10.1006/dbio.1996.0023. [DOI] [PubMed] [Google Scholar]
- Gros J., Scaal M., Marcelle C. A two-step mechanism for myotome formation in chick. Dev. Cell. 2004;6:875–882. doi: 10.1016/j.devcel.2004.05.006. [DOI] [PubMed] [Google Scholar]
- Hayano T., Garg M., Yin D., Sudo M., Kawamata N., Shi S., Chien W., Ding L.-W., Leong G., Mori S. SOX7 is down-regulated in lung cancer. J. Exp. Clin. Canc. Res. 2013;32:17. doi: 10.1186/1756-9966-32-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollway G., Currie P. Vertebrate myotome development. Birth Defects Res. C Embryo Today. 2005;75:172–179. doi: 10.1002/bdrc.20046. [DOI] [PubMed] [Google Scholar]
- Hutcheson D.A., Zhao J., Merrell A., Haldar M., Kardon G. Embryonic and fetal limb myogenic cells are derived from developmentally distinct progenitors and have different requirements for beta-catenin. Genes Dev. 2009;23:997–1013. doi: 10.1101/gad.1769009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikemoto M., Fukada S.-I., Uezumi A., Masuda S., Miyoshi H., Yamamoto H., Wada M.R., Masubuchi N., Miyagoe-Suzuki Y., Takeda S. Autologous transplantation of SM/C-2.6(+) satellite cells transduced with micro-dystrophin CS1 cDNA by lentiviral vector into mdx mice. Mol. Ther. 2007;15:2178–2185. doi: 10.1038/sj.mt.6300295. [DOI] [PubMed] [Google Scholar]
- Kondoh H., Kamachi Y. SOX-partner code for cell specification: regulatory target selection and underlying molecular mechanisms. Int. J. Biochem. Cell Biol. 2010;42:391–399. doi: 10.1016/j.biocel.2009.09.003. [DOI] [PubMed] [Google Scholar]
- Lang K.C., Lin I.H., Teng H.F., Huang Y.C., Li C.L., Tang K.T., Chen S.L. Simultaneous overexpression of Oct4 and Nanog abrogates terminal myogenesis. Am. J. Physiol. Cell Physiol. 2009;297:C43–C54. doi: 10.1152/ajpcell.00468.2008. [DOI] [PubMed] [Google Scholar]
- Lee H.-J., Göring W., Ochs M., Mühlfeld C., Steding G., Paprotta I., Engel W., Adham I.M. Sox15 is required for skeletal muscle regeneration. Mol. Cell. Biol. 2004;24:8428–8436. doi: 10.1128/MCB.24.19.8428-8436.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepper C., Partridge T.A., Fan C.M. An absolute requirement for Pax7-positive satellite cells in acute injury-induced skeletal muscle regeneration. Development. 2011;138:3639–3646. doi: 10.1242/dev.067595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Maltzahn J., Jones A.E., Parks R.J., Rudnicki M.A. Pax7 is critical for the normal function of satellite cells in adult skeletal muscle. Proc. Natl. Acad. Sci. USA. 2013;110:16474–16479. doi: 10.1073/pnas.1307680110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansouri A., Stoykova A., Torres M., Gruss P. Dysgenesis of cephalic neural crest derivatives in Pax7-/- mutant mice. Development. 1996;122:831–838. doi: 10.1242/dev.122.3.831. [DOI] [PubMed] [Google Scholar]
- Marchildon F., Fu D., Lala-Tabbert N., Wiper-Bergeron N. CCAAT/enhancer binding protein beta protects muscle satellite cells from apoptosis after injury and in cancer cachexia. Cell Death Dis. 2016;7:e2109. doi: 10.1038/cddis.2016.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauro A. Satellite cell of skeletal muscle fibers. J. Biophys. Biochem. Cytol. 1961;9:493–495. doi: 10.1083/jcb.9.2.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monsoro-Burq A.H. PAX transcription factors in neural crest development. Semin. Cell Dev. Biol. 2015;44:87–96. doi: 10.1016/j.semcdb.2015.09.015. [DOI] [PubMed] [Google Scholar]
- Motohashi N., Asakura Y., Asakura A. Isolation, culture, and transplantation of muscle satellite cells. J. Vis. Exp. 2014 doi: 10.3791/50846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami A., Shen H., Ishida S., Dickson C. SOX7 and GATA-4 are competitive activators of Fgf-3 transcription. J. Biol. Chem. 2004;279:28564–28573. doi: 10.1074/jbc.M313814200. [DOI] [PubMed] [Google Scholar]
- Murphy M.M., Lawson J.A., Mathew S.J., Hutcheson D.A., Kardon G. Satellite cells, connective tissue fibroblasts and their interactions are crucial for muscle regeneration. Development. 2011;138:3625–3637. doi: 10.1242/dev.064162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niimi T., Hayashi Y., Futaki S., Sekiguchi K. SOX7 and SOX17 regulate the parietal endoderm-specific enhancer activity of mouse laminin alpha1 gene. J. Biol. Chem. 2004;279:38055–38061. doi: 10.1074/jbc.M403724200. [DOI] [PubMed] [Google Scholar]
- Pallafacchina G., Francois S., Regnault B., Czarny B., Dive V., Cumano A., Montarras D., Buckingham M. An adult tissue-specific stem cell in its niche: a gene profiling analysis of in vivo quiescent and activated muscle satellite cells. Stem Cell Res. 2010;4:77–91. doi: 10.1016/j.scr.2009.10.003. [DOI] [PubMed] [Google Scholar]
- Pasut A., Jones A.E., Rudnicki M.A. Isolation and culture of individual myofibers and their satellite cells from adult skeletal muscle. J. Vis. Exp. 2013 doi: 10.3791/50074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Relaix F., Rocancourt D., Mansouri A., Buckingham M. Divergent functions of murine Pax3 and Pax7 in limb muscle development. Genes Dev. 2004;18:1088–1105. doi: 10.1101/gad.301004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Relaix F., Rocancourt D., Mansouri A., Buckingham M. A Pax3/Pax7-dependent population of skeletal muscle progenitor cells. Nature. 2005;435:948–953. doi: 10.1038/nature03594. [DOI] [PubMed] [Google Scholar]
- Relaix F., Montarras D., Zaffran S., Gayraud-Morel B., Rocancourt D., Tajbakhsh S., Mansouri A., Cumano A., Buckingham M. Pax3 and Pax7 have distinct and overlapping functions in adult muscle progenitor cells. J. Cell Biol. 2006;172:91–102. doi: 10.1083/jcb.200508044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambasivan R., Yao R., Kissenpfennig A., Van Wittenberghe L., Paldi A., Gayraud-Morel B., Guenou H., Malissen B., Tajbakhsh S., Galy A. Pax7-expressing satellite cells are indispensable for adult skeletal muscle regeneration. Development. 2011;138:3647–3656. doi: 10.1242/dev.067587. [DOI] [PubMed] [Google Scholar]
- Sarkar A., Hochedlinger K. The sox family of transcription factors: versatile regulators of stem and progenitor cell fate. Cell Stem Cell. 2013;12:15–30. doi: 10.1016/j.stem.2012.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassoon D., Lyons G., Wright W.E., Lin V., Lassar A., Weintraub H., Buckingham M. Expression of two myogenic regulatory factors myogenin and MyoD1 during mouse embryogenesis. Nature. 1989;341:303–307. doi: 10.1038/341303a0. [DOI] [PubMed] [Google Scholar]
- Savage J., Conley A.J., Blais A., Skerjanc I.S. SOX15 and SOX7 differentially regulate the myogenic program in P19 cells. Stem Cells. 2009;27:1231–1243. doi: 10.1002/stem.57. [DOI] [PubMed] [Google Scholar]
- Schefe J.H., Lehmann K.E., Buschmann I.R., Unger T., Funke-Kaiser H. Quantitative real-time RT-PCR data analysis: current concepts and the novel “gene expression’s CT difference” formula. J. Mol. Med. (Berl) 2006;84:901–910. doi: 10.1007/s00109-006-0097-6. [DOI] [PubMed] [Google Scholar]
- Schmidt K., Glaser G., Wernig A., Wegner M., Rosorius O. Sox8 is a specific marker for muscle satellite cells and inhibits myogenesis. J. Biol. Chem. 2003;278:29769–29775. doi: 10.1074/jbc.M301539200. [DOI] [PubMed] [Google Scholar]
- Seale P., Sabourin L.A., Girgis-Gabardo A., Mansouri A., Gruss P., Rudnicki M.A. Pax7 is required for the specification of myogenic satellite cells. Cell. 2000;102:777–786. doi: 10.1016/s0092-8674(00)00066-0. [DOI] [PubMed] [Google Scholar]
- Shimoda M., Kanai-Azuma M., Hara K., Miyazaki S., Kanai Y., Monden M., Miyazaki J. Sox17 plays a substantial role in late-stage differentiation of the extraembryonic endoderm in vitro. J. Cell Sci. 2007;120:3859–3869. doi: 10.1242/jcs.007856. [DOI] [PubMed] [Google Scholar]
- Soullier S., Jay P., Poulat F., Vanacker J.M., Berta P., Laudet V. Diversification pattern of the HMG and SOX family members during evolution. J. Mol. Evol. 1999;48:517–527. doi: 10.1007/pl00006495. [DOI] [PubMed] [Google Scholar]
- Takash W., Cañizares J., Bonneaud N., Poulat F., Mattéi M.G., Jay P., Berta P. SOX7 transcription factor: sequence, chromosomal localisation, expression, transactivation and interference with Wnt signalling. Nucleic Acids Res. 2001;29:4274–4283. doi: 10.1093/nar/29.21.4274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wat M.J., Beck T.F., Hernandez-Garcia A., Yu Z., Veenma D., Garcia M., Holder A.M., Wat J.J., Chen Y., Mohila C.A. Mouse model reveals the role of SOX7 in the development of congenital diaphragmatic hernia associated with recurrent deletions of 8p23.1. Hum. Mol. Genet. 2012;21:4115–4125. doi: 10.1093/hmg/dds241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegner M. From head to toes: the multiple facets of Sox proteins. Nucleic Acids Res. 1999;27:1409–1420. doi: 10.1093/nar/27.6.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H., Price F., Rudnicki M.A. Satellite cells and the muscle stem cell niche. Physiol. Rev. 2013;93:23–67. doi: 10.1152/physrev.00043.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Basta T., Klymkowsky M.W. SOX7 and SOX18 are essential for cardiogenesis in Xenopus. Dev. Dyn. 2005;234:878–891. doi: 10.1002/dvdy.20565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Huang S., Dong W., Li L., Feng Y., Pan L., Han Z., Wang X., Ren G., Su D. SOX7, down-regulated in colorectal cancer, induces apoptosis and inhibits proliferation of colorectal cancer cells. Cancer Lett. 2009;277:29–37. doi: 10.1016/j.canlet.2008.11.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.