Abstract
At least nine neurodegenerative diseases that are caused by the aggregation induced by long tracts of glutamine sequences have been identified. One such polyglutamine-containing protein is huntingtin, which is the primary factor responsible for Huntington’s disease. Sedimentation velocity with fluorescence detection is applied to perform a comparative study of the aggregation of the huntingtin exon 1 protein fragment upon transgenic expression in Drosophila melanogaster and Caenorhabditis elegans. This approach allows the detection of aggregation in complex mixtures under physiologically relevant conditions. Complementary methods used to support this biophysical approach included fluorescence microscopy and semidenaturing detergent agarose gel electrophoresis, as a point of comparison with earlier studies. New analysis tools developed for the analytical ultracentrifuge have made it possible to readily identify a wide range of aggregating species, including the monomer, a set of intermediate aggregates, and insoluble inclusion bodies. Differences in aggregation in the two animal model systems are noted, possibly because of differences in levels of expression of glutamine-rich sequences. An increased level of aggregation is shown to correlate with increased toxicity for both animal models. Co-expression of the human Hsp70 in D. melanogaster showed some mitigation of aggregation and toxicity, correlating best with inclusion body formation. The comparative study emphasizes the value of the analytical ultracentrifuge equipped with fluorescence detection as a useful and rigorous tool for in situ aggregation analysis to assess commonalities in aggregation across animal model systems.
Graphical abstract
Huntington’s disease (HD) is a human hereditary neurodegenerative disease caused by an autosomal dominant mutation involving the expansion of a CAG nucleotide repeat.1 The CAG nucleotide repeat encodes a polyglutamine sequence in the huntingtin (Htt) protein. Hungtingtin polyglutamine stretches containing ≤35 glutamines typically do not cause disease (commonly termed the wild type), whereas the mutant protein contains ≥36 glutamines, resulting in aggregation and disease. At least eight other human diseases share this common disease genotype.1–3 Earlier biochemical studies have elucidated the kinetics and morphology of polyglutamine aggregation, ranging from the formation of soluble oligomers to mesoscale fibrils, the latter being involved in inclusion body formation.4,5 While polyglutamine sequences on their own can aggregate to form toxic species, the sequences flanking the polyglutamine stretch in the Htt protein, containing a nascent helical 17-residue N-terminal domain and a proline-rich C-terminal domain, play a profound role in potentiating protein aggregation. In particular, the N-terminal domain greatly exacerbates the rate of aggregation through the formation of helical bundle intermediates, although the mechanism of this effect is still poorly understood.5,6 The range of aggregation, from oligomers to inclusion bodies, has been recapitulated in vivo in a variety of model systems, using various polyglutamine constructs, although a correlation between the degree of aggregation and toxicity remains elusive.7–16 It is likely that toxicity is not simply a consequence of a gain of function of the aggregated species and is further complicated by a loss of function caused by sequestration and/or mislocalization of other protein factors into polyglutamine-containing inclusion bodies.1,5,17,18
Given the complexity of the protein aggregation pathway, it is critical that new methods that can more precisely map out the size and heterogeneity of aggregation in in vivo model systems be developed. Recent work has shown that the fluorescence detection system in the analytical ultracentrifuge19,20 could be applied to characterize the wide range of polyglutamine protein aggregation seen in yeast cells,21 in cultured murine neuroblastoma cells,22 and in our own work exploring polyglutamine aggregation in Caenorhabditis elegans.23 The analytical ultracentrifuge is unique in offering a rigorous physical picture of particle size and heterogeneity in solution, and we have recently extended this tool to characterize the range of protein aggregation from 1 to 250000 S, including the application of a single experiment that can capture information in the range of 1–3000 S, combining a multispeed method (MSM) with wide distribution analysis (WDA).24,25
The idea of capturing data collected at various rotor speeds in a single experiment is not a new one and was initially described theoretically some time ago as a gravitational sweep approach, in which the rotor speed is increased at a continuous rate.26 However, implementation of this approach for data analysis has only recently been developed in Sedfit, subsequent to the work described here.27 A similar approach that has been implemented in Sedanal some time ago is the MSM approach, which collects data serially over a discrete set of rotor speeds,24,25 and is the method that is used in this study. The analysis algorithm, WDA, plots the data in the form of s*g(s*) as a function of s*. The basic protocol in an MSM approach is to start an ultracentrifuge run at low speed to capture information about the largest particles, followed by increases in the speed in steps during the run, with the final speed chosen so that the smallest species can clear the meniscus, thus accommodating a wide range of sedimentation coefficients from ~1.0 to ~250000 S.
Measuring the sedimentation of fluorescent species in complex mixtures [Biological OnLine Tracer Sedimentation (BOLTS)] has only recently gained traction, and we exploit and improve this new technology in this work. We report here the characterization of polyglutamine aggregation in the context of the Htt exon 1 fragment, using BOLTS combined with fluorescence microscopy and semidenaturing detergent agarose gel electrophoresis (SDD-AGE). The intact Htt protein has rarely been studied because of its size (3144 amino acids in length) and limited expression and solubility in in vivo model systems. Instead, a fragment corresponding to just the first exon of the gene has been commonly used, which recapitulates the aggregation and toxicity of the full-length protein. This truncated construct has been characterized extensively, for example, in neuroblastoma cells.28
The focus of the work described here is on the application of the analytical ultracentrifuge with fluorescence detection as part of a comparative analysis of wild-type and mutant Htt exon 1 aggregation and its effects on toxicity in Drosophila melanogaster29 and C. elegans,30 including an analysis of the role of the human Hsp70 chaperone in affecting aggregation. The range of glutamine lengths studied includes wild-type lengths (25–28 residues in length) and mutant lengths (46–97 residues in length). Such a comparative study will help to distinguish general versus specific effects of an animal model system on aggregate pools within specific size ranges. In addition, the limited ability of Hsp70 to mitigate these specific pools of aggregation in D. melanogaster is reported.
Both animal model systems have been used previously to study the effects of polyglutamine expansions on aggregation and toxicity. In C. elegans, either polyglutamine sequences31,32 or the exon 1 fragment of the huntingtin protein30 has been fused to fluorescent proteins and expressed in transgenic C. elegans. In both fusion constructs, inclusion bodies are readily apparent as fluorescent puncta, as observed in microscopy images. Early work in D. melanogaster has been performed by the Bonini laboratory, studying the aggregation and toxicity of various polyglutamine-containing proteins,18,33,34 including a set of studies exploring the mitigation of aggregation upon expression of Hsp70.35–38 Several reviews describing these findings have been written, along with findings from similar efforts in other laboratories.33,39,40 Specific studies in D. melanogaster using both wild-type and mutant Htt exon 1 fragments have been performed, addressing toxicity and aggregation behavior.29,40 Mutant-length Htt has been shown to accumulate in both nuclei and cytoplasm and is toxic as measured by degeneration in the D. melanogaster eye photoreceptor neurons.41 Pan-neuronal expression is largely lethal after eclosion,42 which encouraged us to use larval models for our studies of aggregation and toxicity. Nevertheless, fertile adults did survive in D. melanogaster expressing various mutant Htt fusion constructs, perhaps reflecting lower expression levels for the pan-tissue driver than for the neuronal drivers, or through mitigating effects of toxicity of the fluorescent protein fusion partner.43 With the genetic power of the D. melanogaster model system, candidate genes (either native genes or human transgenes) can be easily tested for their ability to reduce the level of aggregation and toxicity. For example, as suggested above, Hsp70 overexpression in a D. melanogaster model has been shown to reduce the level of polyglutamine-induced aggregation and toxicity.35,44 In addition, the ability of Hsp70 to reduce the level of mutant Htt aggregation has been tested directly in a cellular model of Huntington’s disease.45 While mitigation of aggregation by Hsp70 is not universal in transgenic systems, such results encouraged us to test the effects of human Hsp70 on Htt aggregation as studied using analytical ultracentrifugation with fluorescence detection.
MATERIALS AND METHODS
D. melanogaster Lines, Maintenance, and Sample Preparation
D. melanogaster lines were maintained on standard medium and grown at room temperature. General D. melanogaster lines were obtained from the Bloomington Drosophila Stock Center. Transgenic lines expressing the Htt exon 1 fragments with lengths of glutamine repeats of 25, 46, 72, and 97 residues each, and fused to green fluorescent protein for detection (UAS-Htt exon1 Qp25-eGFP, UAS-Htt exon1 Qp46-eGFP, UAS-Htt exon1 Qp72-eGFP, and UAS-Htt exon1 Qp97-eGFP), were provided by N. Perrimon.29 The original transgenic line, Qp103-eGFP, was sequenced, and we renamed it Qp97-eGFP, after it was determined to contain 97 sequential glutamines. D. melanogaster lines containing Htt exon 1 constructs with varying length glutamine stretches were crossed with a fly line containing a pan-tissue-expressing promoter (da-Gal4 driver or da-Gal4 GeneSwitch driver). The progeny from this cross were grown at room temperature, and third-instar larvae were collected for each genotype. The GeneSwitch (GS) driver was used to anticipate the co-expression of the HTTQn-eGFP fusion proteins with Hsp70, as this chaperone transgene resides on chromosome 2, making the logistics of the crosses less complex. All experiments were performed using the da-Gal4 constitutive driver unless stated otherwise. Flies involving crosses using the GS system were placed in vials containing approximately 20 μg/mL mifepristone in the medium to induce expression in the larvae immediately upon feeding. Third-instar larvae were collected for analysis, washed briefly, and thrice frozen in liquid nitrogen and thrice thawed on ice to assist in cell lysis. Larvae were then homogenized on ice with a 1 mL glass homogenizer in 2× lysis buffer, containing 100 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) (pH 7.3), 200 mM KCl, 2 mM ethylene glycol tetraacetic acid (EGTA), 2 mM phenylmethanesulfonyl fluoride (PMSF), 2 mM dithiothreitol (DTT), and 2× protease inhibitor (Roche). Samples were allowed to settle on the bench for 1 h, and the supernatant was removed, aliquoted, and flash-frozen in liquid nitrogen. Before the samples were flash-frozen, the total protein concentration was determined using the Coomassie Plus Protein Assay Reagent (Thermo Scientific) in preparation for the various experimental methods used here.
C. elegans Strains, Maintenance, and Sample Preparation
C. elegans were maintained according to standard methods, at 20 °C on nematode growth medium (NGM) seeded with OP50 Escherichia coli.46 The polyglutamine strains [(unc-54)GFP::HttQ28 and (unc-54)GFP::HttQ74], expressing different CAG repeat lengths and fused with green fluorescent protein (GFP) at the N-terminus, have been described elsewhere.30
Synchronized populations for crude extracts were obtained from gravid adults after treatment with a 20% alkaline hypochlorite solution (3.0 mL of bleach, 3.75 mL of 1 M NaOH, and 8.25 mL of doubly distilled H2O) for 5 min and eggs being allowed to hatch in M9 buffer overnight. The next day, the synchronized populations were transferred to fresh NGM plates.
Synchronized 1-day-old adult C. elegans were harvested and washed with M9 buffer (22 mM sodium phosphate, 22 mM potassium phosphate, 85 mM NaCl, and 1 mM MgSO4). Samples were flash-frozen in liquid nitrogen and stored at −80 °C. Protein was extracted after three cycles of freezing and thawing in lysis buffer (same as that used for D. melanogaster). Samples were homogenized in five cycles of 30 s each using a motorized homogenizer (Kontes) followed by a 5 min incubation on ice. Samples were allowed to settle for 1 h on ice, after which the supernatant was removed, flash-frozen, and stored at −80 °C. The total protein concentration was calculated using the Coomassie Plus Protein Assay Reagent (Thermo Scientific).
Western Blot Analysis
Protein extracts were combined with sodium dodecyl sulfate (SDS) sample buffer [125 mM Tris-HCl (pH 6.8), 1% SDS, 10% glycerol, 5% β-mercaptoethanol, and bromophenol blue]. Samples were incubated at 100 °C for 5 min, separated using SDS–PAGE, transferred to nitrocellulose membrane (Thermo Scientific 88018), and blocked with 5% nonfat dry milk in PBS containing 0.001% Tween 20. Membranes were probed with a primary antibody against GFP (Invitrogen A11122) followed by anti-rabbit HRP as a secondary antibody (Cell Signaling 7074S). β-Actin was used as a loading control and was probed with an anti-β-actin antibody (Abcam ab8227). Chemiluminescence was detected using Super SignalWest Femto Maximum Sensitivity Substrate (Thermo Scientific 34095) and imaged using a FluorChem HD2 Imager (Alpha Innotech).
SDD-AGE Analysis
Protein extracts were diluted in SDS sample buffer, incubated at room temperature for 5 min, and loaded onto a 1.5% agarose gel containing 0.1% SDS [in 1× Tris-acetate-EDTA (TAE)]. Using 1× Tris-buffered saline (TBS), capillary action was used to transfer the protein to a nitrocellulose membrane overnight. Blocking and protein detection followed the same procedures that were used for Western blotting.
Analytical Ultracentrifugation
Lysates were prepared in 2× lysis buffer and frozen at −80 °C. For the multispeed method (MSM) runs, samples were adjusted to a total protein concentration of 0.5 mg/mL in 1× lysis buffer. To capture inclusion bodies, SV experiments were performed in a 2 M sucrose solution. A 3 M sucrose stock solution was prepared as previously described.22 Sucrose stock solution concentrations were verified by density measurement (ρ = 0.1257M + 1.00).47
The partial specific volume, density, and viscosity of solutions were calculated using Sednterp version 1.09.48 The temperature-corrected solution densities are 1.00685 g mL−1 for lysis buffer and 1.250 g mL−1 for the 2 M sucrose solution. The viscosities of the solutions are 1.0322 × 10−2 Pa s for the lysis buffer and 0.65 Pa s for 2 M sucrose.
Samples (350 μL each) were loaded into two-channel quartz window charcoal/Epon sedimentation velocity cells with 50 μL of FC43 Fluorinert heavy oil. Cells were placed in an eight-hole AnTi rotor (Beckman-Coulter) and equilibrated to 20 °C in an XL-A analytical ultracentrifuge (Beckman-Coulter) fitted with a fluorescence detection system. For MSM experiments, radial fluorescence scans were collected successively at 3000, 6000, 10000, 20000, 30000, and 50000 rpm using a 488 nm laser for excitation and 520 nm cutoff emission, with a radial step size of 20 μm, and using a constant photomultiplier voltage (voltage of 2193 V, gain of 1, range of 4) in all experiments. The run was continued at 50000 rpm until it was obvious that the meniscus was cleared of the protein.
Data collected from high-speed experiments (in lysis buffer) and low-speed experiments (with 2 M sucrose) were analyzed using Sedfit version 144 (P. Schuck, National Institute of Biomedical Imaging and Bioengineering, National Institutes of Health, Bethesda, MD).49 MSM data were analyzed using SedAnal version 6.01 (P. Sherwood and W. Stafford, Boston Biomedical Research Institute, Watertown, MA).50
High-angular velocity data were fitted using a c(s) distribution. This analytical tool is useful when studying less complex boundaries, as it provides information about the frictional coefficient, thus allowing one to obtain estimates of molar mass.49 In addition, c(s) analysis allows for the removal of systemic time-invariant and radial-invariant noise contributions; this was done using confidence levels (F ratio) set at 0.95. For samples containing 2 M sucrose, an ls-g*(s) distribution was used. This tool allows us to assume a nondiffusing species, which is particularly appropriate for very large particles. The data were also fit including parameters for radial- and time-independent noise, and with the confidence level set at 0.95. The meniscus position was allowed to vary in the fitting procedure while the bottom position was fixed. Sedimentation coefficients for both samples containing lysis buffer and sucrose were corrected to s20, w using Gussi version 1.1.0 (Brautigam C.A. 2015).48 The s20, w values determined from samples containing 2 M sucrose were corrected using the reported values for the density and viscosity of concentrated sucrose solutions using Gussi version 1.1.0.22,51 Data collected using MSM were fitted using dc/dt (concentration profile time-derivative analysis) with the wide distribution analysis option selected, implemented in SedAnal version 6.01.50 To improve the signal-to-noise ratio, data were chosen from 6.40 to 6.59 cm with a step size of 0.01 cm, which gave data from 20 radial positions. An additional 2% smoothing was applied to the distribution.
Confocal Microscopy
Third-instar larvae were isolated from vials and placed on microscope slides in drops of a 70% glycerol solution. Slides were placed on a heating block at 70 °C for 15–20 s to heat kill the larvae. Using a light microscope to visualize the position of the larvae, larvae were placed dorsal side up and topped with cover slides. Larvae were viewed using a Nikon Eclipse 80i confocal microscope, and eGFP-tagged Htt constructs were imaged to obtain Z-stacks (step size of 1.50 μm, medium pinhole size). The gain was adjusted until puncta and diffuse fluorescence were visible with minimal background fluorescence.
One-day-old adult C. elegans were washed three or four times with M9 buffer and placed in a 20 mM levamisole solution. The solution containing the paralyzed worm was then transferred to a 4% (w/v) agar pad on a glass slide. Samples were imaged manually on a Nikon C1 confocal microscope using a 514 nm laser and a 20× objective lens. Images were collected at 70% transmission with a gain set to 4 and a large pinhole.
Quantification of Puncta
Z-Stacks of larval sections were collapsed into single images using ImageJ (version 1.47), and the collapsed images of the four larval sections were opened in Adobe Photoshop. The anterior, precentral, postcentral, and posterior images were pieced together to form a single larva image by cropping out any overlapping sections. To obtain a count of puncta, each larval image was opened in ImageJ (Figure S12A). The image was converted into eight-bit gray scale (Figure S12B), and the threshold was adjusted until each punctum was clearly defined from the background (Figure S12C). The threshold was applied to make the image binary [everything that was determined to be a punctum was turned black, and everything determined to be a part of the background was turned white (Figure S12D)]. In HttQ97 larvae, puncta were distinct enough from the black background to easily determine a threshold value. The high level of diffuse fluorescence in HttQ46 and HttQ72 larvae made the distinction between the larvae and background less clear. To resolve this problem, a high threshold was set to include all puncta, and the threshold was applied to make the image binary. Any background fluorescence that was also included due to the high threshold was manually deleted to prevent ImageJ from counting them as puncta. Once all puncta were successfully separated from the background, the “watershed” function was used to separate any puncta that might have merged together (Figure S12E). Lastly, the “analyze particles” function was used to count the number of black puncta in the binary image, which indicated the total number of aggregates in the larval image. Average counts of puncta were based on three trials (three larvae per trial).
Larval Crawling Assay (D. melanogaster)
The larval crawling assay was adapted from work published by the Littleton laboratory.52 Wandering third-instar larvae grown at room temperature were collected, briefly washed in distilled water, and placed in the middle of a 0.8% agarose 15 cm dish. Larvae were allowed to acclimate for 1 min, and then larval crawling was recorded for 2.5 min. Recordings were taken with a digital camera and analyzed using the MTrack2 plugin for ImageJ, where the distance and time were measured, and the speed was calculated by dividing the distance traveled by the time of recording. At least 20 larvae per genotype were analyzed and compared with a D. melanogaster line containing either the da-Gal4 or daGal4(GS) driver fly line alone as a negative control.
Thrashing Assay (C. elegans)
Synchronized 1-day-old adult C. elegans from strains grown at room temperature were washed briefly in 1× M9 buffer, followed by a 1 min acclimation period. The movement of C. elegans in M9 buffer was recorded with a digital camera on a Nikon Stereoscope (SMZ1500) using NIS Elements software for 1 min. The number of thrashes was counted and analyzed. At least 45 samples per strain were analyzed and compared to N2 C. elegans.
RESULTS
Htt Exon 1 Fragment Expression and Aggregation Assessed Using Western Blotting and SDD-AGE
In preparation for the biophysical analysis of Htt aggregation, Western analysis was performed to determine overall levels of expression, because expression levels themselves can influence the degree of aggregation (Figure S1). The expression levels are similar, when compared to that of β-actin as a standard, even across C. elegans and D. melanogaster model systems, and expression levels for the C. elegans shown here are consistent with that previously reported.29,30 Additional bands for some of these protein fusions have been reported in the literature, presumably because of limited degradation, such as that seen here for HttQ25.29
A more conventional approach used for the analysis of protein aggregation in cell biology is SDD-AGE, a biochemical assay that fractionates protein aggregation by agarose gel electrophoresis using a semidenaturing solution condition. This was performed to make the comparison to the sedimentation velocity experiments, described below. SDD-AGE was performed on the same samples used for Western analysis. At best, only a crude estimate of aggregation is possible using this method. Three categories of aggregates are observed, a tight distribution of lower-molecular weight species, smears representing higher-molecular weight materials, and large-scale aggregates that are caught up in the wells. No evidence of bands running at the molecular weight expected for the monomers is seen in the Htt fusion constructs. For three of the constructs (C. elegans HttQ28, D. melanogaster HttQ25, and D. melanogaster HttQ46), a fairly tight band is seen near the bottom of the gel, in the range of 75–300 kDa, suggesting a minimal structural unit that is resistant to SDS denaturation (Figure 1).
Figure 1.
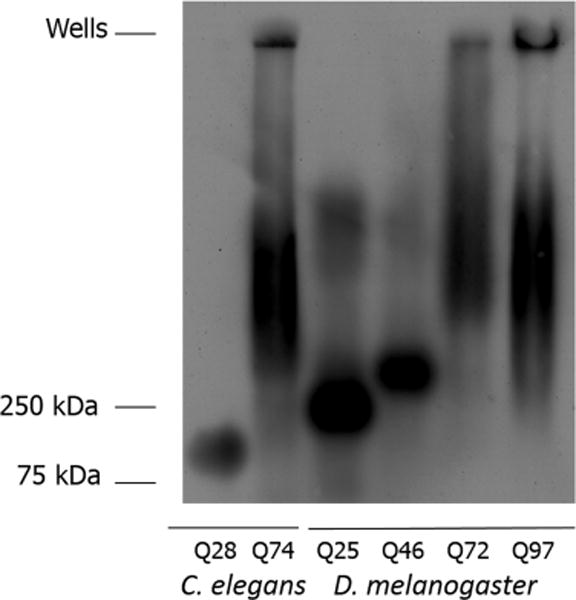
Analysis of protein aggregation using SDD-AGE. Protein from 1-day-old adult C. elegans expressing GFP-HttQ28 and GFP-HttQ74 and third-instar D. melanogaster larvae expressing HttQ25-eGFP, HttQ46-eGFP, HttQ72-eGFP, and HttQ97-eGFP were electrophoresed on an SDD-AGE gel and detected using an anti-GFP antibody. Sample loading is based on total protein determination: 5 μg for HttQ28, 2 μg for HttQ74, and 2 μg each for HttQ25, HttQ46, HttQ72, and HttQ97.
This presumed limited aggregation may be due to aggregation of the GFP itself within the fusion construct, as has been noted in the literature,13,20,23,53–55 or alternatively, sequestration of other factors (see Discussion). Our previous work, studying polyglutamine aggregation in a worm model, showed evidence of significant aggregation of GFP by itself, in both SDD-AGE (~400 kDa) and SV experiments (running at ~80 S), with the sedimentation boundary being present even in the polyglutamine fusion constructs.23 Interestingly, if the discrete bands shown at the bottom of the gel in Figure 1 are indeed due to limited GFP aggregation, the apparent smaller size (when compared to the earlier published work) and increase in size as a function of polyglutamine length may be related to effects of the Htt N-terminal domain. This hypothesis is described below more fully when discussing a similar effect observed in the SV experiments for material sedimenting in the range of 3–5 S.
Absolute magnitudes of individual aggregate bands or smears present cannot be compared across samples, as these intensities can reflect differences in loading amounts. Some smearing is present in the midrange of the gel for both D. melanogaster HttQ25 and HttQ46 samples, suggesting higher-order aggregation. The presence of both the low-molecular weight bands and the higher-molecular weight smearing in these samples likely represents a dynamic partitioning between molecular levels of SDS sensitivity.56 As expected, much more extensive aggregation is seen for the protein fusions with longer polyglutamine sequences (C. elegans HttQ74, D. melanogaster HttQ72, and D. melanogaster HttQ97), including varying degrees of protein caught up in the wells. In these same samples, the lower oligomeric species appear to be absent. On the basis of our previous work,23 the materials in the wells are consistent with inclusion body formation. These results are compared to sedimentation behavior in the SV experiments described below.
Sedimentation Behavior of the Htt Exon 1 Fragment Monomer
Crude extracts for all transgenic animals were prepared in the absence of detergents or denaturants and with no fractionation to minimize solubilization of in vivo aggregation. Appropriate dilutions of crude extracts were loaded into cells to be analyzed by sedimentation velocity methods using an analytical ultracentrifuge equipped with fluorescence detection. Fluorescence detection was enabled through the fusion of the proteins of interest to various fluorescence protein partners as described in Materials and Methods.
In earlier work, characterization of the monomer pool was used to validate the accuracy of the reported s values in crude extracts.23 This validation approach was also performed here. To detect a putative monomer population, data were collected at 50000 rpm and analyzed using a continuous distribution function, c(s) (Figure 2). For all of the D. melanogaster samples, a well-defined distribution is seen in the range of 2.1–2.4 S, consistent with the s value measured for similar fusion constructs in other model systems (Figure 2A, with boundary data shown in Figure S2).22,23 The two C. elegans samples had similar s values, ranging from 2.5 to 2.8 S (Figure 2B, with boundary data shown in Figure S3). Interestingly, in contrast to our previously published results for polyQn-GFP fusions in C. elegans,23 which lacked the Htt exon 1 fragment flanking sequences, some evidence was observed for a small oligomer pool in the range of 3.7–5.5 S for all samples, which may represent intermediates involving flanking sequence interactions, as observed in in vitro biochemical model systems.6,57–59 We also note the apparent increase in the D. melanogaster oligomer in the range of 3.7–5.5 S as a function of polyglutamine length, which could be due to several possibilities: (1) the inhibition of polyglutamine oligomerization or GFP oligomerization through interactions with the N-terminal flanking sequence (these interactions have been suggested to be dependent on polyglutamine length),60 (2) the increased probability of sequestration of other molecular components in the cell,22 or (3) a sedimentation artifact due to Johnston–Ogston drag due to slower-migrating particles.61 The fact that we see this effect in SDD-AGE experiments more strongly supports the first two possibilities. The ability to detect this detailed level of aggregation suggests that experiments could be designed to test for inhibitors or enhancers of flanking sequence interactions.
Figure 2.
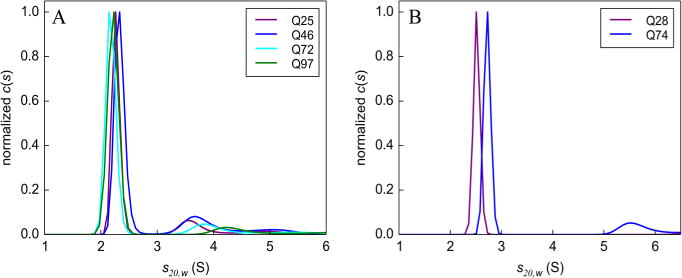
Sedimentation velocity monomer analysis. c(s) sedimentation coefficient distributions in (A) D. melanogaster and (B) C. elegans showing a distinct peak corresponding to the main boundary component. Boundary data used for this analysis are shown in Figures S2 and S3. Data were collected at 50000 rpm.
Characterization of the Htt Exon 1 Fragment Aggregate Pool
To capture the bulk of the aggregates in a single sedimentation velocity experiment, the recently developed multispeed method (MSM) was applied in conjunction with wide distribution analysis (WDA),23,50 identifying species falling in the range of 1–2000 S (Figure 3). A prominent monomer peak is evident for all samples. A sense of the experimental error in loading concentrations can be established from the Western blot results shown in Figure S1, indicating a fairly similar level of expressed recombinant protein in each of the various transgenic animals used. There is a significant decrease in the heights of the monomeric peaks relative to the aggregate distributions for all samples with Htt polyglutamine sequences longer than 28 residues, suggesting an increase in the level of relative aggregation with increasing polyglutamine length. This finding can be quantified by integration under the s*g(s*) distribution curves separately for the 1–20 S pool and the 20–20000 S pool, confirming that the amounts of material in the aggregated pools are indeed significantly larger than that represented in the monomer pool (Table 1). The apparent decrease in the percent aggregate for the D. melanogaster HttQ97 sample may reflect a transfer of material from the aggregate pool into inclusion bodies, as suggested in the SDD-AGE experiment (Figure 1). Such an effect could result in depletion of material in the aggregate pool relative to the monomer pool. This solution experiment provides a unique opportunity to quantify relative amounts of aggregate pools that have not been possible with other biochemical methods.
Figure 3.
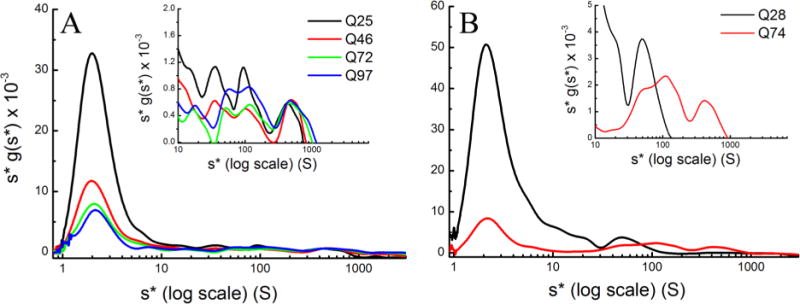
MSM–WDA analysis of (A) HttQ25-eGFP, HttQ46-eGFP, HttQ72-eGFP, and HttQ97-eGFP in D. melanogaster and (B) GFP-HttQ28 and GFP-HttQ74 in C. elegans. All samples prepared in lysis buffer were centrifuged at 3000, 6000, 10000, 20000, 30000, and 50000 rpm until the meniscus was cleared. The log plot of s*g(s*) vs s* shows the complete distribution of s values ranging from 0.8 to 3000 S, and the inset focuses on the 20–3000 S region to better highlight the distribution of the intermediate aggregates.
Table 1.
Analysis of Aggregation Based on the Integration of s* Distributionsa for C. elegans and D. Melanogaster HttQn Samples
| sample | % monomerb | % aggregatesc |
|---|---|---|
| C. elegans HttQ28 | 30 | 70 |
| C. elegans HttQ74 | 3 | 97 |
| D. melanogaster HttQ25 | 34 | 66 |
| D. melanogaster HttQ46 | 24 | 76 |
| D. melanogaster HttQ72 | 7 | 93 |
| D. melanogaster HttQ97 | 18 | 82 |
| D. melanogaster HttQ25d | 35.3 ± 0.8 | 64.7 ± 0.8 |
| D. melanogaster HttQ46d | 16.8 ± 4.8 | 83.2 ± 4.8 |
| D. melanogaster HttQ72d | 6.7 ± 1.7 | 93.3 ± 1.7 |
| D. melanogaster HttQ97d | 14.7 ± 1.9 | 85.6 ± 1.9 |
Expressed as a percentage of the complete integrated areas from 1 to 1000 S in Figure 3.
Based on integration of the 1–20 S area.
Based on integration of the 20–1000 S area.
Average monomer/aggregate percentages, combining all SV runs for each particular HttQn line, including different drivers and levels of Hsp70 expression (see Table 2) because these did not significantly change the distributions.
The insets in Figure 3 show an expansion of the range at which intermediate aggregation is present. In the D. melanogaster samples (Figure 3A), while two major pools of aggregation are quantified here (1–20 and 20–20000 S), recent SV analysis of Htt aggregation in yeast suggests a total of four distinct aggregate pools might be defined on the basis of differential behavior. In yeast, Xi et al.21 show evidence for two differently behaving pools in the range of 20–80 S and above 100 S. They noted a stronger correlation between the level of aggregation and toxicity for the smaller range than for the >100 S range. Thus, the other two pools that we note in our work would encompass the 1–20 S range and a range of >500 S, outside the window of observation of Xi et al. While these pools are empirically defined, they are reproducible, with slight collective x-axis shifts of the pools because of uncertainty in the position of the sample meniscus (compare Figure 3 with the distributions from separate experiments shown in Figure 7). Nevertheless, as suggested above, from a quantitative perspective, we treat all aggregation beyond 20 S as a single intermediate aggregate pool, as presented in Table 1. In addition, this coarser partitioning allows us to demonstrate the reproducibility of the aggregation profiles across samples, and data commingled across samples show a tight distribution of the fraction of monomer and aggregate that is calculated (in Table 1, see the bottom four rows, and see Figure S11). We note that aggregation heterogeneity within each pool is quite complex, with the 20 S pool defined as having the narrowest range. It would be premature to attempt a detailed analysis of this intrapool heterogeneity, although experiments to address this by using multiple fluorescent tags as a way of identifying other molecules that might be contributing to the sedimentation are being developed.
Figure 7.
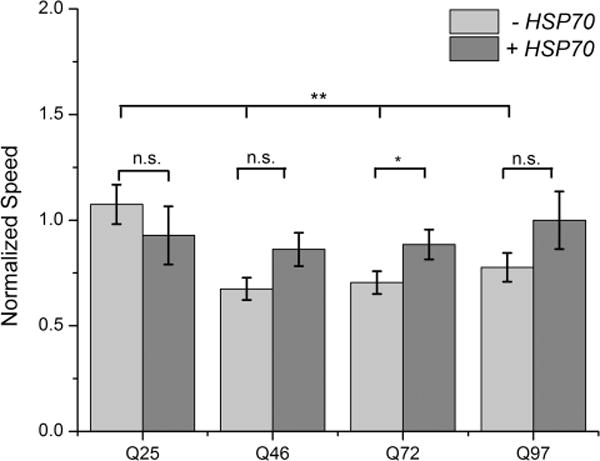
Larval crawling behavior in HttQn-expressing lines with and without expression of Hsp70. HttQ25-eGFP, HttQ46-eGFP, Htt72-eGFP, and Htt97-eGFP average speeds [expressed using the da-Gal4(GS) line] were normalized against the Da-Gal4(GS) driver alone. HttQn-eGFP D. melanogaster lines co-expressed with Hsp70 were normalized against a line expressing Hsp70 alone. Average speeds of Da-Gal4(GS), HttQ25-eGFP with or without Hsp70, and Hsp70 are shown in Figure S10 to make transparent the effects of the normalization calculations. Statistical analysis was performed using a one-way analysis of variance. *P < 0.05; **P < 0.01. n.s., not significant.
The MSM–WDA experiment revealed a significant difference in the distribution of the aggregate pools for the HttQ28 and HttQ74 C. elegans samples (Figure 3B). The single major peak in Q28 (Figure 3B, inset), centered on 45 S, likely reflects GFP aggregation as observed in our previously published work.23 This peak is also seen as a shoulder in the distribution profile for the HttQ74 sample and is likely the same sedimenting species observed in the Drosophila samples consistently seen either as a peak (Q72 and Q97) or as a shoulder (Q25 and Q46) at ~45 S (Figure 3A). However, in contrast to Htt28, the HttQ74 sample has two additional aggregate pools centered around 150 and 450 S, similar in size to the pools seen for the D. melanogaster samples.
Characterization of Htt Exon 1 Fragment Inclusion Bodies
The largest anticipated aggregates would likely be components of inclusion bodies, detected as fluorescent puncta in microscopy images (Figures 4 and S4 for D. melanogaster and Figure 5 for C. elegans). The extent of polyglutamine aggregation in vivo is known to be dependent on glutamine length (as demonstrated in Table 1 for intermediate aggregates) and is characterized here as a general increase in the number of fluorescent puncta for transgenic animals that contain mutant Htt polyglutamine sequences fused to fluorescent proteins. A quantitative analysis of the count of puncta from the D. melanogaster images shown in Figure S4 is provided in Figure 4C and reveals no puncta for HttQ25, a sparse number of puncta for HttQ46 and HttQ72, and a dramatic increase in the number of puncta for HttQ97. A similar increase in the extent of formation of puncta in 1-day-old C. elegans is observed, as expressed in muscle cells (Figure 5), mirroring the findings from the Monteiro laboratory, from where we received these transgenic animals.30
Figure 4.
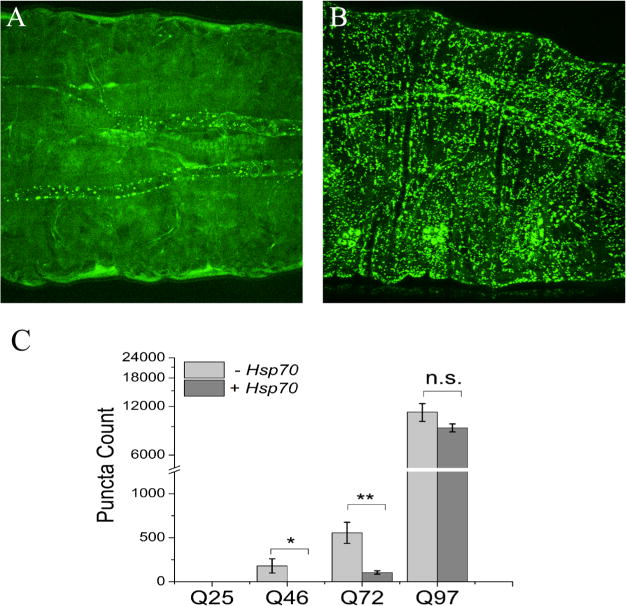
Fluorescence images and analysis of D. melanogaster third-instar larvae. Images showing the expression of polyglutamine expansions in the midsection of third-instar larvae for (A) HttQ46-eGFP and (B) HttQ97-eGFP. (C) Quantitation of puncta in the entire larva with or without Hsp70 expression using the images shown in Figure S4. Image analysis for counts of puncta is described in Materials and Methods (see also Figure S12). The scale bar is 0.1 mm. Statistical analysis was performed using a one-way analysis of variance. *P < 0.05; **P < 0.01. n.s., not significant.
Figure 5.
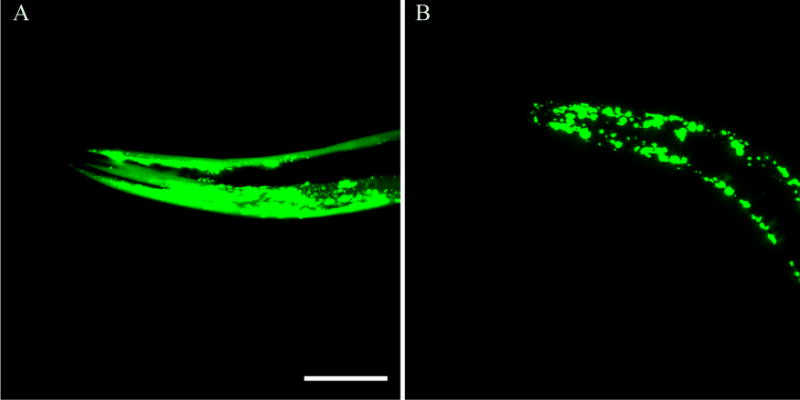
Fluorescence images of C. elegans: (A) GFP-HttQ28 and (B) GFP-HttQ74. Images show the expression of polyglutamine expansions in the body wall of young adult C. elegans. The scale bar is 0.05 mm. Note the increased level of GFP aggregate accumulation in GFP-HttQ74.
To look for evidence of such large-scale aggregates in sedimentation velocity experiments, samples were run at 3000 rpm in the presence of 2 M sucrose. These sample conditions have been shown to be appropriate for revealing inclusion body aggregates.22,23 Three samples were prepared and analyzed by SV: HttQ97 (Figure 6A, B) from D. melanogaster and HttQ74 (Figure 6C, D) and HttQ28 (Figure S5) from C. elegans. The boundary data (Figure 6A, C) show quite a bit of noise, which can be contrasted to that shown in Figures 2 and 3, as is anticipated from Mie scattering effects.27 These effects are similar to the haziness of samples seen by eye for very large particles. Nevertheless, the data are of sufficient quality to extract a set of peaks in a model-free continuous sedimentation distribution, ls-g*(s), resulting in random residuals derived from the fitting of the data. The absence of a clear boundary with a similar rate of movement for the HttQ28 sample (Figure S5) validates the ability to extract meaningful data in the presence of the Mie scattering effect. The aggregation profiles and the weight averages for the ls-g*(s) distributions are very similar to one another for both the D. melanogaster and C. elegans experiments, suggesting that the inclusion bodies are likely to be similar in size. An average of the weighted s values for D. melanogaster HttQ97 is 439120 S, while in C. elegans HttQ74, it is 403900 S. Simple size calculations based on the range of s values reported suggest that they are similar in size to that of the puncta in the corresponding microscopy images (Figures 4 and 5), assuming a spheroidal shape.23
Figure 6.
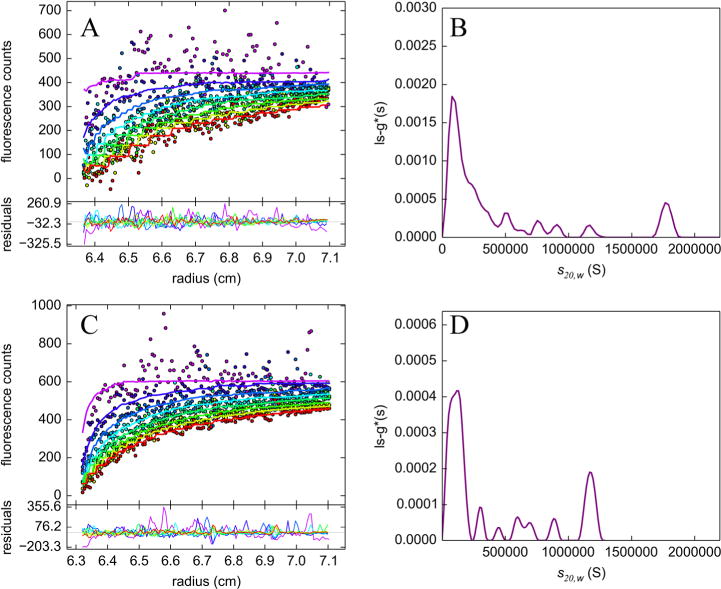
SV analysis in 2 M sucrose. Experimental data and the corresponding ls-g*(s) sedimentation coefficient distributions of (A and B) HttQ97-eGFP in D. melanogaster and (C and D) GFP-HttQ74 in C. elegans. Both time-invariant and radial-invariant noise contributions were removed from the data for the sake of clarity. The weighted s value for HttQ97-EGFP expressed in D. melanogaster is 439120 S, and the weighted s value for GFP-HttQ74 expressed in C. elegans is 403900 S. Data were collected at 3000 rpm.
Correlation of Htt Exon 1 Fragment Aggregation with Toxicity
One of the motivations for this work is to correlate data on protein aggregate profiles with toxicity, as measured using behavioral assays. Motility assays were used to assess behavioral effects, using C. elegans thrashing assays (may be compared to results from the work on these animals by the Monteiro laboratory30), and third-instar crawling assays. Our C. elegans thrashing results (Figure S6) show a significant decrease in the number of thrashes per minute with an increase in polyglutamine length. This result is fully consistent with the C. elegans bending assay performed by the Monteiro laboratory.30 We have also recapitulated the result showing a significant impact on movement upon comparison of wild-type N2 C. elegans with HttQ28 C. elegans. The effect on thrashing (or bending) for HttQ28 probably reflects the presence of puncta, albeit limited in number, in these animals (Figure 5).
The larval crawling motility assay that we used, measuring the speed of travel, is a variant of a behavioral assay studying HttQn constructs published previously,62 and results are shown in Figure 7. The data shown here have been normalized against larval crawling measured using a fly line containing only the da-Gal4 driver as a control. For HttQ46, HttQ72, and HttQ97, larval crawling appears to be equally impaired relative to the HttQ25 larval crawling behavior. In a careful analysis of the videos recorded for the crawling assay (data not presented), the impaired crawling speed is characterized by increased numbers of pausing and turning behaviors. In fact, frames showing such behaviors were counted for each D. melanogaster line, and a histogram of these data recapitulates the results shown in Figure 7. The results from the thrashing assay and the larval crawling assay are compared to the results of protein aggregation in the Discussion.
Effects of Hsp70 on Htt Exon 1 Fragment Aggregation
Having explored the general aggregate profiles of wild-type and mutant Htt constructs in both D. melanogaster and C. elegans, we can now address the question of how specific aggregate pools might respond to potential mitigating factors such as chaperones, particularly as measured by SV methods. We chose to study the effect of Hsp70 on HttQn aggregation. Earlier work from our laboratory showed that human Hsp70 has toxic effects in C. elegans. Furthermore, work from Morimoto’s group showed that yeast Hsp70, when co-expressed with polyglutamine-GFP fusion constructs, showed no effect on aggregation.32 Therefore, no efforts were made to look at crosses of transgenic C. elegans containing Hsp70 and various Htt-containing constructs.
The effects of Hsp70 on aggregation were tested by crossing HttQn D. melanogaster lines with a second line expressing human Hsp70. Because the Hsp70 line and the four HttQn lines all harbor the genes on chromosome 3, we switched to using the Da-Gal4(GS) driver for expression, which resides on chromosome 2. We confirmed Da-Gal4(GS)-driven co- expression of Hsp70 when crossed with two of the four D. melanogaster lines by Western blotting (Figure S7). Aggregation profiles as measured using the MSM SV approach showed no difference regardless of whether the Da-Gal4 or Da-Gal4(GS) drivers were used, as shown for both HttQ25 and HttQ97 (Figure S8 and Tables 1 and 2). Also, no significant difference in overall expression or aggregation was seen upon comparison of SDD-AGE results using the Da-Gal4 driver (Figure 1) and the Da-Gal4(GS) driver (Figure S9, lanes labeled “-Hsp70”).
Table 2.
Analysis of Aggregation Based on Integration of s* Distributionsa for D. melanogaster HttQn Samples Using the Da-Gal4(GS) Driver with or without Hsp70 Expression
| sample | % monomerb | % aggregatesc |
|---|---|---|
| Da-Gal4(GS)/HttQ25 | 35 | 65 |
| Da-Gal4(GS)/HttQ46 | 21 | 79 |
| Da-Gal4(GS)/HttQ72 | 10 | 90 |
| Da-Gal4(GS)/HttQ97 | 14 | 86 |
| Da-Gal4(GS)/HttQ25/Hsp70 | 38 | 62 |
| Da-Gal4(GS)/HttQ46/Hsp70 | 22 | 78 |
| Da-Gal4(GS)/HttQ97/Hsp70 | 9 | 91 |
Expressed as a percentage of the complete integrated areas from 1 to 1000 S in Figure S10.
Based on integration of the 1–20 S area.
Based on integration of the 20–1000 S area.
Observing no significant difference in aggregation profiles using the two different D. melanogaster drivers to express the Htt protein constructs, we analyzed the MSM and SDD-AGE experiments to see if the expression of Hsp70 might affect the intermediate aggregate pools. Samples were prepared for MSM analysis expressing both the HttQn proteins and human Hsp70, and the results are presented in Figure S10 comparing HttQn expression alone (Figure S10A) with that also expressing Hsp70 (Figure S10B). There is no obvious systematic difference in the gross features of the aggregate pools (note insets in Figure S10), suggesting that there is little or no impact of Hsp70 expression on the intermediate aggregate pools. A quantitative treatment of these data shows that the distribution between percent monomer and percent aggregate remains largely the same (Table 2 and Figure S11).
The SDD-AGE analysis of the HttQn D. melanogaster lines was also compared with and without Hsp70 expression (Figure S9). Within the normal variation in signal intensity seen in SDD-AGE, there appears to be little or no impact of Hsp70 on intermediate aggregates with the MSM–WDA analysis. Therefore, the slight, if statistically insignificant, mitigation of toxicity as measured in our crawling assay does not appear to correlate with intermediate aggregate formation.
In contrast to the lack of an impact on intermediate aggregates, co-expression of Hsp70 with the four Htt D. melanogaster lines results in a small decrease in the number of puncta (Figure 4). Such a decrease appears to correlate with a slight increase in crawling speed upon Hsp70 co-expression, as shown in Figure 7 (see also controls in Figure S12; note that Hsp70 itself has an effect on the crawling assay, so the lines co-expressing Hsp70 with the HttQn constructs were normalized against the Hsp70-expressing line to account for this effect).
DISCUSSION
This comparative study takes advantage of technical advances in analytical ultracentrifugation that we employed in recently published work23 and shows that the selective fluorescence detection of individual proteins in crude extracts (BOLTS)19,20,63 can be applied across multiple genetic model systems.
Consistent with the literature, three major Htt exon 1 protein structural subtypes have been identified, using a combination of SV with fluorescence detection and SDD-AGE: (1) monomers, (2) intermediate aggregates, and (3) large particles of the size of inclusion bodies. Furthermore, the intermediate aggregates might be further partitioned among pools determined through SV experiments by us and others,21 encompassing s values of 5–30 S, 40–500 S (with subpartitions of 40–100 and 100–500 S21), and >500 S. Such subpartitioning, while not well-defined in our crude extracts, has been noted recently in SV work on partially purified extracts in yeast, with pool sizes of <100 and >100 S exhibiting different characteristics.21 The latter two pools (40–500 and >500 S) are predicted to have variable SDS sensitivity as judged by the migration of these intermediate pools in the SDD-AGE experiments and may be a reflection of the degree of compaction.64
The low end of the 5–30 S pool (focusing on the shoulder on the right-hand side of the monomer peak), seen most prominently for the D. melanogaster HttQ25 and C. elegans HttQ28 profiles (Figure 2), is particularly intriguing because this is not seen in fusion constructs without the Htt flanking sequences.23 It has been shown by several different groups that the N-terminal flanking sequence of Htt induces limited aggregation (4–12 subunits) through helical pairing interactions in in vitro experiments5 and may be the explanation for this feature in the MSM–WDA experiments. The distinction between the upper end of the 5–30 S pool and the 40–500 S pool is less well-defined, and as suggested in the Results, some of this aggregation may be due to intrinsic aggregation of the fluorescent proteins themselves, as described in our earlier work and as suggested by the low-molecular weight species in the SDD-AGE experiment (Figure 1).23 Above the 40 S limit, the pools seen for both D. melanogaster and C. elegans are similar, suggesting that the properties driving aggregation are intrinsic to the Htt protein fragment, and limitations to clearing of such aggregates could be related to cellular housekeeping principles (e.g., chaperone activity and protein turnover). This general conclusion is further buttressed by noting that the expression in C. elegans is muscle-specific, whereas expression in D. melanogaster larvae is driven by the Da-Ga4 driver, a pan-tissue driver of expression. Both D. melanogaster and C. elegans show significant aggregation in the size range consistent with inclusion body formation.
It is intriguing to note that there are no aggregation pools for the C. elegans HttQ28 sample beyond 50 S in size, whereas the D. melanogaster HttQ25 sample shows aggregation pools not all that dissimilar from those of Htt samples with polyglutamine sequences that are >36 amino acids in length. It has been shown that polyglutamine aggregation can result in sequestration of other proteins,18 particularly those containing polyglut-amine-rich sequences.65 Five percent of the D. melanogaster genome encodes polyglutamine-containing proteins, whereas both humans and C. elegans encode only 0.4%.66 It is worth speculating that the 40–500 S pools in the D. melanogaster HttQ25 sample could involve significant sequestration of other proteins because of this difference. A similar observation has been made in yeast, where aggregates for an HttQ25-GFP fusion construct are noted in the 50–200 S size range;21 yeast also has a higher percentage of polyglutamine-containing proteins, in the 1.0–1.3% range.66
Behavioral assays were performed to look for correlations in toxicity with aggregate pools. For both D. melanogaster and C. elegans, an increase in the level of aggregation and toxicity is seen for polyglutamine lengths above the 36-amino acid threshold. This correlation is observed as monitored either by the ratio of monomer to intermediate aggregate (MSM–WDA experiments) or the number of puncta (confocal images). Our results in the C. elegans experiments mirror the results published by the Morimoto laboratory.67 In the D. melanogaster experiments, there is no apparent glutamine-length dependence of toxicity, which is inconsistent with the clear length dependence for the number of puncta formed. One suggestion is that the toxicity tracks better with the intermediate aggregate pool, which may be more insensitive to glutamine-length dependence.
Finally, mitigation of aggregation and toxicity in D. melanogaster was tested using Hsp70, a chaperone that has been shown in the past to reduce the number of polyglutamine-induced phenotypes in D. melanogaster.35,36 Expression of Hsp70 had only marginal effects on intermediate aggregation relative to the monomer population, as noted in the MSM–WDA (Figure S10 and quantified in Table 2) and SDD-AGE (Figure S9) experiments. In contrast, expression of Hsp70 resulted in a moderate if significant decrease in the number of puncta (Figure 4). This decrease in the number of puncta appears to correlate best with toxicity, which shows some mitigation of crawling speed (Figure 7), albeit with borderline statistical significance.
This work has helped to showcase the power of SV approaches, biophysical technology representing a true solution method, for measuring protein aggregation in complex macromolecular milieus (BOLTS). Technical hurdles for such studies, using fluorescence detection in the analytical ultra-centrifuge, have been discussed in earlier work from our laboratory23 and another.22 The research described here advances the use of this method, in complementation with other biochemical and microscopy methods, to offer a comparative study of polyglutamine aggregation using two different model systems. We believe that this work sets the stage for broader application of BOLTS to other interesting biological and biomedical applications, with particular emphasis on our part on other important protein aggregative diseases implicated in a variety of human neurodegenerative diseases.
Supplementary Material
Acknowledgments
We thank Norbert Perrimon for the D. melanogaster lines harboring the HttQn-eGFP variants and Mervyn Monteiro for the C. elegans strains harboring the GFP-Htt(Q28) and GFP-Htt(Q74) fusion constructs. We thank Glen Ramsay and Aviv Biomedical, Inc., for the use of their facilities to collect SV data and James L. Cole (Department of Molecular and Cell Biology and Department of Chemistry, University of Connecticut, Storrs, CT) and Jeffrey W. Lary (Biophysics Facility Center for Open Research Resources and Equipment, University of Connecticut) for SV data collection. We thank Naomi Chaqueco, Sabrina Kwak, Sydney Hyder, Tim Chaya, and George Neusch for able technical support. Some strains were provided by the CGC, which is funded by the National Institutes of Health Office of Research Infrastructure Programs.
Funding
We gratefully acknowledge funding support from National Institutes of Health Grant 1R15NS081681-01 from the National Institute of Neurological Disorders and Stroke.
ABBREVIATIONS
- BOLTS
biological online tracer sedimentation
- DIC
differential interference contrast
- DTT
dithiothreitol
- eGFP
enhanced green fluorescent protein
- EGTA
ethylene glycol tetraacetic acid
- GFP
green fluorescent protein
- HD
Huntington’s disease
- HEPES
4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid
- HRP
horseradish peroxidase
- Htt
huntingtin
- MSM
multispeed method
- NGM
nematode growth medium
- PMSF
phenylmethanesulfonyl fluoride
- polyQ
polyglutamine
- SDD-AGE
semidenaturating detergent agarose gel electrophoresis
- SDS–PAGE
sodium dodecyl sulfate–polyacrylamide gel electrophoresis
- SV
sedimentation velocity
- TAE
Tris-acetate-EDTA
- TBS
Tris-buffered saline
- WDA
wide distribution analysis
- YFP
yellow fluorescent protein
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.biochem.7b00518.
Western blotting analysis of expressed transgenic proteins (Figure S1), SV boundary data for monomer analysis (Figures S2 and S3), confocal microscopy images of intact D. melanogaster third-instar larvae (Figure S4), SV boundary data for HttQ28 run in 2 M sucrose (Figure S5), behavioral analysis in C. elegans (Figure S6) and D. melanogaster (Figure S12), Western blot analysis of Hsp70 expression (Figure S7), comparison of D. melanogaster drivers on aggregation (Figure S8), effect of Hsp70 expression using SDD-AGE (Figure S9), effect of Hsp70 expression using MSM–WDA (Figure S10), graphical representation of data presented in Table 1 (Figure S11), and processing of confocal images for counts of puncta (Figure S13) (PDF)
ORCID
Robert Fairman: 0000-0002-8103-0783
Notes
The authors declare no competing financial interest.
References
- 1.Arrasate M, Finkbeiner S. Protein aggregates in Huntington’s disease. Exp Neurol. 2012;238:1–11. doi: 10.1016/j.expneurol.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gatchel JR, Zoghbi HY. Diseases of unstable repeat expansion: mechanisms and common principles. Nat Rev Genet. 2005;6:743–755. doi: 10.1038/nrg1691. [DOI] [PubMed] [Google Scholar]
- 3.Mitas M. Trinucleotide repeats associated with human disease. Nucleic Acids Res. 1997;25:2245–2254. doi: 10.1093/nar/25.12.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kodali R, Wetzel R. Polymorphism in the intermediates and products of amyloid assembly. Curr Opin Struct Biol. 2007;17:48–57. doi: 10.1016/j.sbi.2007.01.007. [DOI] [PubMed] [Google Scholar]
- 5.Wetzel R. Physical chemistry of polyglutamine: intriguing tales of a monotonous sequence. J Mol Biol. 2012;421:466–490. doi: 10.1016/j.jmb.2012.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kokona B, Rosenthal ZP, Fairman R. Role of the coiled-coil structural motif in polyglutamine aggregation. Biochemistry. 2014;53:6738–6746. doi: 10.1021/bi500449a. [DOI] [PubMed] [Google Scholar]
- 7.Arrasate M, Mitra S, Schweitzer ES, Segal MR, Finkbeiner S. Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death. Nature. 2004;431:805–810. doi: 10.1038/nature02998. [DOI] [PubMed] [Google Scholar]
- 8.Lee CC, Walters RH, Murphy RM. Reconsidering the mechanism of polyglutamine peptide aggregation. Biochemistry. 2007;46:12810–12820. doi: 10.1021/bi700806c. [DOI] [PubMed] [Google Scholar]
- 9.Miller J, Arrasate M, Brooks E, Libeu CP, Legleiter J, Hatters D, Curtis J, Cheung K, Krishnan P, Mitra S, Widjaja K, Shaby BA, Lotz GP, Newhouse Y, Mitchell EJ, Osmand A, Gray M, Thulasiramin V, Saudou F, Segal M, Yang XW, Masliah E, Thompson LM, Muchowski PJ, Weisgraber KH, Finkbeiner S. Identifying polyglutamine protein species in situ that best predict neurodegeneration. Nat Chem Biol. 2011;7:925–934. doi: 10.1038/nchembio.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mukai H, Isagawa T, Goyama E, Tanaka S, Bence NF, Tamura A, Ono Y, Kopito RR. Formation of morphologically similar globular aggregates from diverse aggregation-prone proteins in mammalian cells. Proc Natl Acad Sci U S A. 2005;102:10887–10892. doi: 10.1073/pnas.0409283102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ossato G, Digman MA, Aiken C, Lukacsovich T, Marsh JL, Gratton E. A two-step path to inclusion formation of huntingtin peptides revealed by number and brightness analysis. Biophys J. 2010;98:3078–3085. doi: 10.1016/j.bpj.2010.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Poirier MA, Li H, Macosko J, Cai S, Amzel M, Ross CA. Huntingtin spheroids and protofibrils as precursors in polyglutamine fibrilization. J Biol Chem. 2002;277:41032–41037. doi: 10.1074/jbc.M205809200. [DOI] [PubMed] [Google Scholar]
- 13.Sathasivam K, Lane A, Legleiter J, Warley A, Woodman B, Finkbeiner S, Paganetti P, Muchowski PJ, Wilson S, Bates GP. Identical oligomeric and fibrillar structures captured from the brains of R6/2 and knock-in mouse models of Huntington’s disease. Hum Mol Genet. 2010;19:65–78. doi: 10.1093/hmg/ddp467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tyedmers J, Mogk A, Bukau B. Cellular strategies for controlling protein aggregation. Nat Rev Mol Cell Biol. 2010;11:777–788. doi: 10.1038/nrm2993. [DOI] [PubMed] [Google Scholar]
- 15.van Ham TJ, Holmberg MA, van der Goot AT, Teuling E, Garcia-Arencibia M, Kim HE, Du D, Thijssen KL, Wiersma M, Burggraaff R, van Bergeijk P, van Rheenen J, Jerre van Veluw G, Hofstra RMW, Rubinsztein DC, Nollen EAA. Identification of MOAG-4/SERF as a Regulator of Age-Related Proteotoxicity. Cell. 2010;142:601–612. doi: 10.1016/j.cell.2010.07.020. [DOI] [PubMed] [Google Scholar]
- 16.Weiss A, Klein C, Woodman B, Sathasivam K, Bibel M, Regulier E, Bates GP, Paganetti P. Sensitive biochemical aggregate detection reveals aggregation onset before symptom development in cellular and murine models of Huntington’s disease. J Neurochem. 2008;104:846–858. doi: 10.1111/j.1471-4159.2007.05032.x. [DOI] [PubMed] [Google Scholar]
- 17.Holmes WM, Klaips CL, Serio TR. Defining the limits: Protein aggregation and toxicity in vivo. Crit Rev Biochem Mol Biol. 2014;49:294–303. doi: 10.3109/10409238.2014.914151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perez MK, Paulson HL, Pendse SJ, Saionz SJ, Bonini NM, Pittman RN. Recruitment and the role of nuclear localization in polyglutamine-mediated aggregation. J Cell Biol. 1998;143:1457–1470. doi: 10.1083/jcb.143.6.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kingsbury JS, Laue TM. Fluorescence-detected sedimentation in dilute and highly concentrated solutions. Methods Enzymol. 2011;492:283–304. doi: 10.1016/B978-0-12-381268-1.00021-5. [DOI] [PubMed] [Google Scholar]
- 20.Kroe RR, Laue TM. NUTS and BOLTS: applications of fluorescence-detected sedimentation. Anal Biochem. 2009;390:1–13. doi: 10.1016/j.ab.2008.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xi W, Wang X, Laue TM, Denis CL. Multiple discrete soluble aggregates influence polyglutamine toxicity in a Huntington’s disease model system. Sci Rep. 2016;6:34916. doi: 10.1038/srep34916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Olshina MA, Angley LM, Ramdzan YM, Tang J, Bailey MF, Hill AF, Hatters DM. Tracking mutant huntingtin aggregation kinetics in cells reveals three major populations that include an invariant oligomer pool. J Biol Chem. 2010;285:21807–21816. doi: 10.1074/jbc.M109.084434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kokona B, May CA, Cunningham NR, Richmond L, Jay Garcia F, Durante JC, Ulrich KM, Roberts CM, Link CD, Stafford WF, Laue TM, Fairman R. Studying polyglutamine aggregation in Caenorhabditis elegans using an analytical ultracentrifuge equipped with fluorescence detection. Protein Sci. 2016;25:605–617. doi: 10.1002/pro.2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Runge MS, Laue TM, Yphantis DA, Lifsics MR, Saito A, Altin M, Reinke K, Williams RC., Jr ATP-induced formation of an associated complex between microtubules and neurofilaments. Proc Natl Acad Sci U S A. 1981;78:1431–1435. doi: 10.1073/pnas.78.3.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stafford WF, Braswell EH. Sedimentation velocity, multi-speed method for analyzing polydisperse solutions. Biophys Chem. 2004;108:273–279. doi: 10.1016/j.bpc.2003.10.027. [DOI] [PubMed] [Google Scholar]
- 26.Machtle W. High-resolution, submicron particle size distribution analysis using gravitational-sweep sedimentation. Biophys J. 1999;76:1080–1091. doi: 10.1016/S0006-3495(99)77273-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ma J, Zhao H, Sandmaier J, Alexander Liddle J, Schuck P. Variable Field Analytical Ultracentrifugation: II. Gravitational Sweep Sedimentation Velocity. Biophys J. 2016;110:103–112. doi: 10.1016/j.bpj.2015.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brignull HR, Morley JF, Morimoto RI. The stress of misfolded proteins: C. elegans models for neurodegenerative disease and aging. Adv Exp Med Biol. 2007;594:167–189. doi: 10.1007/978-0-387-39975-1_15. [DOI] [PubMed] [Google Scholar]
- 29.Zhang S, Binari R, Zhou R, Perrimon N. A genomewide RNA interference screen for modifiers of aggregates formation by mutant Huntingtin in Drosophila. Genetics. 2010;184:1165–1179. doi: 10.1534/genetics.109.112516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang H, Lim PJ, Yin C, Rieckher M, Vogel BE, Monteiro MJ. Suppression of polyglutamine-induced toxicity in cell and animal models of Huntington’s disease by ubiquilin. Hum Mol Genet. 2006;15:1025–1041. doi: 10.1093/hmg/ddl017. [DOI] [PubMed] [Google Scholar]
- 31.Morley JF, Brignull HR, Weyers JJ, Morimoto RI. The threshold for polyglutamine-expansion protein aggregation and cellular toxicity is dynamic and influenced by aging in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2002;99:10417–10422. doi: 10.1073/pnas.152161099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Satyal SH, Schmidt E, Kitagawa K, Sondheimer N, Lindquist S, Kramer JM, Morimoto RI. Polyglutamine aggregates alter protein folding homeostasis in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2000;97:5750–5755. doi: 10.1073/pnas.100107297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bonini NM. A genetic model for human polyglutamine-repeat disease in Drosophila melanogaster. Philos Trans R Soc B. 1999;354:1057–1060. doi: 10.1098/rstb.1999.0458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Warrick JM, Paulson HL, Gray-Board GL, Bui QT, Fischbeck KH, Pittman RN, Bonini NM. Expanded polyglutamine protein forms nuclear inclusions and causes neural degeneration in Drosophila. Cell. 1998;93:939–949. doi: 10.1016/s0092-8674(00)81200-3. [DOI] [PubMed] [Google Scholar]
- 35.Chan HY, Warrick JM, Gray-Board GL, Paulson HL, Bonini NM. Mechanisms of chaperone suppression of polyglutamine disease: selectivity, synergy and modulation of protein solubility in Drosophila. Hum Mol Genet. 2000;9:2811–2820. doi: 10.1093/hmg/9.19.2811. [DOI] [PubMed] [Google Scholar]
- 36.Warrick JM, Chan HY, Gray-Board GL, Chai Y, Paulson HL, Bonini NM. Suppression of polyglutamine-mediated neurodegeneration in Drosophila by the molecular chaperone HSP70. Nat Genet. 1999;23:425–428. doi: 10.1038/70532. [DOI] [PubMed] [Google Scholar]
- 37.Chai Y, Koppenhafer SL, Bonini NM, Paulson HL. Analysis of the role of heat shock protein (Hsp) molecular chaperones in polyglutamine disease. J Neurosci. 1999;19:10338–10347. doi: 10.1523/JNEUROSCI.19-23-10338.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cushman-Nick M, Bonini NM, Shorter J. Hsp104 suppresses polyglutamine-induced degeneration post onset in a drosophila MJD/SCA3 model. PLoS Genet. 2013;9:e1003781. doi: 10.1371/journal.pgen.1003781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chan HY, Bonini NM. Drosophila models of polyglutamine diseases. Methods Mol Biol. 2003;217:241–251. doi: 10.1385/1-59259-330-5:241. [DOI] [PubMed] [Google Scholar]
- 40.Lewis EA, Smith GA. Using Drosophila models of Huntington’s disease as a translatable tool. J Neurosci Methods. 2016;265:89–98. doi: 10.1016/j.jneumeth.2015.07.026. [DOI] [PubMed] [Google Scholar]
- 41.Jackson GR, Salecker I, Dong X, Yao X, Arnheim N, Faber PW, MacDonald ME, Zipursky SL. Polyglutamine-expanded human huntingtin transgenes induce degeneration of Drosophila photoreceptor neurons. Neuron. 1998;21:633–642. doi: 10.1016/s0896-6273(00)80573-5. [DOI] [PubMed] [Google Scholar]
- 42.Steffan JS, Bodai L, Pallos J, Poelman M, McCampbell A, Apostol BL, Kazantsev A, Schmidt E, Zhu YZ, Greenwald M, Kurokawa R, Housman DE, Jackson GR, Marsh JL, Thompson LM. Histone deacetylase inhibitors arrest polyglutamine-dependent neurodegeneration in Drosophila. Nature. 2001;413:739–743. doi: 10.1038/35099568. [DOI] [PubMed] [Google Scholar]
- 43.Doumanis J, Wada K, Kino Y, Moore AW, Nukina N. RNAi screening in Drosophila cells identifies new modifiers of mutant huntingtin aggregation. PLoS One. 2009;4:e7275. doi: 10.1371/journal.pone.0007275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Warrick JM, Morabito LM, Bilen J, Gordesky-Gold B, Faust LZ, Paulson HL, Bonini NM. Ataxin-3 suppresses polyglutamine neurodegeneration in Drosophila by a ubiquitin-associated mechanism. Mol Cell. 2005;18:37–48. doi: 10.1016/j.molcel.2005.02.030. [DOI] [PubMed] [Google Scholar]
- 45.Guzhova IV, Lazarev VF, Kaznacheeva AV, Ippolitova MV, Muronetz VI, Kinev AV, Margulis BA. Novel mechanism of Hsp70 chaperone-mediated prevention of polyglutamine aggregates in a cellular model of huntington disease. Hum Mol Genet. 2011;20:3953–3963. doi: 10.1093/hmg/ddr314. [DOI] [PubMed] [Google Scholar]
- 46.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lide DR, editor. CRC Handbook of Chemistry and Physics. CRC Press Inc; Boca Raton, FL: 1996. [Google Scholar]
- 48.Schuck P, Zhao H. Editorial for the special issue of methods “Modern Analytical Ultracentrifugation”. Methods. 2011;54:1–3. doi: 10.1016/j.ymeth.2011.04.003. [DOI] [PubMed] [Google Scholar]
- 49.Dam J, Velikovsky CA, Mariuzza RA, Urbanke C, Schuck P. Sedimentation velocity analysis of heterogeneous protein-protein interactions: Lamm equation modeling and sedimentation coefficient distributions c(s) Biophys J. 2005;89:619–634. doi: 10.1529/biophysj.105.059568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stafford WF, Sherwood PJ. Analysis of heterologous interacting systems by sedimentation velocity: curve fitting algorithms for estimation of sedimentation coefficients, equilibrium and kinetic constants. Biophys Chem. 2004;108:231–243. doi: 10.1016/j.bpc.2003.10.028. [DOI] [PubMed] [Google Scholar]
- 51.Mok YF, Ryan TM, Yang S, Hatters DM, Howlett GJ, Griffin MD. Sedimentation velocity analysis of amyloid oligomers and fibrils using fluorescence detection. Methods. 2011;54:67–75. doi: 10.1016/j.ymeth.2010.10.004. [DOI] [PubMed] [Google Scholar]
- 52.Lee WC, Yoshihara M, Littleton JT. Cytoplasmic aggregates trap polyglutamine-containing proteins and block axonal transport in a Drosophila model of Huntington’s disease. Proc Natl Acad Sci U S A. 2004;101:3224–3229. doi: 10.1073/pnas.0400243101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jayaraman M, Mishra R, Kodali R, Thakur AK, Koharudin LM, Gronenborn AM, Wetzel R. Kinetically competing huntingtin aggregation pathways control amyloid polymorphism and properties. Biochemistry. 2012;51:2706–2716. doi: 10.1021/bi3000929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Legleiter J, Mitchell E, Lotz GP, Sapp E, Ng C, DiFiglia M, Thompson LM, Muchowski PJ. Mutant huntingtin fragments form oligomers in a polyglutamine length-dependent manner in vitro and in vivo. J Biol Chem. 2010;285:14777–14790. doi: 10.1074/jbc.M109.093708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vitalis A, Pappu RV. Assessing the contribution of heterogeneous distributions of oligomers to aggregation mechanisms of polyglutamine peptides. Biophys Chem. 2011;159:14–23. doi: 10.1016/j.bpc.2011.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Saunders HM, Hughes VA, Cappai R, Bottomley SP. Conformational behavior and aggregation of ataxin-3 in SDS. PLoS One. 2013;8:e69416. doi: 10.1371/journal.pone.0069416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jayaraman M, Kodali R, Sahoo B, Thakur AK, Mayasundari A, Mishra R, Peterson CB, Wetzel R. Slow Amyloid Nucleation via alpha-Helix-Rich Oligomeric Intermediates in Short Polyglutamine-Containing Huntingtin Fragments. J Mol Biol. 2012;415:881–899. doi: 10.1016/j.jmb.2011.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kelley NW, Huang X, Tam S, Spiess C, Frydman J, Pande VS. The predicted structure of the headpiece of the Huntingtin protein and its implications on Huntingtin aggregation. J Mol Biol. 2009;388:919–927. doi: 10.1016/j.jmb.2009.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mishra R, Jayaraman M, Roland BP, Landrum E, Fullam T, Kodali R, Thakur AK, Arduini I, Wetzel R. Inhibiting the Nucleation of Amyloid Structure in a Huntingtin Fragment by Targeting alpha-Helix-Rich Oligomeric Intermediates. J Mol Biol. 2012;415:900–917. doi: 10.1016/j.jmb.2011.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Williamson TE, Vitalis A, Crick SL, Pappu RV. Modulation of polyglutamine conformations and dimer formation by the N-terminus of huntingtin. J Mol Biol. 2010;396:1295–1309. doi: 10.1016/j.jmb.2009.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Correia JJ, Johnson ML, Weiss GH, Yphantis DA. Numerical study of the Johnston-Ogston effect in two-component systems. Biophys Chem. 1976;5:255–264. doi: 10.1016/0301-4622(76)80038-5. [DOI] [PubMed] [Google Scholar]
- 62.Steinert JR, Campesan S, Richards P, Kyriacou CP, Forsythe ID, Giorgini F. Rab11 rescues synaptic dysfunction and behavioural deficits in a Drosophila model of Huntington’s disease. Hum Mol Genet. 2012;21:2912–2922. doi: 10.1093/hmg/dds117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.MacGregor IK, Anderson AL, Laue TM. Fluorescence detection for the XLI analytical ultracentrifuge. Biophys Chem. 2004;108:165–185. doi: 10.1016/j.bpc.2003.10.018. [DOI] [PubMed] [Google Scholar]
- 64.Berger TR, Montie HL, Jain P, Legleiter J, Merry DE. Identification of novel polyglutamine-expanded aggregation species in spinal and bulbar muscular atrophy. Brain Res. 2015;1628:254–264. doi: 10.1016/j.brainres.2015.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Suhr ST, Senut MC, Whitelegge JP, Faull KF, Cuizon DB, Gage FH. Identities of sequestered proteins in aggregates from cells with induced polyglutamine expression. J Cell Biol. 2001;153:283–294. doi: 10.1083/jcb.153.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schaefer MH, Wanker EE, Andrade-Navarro MA. Evolution and function of CAG/polyglutamine repeats in protein-protein interaction networks. Nucleic Acids Res. 2012;40:4273–4287. doi: 10.1093/nar/gks011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Beam M, Silva MC, Morimoto RI. Dynamic imaging by fluorescence correlation spectroscopy identifies diverse populations of polyglutamine oligomers formed in vivo. J Biol Chem. 2012;287:26136–26145. doi: 10.1074/jbc.M112.362764. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


