Abstract
Glioma is the most common primary brain tumor and represents one of the most aggressive and lethal types of human cancer. Recent advances have implicated long noncoding RNAs (lncRNAs) as crucial mediators of cancer development and progression. The present study aimed to investigate the role of a newly-discovered lncRNA, termed eosinophil granule ontogeny transcript (EGOT), in the aggressive abilities of cells in human glioma. It was initially found that the relative transcription level of EGOT in glioma cancerous tissues was significantly lower than that in adjacent non-cancerous tissues. EGOT was differentially expressed in a series of glioma cell lines, with its lowest level in high aggressive U251 and U87 cells. When EGOT was overexpressed by an expression plasmid, cell viability was significantly inhibited in U251 and U87 cells. Furthermore, with EGOT overexpression, the cell cycle was arrested at G0/G1 phase and consequently, cell apoptosis was significantly promoted along with the activities of caspase-3 and caspase-9. The migration abilities of EGOT-overexpressed cells were inhibited by 71.4% in U251 cells and by 69.5% in U87 cells. These data suggest that overexpression of EGOT inhibits cell proliferation and migration, and promotes cell apoptosis in glioma. Therefore, EGOT has potent anticancer activity and may function as a tumor suppressor in human glioma.
Keywords: long noncoding RNA, eosinophil granule ontogeny transcript, glioma, proliferation, metastasis, apoptosis
Introduction
Glioma is the most common brain tumor, accounting for >50% of all brain tumors (1). According to a population-based investigation, the overall median survival duration for patients with malignant glioma is ~15 months (2). Despite significant advances in the therapeutic strategies of glioma, including neurosurgical approaches, chemotherapy and radiotherapy, the prognosis of patients with malignant glioma remains poor, primarily due to the fact that malignant glioma cells are highly aggressive and capable of infiltrating into adjacent normal brain tissue, leading to the eventual failure of complete surgical dissection of the tumor (3,4). Recurrence and resistance to chemotherapy are to be claimed two influential factors for prognosis (5,6). Therefore, developing novel strategies and elucidating innovative therapeutic agents for patients suffering from glioma are imperative and urgent tasks.
Long noncoding RNAs (lncRNAs) are a type of RNAs that widely exist in the nuclei and cytoplasm of eukaryotic cells. These RNAs are non-protein coding and >200 nucleotides in length (7,8). With continuous advances in research approaches, research focused on lncRNAs has recently undergone a rapid expansion. LncRNAs are closely associated with tumor development (9,10) and particularly, have been implicated in glioma progression (11). For example, the lncRNA, MALAT1, has been demonstrated to have crucial roles in the cell proliferation, metastasis and apoptosis processes in glioma (12–14). MALAT1 was proposed as a tumor suppressor in glioma (14). Knockdown of MALAT1 increases the blood-tumor barrier permeability (15), and affects proliferation and the expression of stemness markers in glioma (12). However, only a small proportion of lncRNAs have been functionally characterized in glioma.
LncRNA eosinophil granule ontogeny transcript (EGOT) contains two exons and is nested within an intron of the inositol triphosphate receptor type 1 gene in 3p26.1. EGOT is conserved at the nucleotide level and was initially reported to be involved in eosinophil development and expressed in mature eosinophils (16). Furthermore, EGOT is highly expressed in bone marrow, indicating its role in the development from bone marrow hematopoietic stem cells (17). Recently, an association between aberrant expression of EGOT with malignant status and poor prognosis has been detected in breast cancer (17). EGOT was downregulated in breast cancer and its low levels were significantly associated with larger tumor size, lymph node metastasis and worse prognosis. This pioneering study implied that EGOT may exert potential anticancer activity in human tumorigenesis. However, the functional role of EGOT in other types of cancer remains largely unknown.
Preliminary microarray analysis demonstrated that EGOT was differentially expressed in glioma and non-glioma tissues (data not shown). Based on this finding, the present study aimed to investigate the role of EGOT in cell proliferation and metastasis in glioma. The transcription level of EGOT was initially examined in clinical glioma tissues, followed by a systematic assessment of the biological activities of EGOT in glioma in vitro. The present report represents the first experimental study to elucidate the functional role of EGOT in human cancer.
Materials and methods
Human tissues
A total of 50 glioma tissues and their adjacent non-cancerous tissues were collected from patients (age, 50±5, 27 male:23 female) who underwent neurosurgical dissection at the Department of Oncology, The Second Affiliated Hospital of Zhejiang Chinese Medical University (Hangzhou, China) between May 2015 and May 2016. Infiltrating cases, in which clinicians could not differentiate between cancerous and adjacent non-cancerous tissues, were excluded from this study. These patients had not received chemotherapy nor radiotherapy prior to surgical resection. All cases were diagnosed with glioma by two independent pathologists, without any controversy and the Glioma Diagnosis and Treatment Guidelines of Central Nervous System in China were followed. Written consent was obtained from each patient. Protocols for using these samples for research purposes were approved by the Institutional Review Board at The Second Affiliated Hospital of Zhejiang Chinese Medical University.
Cell lines, cell culture and cell transfection
Normal glial HEB cells were purchased from American Type Culture Collection (ATCC; Manassas, VA, USA). Four glioma cell lines, including A172, U251, U87 and SHG44, were obtained from Shanghai Cell Bank of the Chinese Academy of Sciences (Shanghai, China). All cells were cultured in Dulbecco's modified Eagle's medium (DMEM; ATCC) supplemented with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA). Cells were maintained in an incubator containing 5% CO2 at 37°C. When cells grew to a confluence of 80%, the constructed pcDNA3.1-EGOT plasmid or the vector (pcDNA3.1, an empty plasmid; Addgene, Inc., Cambridge, MA, USA) was transfected into cells using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.). The pcDNA3.1-EGOT recombinant plasmid was constructed according to the protocol outlined in a previous article (18). The whole sequence of EGOT was cloned from the whole genome with the following primers: Forward, 5′-CAAGGCAGAGGTGGGTTTGG-3′ and reverse, 5′-GGCACCTTTTCAGTTGCTCAC-3′ using XhoI and BamHI restriction enzymes (New England BioLabs, Inc., Ipswich, MA, USA). Following digestion, the PCR product was ligated to the linearized pcDNA3.1 vector with T4 ligase (New England BioLabs, Inc.) at 4°C overnight. Trans10-competent cells (Transgen Biotech Co., Ltd., Beijing, China) and Luria-Bertani plates were used to obtain the monoclone via ampicillin selection. The plasmid was sequenced to ensure the plasmid had successfully been implanted.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from fresh frozen samples and cells using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.), as previously described (17). An aliquot of DNase was added to the TRIzol reagent prior to sample preparation. A total of 1 µg RNA was immediately reverse transcribed into cDNA using a Transcriptor First Strand cDNA Synthesis kit (Roche Diagnostics, Basel, Switzerland). Relative levels of EGOT to glyceraldehyde 3-phosphate dehydrogenase (GAPDH) control transcripts were determined by SYBR-Green qPCR (Roche Diagnostics) using the ABI 7900 Fast Real-Time PCR system (Invitrogen; Thermo Fisher Scientific, Inc.). Primer sequences were as follows: EGOT forward, 5′-CACTGCACAGGGAAACACAAA-3′ and reverse, 5′-ACCCTGTTCATAAGCCCTGATG-3′; and GAPDH forward, 5′-ACCACAGTCCATGCCATCAC-3′ and reverse, 5′-TCCACCCTGTTGCTGTA-3′.
RT-qPCR amplification was performed in triplicate. The transcription level of EGOT was normalized to that of GAPDH. The relative level of EGOT transcript was determined via the 2−ΔΔCq method (19).
Cell cycle analysis
Prior to testing, U251 and U87 cells were transfected with vector or EGOT expression plasmid for 48 h, as described. Following this, 1×104 cells were collected and fixed with cold ethanol (70%; 10 min on ice) prior to washing and re-suspension in cold PBS and incubation at 37°C for 30 min with 10 mg/ml RNase and 1 mg/ml propidium iodide (PI; Sigma-Aldrich; Merck KGaA, Darmstadt, Germany). DNA content was analyzed by flow cytometry (BD Biosciences, San Jose, CA, USA). The percentage of cells in each phase of the cell cycle was determined using Cell Quest acquisition software (version 2.9; BD Biosciences).
Flow cytometric analysis of cell apoptosis
Annexin V/PI assay was performed according to the manufacturer's instructions (Invitrogen; Thermo Fisher Scientific, Inc.). Briefly, U251 and U87 cells (1×104 cells) were plated into 6-well plates (Corning, Inc., NY, USA) and transfected with vector or EGOT expression plasmid, as described. Cells were subsequently washed with cold PBS, centrifuged (1,000 × g for 5 min at 4°C), and re-suspended in 100 µl binding buffer containing 2.5 µl fluorescein isothiocyanate-conjugated Annexin V and 1 µl PI (100 µg/ml). Cells were incubated at room temperature for 15 min in the dark. At least 10,000 events were collected and analyzed by flow cytometry (BD Biosciences) and Cell Quest acquisition software (version 2.9; BD Biosciences) was used.
Cell viability determination
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay (Promega Corp., Pittsburgh, PA, USA) was used to measure cell growth abilities according to the manufacturer's protocol. Briefly, U251 and U87 cells were seeded in 96-well plates at an initial density of 5×103/well in DMEM supplemented with 10% FBS in a humidified incubator containing 5% CO2. Each experimental group of cells was spread into six wells and the cultural medium was replaced every other day. Cell viability was assessed for five consecutive days. At each evaluation point (1, 2, 3, 4 and 5 days post-transfection), cell viability was detected using a microplate reader (Tecan Group, Männedorf, Switzerland) at an absorbance of 490 nm. Absorbance of control cells at day 1 was set as 1. Other absorbance values were normalized to the control cells at day 1.
Transwell migration assay
U251 and U87 cells that were transfected with vector or EGOT expression plasmid, respectively, were harvested with serum-free DMEM and 150 µl of cell suspension (6×104 cells) was seeded into the upper chamber (Corning, Inc., Corning, NY, USA), whereas the lower chamber was filled with 600 µl cultural medium supplied with 10% FBS. Following incubation for 12 h at 37°C, cells in each group were fixed with ice-cold methanol for 10 min and stained with crystal violet for 5 min at room temperature. Images were captured under a microscope (magnification, ×200; Nikon Corp., Tokyo, Japan).
Wound-healing assay
Wound-healing assays were performed by creating identical wound areas for anchorage-dependent cells U251 and U87 using 10-µl sterile pipette tips. Briefly, cells were seeded into 6-well plates and co-incubated with the same quantity of plasmids in the presence or absence of EGOT overexpression for 24 h at 37°C. Afterwards, cells were washed with PBS and a cross was scraped in the center of each well, washed with PBS again and immediately replaced with fresh serum-free medium. Images of the cells were captured once the scratch was made. After 24 h growth, cells were observed and images were captured under a Nikon microscope at a magnification of ×200 for each group.
Determination of caspase activities
Activities of caspase-3, caspase-8 and caspase-9 were determined using caspase activity kits according to the instructions provided by Beyotime Institute of Biotechnology (Haimen, China). Briefly, cell lysates were collected from each group of cells. Equal amounts of protein (10 µl) from cell lysates were added into 96-well plates and mixed with an aliquot of 80 µl reaction buffer supplemented with caspase substrate (2 mM). Following incubation for 4 h at 37°C, caspase activities were determined via a TECAN reader at an absorbance of 450 nm.
Western blot analysis
U251 and U87 cells (1×104 cells) were treated with EGOT expressing plasmid and cultured for additional 48 h. Following this, total proteins were extracted with NP40 lysis buffer (Beyotime Institute of Biotechnology) and quantified via the standard BCA method (Thermo Fisher Scientific, Inc., Waltham, MA, USA). Equal amounts of protein from each sample (50 µg) were separated by 10% SDS-PAGE and electroblotted onto nitrocellulose (NC) membranes. Following blocking with TBST supplied with 5% milk, the membranes were incubated with primary antibodies at 4°C overnight. Primary antibodies against caspase-9 (sc-8297), caspase-3 (sc-271028), cytochrome c (cyt c), GAPDH and secondary antibodies were purchased from Santa Cruz Biotechnology, Inc., (Dallas, TX, USA) and was used in a dilution of 1:1,000.
Statistical analysis
All data were expressed as the mean ± standard deviation. Each experiment was repeated at least three times in triplicate. Student's t-test was used to compare the difference between groups under normal distribution. Wilcoxon's test was used for the clinical paired samples and Mann-Whitney test was used to compare values on the cell line experiments. P<0.05 was considered to indicate a statistically significant difference.
Results
EGOT is downregulated in glioma and differentially expressed in glioma cell lines
Initially, the transcription level of EGOT was examined in 50 cases of glioma. It was shown that the mean transcription level of EGOT in the cancerous tissues was only 40% of that in the adjacent non-cancerous tissues, demonstrating a significant decrease (Fig. 1A). Furthermore, compared with the normal glial HEB cells, glioma cell lines consistently exhibited lower transcription levels of EGOT (Fig. 1B). Among the four glioma cell lines, U251 and U87 cells, which are highly aggressive, exhibited the lowest levels of EGOT (Fig. 1B), indicating the negative role of EGOT expression in the aggressiveness of glioma cells. These data suggest that EGOT is downregulated in glioma.
Figure 1.
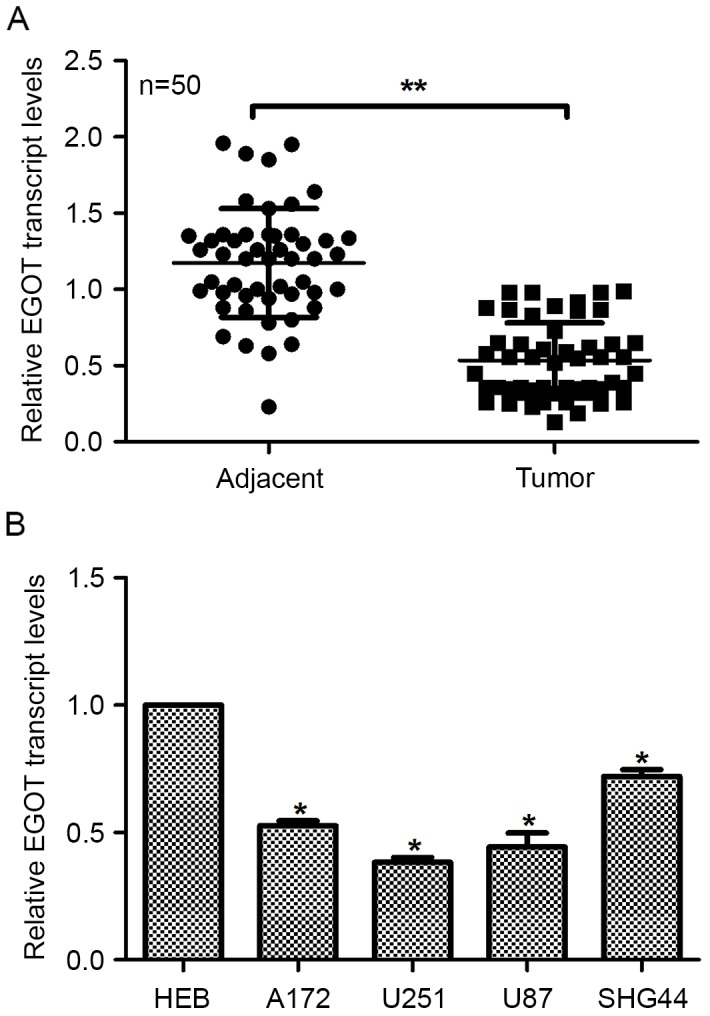
EGOT is downregulated in glioma and differentially expressed in glioma cell lines. (A) Relative transcription levels of EGOT in 50 cases of clinical glioma tissues as well as their matched adjacent non-cancerous tissues were evaluated via reverse transcription-quantitative polymerase chain reaction analysis. (B) Transcription levels of EGOT were analyzed in five cell lines, including the normal glial HEB cells and four glioma cell lines: A172, U251, U87 and SHG44. Glioma cell lines exhibited significantly lower transcription levels of EGOT than the normal HEB cells and among the four glioma cell lines, highly aggressive U251 and U87 cells exhibited the lowest levels of EGOT. All data are expressed as the mean ± standard deviation. *P<0.05 vs. HEB; **P<0.01. EGOT, eosinophil granule ontogeny transcript.
Overexpression of EGOT inhibits the proliferation of U251 and U87 cells
Effects of EGOT overexpression on cell biological activities were assessed in U251 cells and U87 cells. Following transfection of the EGOT expression plasmid, relative EGOT transcription levels significantly increased in U251 cells (4-fold) and U87 cells (3.5-fold; Fig. 2A), as compared with the controls, indicating the successful upregulation of EGOT in glioma cells. Subsequently, cell viability was determined. In U251 cells, while all groups of cells remained constantly proliferative, EGOT-overexpressed cells exhibited a significantly slower proliferative rate, as compared with the controls, from day 4 (Fig. 2B). In U87 cells, it was similarly observed that cell proliferative rates were not significantly different among the three groups of cells in the first 3 days. However, the cell proliferative rate in EGOT-overexpressed cells was significantly inhibited by up to 42% on day 5, as compared with the control cells (Fig. 2C). These data suggest that overexpression of EGOT inhibits cell viability in glioma cells.
Figure 2.
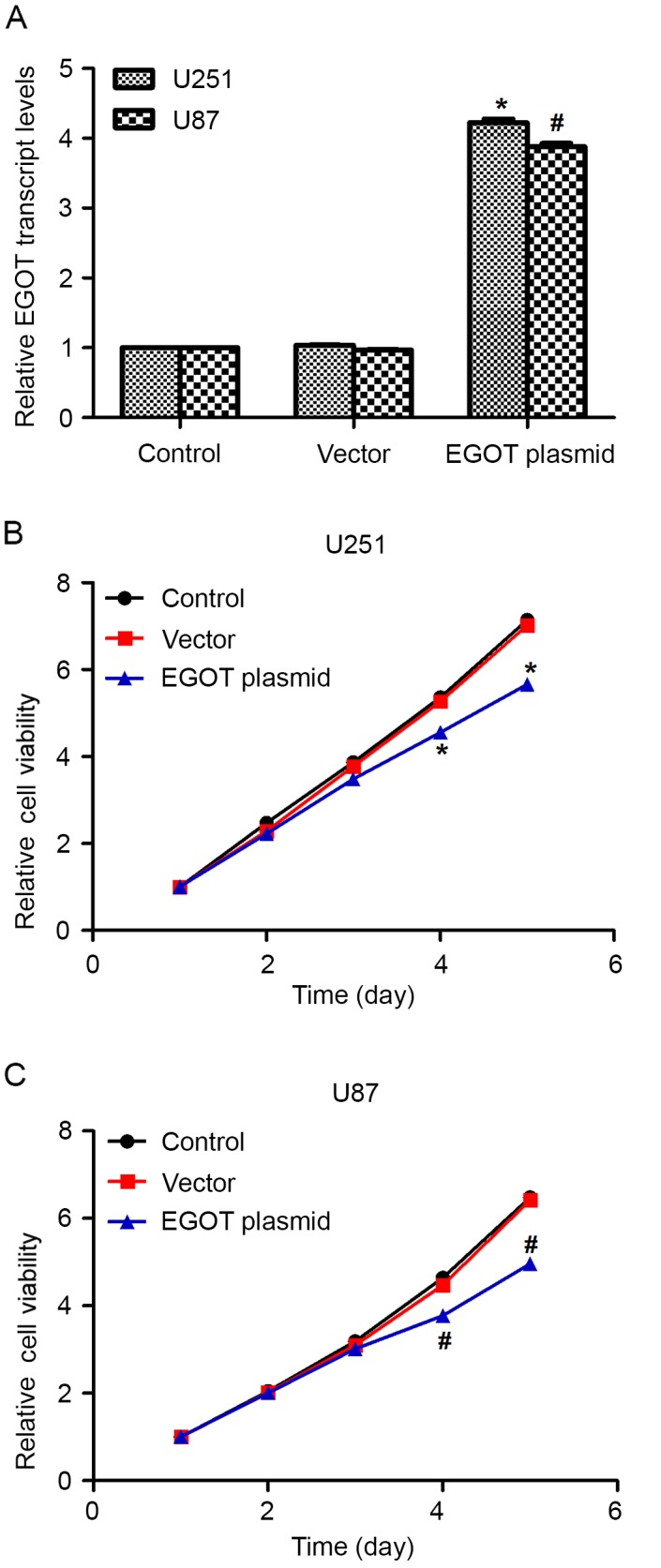
Overexpression of EGOT inhibits cell viability in glioma cells. (A) Relative transcription levels of EGOT in U251 and U87 cells with or without specific expression plasmid of EGOT. MTT assays were conducted to assess the viability of (B) U251 and (C) U87 cells with or without EGOT overexpression. Cell viability was presented relative to the viability of cells on day 1. All data are expressed as the mean ± standard deviation. *P<0.05 vs. control U251 cells; #P<0.05 vs. control U87 cells. EGOT, eosinophil granule ontogeny transcript.
Overexpression of EGOT arrests cell cycle at G0/G1 phase
Cell cycle analysis demonstrated that, following EGOT plasmid transfection, U251 cells were disturbed in each cell cycle phase. The cell percentage in the G0/G1 phase was significantly increased by ~20% after upregulation of EGOT in U251 cells. In turn, the cell percentages in the S phase and G2/M phase were significantly decreased by EGOT overexpression (Fig. 3A). In U87 cells, EGOT-transfected cells significantly accumulated (~16%) in the G0/G1 phase and correspondingly, were significantly less accumulated in the S phase or G2/M phase (Fig. 3B). These observations suggest that overexpression of EGOT promotes cell cycle arrest at G0/G1 phase.
Figure 3.
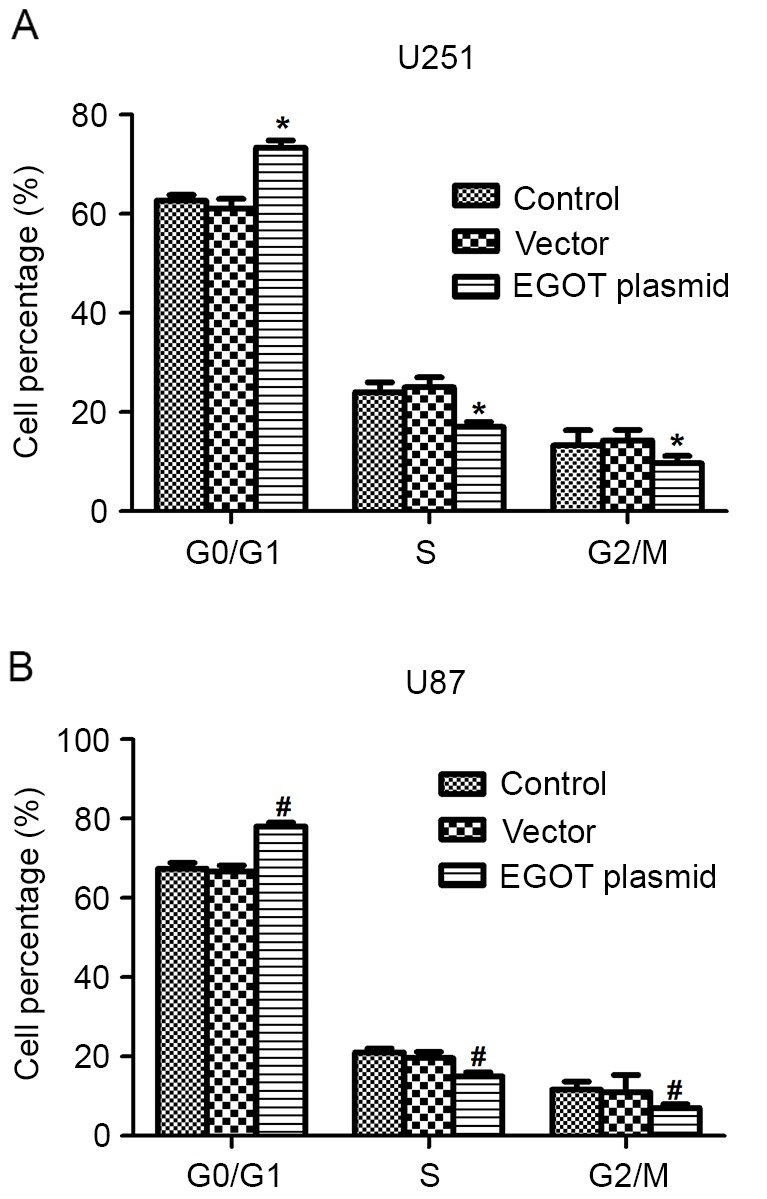
Overexpression of EGOT arrests the cell cycle at G0/G1 phase. (A) In U251 cells, when cells were transfected with EGOT, the cell percentage increased by ~20% in the G0/G1 phase and decreased correspondingly in the S and G2/M phases. (B) U87 cells were also subjected to cell cycle analysis with or without EGOT overexpression. Cell percentages in each phase were indicated. All data are expressed as the mean ± standard deviation. *P<0.05 vs. matched control U251 cells; #P<0.05 vs. matched control U87 cells. EGOT, eosinophil granule ontogeny transcript.
Overexpression of EGOT inhibits cell migration in glioma
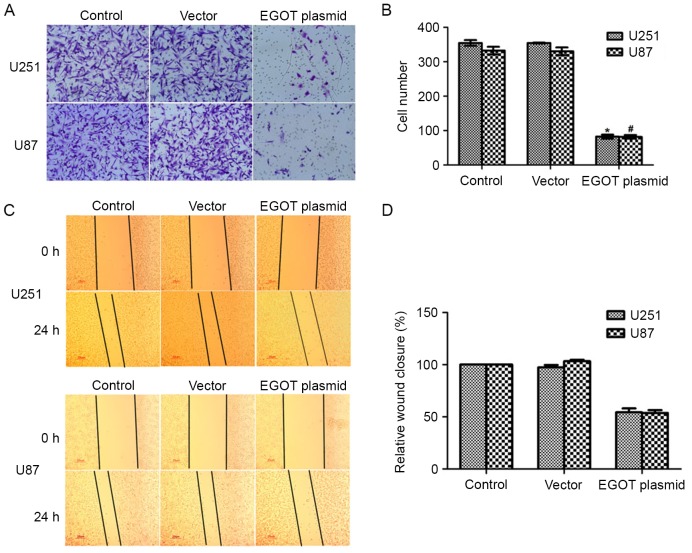
In our preliminary tests, it was found that cell apoptosis was insignificant within 24 h among distinct groups (data not shown). Therefore, to exclude the possible effect of cell apoptosis, cell migration was determined within 24 h in the present study. Cell migration capacity was then assessed using Transwell migration assays. Following transfection with the EGOT plasmid, it was visually observed that migrated cells were markedly less stained in U251 and U87 cells (Fig. 4A). Counting the migrated cells further demonstrated that a mean of ~96 cells had migrated to the lower surface after EGOT overexpression in U251 cells, which was significantly fewer than the ~350 control U251 cells that had successfully migrated (Fig. 4B). In U87 cells, while a mean of 326 control cells migrated, only 84 EGOT-overexpressing cells migrated to the lower surface, demonstrating a significant inhibition of 69.5% in U87 cells (Fig. 4B).
Figure 4.
Overexpression of EGOT inhibits cell migration in glioma. (A) Transwell migration assay was used to assess the migration of cells with or without EGOT overexpression. Cells on the lower surface were stained and images were captured of five random fields of view from each group. (B) Migrated cells in each image were counted and the mean was calculated for each group. (C) Representative images of Transwell assays for U251 and U87 cells upon EGOT plasmid transfection. (D) Quantification of Transwell assays for U251 and U87 cells. All data are expressed as the mean ± standard deviation. *P<0.05 vs. control U251 cells; #P<0.05 vs. control U87 cells. EGOT, eosinophil granule ontogeny transcript.
A wound-healing assay was subsequently performed to further explore the role of EGOT in human glioma. As shown in Fig. 4C and D, overexpression of EGOT in U251 cells retarded cell migration by ~50% compared to the control cells, whereas the migration potential of U87 cells was also inhibited by ~50% when cells were transfected with EGOT-expressing plasmids. These data suggest that overexpression of EGOT inhibits cell migration in glioma.
Overexpression of EGOT induces cell apoptosis and increases the activities of caspase-3 and caspase-9
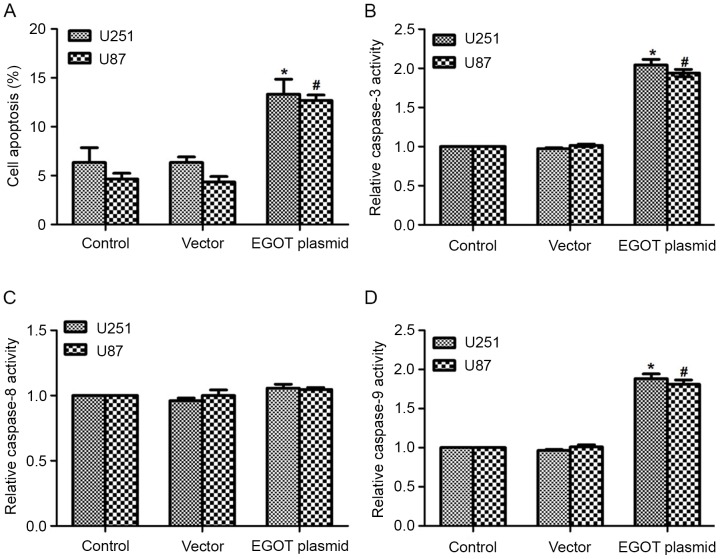
In consideration of the cell cycle arrest detected at G0/G1 phase, cell apoptosis was detected in both cell lines. As expected, it was observed that the EGOT plasmid significantly increased the apoptotic rates in both cell lines (Fig. 5A). Furthermore, with the upregulation of EGOT, the activity of caspase-3 was significantly increased (2-fold) in U251 cells and U87 cells (1.8-fold; Fig. 5B). Caspase-8 activity remained consistent among the different groups (Fig. 5C); whereas the activity of caspase-9 significantly increased following EGOT transfection in both cell lines (Fig. 5D). Subsequent western blot analysis was used to detect the protein expression levels of caspase-3, caspase-9 and cyt c. As shown in Fig. 6, in U251 and U87 cells, transfection of EGOT plasmid increased the protein expression levels of caspase-3, caspase-9 and cyt c. Consistent with the increased activities of caspase-3 and caspase-9, these findings support the hypothesis that EGOT induces cell apoptosis in glioma cells.
Figure 5.
Overexpression of EGOT induces cell apoptosis and increases the activities of caspase-3 and caspase-9. (A) Cell apoptosis was assayed in U251 and U87 cells with or without EGOT overexpression. Activities of (B) caspase-3, (C) caspase-8 and (D) caspase-9 were determined in U251 and U87 cells, respectively. All data are expressed as the mean ± standard deviation. *P<0.05 vs. control U251 cells; #P<0.05 vs. control U87 cells. EGOT, eosinophil granule ontogeny transcript.
Figure 6.
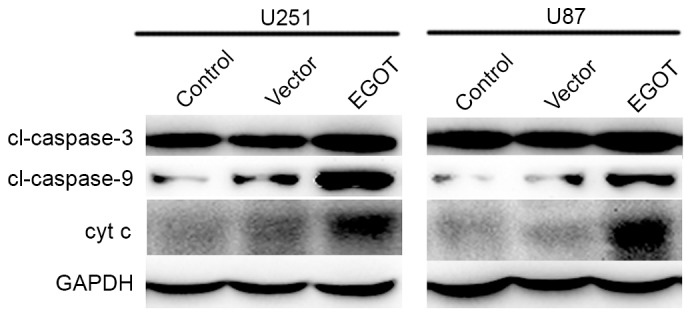
Overexpression of EGOT in glioma cells increased the protein levels of caspase-3, caspase-9 and cyt c. Western blot analysis revealed that when U251 and U87 cells were transfected with EGOT plasmid, the expression of caspase-3, caspase-9 and cyt c was markedly increased. All data are expressed as the mean ± standard deviation. EGOT, eosinophil granule ontogeny transcript; cyt c; cytochrome c.
Discussion
Malignant brain tumors are one of the most lethal types of cancer in adults. As the most common brain tumor, glioma is resistant to the current therapeutic strategies (5). Survival of patients with malignant glioma after surgery and radiotherapy is <1 year (5). Temozolomide chemotherapy may prolong survival; however, research has demonstrated that the majority of patients succumb to their symptoms within two years (6). Therefore, there is an urgent need for novel strategies for the treatment of glioma.
It has previously been demonstrated that ~80% of transcription across the human genome is associated with lncRNAs (8). However, only a small proportion of lncRNAs have been demonstrated to be biologically functional (20). Among the functional lncRNAs, the majority have been implicated in malignant tumors (10,21). The lncRNA, EGOT, was initially thought to be involved in eosinophil development and expressed in mature eosinophils (16); it was subsequently demonstrated that EGOT locus is highly structured, containing several evolutionary conserved regions, and thermodynamic stable secondary structures (22). Notably, the functional roles of EGOT have recently been implicated in ischemic heart diseases (23). Furthermore, another study has found that lncRNA EGOT negatively affects the antiviral response and favors hepatic virus C (HCV) replication (24). These pioneering studies have suggested that EGOT may exert active functional roles in human diseases, including human cancer. For example, the involvement of EGOT into human tumorigenesis has recently been identified in breast cancer (17). It was found that low levels of EGOT were significantly correlated with larger tumor size, lymph node metastasis and a worse prognosis in breast cancer patients.
The present study investigated the role of EGOT in glioma cell activities. EGOT was demonstrated to be downregulated in clinical glioma tissues and, compared with normal glial cells, glioma cells exhibited significantly lower transcription levels of EGOT. These findings were consistent with previous findings, which also revealed the downregulation of EGOT in breast cancer (17). Functionally, overexpression of EGOT in glioma U251 cells and U87 cells significantly inhibited cell proliferation and arrested the cell cycle at G0/G1 phase. The induction of cell apoptosis by EGOT overexpression, in turn, further demonstrated that EGOT functions to inhibit cell proliferation in glioma, since the induction of apoptosis is a good basis for the inhibition of cell proliferation (25). In addition, overexpression of EGOT significantly inhibits cell migration in U251 and U87 cells. These findings strongly suggest that EGOT may inhibit the aggressive activities of glioma cells and function as a tumor suppressor in glioma. However, further investigations are required. In future studies, we will focus on the effects of the depletion of EGOT on glioma. Specific small interfering RNAs or short hairpin RNAs (lentivirus and adenovirus) will be designed and transfected into the cells prior to injection into mice. Furthermore, our group will to collect clinical data in order to elucidate the association between clinicopathological characteristics and the expression of EGOT in human glioma.
It remains unclear how EGOT regulates cell apoptosis in glioma. Cell apoptosis is usually induced by two distinct signal pathways, intrinsic and extrinsic (26). The intrinsic pathway is always initiated by pro-apoptotic factors (B-cell lymphoma-2-associated X protein and Bcl-2-associated death promoter), which leads to increased permeability of the mitochondria membrane, loss of membrane potential and cyt c release into the cytosol. Caspase-3 was thereafter cleaved and activated, followed by caspase-9. By contrast, the extrinsic pathway is associated with the activation of caspase-8. The results of the present study demonstrated that the activity of caspase-8 remained unchanged and the activities of caspase-3 and caspase-9 were significantly increased upon EGOT overexpression. These analyses suggested that the extrinsic pathway may not be critically involved. Instead, the intrinsic pathway may be involved in the apoptosis induction processes induced by EGOT overexpression. However, as a recent report indicated that lncRNA EGOT prefers nuclear localization in HCV-infected cells (24), the subcellular localization of EGOT in human tumor cells remains to be elucidated and merits further investigation into whether the subcellular location is associated with its regulation of cell apoptosis.
In conclusion, the present study is the first to demonstrate the functional role of lncRNA EGOT in human tumorigenesis. Overexpression of EGOT significantly inhibited cell proliferation and migration and induced cell apoptosis in glioma cells. These conclusive findings suggest that EGOT has potent anticancer activity and may function as a tumor suppressor in human glioma.
References
- 1.Taylor LP. Diagnosis, treatment, and prognosis of glioma: Five new things. Neurology. 2010;75(18 Suppl 1):S28–S32. doi: 10.1212/WNL.0b013e3181fb3661. [DOI] [PubMed] [Google Scholar]
- 2.Erdem-Eraslan L, Gravendeel LA, de Rooi J, Eilers PH, Idbaih A, Spliet WG, den Dunnen WF, Teepen JL, Wesseling P, Smitt Sillevis PA, et al. Intrinsic molecular subtypes of glioma are prognostic and predict benefit from adjuvant procarbazine, lomustine, and vincristine chemotherapy in combination with other prognostic factors in anaplastic oligodendroglial brain tumors: A report from EORTC study 26951. J Clin Oncol. 2013;31:328–336. doi: 10.1200/JCO.2012.44.1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sukumari-Ramesh S, Prasad N, Alleyne CH, Vender JR, Dhandapani KM. Overexpression of Nrf2 attenuates Carmustine-induced cytotoxicity in U87MG human glioma cells. BMC Cancer. 2015;15:118. doi: 10.1186/s12885-015-1134-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hu B, Emdad L, Bacolod MD, Kegelman TP, Shen XN, Alzubi MA, Das SK, Sarkar D, Fisher PB. Astrocyte elevated gene-1 interacts with Akt isoform 2 to control glioma growth, survival, and pathogenesis. Cancer Res. 2014;74:7321–7332. doi: 10.1158/0008-5472.CAN-13-2978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Steeg PS, Camphausen KA, Smith QR. Brain metastases as preventive and therapeutic targets. Nat Rev Cancer. 2011;11:352–363. doi: 10.1038/nrc3053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hegi ME, Diserens AC, Gorlia T, Hamou MF, de Tribolet N, Weller M, Kros JM, Hainfellner JA, Mason W, Mariani L, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352:997–1003. doi: 10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- 7.Spizzo R, Almeida MI, Colombatti A, Calin GA. Long non-coding RNAs and cancer: A new frontier of translational research? Oncogene. 2012;31:4577–4587. doi: 10.1038/onc.2011.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chu C, Spitale RC, Chang HY. Technologies to probe functions and mechanisms of long noncoding RNAs. Nat Struct Mol Biol. 2015;22:29–35. doi: 10.1038/nsmb.2921. [DOI] [PubMed] [Google Scholar]
- 9.Kiang KM, Zhang XQ, Leung GK. Long Non-Coding RNAs: The key players in glioma pathogenesis. Cancers (Basel) 2015;7:1406–1424. doi: 10.3390/cancers7030843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li J, Meng H, Bai Y, Wang K. Regulation of lncRNA and Its role in cancer metastasis. Oncol Res. 2016;23:205–217. doi: 10.3727/096504016X14648701447896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ramos AD, Attenello FJ, Lim DA. Uncovering the roles of long noncoding RNAs in neural development and glioma progression. Neurosci Lett. 2016;625:70–79. doi: 10.1016/j.neulet.2015.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Han Y, Zhou L, Wu T, Huang Y, Cheng Z, Li X, Sun T, Zhou Y, Du Z. Downregulation of lncRNA-MALAT1 affects proliferation and the expression of Stemness Markers in glioma stem cell line SHG139S. Cell Mol Neurobiol. 2016;36:1097–1107. doi: 10.1007/s10571-015-0303-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xiang J, Guo S, Jiang S, Xu Y, Li J, Li L, Xiang J. Silencing of long non-coding RNA MALAT1 promotes apoptosis of glioma cells. J Korean Med Sci. 2016;31:688–694. doi: 10.3346/jkms.2016.31.5.688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Han Y, Wu Z, Wu T, Huang Y, Cheng Z, Li X, Sun T, Xie X, Zhou Y, Du Z. Tumor-suppressive function of long noncoding RNA MALAT1 in glioma cells by downregulation of MMP2 and inactivation of ERK/MAPK signaling. Cell Death Dis. 2016;7:e2123. doi: 10.1038/cddis.2015.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ma J, Wang P, Yao Y, Liu Y, Li Z, Liu X, Li Z, Zhao X, Xi Z, Teng H, et al. Knockdown of long non-coding RNA MALAT1 increases the blood-tumor barrier permeability by up-regulating miR-140. Biochim Biophys Acta. 2016;1859:324–338. doi: 10.1016/j.bbagrm.2015.11.008. [DOI] [PubMed] [Google Scholar]
- 16.Wagner LA, Christensen CJ, Dunn DM, Spangrude GJ, Georgelas A, Kelley L, Esplin MS, Weiss RB, Gleich GJ. EGO, a novel, noncoding RNA gene, regulates eosinophil granule protein transcript expression. Blood. 2007;109:5191–5198. doi: 10.1182/blood-2006-06-027987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu SP, Zhang JF, Sui SY, Bai NX, Gao S, Zhang GW, Shi QY, You ZL, Zhan C, Pang D. Downregulation of the long noncoding RNA EGOT correlates with malignant status and poor prognosis in breast cancer. Tumour Biol. 2015;36:9807–9812. doi: 10.1007/s13277-015-3746-y. [DOI] [PubMed] [Google Scholar]
- 18.Liu LX, Deng W, Zhou XT, Chen RP, Xiang MQ, Guo YT, Pu ZJ, Li R, Wang GF, Wu LF. The mechanism of adenosine-mediated activation of lncRNA MEG3 and its antitumor effects in human hepatoma cells. Int J Oncol. 2016;48:421–429. doi: 10.3892/ijo.2015.3248. [DOI] [PubMed] [Google Scholar]
- 19.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 20.Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermüller J, Hofacker IL, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- 21.Zhou J, Li X, Wu M, Lin C, Guo Y, Tian B. Knockdown of long noncoding RNA GHET1 inhibits cell proliferation and invasion of colorectal cancer. Oncol Res. 2016;23:303–309. doi: 10.3727/096504016X14567549091305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rose D, Stadler PF. Molecular evolution of the non-coding eosinophil granule ontogeny transcript. Front Genet. 2011;2:69. doi: 10.3389/fgene.2011.00069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Greco S, Zaccagnini G, Perfetti A, Fuschi P, Valaperta R, Voellenkle C, Castelvecchio S, Gaetano C, Finato N, Beltrami AP, et al. Long noncoding RNA dysregulation in ischemic heart failure. J Transl Med. 2016;14:183. doi: 10.1186/s12967-016-0926-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carnero E, Barriocanal M, Prior C, Unfried Pablo J, Segura V, Guruceaga E, Enguita M, Smerdou C, Gastaminza P, Fortes P. Long noncoding RNA EGOT negatively affects the antiviral response and favors HCV replication. EMBO Rep. 2016;17:1013–1028. doi: 10.15252/embr.201541763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xiang SS, Wang XA, Li HF, Shu YJ, Bao RF, Zhang F, Cao Y, Ye YY, Weng H, Wu WG, et al. Schisandrin B induces apoptosis and cell cycle arrest of gallbladder cancer cells. Molecules. 2014;19:13235–13250. doi: 10.3390/molecules190913235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Spencer SL, Sorger PK. Measuring and modeling apoptosis in single cells. Cell. 2011;144:926–939. doi: 10.1016/j.cell.2011.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]