Summary
Exosomes are membranous nanovesicles of endocytic origin that carry and transfer regulatory bioactive molecules and mediate intercellular communication between cells and tissues. Although seminal exosomes have been identified in human seminal plasma, their exact composition and possible physiologic function remain unknown. The objective of this study was to perform a comprehensive proteomics analysis of exosomes derived from human seminal plasma. Seminal exosomes were isolated and purified from 12 healthy donors using a 30% sucrose cushion‐based exosome‐isolation protocol, followed by characterization by western blot, transmission electron microscopy, and nanoparticle tracking analysis before performing extensive liquid chromatography tandem mass spectrometry proteomics analysis. The identified proteins were analyzed by bioinformatics analysis, and seminal exosomes‐associated proteins were selectively validated by western blot. A total of 1474 proteins were identified in all seminal exosomes samples, with Gene Ontology analysis demonstrating that these identified seminal exosomes‐associated proteins were mostly linked to ‘exosomes,’ ‘cytoplasm,’ and ‘cytosol.’ Bioinformatics analysis indicated that these proteins were mainly involved in biologic processes, including metabolism, energy pathways, protein metabolism, cell growth and maintenance, and transport. Of these identified proteins, PHGDH, LGALS3BP, SEMG1, ACTB, GAPDH, and the exosomal‐marker protein ALIX were validated by western blot. This study provided a more comprehensive description of the seminal exosomes proteome and could also be a resource for further screening of biomarkers and comparative proteomics studies, including those associated with male infertility and prostate cancer.
Keywords: exosomes, proteomics, seminal plasma
Introduction
Exosomes were discovered in 1987 and initially thought to represent non‐specific waste during the maturation of reticulocytes to erythrocytes (Johnstone et al., 1987). In recent years, evidence demonstrated that exosomes are small membranous vesicles (30–150 nm) released by a variety of cells into the extracellular environment and that carry diverse cargo, including proteins, RNA, and lipids, that can be transported and exchanged between cells as a means of intercellular communication (Valadi et al., 2007; Lotvall et al., 2014). Exosomes have been isolated and identified from many cells, such as dendritic cells (Thery et al., 1999), B cells (Raposo et al., 1996), and T cells (Blanchard et al., 2002), with the identified exosomes packaged with diverse cargo reflective of the host cells (Mathivanan et al., 2010). Similarly, exosomes are also detected in virtually all bodily fluids, including seminal plasma (Poliakov et al., 2009), which contains high concentrations and heterogeneous populations of subcellular lipid‐bound seminal prostasomes. Extracellular organelles in seminal fluid were first described in human prostatic fluid in 1977 (Ronquist & Hedstrom, 1977), and the term ‘prostasomes’ was coined in 1982 (Stegmayr & Ronquist, 1982) based on their organ of origin found to be the prostate gland (Aalberts et al., 2014). Although being exocytosed, the generality of prostasomes was unknown at that time. Recently, these seminal prostasomes from seminal plasma were reportedly morphologically and molecularly consistent with exosomes and were appropriately referred to as seminal exosomes (SEs) (Madison et al., 2014; Vojtech et al., 2014; Welch et al., 2017). Due to the difficulties in harvesting sufficient amounts of normal prostatic secretions to get ‘true prostasomes’ for proteomics analysis in the present study, we used SEs purified from seminal plasma.
Previous studies reported that SEs isolated from human seminal plasma carry a distinctive repertoire of small noncoding RNAs with potential regulatory functions (Vojtech et al., 2014). SEs not only exhibit potential anti‐human immunodeficiency virus (HIV)‐1 activity (Madison et al., 2014), but they also have various bioactive functions associated with sperm maturation, capacitation, acrosome reaction, and fertilization (Machtinger et al., 2016). The protein composition of seminal plasma has been previously investigated using proteomics tools, and the seminal‐plasma proteome offers great potential for the discovery of biomarkers to improve diagnosis or classification of a wide range of male reproductive‐system disorders (Gilany et al., 2015). Previously, a systematic attempt to catalogue the SEs proteome was described by Utleg et al. (2003), who identified 139 proteins from five healthy volunteers using microcapillary high‐performance liquid chromatography (HPLC)‐electrospray ionization tandem mass spectrometry (Utleg et al., 2003). In another study, Poliakov et al. (2009) reported for the first time the detailed structure of seminal exosome‐like vesicles. In parallel with the structural analysis, 440 proteins for seminal exosome‐like vesicles were identified from three healthy volunteers using trypsin in‐gel digestion and liquid chromatography tandem mass spectrometry (LC‐MS/MS) (Poliakov et al., 2009). These findings provided great practical value for determining candidate SEs proteins as biomarkers and understanding their biologic functions; however, these studies included small sample sizes and no biologic replicates. Given the relatively high variation in seminal‐plasma protein expression (Yamakawa et al., 2007) between individuals, it is possible that these studies might not have detected authentic differences essential for comparative proteomics studies, including those focusing on male infertility and prostate cancer.
HPLC coupled with LC‐MS/MS analysis allows the direct characterization of SEs associated with the membrane proteins and is more accurate than two‐dimensional electrophoresis (Goodlett & Yi, 2002). Here, we employed high‐resolution HPLC coupled with LC‐MS/MS analysis to perform an in‐depth proteomics analysis of purified human SEs obtained from 12 individual donors with two biologic replicates. We identified a total of 1474 proteins in these two experiments, with Gene Ontology (GO) analysis demonstrating that these SEs‐associated proteins were mostly linked to “exosomes,” “cytoplasm,” and “cytosol.” Bioinformatics analysis indicated that the proteins were mainly involved in biologic processes, including metabolism, energy pathways, protein metabolism, cell growth and maintenance, and transport. This study provided a more comprehensive description of the SEs proteome and could represent a resource for further screening of biomarkers and comparative proteomics studies, including those focused on male infertility and prostate cancer.
Materials and Methods
Ethics statement
This study was approved by the bioethics committees of Nanfang Hospital and the Third Affiliated Hospital of Southern Medical University, Guangzhou, China. Written informed consent was obtained from each healthy donor, and the experimental protocol was established according to the associated national guidelines from the Ministry of Science and Technology of China.
Subjects and semen samples
Semen samples were obtained from 12 healthy donors (mean age: 32.5 years; range: 26–44). These individual donors were randomly assigned to two groups of six (N1–N6 and N7–N12). To avoid inter‐individual variations, equivalent SEs pools from each group under study were analyzed. Mass spectrometry was performed on two biologic replicates. Additionally, SEs samples obtained from three additional healthy individuals (N13–N15) were used for validation of proteomics data by western blot analysis of three biologic replicates. These donors did not present with or have a history of any systemic illnesses, such as cryptorchidism, orchitis, epididymitis, urethritis, testicular atrophy, or sexually transmitted diseases, including HIV. All donors had undergone screening, were non‐smokers, and had a normal body mass index. None had been previously exposed to environmental stressors, including radiation or chemicals. All donors recruited to this study were of proven fertility and exhibited normal semen parameters. Semen samples were obtained from each donor by masturbation following 3 days of sexual abstinence, followed by immediate processing in accordance with 2010 guidelines of the World Health Organization. Routine semen analysis was conducted on a Sperm Class Analyzer (Microptic, Barcelona, Spain) and covered factors, such as liquefaction time, volume, pH, viscosity, agglutination, motility, viability, density, and sperm morphology. Diff‐Quik staining (Microptic) was used to evaluate sperm morphology according to our previous work (Zhou et al., 2015, 2016). The detailed clinical parameters of donors are shown in Table 1.
Table 1.
Clinical parameters of semen samples
| Characteristic | Age (year) | Volume (1.50–5.00 mL) | PH (7.20–8.00) | Count (≥15.0 × 106/mL) | Motility (32.0–100.0%) | Morphology (4.0–100.0%) | BMI (18.5–24.9) |
|---|---|---|---|---|---|---|---|
| N1 | 37 | 3.5 | 7.6 | 156.1 | 48.1 | 11.7 | 21.9 |
| N2 | 27 | 4.0 | 7.6 | 78.0 | 48.8 | 12.0 | 22.2 |
| N3 | 26 | 5.0 | 7.5 | 138.0 | 40.0 | 9.0 | 22.2 |
| N4 | 30 | 2.0 | 7.6 | 69.6 | 43.3 | 10.3 | 22.8 |
| N5 | 28 | 2.0 | 7.6 | 130.6 | 41.1 | 9.8 | 21.4 |
| N6 | 35 | 3.5 | 7.6 | 85.6 | 42.7 | 10.2 | 23.9 |
| N7 | 34 | 3.0 | 7.6 | 110.6 | 39.4 | 9.0 | 23.3 |
| N8 | 30 | 2.5 | 7.6 | 222.8 | 36.5 | 9.0 | 23.0 |
| N9 | 44 | 5.0 | 7.6 | 71.4 | 42.5 | 10.1 | 24.4 |
| N10 | 32 | 5.0 | 7.6 | 25.8 | 71.20 | 20.8 | 23.1 |
| N11 | 38 | 3.0 | 7.6 | 125.2 | 38.5 | 9.1 | 22.1 |
| N12 | 29 | 2.5 | 7.6 | 35.6 | 64.1 | 18.1 | 23.8 |
| N13 | 46 | 2.0 | 7.6 | 87.6 | 44.2 | 11.0 | 22.7 |
| N14 | 28 | 4.0 | 7.6 | 116.9 | 43.9 | 10.0 | 22.5 |
| N15 | 23 | 2.0 | 7.6 | 84.1 | 46.4 | 11.0 | 23.0 |
Preparation of seminal exosomes
SEs were isolated from seminal plasma as described previously (Vojtech et al., 2014), with minor modifications according to Fig. 1A. Briefly, after liquefaction at room temperature for 20 min, semen samples were subjected to sequential centrifugation steps at 1000 g for 10 min, 2400 g for 30 min, and 12,000 g for 30 min, with each step at 4 °C, which removed sperm cell and debris fraction. The supernatant containing the seminal exosomes was diluted in phosphate‐buffered saline (PBS), followed by filtration through 0.45‐μm and 0.22‐μm filters and ultracentrifugation at 100,000 g for 90 min in an SW 28 swinging‐bucker rotor (Beckman Coulter, Brea, CA, USA). Sperm cells and the upper solution of exosome‐depleted seminal plasma were collected and stored at −80 °C for further analysis. Pellets were re‐suspended in PBS and underlayered with a 30% sucrose cushion, followed by centrifugation at 100,000 g for 90 min. The sucrose cushion was washed by re‐suspension in PBS, followed by ultracentrifugation at 100,000 g for 90 min. The pellets were re‐suspended in PBS, and samples were frozen at −80 °C until use.
Figure 1.
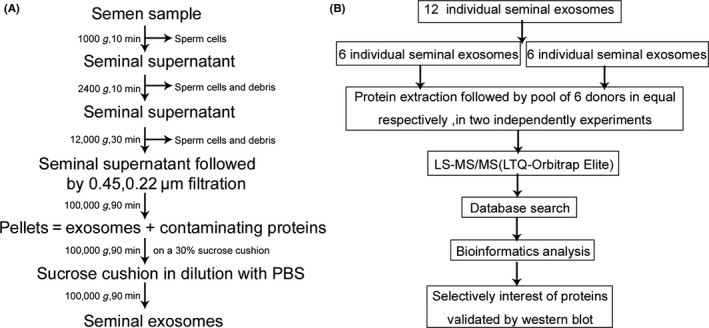
(A) The isolation procedure of exosomes derived from seminal plasma and (B) workflow for the processing of SEs for a comprehensive proteomic analysis.
Nanoparticle tracking analysis (NTA)
Particle size and concentration distribution of the isolated SEs were measured using NTA (v2.3; Malvern Instruments, Malvern, UK) according to manufacturer's instructions. Briefly, SEs samples were vortexed and diluted to a final dilution of 1 : 5000 in filtered molecular‐grade H2O. Blank‐filtered H2O was run as a negative control. Each sample analysis was conducted for 60 s and measured three times using Nanosight automatic analysis settings.
Western blot
Sperm‐cell lysates, SEs‐depleted seminal plasma, and SEs‐protein concentrations were determined using a BCA protein assay kit (P0010S; Beyotime Biotech, Jiangsu, China) according to manufacturer instructions. Subsequently, 30 μg of total protein was heated to 95 °C for 10 min in 1 × DTT‐containing sodium dodecyl sulfate (SDS) sample buffer and separated by 10% SDS‐polyacrylamide gel electrophoresis, followed by transfer onto polyvinylidene fluoride membranes (BioTrace; Bio‐Rad, Hercules, CA, USA). Membranes were blocked with 5% bovine serum albumin for 1 h, and polyclonal antibodies, including anti‐CD81, anti‐HSP‐70, anti‐ALIX, anti‐PHGDH, anti‐LGALS3BP, anti‐SEMG1 (Abclonal, Woburn, MA, USA), anti‐ACTB, anti‐GAPDH (Ray Antibody, Beijing, China), and anti‐Calnexin (Bioworld, Dublin, OH, USA), were used for immunoblotting at 4 °C overnight. Each specific horseradish peroxidase‐conjugated secondary antibody (Ray Antibody) was added accordingly after the membranes were washed in TBS‐Tween solution (TBS‐T) five times for 25 min, and signals were detected by enhanced chemiluminescence (Pierce, Rockford, IL, USA).
Transmission electron microscopy (TEM)
TEM analysis was performed to confirm SEs morphology. Briefly, SEs samples were diluted with PBS to the appropriate concentration, and ~20–40 μL of PBS solution containing SEs was transferred to a copper grid for incubation at room temperature for 5 min. Filter paper was used for absorbing unevaporated solution. SEs samples were negatively stained with 4% phosphotungstic acid solution at room temperature for 5 min and dried at 65 °C for 10 min. Images of SEs samples were obtained using a Hitachi H‐7650 transmission electron microscope (Hitachi, Tokyo, Japan).
Protein extraction
For protein extraction, SEs were transferred to lysis buffer [8m Urea, 2 mm EDTA, 10 mm DTT, 0.5% NP40, 1% Cocktail β] on ice for 30 min. Samples were sonicated for 3 min, and centrifuged at 13,000 g at 4 °C for 10 min to remove debris. Proteins were precipitated with cold 15% trichloroacetic acid for 2 h at −20 °C. After centrifugation at 4 °C at 16,000 g for 5 min, the supernatant was discarded, and the remaining precipitate was washed with cold acetone three times. The protein was re‐suspended in buffer [8m Urea, 100 mm TEAB, 0.5% NP40, PH8.0], and protein concentration was determined using the Bradford kit (C503041, Sangon Biotec, Shanghai, China).
HPLC and LC‐MS/MS analysis
In the study, the proteomics workflow was shown in Fig. 1B. Equal amounts of protein from the individual SEs samples were pooled (GroupA: N1–N6 and GroupB: N7–N12 respectively), then were reduced with a final concentration of 10 mm DTT for 45 min at 37 °C and subsequently alkylated with a final concentration of 25 mm IAM for 55 min at 37 °C under dark conditions. The protein solution was diluted by adding 100 mm TEAB to urea at a concentration of <2 m. Trypsin was added at a 1 : 50 trypsin‐to‐protein mass ratio for the first digestion overnight, followed by addition at a 1 : 100 trypsin‐to‐protein mass ratio for the second 4‐h digestion. The protein digestion was desalted with a Strata X SPE desalting column (Strata X 33‐μm polymeric reversed‐phase column; Phenomenex, Torrance, CA, USA), and peptides were injected into a Waters HPLC e2695/2998 system with a high pH C18 RP column (4.6 × 250 mm, 3.5 μm, 130 Å; Waters Corporation, Milford, MA, USA) at a flow rate of 0.7 mL/min. Peptides were eluted with a gradient from 5% to 30% solvent B over 55 min (solvent A: 2% CAN [pH 10]; solvent B: 98% CAN [pH 10]). A total of 48 fractions were collected every minute from 7 min to 54 min, and the peptides were combined into five fractions and dried by vacuum centrifugation. Peptide fractions were desalted using a Ziptip C18 (Ziptip pipette tips, 10 μL; Merck Millipore, Billerica, MA, USA) according to manufacturer instructions. Samples were dried under vacuum and stored at −20 °C until performance of MS analyses.
LC‐MS/MS was performed using a NanoLC 1000 LC‐MS/MS on an Easy‐nanoLC 1000 (Thermo Scientific, Odense, Denmark) with an analytical column heater (40 °C) coupled to an LTQ‐Orbitrap Elite. Trypsin‐digested fractions were dried, reconstituted in Solvent A (2% ACN, 0.1% FA) and delivered to a trap column [Acclaim PepMap 100C18, 3 μm, 100 Å (75 μm × 2 cm) at 5 μL/min in 100% solvent A (2% ACN, 0.1% FA)]. Peptides eluted from the trap column were loaded onto an analytical column (Acclaim PepMap RSLC C18, 2 μm, 100 Å [50 μm × 15 cm] at ~300 nL/min in a 60‐min gradient from 0% to 35% solvent B [98% ACN, 0.1% FA]). The eluent was sprayed via distal coated‐emitter tips butt‐connected to the analytical column. The mass spectrometer was operated in data‐dependent mode, automatically switching between MS and MS/MS. Full‐scan MS spectra (from m/z 350–1500) were acquired in the Orbitrap at a resolution of 60,000 full width at half maximum at 400 m/z using an automatic gain control setting of 1E6 ions. After the survey scans, the top 10 most intense precursors were selected for collision‐induced dissociation in the liner ion trap at a normalized‐collision energy of 33%. Dynamic exclusion was set at 60 s.
Data processing
The resulting MS/MS raw data were processed with Proteome Discoverer (v1.3; Thermo Scientific). Generated peak lists were searched against the Swiss‐Prot database containing human proteins and common contaminants using SEQUEST. Trypsin was chosen as the enzyme, and two missed cleavages were allowed. Carbamidomethylation (C) was set as a fixed modification; oxidation (M) and acetylation (N‐term) were set as variable modifications. The searches were performed with the precursor mass tolerance of 20 ppm and the maximum fragment mass error 0.8 Da. The criteria for protein identifications were accepted if a minimum of two peptides were detected or a unique peptide was detected with a false‐discovery rate <1%.
Statistical analysis
Functional‐enrichment analysis for GO terms was performed using FunRich (v3.0; released on December 2016) (Pathan et al., 2015). GO analysis of annotated proteins was performed for cellular components, molecular functions and biologic processes as previously described (van Herwijnen et al., 2016). Enriched terms were ranked by p‐value (hypergeometric test) using FunRich. A p < 0.05 was considered significant. Conversion from Uniprot identifier to Uniprot accession number was performed to compare proteins in different proteomic studies by Uniprot Retrieve/ID mapping (http://www.uniprot.org/uploadlists/). Comparison of identified proteins with the previously published data was performed using the Venn Diagrams web tool (http://bioinformatics.psb.ugent.be/webtools/Venn/).
Results
SEs characterization
To confirm the presence of exosomes, exosome‐marker proteins, including HSP70 and CD81, were validated by western blot (Fig. 2A). In contrast, these exosomal‐specific proteins were absent from collected exosome‐depleted fractions, whereas the endoplasmic reticulum marker calnexin was detected in sperm‐cell lysates. TEM results showed that SEs contained lipid bilayer‐bound membranes, with size distributions peaking at 105 nm diameter according to NTA (Fig. 2B and C). These data indicated that the successful isolation and purification of SEs are from seminal plasma.
Figure 2.
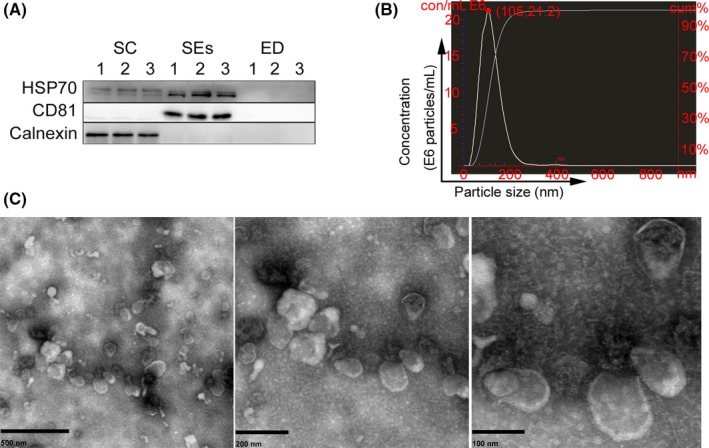
Characterization of the isolated SEs: (A) Western blot analysis of the exosomal‐marker proteins CD81, HSP70, and the endoplasmic reticulum marker calnexin. Sperm cell lysates (SC) and exosomes‐depleted seminal plasma (ED) were used as control. (B) After SEs samples with a final dilution of 1 : 5000, size distribution of SEs were determined by nanoparticle tracking analysis. The red dot indicated particle size (105 nm) and concentrations (21.2 E6 particles/mL) and finally the concentrations of SEs is 1.06 E11 particles/mL. (C) Transmission electron micrographs of the isolated SEs, representative image of SEs under different magnifications from left to right. [Colour figure can be viewed at wileyonlinelibrary.com].
Comprehensive proteomics analysis of SEs proteome
To determine SEs protein composition, extensive LC‐MS/MS proteomics analyses were performed, resulting in the identification of a total of 1474 proteins in both groups. We determined the presence of 354 overlapping proteins (24% of the total) between these two groups (897 proteins and 931 proteins respectively) (Fig. 3A, Tables S1 and S2). Importantly, from the proteins shared between these two groups, several SEs‐marker proteins, including ALIX and HSP70 and cytosolic proteins (annexins and Ras‐related proteins), were identified, whereas intracellular proteins (Calnexin) not expected to be enriched in SEs were indeed lacking, which was consistent with western blot results. Furthermore, western blot analysis of these shared proteins was performed to confirm their identities (Fig. 3B), including the exosomal protein ALIX, HSP70, and the SEs‐associated proteins ACTB, GAPDH, PHGDH, LGALS3BP, and SEMG1. Although relatively high individual variation existed, with some proteins uniquely identified in specific donors, portions of the shared proteins among all SEs samples suggested the presence of a common SEs proteome.
Figure 3.
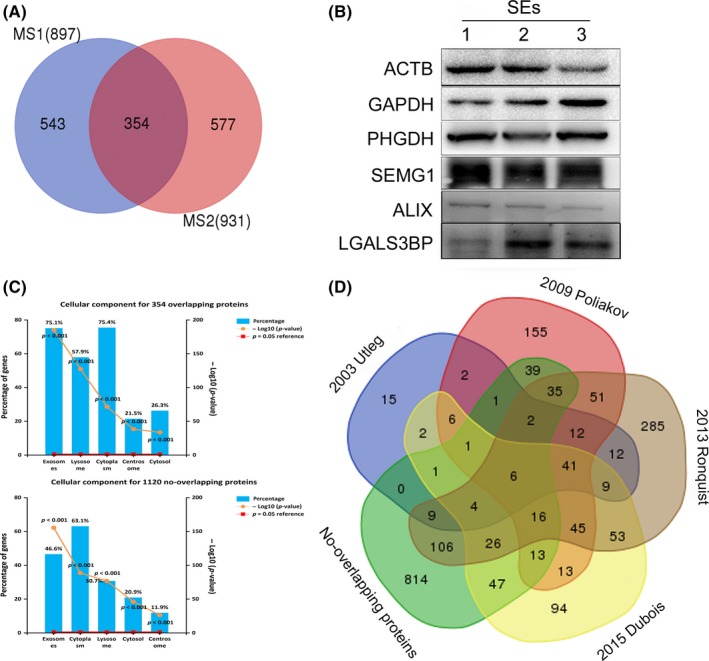
(A) Venn diagram illustrating overlapping proteins in the two independent MS experiments. (B) Selectively SEs‐associated proteins were validated by western blot. (C) The GO analysis (cellular component) of the overlapping proteins and no‐overlapping proteins separately. (D) Comparison of the 1474 proteins identified in the present study with previously published data from four studies. [Colour figure can be viewed at wileyonlinelibrary.com].
To investigate whether non‐overlapping proteins were present in the SEs proteome, but not in the contaminants, we determined the subcellular origins of these non‐overlapping proteins by functional‐enrichment analysis to determine cellular components. Our results revealed a high percentage of proteins linked to GO terms, such as “exosomes,” “lysosome,” and “cytoplasm,” which was similar to these 354 overlapping proteins (Fig. 3C) and suggested that the non‐overlapping proteins were likely derived from SEs. Additionally, comparison of the non‐overlapping proteins identified in SEs with previously published raw data obtained from four SEs studies (Utleg et al., 2003; Poliakov et al., 2009; Ronquist et al., 2013; Dubois et al., 2015) indicated that 306 proteins accounting for 27.3% of the 1120 non‐overlapping proteins had been previously identified in at least one study. Moreover, some of these 306 proteins, including CD81, LGALS3, and TSG101, were validated by western blot in other SEs studies (Kovak et al., 2013; Chiasserini et al., 2015). Remarkably, 814 proteins identified in this study had not been previously identified in SEs studies (Fig. 3D), indicating that they constituted novel SEs proteins. These results indicated that the non‐overlapping proteins identified in this study were most probably derived from SEs. Therefore, we selected all proteins (1474 proteins) identified in the SEs samples for further analysis.
To investigate the functional basis of the identified SEs‐associated proteins, GO analysis was conducted to analyze and classify their molecular functions. Proteins were sorted into categories according to their ontology as determined from their GO annotation terms (Fig. 4). The annotated biologic processes of the proteins revealed enrichment of SEs‐associated proteins related to “metabolism,” “energy pathways,” “protein metabolism,” “cell growth and maintenance,” and “transport.”
Figure 4.
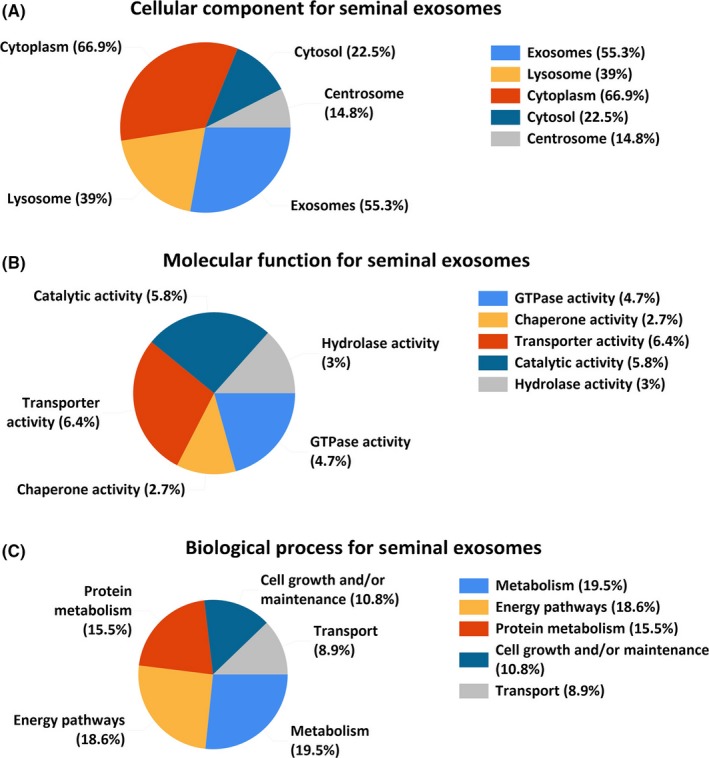
The GO analysis of the 1474 proteins identified in the study. (A) cellular component, (B) molecular function, and (C) Biologic process. [Colour figure can be viewed at wileyonlinelibrary.com].
SEs proteins compared with published exosomal databases and previously seminal‐plasma proteomic studies
To get more insight into these SEs proteins identified in human seminal plasma, we compared the 1474 proteins identified in the SEs with previously published exosomal studies included in the ExoCarta (Mathivanan & Simpson, 2009; Mathivanan et al., 2012; Keerthikumar et al., 2016) and Vesiclepedia (Kalra et al., 2012) databases. Our results indicated overlaps of 83% and 95% of proteins among the top 100 identified exosomal proteins in the ExoCarta and Vesiclepedia database, respectively, indicating a highly sensitive and comprehensive profiling of the SEs proteome (Fig. 5A). Additionally, according to Rolland et al. (2013), who summarized nine previous proteomic studies and obtained a list of 2545 unique proteins in human seminal plasma, 864 proteins (58.6%) were found to be overlapped in the SEs proteome. Moreover, some of these overlapped proteins between seminal plasma and the SEs proteome, such as PHGDH and LGALS3BP, were validated by western blot in the SEs samples (Fig. 3B). This suggests that the overlapping proteins most likely are derived from seminal plasma, while the no‐overlapping proteins unique in the SEs are most likely novel seminal‐plasma proteins (Fig. 5B).
Figure 5.
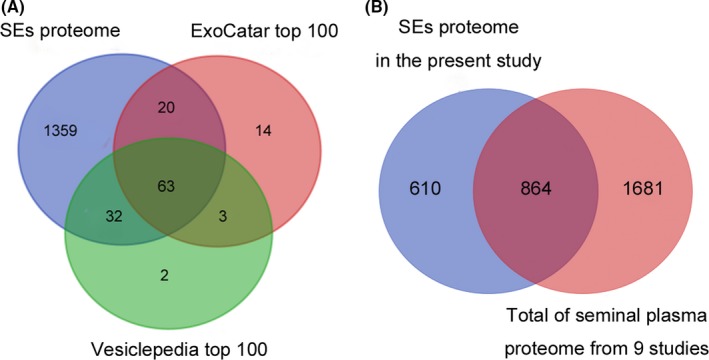
(A) Venn diagram describing common proteins in the all identified proteins between the ExoCarta database and EV database Vesiclepedia top 100. (B) Venn diagram representing shared proteins and non‐SEs proteins compared the published studies that Rolland et al. summarized total of seminal‐plasma proteome from nine studies in 2013. [Colour figure can be viewed at wileyonlinelibrary.com].
Discussion
To determine the composition of SEs as a reference dataset for comparative proteomics analysis of SEs between physiologic and pathologic states, including male infertility and prostate cancer, we performed comprehensive proteomics analysis of purified SEs isolated from the seminal plasma of 12 healthy donors using high‐resolution HPLC combined with LC‐MS/MS. A total of 1474 proteins were identified in all the SEs samples, whereas the low number of overlapping 354 proteins identified in the 2 pools (Tables S1 and S2). In this study, despite equivalent amount of total proteins from each individual in the two pools (Batruch et al., 2011; Rolland et al., 2013), large variations were observed in the SEs proteome (39.5% and 38.0% of the total 1474 proteins respectively). Body fluids, such as milk and seminal plasma, exhibit high degrees of variation in their composition between individuals. A previous study demonstrated coefficients of variation in protein spots ranging from 24.5% to 129.9%, with a median value of 63.1%, according to an analysis of the seminal‐plasma proteome from 10 fertile men (Yamakawa et al., 2007). Another study involving proteomics analysis of human milk‐derived extracellular vesicles revealed that 367 proteins were shared among seven donors, which was only 19% of the 1963 unique proteins in the milk‐derived extracellular vesicle proteome (van Herwijnen et al., 2016). Our results were consistent with the findings of these studies, demonstrating that SEs‐protein expression among different individuals is relatively variable. Additionally, in this study, proteomics analysis of these donors showed variations in sperm concentration from 25.8 × 106/mL to 222.8 × 106/mL, with progressive sperm motility ranging from 36.5% to 71.2%. Moreover, almost all of the SEs proteins identified were extracellular proteins, with high variability in proteolysis results (Yamakawa et al., 2007). It remains unknown whether different conditions dictate changes in SEs‐protein composition; however, the findings of our study and those of previous studies illustrate several properties of the SEs proteome, including that these SEs proteins behave over a broader dynamic range in the normal physiologic state and provide us with a basic reference to distinguish authentic differences from individual variations observed following comparison of the results of different proteomics studies.
SEs proteomes from human seminal plasma have been well documented. The previous study reported that 139 SEs proteins could be categorized as enzymes, transport/structural proteins, GTP proteins, chaperone proteins, and signal‐transduction proteins (Utleg et al., 2003). In an another study, Poliakov et al. (2009) reported that 440 proteins were mainly involved in metabolic processes, transport, protease activity, ion binding, and signal transduction. The 1474 proteins identified in this study were subjected to GO analysis, which revealed that these SEs proteins were involved in biologic processes, including metabolism, energy pathways, protein metabolism, cell growth and maintenance, and transport. A recent study (Ronquist et al., 2013) reported that SEs from four different species, including humans, are able to produce extracellular adenosine triphosphate, which suggests an energy metabolic function associated with SEs. SEs play an important role in cell responses and the ability of sperm cells to fertilize the ovum (Machtinger et al., 2016). Comparison of the 1474 proteins identified here with previously published data from four studies (Utleg et al., 2003; Poliakov et al., 2009; Ronquist et al., 2013; Dubois et al., 2015) indicated that nearly 54% of the proteins had been previously identified, and 56% were previously identified in our quantitative proteomics data associated with male infertility (data not shown). These results indicated the accuracy and reliability of our results and confirmed the acquisition of an SEs‐specific proteome.
The SEs from seminal plasma are originate from several cell types in the male genital tract (Madison et al., 2014; Vojtech et al., 2014; Machtinger et al., 2016) and play vital roles in sperm maturation and fertilization (Machtinger et al., 2016). However, SEs are not recognized as a major component of seminal plasma. To compare and assess the relative contribution of the SEs proteome to proteins previously identified in seminal plasma, we compared with previous proteomics studies of seminal plasma that Rolland et al. summarized (Rolland et al., 2013). Our analyses revealed that 864 proteins identified in our study overlapped with those found in the seminal‐plasma proteome, accounting for 33.9% of the seminal‐plasma proteome consisting of 2545 unique proteins. The outcome was expected, given that seminal plasma contains SEs. Here, SEs samples were isolated from seminal plasma for purification and concentration. The concentrations of SEs determined by NTA analysis demonstrated the presence of 1012 SEs/mL of ejaculate, which was consistent with previous studies (Vojtech et al., 2014; Yang et al., 2016; Welch et al., 2017) and suggested that a substantial number of SEs proteins are present in the seminal‐plasma proteome. Remarkably, 610 proteins identified in SEs were not identified in the seminal‐plasma proteome, likely because proteins are more abundant in seminal plasma, including semenogelins I and II (SEMG1 and SEMG2) represent 80% of all proteins in seminal plasma, thereby hindering the identification of less abundant proteins. In contrasted with results from a previous study (Utleg et al., 2003) showing that these proteins were contaminants rather than components of SEs, our results showed that these proteins were also identified in SEs, with SEMG1 was validated by western blot. A possible explanation is that the SEs proteins were not derived from soluble contaminants, but were indeed present in SEs.
Seminal plasma is an excellent source of protein biomarkers because it circulates through and comes in contact with the male reproductive system (Gilany et al., 2015). Consequently, seminal plasma as well as the SEs proteomics has great potential for the discovery of biomarkers for male diseases including male infertility and prostate cancer (Drake & Kislinger, 2014; Machtinger et al., 2016). Among the shared proteins identified in the SEs proteome, the following proteins have been successfully used for biomarker discovery of male infertility and male reproduction disorder in previous studies: semenogelins1 (SEMG1), semenogelins2 (SEMG2), prolactin‐inducible protein (PIP), fibronectin (FN1), prostatic acid phosphatase (ACPP), kallikrein3 (KLK3) (Wang et al., 2009; Drabovich et al., 2011; Batruch et al., 2012). Interestingly, the no‐overlapping SEs proteins identified in this study were almost not reported in the seminal‐plasma proteomics studies (Intasqui et al., 2015, 2016). This study demonstrated that seminal plasma contains a more varied population of proteins, but that a substantial number of these proteins are associated with SEs. These novel SEs‐associated proteins may also serve as potential biomarkers and provide insight into the possible functions performed by these proteins in male diseases and cancers.
Conclusion
This study involving proteomics analysis of SEs provides a platform for further studies in comparative proteomics, and it could also be a resource for further screening of biomarkers for male diseases including male infertility and prostate cancer.
Conflict of interest
The authors declare that they have no competing interests.
Authors’ contributions
CDL designed the study. TQ and CYW participated in collection of semen samples; WSZ and PW performed the western blot. JKY, QZZ, and MKC analyzed the experimental data. CY drafted the manuscript. WBG and JB participated in the revising of the manuscript. All authors read and approved the final manuscript.
Supporting information
Table S1. The list of 897 proteins identified in the MS1 (N1‐N6).
Table S2. The list of 931 proteins identified in the MS2 (N7‐N12).
AcknowledgEments
The authors thank Prof. Rangke Wu for his contributions to the manuscript revision. This study was supported in part by the Guangdong Provincial Science and Technology Program (Nos. 2014A020212204, 2013B021800317), Guangdong Provincial Natural Science Foundation of China (Nos. 2014A030313291, 2015A030310027, 2016A030310393), Science and Technology Innovation Project of Southern Medical University (QD2014N005), and Medical Scientific Research Foundation of Guangdong Province (A2014430).
References
- Aalberts M, Stout TA & Stoorvogel W. (2014) Prostasomes: extracellular vesicles from the prostate. Reproduction 147, R1–R14. [DOI] [PubMed] [Google Scholar]
- Batruch I, Lecker I, Kagedan D, Smith CR, Mullen BJ, Grober E, Lo KC, Diamandis EP & Jarvi KA. (2011) Proteomic analysis of seminal plasma from normal volunteers and post‐vasectomy patients identifies over 2000 proteins and candidate biomarkers of the urogenital system. J Proteome Res 10, 941–953. [DOI] [PubMed] [Google Scholar]
- Batruch I, Smith CR, Mullen BJ, Grober E, Lo KC, Diamandis EP & Jarvi KA. (2012) Analysis of seminal plasma from patients with non‐obstructive azoospermia and identification of candidate biomarkers of male infertility. J Proteome Res 11, 1503–1511. [DOI] [PubMed] [Google Scholar]
- Blanchard N, Lankar D, Faure F, Regnault A, Dumont C, Raposo G & Hivroz C. (2002) TCR activation of human T cells induces the production of exosomes bearing the TCR/CD3/zeta complex. J Immunol 168, 3235–3241. [DOI] [PubMed] [Google Scholar]
- Chiasserini D, Mazzoni M, Bordi F, Sennato S, Susta F, Orvietani PL, Binaglia L & Palmerini CA. (2015) Identification and partial characterization of two populations of prostasomes by a combination of dynamic light scattering and proteomic analysis. J Membr Biol 248, 991–1004. [DOI] [PubMed] [Google Scholar]
- Drabovich AP, Jarvi K & Diamandis EP. (2011) Verification of male infertility biomarkers in seminal plasma by multiplex selected reaction monitoring assay. Mol Cell Proteomics 10, M110.004127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake RR & Kislinger T. (2014) The proteomics of prostate cancer exosomes. Expert Rev Proteomics 11, 167–177. [DOI] [PubMed] [Google Scholar]
- Dubois L, Ronquist KK, Ek B, Ronquist G & Larsson A. (2015) Proteomic profiling of detergent resistant membranes (lipid rafts) of prostasomes. Mol Cell Proteomics 14, 3015–3022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilany K, Minai‐Tehrani A, Savadi‐Shiraz E, Rezadoost H & Lakpour N. (2015) Exploring the human seminal plasma proteome: an unexplored gold mine of biomarker for male infertility and male reproduction disorder. J Reprod Infertil 16, 61–71. [PMC free article] [PubMed] [Google Scholar]
- Goodlett DR & Yi EC. (2002) Proteomics without polyacrylamide: qualitative and quantitative uses of tandem mass spectrometry in proteome analysis. Funct Integr Genomics 2, 138–153. [DOI] [PubMed] [Google Scholar]
- van Herwijnen MJ, Zonneveld MI, Goerdayal S, Nolte‐’t Hoen EN, Garssen J, Stahl B, Maarten AA, Redegeld FA & Wauben MH. Comprehensive proteomic analysis of human milk‐derived extracellular vesicles unveils a novel functional proteome distinct from other milk components. Mol Cell Proteomics 2016;15:3412–3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Intasqui P, Antoniassi MP, Camargo M, Nichi M, Carvalho VM, Cardozo KH, Zylbersztejn DS & Bertolla RP. (2015) Differences in the seminal plasma proteome are associated with oxidative stress levels in men with normal semen parameters. Fertil Steril 104, 292–301. [DOI] [PubMed] [Google Scholar]
- Intasqui P, Camargo M, Antoniassi MP, Cedenho AP, Carvalho VM, Cardozo KH, Zylbersztejn DS & Bertolla RP. (2016) Association between the seminal plasma proteome and sperm functional traits. Fertil Steril 105, 617–628. [DOI] [PubMed] [Google Scholar]
- Johnstone RM, Adam M, Hammond JR, Orr L & Turbide C. (1987) Vesicle formation during reticulocyte maturation. Association of plasma membrane activities with released vesicles (exosomes). J Biol Chem 262, 9412–9420. [PubMed] [Google Scholar]
- Kalra H, Simpson RJ, Ji H, Aikawa E, Altevogt P, Askenase P, Bond VC, Borras FE, Breakefield X, Budnik V, Buzas E, Camussi G, Clayton A, Cocucci E, Falcon‐Perez JM, Gabrielsson S, Gho YS, Gupta D, Harsha HC, Hendrix A, Hill AF, Inal JM, Jenster G, Kramer‐Albers EM, Lim SK, Llorente A, Lotvall J, Marcilla A, Mincheva‐Nilsson L, Nazarenko I, Nieuwland R, Nolte‐’t Hoen HN, Pandey A, Patel T, Piper MG, Pluchino S, Prasad TS, Rajendran L, Raposo G, Record M, Reid GE, Sanchez‐Madrid F, Schiffelers RM, Siljander P, Stensballe A, Stoorvogel W, Taylor D, Thery C, Valadi H, van Balkom BW, Vazquez J, Vidal M, Wauben MH, Yanez‐Mo M, Zoeller M & Mathivanan S (2012) Vesiclepedia: a compendium for extracellular vesicles with continuous community annotation. PLoS Biol, 10, e1001450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keerthikumar S, Chisanga D, Ariyaratne D, Al SH, Anand S, Zhao K, Samuel M, Pathan M, Jois M, Chilamkurti N, Gangoda L & Mathivanan S. (2016) ExoCarta: a web‐based compendium of exosomal cargo. J Mol Biol 428, 688–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovak MR, Saraswati S, Goddard SD & Diekman AB. (2013) Proteomic identification of galectin‐3 binding ligands and characterization of galectin‐3 proteolytic cleavage in human prostasomes. Andrology 1, 682–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotvall J, Hill AF, Hochberg F, Buzas EI, Di Vizio D, Gardiner C, Gho YS, Kurochkin IV, Mathivanan S, Quesenberry P, Sahoo S, Tahara H, Wauben MH, Witwer KW & Thery C. (2014) Minimal experimental requirements for definition of extracellular vesicles and their functions: a position statement from the International Society for Extracellular Vesicles. J Extracell Vesicles 3, 26913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machtinger R, Laurent LC & Baccarelli AA. (2016) Extracellular vesicles: roles in gamete maturation, fertilization and embryo implantation. Hum Reprod Update 22, 182–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madison MN, Roller RJ & Okeoma CM. (2014) Human semen contains exosomes with potent anti‐HIV‐1 activity. Retrovirology 11, 102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathivanan S & Simpson RJ. (2009) ExoCarta: a compendium of exosomal proteins and RNA. Proteomics 9, 4997–5000. [DOI] [PubMed] [Google Scholar]
- Mathivanan S, Lim JW, Tauro BJ, Ji H, Moritz RL & Simpson RJ. (2010) Proteomics analysis of A33 immunoaffinity‐purified exosomes released from the human colon tumor cell line LIM1215 reveals a tissue‐specific protein signature. Mol Cell Proteomics 9, 197–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathivanan S, Fahner CJ, Reid GE & Simpson RJ. (2012) ExoCarta 2012: database of exosomal proteins, RNA and lipids. Nucleic Acids Res 40, D1241–D1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathan M, Keerthikumar S, Ang CS, Gangoda L, Quek CY, Williamson NA, Mouradov D, Sieber OM, Simpson RJ, Salim A, Bacic A, Hill AF, Stroud DA, Ryan MT, Agbinya JI, Mariadason JM, Burgess AW & Mathivanan S. (2015) FunRich: an open access standalone functional enrichment and interaction network analysis tool. Proteomics 15, 2597–2601. [DOI] [PubMed] [Google Scholar]
- Poliakov A, Spilman M, Dokland T, Amling CL & Mobley JA. (2009) Structural heterogeneity and protein composition of exosome‐like vesicles (prostasomes) in human semen. Prostate 69, 159–167. [DOI] [PubMed] [Google Scholar]
- Raposo G, Nijman HW, Stoorvogel W, Liejendekker R, Harding CV, Melief CJ & Geuze HJ. (1996) B lymphocytes secrete antigen‐presenting vesicles. J Exp Med 183, 1161–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolland AD, Lavigne R, Dauly C, Calvel P, Kervarrec C, Freour T, Evrard B, Rioux‐Leclercq N, Auger J & Pineau C. (2013) Identification of genital tract markers in the human seminal plasma using an integrative genomics approach. Hum Reprod 28, 199–209. [DOI] [PubMed] [Google Scholar]
- Ronquist G & Hedstrom M. (1977) Restoration of detergent‐inactivated adenosine triphosphatase activity of human prostatic fluid with concanavalin A. Biochim Biophys Acta 483, 483–486. [DOI] [PubMed] [Google Scholar]
- Ronquist KG, Ek B, Morrell J, Stavreus‐Evers A, Strom HB, Humblot P, Ronquist G & Larsson A. (2013) Prostasomes from four different species are able to produce extracellular adenosine triphosphate (ATP). Biochim Biophys Acta 1830, 4604–4610. [DOI] [PubMed] [Google Scholar]
- Stegmayr B & Ronquist G. (1982) Promotive effect on human sperm progressive motility by prostasomes. Urol Res 10, 253–257. [DOI] [PubMed] [Google Scholar]
- Thery C, Regnault A, Garin J, Wolfers J, Zitvogel L, Ricciardi‐Castagnoli P, Raposo G & Amigorena S. (1999) Molecular characterization of dendritic cell‐derived exosomes. Selective accumulation of the heat shock protein hsc73. J Cell Biol 147, 599–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utleg AG, Yi EC, Xie T, Shannon P, White JT, Goodlett DR, Hood L & Lin B. (2003) Proteomic analysis of human prostasomes. Prostate 56, 150–161. [DOI] [PubMed] [Google Scholar]
- Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ & Lotvall JO. (2007) Exosome‐mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol 9, 654–659. [DOI] [PubMed] [Google Scholar]
- Vojtech L, Woo S, Hughes S, Levy C, Ballweber L, Sauteraud RP, Strobl J, Westerberg K, Gottardo R, Tewari M & Hladik F. (2014) Exosomes in human semen carry a distinctive repertoire of small non‐coding RNAs with potential regulatory functions. Nucleic Acids Res 42, 7290–7304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Wang J, Zhang HR, Shi HJ, Ma D, Zhao HX, Lin B & Li RS. (2009) Proteomic analysis of seminal plasma from asthenozoospermia patients reveals proteins that affect oxidative stress responses and semen quality. Asian J Androl 11, 484–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch JL, Madison MN, Margolick JB, Galvin S, Gupta P, Martinez‐Maza O, Dash C & Okeoma CM. (2017) Effect of prolonged freezing of semen on exosome recovery and biologic activity. Sci Rep 7, 45034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization . (2010) WHO Laboratory Manual for the Examination and Processing of Human Semen, 5th edn WHO, Geneva. [Google Scholar]
- Yamakawa K, Yoshida K, Nishikawa H, Kato T & Iwamoto T. (2007) Comparative analysis of interindividual variations in the seminal plasma proteome of fertile men with identification of potential markers for azoospermia in infertile patients. J Androl 28, 858–865. [DOI] [PubMed] [Google Scholar]
- Yang C, Guo WB, Zhang WS, Bian J, Yang JK, Qi T, Wang CY & Liu CD (2016) [Extraction and identification of semen‐derived exosomes using PEG6000]. Nan Fang Yi Ke Da Xue Xue Bao 36, 1531–1535. [PubMed] [Google Scholar]
- Zhou JH, Zhou QZ, Lyu XM, Zhu T, Chen ZJ, Chen MK, Xia H, Wang CY, Qi T, Li X & Liu CD (2015) The expression of cysteine‐rich secretory protein 2 (CRISP2) and its specific regulator miR‐27b in the spermatozoa of patients with asthenozoospermia. Biol Reprod 92, 28. [DOI] [PubMed] [Google Scholar]
- Zhou JH, Zhou QZ, Yang JK, Lyu XM, Bian J, Guo WB, Chen ZJ, Xia M, Xia H, Qi T, Li X & Liu CD (2016) MicroRNA‐27a‐mediated repression of cysteine‐rich secretory protein 2 translation in asthenoteratozoospermic patients. Asian J Androl https://doi.org/10.4103/1008-682X.185001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. The list of 897 proteins identified in the MS1 (N1‐N6).
Table S2. The list of 931 proteins identified in the MS2 (N7‐N12).


