Abstract
Chromatin is reprogrammed after fertilization to produce a totipotent zygote with the potential to generate a new organism1. The maternal genome inherited through the oocyte and the paternal genome provided by sperm coexist as separate haploid nuclei in the zygote. How these two epigenetically distinct genomes are spatially organized is poorly understood. Existing chromosome conformation capture-based methods2–5 are inapplicable to oocytes and zygotes due to a paucity of material. To study the 3D chromatin organization in rare cell types, we developed a single-nucleus Hi-C (snHi-C) protocol that provides >10-fold more contacts per cell than the previous method2. Here we show that chromatin architecture is uniquely reorganized during the mouse oocyte-to-zygote transition and is distinct in paternal and maternal nuclei within single-cell zygotes. Features of genomic organization including compartments, topologically associating domains (TADs) and loops are present in individual oocytes when averaged over the genome; each feature at a locus is variable between cells. At the sub-megabase level, we observe stochastic clusters of contacts that violate TAD boundaries but average into TADs. Strikingly, we found that TADs and loops but not compartments are present in zygotic maternal chromatin, suggesting that these are generated by different mechanisms. Our results demonstrate that the global chromatin organization of zygote nuclei is fundamentally different from other interphase cells. An understanding of this zygotic chromatin “ground state” has the potential to provide insights into reprogramming to totipotency.
To investigate 3D genome organization in nuclei of single cells, we developed a genome-wide high-resolution in situ Hi-C approach. Conventional Hi-C methods include biotin incorporation and enrichment for ligated fragments6, which might limit fragment retrieval. We simplified the protocol by omitting these steps, similarly to genome conformation capture7 (Fig. 1a, Extended Data Fig. 1, see Methods). To verify the protocol, we compared data from population and single-cell data from K562 (human chronic myelogenous leukemia) cells and obtained a dependence of contact probability Pc(s), on genomic distance s, that matched conventional in situ Hi-C on bulk K562 cells8 (Fig. 1b). When applied to oocytes (Fig. 1c), our method was remarkably efficient at capturing chromosomal interactions: snHi-C revealed up to 1.9 × 106 contacts per cell after filtering, yielding 1–2 orders of magnitude more contacts than published single-cell Hi-C data2 and exceeding contact frequencies in single-cell Hi-C preprints9,10. Half of the cells had >3.39 × 105 contacts per cell and 7.1% had >1 × 106 contacts per cell (Supplementary Table 1). These high-density snHi-C data enabled us to examine chromatin features directly in single-cell maps.
Figure 1. Genome-wide high-resolution single-nucleus Hi-C approach.
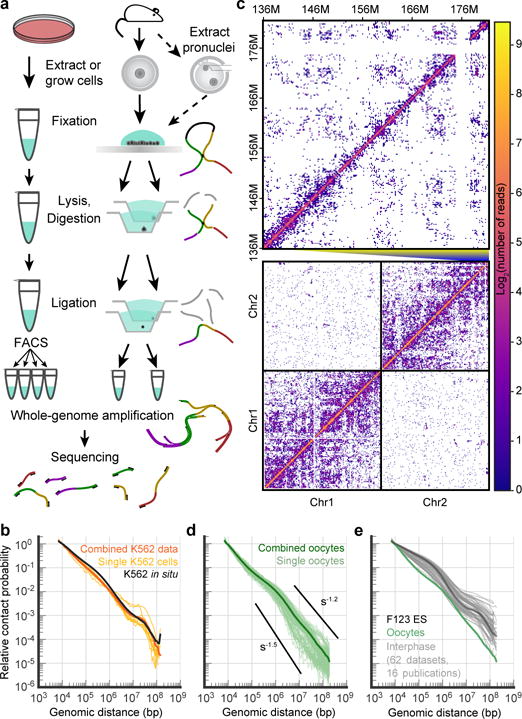
a, snHi-C workflow for cell culture and oocytes/zygotes. b, Dependence of contact probability on genomic separation, Pc(s), for single K562 cells (n=9) with >30K total contacts (yellow), combined single-cell K562 data (orange) and published population K562 data8 (black). P(s) here and below were normalized to be 1 at 10 kb. c, Example contact map from a single oocyte (cell 1). Below: chromosomes 1 and 2 at 1 Mb resolution. Above: fragment of chromosome 2 at 200-kb resolution. d, Pc(s) in single oocytes with >30K total contacts (n=84) and in combined data. Black lines show slopes for Pc(s)=s−1.5 and Pc(s)=s−1.2. e, Pc(s) of oocytes (green, combined data) compared to published interphase cells (grey) with highlighted curve for ES cells30 (black).
To investigate higher-level chromatin organization in oocytes, we examined how contact probability Pc(s)6,11 depends on genomic distance in individual cells and pooled data. In oocytes, Pc(s) were consistent between individual cells (Fig. 1d) but strikingly different from the characteristic shape in other mammalian interphase cells (Fig. 1e). For s>1 Mb, we observed a steeper (~s−1.5) decay in oocyte Pc(s), closer to the random walks of yeast chromosomes12–14. Our simulations showed that steeper Pc(s) can be attributed to the larger volume of oocyte nuclei (~25 μm versus ~6 μm diameter in somatic cells, Extended Data Fig. 2a).
Another major feature of mammalian chromosomes is segregation into A/B (active/inactive) compartments6. Although assignment of compartments from snHi-C data was impossible due to its sparsity, an enrichment of interactions between the same compartment type and depletion between different types became evident in individual cells when compartments were assigned using GC content (Fig. 2a) or population Hi-C from other cell types (Extended Data Fig. 3, 9). We also examined whether loops8 and TADs15,16, prominent functional features of chromatin organization17–19, are present in individual cells. Averaging over all genomic positions of loops and TADs identified in population Hi-C8 (for CH12-LX cells, see Methods) revealed that both are present in individual oocytes as average enrichments (Fig. 2a, Extended Data Fig. 3b) but vary between cells (Extended Data Fig. 3c), reflecting both inter-cell variability and variations in experimental conditions. We conclude that a single nucleus shows enrichment of interactions between regions of the same compartment type, within TADs and loops.
Figure 2. SnHi-C identifies TADs and chromatin loops in individual cells.
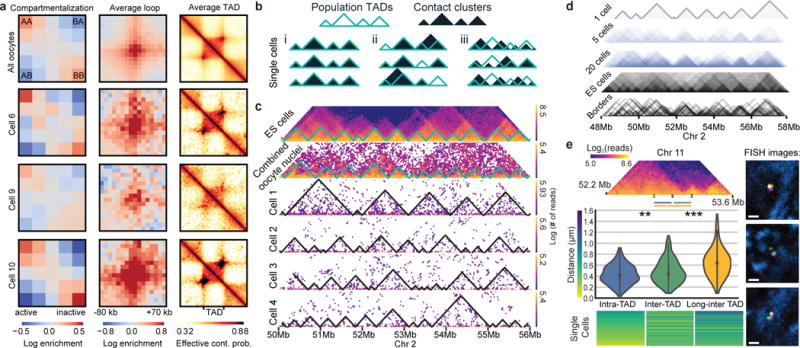
a, Contact enrichment of A/B compartments, averaging over loop and TAD positions annotated in CH12-LX cells8 in combined and single oocyte data. b, Scenarios leading to TADs in combined data. c, Variable contact clusters of top 4 single oocytes; first row - mESCs data30, second row - combined data from all oocytes (n=120). Resolution of all maps is 40 kb. d, Superimposing contact cluster annotation from cells (n=20) compared to population Hi-C TAD annotations. e, 3D FISH in mES cells quantifies TAD boundary violations in single cells (n=211). Top: Hi-C map of tested region with FISH probe locations. Middle: Distribution of measured distances, average distances (left to right) 0.428, 0.484, and 0.646 μm, relative contact probabilities from F123 cells at 20 kb resolution are 0.0095, 0.0037, 0.0024. Wilcoxon test p-values: ** − 0.007, *** − 2.5×10−16. Bottom: Heatmap of FISH measurements with colour-coded distances. Right: representative FISH images with adjusted gamma values. Scale bar − 1 μm. Probes (yellow, magenta, green) and DAPI (blue).
Using snHi-C data, we asked if TADs constitute physically isolated domains in individual cells or result from a tendency of chromosomes in individual cells to interact more within and less outside of a domain. We envisioned three scenarios (Fig. 2b): (i) all population-identified TADs are present in every individual cell; (ii) only population TAD boundaries are present, but individual TADs can be missing or fused in single cells; (iii) contacts may be clustered in individual cells, but clusters do not always match population TADs, revealing them only as an average feature. To distinguish between these scenarios, we examined high-coverage regions in the top snHi-C maps (Fig. 2c–d) and segmented chromosomes into domains of enriched contact frequency (contact clusters) using an exact segmentation algorithm that maximizes modularity (see Methods; comparable results for modularity segmentation of population TADs were obtained using an algorithm from ref. 20, see Extended Data Fig. 4a). We found that single-cell contact clusters do not always match population TADs, as contacts clusters are highly variable and frequently violate TAD borders. Nevertheless, variable contact clusters averaged into TADs when pooled together (Fig. 2d). The high cell-to-cell variations in contact patterns cannot be solely explained by experimental DNA loss because first, we often observe a presence of border-violating clusters rather than absence, and second, such patterns are also observed in regions of high read coverage (Extended Data Fig. 4b–c).
We validated frequent violations of TAD boundaries using 3D DNA FISH for equidistant pairs of probes located within a TAD and across a TAD border. Scenarios (i) and (ii) with TADs present in single cells are expected to yield intra-TAD distances mostly shorter than inter-TAD distances. However, imaging of ES cells showed that inter-TAD distances are shorter than intra-TAD in 42% of cases, although the average inter-TAD distance is larger than average intra-TAD (Wilcoxon test p-value=0.007). This indicates that although TAD borders affect the average distance, they do not always insulate in single cells (Fig. 2e). Surprisingly, even the long inter-TAD pair was closer than the intra-TAD pair in 18% of cells, despite having twice the linear genome separation, having 50% larger average distance and 4x lower contact probability. Together, both imaging data and snHi-C support scenario (iii), where TADs reflect a tendency for contact enrichment arising from a diverse conformational ensemble, rather than being isolated blocks of DNA present in individual cells.
The mechanism of TAD formation by loop extrusion and boundary insulation21,22 provides a rationale for TAD stochasticity and frequent boundary violations. Although insulation prevents extruded loops from crossing boundaries, contact clusters naturally emerge from the 3D spatial proximity of DNA in a confined volume22 (Extended Data Fig. 5, Extended Data Fig. 6, and ref. 11). Since contact clusters are also detected in K562 snHi-C (Extended Data Fig. 4d), stochastic cluster formation is likely a universal property of chromatin in single cells.
Next, we investigated chromatin rearrangement during the transition from transcriptionally active immature oocytes (non-surrounded nucleolus, NSN) into transcriptionally inactive mature (surrounded-nucleolus, SN) oocytes23,24 (Fig. 3a). We observed a significant decrease in loop, TAD and compartment strengths, (Fig. 3b–d and Extended Data Fig. 7; all Mann-Whitney p<0.005) during maturation, which may be related to transcriptional silencing and visual detachment of chromatin from the nuclear envelope (Fig. 3a)23,24. While Pc(s) scalings are similar, mature oocytes display more long-range (>400 kb) contacts (Mood’s equal median test p=0.02), and significantly less cell-to-cell variation in scalings, (Levene’s test p=0.007) (Fig. 3e–g). These findings are consistent with progressive chromatin reorganization during oocyte maturation.
Figure 3. Chromatin reorganization during oocyte maturation.
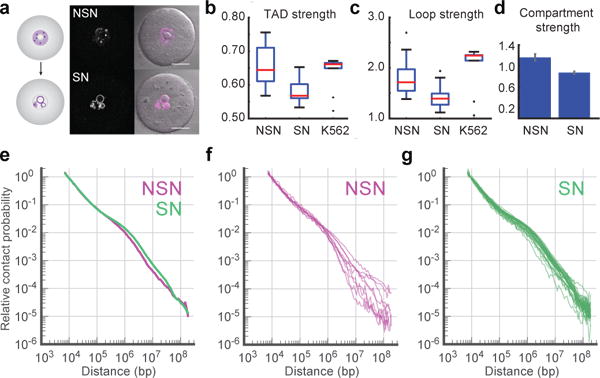
a, Immature (Non-Surrounded Nucleolus, NSN) and mature (Surrounded Nucleolus, SN) oocytes stained with Hoechst (magenta). Scale bar, 25 μm. Images were adjusted in brightness/contrast settings in the individual channels using ImageJ. b–d, Comparison of average TAD strength (b), loop strength (c) and compartment strength (d) in NSN and SN oocytes with Hoechst staining (n(NSN)=15, n(SN)=30). Error bars in (d) show standard deviation, obtained by bootstrapping. e, Pc(s) in data from combined NSN and SN oocytes, scored by Hoechst staining. f–g, Pc(s) in single Hoechst-stained NSN (f, n=9) and SN oocytes (g, n=27) with >30K total contacts.
We next addressed the key question, namely if and how chromatin is reorganized during the oocyte-to-zygote transition, and whether it is different between the maternal and paternal genomes that have different biological histories and epigenetic modifications1. Oocyte chromosomes decondense after two meiotic divisions into the maternal nucleus while in the compacted sperm chromatin protamines are replaced by histones to form the paternal nucleus25,26. To determine whether chromatin architecture is inherited or established de novo after fertilization, maternal and paternal nuclei extracted from predominantly G1 phase zygotes were subjected to snHi-C (Fig. 1a and Fig. 4a; similar results were obtained without extracting nuclei, see Extended Data Fig. 8a–b). In the best nuclei we detected 6 × 105 contacts, which is two-fold higher than for somatic cells and three-fold lower than for best oocytes. Averaging over TADs and loops identified previously8 indicated these features are present at similar strengths in maternal and paternal nuclei (Fig. 4b, Extended Data Fig. 3, 9). Although A and B compartmentalization is observed in paternal nuclei, it is strikingly absent from maternal nuclei (Fig. 4b–c, Extended Data Fig. 9a–b). To our knowledge, this is the first example of mammalian interphase nuclei presenting essentially no A/B compartmentalization. To corroborate this novel finding, we imaged 25 loci across chromosome 11 simultaneously using 3D FISH and measured nearest neighbour distances of A and B compartment probes to each other (Fig. 4d, Extended Data Fig. 10a–b). In agreement with Hi-C, we found that compartmentalization is most pronounced in ES cells (p<10−16 one-sided Mann-Whitney U-test, Extended Data Fig. 10c); paternal nuclei display weak but significant compartmentalization (p<0.01); and compartmentalization in maternal nuclei is undetectable as compared to a randomized control (p=0.08, Fig. 4d, see Methods). The lack of compartmentalization suggests that compartments are established de novo in the maternal genome, which may be due to a transcriptionally inactive extended G1 phase after fertilization1. Paternal genome compartmentalization is either inherited from sperm chromatin or established with faster kinetics. The weak compartmentalization aligns with detection of hyperacetylated histone H4, a hallmark of active chromatin, in early G1 phase paternal nuclei and earlier transcriptional activation27. These results further suggest that the mechanisms forming compartments are distinct from those forming TADs and loops, in agreement with a recent preprint28. We propose that the chromatin organization of zygotic nuclei denotes a “ground state” produced by transcriptional silencing, chromosome condensation and an exchange in cohesin complex composition at fertilization29.
Figure 4. Distinct chromatin architecture in haploid nuclei of totipotent zygotes.
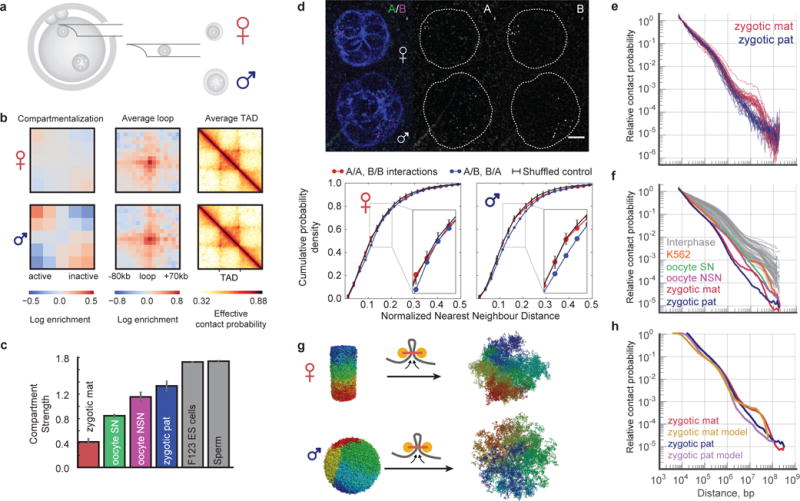
a, Extraction of nuclei from zygotes. b, Same as Fig. 2a but for combined data from zygote nuclei. Top: maternal (n=31), Bottom: paternal (n=24). c, Comparison of compartment signal strength in combined maternal (mat) and paternal (pat) zygote nuclei, oocytes, ES cell30 and sperm25 data. Error bars as in Fig. 3d. d, 3D FISH compartment quantification in zygote nuclei. Top: Deconvolved zygote FISH image, adjusted in brightness/contrast settings, background subtracted using ImageJ. Scale bar, 5 μm. Bottom: cumulative probability density plots for nearest-neighbour distances between same compartment and different compartment probe pairs for maternal (n=33) and paternal (n=37) nuclei. Black denotes average reshuffled control with random compartment assignment; error bars show 5% and 95% percentiles. e, Pc(s) in single maternal and paternal nuclei with >30K total contacts (mat=24, pat=20). f, Pc(s) from all combined datasets with all previously published data from mammalian interphase nuclei. g, In models of maternal and paternal nuclei, loop extrusion is initialized with a model of metaphase chromosome11 or compact fractal globule. Colour denotes position along the chromosome. h, Model Pc(s) compared to experimental Pc(s).
Finally, we used polymer modelling to understand differences in contact probability Pc(s) between the three analyzed cell types and somatic cells. Pc(s) scale similarly for s<3 Mb in zygotic paternal and maternal genomes (Fig. 4e), followed by a plateau between 3–12 Mb for the maternal genome, while Pc(s) continues to decrease for the paternal genome. Both curves show a remarkable 105-fold drop in contact probability (for s=10 kb to 100 Mb) and differ from somatic cells (Fig. 4f and Extended Data Fig. 8c). Polymer modelling demonstrated that the steeper drop of Pc(s) in oocytes and zygotes can be explained by their larger nuclear volumes (Extended Data Fig. 2), while the shallow scaling at <0.5 Mb may reflect local compaction by loop extrusion that was suggested to form TADs and loops21,22. Simulations suggested that differences in chromatin organization of paternal and maternal genomes at the long range (>3 Mb) can reflect their different histories. Simulations of decondensation subject to loop extrusion that start from a metaphase chromosome11 (Fig. 4g and Extended Data Fig. 6) result in Pc(s) resembling those of maternal nuclei (Fig. 4h, Extended Data Fig. 2c), while simulations that start with the compact fractal globule6 (as a model of protamine-compacted state) (Fig. 4g and Extended Data Fig. 2b and 5) can reproduce paternal Pc(s). Taken together, these results suggest that the factors influencing Pc(s) are nuclear density, cell cycle stage, and memory of the previous chromosome state. Maternal nuclei and somatic cells are both predominantly in G1 phase and experienced recent chromosome decondensation making their global genome organization most similar. Paternal nuclei have a different cell biological history and are thus different from somatic cells. Oocytes experienced the last mitosis weeks or months ago and are arrested in prophase I; they therefore differ the most from somatic cells.
In summary, our work provides insights into general principles of chromosome organization and specific biological aspects of oocyte and zygote genomes. We find that all known levels of chromosomal organization only exist as population averages, and in single cells are overshadowed by stochasticity of single-cell conformations. The unanticipated finding that zygotic maternal chromatin contains TADs and loops but no compartments suggests that it represents a transition state towards building the embryonic chromatin organization of a totipotent cell and raises the question of how the paternal chromatin establishes or maintains compartments after fertilization. These distinct states of higher-order structure also suggest that loops and compartments are formed by distinct mechanisms. Lastly, snHi-C will enable the study of chromatin organization during development and in rare cell types, like stem cells and distinct cells within highly heterogeneous tumours. By combining snHi-C with other single-cell approaches including single-cell transcriptome and methylome analyses, it will be possible to build a comprehensive picture of the interplay between genome folding and transcription in generating identities of individual cells.
Extended Data
Extended Data Figure 1. Comparison of conventional and strong fixation conditions for Hi-C.
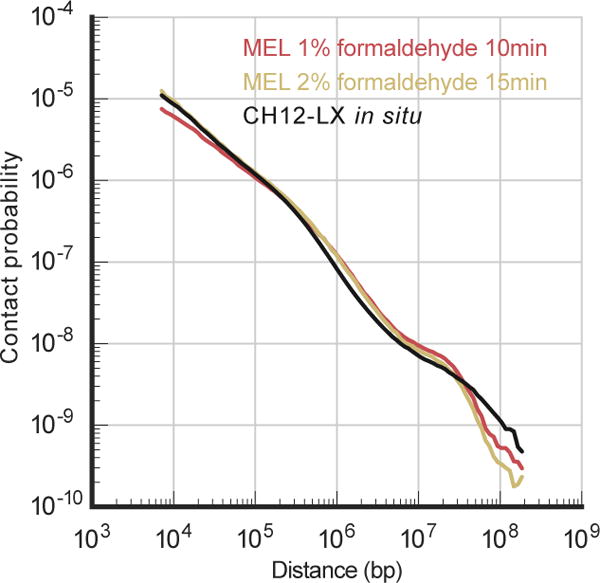
Pc(s) of contact probability over genomic separation has similar shape under conventional (1% of formaldehyde for 10 minutes) and strong (2% of formaldehyde for 15 minutes) fixation conditions. Pc(s) plot for the CH12-LX cell line is constructed using previously published in situ Hi-C data8 and is normalized to integrate to 1.
Extended Data Figure 2. Simulations of Pc(s) of oocytes, maternal and paternal nuclei.
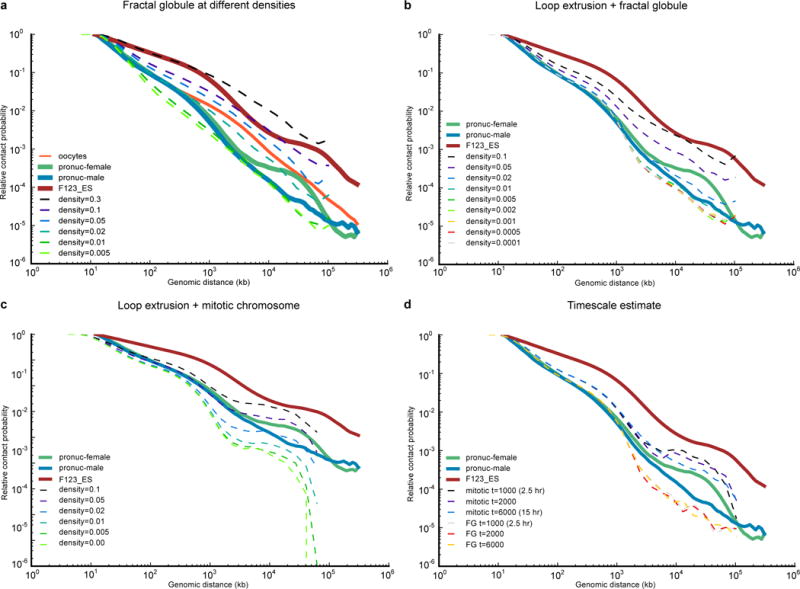
a–c, Pc(s) for various polymer models. All simulated Pc(s) curves were calculated using contact radius of 10 monomer diameters (100 nm). a, decondensed fractal globule, b, loop extrusion model starting with fractal globule, c, loop extrusion model starting with mitotic chromosome. d, Simulations in a–c and in Fig. 4h were run for 2000 loop extrusion steps, which represents around 5 hours of real time (see Methods). In reality, zygotes spent 7–10 hours post fertilization. To ensure that Pc(s) does not change significantly over this timescale, we simulated one run for 3 times longer (6000 loop extrusion steps). Note that since this figure was obtained from only two simulations, and not an average of many, and therefore the Pc(s) does not exactly match Fig. 4h. Even after 6000 loop extrusion steps, the Pc(s) curves starting with the fractal globule and with mitotic chromosome model are very distinct, and different by almost two orders of magnitude at 10 Mb.
Extended Data Figure 3. Quantification of average features in Hi-C maps.
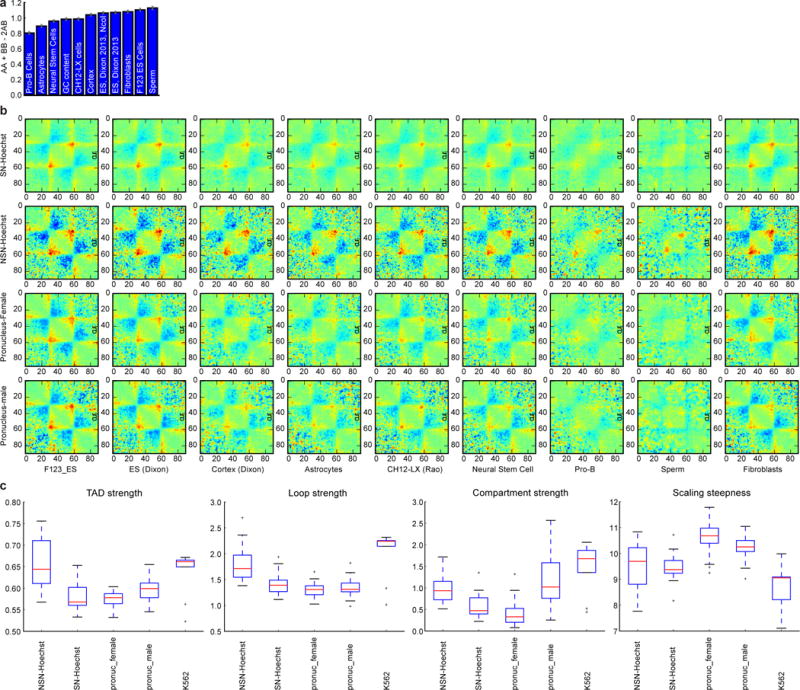
a, Using compartment annotation from different published datasets and quantification of compartment strength indicates highest similarity of oocyte compartments to sperm, mESCs and fibroblasts chromatin. Errorbars as in Fig. 3d. b, Average TADs calculated over TADs computed from various cell types. Note that high-resolution TAD calling is only available in CH12-LX cells8. For this figure, all TADs were all computed using lavaburst algorithm described in the methods. The value plotted here is natural log of observed-over-expected of the TAD enrichment. Unlike plots in the main figures, these are true observed-over-expected probabilities, not “effective contact probability”. Colourmap is jet, with range from −0.5 to 0.5. c, TAD, loop and compartment strength as well as scaling steepness (definitions are in Methods) in different classes of cells. Boxplots were generated using matplotlib (version 1.5.1) library for Python with default parameters.
Extended Data Figure 4. Stochasticity of contact clusters and validation of contact cluster annotation algorithm.
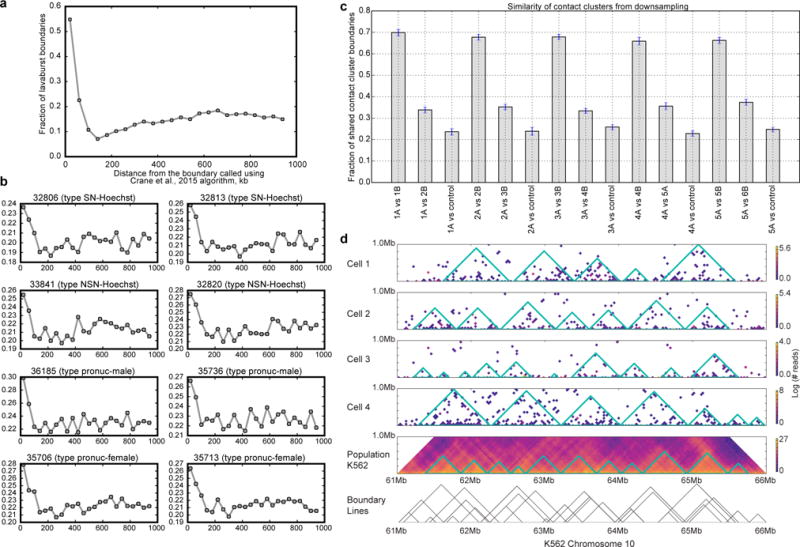
a, For this figure, boundaries were called on population data from CH12 cells (Rao, 2014) at 20 kb resolution using two different methods: lavaburst with modularity score, with an average domain size of 25 bins (500 kb), and a method from ref. 20, downloaded from https://github.com/dekkerlab/crane-nature-2015 (most recent commit, August 28, 2016). The latter method was used with default parameters, on whole-chromosome heatmaps. The plot shows fraction of lavaburst boundaries that are located within a certain distance of Crane et al. boundaries; step is 40 kb. Modularity score boundaries align very well with boundaries called using (Crane et al., 2015) as seen from the figure. For example, 77% of boundaries called using modularity score were within an 80 kb of (Crane et al.,) algorithm boundaries (32% expected if boundaries were randomized by offsetting them by 1 Mb). b, same as (a), but for top two single cell in each set. c, Contact cluster calling is robust to downsampling. From each of the top 5 single-cell oocytes, we obtained two maps down-sampled by 50% (1A and 1B from cell 1, etc.). We then compared contact clusters called in the two same-cell downsampled maps to each other (1A vs 1B), two maps from different cells (1A vs 2B), and each map to its randomly-shuffled control (1A vs control). Two maps from the same cell overlap by 65–70% of domain boundaries with 80 kb error margin. Overlap between different cells is about 1.5 times less (30–40%), and overlap with the reshuffled control is about 20–30%. Displayed are the average over all chromosomes and 95% confidence intervals of the fraction of overlap. d, The Hi-C contact cluster annotation of the top four single cell K562 cells is compared with the published population Hi-C map8.
Extended Data Figure 5. Contact clusters can form in polymer models with no average structure.
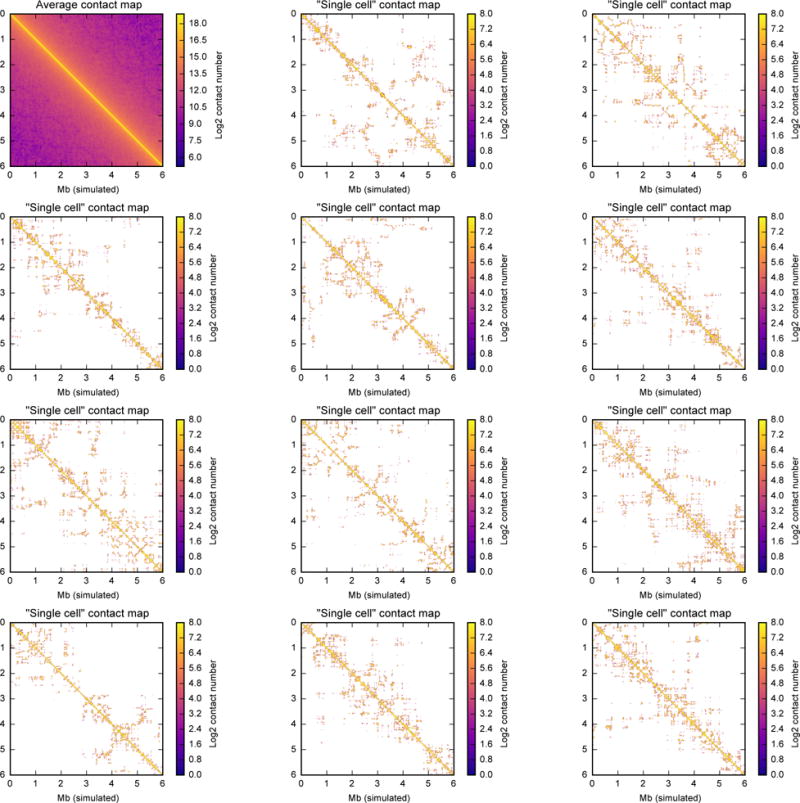
This figure shows contact maps of a 10,000 monomer region in fractal globules at density 0.05 (see Methods for the fractal globule creation descriptions). Each contact map was calculated with a contact radius of 10, at bin size of 16 monomers (approximately 10 kb, if we assume 600-bp monomers as in the other models or in ref. 11,22. First map (top left) shows a population average contact map calculated from 2000 independent realizations. Fractal globule is a model in which monomers are all treated equally and have no specific organization; therefore, a population average contact map of the fractal globule would be completely uniform (e.g. contact probability only depends on the distance between the two regions, Pc(i,j) = Pc(abs(i–j))). Each of the remaining 11 maps shows a “single-cell” contact map from 11 single conformations. Note the high degree of variability between single-conformation contact map, despite the complete homogeneity of the average contact map. See Figure S17 in ref. 11 for similar maps from our model of mitotic chromosomes. Note that, unlike in Hi-C, where each fragment end can form only one contact, in our simulations we record all contacts happening within the contact radius 10, and each monomer can form many contacts. Thus, this map shows more contacts than a single-cell Hi-C map would, even if Hi-C had the same capture radius. The map thus shows all potential contacts that could be extracted from a single conformation if sn-HiC was “performed” on the same conformation many times, each time choosing one neighbour within the contact radius of 10.
Extended Data Figure 6. TADs are not visible in single polymers undergoing loop extrusion.
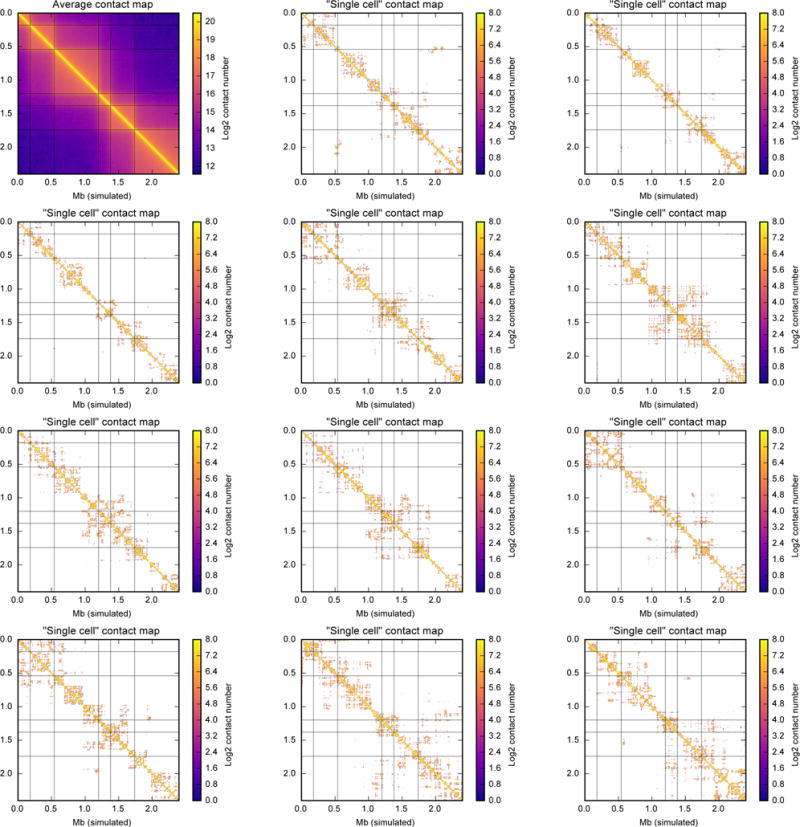
Similar to Extended Data Fig. 5, but for our model of loop extrusion starting with mitotic-like conformation (maternal nuclei). In this model, a 77-Mb chromosome (600-bp monomers; 128,000 monomers) is divided into 64 blocks of 3 TADs each. TAD sizes are 300, 600, and 1,100 monomers (180 kb, 360 kb and 660 kb). See ref. 22 and Methods for the description of the model. Thin gray lines denote TAD boundaries on all heatmaps. Each panel shows a block of 6 consecutive TADs, 4,000 monomers, or 2.4 Mb. Contact map is calculated at contact radius 10, and for bin size of 6 kb (10 monomers). For a population average map, 15,000 conformations were used. From each of 50 independent runs, we sampled 10 conformation at block numbers 1,100, 1,200 ..., 2,000. From each conformation, we sampled 30 non-overlapping blocks of 6 TADs (excluding first and last out of 32 blocks 0...4k, 4k..8k, 124k...128k) totalling 15,000 blocks. Single-cell map was calculated from a single randomly chosen block.
Extended Data Figure 7. Sn-HiC results for NSN and SN oocytes sorted by DIC scoring.
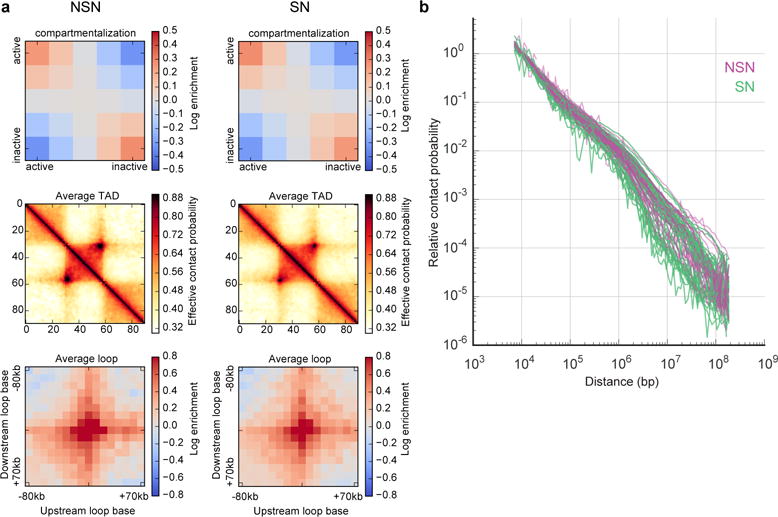
Note that Hoechst staining (see Fig. 3) is necessary for proper sorting of NSN and SN oocyte populations. a, Compartment signal, average TAD, average loop in oocytes staged by DIC with no DNA staining (n(NSN)=29, n(SN)=40). b, Pc(s) (for cells with >30K contacts, n(NSN)=25, n(SN)=30) for oocytes staged by DIC with no DNA staining.
Extended Data Figure 8. Pc(s) and compartment strength in zygote nuclei in comparison to other cell types.
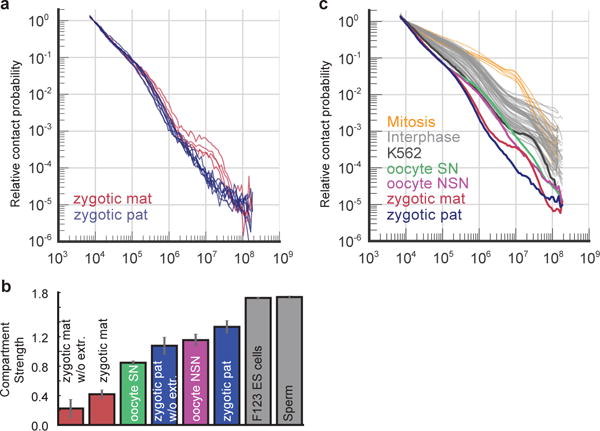
a, Pc(s) for maternal (mat) and paternal (pat) zygotic nuclei with >30K total contacts analysed without nuclear extraction (n(maternal)=4, n(paternal)=7). b, Comparison of compartment signal strength in combined maternal and paternal zygote nuclei with or without using nuclear extraction, with NSN and SN oocytes (staged with Hoechst staining), and published ES cell30 and sperm25 data. c, Pc(s) for K562 cells, paternal and maternal nuclei, NSN and SN oocytes (this work), interphase cells6,8,11,15,30,36–45 and mitotic chromosomes11.
Extended Data Figure 9. Comparison of all mm9 datasets.
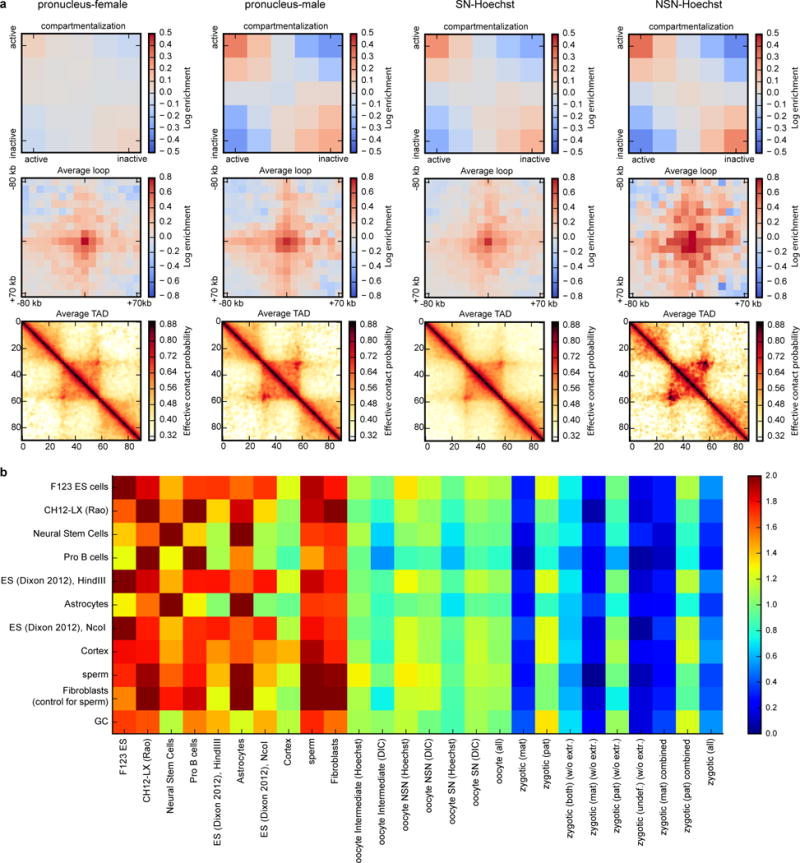
a, Same as Fig. 4b, but for oocytes and zygotic nuclei together. b, Compartment strength quantified in different datasets (columns, both published and ours) based on compartment annotation from published datasets (rows). The highest values in each column represent cell types most similar to the data of interest. Note that the first 9 columns have the highest value on the main diagonal, which correspond to compartment strength evaluated using eigenvector from the same dataset. Also note that Cortex cells have similar compartment strength to oocytes and paternal zygotic nuclei.
Extended Data Figure 10. Design and validation of FISH probes for quantification of compartments.
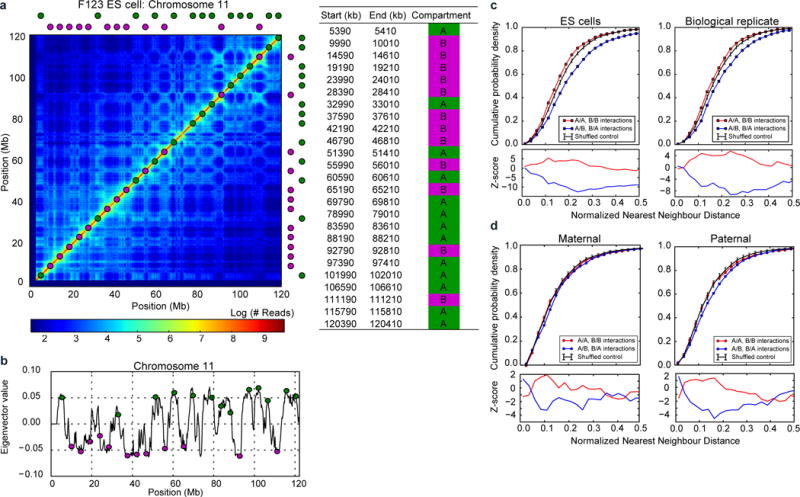
a, FISH probe design for quantifying compartment segregation; left: exact locations of designed probes, right: probe locations are shown superimposed on the Hi-C data (200 kb resolution) from F123 ES cells30 b, probe locations shown relative to the profile of compartment strength (200 kb resolution) as measured by the first eigenvector of the Hi-C map’s eigenvector decomposition. c–d, top: nearest neighbour FISH distances - the same as curves in Figure 4d, but shown for (c) ES cells (n(replicate1)=87, n(replicate2)=78) and (d) maternal (n=33) and paternal (n=37) zygotic nuclei; bottom: z-scores showing the number of standard deviations from the expected minimum distance distribution of the control data; the control distribution was obtained from randomly reshuffling probe colours as described in the methods.
Supplementary Material
Acknowledgments
We thank Christian Theußl for help with pronuclear extraction procedure, Sabrina Ladstätter for assistance in scoring oocyte stages and Kerstin Klien for experimental support and mouse colony management. We are grateful to Ian Adams, Shelagh Boyle, Isabelle Vassias-Jossic, Genevieve Almouzni and Wendy Bickmore for advice and help with FISH experiments. lllumina sequencing was performed at the VBCF NGS Unit (www.vbcf.ac.at) except Hi-C libraries from MEL cells, which were sequenced in the Laboratory of Evolutionary Genomics of the Faculty of Bioengineering and Bioinformatics, Moscow State University, by Maria Logacheva. K562 cells were a gift from Alexander Stark lab. We thank the staff of the Institute of Genetics and Molecular Medicine imaging facility and Vienna Biocenter BioOptics facility for assistance with imaging and analysis. We thank all members of the K.T.-K. lab for fruitful discussions, Life Science Editors for editorial assistance and Rob Illingworth for critically reading the manuscript. J.G. is an associated student of the DK Chromosome Dynamics supported by the grant W1238-B20 from the Austrian Science Fund (FWF). H.B.B. was partly supported by the Natural Sciences and Engineering Research Council of Canada, PGS-D. This work was funded by the Austrian Academy of Sciences and by the European Research Council (ERC-StG-336460 ChromHeritance) to K.T.-K. as well as by a grant from the Russian Science Foundation (14-24-00022) to S.V.U. and S.V.R. The work in the Mirny lab is supported by R01 GM114190, U54 DK107980 from the National Institute of Health, and 1504942 from the National Science Foundation.
Data availability
All sequencing data in support of the findings of this study have been deposited in the Gene Expression Omnibus (GEO) under accession number GSE80006. Source data for figures (Fig. 1b, d, e, Fig. 2c–e, Fig. 3b–g, Fig. 4c–f, Extended Data: Fig. 1, Fig. 2, Fig. 3a, c, Fig. 4a–d, Fig. 7b, Fig. 8a–c, Fig. 10a–c) are provided with the paper.
Footnotes
Supplementary Information
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author contributions
K.T.-K. conceived the project. I.M.F., M.I., S.V.U. and K.T.-K. conceived the method. I.M.F. developed the method. I.M.F. and J.G., supervised by K.T.-K., performed snHi-C on oocytes and zygotes. S.V.U. supervised by S.V.R. and K.T.-K. performed scHi-C on K562 cells. I.M.F. supervised by S.V.R. performed in situ Hi-C on MEL cells. I.M.F. supervised by K.T-K performed 3D FISH on ES cells. J.G. supervised by K.T-K performed 3D FISH on zygote. N.A. developed and maintains the library lavaburst for TAD calling. M.I. and H.B.B. supervised by L.A.M. developed and performed snHi-C data analysis. H.B.B. led FISH data analysis and performed contact cluster analysis. M.I. performed simulations, processed the data, and performed genome-wide averaging analyses. M.I., H.B.B., I.M.F., and J.G. prepared the figures. M.I., I.M.F., J.G., H.B.B., L.A.M., and K.T.-K. wrote the manuscript with input from all authors.
Author information
Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests. Correspondence and requests for materials should be addressed to K.T.-K. (kikue.tachibana@imba.oeaw.ac.at) and L.A.M. (leonid@mit.edu).
References
- 1.Zhou L, Dean J. Reprogramming the genome to totipotency in mouse embryos. Trends Cell Biol. 2015;25:82–91. doi: 10.1016/j.tcb.2014.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nagano T, et al. Single-cell Hi-C reveals cell-to-cell variability in chromosome structure. Nature. 2013;502:59–64. doi: 10.1038/nature12593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ma W, et al. Fine-scale chromatin interaction maps reveal the cis-regulatory landscape of human lincRNA genes. Nat Methods. 2015;12:71–78. doi: 10.1038/nmeth.3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hsieh THS, et al. Mapping Nucleosome Resolution Chromosome Folding in Yeast by Micro-C. Cell. 2015;162:108–119. doi: 10.1016/j.cell.2015.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barutcu AR, et al. C-ing the Genome: A Compendium of Chromosome Conformation Capture Methods to Study Higher-Order Chromatin Organization. J Cell Physiol. 2016;231:31–35. doi: 10.1002/jcp.25062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lieberman-Aiden E, et al. Comprehensive mapping of long range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rodley CDM, Bertels F, Jones B, O’Sullivan JM. Global identification of yeast chromosome interactions using Genome conformation capture. Fungal Genet Biol. 2009;46:879–886. doi: 10.1016/j.fgb.2009.07.006. [DOI] [PubMed] [Google Scholar]
- 8.Rao SSP, et al. A 3D Map of the Human Genome at Kilobase Resolution Reveals Principles of Chromatin Looping. Cell. 2014;159:1665–1680. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nagano T, et al. Cell cycle dynamics of chromosomal organisation at single-cell resolution. 2016 doi: 10.1101/094466. Preprint at http://biorxiv.org/content/early/2016/12/15/094466. [DOI] [PMC free article] [PubMed]
- 10.Ramani V, et al. Massively multiplex single-cell Hi-C. 2016 doi: 10.1101/065052. Preprint at http://biorxiv.org/content/early/2016/07/23/065052. [DOI] [PMC free article] [PubMed]
- 11.Naumova N, et al. Organization of the Mitotic Chromosome. Science. 2013;342:948–953. doi: 10.1126/science.1236083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mizuguchi T, et al. Cohesin-dependent globules and heterochromatin shape 3D genome architecture in S. pombe. Nature. 2014;516:432–435. doi: 10.1038/nature13833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tjong H, Gong K, Chen L, Alber F. Physical tethering and volume exclusion determine higher-order genome organization in budding yeast. Genome Res. 2012;22:1295–1305. doi: 10.1101/gr.129437.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Halverson JD, Smrek J, Kremer K, Grosberg AY. From a melt of rings to chromosome territories: the role of topological constraints in genome folding. Rep Prog Phys. 2014;77:022601. doi: 10.1088/0034-4885/77/2/022601. [DOI] [PubMed] [Google Scholar]
- 15.Dixon JR, et al. Topological Domains in Mammalian Genomes Identified by Analysis of Chromatin Interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nora EP, et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature. 2012;485:381–385. doi: 10.1038/nature11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Flavahan WA, et al. Insulator dysfunction and oncogene activation in IDH mutant gliomas. Nature. 2016;529:110–114. doi: 10.1038/nature16490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Franke M, et al. Formation of new chromatin domains determines pathogenicity of genomic duplications. Nature. 2016;538:265–269. doi: 10.1038/nature19800. [DOI] [PubMed] [Google Scholar]
- 19.Lupiáñez DG, et al. Disruptions of Topological Chromatin Domains Cause Pathogenic Rewiring of Gene-Enhancer Interactions. Cell. 2015;161:1012–1025. doi: 10.1016/j.cell.2015.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Crane E, et al. Condensin-driven remodelling of X chromosome topology during dosage compensation. Nature. 2015;523:240–244. doi: 10.1038/nature14450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sanborn AL, et al. Chromatin extrusion explains key features of loop and domain formation in wild-type and engineered genomes. Proc Natl Acad Sci. 2015;112:E6456–E6465. doi: 10.1073/pnas.1518552112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fudenberg G, et al. Formation of Chromosomal Domains by Loop Extrusion. Cell Rep. 2016;15:2038–2049. doi: 10.1016/j.celrep.2016.04.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miyara F, et al. Chromatin configuration and transcriptional control in human and mouse oocytes. Mol Reprod Dev. 2003;64:458–470. doi: 10.1002/mrd.10233. [DOI] [PubMed] [Google Scholar]
- 24.Bouniol-Baly C, et al. Differential Transcriptional Activity Associated with Chromatin Configuration in Fully Grown Mouse Germinal Vesicle Oocytes. Biol Reprod. 1999;60:580–587. doi: 10.1095/biolreprod60.3.580. [DOI] [PubMed] [Google Scholar]
- 25.Battulin N, et al. Comparison of the three-dimensional organization of sperm and fibroblast genomes using the Hi-C approach. Genome Biol. 2015;16 doi: 10.1186/s13059-015-0642-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ward WS, Coffey DS. DNA packaging and organization in mammalian spermatozoa: comparison with somatic cells. Biol Reprod. 1991;44:569–574. doi: 10.1095/biolreprod44.4.569. [DOI] [PubMed] [Google Scholar]
- 27.Adenot PG, Mercier Y, Renard JP, Thompson EM. Differential H4 acetylation of paternal and maternal chromatin precedes DNA replication and differential transcriptional activity in pronuclei of 1-cell mouse embryos. Dev Camb Engl. 1997;124:4615–4625. doi: 10.1242/dev.124.22.4615. [DOI] [PubMed] [Google Scholar]
- 28.Schwarzer W, et al. Two independent modes of chromosome organization are revealed by cohesin removal. 2016 doi: 10.1101/094185. Preprint at http://biorxiv.org/content/early/2016/12/15/094185. [DOI] [PMC free article] [PubMed]
- 29.Tachibana-Konwalski K, et al. Rec8-containing cohesin maintains bivalents without turnover during the growing phase of mouse oocytes. Genes Dev. 2010;24:2505–2516. doi: 10.1101/gad.605910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Selvaraj S, Dixon JR, Bansal V, Ren B. Whole-genome haplotype reconstruction using proximity-ligation and shotgun sequencing. Nat Biotechnol. 2013;31:1111–1118. doi: 10.1038/nbt.2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


